Abstract
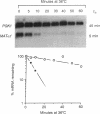
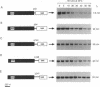
In yeast, the mRNA encoded by the MAT alpha 1 gene is unstable (t1/2 = 5 min) and the mRNAs encoded by the ACT1 gene (t1/2 = 30 min) and the PGK1 gene (t1/2 = 45 min) are stable. To understand the RNA structural features that dictate mRNA decay rates in yeast, we have constructed PGK1/MAT alpha 1 and ACT1/MAT alpha 1 gene fusions and analyzed the decay rates of the resultant chimeric transcripts. Fusion of a MAT alpha 1 segment containing 73% of the coding region and the 3' untranslated region to either of the stable genes is sufficient to cause rapid decay of the chimeric mRNAs (t1/2 = 6-7.5 min). Sequences required for this rapid decay are not found in the MAT alpha 1 3' untranslated region but are located within a 42-nucleotide segment of the coding region that has a high content (8 out of 14) of rare codons. Introduction of a translational stop codon upstream of this region stabilizes the hybrid mRNAs, indicating that the rapid decay promoted by these sequences is dependent on ribosomal translocation.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aota S., Gojobori T., Ishibashi F., Maruyama T., Ikemura T. Codon usage tabulated from the GenBank Genetic Sequence Data. Nucleic Acids Res. 1988;16 (Suppl):r315–r402. doi: 10.1093/nar/16.suppl.r315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astell C. R., Ahlstrom-Jonasson L., Smith M., Tatchell K., Nasmyth K. A., Hall B. D. The sequence of the DNAs coding for the mating-type loci of Saccharomyces cerevisiae. Cell. 1981 Nov;27(1 Pt 2):15–23. doi: 10.1016/0092-8674(81)90356-1. [DOI] [PubMed] [Google Scholar]
- Casey J. L., Hentze M. W., Koeller D. M., Caughman S. W., Rouault T. A., Klausner R. D., Harford J. B. Iron-responsive elements: regulatory RNA sequences that control mRNA levels and translation. Science. 1988 May 13;240(4854):924–928. doi: 10.1126/science.2452485. [DOI] [PubMed] [Google Scholar]
- Cleveland D. W. Autoregulated instability of tubulin mRNAs: a novel eukaryotic regulatory mechanism. Trends Biochem Sci. 1988 Sep;13(9):339–343. doi: 10.1016/0968-0004(88)90103-x. [DOI] [PubMed] [Google Scholar]
- Elledge S. J., Davis R. W. A family of versatile centromeric vectors designed for use in the sectoring-shuffle mutagenesis assay in Saccharomyces cerevisiae. Gene. 1988 Oct 30;70(2):303–312. doi: 10.1016/0378-1119(88)90202-8. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Gay D. A., Sisodia S. S., Cleveland D. W. Autoregulatory control of beta-tubulin mRNA stability is linked to translation elongation. Proc Natl Acad Sci U S A. 1989 Aug;86(15):5763–5767. doi: 10.1073/pnas.86.15.5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay D. A., Yen T. J., Lau J. T., Cleveland D. W. Sequences that confer beta-tubulin autoregulation through modulated mRNA stability reside within exon 1 of a beta-tubulin mRNA. Cell. 1987 Aug 28;50(5):671–679. doi: 10.1016/0092-8674(87)90325-4. [DOI] [PubMed] [Google Scholar]
- Graves R. A., Pandey N. B., Chodchoy N., Marzluff W. F. Translation is required for regulation of histone mRNA degradation. Cell. 1987 Feb 27;48(4):615–626. doi: 10.1016/0092-8674(87)90240-6. [DOI] [PubMed] [Google Scholar]
- Harland R., Misher L. Stability of RNA in developing Xenopus embryos and identification of a destabilizing sequence in TFIIIA messenger RNA. Development. 1988 Apr;102(4):837–852. doi: 10.1242/dev.102.4.837. [DOI] [PubMed] [Google Scholar]
- Hitzeman R. A., Hagie F. E., Hayflick J. S., Chen C. Y., Seeburg P. H., Derynck R. The primary structure of the Saccharomyces cerevisiae gene for 3-phosphoglycerate kinase. Nucleic Acids Res. 1982 Dec 11;10(23):7791–7808. doi: 10.1093/nar/10.23.7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekema A., Kastelein R. A., Vasser M., de Boer H. A. Codon replacement in the PGK1 gene of Saccharomyces cerevisiae: experimental approach to study the role of biased codon usage in gene expression. Mol Cell Biol. 1987 Aug;7(8):2914–2924. doi: 10.1128/mcb.7.8.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabnick K. S., Housman D. E. Determinants that contribute to cytoplasmic stability of human c-fos and beta-globin mRNAs are located at several sites in each mRNA. Mol Cell Biol. 1988 Aug;8(8):3244–3250. doi: 10.1128/mcb.8.8.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losson R., Lacroute F. Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5134–5137. doi: 10.1073/pnas.76.10.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüscher B., Stauber C., Schindler R., Schümperli D. Faithful cell-cycle regulation of a recombinant mouse histone H4 gene is controlled by sequences in the 3'-terminal part of the gene. Proc Natl Acad Sci U S A. 1985 Jul;82(13):4389–4393. doi: 10.1073/pnas.82.13.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat L. E., Kinniburgh A. J., Rachmilewitz E. A., Ross J. Unstable beta-globin mRNA in mRNA-deficient beta o thalassemia. Cell. 1981 Dec;27(3 Pt 2):543–553. doi: 10.1016/0092-8674(81)90396-2. [DOI] [PubMed] [Google Scholar]
- Mellor J., Dobson M. J., Kingsman A. J., Kingsman S. M. A transcriptional activator is located in the coding region of the yeast PGK gene. Nucleic Acids Res. 1987 Aug 11;15(15):6243–6259. doi: 10.1093/nar/15.15.6243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyuhas O., Perry R. P. Relationship between size, stability and abundance of the messenger RNA of mouse L cells. Cell. 1979 Jan;16(1):139–148. doi: 10.1016/0092-8674(79)90195-8. [DOI] [PubMed] [Google Scholar]
- Müllner E. W., Kühn L. C. A stem-loop in the 3' untranslated region mediates iron-dependent regulation of transferrin receptor mRNA stability in the cytoplasm. Cell. 1988 Jun 3;53(5):815–825. doi: 10.1016/0092-8674(88)90098-0. [DOI] [PubMed] [Google Scholar]
- Ng R., Abelson J. Isolation and sequence of the gene for actin in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3912–3916. doi: 10.1073/pnas.77.7.3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet M., Scafe C., Sexton J., Young R. Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol Cell Biol. 1987 May;7(5):1602–1611. doi: 10.1128/mcb.7.5.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudel U., Soreq H., Littauer U. Z. Globin mRNA species containing poly(A) segments of different lengths. Their functional stability in Xenopus oocytes. Eur J Biochem. 1976 Apr 15;64(1):115–121. doi: 10.1111/j.1432-1033.1976.tb10279.x. [DOI] [PubMed] [Google Scholar]
- Pandey N. B., Marzluff W. F. The stem-loop structure at the 3' end of histone mRNA is necessary and sufficient for regulation of histone mRNA stability. Mol Cell Biol. 1987 Dec;7(12):4557–4559. doi: 10.1128/mcb.7.12.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J., Kobs G. H4 histone messenger RNA decay in cell-free extracts initiates at or near the 3' terminus and proceeds 3' to 5'. J Mol Biol. 1986 Apr 20;188(4):579–593. doi: 10.1016/s0022-2836(86)80008-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago T. C., Bettany A. J., Purvis I. J., Brown A. J. Messenger RNA stability in Saccharomyces cerevisiae: the influence of translation and poly(A) tail length. Nucleic Acids Res. 1987 Mar 25;15(6):2417–2429. doi: 10.1093/nar/15.6.2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro R. A., Herrick D., Manrow R. E., Blinder D., Jacobson A. Determinants of mRNA stability in Dictyostelium discoideum amoebae: differences in poly(A) tail length, ribosome loading, and mRNA size cannot account for the heterogeneity of mRNA decay rates. Mol Cell Biol. 1988 May;8(5):1957–1969. doi: 10.1128/mcb.8.5.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw G., Kamen R. A conserved AU sequence from the 3' untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986 Aug 29;46(5):659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- Shyu A. B., Greenberg M. E., Belasco J. G. The c-fos transcript is targeted for rapid decay by two distinct mRNA degradation pathways. Genes Dev. 1989 Jan;3(1):60–72. doi: 10.1101/gad.3.1.60. [DOI] [PubMed] [Google Scholar]
- Singer R. H., Penman S. Messenger RNA in HeLa cells: kinetics of formation and decay. J Mol Biol. 1973 Aug 5;78(2):321–334. doi: 10.1016/0022-2836(73)90119-8. [DOI] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varenne S., Buc J., Lloubes R., Lazdunski C. Translation is a non-uniform process. Effect of tRNA availability on the rate of elongation of nascent polypeptide chains. J Mol Biol. 1984 Dec 15;180(3):549–576. doi: 10.1016/0022-2836(84)90027-5. [DOI] [PubMed] [Google Scholar]
- Wolin S. L., Walter P. Ribosome pausing and stacking during translation of a eukaryotic mRNA. EMBO J. 1988 Nov;7(11):3559–3569. doi: 10.1002/j.1460-2075.1988.tb03233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen T. J., Machlin P. S., Cleveland D. W. Autoregulated instability of beta-tubulin mRNAs by recognition of the nascent amino terminus of beta-tubulin. Nature. 1988 Aug 18;334(6183):580–585. doi: 10.1038/334580a0. [DOI] [PubMed] [Google Scholar]