Abstract
We report here the main characteristics of five new species ‘Urmitella timonensis’ strain Marseille-P2918T (CSUR P2918), ‘Blautia marasmi’ strain Marseille-P2377T (CSUR P2377), ‘Lachnoclostridium pacaense’ strain Marseille-P3100T (CSUR P3100), ‘Bacillus marasmi’ strain Marseille-P3556T (CSUR P3556) and ‘Anaerotruncus rubiinfantis’ strain MT15T (CSUR P2276), which were isolated recently from stool samples taken from undernourished children in Niger and Senegal using microbial culturomics.
Keywords: Culturomics, human gut microbiota, malnutrition, new species, taxonogenomics
In 2016, as a part of culturomics study of the human microbiome, we isolated five bacterial strains from stool samples of patients in Senegal and Niger with malnutrition (marasmus and kwashiorkor) [6], [7] that could not be identified by our systematic matrix-assisted laser desorption–ionization time-of-flight mass spectrometry (MALDI-TOF MS) screening on a Microflex spectrometer (Bruker Daltonics, Bremen, Germany) [1], [2], [4]. The patients signed informed consent; the study was validated by the ethics committee of the Institut Federatif de Recherche IFR48 under number 09-022.
In this study, we will describe these five bacterial strains, which were deposited in the open Collection de Souches de l’Unité des Rickettsies (CSUR, WDCM 875) under reference numbers P2918 (‘Urmitella timonensis’ strain Marseille-P2918T) [8], P2377 (‘Blautia marasmi’ strain Marseille-P2377T), P3100 (‘Lachnoclostridium pacaense’ strain Marseille-P3100T), P3556 (‘Bacillus marasmi’ strain Marseille-P3556T) and P2276 (‘Anaerotruncus rubiinfantis’ strain MT15T), respectively.
Two strains, Marseille-P2918T and Marseille-P3100T, were isolated from a 3.3-month-old Senegalese girl with clinical aspects of marasmus (70 cm, 7 kg, weight-for-height Z score −1.75). Strain Marseille-P2377T was isolated from a 2.5-month-old Senegalese boy with severe acute malnutrition (64 cm, 3 kg, weight-for-height Z score −8.9). Meanwhile, strain Marseille-P3556T was isolated from a Nigerian child with clinical aspects of marasmus, but anthropometric criteria were not available for this child. Strain MT15T was isolated from a 13-month-old Nigerian girl with oedematous severe acute malnutrition, also known as kwashiorkor (72 cm, 8 kg, presence of oedema).
The following conditions for initial growth existed for each strain: Strain Marseille-P2918T was isolated after 7 days in blood culture bottle + sheep’s blood, in anaerobe condition, 37°C; strain Marseille-P2377T was isolated after 3 days in blood culture bottle, in anaerobe condition, 37°C; strain Marseille-P3100T was isolated after 3 days of culture in COS medium (Columbia blood agar with sheep’s blood medium), anaerobe condition, 37°C; strain Marseille-P3556T was isolated after 1 day in blood culture bottle + sheep’s blood + rumen, aerobic condition, 37°Cand strain MT15T was isolated after 10 days in blood culture + sheep’s blood, anaerobe condition, 37°C.
In this study, we described the colony morphologies of these five bacterial species. Colonies of strain Marseille-P2918T were circular, smooth, very small and pale grey with a mean diameter of 0.2 to 0.5 mm. Colonies of the strain Marseille-P2377T were white, circular, smooth, convex with intact edges with a larger diameter of 0.5 to 3 mm. Colonies of the strain Marseille-P3100T were white, circular, smooth with intact edges and a mean diameter of 1 to 3 mm. Colonies of the strain Marseille-P3556T were flat, smooth, small, circular and pale grey with a mean diameter of 0.5 to 2 mm. Colonies of the strain MT15T were small, smooth with intact edges and a mean diameter of 1 to 3 mm.
All five bacterial strains were Gram-positive, rod-shaped for strain Marseille-P2918T, Marseille-P3100T, Marseille-P3556T, MT15T; and coccus-shaped for strain Marseille-P2377T; polymorphic.
The 16S rRNA gene of these five strains were sequenced using fD1-rP2 primers as previously described using a 3130-XL sequencer (Applied Biosciences, Saint Aubin, France).
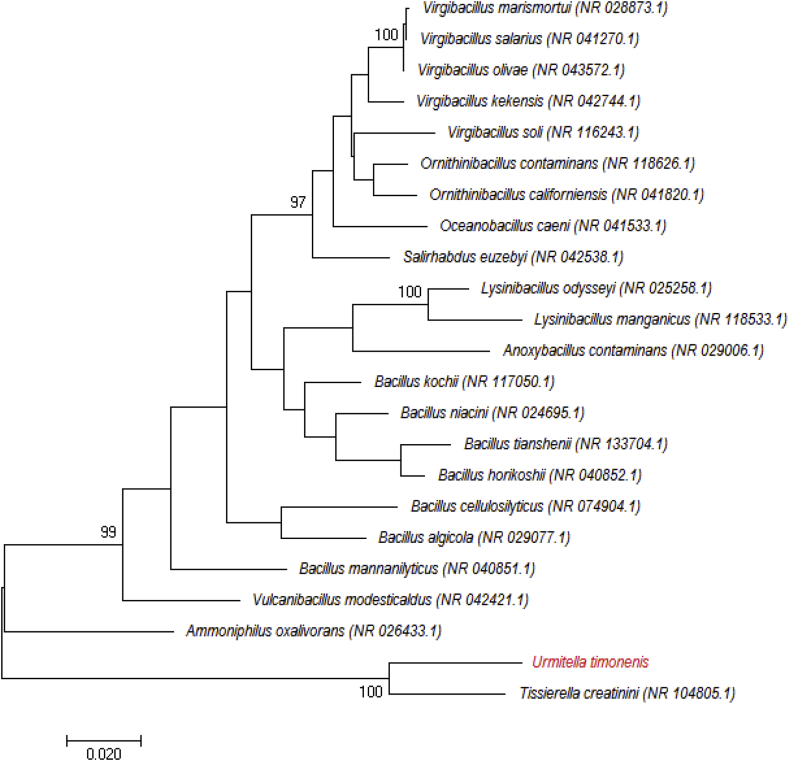
Strain Marseille-P2918T exhibited a 93.25% 16S rRNA gene sequence identity with Tissierella creatinini strain BN11 (GenBank accession no. NR_104805), the phylogenetically closest species with standing in nomenclature (Fig. 1), which putatively classifies it as a member of the family Tissierellaceae in the phylum Firmicutes. Strain Marseille-P2918T exhibits a 16S rRNA gene sequence divergence of >5% with its phylogenetically closest species with standing in nomenclature [3], [5]. Thus, we propose the creation of the new genus ‘Urmitella’ (ur.mit.tel′la, NL, gen. fem, ‘URMITE,’ the name of the laboratory where the strain was isolated in Marseille, France). ‘Urmitella timonensis’ is proposed as the type species of the ‘Urmitella’ genus. Strain Marseille-P2918T is the type strain of the new species ‘Urmitella timonensis’ (ti.mo.nen′sis, L adj. fem., ‘Timone,’ the name of the main hospital of Marseille, France, where the strain was isolated).
Fig. 1.
Phylogenetic tree showing position of ‘Urmitella timonensis’ strain Marseille-P2918T relative to other phylogenetically close neighbours. Sequences were aligned using CLUSTALW, and phylogenetic inferences were obtained using maximum-likelihood method within MEGA software. Numbers at nodes are percentages of bootstrap values obtained by repeating analysis 500 times to generate majority consensus tree. Only bootstraps scores of at least 90% were retained. Scale bar indicates 2% nucleotide sequence divergence.
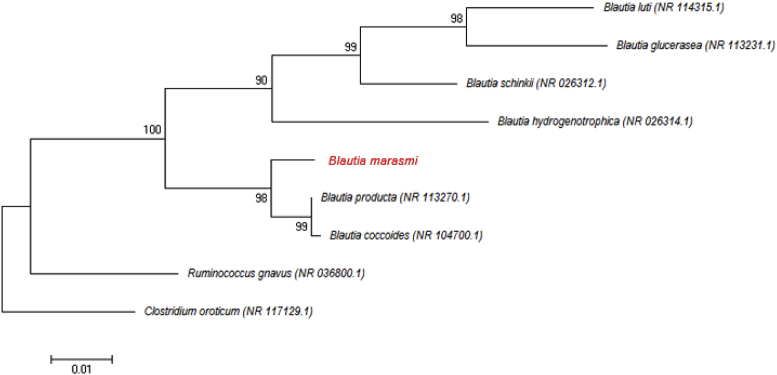
Strain Marseille-P2377T exhibited a 98.39% 16S rRNA gene sequence identity with Blautia producta strain JCM1471 (GenBank accession no. NR_113270), the phylogenetically closest species with standing in nomenclature (Fig. 2), which putatively classifies it as a member of the genus Blautia within the family Lachnospiraceae in the phylum Firmicutes. Strain Marseille-P2377T exhibited a 16S rRNA sequence divergence of >1.3% with its phylogenetically closest species with standing in nomenclature [3], [5]. We thus propose the creation of a new species, ‘Blautia marasmi’ (ma.ras.mi, L. adj. fem., referring to the marasmus disease, with which patients were afflicted). Strain Marseille-P2377T is the type strain of the new species ‘Blautia marasmi.’
Fig. 2.
Phylogenetic tree showing position of ‘Blautia marasmi’ Marseille-P2377T relative to other phylogenetically close neighbours. Sequences were aligned using CLUSTALW, and phylogenetic inferences were obtained using maximum-likelihood method within MEGA software. Numbers at nodes are percentages of bootstrap values obtained by repeating analysis 500 times to generate majority consensus tree. Only bootstraps scores of at least 90% were retained. Scale bar indicates 1% nucleotide sequence divergence.
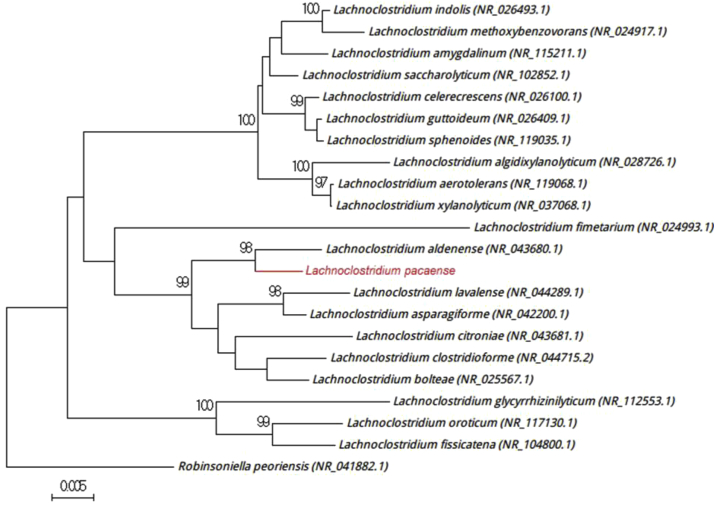
Strain Marseille-P3100T exhibited a 98.55% sequence identity with Lachnoclostridium aldenense strain RMA 9741 (GenBank accession no. NR_043680), the phylogenetically closest species with standing in nomenclature (Fig. 3), which putatively classifies it as a member of the genus Lachnoclostridium within the family Lachnospiraceae in the phylum Firmicutes. Strain Marseille-P3100T exhibited a 16S rRNA sequence divergence of >1.3% with its phylogenetically closest species with standing in nomenclature [3], [5]. We thus propose the creation of the new species, ‘Lachnoclostridium pacaense’ (pa.ca.en′se, L. adj. fem., ‘PACA,’ the name of the region Provence-Alpes-Côte d’Azur, France, where the strain was isolated). Strain Marseille-P3100T is the type strain of the new species ‘Lachnoclostridium pacaense.’
Fig. 3.
Phylogenetic tree showing position of ‘Lachnoclostridium pacaense’ Marseille-P3100T relative to other phylogenetically close neighbours. Sequences were aligned using CLUSTALW, and phylogenetic inferences were obtained using maximum-likelihood method within MEGA software. Numbers at nodes are percentages of bootstrap values obtained by repeating analysis 500 times to generate majority consensus tree. Only bootstraps scores of at least 90% were retained. Scale bar indicates 0.5% nucleotide sequence divergence.
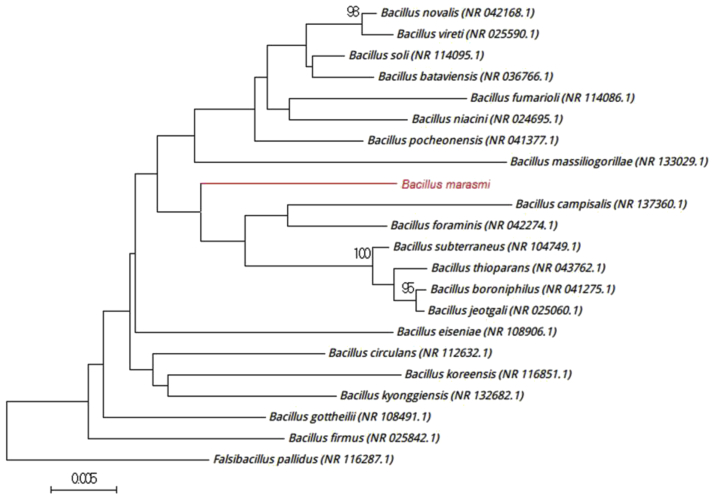
Strain Marseille-P3556T exhibited a 97.28% 16S rRNA gene sequence identity with Bacillus subterraneus strain COOI3B (NR_104749), the phylogenetically closest species with standing in nomenclature (Fig. 4), which putatively classifies it as a member of the genus Bacillus within the family Bacillaceae in the phylum Firmicutes. Strain Marseille-P3556T exhibits a 16S rRNA gene sequence divergence of >1.3% with its phylogenetically closest species with standing in nomenclature [3], [5]. Thus, we propose the creation of the new species ‘Bacillus marasmi’ (ma.ras.mi, L. adj. fem, ‘marasmus,’ the disease of with which patients were afflicted). Strain Marseille-P3556T is the type strain of the new species Bacillus marasmi.
Fig. 4.
Phylogenetic tree showing position of ‘Bacillus marasmi’ Marseille-P3556T relative to other phylogenetically close neighbours. Sequences were aligned using CLUSTALW, and phylogenetic inferences were obtained using maximum-likelihood method within MEGA software. Numbers at nodes are percentages of bootstrap values were obtained by repeating analysis 500 times to generate majority consensus tree. Only bootstraps score at least 90% were retained. Scale bar indicates 0.5% nucleotide sequence divergence.
Strain MT15T exhibited a 94.07% 16S rRNA gene sequence identity with Anaerotruncus colihominis strain WAL 14565 (NR_027558), the phylogenetically closest species with standing in nomenclature (Fig. 5), which putatively classifies it as a member of the genus Anaerotruncus within the family Ruminococcaceae in the phylum Firmicutes. Strain MT15T, exhibiting a 16S rRNA gene sequence, had a divergence of >1.3% with its phylogenetically closest species with standing in nomenclature [3], [5]. Thus, we propose the creation of the new species ‘Anaerotruncus rubiinfantis,’ from ru.be.us, adj., from infantis (L. gen. n.), ‘red infant,’ a reference to the hair discoloration observed in kwashiorkor patients. Strain MT15T is the type strain of the new species ‘Anaerotruncus rubiinfantis.’
Fig. 5.
Phylogenetic tree showing position of ‘Anaerotruncus rubiinfantis’ MT15T relative to other phylogenetically close neighbours. Sequences were aligned using CLUSTALW, and phylogenetic inferences were obtained using maximum-likelihood method within MEGA software. Numbers at nodes are percentages of bootstrap values obtained by repeating analysis 500 times to generate majority consensus tree. Only bootstraps score at least 90% were retained. Scale bar indicates 1% nucleotide sequence divergence.
Nucleotide sequence accession number
All the 16S rRNA gene sequences were deposited in GenBank under accession numbers LT598554 (‘Urmitella timonensis’ strain Marseille-P2918T), LT631508 (‘Blautia marasmi’ strain Marseille-P2377T), LT631510 (‘Lachnoclostridium pacaense’ strain Marseille-P3100T), LT671590 (‘Bacillus marasmi’ strain Marseille-P3556T) and LN881593 (‘Anaerotruncus rubiinfantis’ strain MT15T), respectively.
Deposit in a culture collection
All strains were deposited in the open Collection de Souches de l’Unité des Rickettsies (CSUR, WDCM 875) under reference numbers P2918 (‘Urmitella timonensis’ strain Marseille-P2918T), P2377 (‘Blautia marasmi’ strain Marseille-P2377T), P3100 (‘Lachnoclostridium pacaense’ strain Marseille-P3100T), P3556 (‘Bacillus marasmi’ strain Marseille-P3556T) and P2276 (‘Anaerotruncus rubiinfantis’ strain MT15T), respectively.
Conflict of Interest
None declared.
Acknowledgement
This study was funded by the Fondation Méditerranée Infection.
References
- 1.Lagier J.C., Hugon P., Khelaifia S., Fournier P.E., La Scola B., Raoult D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev. 2015;28:237–264. doi: 10.1128/CMR.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lagier J.C., Armougom F., Million M., Hugon P., Pagnier I., Bittar F. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect. 2012;18:1185–1193. doi: 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
- 3.Huson D.H., Auch A.F., Qi J., Schuster S.C. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seng P., Drancourt M., Gouriet F., La Scola B., Fournier P.E., Rolain J.M. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis. 2009;49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 5.Stackebrandt E., Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today. 2006;33:152. [Google Scholar]
- 6.Million M., Tidjani Alou M., Khelaifia S., Bachar D., Lagier J.C., Dione N. Increased gut redox and depletion of anaerobic and methanogenic prokaryotes in severe acute malnutrition. Sci Rep. 2016;6:26051. doi: 10.1038/srep26051. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Million M., Diallo A., Raoult D. Gut microbiota and malnutrition. Microb Pathog. 2016 Feb 4 doi: 10.1016/j.micpath.2016.02.003. (pii: S0882-4010(15)30212-6) [DOI] [PubMed] [Google Scholar]
- 8.Lagier J.C., Khelaifia S., Alou M.T., Ndongo S., Dione N., Hugon P. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol. 2016;1:16203. doi: 10.1038/nmicrobiol.2016.203. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]