Abstract
We recently reported the genome of Orientia tsutsugamushi (OT) strain Karp (GenBank Accession #: NZ_LYMA00000000.2, https://www.ncbi.nlm.nih.gov/nuccore/NZ_LYMA00000000.2) with > 2 Mb in size through clone-based sequencing and high throughput genomic shotgun sequencing (HTS). The genomes of OT strains AFSC4 and AFSC7 were similarly sequenced by HTS Since strains AFSC4 (GenBank Accession #: NZ_LYMT00000000.1, https://www.ncbi.nlm.nih.gov/nuccore/1035784408) and AFSC7 (GenBank Accession #: NZ_LYMB00000000.1, https://www.ncbi.nlm.nih.gov/nuccore/1035854767) were more resistant to antibiotics than strain Karp, we conducted comparative analysis of the three draft genomes annotated by RAST server aimed to identify possible genetic bases of difference in microbial antibiotic sensitivity. Intraspecies comparative genomics analysis of the three OT strains revealed that two ORFs encoding hypothetical proteins in both strains AFSC4 and AFSC7 are absent in strain Karp.
Keywords: Orientia tsutsugamushi, Comparative genomic analysis, Antibiotic sensitivity, RAST genome annotation, Gene function comparison, Genome sequence comparison
| Specifications | |
|---|---|
| Organism/cell line/tissue | Orientia tsutsugamushi strain Karp, AFSC4 and AFSC7 |
| Sex | N/A |
| Sequencer or array type | Sanger, Illumina MiSeq V2 250 × 2 PE |
| Data format | Analyzed |
| Experimental factors | Bacteria isolated from different geographic areas with distinguished sensitivities to antibiotics |
| Experimental features | The genome sequences of three Orientia tsutsugamushi strains were annotated and compared. |
| Consent | N/A |
| Sample source location | The strains were originally collected from Papua New Guinea and Thailand. |
1. Direct link to deposited data
Orientia tsutsugamushi, Karp https://www.ncbi.nlm.nih.gov/genome/710?genome_assembly_id=290025.
Orientia tsutsugamushi, AFSC4 https://www.ncbi.nlm.nih.gov/genome/710?genome_assembly_id=276762.
Orientia tsutsugamushi, AFSC7 http://www.ncbi.nlm.nih.gov/genome/710?genome_assembly_id=276760.
2. Experimental design, materials and methods
Orientia tsutsugamushi (OT), the etiological agent of scrub typhus infects human via chigger bite, is an obligate intracellular organism. The disease is endemic in the scrub typhus triangle or so called “tsutsugamushi triangle” which extends from northern Japan and far-eastern Russia in the north, to northern Australia in the south, and to Pakistan in the west with a million cases reported yearly and a billion people under the risk [1]. Many OT isolates have been obtained from endemic areas but also from the areas outside of the scrub typhus triangle [2], [3]. Clinical antibiotic resistant infections have been reported in Southern India [4] and Southern Thailand [5], [6], [7]. In vitro studies and animal models showed that Karp strain was sensitive to doxycycline and azithromycin whereas AFSC4 and AFSC7 were much less sensitive [8]. We recently reported the 2 Mb genome of strain Karp using both clone-based sequencing and high-throughput genomic shotgun sequencing [9]. In this study, we conducted genomic sequencing of strains AFSC4 and AFSC7 isolated from Thailand and made genomic comparison among the three strains of OT.
All Orientia tsutsugamushi strains were cultured in irradiated L929 cells. Purified bacteria with minimum amount of host material was prepared by serious ultracentrifugation in renografin density gradient [10]. The DNAs extracted from the purified bacteria of strain AFSC4 and AFSC7 were subjected to whole genome shotgun sequencing (WGS) using Illumina MiSeq platform with the 2 × 250 bp pair-end mode. Briefly, 50 ng of purified DNA was subjected to DNA library construction using the Nextera DNA Sample Prep Kit (Illumina), and the generated fastq files were used in the downstream sequencing data analysis, which was performed by CLC Genomics Workbench Version 9.0.1. The raw reads were trimmed and filtered under the parameters: reads phred score > Q30, base error probability score > 0.05, base ambiguities < 2 and read-length < 15 bp. The trimmed raw reads of strain AFSC4 and AFSC7 were de novo assembled into original 1201 and 1803 contigs, with the average coverage 120 and 179, respectively. The parameters of de novo assembly in CLC Genomics Workbench v.8.5 were minimum contig length 300 bp, no scaffolding, mismatch cost = 2, insertion cost = 3, deletion cost = 3, length fraction = 5, similarity fraction = 0.8. The original contigs of two strains obtained from WGS were first mapped to genomes of three reference strains of OT: 1) strain Karp (GenBank Accession#: LYMA00000000), 2) strain Boryong (NC_009488.1) and 3) strain Ikeda (NC_010793.1). The original contigs were also blasted in parallel against nt/nr database to remove those with sequence homology to mouse and other environmental organisms as well as those with only very low coverage. The reads from the contigs, which passed the filtering, were retrieved and subjected to a second round of de novo assembly.
All retained WGS contigs were able to be mapped to at least one of the three OT genome references, the final contig numbers of AFSC4 and AFSC7 were 452 and 485, respectively. The draft genome of AFSC4 has 1,295,323 bp (29.9% G + C) and AFSC7 has 1,437,569 bp (30.0% G + C). The contigs of AFSC4 and AFSC7 were aligned to the Ikeda reference genome using CONTIGuator genome finishing tool to obtain the oriented and arranged contig series as the draft genomes [11]. In comparison with the Karp draft genome 2,026,724 bp in length with 30.41% G + C content [9], both of AFSC 4 and AFSC 7 draft genome contributed totally from the contigs are significantly shorter. The smaller sizes of draft genome with higher numbers of contigs were evidently contributed by the high content (about 40%) of repetitive sequence in genomes of OT. The ambiguity and abundance of the repeats prevented generation of long contigs in de novo assembly. Many junctions between contigs of AFSC4 and AFSC7 could only be possibly closed with the long reads generated by other sequencing approaches or methods in the future.
All three genomes were annotated using RAST server Version 2.0 [12]. The numbers of the contigs would not influence the gene annotation. The strain Karp was annotated with 2089 functional elements, including 2052 coding sequences (CDSs) and 37 RNAs. Total 1090 CDSs transcribed from the positive strand and 999 transcribed from negative strand. 32% (656) of the CDSs were categorized into 185 subsystems (Fig. 1A). The strain AFSC4 has 1149 annotated CDS and 36 RNAs, 560 CDSs were transcribed from the positive strand and 625 were transcribed from negative strand. 44% (502) of the CDSs were categorized into 186 subsystems (Fig. 1B). In the study, the strain AFSC7 was annotated with 1376 CDSs and 37 RNAs. In that, 652 CDSs were transcribed from the positive strand and 761 were transcribed from negative strand; 41% (557) of the CDSs were categorized into 185 subsystems (Fig. 1C). AFSC4 and AFSC7 have similar subsystem distribution, while Karp has more CDSs categorized into the membrane transporter, the most abundant subsystems. Protein metabolism and RNA metabolism are the second and third abundant subsystems of these three strains, respectively.
Fig. 1.
The gene subsystem distribution of OT strain KARP, AFSC4 and AFSC7. The analysis was performed using RAST server Version 2.0. The pie charts show the distribution of OT strain (A) KARP (B) AFSC4 and (C) AFSC7.
The SEED Viewer functional comparison [13] compared the genes encompassed in functional groups or subsystems in genomes, pairwisely, which identified 682 CDSs of AFSC4 and 677 CDSs of AFSC7 with known functions and are comparable to those of CDS identified in Karp genome. The Seed Viewer functional- based comparative analysis revealed five protein-coding genes that are present in the genomes of AFSC4 and AFSC7, but absent in the genome of Karp. The sequence length of these five genes ranged from 135 bp to 543 bp. However, all of these five genes could be found in the Karp genome, when the gene sequences were blasted against the draft genome. The findings indicated that the functional comparison between genomes by the computational tool may not be optimal for some of the shorter genes, and shown as false positive findings.
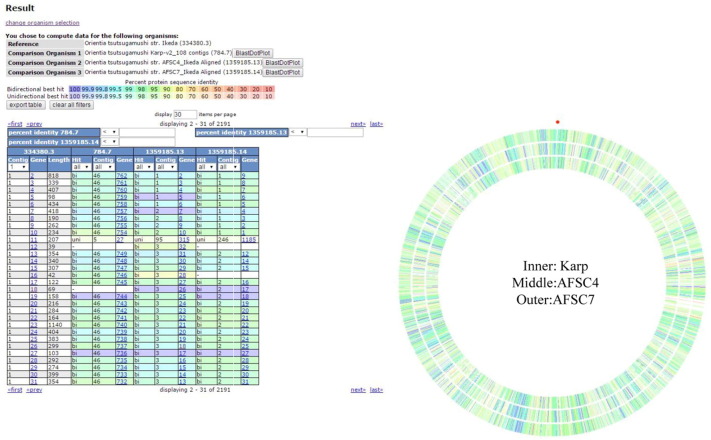
To identify genomic differences or possible genes that might contribute to the differential of the antibiotic sensitivity among the closely related strains of OT, we further analyzed the CDSs of the genomes using the SEED Viewer computational tool for sequence comparison. The genome sequences of the compared strains were aligned to a reference genome. The output listed the genes on the reference genome following the chromosomal order and displayed the gene hits on the compared strains without concerning the functions of the listed genes. The genome of OT strain Ikeda was used as the reference strain for the detail sequence comparison (Fig. 2). The comparative analysis revealed 28 CDSs that are present in AFSC4 and AFSC7 but are absent in Karp. However, specific Blast verification for gene sequences of each identified CDS showed only two CDSs, number 1436 and 2073, were truly missing in the Karp genome (Table 1). Fig. 3, Fig. 4 show the sequences of these two genes were blasted against the draft genomes of the three OT strains. The proteins encoded by these two genes are hypothetical without known biological function. Although the two genes identified are coding for hypothetical proteins without known biological function, the close genomic comparisons of three OT strains with evident differences in antibiotic tolerance has provided the possible targets for investigation of OT drug-resistance mechanism(s).
Fig. 2.
The results of the SEED Viewer sequence compare.
Table 1.
The summary of the two coding sequences present in AFSC4 and AFSC7 but absent in Karp.
| Reference Orientia tsutsugamushi str. Ikeda |
Blast against Karp, AFSC4 and AFSC7 draft genome |
||||
|---|---|---|---|---|---|
| Gene ID | Function | Length (bp) | Sequence | Identity | Hit notes |
| 1436 | Hypothetical protein | 1032 | atgcaacaagaagaatatattaaaatttcttttttaggagaagatagtcctacattaattaaaaacttttttttagcattaaatcaactacagcatgatatatcttgtcttgctattactagctattataatagcaagcctcatcaaaagcaccttttgagctatcttaaaaatcaacaaattgagtcagtttttagaaaggtttgtgaaaataaaggatacttcaagcaagttattaaaaaagttcacaaaaataaaaacaaagactttaggcaagtggttaatggacttcttgaatatgaaaataagcacgatcaacaaaaatcttacaaagttgaagatgataaatcgttactgcaagctattaatatagcaaataaaaactacaagtactttatgctagatcttgatgatgctcaggatgatctcttgaaggatggacaggttgaaatagccttctcggatcaattctcagagttattatctaagattaaaacgcttatgtttgttatggcagatgaaagaatggggcaacgtattattaattgtataccagatgggtctatattacctaatttagcatattttcattttgatatggaatttggtgcgtttgataaatttagcaaaggaactttatctaagtttactcaaaaaggatctataaaggtagatgatgtcttttttattcattgcgatccagagtttattggttataattcaggccatagttcaagaaaacatccaagtccagcaaaaaattcagatttagcatactatatcagcaaggatgcagcattacactgcttcaacattgattgtaaaacattagttttaagcgctaacattgaggtaggagatgatgaagatttgcaaacaaaaatatctgaaatttctcaaaataataccactaaacggttaattttgccgtatgaattctatgatgaaccaatttgtgatagtaatggcgagatagctaagttattaggaatagaagaaagctcttttgattgttgctgctctatttcttag | 24/26 (92%) | One full length hit was found in AFSC4 and AFSC7, but not in Karp. (a 24 nt fragment was blast matched) |
| 2073 | Hypothetical protein | 192 | ttgaagcaatgtacaccagtaatagaagaagtagatgtacaaaaaattagtcaatgtataactagatcatacaatatatcaaaatattcagttgttacattgcaaaaaattaccgatattactaaagtaaataaggaaataaaaaaagttaattttttcttaacctataagctatctacaaagtgccattaa | 75/77 (97%), 14/14 (100%), 14/14 (100%) | One full length was found in AFSC4 and AFSC7 genomes, but not found in Karp. (three fragments found with 75, 14, and 14 nt blast match, respectively) |
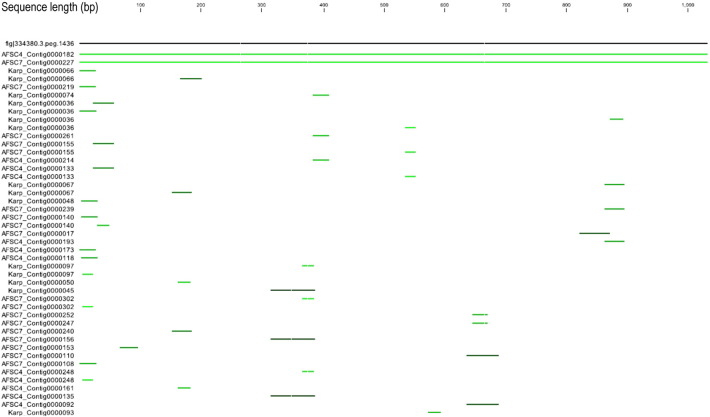
Fig. 3.
The blast hits of the CDS#1436 sequence against the draft genomes of OT strain Karp, AFSC4, AFSC7.
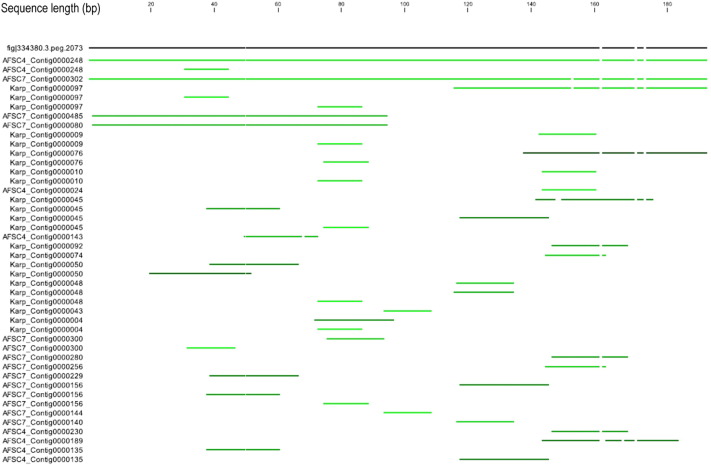
Fig. 4.
The blast hits of the CDS# 2073 sequence against the draft genomes of OT strain Karp, AFSC4, AFSC7.
Four genome sequences were chosen as the inputs for sequence compare. The database available complete genome of OT strain Ikeda was used as reference for comparison. The circles represent the blast identity of the compared strains against the reference strain, the red dot indicates the first based of the reference sequence, and the continuous bases were extended clockwise. The inner circle indicates the blast hit in Karp, the middle indicates the blast hit in AFSC4 and the outer circle indicates the blast hit in AFSC7. The rainbow spectrum of color was used to indicate the identity (purple: high to red: low) of the sequence hit on the corresponding position of the reference genome. Most of the blast hits on the three strains have high identity comparing to the reference sequences.
On the top, the black line represents the sequence of the coding sequence Fig | 334380.3.peg.1436 (CDS#1436 in short), the numbers above indicate the length of the sequence. The other lines (green) below the top line indicate the sequences of the contigs of the three draft genomes that can be matched to the CDS#1436. The brighter green indicates the higher identity. Only the FASC4_contig0000162 and AFSC7_Contig000227 encompass the full-length sequence of CDS#1436.
On the top, the black line represents the sequence of the coding sequence Fig | 334380.3.peg.2073 (CDS#2073 in short), the numbers above indicate the length of the sequence. The other lines (green) below the top line indicate the sequences of the contigs of the three draft genomes that can be matched to CDS#2073. The brighter green indicates the higher identity. Only the AFSC4_contig0000248 and AFSC7_Contig000302 encompass the full-length sequence of CDS#2073. Karp_contig0000097 has partial sequence matched to the CDS#2073.
3. Nucleotide sequence accession number
The first versions of draft genome sequences of Orientia tsutsugamushi strain AFSC4 and AFSC7 were deposited in GenBank under accession number LYMT00000000 and LYMB00000000, respectively.
Acknowledgement
The authors wish to thank Dr. Gregory Dasch for purifying Karp, AFSC4 and AFSC7 strains of Orientia tsutsugamushi from L929 cells and Ms. Zhiwen Zhang and Ms. Tatyana Belinskaya for subsequent DNA extraction for WGS. This work was supported in part by Work Unit Number (WUN) 6000.RAD1.J.A0310 and an FDA Modernizing Science grant. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. The opinions and assertions contained herein are the private ones of the authors and are not to be construed as official or as reflecting the views of the Department of the Navy, the Naval service at large, the Department of Defense, or the U. S. Government. Authors Chien-Chung Chao and Wei-Mei Ching are employees of the U. S. Government. This work was prepared as part of official duties. Title 17 U.S.C. §105 provides that ‘Copyright protection under this title is not available for any work of the United States Government.’ Title 17 U.S.C. §101 defines a U.S. Government work as a work prepared by employee of the U.S. Government as part of that person's official duties.
References
- 1.Kelly D.J., Fuerst P.A., Ching W.M., Richards A.L. Scrub typhus: the geographic distribution of phenotypic and genotypic variants of Orientia tsutsugamushi. Clinical Infectious Diseases: An Official Publication of the Infectious Diseases Society of America. 2009;48(Suppl. 3):S203–S230. doi: 10.1086/596576. PubMed PMID: 19220144. [DOI] [PubMed] [Google Scholar]
- 2.Balcells M.E., Rabagliati R., Garcia P., Poggi H., Oddo D., Concha M. Endemic scrub typhus-like illness, Chile. Emerg. Infect. Dis. 2011;17(9):1659–1663. doi: 10.3201/eid1709.100960. PubMed PMID: 21888791; PubMed Central PMCID: PMCPMC3322051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Izzard L., Fuller A., Blacksell S.D., Paris D.H., Richards A.L., Aukkanit N. Isolation of a novel Orientia species (O. chuto sp. nov.) from a patient infected in Dubai. J. Clin. Microbiol. 2010;48(12):4404–4409. doi: 10.1128/JCM.01526-10. PubMed PMID: 20926708; PubMed Central PMCID: PMCPMC3008486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mathai E., Rolain J.M., Verghese G.M., Abraham O.C., Mathai D., Mathai M. Outbreak of scrub typhus in southern India during the cooler months. Ann. N. Y. Acad. Sci. 2003;990:359–364. doi: 10.1111/j.1749-6632.2003.tb07391.x. PubMed PMID: 12860654. [DOI] [PubMed] [Google Scholar]
- 5.Watt G., Chouriyagune C., Ruangweerayud R., Watcharapichat P., Phulsuksombati D., Jongsakul K. Scrub typhus infections poorly responsive to antibiotics in northern Thailand. Lancet. 1996;348(9020):86–89. doi: 10.1016/s0140-6736(96)02501-9. PubMed PMID: 8676722. [DOI] [PubMed] [Google Scholar]
- 6.Watt G., Kantipong P., Jongsakul K., Watcharapichat P., Phulsuksombati D. Azithromycin activities against Orientia tsutsugamushi strains isolated in cases of scrub typhus in Northern Thailand. Antimicrob. Agents Chemother. 1999;43(11):2817–2818. doi: 10.1128/aac.43.11.2817. PubMed PMID: 10543774; PubMed Central PMCID: PMCPMC89570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watt G., Kantipong P., Jongsakul K., Watcharapichat P., Phulsuksombati D., Strickman D. Doxycycline and rifampicin for mild scrub-typhus infections in northern Thailand: a randomised trial. Lancet. 2000;356(9235):1057–1061. doi: 10.1016/S0140-6736(00)02728-8. PubMed PMID: 11009140. [DOI] [PubMed] [Google Scholar]
- 8.Strickman D., Sheer T., Salata K., Hershey J., Dasch G., Kelly D. In vitro effectiveness of azithromycin against doxycycline-resistant and -susceptible strains of Rickettsia tsutsugamushi, etiologic agent of scrub typhus. Antimicrob. Agents Chemother. 1995;39(11):2406–2410. doi: 10.1128/aac.39.11.2406. PubMed PMID: 8585717; PubMed Central PMCID: PMCPMC162956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liao H.M., Chao C.C., Lei H., Li B., Tsai S., Hung G.C. Genomic sequencing of Orientia tsutsugamushi strain Karp, an assembly comparable to the genome size of the strain Ikeda. Genome Announc. 2016;4(4) doi: 10.1128/genomeA.00702-16. PubMed PMID: 27540052; PubMed Central PMCID: PMCPMC4991697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dasch G.A., Halle S., Bourgeois A.L. Sensitive microplate enzyme-linked immunosorbent assay for detection of antibodies against the scrub typhus rickettsia, Rickettsia tsutsugamushi. J. Clin. Microbiol. 1979;9(1):38–48. doi: 10.1128/jcm.9.1.38-48.1979. PubMed PMID: 107185; PubMed Central PMCID: PMCPMC272954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galardini M., Biondi E.G., Bazzicalupo M., Mengoni A. CONTIGuator: a bacterial genomes finishing tool for structural insights on draft genomes. Source Code Biol Med. 2011;6:11. doi: 10.1186/1751-0473-6-11. PubMed PMID: 21693004; PubMed Central PMCID: PMCPMC3133546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Overbeek R., Olson R., Pusch G.D., Olsen G.J., Davis J.J., Disz T. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST) Nucleic Acids Res. 2014;42(Database issue):D206–D214. doi: 10.1093/nar/gkt1226. PubMed PMID: 24293654; PubMed Central PMCID: PMCPMC3965101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Overbeek R., Begley T., Butler R.M., Choudhuri J.V., Chuang H.Y., Cohoon M. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33(17):5691–5702. doi: 10.1093/nar/gki866. PubMed PMID: 16214803; PubMed Central PMCID: PMCPMC1251668. [DOI] [PMC free article] [PubMed] [Google Scholar]