Abstract
One of the most fundamental processes of life is protein synthesis by the ribosome. Although much is known about the function and structure of this macromolecular complex, our understanding on its assembly is still vague. In this issue of The EMBO Journal, Malyutin et al (2017) provide a detailed picture of one of the latest assembly stages of the yeast 60S ribosomal subunit. The cryo‐EM map of the 60S‐Nmd3‐Lsg1‐Tif6 complex sheds new light on the function of Nmd3, Lsg1 and Tif6—and their release mechanisms—right before the 60S subunit joins the pool of actively translating ribosomes.
Subject Categories: Protein Biosynthesis & Quality Control, RNA Biology, Structural Biology
The eukaryotic ribosome consists of four ribosomal RNA molecules (rRNA) and ~80 ribosomal proteins (r‐proteins). These components are organized into the 40S small and 60S large subunits that associate forming the 80S ribosome. Ribosome assembly starts in the nucleus and is completed in the cytoplasm, with immature particles exported from the nucleus to the cytoplasm during this process. Assembly of the eukaryotic ribosome involves more than 200 biogenesis factors that support folding, processing and modifications of the rRNA, enable nuclear export of the assembling ribosomal particles or functionally test the active sites of the subunits (Karbstein, 2013; Woolford & Baserga, 2013; Greber, 2016). The general mechanisms by which these factors perform their functions remain poorly understood.
Pioneering work from the Hurt and Beckman's laboratories (Bradatsch et al, 2012) provided the first moderate resolution (11.9 Å) cryo‐EM reconstruction of a pre‐60S particle revealing how multiple assembly factors bind immature ribosomal particles. The recent introduction of direct electron detectors in cryo‐electron microscopy (cryo‐EM) is transforming our understanding of the role of assembly factors (Razi et al, 2017). We are now able to capture high‐resolution snapshots of the assembly process that shows multiple maturation factors simultaneously bound to pre‐ribosomal particles. In this issue, Malyutin et al (2017) present the cryo‐EM structure of the 60S subunit in complex with Nmd3, Lsg1 and Tif6 (Fig 1), providing a 3D view of one of the latest stages of 60S maturation.
Figure 1. Yeast 60S subunit bound to assembly factors.
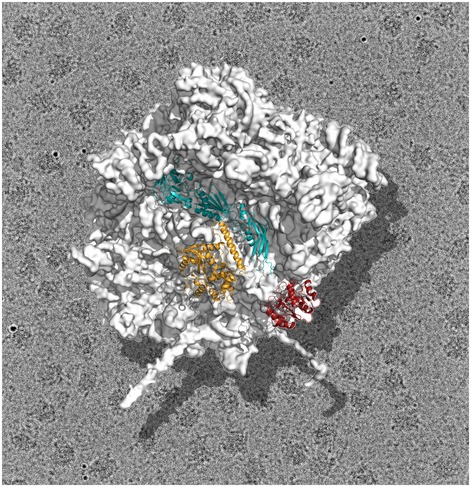
Cryo‐EM structure of the yeast 60S subunit in complex with assembly factors Nmd3 (teal), Lsg1 (gold) and Tif6 (red). This structure provides a 3D view of one of the latest stages of 60S maturation capturing the complete subunit right before the release of Nmd3, Lsg1 and Tif6 in the cytoplasm of yeast cells. This final step allows the 60S subunit to join the pool of actively translating ribosomes.
Assembly factor Nmd3 associates with the pre‐60S particle in the nucleus and participates in the nuclear export process along with a myriad of transiently interacting factors. Most of these factors are released prior to nuclear export. However, Nmd3 remains associated with the ribosomal particle until late during cytoplasmic maturation, where it is ultimately released in a process assisted by the GTPase Lsg1 (Hedges et al, 2005). Tif6 is a ribosome anti‐association factor that is released at the end of the maturation process assisted by the Efl1 GTPase and protein Sdo1. The reconstitution of the 60S‐Nmd3‐Lsg1‐Tif6 complex by Malyutin et al (2017) was done in vitro using purified components and mature 60S subunits, and it likely captures the complete subunit right before the release of Nmd3 and Tif6 in the cytoplasm of yeast cells.
The cryo‐EM map reveals that Nmd3 spans the joining face of the 60S subunit from the uL1 protein on the L1 stalk, covers the E and P sites and ends up contacting Tif6, which is bound at the sarcin‐ricin loop (SRL). The cryo‐EM map is of sufficient resolution to build an atomic model of all three domains of Nmd3. The N‐terminal domain of Nmd3 contacts Tif6, the second domain adopts a fold similar to that of r‐protein eL22 and occupies the P site, while the third domain contacts the L1 stalk, which is seen to adopt a closed conformation. Density for GTPase Lsg1 is also apparent in the cryo‐EM map. This protein binds at the intersubunit surface of the 60S subunit contacting helix 69, essential for association with the smaller subunit. A striking finding of the structure is that binding of Lsg1 induces a shift of rRNA helix 69 that causes guanosine 2261 to flip out towards residues near switch I in the GTPase domain of Lsg1, possibly triggering GTP hydrolysis. This observation suggests that helix 69 could act as a general activator of GTPases involved in large subunit biogenesis. The structure also suggests a role for Nmd3 in promoting the loading of uL16, a step that causes drastic conformational changes leading to the formation of the mature subunit.
The cryo‐EM map of the 60S‐Nmd3‐Lsg1‐Tif6 complex provides new examples of how assembly factors act as checkpoint proteins in the mature subunits. It reveals that the domain of Nmd3 bound to the E site adopts a topology remarkably similar to eIF5A, a protein factor necessary for the rescue of ribosomes stalled on polyproline‐containing sequences. One of the domains of Nmd3 also occupies the P site, thus adding to the list of assembly factors that perform quality control checks and test the functionality and conformation of specific ribosome sites.
Finally, this study deepens our understanding of how late‐stage assembly factors are released right before the active 60S subunit joins the pool of translating ribosomes. Cryo‐EM analysis of several partial sub‐complexes undertaken in this study, in addition to the full 60S‐Nmd3‐Lsg1‐Tif6 complex, allowed the authors to propose a sequence of events leading to the release of Nmd3, Lsg1 and Tif6. They suggest that breakage of the interaction between Nmd3 and Tif6 constitutes the initial event that destabilizes the complex. The L1 stalk is then free to move from a closed to an open conformation, thereby pulling out Nmd3 from the P and E sites. The absence of direct contacts between Lsg1 and Nmd3 suggests that another factor, possibly Efl1, may be responsible for disengaging Nmd3 from Tif6 and initiate the release. Subsequent entry of Sdo1, recruited by Efl1, releases Tif6.
In a related publication (Ma et al, 2017), the Woolford and Gao's groups reported the cryo‐EM structure of a pre‐60S particle purified with epitope‐tagged Nmd3. They found that these particles are depleted for r‐proteins uL16, uL10, uL11, eL40 and eL41 and are bound to the Nmd3, Lsg1, Tif6 and Reh1 assembly factors. The structure reported in that study likely typifies the maturation stage of the 60S subunit just prior to that shown in the Malyutin et al (2017) article. Therefore, it brings additional insights on the maturation events occurring at these late stages. A noticeable difference between the two structures is that the density for Lsg1 appears fragmented in the pre‐60S particle. The N‐terminal region of Nmd3 is also not well ordered, suggesting that Nmd3 and Lsg1 bind in a flexible manner. These observations are consistent with these pre‐60S particles representing a late cytoplasmic stage just prior to the incorporation of uL16. This stage of maturation is earlier than that described in the structure of the in vitro reconstituted 60S‐Nmd3‐Lsg1‐Tif6 complex and suggests a different order of release for Nmd3 and Tif6 from that proposed in Malyutin et al (2017). The structure by Ma et al (2017) implies that Tif6 is the last remaining factor released from the maturing particle and that Nmd3 release precedes this event. Additional structures and genetic analysis will need to be done to resolve these conflicting models.
Overall, the structures presented by Malyutin et al (2017) and Ma et al (2017) provide two important snapshots of a cellular pathway essential to sustain life. We see cryo‐EM with the incorporation of direct electron detectors as a technique of great promise that is currently transforming our understanding of the ribosome biogenesis process and the role played by assembly factors. Studies in the last decade linking defects in ribosome assembly with cancer development make these studies of general importance and have the potential to translate into clinical benefits.
See also: AG Malyutin et al (April 2017) and C Ma et al (March 2017)
References
- Bradatsch B, Leidig C, Granneman S, Gnadig M, Tollervey D, Bottcher B, Beckmann R, Hurt E (2012) Structure of the pre‐60S ribosomal subunit with nuclear export factor Arx1 bound at the exit tunnel. Nat Struct Mol Biol 19: 1234–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber BJ (2016) Mechanistic insight into eukaryotic 60S ribosomal subunit biogenesis by cryo‐electron microscopy. RNA 22: 1643–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges J, West M, Johnson AW (2005) Release of the export adapter, Nmd3p, from the 60S ribosomal subunit requires Rpl10p and the cytoplasmic GTPase Lsg1p. EMBO J 24: 567–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbstein K (2013) Quality control mechanisms during ribosome maturation. Trends Cell Biol 23: 242–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Wu S, Li N, Chen Y, Yan K, Li Z, Zheng L, Lei J, Woolford JL Jr, Gao N (2017) Structural snapshot of cytoplasmic pre‐60S ribosomal particles bound by Nmd3, Lsg1, Tif6 and Reh1. Nat Struct Mol Biol 24: 214–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malyutin AG, Musalgaonkar S, Patchett S, Frank J, Johnson AW (2017) Nmd3 is a structural mimic of eIF5A, and activates the cpGTPase Lsg1 during 60S ribosome biogenesis. EMBO J 36: 854–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razi A, Britton RA, Ortega J (2017) The impact of recent improvements in cryo‐electron microscopy technology on the understanding of bacterial ribosome assembly. Nucleic Acids Res 45: 1027–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford JL Jr, Baserga SJ (2013) Ribosome biogenesis in the yeast Saccharomyces cerevisiae . Genetics 195: 643–681 [DOI] [PMC free article] [PubMed] [Google Scholar]


