Abstract
We propose the main characteristics of a new bacterium species named Nissabacter archeti strain 2134 (CSURP3445 = LT631518), isolated from pustule scalp of a 29-year-old man at hospital Archet 2, Nice, south of France.
Keywords: Enterobacteria, human infection, Nissabacter archeti, pustule, scalp, taxonomy
A Gram-negative bacterial strain 2134, CSURP3445, Enterobacteria, aeroanaerobic, isolated from pustule scalp of a 29-year-old man from Archet 2 Hospital, Nice, south of France, was studied for its taxonomy allocation. Initially this bacterium grew on 5% blood agar plate in an aerobic atmosphere at 37°C after 24 hours' incubation. The strain also grew on Luria-Bertani agar to 37°C after 24 hours' incubation. The colonies were whitish, circular and smooth, with a diameter of 0.5 to 1.0 mm. Identification by matrix-assisted laser desorption–ionization time-of-flight mass spectrometry (MALDI-TOF MS) screening on a MicroFlex spectrometer protein analysis (Bruker Daltonics, Leipzig, Germany) did not allow the identification of the strain 2134 to the genus level [1]. Consequently, the sequence of 16S rRNA gene was determined to specify its phylogenetic position. Briefly, PCR of 16S rRNA gene was performed using the fD1-rP2 primers as previously described [2] using Quantitec PCR System 2720 thermal cyclers (Applied Biosystems, Bedford, MA, USA). The PCR product was purified and sequenced using ABI Prism 3130xl Genetic Analyzer capillary sequencer (Applied Biosystems), which generated 1507 bp.
Under electron microscopy, individual cells have a slightly oval form with flagella and were 3.5 μm in length and 1.0 μm in diameter.
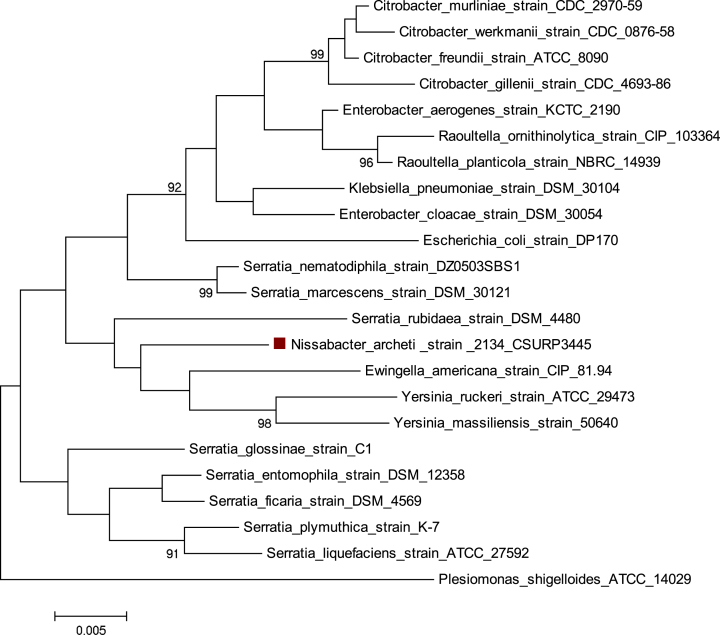
The 16S rRNA gene of strain 2134 exhibited a 97.67% 16S rRNA gene sequence similarity with Ewingella americana strain CIP 81.94. Strain 2134 is closely related to two standing nomenclature species, Serratia rubidae DSM 4480 and Ewingella americana strain CIP81.94 (Fig. 1). Because the branches grouping strain 2134 with these two species are not supported by significant bootstrap values (>90%), we propose that the strain 2134 is representative of a new genus within the Enterobacteriaceae family.
Fig. 1.
Phylogenetic position of strain 2134 within Enterobacteriaceae family by 16S rDNA sequence analysis. Unrooted neighbour-joining tree of concatenated 16S rDNA of 22 reference strains of representative members of Enterobacteriaceae family closely related to strain 2134 [3]. Pleisomonas shigelloides ATCC 14029 was used as outgroup. Values below lines are bootstrap values (1000 replicates) expressed in percent (only values greater than 90% are shown) [4]. Scale bar = accumulated changes per nucleotide. Analyses were conducted in MEGA7 [5].
Taxonomically, the bacterial family Enterobacteriaceae currently has 53 genera (and over 170 named species). Of these, 26 genera are known to be associated with infections in humans [3], [4]. Members of the Enterobacteriaceae are small, Gram-negative, nonsporing straight rods. They are facultatively anaerobic, and most species grow well at 37°C, although some species grow better at 25 to 30°C [6]. Strain 2134 exhibited a 16S rRNA gene sequence similarity of ≤98.65 % with a validly published name with standing in nomenclature [7]. We propose the new genus Nissabacter as a new member of Enterobacteriaceae family for the fact that it was first isolated from Nice a south-east city of France, and the description of the first species of this genus Nissabacter archeti for the fact that it was isolated from human pustule scalp in the bacteriology laboratory of the Archet 2 Hospital.
Deposit in a culture collection
Strain Nissabacter archeti strain 2134 was deposited in the Collection de Souches de l'Unité des Rickettsies (CSUR, WDCM 875) under number P3445.
Nucleotide sequence accession number
The 16S rRNA gene sequence of the strain 2134 was deposited in GenBank under accession number LT631518 under the name Nissabacter archeti strain 2134.
Acknowledgements
This study was funded by the Fondation Méditerranée Infection. The authors thank F. Cadoret for administrative tasks.
Conflict of Interest
None declared.
References
- 1.Seng P., Rolain J.M., Fournier P.E., La Scola B., Drancourt M., Raoult D. MALDI-TOF-mass spectrometry applications in clinical microbiology. Future Microbiol. 2010;5:1733–1754. doi: 10.2217/fmb.10.127. [DOI] [PubMed] [Google Scholar]
- 2.Drancourt M., Bollet C., Carlioz A., Martelin R., Gayral J.P., Raoult D. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J Clin Microbiol. 2000;38:3623–3630. doi: 10.1128/jcm.38.10.3623-3630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khan F., Rizvi M., Shukla I., Malik A. A novel approach for identification of members of Enterobacteriaceae isolated from clinical samples. Biol Med. 2011;3(2 special issue):313–319. [Google Scholar]
- 4.Borman E.K., Stuart C.A., Wheeler K.M. Taxonomy of the family Enterobacteriaceae. J Bacteriol. 1944;48:351–367. doi: 10.1128/jb.48.3.351-367.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Health Protection Agency . 2013. UK standards for microbiology investigations identification of Campylobacter species; pp. 1–22. ID23. [Google Scholar]
- 7.Kim M., Oh H.S., Park S.C., Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64(pt 2):346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]