Abstract
During ribosome biogenesis in eukaryotes, nascent subunits are exported to the cytoplasm in a functionally inactive state. 60S subunits are activated through a series of cytoplasmic maturation events. The last known events in the cytoplasm are the release of Tif6 by Efl1 and Sdo1 and the release of the export adapter, Nmd3, by the GTPase Lsg1. Here, we have used cryo‐electron microscopy to determine the structure of the 60S subunit bound by Nmd3, Lsg1, and Tif6. We find that a central domain of Nmd3 mimics the translation elongation factor eIF5A, inserting into the E site of the ribosome and pulling the L1 stalk into a closed position. Additional domains occupy the P site and extend toward the sarcin–ricin loop to interact with Tif6. Nmd3 and Lsg1 together embrace helix 69 of the B2a intersubunit bridge, inducing base flipping that we suggest may activate the GTPase activity of Lsg1.
Keywords: eIF5A, LSG1, NMD3, ribosome biogenesis
Subject Categories: Protein Biosynthesis & Quality Control, RNA Biology, Structural Biology
Introduction
The ribosome is a remarkably intricate and dynamic machine whose assembly requires the precise processing and folding of its RNA along with the incorporation of numerous ribosomal proteins. Because the mature ribosome and its attendant translation factors are responsible for decoding the cell's genome during mRNA translation, the correct assembly of this complex machine is critical for faithful gene expression.
The eukaryotic ribosome comprises a large (60S) and a small (40S) subunit. Their synthesis begins with rRNA transcription in the nucleolus where initial assembly of the SSU processome promotes co‐transcriptional processing and folding to form the 90S particle (Dragon et al, 2002; Grandi et al, 2002; Woolford & Baserga, 2013; Gerhardy et al, 2014; Kornprobst et al, 2016). An early cleavage event releases the pre‐40S from the assembling pre‐60S subunit (Osheim et al, 2004; Koš & Tollervey, 2010; Karbstein, 2013; Woolford & Baserga, 2013; Gerhardy et al, 2014; Henras et al, 2015). The two subunits then follow independent paths of additional assembly events, nuclear export, and final maturation in the cytoplasm (Tschochner & Hurt, 2003; Zemp & Kutay, 2007; Panse & Johnson, 2010; Thomson et al, 2013). Export of the 60S subunit in eukaryotes is strictly dependent on Nmd3, a highly conserved protein whose C‐terminal leucine‐rich nuclear export signal recruits the export receptor Crm1 (Ho et al, 2000; Gadal et al, 2001; Thomas & Kutay, 2003; Trotta et al, 2003). Additional factors, including Arx1 and the mRNA export factor Mex67‐Mtr2, also contribute to export in yeast (Bradatsch et al, 2007; Yao et al, 2007; Hung et al, 2008). Although hundreds of transiently interacting factors, many of which are essential proteins, orchestrate the cascade of assembly events from the nucleolus to the cytoplasm (Nissan et al, 2002; Fromont‐Racine et al, 2003; Strunk & Karbstein, 2009; Kressler et al, 2010; Thomson et al, 2013; Woolford & Baserga, 2013), the majority of these factors are released prior to nuclear export (Gerhardy et al, 2014). Thus, the complexity of the pre‐ribosomal particles that enter the cytoplasm is significantly reduced.
The final maturation of 60S subunits in the cytoplasm involves the release and recycling of trans‐acting factors and the incorporation of the remaining ribosomal proteins (reviewed in Zemp & Kutay, 2007; Panse & Johnson, 2010; Karbstein, 2013), resulting ultimately in their functional activation. These events can be ordered into a pathway that reveals a hierarchical dependence on ATPase‐ and GTPase‐driven maturation steps (Lo et al, 2010). The final known steps in cytoplasmic maturation include assembly of the P stalk (Kemmler et al, 2009; Lo et al, 2009) and subsequent release of the ribosome anti‐association factor, eIF6 (Tif6 in yeast), and the nuclear export factor Nmd3. The GTPase Efl1, a paralog of the translation elongation factor EF2, together with the protein Sdo1, promote the release of Tif6 (Senger et al, 2001; Weis et al, 2015). Because Tif6 sterically blocks association of the 40S subunit (Russell & Spremulli, 1980; Gartmann et al, 2010), it must be removed to allow assembly of translationally active 80S ribosomes (Raychaudhuri et al, 1984). We and others have proposed that this release event represents a quasi‐functional “test drive” of the nascent 60S subunit in which protein mimics of translation factors assess functionality of newly made subunits prior to their release into the translating pool (Bussiere et al, 2012; Weis et al, 2015). Although Nmd3 functions in nuclear export, it remains associated with pre‐60S particles until late in cytoplasmic maturation. Its release depends on the assembly of uL16 (Rpl10) into the subunit to complete the peptidyl‐transferase center (Bussiere et al, 2012) and assembly of eL40 (Fernández‐Pevida et al, 2012). In addition, depletion of Sdo1 or Efl1 leads to accumulation of both Tif6 and Nmd3 on subunits (Lo et al, 2010; Finch et al, 2011), suggesting that Nmd3 is released in concert with or after Tif6. Thus, at the time of release of Nmd3 by the GTPase Lsg1 (Hedges et al, 2005), all known assembly events on the 60S subunit have been completed.
Although molecular genetic studies have identified the functions of many individual ribosome biogenesis factors and the order of events, the way these factors collaborate to assemble ribosomes has only begun to be unraveled by recent structural studies. High‐resolution cryo‐electron microscopy (EM) of native nuclear pre‐ribosomal particles has revealed the architecture of several assembly intermediates, delineating the structures and binding sites for numerous trans‐acting factors (Bradatsch et al, 2012; Bussiere et al, 2012; Greber et al, 2012; Leidig et al, 2014; Kornprobst et al, 2016; Murray et al, 2016; Wu et al, 2016). These structures display remarkable complexity of interactions among transacting factors and the pre‐60S subunit, as well as unexpected insights to RNA rearrangements that must occur during ribosome assembly (reviewed in Greber, 2016). Among the factors on the nuclear pre‐60S subunit is the essential nuclear GTPase Nog2 (also named Nug2) as well as numerous shuttling factors that are exported with the pre‐60S subunit, including eIF6 and Arx1. Structures have also been determined for complexes of eIF6 and Arx1 reconstituted with mature 60S subunits (Klinge et al, 2011; Greber et al, 2016). However, thus far, these structures have lacked the export adaptor, Nmd3. UV cross‐linking studies have suggested that Nmd3‐rRNA contacts overlap with the Nog2 binding site (Matsuo et al, 2014). Thus, recruitment of Nmd3 is thought to be regulated by the release of Nog2 as a checkpoint for nuclear export (Matsuo et al, 2014). While the structures of these nuclear pre‐ribosomal intermediates have provided important insights into assembly mechanisms and structural transitions that take place prior to export, less is known about the events of cytoplasmic maturation of pre‐60S particles. However, recent cryo‐EM of partially reconstituted cytoplasmic 60S with eIF6, Sdo1, and Efl1 identified Sdo1 as a ribosome recycling factor‐like protein that binds in the P site to engage and activate the GTPase activity of Efl1 (Weis et al, 2015).
Here, we have focused on the late cytoplasmic 60S‐Nmd3‐Lsg1 complex. Whereas many earlier ribosome biogenesis steps involve rearrangements of RNA and proteins that are not amenable to reconstitution, we reasoned that the 60S‐Nmd3‐Lsg1 complex, representing the completed subunit before release of Nmd3, could be faithfully reconstituted in vitro. We present a three‐dimensional (3D) reconstruction of the yeast proteins Nmd3, Lsg1, and Tif6 in complex with the 60S subunit, determined by single‐particle cryo‐electron microscopy. 60S‐Nmd3 and 60S‐Nmd3‐Lsg1‐Tif6 complexes were resolved to 3.1 and 3.3 Å, respectively. Lsg1 is an active GTPase in this complex and requires the presence of both the 60S subunit and Nmd3. Thus, this reconstituted complex likely captures the salient features of Nmd3 and Lsg1 binding.
Results
Reconstitution of a 60S‐Nmd3‐Lsg1‐Tif6 complex
We reconstituted complexes of yeast 60S subunits with combinations of Nmd3, Lsg1, and Tif6 from purified components in vitro. Nmd3 was purified from yeast as a fusion to maltose‐binding protein (MBP). Lsg1 was purified from E. coli with a C‐terminal 6xHistidine tag. Lsg1 belongs to the family of circularly permuted GTPases (cpGTPases) in which the G motifs are reordered (Reynaud et al, 2005; Anand et al, 2006). Although it has previously been reported that free human Lsg1 is an active GTPase in vitro (Reynaud et al, 2005), this had not been shown for the yeast protein. We found that free yeast Lsg1 protein had no detectable GTP hydrolysis activity (Fig 1A). The GTPase activity was not stimulated by Nmd3 alone and only modestly stimulated by 60S subunits. However, in the presence of both 60S subunits and Nmd3, we observed robust GTPase activity (Fig 1A). Neither Nmd3 nor free 60S subunits had significant GTPase activity themselves, demonstrating that the increased GTP hydrolysis by Lsg1 resulted from the stimulation of the Lsg1 GTPase center by the 60S subunit and Nmd3 together, and was not an additive effect of the individual components. In the presence of Nmd3 and 60S subunits, a minimum estimate of the rate of GTP hydrolysis by Lsg1 was 190 min−1 (Appendix Fig S1), comparable to, but higher than, the turnover rate of 14 min−1 reported for the bacterial cpGTPase RbgA in the presence of 50S subunits (Achila et al, 2012). Unlike previously characterized cpGTPases (Achila et al, 2012; Ash et al, 2012; Matsuo et al, 2014), Lsg1 was also not stimulated by increased concentrations of potassium, either as free protein or in the presence of 60S and Nmd3 (Appendix Fig S1B). Together, these results demonstrate that the reconstituted 60S‐Nmd3‐Lsg1 complex is active.
Figure 1. Lsg1 and Nmd3 form an active complex with the 60S subunit.
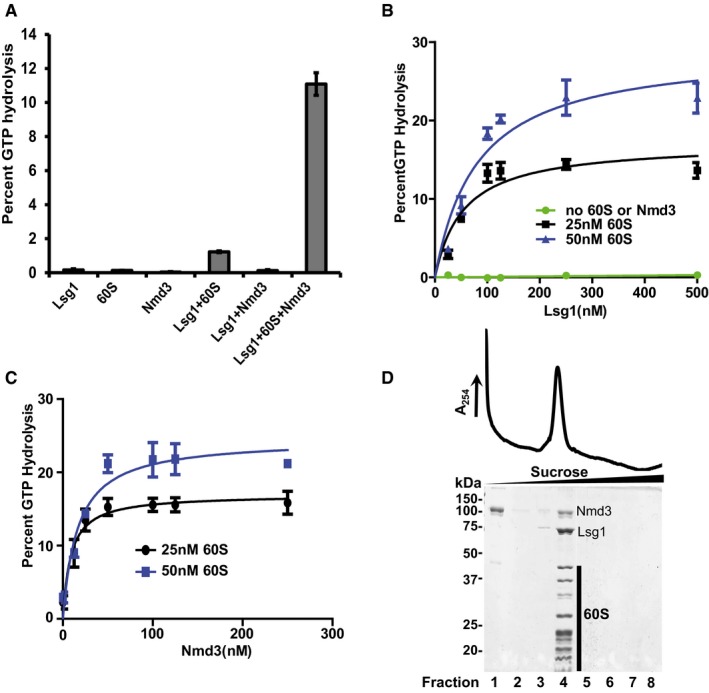
- Percent GTP hydrolysis in reactions containing 125 nM Lsg1, 100 nM Nmd3, or 25 nM 60S alone, or in the indicated combinations, was determined by monitoring the release of free phosphate as described in Materials and Methods.
- Representative curves for percent GTP hydrolysis by increasing concentrations of Lsg1, as indicated in the figure, without Nmd3 or 60S subunit (green) or reactions containing 100 nM Nmd3 with 25 nM (black) and 50 nM (blue) 60S.
- Representative curves for percent GTP hydrolysis by 125 nM Lsg1 in reactions containing 25 nM (black) and 50 nM (blue) 60S subunits, and increasing concentrations of Nmd3.
- Migration of 60S‐Nmd3‐Lsg1 complex (stoichiometry of 1:4:5) in the presence of GMPPNP after sedimentation through 10–30% sucrose. Fractions were precipitated with 10% TCA and analyzed on 10% SDS–PAGE. MBP‐(TEV)‐HIS6‐Nmd3 and Lsg1‐6His and 60S subunit proteins are indicated.
To ensure sufficient occupancy of Lsg1 and Nmd3 on 60S subunits for 3D reconstruction, we performed titration experiments, using the dependence of the Lsg1 GTPase on both Nmd3 and 60S subunits as a proxy for protein occupancy. We reasoned that monitoring Lsg1 GTPase activity would report on Lsg1 binding to a specific site on the 60S subunit and avoid issues that could arise from non‐specific binding. We titrated Lsg1 at two different 60S subunit concentrations, keeping Nmd3 constant (Fig 1B). At each 60S concentration, GTP hydrolysis approached a maximum as Lsg1 concentration was increased, suggesting that Lsg1 approached saturation. Near‐saturation with Lsg1 was achieved at ~10 Lsg1 molecules per 60S subunit. A similar titration of Nmd3 at constant 60S subunit concentration and two concentrations of Lsg1 showed saturation with Nmd3 at a ratio of ~4 Nmd3 molecules per 60S subunit (Fig 1C). GTP hydrolysis data in Fig 1B and C were fitted to saturating‐specific single‐site binding curves. Parabolic standard slope saturating curves indicated that both Lsg1 and Nmd3 bind to single sites in the 60S‐Nmd3‐Lsg1 complex. Sucrose density gradient sedimentation of 60S‐Nmd3‐Lsg1 complexes in the presence of the non‐hydrolyzable GTP analog guanosine 5′‐[β,γ‐imido]triphosphate (GMPPNP) confirmed that 60S, Nmd3 and Lsg1 co‐sedimented as a stable complex (Fig 1D).
Structure determination of Nmd3‐containing complexes
To better understand the function of Nmd3 and Lsg1 in the assembly of the 60S subunit, we utilized cryo‐EM to characterize the structures of 60S‐Nmd3, 60S‐Nmd3‐Lsg1, and 60S‐Lsg1 complexes. Lsg1‐containing complexes were prepared in the presence of GMPPNP. To improve the distribution of particle orientations in ice, 0.5% w/v of glutaraldehyde was added to complexes prior to freezing.
Refined maps for 60S‐Nmd3 and 60S‐Nmd3‐Lsg1 complexes contained densities not present on the mature 60S subunits alone. However, 60S‐Lsg1 maps showed no extra density compared to the mature 60S subunit. Consequently, we focused our effort on the 60S‐Nmd3 and 60S‐Nmd3‐Lsg1 complexes. In the 60S‐Nmd3 complex, a mass of density could be seen between the A site and the SRL, but only at low thresholds, suggesting a high degree of flexibility of this domain of Nmd3. This region was better defined in the presence of Lsg1 and appeared to project toward the position that Tif6 would occupy on the 60S subunit. Because Tif6 and Nmd3 are both present on the same pre‐60S particles in vivo (Lo et al, 2010), we prepared and characterized an additional 60S‐Nmd3‐Lsg1‐Tif6 (60SNLT) complex in order to determine if Nmd3 made contacts with Tif6.
It was apparent that both Nmd3 and the L1 stalk existed in multiple conformations in these complexes, with the L1 stalk ranging from open to closed. In the closed and intermediate states, additional density could be seen in contact with the L1 stalk. To separate the various states and improve resolution of Nmd3 and Lsg1, we used 3D classification with signal subtraction (Bai et al, 2015). The best maps were achieved with the stalk in the closed state, primarily due to high representation of this state in our particles. A 60SNLT map was refined to 3.3 Å resolution from ~19,000 particles (Appendix Table S1).
To further improve the quality of the Nmd3 density, datasets from 60S‐Nmd3, 60S‐Nmd3‐Lsg1, and 60SNLT were combined into a single set after 3D classification of individual runs, resulting in a total of 226,516 particles. The combined 60S‐Nmd3 dataset resulted in an overall map at 3.1 Å resolution, with interior 60S subunit regions reaching resolutions better than 3 Å (Appendix Fig S2). See Appendix Figs S3–S8 and Appendix Table S1 for classification and refinement strategies.
Comparing the maps for 60S‐Nmd3 and 60SNLT to that of a mature 60S subunit, we were able to assign densities not present on the mature 60S subunit to Nmd3, Lsg1, and Tif6 (Fig 2A). Although we reconstituted complexes using Nmd3 with an N‐terminal MBP tag, we did not observe any density for MBP, indicating that MBP did not adopt any specific position in the reconstituted particle. To gain greater insight into the function of Nmd3 and its interaction with the subunit, we sought to assign amino acid sequence to the density. However, there were no structures for Nmd3‐family proteins, and computer‐generated models did not predict structures that corresponded to the observed density. Fortunately, the majority of Nmd3 density in the combined 60S‐Nmd3 map that we observed between uL1 and the A site was of sufficiently high resolution to allow assignment of 254 amino acids, A147 to K401 (Figs 2A and C, and 3A). The 60SNLT complex was used to build a portion of the remaining N‐terminal residues from 40 to 146. The C‐terminal 117 amino acids of Nmd3, containing the nuclear import and export signals (Hedges et al, 2006), were not resolved in our structure.
Figure 2. Structure of the 60S‐Nmd3‐Lsg1‐Tif6‐GMPPNP complex.
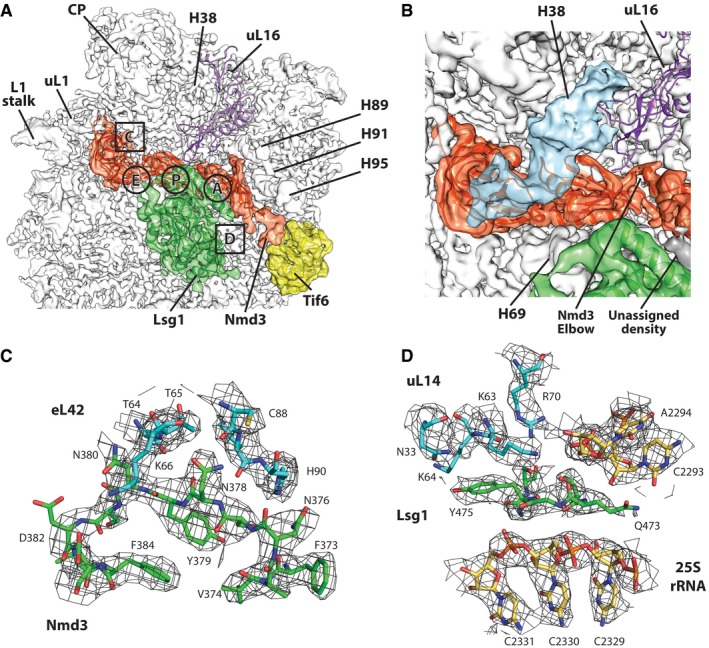
- Cryo‐EM reconstruction of yeast 60S subunit (white) in complex with Nmd3 (orange), Lsg1 (green), and Tif6 (yellow). A site, P site, and E site and regions of the density map shown in detail in panels (C) and (D) are indicated.
- Zoomed view of reconstruction from a class of particles with a deformed H38 (shown in blue). The elbow region of Nmd3 connecting the eL22‐like and N‐terminal domains and an unassigned density connecting Lsg1 and Nmd3 are also shown.
- Region extracted from density map in (A) showing the interface region of Nmd3 and eL42.
- Region extracted from density map in (A) showing density for Lsg1 wedged underneath uL14.
Figure 3. The structure of Nmd3.
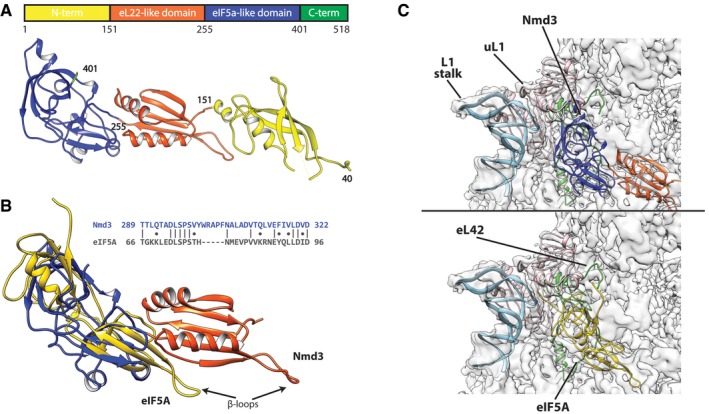
- Linear map of Nmd3 and atomic structure colored on proposed domains. Amino acid positions are given by numbers.
- Sequence and structural alignment of eIF5a (golden) with eIF5a and eL22‐like domains of Nmd3.
- Comparison of Nmd3 from the 60S‐Nmd3‐Lsg1 complex (upper panel, colored as in A) and eIF5A (PDB: 5gak) (lower panel, golden) bound to the 60S subunit. The L1 stalk (light blue), uL1 (pink), and eL42 (green) are highlighted.
Nmd3 spans the joining face of the 60S subunit from Tif6 to the L1 stalk
In both 60S‐Nmd3 and 60SNLT complexes, the primary state for the 60S subunit was with the L1 stalk in the closed conformation and Nmd3 spanning the 60S subunit from the uL1 protein on the L1 stalk through the E site, the P site and extending toward Tif6 at the sarcin–ricin loop (SRL). The position of Nmd3 in our reconstituted particles was remarkably similar to that of unidentified densities in the recently described structure of a Yvh1‐containing pre‐60S particle (Sarkar et al, 2016). In that work, density attributed to Nmd3 was observed spanning from Tif6 to the P site, while additional density between the L1 stalk and the P site was tentatively attributed to Lsg1. Based on the structure presented here, we can attribute both of the unassigned densities in the Yvh1‐particle to Nmd3 alone.
Overall, the assigned Nmd3 sequence can be separated into three domains (Fig 3A). The N‐terminal domain is composed of a long four‐stranded beta sheet and two alpha helices, and spans from Tif6 toward the P site (SRL; helices 89, 91, 92, and 95). Although the extreme N‐terminus of Nmd3 (residues 1–39) was not of sufficient resolution to assign amino acids, this region of Nmd3 appears to interact directly with Tif6 (Fig 2A). Possibly, the MBP fusion destabilizes the Nmd3‐Tif6 interaction, leading to reduced resolution of the extreme N‐terminus of Nmd3. Because we could resolve the N‐terminal domain of Nmd3 only in the presence of Tif6, the interaction of Tif6 appears to stabilize the N‐terminus of Nmd3. Consistent with this stabilization, the addition of Tif6 modestly enhanced the Nmd3‐stimulated Lsg1 GTPase activity (Appendix Fig S1C).
The second domain of Nmd3, spanning residues 152 through 255, is composed of a five‐stranded beta sheet and two alpha helices, and contains a long flexible loop (Fig 3B). This domain occupies the ribosomal P site in the closed state of the L1 stalk. In this state, the flexible loop (V229–V240) is positioned directly above and covering the peptide exit tunnel. We compared the fold of this domain to known structures using the DALI server and found that it adopts a similar fold as that of ribosomal protein eL22 (Z = 5.5, r.m.s.d. 3.0 Å, PDB: 4uk1, chain H). Thus, we refer to this domain as eL22‐like. Despite the similarity of folds, we do not suggest functional relatedness between eL22 and Nmd3.
At the elbow‐like junction of the N‐terminal and eL22‐like domains, Nmd3 closely approaches the peptidyl (P) site loop of uL16 (Fig 2B). This flexible loop of uL16 is required for the release of Nmd3 from the nascent subunit (Bussiere et al, 2012). Mutations at the base of this loop (R98S, R98C, H123P) are associated with T‐cell acute lymphoblastic leukemia in humans, and they also impair the release of Nmd3 and Tif6 (De Keersmaecker et al, 2013). This loop of uL16 is stabilized by P site ligands but is unstructured in our model. Extension of the loop into the P site, as observed for mature ribosomes in the presence of tRNA or eIF5A (Schmidt et al, 2016), would be precluded in our structure due to steric clash with Nmd3. As a consequence, the P site loop of uL16 is not resolved and must be displaced above the elbow region at A147 of Nmd3.
The third domain of Nmd3 (aa 256–401) interacts with uL1 of the L1 stalk and eL42. Thirty of the assigned amino acids of Nmd3 are at the interface with uL1, resulting in a buried surface area of ~577 Å2 (Fig 3C). However, due to poor resolution of uL1 density, assignment of amino acids of uL1 could not be performed reliably. Nmd3 shares an interface of 744 Å2 with eL42, composed of 24 residues of Nmd3 and 21 residues of eL42 (Figs 2C and 3C). This domain of Nmd3 interface is stabilized by a number of predicted hydrogen bonds and salt bridges (Appendix Table S2; PDBePISA server).
A domain of NMD3 resembles eIF5A in both structure and mode of binding
Euryarchaeal Nmd3 proteins have previously been annotated as containing an eIF5A‐like domain, based on sequence comparison (Aravind & Koonin, 2000). However, no sequence similarity between eukaryotic Nmd3 and eIF5A proteins had been reported. Our structure shows that the third domain of Nmd3 (aa 256–401) adopts the same topology as that of eIF5A (Dali: Z = 3.5, r.m.s.d 4.0 Å, PDB: 5gak‐q). In addition, we detected limited sequence similarity between residues 289–326 of Nmd3 and 66–97 of eIF5A (Fig 3B). We will refer to this domain as the eIF5A‐like domain of Nmd3.
eIF5A and its bacterial homolog EF‐P are necessary for the rescue of ribosomes stalled on polyproline‐containing sequences and function by recognizing an empty E site (Doerfel et al, 2013; Gutierrez et al, 2013; Ude et al, 2013). eIF5A is located in the tRNA path of the 60S subunit between the E and P sites (Melnikov et al, 2016). eIF5A interacts with uL1, inducing a closed conformation of the L1 stalk (Schmidt et al, 2016). This closed conformation is additionally stabilized by interactions between the N‐terminal extension of eIF5A and eL42 (Fig 3C, lower panel). The position of Nmd3 in the tRNA path, its interaction with eL42 and uL1, and its influence on inducing a closed position of the L1 stalk are remarkably similar to the interaction of eIF5A with the 60S subunit (Fig 3C, Movie EV1).
An essential element of eIF5A is the extended β3–β4 loop that carries hypusine and interacts with the acceptor stem of P site tRNAs (Wolff et al, 2007). The corresponding loop in Nmd3 (determined by structural alignment of eIF5a‐like domain and eIF5A) is severely truncated, lacks the corresponding lysine of eIF5A that is modified to hypusine, and does not protrude beyond the core of the domain. eIF5A contains an N‐terminal extension that emanates from a tight turn and folds back along the protein toward the L1 stalk (Melnikov et al, 2016; Schmidt et al, 2016). In Nmd3, the corresponding strand projects forward, toward the P site, and contributes to the five‐stranded beta sheet in the eL22‐like domain. Interestingly, this strand supports a flexible loop (aa 229–239) that extends beyond the core of the eL22‐like domain, reminiscent of the hypusine‐containing β3–β4 loop of eIF5A. However, this loop of Nmd3 does not contain a lysine that could be a substrate for modification.
Nmd3 and the L1 stalk exhibit multiple conformations
Although our highest‐resolution maps were from particles with the L1 stalk in the closed conformation, we observed additional states of the L1 stalk. We refined several small subsets into low‐resolution densities displaying various conformations of L1 and Nmd3 using focused classification with signal subtraction to improve the EM densities by removing particles of alternative conformations (see Appendix Figs S3–S8 for classification strategies, Appendix Table S1 for description of classes). Four primary states were observed for the L1 stalk: closed, open, and two intermediate positions (Fig 4A). Nmd3 density becomes progressively less defined as the L1 stalk moves from the closed to the open state and completely disappears in the open state. The N‐terminal domain of Nmd3 can only be observed in maps with L1 in the fully closed state and disappears completely as the L1 stalk transitions toward the open state. In the intermediate positions of the L1 stalk, we were able to rigid body‐fit the uL1‐Nmd3 structure of the closed complex into the densities. In the different states, the L1 stalk appeared to lift Nmd3 as a single, rigid unit, as both uL1 and Nmd3 maintained the same relative configuration (Fig 4B). However, it is possible that the eL22‐like domain of Nmd3 could adopt a different relative conformation in the presence of a P‐site ligand.
Figure 4. The L1 stalk and Nmd3 adopt multiple states.
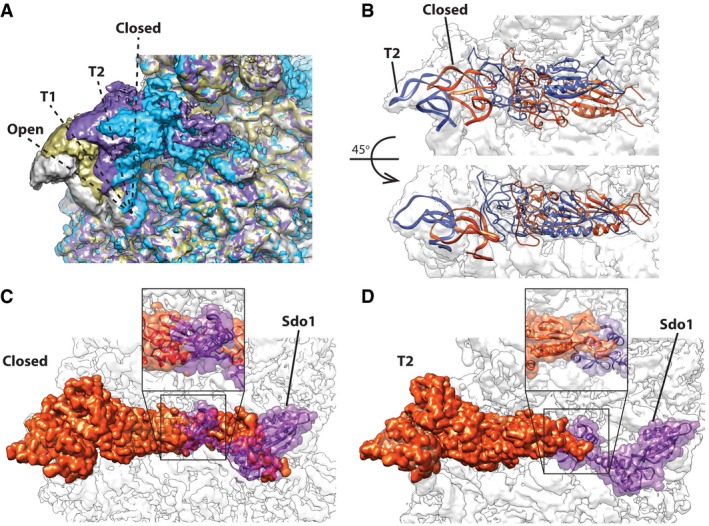
-
AOverlap of the four observed states for L1 stalk/Nmd3. T1 (tan) and T2 (purple) are transition states between the fully open (gray) and fully closed (blue).
-
BRigid‐body docking of uL1‐L1‐Nmd3 into T2 state, showing the atomic model overlap between the closed (orange) and T2 position (blue).
-
C, DModel for coordination of Nmd3 action with Sdo1. (C) In the closed conformation of L1 stalk/Nmd3, the eL22‐like domain of Nmd3 occupies the P site where it would clash with domain I of Sdo1. (D) In transition state T2, the clash between Nmd3 and Sdo1 in the P site is greatly reduced, potentially allowing for the accommodation of Sdo1.
During 60S subunit maturation, Sdo1 binds in the P site to activate Efl1 for the release of Tif6. We note that in the closed state of the L1 stalk, the eL22 domain of Nmd3 is fully engaged with the P site where it would sterically clash with the N‐terminal domain of Sdo1 (Fig 4C). However, the N‐terminal domain of Sdo1 could be accommodated in the P site in the T2 state (Fig 4D). These observations suggest dynamic movement of Nmd3 and the L1 stalk to allow Sdo1 binding (see Discussion).
Helix 38 exhibits multiple conformations in the presence of Nmd3
Density between the eIF5A‐like domain of Nmd3 and the central protuberance (CP) of the large subunit (Fig 2B) was initially attributed to the C‐terminal domain of Nmd3, as the maps showed no obvious changes in conformation of the Nmd3‐bound 60S subunit relative to the mature subunit. However, after several rounds of classification (Appendix Fig S8), a subset of particles was isolated showing a clearly distorted H38 merging into the unassigned density with a well‐resolved RNA‐helical motif (Fig 2B). This interaction between H38 and Nmd3 is consistent with UV cross‐linking data (Appendix Fig S9; Matsuo et al, 2014). In addition to the distortion of H38, which contributes to the binding pocket of uL16, the density for uL16 residues 1–32 and 95–124 is poorly resolved, suggesting destabilization of uL16 due to the movement of H38 in the presence of Nmd3.
Lsg1 binds to the joining face of the 60S subunit
In 60S‐Nmd3‐Lsg1 and 60SNLT complexes, Lsg1 density is found on the intersubunit surface of the 60S subunit, in close contact with helices 61, 62, 64, 69, 71, and 92 (Fig 2A). The Lsg1 density showed little direct interaction with biogenesis factors or ribosomal proteins, reflecting the paucity of ribosomal proteins on the joining face of the subunit. Despite the role of Lsg1 in Nmd3 release, we did not observe a direct connection between the resolved densities of Lsg1 and Nmd3 in our maps. However, an unidentified density above helix 71 appears to originate within Lsg1 and joins the N‐terminal domain of Nmd3 near the SRL (Fig 2B). Whereas no direct connection between Lsg1 and Tif6 was observed, residues 473–477 of Lsg1 wedge between the tip of helix 71 and K64 of uL14 (Fig 2D), a binding partner of Tif6. In our structure, we noticed the absence of eL41, a short 25‐residue‐long, helical protein that binds adjacent to H69 and would be positioned beneath Lsg1. eL41 is a non‐essential protein that makes only modest contacts with the 60S subunit. eL41 peptide did not stimulate the GTPase activity of Lsg1 (Appendix Fig S10A), deletion of RPL41A and RPL41B had no apparent growth defect, and we did not observe synthetic genetic interaction between a lsg‐R208G, which has a mild temperature‐sensitive defect, and deletion of RPL41A and RPL41B (Appendix Fig S10B). Together, these results suggest that eL41 does not play a role in Lsg1 function.
The Lsg1 protein density was resolved in the range of 3.3–5 Å (Appendix Fig S2D), which allowed us to identify secondary structure elements, as well as to fit and refine an in silico model generated using I‐TASSER server (Fig 5). The model accounts for residues 135 through 503 of Lsg1. Although the N‐ and C‐termini of Lsg1 are predicted to be largely unstructured and flexible and the densities are absent in our map, most of the observed density is of sufficient resolution to confirm the fits of large amino acids. Lsg1 is a circularly permuted GTPase, with the sequence motifs appearing in the order of G4‐G5‐G1‐G2‐G3 (Fig 5A). Nevertheless, the overall structure of the active site is that of a classical GTPase (Fig 5A and B). Additionally, the map contains density attributable to GMPPNP, located in the same pocket as would be expected from other GTPases. Structurally, the GTPase domain of Lsg1 is closely related to GTP‐bound YlqF (Kim et al, 2008) as determined by the PDBeFold server (Appendix Fig S11). The Q score for the alignment of residues 184–503 of Lsg1 to GTP‐bound states of YlqF (PDB: 3cnn) is 0.39, the Z‐score is 5.6, and the root mean square deviation is 2.75 Å. DALI server structural comparison also confirmed structural similarity with YlqF (Z = 12.3) and, to a lesser degree, with GTPases YqeH (Z = 7.4) and EngC (YjeQ) (Z = 6.3).
Figure 5. The structure of Lsg1.
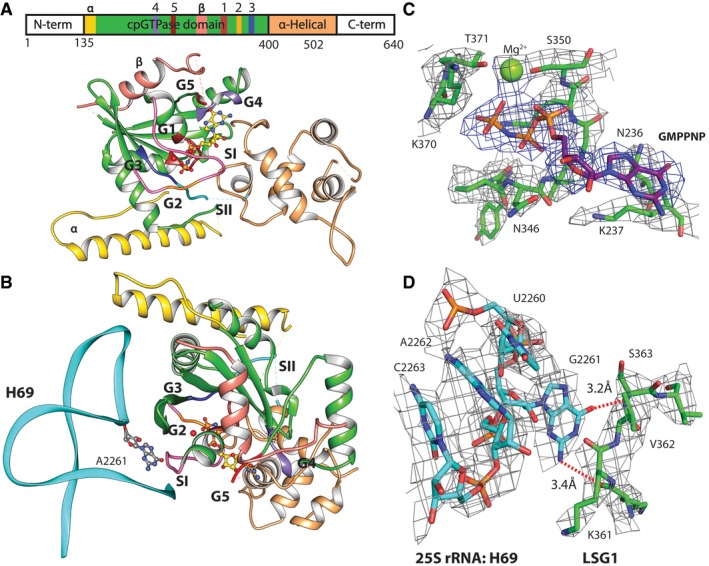
- Domain view of Lsg1 in the presence of GMPPNP. Sequence motifs G1–G5 and Switch regions are shown in different colors. G1, red; G2, orange among the pink of Switch I; G3, blue; G4, purple; G5, brown; and Switch II in teal. Residues 135–175 are shown in yellow and marked with “α”. Extra insertion in the cpGTPase domain is shown in salmon and marked with “β”, and the α‐helical domain is shown in tan. Linear sequence map shows all of the domains and G motifs in the corresponding colors.
- Lsg1 as in (A), but rotated around the horizontal axis and with helix 69 showing flipped‐out G2261.
- Density of the GMPNP pocket shown at 4 sigma and Pymol carve level of 2.4 Å.
- Map density around G2261 and opposing residues of Switch I, distances to possible backbone contacts are shown with red dashed lines. Density shown at 4 sigma and Pymol carve level of 2 Å.
The G4 motif spans residues N236–L240 and is located near the guanine base of the ligand (Fig 5C and D). Nucleotide‐binding is stabilized via hydrogen bonding with N236 and D239 of the G4 box. The G5 motif (SAL) is within the vicinity of the guanosine of the GMPPNP, but does not appear to interact directly with it. L266 of the G5 motif and K237 of G4 appear to form a hydrophobic pocket around the guanine nucleotide, while S264 appears to interact with O6 of the GNPPNP and D239 of the G4 motif. The G1 motif consists of residues 343–350 (GYPNVGKS) and creates a pocket for the phosphate groups of GMPPNP. We have previously found that mutations in K349 or S350 inactivate the GTPase in vivo (Hedges et al, 2005). The main‐chain nitrogen and oxygen atoms of the G1 motif make hydrogen bonds with the phosphate atoms of the ligand. K349 interacts with the nitrogen between β‐ and γ‐phosphates and the oxygen of the γ‐phosphate. The G2 motif corresponds to residues 369–371 (GKT) within the Switch I region 368–375. The backbone oxygen of G369 appears to be stabilizing the γ‐phosphate. We also observe extra density located above the γ‐phosphate of GMPPNP, likely corresponding to a coordinating Mg2+ ion (Anand et al, 2006). Threonine 371 of G2 interacts with this ion, presumably stabilizing it above the O3 of the γ‐phosphate (Fig 5D). The G3 motif DCPG (aa 387–390) covers the Switch II region (389–391). No direct interaction of Switch II with the GMPPNP is observed. We also do not observe density corresponding to the expected K+ ion that stabilizes Switch I in this family of GTPases (Anand et al, 2013); however, the absence of K+ may be due to the use of the GTP analog GMPPNP, which has been reported to destabilize the coordination shell of the monovalent ion (Kuhle & Ficner, 2014).
Lsg1 distorts Helix 69
Unlike in 60S‐Nmd3 complexes without Lsg1, helix 69 is distorted in the presence of Lsg1, having undergone a shift of about 6–12 Å from its canonical position (PDB: 4v88), toward the L1 stalk, and a bend toward Nmd3. In addition to the distorted position of H69, another striking feature of Lsg1‐containing complexes is the displacement of guanosine 2261 of helix 69, which is flipped out in the presence of Lsg1 and makes backbone contacts to residues 361 and 362 between the Switch I and G1 box of Lsg1 (Fig 5B and D). The binding of Lsg1 distorts H69 by pushing U2260 into the helix where it stacks on A2262, thereby evicting G2261. This interaction may help anchor Switch I in a GTP‐binding conformation. Switch I and the loop connecting it to the G1 motif are in direct contact with the ribosomal RNA and are located above helices 69 and 71, making these regions likely sensors of the correct positioning of Lsg1 on the subunit for activation of its GTPase.
Relative to YlqF and other cpGTPases, Lsg1 displays an insertion in the GTPase domain spanning residues 266–338. This is a flexible subdomain that is not well resolved in our density, located within close proximity of the tip of helix 69, and potentially helping to anchor Lsg1 on the 60S subunit (Fig 5A and B). The general shape of this subdomain can be traced, but the long flexible loops prevent accurate sequence assignment to this region. Similarly, a beta‐strand and an alpha‐helical density, putatively assigned to residues 135–175, make contact with the tip of helix 69. However, the resolution of this segment is not sufficient to assign the sequence to the residues (Fig 5A and B).
The C‐terminal α‐helical domain of Lsg1 is structurally similar to the C‐terminal domain of YlqF (Appendix Fig S11; Kim et al, 2008). Many members of cpGTPases families are predicted to contain additional C‐terminal domains, such as α‐helical or zinc‐binding, that potentially assist in anchoring Switch II in correct configuration (Anand et al, 2006). Residues 491–497 of Lsg1 are in close proximity to the guanine nucleotide and might participate in stabilizing GTP binding. Residues 410 through 417, 436 through 442, and 467 through 492 are located above the rRNA and likely help in the binding and stability of Lsg1 on the 60S subunit, as was predicted for the C‐terminal domain of YlqF family proteins.
Discussion
Nmd3 is a molecular mimic of eIF5A
Recent structural analyses have revealed that several ribosome biogenesis factors interrogate the ligand binding sites of the pre‐60S subunit. These include Nog1 and Rei1, which sequentially probe the polypeptide exit tunnel from the solvent surface of the subunit (Weis et al, 2015; Greber et al, 2016; Wu et al, 2016); Sdo1, which interrogates the P site and inserts into the exit tunnel from the joining face of the subunit; and Efl1, which samples the GTPase‐activating center (Weis et al, 2015; Greber et al, 2016; Wu et al, 2016). Among these, Efl1 is a close paralog of a bona fide translation factor, suggesting the evolution of molecular mimicry of translation factors in the biogenesis pathway. This use of molecular mimics during ribosome biogenesis may allow functional partitioning of quality control checks during ribosome assembly from translation, by using biogenesis factors in place of true translation factors. Here, we have extended our understanding of molecular mimicry in ribosome biogenesis by showing that Nmd3 comprises domains that are related to the elongation factor eIF5A.
Nmd3 is a highly conserved protein found throughout archaea and eukarya. It is essential in eukaryotes for its role in nuclear export of the nascent 60S subunit. However, its presence in archaea, which lack nuclei, suggests that it has an additional function more basic to ribosome biology. Sequence analysis revealed that euryarchaeal Nmd3 proteins are fusions of an N‐terminal zinc‐binding domain to a C‐terminal eIF5A‐like domain (Aravind & Koonin, 2000). However, a function for Nmd3 in archaea has not been identified and structures for Nmd3 protein family members have not been reported previously. Eukaryotic Nmd3 proteins contain an N‐terminal zinc‐binding domain, but no sequence homology to eIF5A had been reported. Remarkably, we find that Nmd3 not only contains a domain corresponding to the entirety of eIF5A, but this domain also interacts with the ribosome in a manner similar to eIF5A. The eIF5A‐like domain of Nmd3 occupies the tRNA E site, bound to the L1 stalk and an additional domain of Nmd3 occupies the P site. Thus, while Sdo1 and Efl1 probe the P site, the GTPase‐activating center and the P stalk (Weis et al, 2015), Nmd3 probes the L1 stalk, the E site, and the P site.
In a previous low‐resolution cryo‐EM reconstruction of a Nmd3‐60S subunit complex (Sengupta et al, 2010), we identified MBP‐Nmd3 as a mass on the joining face of the subunit, adjacent to H69, a position where we now find Lsg1. The 60S‐Nmd3 complex is quite flexible, existing in a number of states with the N‐terminal domain apparently stabilized by Tif6, which was not present in the previous work. The conformational heterogeneity of the 60S‐Nmd3 particles used in that work and the lack of robust classification methods at the time could have influenced the reconstruction. Nevertheless, the current position of Nmd3 is consistent with recent UV cross‐linking data for Nmd3 (Appendix Fig S9; Matsuo et al, 2014). In addition, this position of Nmd3 corresponds exactly to unidentified densities in a recent native pre‐60S particle affinity purified with the biogenesis factor Yvh1 (Sarkar et al, 2016), densities that we can now attribute to Nmd3. Lastly, we find that Nmd3 and the 60S subunit together stimulate the GTPase activity of Lsg1. Together, these results strongly suggest that the present structure of a reconstituted 60S‐Nmd3 complex is a faithful representation of the native complex.
Implications for uL16 loading and Nog1 binding
In one class of particles, we observed a severely distorted H38 such that the tip of the helix is unwound and interacts with Nmd3 at the junction of the L22 and eIF5A domains. In the reconstruction from these particles, the density for uL16 was also less well resolved. A similar distortion of H38 and loss of resolution of uL16 was observed in the native Yvh1‐pre‐60S particle (Sarkar et al, 2016). Because Nmd3 assembles onto the ribosome prior to the loading of uL16 (West et al, 2005), the presence of this class of particles raises the interesting possibility that Nmd3 holds open H38 to facilitate uL16 loading. Extensive genetic interactions between NMD3 and RPL10 (encoding uL16) have been noted previously (Karl et al, 1999; Zuk et al, 1999; Hedges et al, 2005). In particular, some mutations in RPL10 can be suppressed by mutations in Nmd3, often in highly allele‐specific interactions. While these interactions have implicated uL16 as being required for the release of Nmd3 (Hedges et al, 2005), the mechanism of suppression has not been fully explored. rpl10‐G161D is a temperature‐sensitive allele that is suppressed by nmd3 mutations L291F, A336P, or I279F (Karl et al, 1999). Aspartate at position 161 would introduce a destabilizing negative charge at the interface of uL16 and 5S rRNA. L291, A336, and I279 are all on the solvent surface of Nmd3 at the interface with the distorted H38 (Fig 2B). Weakening the interaction between Nmd3 and H38 could facilitate the release of H38, allowing it to adopt its mature position in the subunit and promote the accommodation of mutant uL16. Thus, we suggest that Nmd3 not only requires the loading of uL16 for its release but that it may also promote the loading of uL16.
Interestingly, the position of the N‐terminal domain of Nmd3 in the 60SNLT complex is incompatible with the binding of the GTPase Nog1, which binds to pre‐60S particles in the nucleus prior to Nmd3 loading. Nog1 inserts a helical bundle into the A site of the ribosome (Leidig et al, 2014; Wu et al, 2016) which would sterically clash with Nmd3. However, both proteins coexist on late nuclear pre‐60S particles (Kallstrom et al, 2003; Altvater et al, 2012). Thus, the N‐terminal domain of Nmd3 must be highly dynamic, undergoing large‐scale movements during ribosome maturation.
Stimulation of the GTPase activity of Lsg1
The release of Nmd3 from pre‐60S subunits in the cytoplasm is necessary for the production of mature 60S subunits and for the recycling of Nmd3 to the nucleus, to support continued export of ribosomes. The release of Nmd3 requires the GTPase Lsg1, but the mechanism of release and the way in which Lsg1 is activated have not been described. We found that Nmd3 and mature 60S subunits together, but not 60S alone, were sufficient to stimulate the GTPase activity of Lsg1, allowing us to begin to speculate about the mechanism of activation of the GTPase. H69 is a critical element of the 60S joining face that interacts with h44 of the small subunit and forms the B2a intersubunit bridge. Lsg1 binds the joining face of the 60S subunit and embraces H69 to flip out G2261 toward residues of Lsg1 immediately upstream of Switch I. Because Switch I is expected to coordinate a monovalent cation as a structural cofactor in the active site, coupling G2261 to Switch I could provide a means of regulating activation of the GTPase only in the presence of Nmd3 and the 60S subunit. In cpGTPases Switch II, whose positioning is needed for stable interaction of GTP in its binding pocket, is directly connected to the C‐terminal domain of the protein. This arrangement is thought to couple Switch II positioning to proper engagement of the C‐terminus with the ribosome (Anand et al, 2006). Although the C‐terminal regions of Lsg1 resolved in our structure do contact RNA, no obvious rearrangements of these RNA elements were observed, focusing attention on helix 69. While a detailed mutational analysis will be necessary to fully understand activation of Lsg1, it is possible that H69 is a general activation site for other cpGTPases involved in large subunit biogenesis. Previously, it was shown that the GTPase Nog2 is bound in a GTPase‐inactive state in pre‐ribosomes before H69 undergoes a 180° rotation, possibly to engage with Nog2 (Wu et al, 2016). It will be interesting to know whether RbgA, a homolog of Lsg1 that is required for the loading of uL16, has a similar binding site to Lsg1 on the pre‐50S subunit (Britton, 2009; Achila et al, 2012).
The order of release of Nmd3 and Tif6
Previous genetic analysis led us to conclude that the release of Nmd3 by Lsg1 follows the release of Tif6 by Sdo1 and Efl1 (Lo et al, 2010). This conclusion was based on the observations that depletion of Efl1 or Sdo1 traps both Nmd3 and Tif6 on subunits, a block that can be bypassed by mutations in Tif6 (Lo et al, 2010; Finch et al, 2011). In contrast, depletion of Lsg1 traps only Nmd3 and not Tif6 (Appendix Fig S12), and Lsg1 mutants can be bypassed by mutations in Nmd3, but not Tif6. Thus, our genetic analysis places the release of Nmd3 after the release of Tif6 (Lo et al, 2010). However, recent structural analysis of 60S subunits isolated from dominant negative Sdo1‐expressing Dictyostelium revealed a population of particles containing Tif6 but not Nmd3, suggesting that Nmd3 was released prior to Tif6 (Weis et al, 2015). While this may be the case, it is also possible that under some conditions Tif6 rebinds to mature subunits, as it was found to be highly enriched in mature 60S subunits containing the ribosome quality control complex after dissociation of 40S subunits (Shen et al, 2015). Thus, the presence of Tif6 on 60S may reflect a role in subunit recycling separate from biogenesis.
How does the 60S‐Nmd3‐Lsg1‐Tif6 complex inform us about the coordination of the release of Tif6 and Nmd3? In our structures, we observed the L1 stalk in multiple different positions, ranging from open to partially closed to fully closed. Nmd3 interaction with Tif6 appears to stabilize Nmd3 and the L1 stalk in the fully closed state. In this position, the eIF5A domain of Nmd3 is fully engaged with the E site, and the eL22‐like domain of Nmd3 occupies the P site, in a conformation that is incompatible with Sdo1 binding to that site (Fig 5C). In addition, this closed state of the L1 stalk is likely stabilized through Nmd3 interaction with Tif6. Thus, we suggest that the linkage between Nmd3 and Tif6 must be broken to permit the L1 stalk to retract Nmd3 from the P site, allowing Sdo1 to bind (Fig 6). What breaks the linkage between Nmd3 and Tif6? Because we have not observed a role for Lsg1 in the release of Tif6, we suspect that Efl1 may disengage Nmd3 from Tif6 to initiate retraction of Nmd3 and the L1 stalk. Recruitment of Sdo1 to activate Efl1 would then release Tif6, whereas Nmd3 could be released independently, once the linkage with Tif6 is broken. In this model, the release of Nmd3 and Tif6 is coupled rather than linear dependence of Nmd3 release on the release of Tif6.
Figure 6. Model for coordination of Nmd3 and Sdo1 binding.
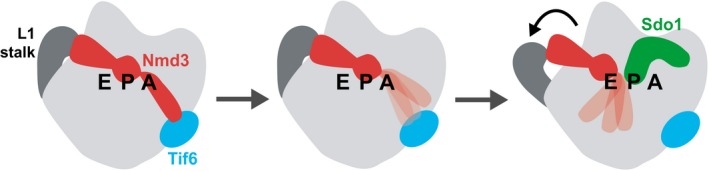
In the fully closed L1 stalk position (left), the eIF5A domain of Nmd3 is engaged with the E site, while the eL22‐like domain occupies the P site. This position is stabilized by the interaction between the N‐terminus of Nmd3 with Tif6. We propose that breaking the linkage between Nmd3 and Tif6 destabilizes the N‐terminus of Nmd3 (middle), allowing the L1 stalk to retract. Nmd3 is moved out of the P site in the retracted position, allowing Sdo1 to bind (right).
Materials and Methods
Strains and plasmids
Yeast strains and plasmids used in this work are listed in Appendix Tables S3 and S4, respectively. AJY3842 was made by amplifying the GFP‐His3MX cassette (Longtine et al, 1998) and integrating into AJY3827. AJY3845 was made by crossing AJY1705 (Hedges et al, 2005) and AJY3827. AJYAJY4073 was made by sequential integrations into rpl41b∆::KanMX (Research Genetics). The His3MX‐P GAL1 ‐3xHA cassette (Longtine et al, 1998) was amplified with homology to the LSG1 locus and integrated into the rpl41b∆ strain. Because the rpl41a∆::KanMX strain was incorrect in our knockout collection from Research Genetics, it was remade in BY4741 using the KanMX cassette (Longtine et al, 1998). The KanMX marker was then replaced with clonNAT resistance (Nat) (Tong & Boone, 2006) to give AJY4066. Finally, rpl41a∆::Nat was amplified from AJY4066 and integrated into the rpl41b∆::KanMX His3MX‐P GAL1 ‐3xHA‐LSG1 strain to give AJY4073. pAJ2229 contained wild‐type LSG1 under control of its native promoter in pRS315. pAJ3021 was made by inverse PCR to introduce R208G, a mildly temperature‐sensitive allele of Lsg1. pAJ3283 was made by cloning TIF6 into pET28b and the amino acid sequence MGSSHHHHHHSLRRASLGS was added to the N‐terminus for purification. pAJ3420 was made by cloning LSG1 into pET21a, and the amino acid sequence AAALEHHHHHH was added to the C‐terminus for purification. Vector sequences will be provided upon request.
Purification of 60S subunits from yeast
Yeast strain AJY2781 was grown in 6 l of YPD to OD600 of 0.6. Cells were washed with Ribo Buffer A (100 mM KCl, 20 mM HEPES‐KOH, pH 7.4, 2.5 mM Mg(OAc)2) and pelleted. Cell pellet was resuspended in 1/3rd pellet weight of lysis buffer (Ribo buffer A with freshly added 1 mg/ml heparin, 2 mM DTT, and protease inhibitors (PIs) (1 mM PMSF and 1 μM each leupeptin and pepstatin)), and dispersed in liquid N2. Cells were lysed in Retsch mixer‐mill, cooled with liquid nitrogen. Frozen grindate was thawed and reconstituted in 2/3rd pellet weight of lysis buffer and cleared by centrifugation at 25,000 g for 25 min. Extract was layered over 2.5‐ml sucrose cushions (Ribo Buffer A supplemented with 500 mM KCl, 1 M sucrose, and 2 mM DTT) and centrifuged for 106 min at 370,000 g in Type 70Ti rotor (Beckman Coulter). The pellet was resuspended in high salt wash buffer (Ribo Buffer A with 500 mM KCl, 1 mg/ml heparin, 2 mM DTT). Insoluble material was removed by centrifugation, and the soluble fraction was layered onto 250 μl sucrose cushions, as above, and centrifuged for 30 min at 541,000 g in a TLA 100.3 rotor (Beckman Coulter). The pellets were resuspended in subunit separation buffer (50 mM HEPES‐KOH, pH 7.4, 2 mM MgCl2, 500 mM KCl, and 2 mM DTT). The suspension was incubated on ice for 15 min and then at 37°C for 10 min after addition of puromycin to 1 mM final concentration. The sample was then centrifuged through 5–20% sucrose gradients (5–20% sucrose, 50 mM HEPES‐KOH, pH 7.4, 5 mM MgCl2, 500 mM KCl, and 2 mM DTT) for 15 h at 71,800 g in an SW32 Ti rotor (Beckman Coulter). Fractions containing the 60S and 40S peaks were pooled separately and concentrated using an Amicon Ultra 15 100 K (Millipore), and buffer was changed to Ribosome storage buffer (Ribo buffer A with 250 mM sucrose and 2 mM DTT). 20 μl of aliquots of subunits was stored at 1 μM each at −80°C.
Protein purification
Lsg1‐6His
One liter of bacterial culture of Codon plus (RIL) cells (Stratagene) with pAJ3420 was grown to OD600 of 0.5 and induced with 1 mM IPTG for 4 h at 30°C. Cells were harvested and washed with Lysis buffer (40 mM Tris, pH 8.0, 500 mM NaCl, and 10% glycerol). The cell pellet was resuspended in 40 ml Lysis buffer (supplemented with 5 mM β‐mercaptoethanol (BME) and 1 mM PMSF and 1 μM each leupeptin and pepstatin) and disrupted by sonication. Lysates were cleared by centrifugation at 20,000 g for 20 min. Imidazole was added to 10 mM. Clarified lysate was bound to a 1 ml Ni‐NTA column (HisTrap HP, GE Healthcare) by pumping at 1 ml/min. The column was first washed with 10 ml lysis buffer supplemented with 10 m imidazole and 5 mM BME and then with low salt buffer (40 mM Tris, pH 8.0, 50 mM NaCl, 10% glycerol, 20 mM imidazole, and 5 mM BME). Bound protein was eluted with low salt buffer with 250 mM imidazole. Fractions containing protein were pooled and bound to 2 ml Source S (GE Healthcare) column pre‐equilibrated with S buffer (40 mM Tris, pH 7.5, 50 mM NaCl, 10% glycerol, and 5 mM BME). Column was washed with 10 ml S buffer. Bound proteins were eluted with a 20 ml linear NaCl gradient (50 mM to 1 M) in S buffer. Fractions containing Lsg1 were pooled and dialyzed in 20 mM Tris, pH 7.5, 150 mM KOAc, 1 mM DTT, and 10% glycerol. Dialyzed protein was stored at −80°C.
MBP‐(TEV)‐HIS6‐Nmd3
One liter of BJ5464 with pAJ1381 was grown to OD600 of 0.6 in selective medium containing 2% glucose. Cells were harvested, washed, and resuspended in two volumes of extract buffer: 50 mM Tris, pH 8, 450 mM NaCl, 100 mM KCl, and 10% glycerol plus PIs. Cells were disrupted by vortexing with glass beads. The crude extract was clarified by centrifugation first for 10 min at 10,000 g and then for 20 min at 25,000 g at 4°C. Imidazole was added to 10 mM, and clarified lysate was bound to 1 ml Ni‐NTA column (HisTrap HP, GE Healthcare) by pumping at 1 ml/min. The column was washed twice with 10 ml of extract buffer containing 10 and 30 mM imidazole, respectively, and the protein was eluted in 0.330 ml fractions of extract buffer supplemented with 250 mM imidazole. Fractions containing protein were pooled and incubated for 2 h at 4°C with 1 ml of amylose resin equilibrated in amylose buffer (20 mM Tris, pH 7.5, 50 mM NaCl, 10% glycerol plus PIs). The beads were washed with 5 ml extract buffer and 5 ml amylose buffer. Bound protein was eluted with amylose buffer supplemented with 50 mM maltose in 300 μl fractions. Concentration was determined by Bradford, and protein was stored at −80°C.
6His‐Kemptide‐Tif6
200 milliliters of bacterial culture of Codon plus (RIL) cells (Stratagene) with pAJ3283 was grown to OD600 of 0.5 and induced with 1 mM IPTG for 4 h at 30°C. Cells were harvested and washed with Lysis buffer (50 mM NaPO4 pH 8.0, 500 mM NaCl, 10% glycerol, 6 mM BME). The cell pellet was resuspended in 10 ml Lysis buffer (supplemented with PIs) and disrupted by sonication. Lysates were cleared by centrifugation at 20,000 g for 20 min. Imidazole was added to 10 mM. Clarified lysate was bound to a 400 μl NiNTA resin bed (Qiagen) equilibrated with lysis buffer. The column was first washed with 10 ml lysis buffer supplemented with 10 mM imidazole and then with 10 ml supplemented with 30 mM imidazole. Bound protein was eluted in 300 μl fractions of lysis buffer supplemented with 200 mM imidazole. Fractions containing eluted protein were further purified by gel filtration on 170 ml S200 column in S200 buffer (20 mM Tris–HCl pH 7.5, 150 mM NaCl, 10% glycerol, and 6 mM BME). Fractions containing 6His‐Kemptide‐Tif6 were pooled and concentrated using Amicon Ultra‐15 10K device. Chemically synthesized Rpl41 peptide was purchased from GenScript.
GTPase assays
Specified amounts of Lsg1, Nmd3, 60S, as indicated in the figure legends, were mixed in a 20 μl volume in 1× GTPase buffer (20 mM HEPES‐KOH, pH 7.4, 2 mM MgOAc, 50 mM KOAc, 1 mM DTT) and containing 50 μM GTP. Reactions were spiked with ~1 × 105 cpm of [γ‐32P]‐GTP to trace the hydrolysis of gamma phosphate. Reactions were incubated at 30°C for 10 min and stopped by addition of 5 μl of 0.5 M EDTA. 1 μl of each reaction was spotted on a TLC plate (PEI‐cellulose, Sigma‐Aldrich). Free phosphate was separated from GTP by developing the TLC plate for 10 min in 0.8 M LiCl, 0.8 M CH3COOH. Plates were imaged on a phosphoimager screen and signal intensities for free phosphate and GTP were analyzed using ImageJ software. All samples were corrected for non‐enzymatic background hydrolysis. Percent GTP hydrolysis was calculated as (free phosphate/total phosphate) × 100. Percent GTP hydrolysis values from Lsg1 or Nmd3 titration assays were fitted to saturating‐specific single‐site binding curves using GraphPad Prism software.
Assembly of complexes for cryo‐EM
60S subunits (0.1 μM) were mixed with Nmd3 (0.4 μM) alone or with Lsg1 (1 μM), Tif6 (0.5 μM), and 200 μM GMPPNP in a total volume of 20 μl in GTPase buffer. The samples were incubated at 30°C for 10 min. Glutaraldehyde was added to a final concentration of 0.5% w/v, and the samples were incubated for 10 min at 0°C. Unreacted glutaraldehyde was quenched with addition of Tris, pH 7.4 to final concentration of 75 mM, and incubation at 0°C for 10 min. The samples were immediately flash‐frozen and stored in liquid N2 until needed.
Electron microscopy
Three microliters of each sample was applied to 300 mesh, holey R1.2/1.3 gold grids that were either prepared in laboratory (Russo & Passmore, 2014) from Quantifoil carbon grids or acquired commercially (UltrAuFoil R 1.2/1.3 Quantifoil). Prior to sample application, the grids were plasma‐cleaned for 25 s at 10 W and 4.3 to 1 O2:H2 ratio. 3 μl of aliquots of prepared complexes was applied to the prepared grids. Grids were blotted for 3–6 s at 4°C and 100% relative humidity and flash‐frozen in liquid ethane on a Vitrobot Mark IV (FEI, Hillsboro). Images for all but one dataset were acquired on an FEI Tecnai F30 Polara operating at 300 kV utilizing Leginon for automated data collection (Suloway et al, 2005). The movies were taken at nominal magnification of 32,000 on a Gatan K2 Summit direct electron detector (Gatan, Pleasanton) with a pixel size of 1.26 Å. The remaining collection was on a Titan Krios (FEI, Hillsboro) operating at 300 kV. In this case, the movies were taken at nominal magnification of 22,500 on a Gatan K2 Summit direct electron detector with a pixel size of 1.1 Å. For all images, the dose rate was set at eight counts per physical pixel per second. A total of 40 frames per stack were recorded at 0.2 s per frame, for a total exposure time of 8 s and a total exposure dose of ~40 e− Å−2.
Image processing
Movie stacks were aligned using dosefgpu_driftcorr summing all of the frames in each stack (Li et al, 2013). CTF estimation was done using CTFFIND3 (Mindell & Grigorieff, 2003). The remaining processing steps were done with RELION (Scheres, 2012a,b). Reported overall resolutions were calculated following the “gold‐standard” protocol employing the FSC = 0.143 criterion (Scheres, 2012a). Appendix Table S5 summarizes the number of images collected, particles extracted, number of particles used for classification and refinement, as well as the final resolution for the refined maps. For model building, maps were sharpened by applying a negative B‐factor (Appendix Table S5) that was estimated using the automated procedure in RELION.
Subset of data (Appendix Fig S8) was reprocessed with MotionCor2 (Zheng et al, 2016) (Patch 5 5, Iter = 10, Tol = 0.5, FmDose = 1.32). The aligned, non‐dose weighted images were used for CTF estimation by Gctf (Zhang, 2016). The remaining processing steps were done with RELION 2.0 (open beta) (Kimanius et al, 2016). Following the first refinement step, dose weighted particles were used for further processing.
Model building
To generate an initial model for the 60S‐Nmd3‐Lsg1‐Tfi6 complex, UCSF Chimera (Pettersen et al, 2004) was used for initial rigid‐body fitting of a 60S map (PDB: 4v88) into the refined volume. Further model building and correction was done in Coot (Emsley et al, 2010). In silico model generation by homology for Nmd3 and Lsg1 was attempted with I‐TASSER server (Zhang, 2008; Roy et al, 2010; Yang & Zhang, 2015; Yang et al, 2015). An Lsg1 model that fit best to the density attributed to Lsg1 was used to build the final structure. Residues 135 through 501 could be assigned to density. The final Lsg1 model was refined using MODREFINER (Xu & Zhang, 2011). Initial chain trace for Nmd3 was done in Coot on skeletonized map with baton mode. Density for residues 371–373 was used as starting point for sequence assignment.
Sharpened maps used in model building were also used for model refinement in REFMAC v5.8. MolProbity was used for validation of the refined models.
DALI and PDBeFold servers were used to search the PDB database for structural matches with Nmd3 and Lsg1 (Krissinel & Henrick, 2004, 2005; Holm & Rosenström, 2010). PDBePISA was employed to analyze the interacting regions between Nmd3 and other 60S proteins (Krissinel & Henrick, 2007).
Sucrose density gradient sedimentation of 60S‐Nmd3‐Lsg1·GMPPNP complex
50 μl of binding reaction containing 5 pmol 60S, 25 pmol Lsg1, 10 pmol Nmd3, and 200 μM GMP‐PNP in binding buffer (20 mM HEPES‐KOH pH 7.5, 2 mM Mg(OAc)2, 250 mM KOAc and 1 mM DTT) was incubated at 30°C for 15 min. The reaction mixture was sedimented through 10–30% sucrose gradient in SW55 Ti rotor for 75 min at 280,000 g. The gradients were fractionated into 600 μl fractions. Contents of the fractions were precipitated in 10% TCA and analyzed on 10% SDS–PAGE.
Accession codes
The cryo‐EM maps have been deposited in the EMDB with accession codes of EMDB‐8368 (60S‐Nmd3 only, not combined) and EMDB‐8362 (60S‐Nmd3‐Lsg1‐Tif6). Corresponding PDB codes are 5T6R and 5T62, respectively.
Author contributions
JF and AWJ designed the study. SM purified reagents, carried out in vitro complex assembly and assays, and contributed to writing. SP provided critical genetic insights and contributed to writing. AWJ directed the biochemical work and JF directed the cryo‐EM experiments. AM performed the cryo‐EM experiments and built the atomic models. AGM, JF, and AWJ interpreted the structures and wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Movie EV1
Review Process File
Source Data for Figure 1
Acknowledgements
We thank Bob Grassucci for his help in making gold grids and supporting microscopy on the F30 electron microscope. We thank the Simons Electron Microscopy Center at the New York Structural Biology Center (NYSBC) for providing us access to and support on the Titan Krios electron microscope, and Jeff Recchia‐Rife for support in protein purification. This work was supported by HHMI and grant NIH GM29169 (to JF) and grant NIH GM53655 (to AWJ). The Simons Electron Microscopy Center is supported by a grant from the Simons Foundation (Grant Number: 349247) with additional support from: NIH S10 OD019994‐01, the Agouron Institute (Grant Number: F00316), NIH S10 RR029300‐01, NIH S10 RR017291‐01, NYSTAR, and NIH C06 RR017528‐01‐CEM.
Note added in proof
Related work published while this work was in press reports a similar structure for Nmd3 (Ma C, Wu S, Li N, Chen Y, Yan K, Li Z, Zheng L, Lei J, Woolford JL Jr, Gao N (2017) Structural snapshot of cytoplasmic pre‐60S ribosomal particles bound by Nmd3, Lsg1, Tif6 and Reh1. Nat Struct Mol Biol doi:10.1038/nsmb.3364).
The EMBO Journal (2017) 36: 854–868
See also: A Razi & J Ortega (April 2017)
Contributor Information
Joachim Frank, Email: jf2192@cumc.columbia.edu.
Arlen W Johnson, Email: arlen@mail.utexas.edu.
References
- Achila D, Gulati M, Jain N, Britton RA (2012) Biochemical characterization of ribosome assembly GTPase RbgA in Bacillus subtilis . J Biol Chem 287: 8417–8423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altvater M, Chang Y, Melnik A, Occhipinti L, Schutz S, Rothenbusch U, Picotti P, Panse VG (2012) Targeted proteomics reveals compositional dynamics of 60S pre‐ribosomes after nuclear export. Mol Syst Biol 8: 628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand B, Verma SK, Prakash B (2006) Structural stabilization of GTP‐binding domains in circularly permuted GTPases: implications for RNA binding. Nucleic Acids Res 34: 2196–2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand B, Majumdar S, Prakash B (2013) Structural basis unifying diverse GTP hydrolysis mechanisms. Biochemistry 52: 1122–1130 [DOI] [PubMed] [Google Scholar]
- Aravind L, Koonin EV (2000) Eukaryote‐specific domains in translation initiation factors: implications for translation regulation and evolution of the translation system. Genome Res 10: 1172–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ash M‐R, Maher MJ, Mitchell Guss J, Jormakka M (2012) The cation‐dependent G‐proteins: in a class of their own. FEBS Lett 586: 2218–2224 [DOI] [PubMed] [Google Scholar]
- Bai X‐C, Rajendra E, Yang G, Shi Y, Scheres SHW (2015) Sampling the conformational space of the catalytic subunit of human γ‐secretase. eLife 4: e11182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradatsch B, Katahira J, Kowalinski E, Bange G, Yao W, Sekimoto T, Baumgartel V, Boese G, Bassler J, Wild K, Peters R, Yoneda Y, Sinning I, Hurt E (2007) Arx1 functions as an unorthodox nuclear export receptor for the 60S preribosomal subunit. Mol Cell 27: 767–779 [DOI] [PubMed] [Google Scholar]
- Bradatsch B, Leidig C, Granneman S, Gnadig M, Tollervey D, Bottcher B, Beckmann R, Hurt E (2012) Structure of the pre‐60S ribosomal subunit with nuclear export factor Arx1 bound at the exit tunnel. Nat Struct Mol Biol 19: 1234–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton RA (2009) Role of GTPases in bacterial ribosome assembly. Annu Rev Microbiol 63: 155–176 [DOI] [PubMed] [Google Scholar]
- Bussiere C, Hashem Y, Arora S, Frank J, Johnson AW (2012) Integrity of the P‐site is probed during maturation of the 60S ribosomal subunit. J Cell Biol 197: 747–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Keersmaecker K, Atak ZK, Li N, Vicente C, Patchett S, Girardi T, Gianfelici V, Geerdens E, Clappier E, Porcu M, Lahortiga I, Luca R, Yan J, Hulselmans G, Vranckx H, Vandepoel R, Sweron B, Jacobs K, Mentens N, Wlodarska I et al (2013) Exome sequencing identifies mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T‐cell acute lymphoblastic leukemia. Nat Genet 45: 186–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerfel LK, Wohlgemuth I, Kothe C, Peske F, Urlaub H, Rodnina MV (2013) EF‐P is essential for rapid synthesis of proteins containing consecutive proline residues. Science 339: 85–88 [DOI] [PubMed] [Google Scholar]
- Dragon F, Gallagher JE, Compagnone‐Post PA, Mitchell BM, Porwancher KA, Wehner KA, Wormsley S, Settlage RE, Shabanowitz J, Osheim Y, Beyer AL, Hunt DF, Baserga SJ (2002) A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417: 967–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of coot. Acta Crystallogr D Biol Crystallogr 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández‐Pevida A, Rodríguez‐Galán O, Díaz‐Quintana A, Kressler D, de la Cruz J (2012) Yeast ribosomal protein L40 assembles late into precursor 60 S ribosomes and is required for their cytoplasmic maturation. J Biol Chem 287: 38390–38407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch AJ, Hilcenko C, Basse N, Drynan LF, Goyenechea B, Menne TF, Gonzalez Fernandez A, Simpson P, D'Santos CS, Arends MJ, Donadieu J, Bellanne‐Chantelot C, Costanzo M, Boone C, McKenzie AN, Freund SM, Warren AJ (2011) Uncoupling of GTP hydrolysis from eIF6 release on the ribosome causes Shwachman‐diamond syndrome. Genes Dev 25: 917–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont‐Racine M, Senger B, Saveanu C, Fasiolo F (2003) Ribosome assembly in eukaryotes. Gene 313: 17–42 [DOI] [PubMed] [Google Scholar]
- Gadal O, Strauss D, Kessl J, Trumpower B, Tollervey D, Hurt E (2001) Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence‐containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol Cell Biol 21: 3405–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartmann M, Blau M, Armache JP, Mielke T, Topf M, Beckmann R (2010) Mechanism of eIF6‐mediated inhibition of ribosomal subunit joining. J Biol Chem 285: 14848–14851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardy S, Menet AM, Pena C, Petkowski JJ, Panse VG (2014) Assembly and nuclear export of pre‐ribosomal particles in budding yeast. Chromosoma 123: 327–344 [DOI] [PubMed] [Google Scholar]
- Grandi P, Rybin V, Bassler J, Petfalski E, Strauss D, Marzioch M, Schafer T, Kuster B, Tschochner H, Tollervey D, Gavin AC, Hurt E (2002) 90S pre‐ribosomes include the 35S pre‐rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol Cell 10: 105–115 [DOI] [PubMed] [Google Scholar]
- Greber BJ, Boehringer D, Montellese C, Ban N (2012) Cryo‐EM structures of Arx1 and maturation factors Rei1 and Jjj1 bound to the 60S ribosomal subunit. Nat Struct Mol Biol 19: 1228–1233 [DOI] [PubMed] [Google Scholar]
- Greber BJ (2016) Mechanistic insight into eukaryotic 60S ribosomal subunit biogenesis by cryo‐electron microscopy. RNA 22: 1643–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber BJ, Gerhardy S, Leitner A, Leibundgut M, Salem M, Boehringer D, Leulliot N, Aebersold R, Panse VG, Ban N (2016) Insertion of the biogenesis factor Rei1 probes the ribosomal tunnel during 60S maturation. Cell 164: 91–102 [DOI] [PubMed] [Google Scholar]
- Gutierrez E, Shin BS, Woolstenhulme CJ, Kim JR, Saini P, Buskirk AR, Dever TE (2013) eIF5A promotes translation of polyproline motifs. Mol Cell 51: 35–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges J, West M, Johnson AW (2005) Release of the export adapter, Nmd3p, from the 60S ribosomal subunit requires Rpl10p and the cytoplasmic GTPase Lsg1p. EMBO J 24: 567–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges J, Chen YI, West M, Bussiere C, Johnson AW (2006) Mapping the functional domains of yeast NMD3, the nuclear export adapter for the 60 S ribosomal subunit. J Biol Chem 281: 36579–36587 [DOI] [PubMed] [Google Scholar]
- Henras AK, Plisson‐Chastang C, O'Donohue MF, Chakraborty A, Gleizes PE (2015) An overview of pre‐ribosomal RNA processing in eukaryotes. Wiley Interdiscip Rev RNA 6: 225–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho JH, Kallstrom G, Johnson AW (2000) Nmd3p is a Crm1p‐dependent adapter protein for nuclear export of the large ribosomal subunit. J Cell Biol 151: 1057–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Rosenström P (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res 38: W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung NJ, Lo KY, Patel SS, Helmke K, Johnson AW (2008) Arx1 is a nuclear export receptor for the 60S ribosomal subunit in yeast. Mol Biol Cell 19: 735–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallstrom G, Hedges J, Johnson A (2003) The putative GTPases Nog1p and Lsg1p are required for 60S ribosomal subunit biogenesis and are localized to the nucleus and cytoplasm, respectively. Mol Cell Biol 23: 4344–4355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbstein K (2013) Quality control mechanisms during ribosome maturation. Trends Cell Biol 23: 242–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl T, Onder K, Kodzius R, Pichova A, Wimmer H, Thr A, Hundsberger H, Loffler M, Klade T, Beyer A, Breitenbach M, Koller L (1999) GRC5 and NMD3 function in translational control of gene expression and interact genetically. Curr Genet 34: 419–429 [DOI] [PubMed] [Google Scholar]
- Kemmler S, Occhipinti L, Veisu M, Panse VG (2009) Yvh1 is required for a late maturation step in the 60S biogenesis pathway. J Cell Biol 186: 863–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DJ, Jang JY, Yoon H‐J, Suh SW (2008) Crystal structure of YlqF, a circularly permuted GTPase: implications for its GTPase activation in 50 S ribosomal subunit assembly. Proteins 72: 1363–1370 [DOI] [PubMed] [Google Scholar]
- Kimanius D, Forsberg BO, Scheres SHW, Lindahl E (2016) Accelerated cryo‐EM structure determination with parallelisation using GPUs in RELION‐2. Elife 5: e18722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinge S, Voigts‐Hoffmann F, Leibundgut M, Arpagaus S, Ban N (2011) Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science 334: 941–948 [DOI] [PubMed] [Google Scholar]
- Kornprobst M, Turk M, Kellner N, Cheng J, Flemming D, Kos‐Braun I, Kos M, Thoms M, Berninghausen O, Beckmann R, Hurt E (2016) Architecture of the 90S pre‐ribosome: a structural view on the birth of the eukaryotic ribosome. Cell 166: 380–393 [DOI] [PubMed] [Google Scholar]
- Koš M, Tollervey D (2010) Yeast pre‐rRNA processing and modification occur cotranscriptionally. Mol Cell 37: 809–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D, Hurt E, Baβler J (2010) Driving ribosome assembly. Biochim Biophys Acta 1803: 673–683 [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K (2004) Secondary‐structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr 60: 2256–2268 [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K (2005) Multiple alignment of protein structures in three dimensions In Computational life sciences, Berthold MR, Glen RC, Diederichs K, Kohlbacher O, Fischer I. (eds), pp 67–78. Berlin, Heidelberg: Springer; [Google Scholar]
- Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372: 774–797 [DOI] [PubMed] [Google Scholar]
- Kuhle B, Ficner R (2014) A monovalent cation acts as structural and catalytic cofactor in translational GTPases. EMBO J 33: 2547–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidig C, Thoms M, Holdermann I, Bradatsch B, Berninghausen O, Bange G, Sinning I, Hurt E, Beckmann R (2014) 60S ribosome biogenesis requires rotation of the 5S ribonucleoprotein particle. Nat Commun 5: 3491 [DOI] [PubMed] [Google Scholar]
- Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng Y (2013) Electron counting and beam‐induced motion correction enable near‐atomic‐resolution single‐particle cryo‐EM. Nat Methods 10: 584–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo KY, Li Z, Wang F, Marcotte EM, Johnson AW (2009) Ribosome stalk assembly requires the dual‐specificity phosphatase Yvh1 for the exchange of Mrt4 with P0. J Cell Biol 186: 849–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo KY, Li Z, Bussiere C, Bresson S, Marcotte EM, Johnson AW (2010) Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit. Mol Cell 39: 196–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR (1998) Additional modules for versatile and economical PCR‐based gene deletion and modification in Saccharomyces cerevisiae . Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Matsuo Y, Granneman S, Thoms M, Manikas R‐G, Tollervey D, Hurt E (2014) Coupled GTPase and remodelling ATPase activities form a checkpoint for ribosome export. Nature 505: 112–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikov S, Mailliot J, Shin BS, Rigger L, Yusupova G, Micura R, Dever TE, Yusupov M (2016) Crystal structure of hypusine‐containing translation factor eIF5A bound to a rotated eukaryotic ribosome. J Mol Biol 428: 3570–3576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N (2003) Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol 142: 334–347 [DOI] [PubMed] [Google Scholar]
- Murray J, Savva CG, Shin B‐S, Dever TE, Ramakrishnan V, Fernández IS (2016) Structural characterization of ribosome recruitment and translocation by type IV IRES. eLife 5: e13567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissan TA, Bassler J, Petfalski E, Tollervey D, Hurt E (2002) 60S pre‐ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J 21: 5539–5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osheim YN, French SL, Keck KM, Champion EA, Spasov K, Dragon F, Baserga SJ, Beyer AL (2004) Pre‐18S ribosomal rna is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae . Mol Cell 16: 943–954 [DOI] [PubMed] [Google Scholar]
- Panse VG, Johnson AW (2010) Maturation of eukaryotic ribosomes: acquisition of functionality. Trends Biochem Sci 35: 260–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Raychaudhuri P, Stringer EA, Valenzuela DM, Maitra U (1984) Ribosomal subunit antiassociation activity in rabbit reticulocyte lysates. Evidence for a low molecular weight ribosomal subunit antiassociation protein factor (Mr = 25,000). J Biol Chem 259: 11930–11935 [PubMed] [Google Scholar]
- Reynaud EG, Andrade MA, Bonneau F, Ly TB, Knop M, Scheffzek K, Pepperkok R (2005) Human Lsg1 defines a family of essential GTPases that correlates with the evolution of compartmentalization. BMC Biol 3: 21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y (2010) I‐TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5: 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell DW, Spremulli LL (1980) Mechanism of action of the wheat germ ribosome dissociation factor: interaction with the 60 S subunit. Arch Biochem Biophys 201: 518–526 [DOI] [PubMed] [Google Scholar]
- Russo CJ, Passmore LA (2014) Ultrastable gold substrates for electron cryomicroscopy. Science 346: 1377–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A, Pech M, Thoms M, Beckmann R, Hurt E (2016) Ribosome‐stalk biogenesis is coupled with recruitment of nuclear‐export factor to the nascent 60S subunit. Nat Struct Mol Biol 23: 1074–1082 [DOI] [PubMed] [Google Scholar]
- Scheres SHW (2012a) A bayesian view on cryo‐EM structure determination. J Mol Biol 415: 406–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SHW (2012b) RELION: implementation of a Bayesian approach to cryo‐EM structure determination. J Struct Biol 180: 519–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt C, Becker T, Heuer A, Braunger K, Shanmuganathan V, Pech M, Berninghausen O, Wilson DN, Beckmann R (2016) Structure of the hypusinylated eukaryotic translation factor eIF‐5A bound to the ribosome. Nucleic Acids Res 44: 1944–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senger B, Lafontaine DL, Graindorge JS, Gadal O, Camasses A, Sanni A, Garnier JM, Breitenbach M, Hurt E, Fasiolo F (2001) The nucle(ol)ar Tif6p and Efl1p are required for a late cytoplasmic step of ribosome synthesis. Mol Cell 8: 1363–1373 [DOI] [PubMed] [Google Scholar]
- Sengupta J, Bussiere C, Pallesen J, West M, Johnson AW, Frank J (2010) Characterization of the nuclear export adaptor protein Nmd3 in association with the 60S ribosomal subunit. J Cell Biol 189: 1079–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen PS, Park J, Qin Y, Li X, Parsawar K, Larson MH, Cox J, Cheng Y, Lambowitz AM, Weissman JS, Brandman O, Frost A (2015) Rqc2p and 60S ribosomal subunits mediate mRNA‐independent elongation of nascent chains. Science 347: 75–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunk BS, Karbstein K (2009) Powering through ribosome assembly. RNA 15: 2083–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B (2005) Automated molecular microscopy: the new Leginon system. J Struct Biol 151: 41–60 [DOI] [PubMed] [Google Scholar]
- Thomas F, Kutay U (2003) Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway. J Cell Sci 116: 2409–2419 [DOI] [PubMed] [Google Scholar]
- Thomson E, Ferreira‐Cerca S, Hurt E (2013) Eukaryotic ribosome biogenesis at a glance. J Cell Sci 126: 4815–4821 [DOI] [PubMed] [Google Scholar]
- Tong AH, Boone C (2006) Synthetic genetic array analysis in Saccharomyces cerevisiae . Methods Mol Biol 313: 171–192 [DOI] [PubMed] [Google Scholar]
- Trotta CR, Lund E, Kahan L, Johnson AW, Dahlberg JE (2003) Coordinated nuclear export of 60S ribosomal subunits and NMD3 in vertebrates. EMBO J 22: 2841–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschochner H, Hurt E (2003) Pre‐ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol 13: 255–263 [DOI] [PubMed] [Google Scholar]
- Ude S, Lassak J, Starosta AL, Kraxenberger T, Wilson DN, Jung K (2013) Translation elongation factor EF‐P alleviates ribosome stalling at polyproline stretches. Science 339: 82–85 [DOI] [PubMed] [Google Scholar]
- Weis F, Giudice E, Churcher M, Jin L, Hilcenko C, Wong CC, Traynor D, Kay RR, Warren AJ (2015) Mechanism of eIF6 release from the nascent 60S ribosomal subunit. Nat Struct Mol Biol 22: 914–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West M, Hedges JB, Chen A, Johnson AW (2005) Defining the order in which Nmd3p and Rpl10p load onto nascent 60S ribosomal subunits. Mol Cell Biol 25: 3802–3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff EC, Kang KR, Kim YS, Park MH (2007) Posttranslational synthesis of hypusine: evolutionary progression and specificity of the hypusine modification. Amino Acids 33: 341–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford JL, Baserga SJ (2013) Ribosome biogenesis in the yeast Saccharomyces cerevisiae . Genetics 195: 643–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Tutuncuoglu B, Yan K, Brown H, Zhang Y, Tan D, Gamalinda M, Yuan Y, Li Z, Jakovljevic J, Ma C, Lei J, Dong MQ, Woolford JL Jr, Gao N (2016) Diverse roles of assembly factors revealed by structures of late nuclear pre‐60S ribosomes. Nature 534: 133–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Zhang Y (2011) Improving the physical realism and structural accuracy of protein models by a two‐step atomic‐level energy minimization. Biophys J 101: 2525–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zhang Y (2015) I‐TASSER server: new development for protein structure and function predictions. Nucleic Acids Res 43: W174–W181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y (2015) The I‐TASSER Suite: protein structure and function prediction. Nat Methods 12: 7–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao W, Roser D, Kohler A, Bradatsch B, Bassler J, Hurt E (2007) Nuclear export of ribosomal 60S subunits by the general mRNA export receptor Mex67‐Mtr2. Mol Cell 26: 51–62 [DOI] [PubMed] [Google Scholar]
- Zemp I, Kutay U (2007) Nuclear export and cytoplasmic maturation of ribosomal subunits. FEBS Lett 581: 2783–2793 [DOI] [PubMed] [Google Scholar]
- Zhang Y (2008) I‐TASSER server for protein 3D structure prediction. BMC Bioinformatics 9: 40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K (2016) Gctf: real‐time CTF determination and correction. J Struct Biol 193: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Palovcak E, Armache J‐P, Cheng Y, Agard D (2016) Anisotropic correction of beam‐induced motion for improved single‐particle electron cryo‐microscopy. bioRxiv doi: 10.1101/061960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk D, Belk JP, Jacobson A (1999) Temperature‐sensitive mutations in the Saccharomyces cerevisiae MRT4, GRC5, SLA2 and THS1 genes result in defects in mRNA turnover. Genetics 153: 35–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Movie EV1
Review Process File
Source Data for Figure 1


