ABSTRACT
Burkholderia contaminans MS14 was isolated from soil in Mississippi. When it is cultivated on nutrient broth-yeast extract agar, the colonies exhibit bactericidal activity against a wide range of plant-pathogenic bacteria. A bacteriostatic compound with siderophore activity was successfully purified and was determined by nuclear magnetic resonance spectroscopy to be ornibactin. Isolation of the bactericidal compound has not yet been achieved; therefore, the exact nature of the bactericidal compound is still unknown. During an attempt to isolate the bactericidal compound, an interesting relationship between the production of ornibactin and the bactericidal activity of MS14 was characterized. Transposon mutagenesis resulted in two strains that lost bactericidal activity, with insertional mutations in a nonribosomal peptide synthetase (NRPS) gene for ornibactin biosynthesis and a luxR family transcriptional regulatory gene. Coculture of these two mutant strains resulted in restoration of the bactericidal activity. Furthermore, the addition of ornibactin to the NRPS mutant restored the bactericidal phenotype. It has been demonstrated that, in MS14, ornibactin has an alternative function, aside from iron sequestration. Comparison of the ornibactin biosynthesis genes in Burkholderia species shows diversity among the regulatory elements, while the gene products for ornibactin synthesis are conserved. This is an interesting observation, given that ornibactin is thought to have the same defined function within Burkholderia species. Ornibactin is produced by most Burkholderia species, and its role in regulating the production of secondary metabolites should be investigated.
IMPORTANCE Identification of the antibacterial product from strain MS14 is not the key feature of this study. We present a series of experiments that demonstrate that ornibactin is directly involved in the bactericidal phenotype of MS14. This observation provides evidence for an alternative function for ornibactin, aside from iron sequestration. Ornibactin should be further evaluated for its role in regulating the biosynthesis of secondary metabolites in other Burkholderia species.
KEYWORDS: Burkholderia contaminans MS14, antibacterial activity, ornibactin
INTRODUCTION
The genus Burkholderia is composed of Gram-negative, rod-shaped, motile, environmental, versatile, non-spore-forming bacteria that have been identified in many diverse ecological niches (1). Currently, 88 species have been recognized in the genus Burkholderia (2). The bacteria have the ability to use a large array of carbon sources to synthesize secondary metabolites (3, 4). The Burkholderia cepacia complex is a group of Burkholderia species that includes soil isolates and opportunistic bacteria that cause lung disease in immunocompromised individuals (5). The B. cepacia complex group is composed of 9 different genomovars and at least 18 different species (6). Conversely, some strains of Burkholderia cepacia are related to the promotion of plant growth and are considered to be plant-growth-promoting bacteria (PGPB). For example, B. cepacia strains could protect crops from the damping-off diseases caused by Pythium species and Rhizoctonia solani (3). Interest in the use of Burkholderia species or their secondary metabolites in agriculture has increased. In addition, multiple antimicrobials produced by Burkholderia species, such as occidiofungin (7), pyrrolnitrin (4), pyoluteorin (8), and AFC-BC11 (9), have been identified.
Siderophores are small-molecule, ferric-ion-specific, chelating agents secreted by bacteria and fungi growing under low-iron stress. They scavenge iron from the environment and make it available to the microbial cells (10). Siderophores are bacteriostatic agents that can inhibit the growth of pathogenic microorganisms by depleting iron in the soil (11). Many siderophores are synthesized by nonribosomal peptide synthetases (NRPSs) and polyketide synthases (PKSs). NRPSs and PKSs are large multimodular enzymes that are involved in natural product synthesis in many microorganisms (12). NRPSs, which are involved in the biosynthesis of oligopeptides, are grouped by active sites termed modules, with each module being required to catalyze one single cycle of product length elongation. The number and order of the modules of a NRPS protein mainly follow the “collinearity rule” (13).
Ornibactin is a product of nonribosomal peptide synthesis with siderophore activity. Ornibactin production in Burkholderia cepacia was shown to be critical for establishing an infection in a murine chronic respiratory infection model (14). In the same report, however, the authors noted that ornibactin appeared to be important for bacterial adherence or colonization. Furthermore, the report showed that the absence of ornibactin production led to a significant increase in the production of salicylic acid, suggesting that ornibactin production represses salicylic acid biosynthesis. These are interesting observations, but no direct link to an alternative function for ornibactin could be made. There is a long-term understanding that the role of ornibactin in virulence involves providing a source of iron in iron-restricted environments. To date, the best described function for ornibactin is its ability to sequester iron within the lung from iron-binding proteins, such as lactoferrin. This activity is crucial for survival within the respiratory mucus (15, 16).
In this study, we showed that B. contaminans MS14 produces a bactericidal compound that has a broad spectrum of activity toward Gram-negative bacterial plant pathogens. In addition, the bacteriostatic compound ornibactin was isolated and was shown to be an important component for the bactericidal phenotype of MS14. It was demonstrated that growing an ornibactin synthesis mutant in proximity to a luxR mutant restored MS14 bactericidal activity. We also showed that the addition of ornibactin to colonies of the ornibactin synthesis mutant restored the bactericidal phenotype, thus providing a direct observation of an alternative function for ornibactin. The genes involved in the initiation and regulation of biosynthesis, as well as regulatory elements, have significant diversity among Burkholderia species, suggesting that ornibactin has other functions within Burkholderia. The findings suggest that ornibactin is an important component for the production, or possibly the function, of the bactericidal secondary metabolite produced by B. contaminans MS14, and they support studies looking into its alternative functions within other bacterial species.
RESULTS
Antibacterial activity of MS14.
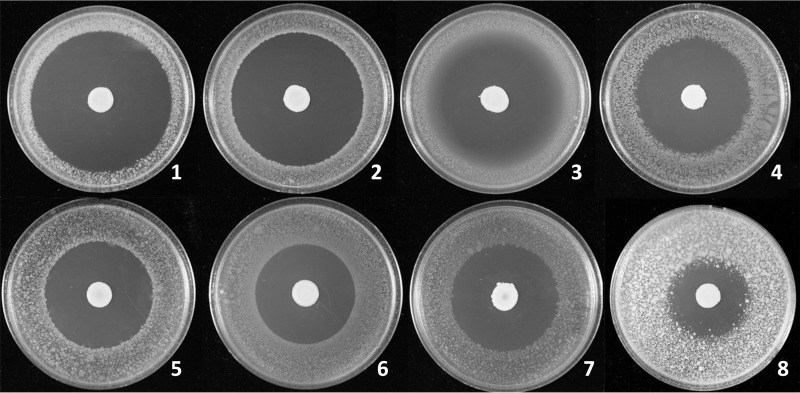
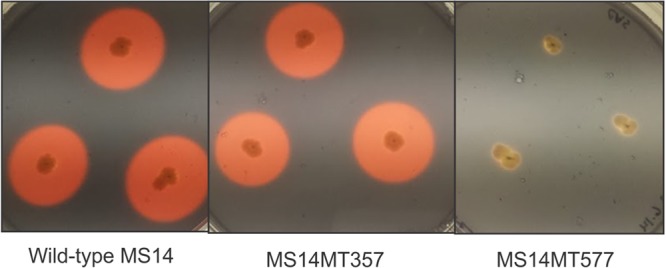
Burkholderia contaminans MS14 was shown previously to produce a potent antifungal named occidiofungin (17). In this study, zone-of-inhibition plate assays of strain MS14 grown on nutrient broth-yeast extract (NBY) demonstrated significant bactericidal activity against a broad array of plant bacterial pathogens (Fig. 1 and Table 1). Culture of agar plugs within the zones of inhibition from the indicator bacterial strain Erwinia amylovora did not yield any viable colonies, supporting the classification of the MS14 antibacterial product as a bactericidal compound. Xanthomonas citri pv. malvacearum, one of the most destructive pathogens on cotton (18), was best inhibited by strain MS14, with a 36-mm-radius bactericidal zone on the plate. Pectobacterium carotovorum EC101, which is the pathogen of bacterial soft rot on potatoes and other vegetables (19), and Ralstonia solanacearum, which causes bacterial wilt of tomatoes and potatoes (20), were also significantly inhibited by strain MS14. The apple and pear fire blight pathogen Erwinia amylovora (21) and the bacterial panicle blight pathogen Burkholderia glumae (22) were also highly sensitive to MS14, with inhibition zone radii of 23 mm and 22 mm, respectively. Plate assays revealed that strain MS14 could significantly inhibit the Gram-positive bacterium Clavibacter michiganensis subsp. michiganensis, which is the pathogen of a major tomato disease, namely, tomato wilt and canker (23). However, another Gram-positive bacterium, Bacillus megaterium, was not very sensitive to the growth of strain MS14, compared to the other pathogenic bacteria tested. Overall, these data indicate that the cell metabolites of strain MS14 have possible applications as potent broad-spectrum antibacterial agents against plant pathogens. Mutagenesis analysis of MS14 generated the antibacterial-defective mutants MS14MT357 and MS14MT577, which retained similar antifungal patterns, compared to the wild-type strain (Fig. 2). These data indicate that the antibacterial mechanism is independent of production of the antifungal occidiofungin.
FIG 1.
Antibacterial activity of Burkholderia contaminans MS14 against Xanthomonas citri pv. malvacearum MSCT1 (1), Pectobacterium carotovorum EC101 (2), Ralstonia solanacearum (3), Pseudomonas syringae pv. syringae B301D (4), Erwinia amylovora 2029 (5), Burkholderia glumae 291 (6), Escherichia coli (7), and Clavibacter michiganensis subsp. michiganensis Lu-01 (8). Aliquots (5 μl) of bacterial suspensions (optical density at 420 nm [OD420] values of 0.3) were inoculated onto the centers of NBY plates. After the plates had been incubated for 3 days at 28°C, the NBY plates were oversprayed with a suspension of indicator pathogenic bacteria (OD420 values of 0.3). Inhibition zones were measured from the margins of bacterial colonies 24 h later. The results show that MS14 has broad-spectrum antibacterial activities against the tested pathogens.
TABLE 1.
Antibacterial activities of Burkholderia contaminans MS14
| Indicator pathogenic bacterium | Inhibition zone radius (mm)a |
||
|---|---|---|---|
| MS14 | MT357 | MT577 | |
| Xanthomonas citri pv. malvacearum MSCT1 | 36 ± 1.66 | 0 | 0 |
| Pectobacterium carotovorum subsp. carotovora EC101 | 33 ± 0.86 | 0 | 1 ± 0.5 |
| Ralstonia solanacearum 102 | 31 ± 0.08 | 0 | 0 |
| Pseudomonas syringae pv. syringae B301D | 29 ± 1.00 | 0 | 0 |
| Erwinia amylovora 2029 | 23 ± 0.85 | 0 | 0 |
| Burkholderia glumae 291 | 22 ± 0.08 | 4 ± 1.55 | 5 ± 1.30 |
| Escherichia coli JM109 | 22 ± 0.77 | 0 | 0 |
| Clavibacter michiganensis subsp. michiganensis 1-07 | 17 ± 0.07 | 0 | 0 |
| Bacillus megaterium KM | 2.5 ± 0.04 | 0 | 0 |
Mean ± standard deviation.
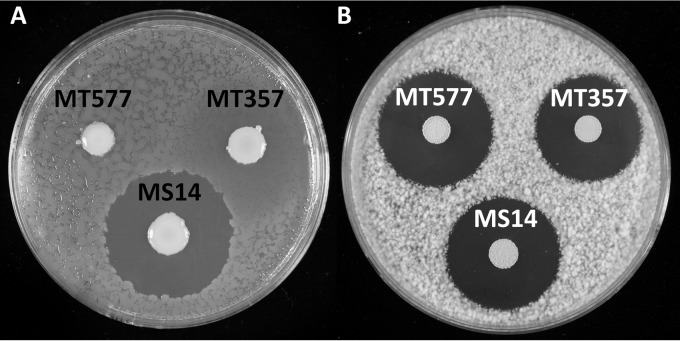
FIG 2.
Plate bioassays of antimicrobial activities of Burkholderia contaminans strain MS14 and the mutants MS14MT357 (luxR::Tn5) and MS14MT577 (orbI::Tn5). (A) Loss of antibacterial activity of strain MS14 against the bacterial indicator Erwinia amylovora in MS14MT357 and MS14MT577. (B) Antifungal activities of strains MS14, MT357, and MT577 against the fungal indicator Geotrichum candidum. The results show that the genes disrupted in the mutants are not related to production of the antifungal activity of strain MS14.
Identification of genes involved in production of the antibacterial product.
Mutants of strain MS14 were created by EZ-Tn5 transposon insertion and were tested for antibacterial activity against our indicator strain of Erwinia amylovora. Two mutants that lost activity in the overlay assay were named MS14MT357 and MS14MT577; the plasmid rescue method was used to obtain plasmids pDP357 and pDP577, respectively, from the genomes of the mutants. Plasmid details are shown in Table 2. BLAST analysis using the DNA sequence generated from plasmid pDP357 (rescued from the mutant MS14MT357) against the MS14 genome showed that the disrupted gene NL30_RS14390 is 672 bp in size and is a luxR family transcriptional regulator (see Fig. S1 in the supplemental material). Sequence analysis of pDP577 revealed that the disrupted gene in mutant MS14MT577 is at locus NL30_RS14890, which is 9,663 bp in size (Fig. S2) and encodes a 3,219-amino-acid peptide. The deduced peptide of NL30_RS14895 shares 93% identity with the product of the orbI gene in Burkholderia cenocepacia J2315, which is one of the two NRPS genes for siderophore ornibactin biosynthesis (24). Given the size of the gene product in the MS14MT577 mutant, complementation cannot be achieved. The downstream genes are also involved in ornibactin biosynthesis; therefore, any possible polar effects would be on genes involved in the synthesis of the same product.
TABLE 2.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Source |
|---|---|---|
| Escherichia coli | ||
| Ec100D | F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80dlacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara leu)7697 galU galK λ− rpsL (Strr) nupG pir+(DHFR) | Epicentre Corp. |
| Burkholderia contaminans | ||
| MS14 | Wild-type strain | |
| MS14MT357 | luxR::Tn5 derivative of MS14; Kmr | This study |
| MS14MT577 | NRPS gene::Tn5 derivative of MS14; Kmr | This study |
| Plasmids | ||
| pDP357 | EZ-Tn5 carrying 1.2-kb genomic DNA of MS14MT357; Kmr | This study |
| pDP577 | EZ-Tn5 carrying 0.9-kb genomic DNA of MS14MT577; Kmr | This study |
| pMLS7 | Expression vector of Burkholderia; Tpr | Lefebre and Valvano (25) |
| pDP357-2 | pMLS7 carrying 828-bp BamHI and HindIII fragment containing intact luxR gene; Tpr | This study |
Kmr, kanamycin resistant; Tpr, trimethoprim resistant.
Complementation of the mutated luxR-type gene.
The intact luxR-type transcriptional regulator gene was cloned into the Burkholderia expression vector pMLS7 by using the primer pair LuxRF (5′-CTGAGGATCCATTCAAACTAAACGAACGGGG-3′) and LuxRR (5′-GACGAAGCTTTGGCTCAGCGCGTTTC-3′), with the addition of BamHI and HindIII enzyme-digesting sequences, respectively. The cloned luxR genes were regulated by the S7 ribosomal protein promoter (25). The generated plasmid, pDP357-2, was transformed into the mutant MS14MT357 to be expressed constitutively. Plate bioassays revealed that the antibacterial activities of the mutants against Erwinia amylovora had fully been restored to the wild-type level, compared with the strain MS14 (Fig. S3). Considering that the EZ-Tn5 transposome was reported previously to have no polar effects leading to the inactivation of downstream genes (26) and that luxR gene complementation could fully restore MS14 antibacterial activities, we think that downstream genes were unlikely to have been affected by the insertional mutagenesis. The results demonstrated that the LuxR family transcriptional regulator is essential for the observed bactericidal activity in strain MS14.
Isolation and characterization of products from MS14MT357 and MS14MT577.
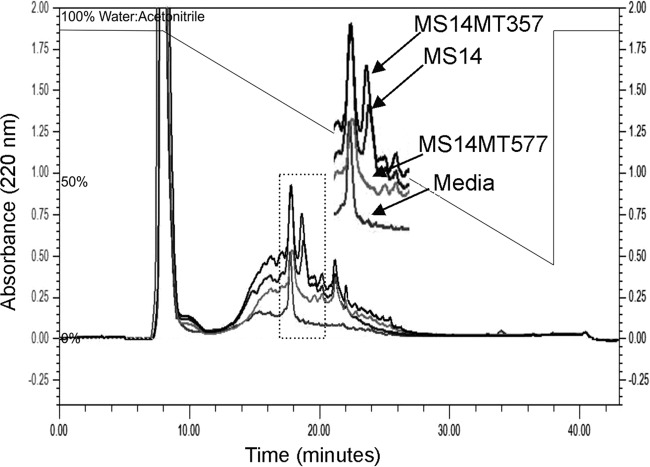
The wild-type strain MS14 and the mutant strains MS14MT357 and MS14MT577 were cultured and antimicrobial compounds were extracted following an identical procedure. Extracts were analyzed on a reverse-phase high-performance liquid chromatography (RP-HPLC) column to determine differences among the isolated products. Wild-type MS14 and MS14MT357 had comparable peaks at the retention time of 18 min, eluting in 64:36 water/acetonitrile (Fig. 3). The mutant strain MS14MT577 did not produce a similar product at this retention time. In a MIC assay, the fraction at 18 min was the only product that exhibited any inhibitory activity against E. amylovora, but this activity was clearly not bactericidal. The initial Diaion HP-20 extracts had bactericidal activity in a zone-of-inhibition plate assay, but the bactericidal activity was not recovered from any RP-HPLC fraction. Isolation of the bactericidal compound has not yet been achieved.
FIG 3.
RP-HPLC chromatograms. An overlay of the chromatograms at 220 nm of the final purification step of the wild-type MS14 fraction, MS14MT357, and MS14MT577, using a C18 column (4.6 by 250 mm), is shown. Extracted medium was run as a negative control. The dotted box represents the region that was enlarged.
The bacteriostatic product was isolated from wild-type MS14, and the structure was characterized by correlation spectroscopy (COSY), total correlation spectroscopy (TOCSY), nuclear Overhauser effect spectroscopy (NOESY), and heteronuclear single quantum coherence (HSQC) nuclear magnetic resonance (NMR) (Fig. S4, S5, S6, and S7) and mass spectrometry. NMR analysis revealed that the purified product contained TOCSY spin systems for 3-hydroxyoctanoic acid (HOA), ornithine (Orn), aspartic acid (Asp), serine (Ser), and putrescine (Put) (Fig. S8 and Table S1). Furthermore, nuclear Overhauser effects (NOEs) were observed in the NOESY experiment, confirming the assigned position of each residue within the structure. NOEs were observed between Orn(Nδ-OH)1λ and Asp(β-OH)2NH, Orn(Nδ-OH)1α and Asp(β-OH)2NH, Asp(β-OH)2α and Ser3NH, Ser3α and Orn(Nδ-OH)4NH, Orn(Nδ-OH)4δ and formyl, Orn(Nδ-OH)4α and PutNH, and Ser3α and PutNH3+ (Fig. S9 and S10). The isolated product was structurally determined to be ornibactin-F, with a mass of 737 Da. In addition, a chrome azurol S (CAS) plate assay was used to demonstrate that the isolated product had siderophore activity (Fig. 4). The observed lack of the product in the MS14MT357 strain is to be expected, given that the mutation is within the biosynthesis pathway for ornibactin.
FIG 4.
Chrome azurol S (CAS) plate assay. The wild-type MS14 and MS14MT357 strains have clear zones of siderophore activity, while the MS14MT577 strain has lost this activity due to the absence of ornibactin production.
Restoration of bactericidal activity through coculture of MS14MT357 and MS14MT577.
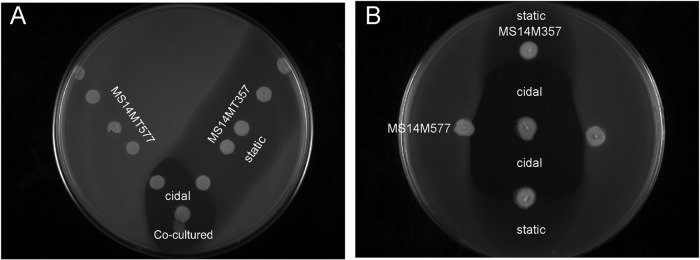
The isolated ornibactin-F product does not account for the bactericidal activity observed in the wild-type strain. This is supported by the lack of bactericidal activity in the MS14MT357 strain, which is capable of producing the same ornibactin product as wild-type MS14. Therefore, ornibactin is not directly responsible for the observed bactericidal activity. The relationship between ornibactin production and the LuxR family transcriptional regulator for synthesis of the bactericidal compound was further evaluated using plate overlay assays (Fig. 5). The mutant strains MS14MT357 and MS14MT577 were spotted on a plate in the shape of a V, with the colonies at the bottom being mixed cultures. Siderophore bacteriostatic activity was observed around each colony of the MS14MT357 strain along the right side of the V, while MS14MT577 did not inhibit the growth of the indicator strain along the left side of the V. The bactericidal activity, which was evident in the clear zone of inhibition at the bottom of the V, was present only when the MS14MT357 and MS14MT577 mutants were grown in close proximity. Similarly, when the strains were grown perpendicular to each other, a clear bactericidal zone of inhibition was present when cultures were mixed at the center of the plate (Fig. 5B). Synthesis of both the LuxR family transcriptional regulator and ornibactin is required for production of the bactericidal compound. To test this observation, the bactericidal and siderophore activities were tested with elevated concentrations of ferric iron. Increasing concentrations of ferric iron have been shown to regulate the synthesis of ornibactin (27). If ornibactin is directly involved in the regulation of the bactericidal product, then bactericidal activity should be absent at the same ferric iron concentrations that inhibit ornibactin biosynthesis. A concomitant loss of siderophore activity and bactericidal activity was observed for Diaion HP-20 extracts with increasing concentrations of available ferric iron (Fig. 6A and B). The loss of siderophore and bactericidal activities corresponded to loss of the ornibactin product observed by RP-HPLC (Fig. 6C). The data support the observation that ornibactin is directly required for the bactericidal activity of MS14. One possibility is that an ornibactin by-product is responsible for the observed bactericidal activity. This would suggest that the luxR gene NL30_RS14395 product is involved in the regulation of a product that modifies ornibactin. This scenario is unlikely, given that this product would presumably be isolated by the same extraction method as used to isolate ornibactin. It is more likely that ornibactin has a regulatory role in MS14 and promotes the synthesis of a bactericidal secondary product. This is supported by the observation that bactericidal activity was restored only in the mutant deficient in ornibactin production and not in the luxR regulatory mutant strain. When the ornibactin-deficient mutant MS14MT577 was grown on plates made from the culture broth of the MS14MT357 strain, the clear bactericidal zone of inhibition was restored (Fig. 7A). In a related assay, 100 μg of ornibactin was added to the center of a plate that had been previously stabbed with the MS14MT577 mutant (Fig. 7B and C). The addition of ornibactin restored the bactericidal activity of the MS14MT577 mutant strain, demonstrating that ornibactin has an activity aside from just sequestering iron in B. contaminans MS14.
FIG 5.
Bioassay for antibacterial activity. The mutant strains MS14MT357 and MS14MT577 were grown in proximity in a V assay (A) and a vertical assay (B). Mixed cultures are present at the bottom of the V in the V assay and in the center in the vertical assay. Both assays show that the mixed cultures produce bactericidal activity and that the bactericidal activity is also present in stabs of the two strains grown in close proximity (“cidal” indicates the bactericidal region on the plate). The bacteriostatic activity of ornibactin production in the MS14MT357 strain is visible in the observed growth reduction of the indicator strain (“static” indicates the bacteriostatic region on the plate).
FIG 6.
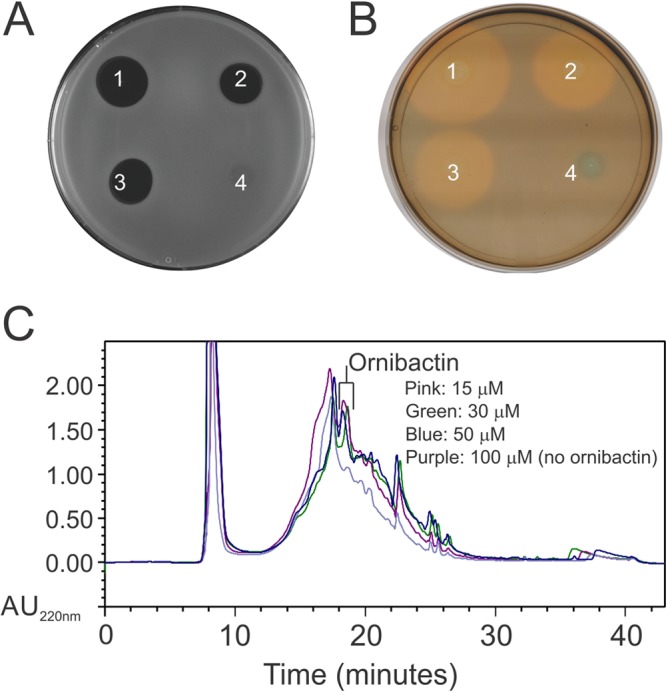
Bioactivity with supplemented ferric iron. (A) Antibacterial activity against E. amylovora of Diaion HP-20 extracts of cultures grown with ferric iron at 15 (1), 30 (2), 50 (3), and 100 (4) μM. (B) Chrome azurol S (CAS) plate assay of Diaion HP-20 extracts of cultures grown with ferric iron at 15 (1), 30 (2), 50 (3), and 100 (4) μM. (C) RP-HPLC chromatograms of Diaion HP-20 extracts of cultures grown with ferric iron at 15, 30, 50, and 100 μM. No siderophore or bactericidal activity is observed in cultures grown with 100 μM ferric iron.
FIG 7.
Bioassays of MS14MT577. (A) MS14MT577 strain grown on a plate made from a 3-day-old culture broth of the MS14MT357 strain and overlaid with the E. amylovora indicator strain after 3 days of growth. (B) Negative control in which the indicator strain was overlaid 3 days after 100 μg of ornibactin was spotted in the center of the plate. (C) Four inoculated colonies of MS14MT577 with ornibactin spotted in the center of the plate and overlaid with the indicator strain 3 days later. Bactericidal activity of the MS14MT577 strain was restored when MS14MT577 was grown on the culture broth of the MS14MT357 strain or when ornibactin was added directly.
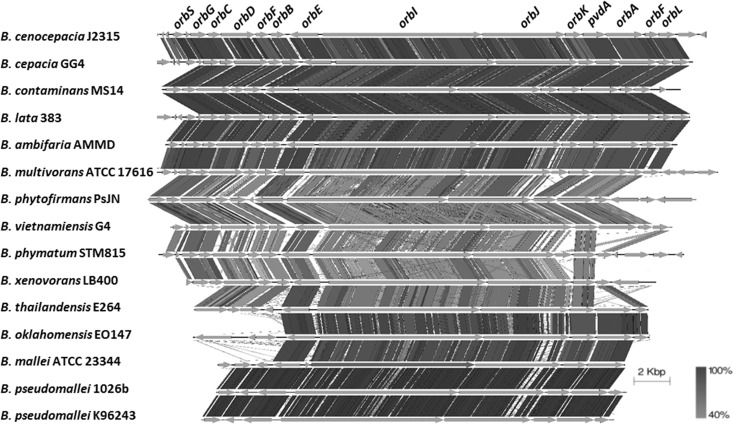
Genetic architecture of ornibactin biosynthesis loci among Burkholderia species.
The relationship between the production of ornibactin and B. cepacia complex virulence remains unclear (14). Given the demonstrated function of ornibactin in MS14 bactericidal activity, ornibactin biosynthesis loci were compared among Burkholderia species (Fig. 8). B. cenocepacia was selected as the reference because the ornibactin biosynthesis locus is best described in strain J2315 (24, 28). Using this reference, the ornibactin loci were identified for 13 Burkholderia species, including the pathogenic species Burkholderia multivorans ATCC 17616, Burkholderia mallei ATCC 23344, Burkholderia thailandensis E264, Burkholderia oklahomensis EO147, Burkholderia pseudomallei 1026b, and B. pseudomallei K96243 (29–32), the PGPB Burkholderia lata 383, Burkholderia ambifaria AMMD, and Burkholderia phytofirmans PsJN (33–35), and the soil isolates B. cepacia GG4, Burkholderia vietnamiensis G4, Burkholderia phymatum STM815, and Burkholderia xenovorans LB400 (36–38). We also analyzed the plant pathogens B. glumae BGR1 and Burkholderia gladioli BSR3 (39, 40); however, the ornibactin biosynthesis loci were not identified for the two species. The comparison of ornibactin loci demonstrated a high degree of conservation of the orbI, orbJ, orbE, and pvdA genes, which are responsible for ornibactin biosynthesis and ornibactin export across cytoplasmic membranes. The MS14 NRPS genes NL30_RS14890 and NL30_RS14895 share 90% nucleotide identity with the orbJ and orbI genes, respectively, in B. cenocepacia J2315, and the deduced peptides share 93% identity with those in J2315. In contrast, the genes from orbS to orbB and from orbA to orbL, which are involved in ornibactin biosynthesis initiation, regulation, transportation, and modification, show significant diversity among the studied Burkholderia genomes. In this analysis, the ornibactin products appear to be fairly consistent across Burkholderia species. The main question is why the regulatory elements for ornibactin production are so diverse, for a product that is thought to have a limited function in sequestering iron. The genomic comparison further illustrates the need to evaluate the possible functions ornibactin may have within Burkholderia.
FIG 8.
Ornibactin biosynthesis locus genetics of Burkholderia species. The ornibactin loci showed a high degree of conservation of the orbL, orbJ, orbE, and pvdA genes among the Burkholderia species compared; those genes are responsible for the biosynthesis and secretion of ornibactin. The bar at the right bottom indicates the degrees of similarity (40 to 100%).
DISCUSSION
The findings from this study demonstrate that MS14 has antibacterial activities against a wide range of plant-pathogenic bacteria, including Erwinia amylovora, Xanthomonas citri pv. malvacearum, and Clavibacter michiganensis subsp. michiganensis. Random mutagenesis studies resulted in the identification of two MS14 mutants with losses in antibacterial production. Both of the mutations occurred within regions that are not directly involved in the biosynthesis of the bactericidal product. Bactericidal activity could be restored by growing the ornibactin NRPS mutant and the LuxR family transcriptional regulatory mutant in proximity, suggesting that ornibactin production is essential for production of the antibacterial compound. We also showed a significant amount of diversity among Burkholderia species for the initiation and regulation of ornibactin biosynthesis. Given the genomic diversity of these regions, ornibactin presumably has evolved to have functional roles in addition to iron sequestration and iron uptake.
A LuxR family transcriptional regulator and the synthesis of ornibactin were essential for the production of a bactericidal compound in B. contaminans MS14 under the culture conditions tested. Mutations in each of the luxR and orbI genes resulted in a loss of production of the bactericidal activity. The LuxR bacterial protein family is one of the largest groups involved in the regulation of cell functions to control a variety of phenotypes (41). The results of this research suggest that production of the bactericidal activity in MS14 is regulated by the luxR gene (NL30_RS14390) via quorum-sensing communication. Currently, there are multiple mechanisms for the LuxR proteins to regulate cell functions in nearly all known Gram-negative quorum-sensing systems (42). Further studies are needed to understand the details of the regulation of bacterial activity. As shown previously, the genetic locus NL30_RS14890 is a homolog of the orbI gene of B. cenocepacia J2315, and the deduced peptide of NL30_RS14890 shares 93% identity with the OrbI protein, which is one of the two synthetases for production of the siderophore ornibactin (24). Mutation of the orbI gene in B. contaminans MS14 led to the ornibactin-negative phenotype, which is consistent with the result obtained in B. cenocepacia J2315 (24). Therefore, current data support the idea that luxR gene transcriptional regulation of the biosynthesis genes for the unknown bactericidal compound(s) and the orbI gene is involved in the biosynthesis of the siderophore ornibactin, which is associated with the production of bactericidal activity.
The structure of the isolated siderophore determined from our analysis is the same as that reported previously for ornibactin-F (43). The mass for this product also is similar to the reported mass for ornibactin-F, i.e., 737 Da (43). Ornibactin (27) is a tetrapeptide siderophore that was reported to be produced first by Pseudomonas (44) and then by several B. cenocepacia strains (27, 45). The ornibactin gene cluster contains two core NRPS genes (24), each of which is composed of an amino acid adenylation domain and a condensation domain. The two domains are core components of the NRPS mechanism (46). These domains for ornibactin biosynthesis were conserved among the Burkholderia species that were compared (Fig. 8), suggesting that there is little deviation from the core structure of the ornibactin product.
Bactericidal activity is associated with the presence of ornibactin and is absent when ornibactin production is inhibited with higher iron concentrations. Concentrations of ≥15 μM ferric iron were enough to suppress ornibactin biosynthesis by members of the Burkholderia cepacia complex. Ornibactin production in MS14 was shown to be suppressed with the addition of 100 μM ferric iron. At that iron concentration, there was a concomitant loss of bactericidal activity (Fig. 6). Comparison of the ornibactin gene cluster with previously sequenced genomes of 14 Burkholderia species, including plant-growth-promoting strains and mammal-pathogenic strains, indicated that the ornibactin biosynthesis gene cluster commonly exists among other Burkholderia species. The diversity in the regulatory and transport regions within the gene cluster for ornibactin biosynthesis supports additional regulatory roles for the compound in Burkholderia species. In the case of MS14, ornibactin does have a clear role in promoting the bactericidal activity of the strain. Given the requirement of ornibactin for production of the bactericidal compound in MS14, ornibactin may also be crucial for the synthesis of other secondary metabolites in other bacterial systems. Ornibactin possibly has a broader function for virulence, aside from just sequestering iron. It is clear that ornibactin has an alternative function aside from iron sequestration in MS14, and a better understanding of this alternative activity could possibly promote the isolation of novel secondary metabolites or yield a more complete picture of the role of ornibactin in bacterial virulence.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture medium.
Bacterial strains and plasmids used in this study are listed in Table 2. Escherichia coli strain TransforMax EC100DTM pir+ (Epicentre Biotechnologies, Madison, WI) was used for plasmid rescue cloning and was cultured in Luria-Bertani (LB) medium at 37°C. Nutrient broth-yeast extract (NBY) agar (47) was used for cultures of Burkholderia strains and for plate bioassays of antimicrobial activities. Potato dextrose agar (PDA) (Difco, Detroit, MI) was used for plate bioassays to evaluate antifungal activities. Antibiotics (Sigma Chemical Co., St. Louis, MO), if applicable, were added to media at the following concentrations: trimethoprim, 100 mg/ml; kanamycin, 100 mg/ml for Escherichia coli and 300 mg/ml for Burkholderia strains.
Bioassay for antimicrobial activities.
B. contaminans MS14 and its mutants used in this study were evaluated for antibacterial activities against Erwinia amylovora 2029 and other pathogenic indicators by using NBY plate bioassays. The bioassay was similar to that described by Scholz-Schroeder and colleagues (48). Briefly, MS14 and mutants were grown overnight in 5 ml of NBY liquid medium at 28°C. The bacterial cells were then collected by centrifugation and suspended in sterile distilled water to an optical density (OD) of 0.3 (approximately 2 ×108 CFU/ml). Aliquots (5 μl) of bacterial suspensions were inoculated onto the centers of NBY plates. After the plates had been incubated for 2 days at 28°C, NBY plates were oversprayed with a suspension of the indicator bacterial strains (OD at 420 nm [OD420] of 0.3) and PDA plates were oversprayed with the indicator fungus Geotrichum candidum F-260 (OD420 of 0.3). Inhibition zones were measured from the margins of bacterial colonies 12 to 24 h later, and the sizes of the zones were compared between MS14 and the mutants. MS14MT577 was grown on plates made from the culture supernatants of extracted media from soft agar stabs. Supernatants were collected as described below and were used to make 1.5% agar plates. The MS14MT577 strain was stabbed into the culture supernatant agar plates and grown for 3 days at 28°C. The plates were placed in an oven at 60°C for 1 h, to kill the MS14MT577 strain, before being overlaid with E. amylovora 2029. E. amylovora was grown to an OD600 of 0.2 and was diluted 20-fold in NBY agar (0.75%). Five milliliters of the soft agar suspension was poured over the plates, and the plates were placed at 28°C for 1 day. In another assay, four colonies were stabbed into the center of NBY agar plates before 100 μg of ornibactin (10 μg/μl stock solution in 35% acetonitrile) was added to the center of the colonies. The plates were incubated for 3 days before being overlaid with the indicator bacterial strain embedded in the top agar, as described above. Three replicates of the plate bioassays were performed independently, and standard deviations of the means were calculated. To differentiate bactericidal activity from bacteriostatic activity, agar plugs (1 by 1 cm each) within the zones of inhibition of the indicator bacterial strain Erwinia amylovora were picked up and suspended in 1 ml of NBY medium; 100 μl of the suspension was plated. Bactericidal activity was indicated by no growth of the indicator bacterial strain from the agar plugs cut from the clear zones of inhibition. The initial CFU (time zero) from an agar plug (1 by 1 cm) suspended in 1 ml of medium was ∼8 × 104 CFU.
Random mutagenesis.
The EZ-Tn5 <R6Kγori/KAN-2>Tnp transposome kit was used, as recommended by the manufacturer (Epicentre Biotechnologies, Madison, WI), to characterize the genes dedicated to the antibacterial activity of MS14. MS14 acquired kanamycin resistance on NBY plates with the EZ-Tn5 transposon insertion into the genome, and mutants could be selected on NBY plates supplemented with 300 μg/ml kanamycin. The mutants that exhibited reduced or no antibacterial activity against Erwinia amylovora were isolated; 16S rRNA and recA genes were cloned and sequenced to confirm that the resulting mutants were derivatives of strain MS14. Plasmid rescue cloning was performed according to the transposome kit instructions, to generate the plasmids pPD357 and pPD577 (Table 2). To confirm that the rescue plasmid contained the transposon sequence, a portion of the Tn5 transposon sequence was amplified by PCR with the primers R6kF1 (5′-GGGTAGCCAGCAGCATCCT-3′) and R6kR1 (5′-CATGATCGTGCTCCTGTCGTT-3′). The positive rescue clones were sequenced for further analysis. Sequence analysis was accomplished using the Lasergene Cloning Suite (version 12; DNASTAR, Inc., Madison, WI). Genes were searched against the B. contaminans MS14 reference genome (49). The BLASTn comparison of genomes was visualized with the BLAST Ring Image Generator (BRIG) (50).
Analysis and isolation of the siderophore product.
The wild-type strain MS14 was grown overnight at 28°C on modified NBY (487 ml distilled water, 2.5 g peptone, 1.5 g Todd-Hewitt broth, 1.0 g yeast extract, 1.0 g anhydrous K2HPO4, 0.25 g KH2PO4, and 1.5% agar, with 12.5 ml of 20% glucose and 0.5 ml of 1 M MgSO4 added after autoclaving) agar plates. Colonies from the overnight NBY agar plates were stabbed into 500 ml of modified NBY soft agar (NBY medium with only 0.75% agar). The inoculum in soft agar was placed at 28°C for 3 days and then frozen at −80°C. The medium was thawed in a 65°C water bath for 1 h. The inoculum was then placed in 250-ml centrifuge bottles and centrifuged at 20,000 × g for 30 min. The collected supernatant was pooled, mixed with 1 g of the polyaromatic absorbance resin Diaion HP-20, and shaken for 1 h. The resin was allowed to settle before the supernatant was decanted, and the resin was suspended following the decantation of the medium in 10 ml of 50% acetonitrile in water. The extract was dried by lyophilization and suspended in 1 ml of 35% acetonitrile in water. The extracts were tested for siderophore and antibacterial activities by spotting 10 μl of the extract on a chrome azurol S (CAS) plate or on an NBY plate overlaid with Erwinia amylovora 2029. RP-HPLC was performed using a C18 column (4.6 by 250 mm; Grace-Vydac catalog no. 201TP54) on a Bio-Rad BioLogic DuoFlow F10 system with a QuadTec UV-visible detector. Fractions were separated by using a 30-min gradient from 90:10 to 20:80 (water with 0.1% trifluoroacetic acid/acetonitrile with 0.1% trifluoroacetic acid).
Structural determination of the siderophore product by NMR.
A 2 mM sample of the purified bacteriostatic compound was prepared in acetonitrile-D3 (Cambridge Isotopes)/H2O (50:50). The NMR data were collected with a Bruker Avance III HD 600-MHz spectrometer and a Bruker Avance III HD 850-MHz spectrometer, using TCI CryoProbes for each spectrometer. The 1H resonances were assigned according to standard methods (51), using COSY, TOCSY, NOESY, and 13C-HSQC NMR experiments. NMR data were collected at 10°C. The carrier frequency was centered on the water resonance, which was suppressed minimally by using standard presaturation methods. A 2.0-s relaxation delay between scans was used. The TOCSY experiment was performed with a 60-ms mixing time, using the Bruker DIPSI-2 spinlock sequence. The NOESY experiment was performed with a 400-ms mixing time. The parameters for collecting the HSQC spectrum were optimized for the observation of aliphatic and aromatic CH groups. The spectral sweep widths for the TOCSY and NOESY experiments were 11.35 ppm in both dimensions. The spectral sweep widths for the HSQC experiment were 11.35 ppm in the proton dimension and 0 to 85 ppm in the carbon dimension. All two-dimensional data were collected with 2,048 complex points for the acquisition dimension and 256 complex points for the indirect dimensions, except for the HSQC data, which were collected with 2,048 and 128 complex points for the direct and indirect dimensions, respectively. Phase-sensitive indirect detection for the NOESY, TOCSY, and COSY experiments was achieved using standard Bruker pulse sequences. 1H chemical shifts were referenced to the residual water peak (3.33 ppm). Data were processed with NMRPipe (52) by removing the residual water signal by deconvolution, multiplying the data in both dimensions by a squared sine-bell function with 45° or 60° shifts (for the 1H dimension of HSQC), zero-filling once, and performing Fourier transformation and baseline correction. Data were analyzed with the interactive computer program NMRView (53).
Mass spectrometry of the siderophore product.
The mass of the purified bacteriostatic product was confirmed by matrix-assisted laser desorption ionization (MALDI) using a Shimadzu/Kratos MALDI-TOF mass spectrometer in both the linear and reflectron modes. The isolated compound was further analyzed by an electrospray mass spectrometer using a Thermo Fisher Deca XP ion trap mass spectrometer. The compound was dissolved in acetonitrile/water (50:50 [vol/vol]) with 0.1% formic acid and injected into a 1-μl/min flow of the same solvent by using a Harvard syringe pump. The flow was sprayed using the nano-liquid chromatography interface. Tandem mass spectrometry was performed with singly charged ions using standard collision energy (34 V) and higher collision energy (50 V).
Plasmid construction for luxR gene complementation.
The intact luxR gene was amplified using the primer pair LuxRF (5′-CTGAGGATCCATTCAAACTAAACGAACGGGG-3′) and LuxRR (5′-GACGAAGCTTTGGCTCAGCGCGTTTC-3′), in which restriction endonuclease cutting sites (underlined; for BamHI and HindIII, respectively) were added. The resulting PCR product, containing the intact wild-type luxR gene, was digested with BamHI and HindIII and then cloned into the expression vector pMLS7 to generate the plasmid pDP357-2, as described previously (54). The plasmid pDP357-2 was electroporated into competent MT357 cells to recover the wild-type characteristics. Empty vector was used as a negative control. Single colonies were picked from NBY plates supplemented with trimethoprim (100 μg/ml) and kanamycin (300 μg/ml). Plasmid was extracted from the colonies, and sequencing confirmed the existence of the resultant plasmid pDP357-2. Plate bioassays were used to evaluate the antibacterial activity of the resulting cells.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Rama Krishna and the support of the UAB Cancer Center NMR Shared Facility.
This research was funded in part by the USDA NIFA (grant MIS-401170 to S.-E.L.). The UAB Cancer Center NMR Shared Facility is funded by CCSG 1P30 CA-13148, NCI grant 1P30 CA-13148, and NCRR grant 1S10 RR022994-01A1.
We have no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00051-17.
REFERENCES
- 1.Francis F, Kim J, Ramaraj T, Farmer A, Rush MC, Ham JH. 2013. Comparative genomic analysis of two Burkholderia glumae strains from different geographic origins reveals a high degree of plasticity in genome structure associated with genomic islands. Mol Genet Genomics 288:195–203. doi: 10.1007/s00438-013-0744-x. [DOI] [PubMed] [Google Scholar]
- 2.De Meyer SE, Cnockaert M, Ardley JK, Maker G, Yates R, Howieson JG, Vandamme P. 2013. Burkholderia sprentiae sp. nov., isolated from Lebeckia ambigua root nodules. Int J Syst Evol Microbiol 63:3950–3957. doi: 10.1099/ijs.0.048777-0. [DOI] [PubMed] [Google Scholar]
- 3.Parke JL, Gurian-Sherman D. 2001. Diversity of the Burkholderia cepacia complex and implications for risk assessment of biological control strains. Annu Rev Phytopathol 39:225–258. doi: 10.1146/annurev.phyto.39.1.225. [DOI] [PubMed] [Google Scholar]
- 4.el-Banna N, Winkelmann G. 1998. Pyrrolnitrin from Burkholderia cepacia: antibiotic activity against fungi and novel activities against streptomycetes. J Appl Microbiol 85:69–78. doi: 10.1046/j.1365-2672.1998.00473.x. [DOI] [PubMed] [Google Scholar]
- 5.Mahenthiralingam E, Urban TA, Goldberg JB. 2005. The multifarious, multireplicon Burkholderia cepacia complex. Nat Rev Microbiol 3:144–156. doi: 10.1038/nrmicro1085. [DOI] [PubMed] [Google Scholar]
- 6.Lipuma JJ. 2005. Update on the Burkholderia cepacia complex. Curr Opin Pulm Med 11:528–533. doi: 10.1097/01.mcp.0000181475.85187.ed. [DOI] [PubMed] [Google Scholar]
- 7.Lu S-E, Novak J, Austin FW, Gu G, Ellis D, Kirk M, Wilson-Stanford S, Tonelli M, Smith L. 2009. Occidiofungin, a unique antifungal glycopeptide produced by a strain of Burkholderia contaminans. Biochemistry 48:8312–8321. doi: 10.1021/bi900814c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birchall GR, Hughes CG, Rees AH. 1970. Newer synthes of the pyoluteorin antibiotics. Tetrahedron Lett (56):4879–4882. [DOI] [PubMed] [Google Scholar]
- 9.Kang Y, Carlson R, Tharpe W, Schell MA. 1998. Characterization of genes involved in biosynthesis of a novel antibiotic from Burkholderia cepacia BC11 and their role in biological control of Rhizoctonia solani. Appl Environ Microbiol 64:3939–3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neilands JB. 1995. Siderophores: structure and function of microbial iron transport compounds. J Biol Chem 270:26723–26726. doi: 10.1074/jbc.270.45.26723. [DOI] [PubMed] [Google Scholar]
- 11.Hider RC, Kong X. 2010. Chemistry and biology of siderophores. Nat Prod Rep 27:637–657. doi: 10.1039/b906679a. [DOI] [PubMed] [Google Scholar]
- 12.Wang H, Fewer DP, Holm L, Rouhiainen L, Sivonen K. 2014. Atlas of nonribosomal peptide and polyketide biosynthetic pathways reveals common occurrence of nonmodular enzymes. Proc Natl Acad Sci U S A 111:9259–9264. doi: 10.1073/pnas.1401734111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marahiel MA, Stachelhaus T, Mootz HD. 1997. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem Rev 97:2651–2674. doi: 10.1021/cr960029e. [DOI] [PubMed] [Google Scholar]
- 14.Sokol P, Darling P, Woods D, Mahenthiralingam E, Kooi C. 1999. Role of ornibactin biosynthesis in the virulence of Burkholderia cepacia: characterization of pvdA, the gene encoding l-ornithine N5-oxygenase. Infect Immun 67:4443–4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, Lory S, Ramphal R, Jin S. 1996. Isolation and characterization of Pseudomonas aeruginosa genes inducible by respiratory mucus derived from cystic fibrosis patients. Mol Microbiol 22:1005–1012. doi: 10.1046/j.1365-2958.1996.01533.x. [DOI] [PubMed] [Google Scholar]
- 16.Gray-Owen SD, Schyvers AB. 1996. Bacterial transferrin and lactoferrin receptors. Trends Microbiol 4:185–191. doi: 10.1016/0966-842X(96)10025-1. [DOI] [PubMed] [Google Scholar]
- 17.Wang X, Liu A, Guerrero A, Liu J, Yu X, Deng P, Ma L, Baird S, Smith L, Li X. 2016. Occidiofungin is an important component responsible for the antifungal activity of Burkholderia pyrrocinia strain Lyc2. J Appl Microbiol 120:607–618. doi: 10.1111/jam.13036. [DOI] [PubMed] [Google Scholar]
- 18.Schaad NW, Postnikova E, Lacy G, Sechler A, Agarkova I, Stromberg PE, Stromberg VK, Vidaver AK. 2006. Emended classification of xanthomonad pathogens on citrus. Syst Appl Microbiol 29:690–695. doi: 10.1016/j.syapm.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 19.Toth IK, Bell KS, Holeva MC, Birch PR. 2003. Soft rot erwiniae: from genes to genomes. Mol Plant Pathol 4:17–30. doi: 10.1046/j.1364-3703.2003.00149.x. [DOI] [PubMed] [Google Scholar]
- 20.Prior P, Allen C, Elphinstone J (ed). 1998. Bacterial wilt disease: molecular and ecological aspects. Springer Verlag, Berlin, Germany. [Google Scholar]
- 21.Billing E. 1974. The effect of temperature on the growth of the fireblight pathogen, Erwinia amylovora. J Appl Bacteriol 37:643–648. doi: 10.1111/j.1365-2672.1974.tb00488.x. [DOI] [PubMed] [Google Scholar]
- 22.Ham JH, Melanson RA, Rush MC. 2011. Burkholderia glumae: next major pathogen of rice? Mol Plant Pathol 12:329–339. doi: 10.1111/j.1364-3703.2010.00676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gartemann KH, Kirchner O, Engemann J, Grafen I, Eichenlaub R, Burger A. 2003. Clavibacter michiganensis subsp. michiganensis: first steps in the understanding of virulence of a Gram-positive phytopathogenic bacterium. J Biotechnol 106:179–191. [DOI] [PubMed] [Google Scholar]
- 24.Agnoli K, Lowe CA, Farmer KL, Husnain SI, Thomas MS. 2006. The ornibactin biosynthesis and transport genes of Burkholderia cenocepacia are regulated by an extracytoplasmic function σ factor which is a part of the Fur regulon. J Bacteriol 188:3631–3644. doi: 10.1128/JB.188.10.3631-3644.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lefebre MD, Valvano MA. 2002. Construction and evaluation of plasmid vectors optimized for constitutive and regulated gene expression in Burkholderia cepacia complex isolates. Appl Environ Microbiol 68:5956–5964. doi: 10.1128/AEM.68.12.5956-5964.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barquist L, Langridge GC, Turner DJ, Phan M-D, Turner AK, Bateman A, Parkhill J, Wain J, Gardner PP. 2013. A comparison of dense transposon insertion libraries in the Salmonella serovars Typhi and Typhimurium. Nucleic Acids Res 41:4549–4564. doi: 10.1093/nar/gkt148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyer J-M, Van VT, Stintzi A, Berge O, Winkelmann G. 1995. Ornibactin production and transport properties in strains of Burkholderia vietnamiensis and Burkholderia cepacia (formerly Pseudomonas cepacia). Biometals 8:309–317. doi: 10.1007/BF00141604. [DOI] [PubMed] [Google Scholar]
- 28.Holden MT, Seth-Smith HM, Crossman LC, Sebaihia M, Bentley SD, Cerdeno-Tarraga AM, Thomson NR, Bason N, Quail MA, Sharp S, Cherevach I, Churcher C, Goodhead I, Hauser H, Holroyd N, Mungall K, Scott P, Walker D, White B, Rose H, Iversen P, Mil-Homens D, Rocha EP, Fialho AM, Baldwin A, Dowson C, Barrell BG, Govan JR, Vandamme P, Hart CA, Mahenthiralingam E, Parkhill J. 2009. The genome of Burkholderia cenocepacia J2315, an epidemic pathogen of cystic fibrosis patients. J Bacteriol 191:261–277. doi: 10.1128/JB.01230-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeShazer D, Waag DM, Fritz DL, Woods DE. 2001. Identification of a Burkholderia mallei polysaccharide gene cluster by subtractive hybridization and demonstration that the encoded capsule is an essential virulence determinant. Microb Pathog 30:253–269. doi: 10.1006/mpat.2000.0430. [DOI] [PubMed] [Google Scholar]
- 30.Biddick R, Spilker T, Martin A, LiPuma JJ. 2003. Evidence of transmission of Burkholderia cepacia, Burkholderia multivorans and Burkholderia dolosa among persons with cystic fibrosis. FEMS Microbiol Lett 228:57–62. doi: 10.1016/S0378-1097(03)00724-9. [DOI] [PubMed] [Google Scholar]
- 31.Glass MB, Steigerwalt AG, Jordan JG, Wilkins PP, Gee JE. 2006. Burkholderia oklahomensis sp. nov., a Burkholderia pseudomallei-like species formerly known as the Oklahoma strain of Pseudomonas pseudomallei. Int J Syst Evol Microbiol 56:2171–2176. doi: 10.1099/ijs.0.63991-0. [DOI] [PubMed] [Google Scholar]
- 32.Knappe TA, Linne U, Zirah S, Rebuffat S, Xie X, Marahiel MA. 2008. Isolation and structural characterization of capistruin, a lasso peptide predicted from the genome sequence of Burkholderia thailandensis E264. J Am Chem Soc 130:11446–11454. doi: 10.1021/ja802966g. [DOI] [PubMed] [Google Scholar]
- 33.Coenye T, Mahenthiralingam E, Henry D, LiPuma JJ, Laevens S, Gillis M, Speert DP, Vandamme P. 2001. Burkholderia ambifaria sp. nov., a novel member of the Burkholderia cepacia complex including biocontrol and cystic fibrosis-related isolates. Int J Syst Evol Microbiol 51:1481–1490. doi: 10.1099/00207713-51-4-1481. [DOI] [PubMed] [Google Scholar]
- 34.Vanlaere E, Baldwin A, Gevers D, Henry D, De Brandt E, LiPuma JJ, Mahenthiralingam E, Speert DP, Dowson C, Vandamme P. 2009. Taxon K, a complex within the Burkholderia cepacia complex, comprises at least two novel species, Burkholderia contaminans sp. nov. and Burkholderia lata sp. nov. Int J Syst Evol Microbiol 59:102–111. doi: 10.1099/ijs.0.001123-0. [DOI] [PubMed] [Google Scholar]
- 35.Weilharter A, Mitter B, Shin MV, Chain PS, Nowak J, Sessitsch A. 2011. Complete genome sequence of the plant growth-promoting endophyte Burkholderia phytofirmans strain PsJN. J Bacteriol 193:3383–3384. doi: 10.1128/JB.05055-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hong KW, Koh CL, Sam CK, Yin WF, Chan KG. 2012. Complete genome sequence of Burkholderia sp. strain GG4, a betaproteobacterium that reduces 3-oxo-N-acylhomoserine lactones and produces different N-acylhomoserine lactones. J Bacteriol 194:6317. doi: 10.1128/JB.01578-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vandamme P, Goris J, Chen WM, de Vos P, Willems A. 2002. Burkholderia tuberum sp. nov. and Burkholderia phymatum sp. nov., nodulate the roots of tropical legumes. Syst Appl Microbiol 25:507–512. doi: 10.1078/07232020260517634. [DOI] [PubMed] [Google Scholar]
- 38.Fries MR, Forney LJ, Tiedje JM. 1997. Phenol- and toluene-degrading microbial populations from an aquifer in which successful trichloroethene cometabolism occurred. Appl Environ Microbiol 63:1523–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lim J, Lee TH, Nahm BH, Choi YD, Kim M, Hwang I. 2009. Complete genome sequence of Burkholderia glumae BGR1. J Bacteriol 191:3758–3759. doi: 10.1128/JB.00349-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seo YS, Lim J, Choi BS, Kim H, Goo E, Lee B, Lim JS, Choi IY, Moon JS, Kim J, Hwang I. 2011. Complete genome sequence of Burkholderia gladioli BSR3. J Bacteriol 193:3149. doi: 10.1128/JB.00420-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fuqua C, Winans SC, Greenberg EP. 1996. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu Rev Microbiol 50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 42.Ng W-L, Bassler BL. 2009. Bacterial quorum-sensing network architectures. Annu Rev Genet 43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stephan H, Freund S, Meyer JM, Winkelmann G, Jung G. 1993. Structure elucidation of the gallium-ornibactin complex by 2D-NMR spectroscopy. Liebigs Ann Chem 1993:43–48. doi: 10.1002/jlac.199319930108. [DOI] [Google Scholar]
- 44.Stephan H, Freund S, Beck W, Jung G, Meyer JM, Winkelmann G. 1993. Ornibactins: a new family of siderophores from Pseudomonas. Biometals 6:93–100. [DOI] [PubMed] [Google Scholar]
- 45.Darling P, Chan M, Cox AD, Sokol PA. 1998. Siderophore production by cystic fibrosis isolates of Burkholderia cepacia. Infect Immun 66:874–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cane DE, Walsh CT. 1999. The parallel and convergent universes of polyketide synthases and nonribosomal peptide synthetases. Chem Biol 6:R319–R325. doi: 10.1016/S1074-5521(00)80001-0. [DOI] [PubMed] [Google Scholar]
- 47.Vidaver AK. 1967. Synthetic and complex media for the rapid detection of fluorescence of phytopathogenic pseudomonads: effect of the carbon source. Appl Microbiol 15:1523–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scholz-Schroeder BK, Hutchison ML, Grgurina I, Gross DC. 2001. The contribution of syringopeptin and syringomycin to virulence of Pseudomonas syringae pv. syringae strain B301D on the basis of sypA and syrB1 biosynthesis mutant analysis. Mol Plant Microbe Interact 14:336–348. doi: 10.1094/MPMI.2001.14.3.336. [DOI] [PubMed] [Google Scholar]
- 49.Deng P, Wang X, Baird SM, Showmaker KC, Smith L, Peterson DG, Lu S. 2016. Comparative genome-wide analysis reveals that Burkholderia contaminans MS14 possesses multiple antimicrobial biosynthesis genes but not major genetic loci required for pathogenesis. Microbiologyopen 5:353–369. doi: 10.1002/mbo3.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wüthrich K. 1986. NMR of proteins and nucleic acids. Wiley, New York, NY. [Google Scholar]
- 52.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293. [DOI] [PubMed] [Google Scholar]
- 53.Johnson BA, Blevins RA. 1994. NMRView: a computer program for the visualization and analysis of NMR data. J Biomol NMR 4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 54.Gu G, Wang N, Chaney N, Smith L, Lu SE. 2009. AmbR1 is a key transcriptional regulator for production of antifungal activity of Burkholderia contaminans strain MS14. FEMS Microbiol Lett 297:54–60. doi: 10.1111/j.1574-6968.2009.01653.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.