ABSTRACT
In Streptococcus thermophilus, gene transfer events and loss of ancestral traits over the years contribute to its high level of adaptation to milk environments. Biofilm formation capacity, a phenotype that is lost in the majority of strains, plays a role in persistence in dairy environments, such as milk pasteurization and cheese manufacturing plants. To investigate this property, we have studied S. thermophilus UC8547, a fast-acidifying dairy starter culture selected for its high capacity to form biofilm on stainless steel under environmental conditions resembling the dairy environment. Using a dynamic flow cell apparatus, it was shown that S. thermophilus UC8547 biofilm formation on stainless steel depends on the presence of milk proteins. From this strain, which harbors the prtS gene for the cell wall protease and shows an aggregative phenotype, spontaneous mutants with impaired biofilm capacity can be isolated at high frequency. These mutants lack the PrtS expendable island, as confirmed by comparison of the genome sequence of UC8547Δ3 with that of the parent strain. The prtS island excision occurs between two 26-bp direct repeats located in the two copies of the ISSth1 flanking this genomic island. The central role of PrtS was confirmed by analyzing the derivative strain UC8547Δ16, whose prtS gene was interrupted by an insertional mutation, thereby making it incapable of biofilm formation. PrtS, acting as a binding substance between the milk proteins adhered to stainless steel and S. thermophilus cell envelopes, mediates biofilm formation in dairy environments. This feature provides S. thermophilus with an ecological benefit for its survival and persistence in this environment.
IMPORTANCE The increased persistence of S. thermophilus biofilm has consequences in the dairy environment: if, on the one hand, the release of this microorganism from biofilm can promote the fermentation of artisanal cheeses, under industrial conditions it may lead to undesirable contamination of dairy products. The study of the molecular mechanism driving S. thermophilus biofilm formation provides increased knowledge on how an ancestral trait affects relevant phenotypes, such as persistence in the environment and efficiency of growth in milk. This study provides insight into the genetic factors affecting biofilm formation at dairy plants.
KEYWORDS: biofilm, genome, milk, PrtS, stainless steel, Streptococcus thermophilus
INTRODUCTION
For many years, the dairy industry has benefited from Streptococcus thermophilus natural activity, as that species is the most used thermophilic starter culture for yogurt and cheese manufacture (1). Complete genome analysis revealed that this microorganism harbors genes that well describe its adaptation to milk environments, seen after experiencing a series of horizontal gene transfer (HGT) episodes and loss-of-function events that differentiated this bacterium from its original commensal nature (2–4). Even if the exact source is not well known and its presence was originally correlated to Lactococcus lactis (5), the cell envelope proteinase PrtS in S. thermophilus could be indicated as an example of a gene acquired by HGT that recently appears to be highly conserved and therefore selected in the industrial strains (6). This is supported by the fact that in S. thermophilus, the prtS gene, coding for an LPXTG-containing serine proteinase of the subtilisin family (5), is part of a genomic island flanked by conserved insertion sequence (IS) elements (6–8). This mobile island has been shown to be involved in recombination events involving the flanking IS elements that lead to the loss of the prtS genomic island (6–8), therefore influencing gene homeostasis and species heterogeneity (7). The close proximity of these IS elements to genes involved in adaptation to milk environments reveals that their acquisition in this bacterium's genome could represent a relatively recent excision event (8). prtS genetic adoption generated beneficial properties in S. thermophilus, like fast growth in milk substrate and the ability to hydrolyze casein with a consequent high acidification rate (6); moreover, additional unknown functions could be conferred by these horizontal genomic rearrangements compared to prtS-deficient strains.
Another trait that S. thermophilus shows prior to adaptation to the milk environment and that is maintained in the domestication process in milk is the ability to form biofilm (9). High levels of contamination by biofilms formed by thermophilic bacteria have been detected in the heating sections of cheese-milk pasteurization equipment, where the temperature ranges from 30 to 73°C, and these contaminated sections are consequently released in the pasteurized milk (10, 11). This can cause defects in milk and cheese quality, such as acidic flavor and undesirable texture (11, 12). Even if further studies are needed, biofilm formation by streptococci seems to be influenced by the different milk components and by the surface material composition of the dairy equipment as well (13, 14). When cheeses such as Sicilian PDO Ragusano (15) and Caciocavallo Palermitano (16) are produced using Tina wooden vats, S. thermophilus appeared to be one of the dominant species in the biofilm isolate analysis. Due to the variabilities in their life environments, many streptococcal species develop the capacity to form a biofilm in order to survive unfavorable conditions, thereby obtaining an ecological advantage.
Recently, Couvigny et al. (9) provided new information about the biofilm-associated genes that S. thermophilus can activate and express for biofilm formation. When a collection of S. thermophilus strains obtained from different dairy products and with diverse geographical origins were studied for their capacity to produce biofilm, their varied behavior raised interest, as it was observed that a few strains acted as strong biofilm producers while the majority were poor biofilm producers. Three niche-specific genes involved in adhesion to and biofilm formation on polystyrene can justify this peculiarity (9). One of these, STH8232_017, is located in a genomic island which harbors open reading frames (ORFs) encoding transposases and appears to be acquired by HGT. This gene is heterogeneously found in S. thermophilus strains. Orthologs of the other two ORFs, STH8232_0714 and STH8232_1361, are detected in many host-associated bacteria, being involved in adhesion to mammalian cells, but they rarely are found in S. thermophilus. These observations led to the hypothesis that these genes code for ancestral functions that have been lost in the majority of S. thermophilus strains (9).
The aim of this study was therefore to investigate the mechanism behind biofilm formation by S. thermophilus strains in milk environments and to understand in more detail the molecular basis of and the gene exchange episodes driving adhesion and persistence on stainless steel surfaces.
RESULTS
Biofilm formation capacity of S. thermophilus UC8547.
We investigated the capacity to form biofilm on abiotic surfaces in the presence of milk, under conditions resembling the natural environment of S. thermophilus species, by analyzing 25 strains (Fig. S1). Since a static biofilm assay showed that S. thermophilus UC8547 had the highest ability to produce biofilm (Fig. S1), this strain was selected for further studies. UC8547 is a fast acidifier, dropping the milk pH below 4.8 after 6 h (Fig. 1). PCR and reverse transcription-PCR (RT-PCR) experiments showed that it harbors the gene coding for the S. thermophilus serine protease PrtS and expresses it when grown in LM17 (data not shown) (5). Moreover, this strain showed an aggregation phenotype when grown in liquid medium and a hydrophobic phenotype when tested with the microbial adhesion to solvents (MATS) assay (Fig. 2).
FIG 1.
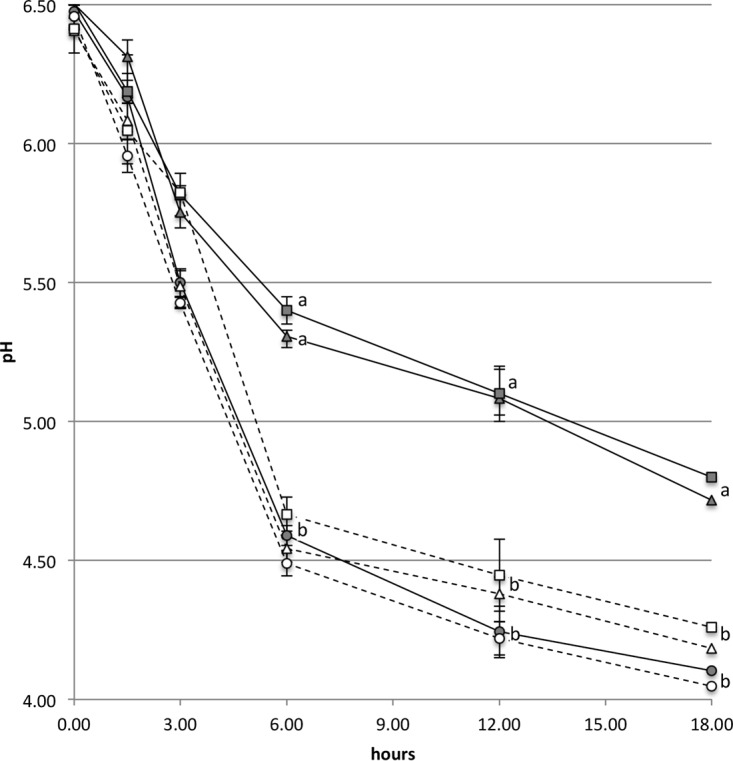
Acidification rate of S. thermophilus UC8547 (○), its spontaneous mutant UC8547Δ3 (△), and the prtS-null mutant UC8547Δ16 (□). Strains were grown in reconstituted skim milk (gray symbols and continuous line) and in reconstituted skim milk supplemented with 5 g/liter of hydrolyzed casein (white symbols and dotted line). The two derivative strains show a reduced acidification rate, which is restored by the addition of hydrolyzed casein. Data represent the means of the results from three independent experiments. Standard deviations are indicated by bars. A statistically significant difference (P < 0.05, one-way analysis of variance [ANOVA]) was observed at 6 h and thereafter between the two mutants (UC8547Δ3 and UC8547Δ16) grown in milk and the other analyzed samples, as indicated by different letters.
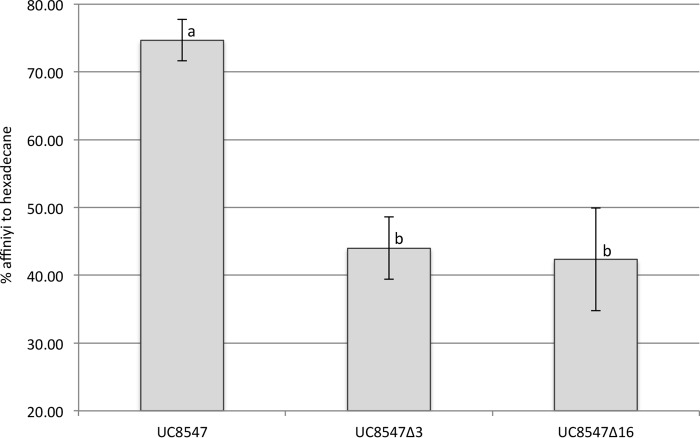
FIG 2.
Cell surface hydrophobicity of S. thermophilus UC8547 and its prtS-null derivatives UC8547Δ3 and UC8547Δ16. The lack of the prtS gene reduced the cell affinity to hexadecane solvent. Data represent the means of the results from three independent experiments. Standard deviations are indicated by error bars. Values with different letters are significantly different (P < 0.05, one-way analysis of variance [ANOVA]).
When tested in the flow cell apparatus using dRSM as the medium, the counts of S. thermophilus UC8547 sessile and planktonic cells (Fig. 3) show that adhered cells increased over time. At 3 h, the count was 6.2 × 103 CFU/cm2; at 6 h, the count was 6.3 × 106 CFU/cm2, and the highest cell density was reached at 18 h, with a count of 5.0 × 107 CFU/cm2. The density of planktonic cells increased during the first 6 h and then remained constant over the 36 h of the experiments. When the same samples were analyzed by scanning electron microscopy (SEM) (Fig. 4), it was observed that after 3 h, the stainless steel surface was covered by a matrix, presumably composed of adhered milk proteins, in which are enclosed S. thermophilus cells (Fig. S2). The thickness of the biofilm increased over time, and after 30 h, the wire was covered by a multilayer biofilm (up to 20 μm) of S. thermophilus cells. When LM17 was used as the medium, the cell density in the biofilm on stainless steel was significantly lower, reaching the highest value of 1.5 × 103 CFU/cm2 after 24 h, while the counts of planktonic cells were comparable to those observed in the experiment with milk (Fig. 3).
FIG 3.
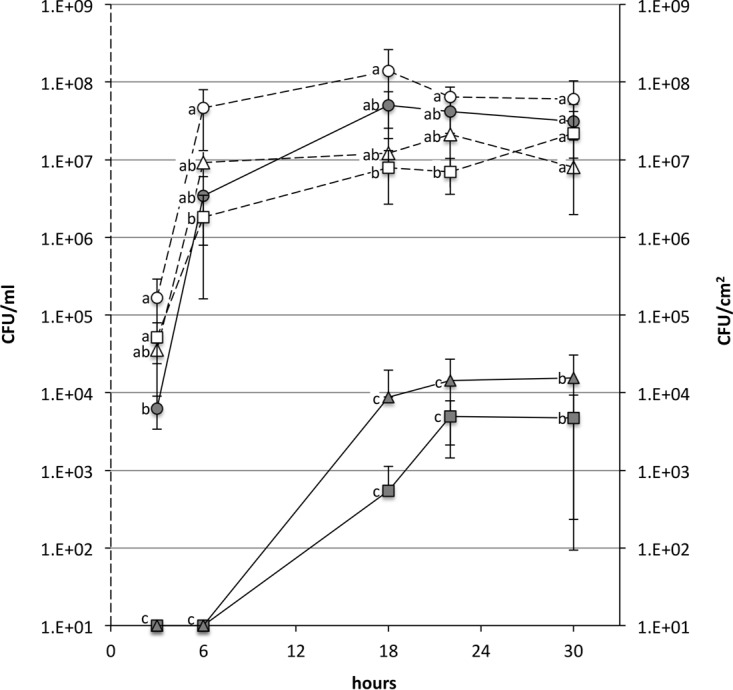
Biofilm formation in dynamic milk flow by S. thermophilus UC8547 (○), its spontaneous mutant UC8547Δ3 (△), and the prtS-null mutant UC8547Δ16 (□) over 36 h. Planktonic cells (white symbols and dotted line) are expressed as CFU per milliliter, while sessile cells (gray symbols and continuous line) are expressed as CFU per square centimeter. The reported data are the averages of the results from three independent experiments. Standard deviations are indicated by bars. No significant differences were observed in the planktonic cells counts between the three isogenic strains. The numbers of sessile cells adhered to stainless steel indicate that UC8547Δ3 and UC8547Δ16 have impaired biofilm formation. Values with different letters are significantly different (P < 0.05, one-way ANOVA).
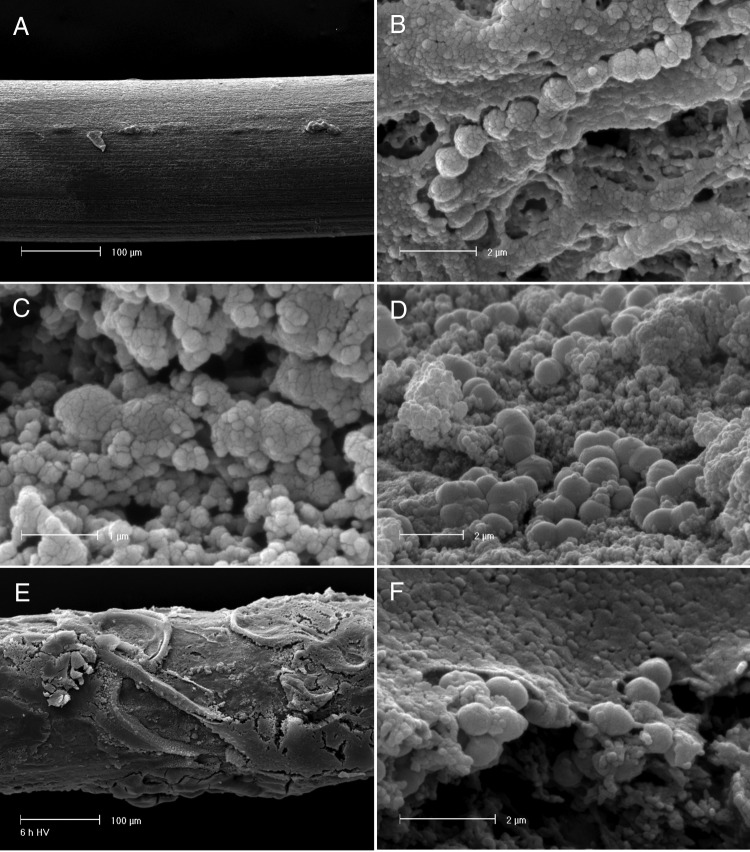
FIG 4.
SEM observation of S. thermophilus UC8547 biofilm on stainless steel. (A) The stainless steel wire at the beginning of the experiment. (B) S. thermophilus cells adhered to the surface after 3 h. (C) Sessile cells at 6 h and 18 h. (D) After 30 h, an evident biofilm is present over the stainless steel wire (E and F) and cells are included in a multilayer structure.
prtS mutants have impaired biofilm formation ability.
To identify the cellular mechanisms at the basis of biofilm formation, isogenic spontaneous mutants of the parent strain S. thermophilus UC8547 were selected after subculturing for 30 days. After this period, 6% of the analyzed colonies lost the aggregative phenotypes. Five nonaggregative derivatives were isolated and when tested showed poor biofilm formation in static-mode experiments. When grown in milk, all five derivatives had a reduced acidification compared to the parent strains, as shown in Fig. 1 for one representative of them, named UC8547Δ3. All the nonaggregative mutants investigated by PCR lacked the gene coding for PrtS. Additionally, all the studied spontaneous mutants presented lower cell hydrophobicity than S. thermophilus UC8547 (Fig. 2). As shown in Fig. 3, a selected spontaneous mutant, named UC8547Δ3, showed a limited ability to form biofilm on stainless steel in milk, reaching the highest counts of sessile cells (1.5 × 104 CFU/cm2) after 30 h. The counts of planktonic cells were substantially equal to those of the parent strain. The addition of hydrolyzed casein to RSM did not significantly affect the numbers of planktonic and sessile S. thermophilus UC8547Δ3 cells (data not shown). The SEM observation confirmed that a limited biofilm structure is formed over the stainless steel surface compared to that with the parent strain (Fig. 5).
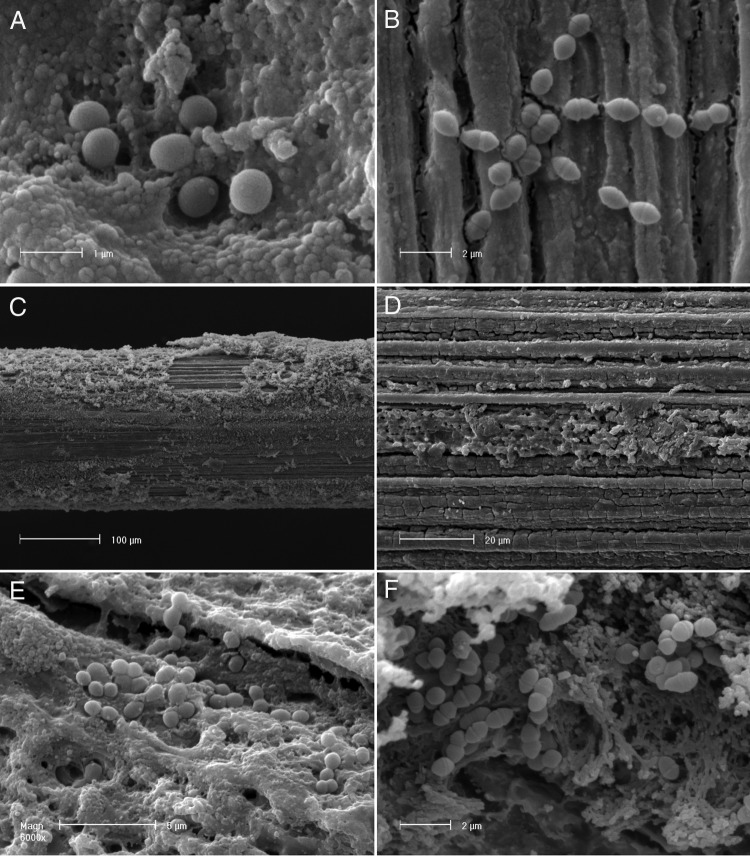
FIG 5.
(A and B) SEM observations of UC8547Δ3 and UC8547Δ16 biofilm on stainless steel. S. thermophilus UC8547Δ3 (A) and UC8547Δ16 (B) cells adhered to the surface after 6 h. (C and D) Low-magnification observation of the stainless steel wire at 30 h for S. thermophilus UC8547Δ3 (C) and UC8547Δ16 (D). The biofilm matrix is substantially limited compared to that of the parent strain UC8547. (E and F) Higher magnification of the same sample shows that adhered cells of UC8547Δ3 (E) and UC8547Δ16 (F) are not included in a multilayer biofilm structure.
To confirm the role of the cell wall-associated proteinase in biofilm formation, a mutant was constructed by a single-crossover insertional mutation. The resulting knockout mutant, named S. thermophilus UC8547Δ16, showed impaired biofilm formation in the presence of milk or casein, similar to that of the spontaneous mutant UC8547Δ3 (Fig. 3). While the counts of planktonic cells of the two mutants were comparable to those of the parent strain UC8547, the cell density on the stainless steel was substantially lower. Electron microscopy observations confirmed the limited capacity to form biofilm under the tested conditions, as shown in Fig. 5. Differently from the parent strain UC8547, but as observed in the UC8547Δ3 experiments, few cells of UC8547Δ16, mainly in a monolayer, were detected over the abiotic surface. As expected, inactivation of the gene coding for the cell wall-associated protease resulted in a reduced acidification rate of S. thermophilus UC8547Δ16 in milk (Fig. 1). A reduction in cell hydrophobicity (Fig. 2) and the loss of the aggregative phenotype were also observed, indicating that, as observed by Couvigny et al. (9), cell hydrophobicity has a central role in the biofilm formation.
PrtS affects the conjugal transfer of pAMβ1 plasmid.
The observation that both the spontaneous and the knockout mutants lost the aggregative phenotype when grown in LM17 led us to assess if the loss of PrtS affects horizontal gene transfer. For this purpose, we used as a donor L. lactis SH4174 harboring pAMβ1 (17), an erythromycin resistance plasmid with a broad host range in Firmicutes (18). The data from three independent plate mating experiments demonstrated that the transfer of pAMβ1 occurred at a higher frequency in S. thermophilus UC8547 (3.7 × 10−4) than in the PrtS-negative mutants UC8547Δ3 (8.5 × 10−8) and UC8547Δ16 (9.2 × 10−8), indicating that in S. thermophilus UC8547, the aggregative phenotype associated with the cell wall proteinase PrtS positively affects horizontal gene transfer.
Genome analysis of S. thermophilus UC8547 and UC8547Δ3.
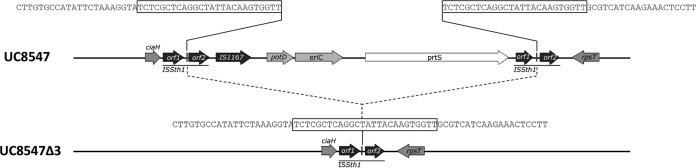
De novo shotgun sequencing of S. thermophilus UC8547 and its spontaneous mutant UC8547Δ3 was performed. A 1,795,194-bp assembly was obtained from the genome analysis of the parent strain UC8547, consisting of a total of 77 contigs. These annotated contigs contain 1,960 putative coding sequences (CDSs) and 55 predicted RNAs. In the case of S. thermophilus UC8547Δ3, a slightly smaller assembly of 1,779,153 bp was obtained, consisting of a total of 66 contigs. The annotated contigs of the mutant contain 1,954 putative CDSs and 55 predicted RNAs. The whole-genome sequence analysis of S. thermophilus UC8547 and UC8547Δ3 made it possible to search for the genes involved in the biofilm formation on polystyrene plates and responsible for adhesion to epithelial cells (9). Both S. thermophilus UC8547 and its isogenic mutant harbor the STH8232_1361 gene, coding for a transmembrane protein of the polysaccharide transporter (PST) family, and lack the STH8232_017 gene, coding for a predicted cytoplasmic protein of unknown function. Both strains harbor an identical copy of a gene that shows 68% identity with STH8232_0714, a gene that codes for a putative surface protein that contains the mucin binding domain MucBP. The genome analysis showed that S. thermophilus UC8547 contains the prtS gene on the mobile genetic element previously identified in the fast-acidifying S. thermophilus strains. This genomic island is located in the ciaH-rpsT chromosomal region, as observed in other S. thermophilus strains (6, 8), downstream of the ciaH pseudogene and between two tandem ISSth1 elements (Fig. 6). The comparative genomic analysis of the parent and derivative strains revealed that UC8547Δ3 has encountered a major deletion event involving the 11,677-bp prtS island. Figure 6 shows that the deletion involved orf2 of ISSth1, IS1167, the eriC gene encoding a chloride channel protein, the potD gene coding for the ABC transporter spermidine putrescine-binding protein PotD, the prtS gene, and orf1 of the second ISSth1 (Fig. 6). In S. thermophilus UC8547Δ3, the excision event involved two 26-bp direct repeat sequences (TCTCGCTCAGGCTATTACAAGTGGTT) located between the two open reading frames of ISSth1 (Fig. 6). The excision event occurred through recombination between the two copies of ISSth1, resulting in the reconstruction of a complete copy of this insertion sequence. The excision site identified by genome comparison of S. thermophilus UC8547 and UC8547Δ3 differs from that identified in other S. thermophilus strains, with the left junction 769 bp downstream and the right junction 883 bp upstream of two direct repeat sequences homologous to those described in S. thermophilus LMD-9 (8). The comparison of the two genomes did not show other relevant mutations.
FIG 6.
Schematic representation of the prtS genomic island of S. thermophilus UC8547. Genes are represented by arrows. In black are the IS elements ISSth1, composed of two ORFs, and IS1167. In the spontaneous mutant UC8547Δ3, the excision event occurred via recombination between the two flanking ISSth1 elements. The nucleotide sequences of direct repeats of the deletion junctions are boxed.
DISCUSSION
The scientific hypothesis we have investigated is that the cell wall-associated proteinase PrtS has a central role in adhesion to stainless steel in milk environments. The rationale behind this hypothesis is that biofilm formation is based on two major stages: biofilm starts with the adsorption of milk proteins to stainless steel, which is followed by S. thermophilus adhesion and colonization of this preconditioned surface. The adsorption of milk proteins to stainless steel is a multistep process that has been extensively studied. The first step is the electrostatic adsorption of β-lactoglobulin to negatively charged surfaces (19), such as that of stainless steel at neutral pH (20), and that is followed by the adsorption of κ-casein to the β-lactoglobulin-saturated layer. This adsorption process is temperature dependent, increasing when the temperature rises (21). The adsorbed layer of milk protein has been demonstrated to affect bacterial adherence and biofilm formation: Barnes et al. (22) observed that preconditioning of stainless steel surfaces with milk or individual milk proteins reduced the adhesion of Staphylococcus aureus, Pseudomonas fragi, Escherichia coli, Listeria monocytogenes, and Serratia marcescens to stainless steel after 2 h of incubation. In contrast, in flow cell experiments, Al-Makhlafi et al. (23) showed that β-lactoglobulin increases the initial adhesion of Listeria monocytogenes and the bacterial colonization over time. The hypothesis that PrtS protein is the binding force between milk-conditioned stainless steel and S. thermophilus cells is based on the presence in this proteinase of a substrate binding site, recognizing caseins, and of the sortase target site LPXTG motif, which allows a covalent bond to the cell wall (5). Moreover, PrtS lacks the B domain involved in the autoproteolytic process, which results in the release from the cell wall of other proteinases of lactic acid bacteria (5). To investigate if the ability to form biofilm under environmental conditions resembling the dairy environment is mediated by PrtS, we have selected a strain of S. thermophilus with a high biofilm formation capacity, isolated a spontaneous mutant, made the prtS gene-null mutant, assessed the biofilm formation in a flow cell apparatus, and compared the whole-genome sequences of the parent and derivative strains. The following observations support the hypothesis of a PrtS-mediated biofilm formation in dairy environment: (i) S. thermophilus UC8547 adheres to and forms a biofilm on stainless steel in milk; after 3 h of milk flow, cells attached to the conditioned surface are visible, and then a multilayer biofilm in which cells are embedded in an extracellular matrix is formed. Since the S. thermophilus exopolysaccharide gene cluster (24) is not present in the UC8547 genome and this strain does not produce a capsule, the exopolysaccharide (EPS) cannot be involved in the extracellular matrix formation and adhesion. In contrast, when the same experiments were performed in LM17, a very limited biofilm was observed, indicating the fundamental role of milk proteins in this process. (ii) PrtS is involved in the adhesion process. When this gene was interrupted by a single-crossover insertional mutation, the derivative strain UC8547Δ16 lost the proteolytic activity and failed to form biofilm. Moreover, when the whole-genome sequence of the spontaneous mutant UC8547Δ3 with impaired biofilm formation was compared with that of the wild-type strain, a deletion involving the expendable prtS island was detected. No differences in the genes involved in biofilm formation onto polystyrene genes, recently described by Couvigny et al. (9), were observed. The results of this study strongly indicate that the cell wall-associated proteinase PrtS, besides improving the growth and acidification rate in milk, mediates the adhesion of S. thermophilus cells to surfaces conditioned by milk. Two potential mechanisms at the basis of adhesion can be hypothesized: PrtS acts as a binding substance between the cell envelope and the milk proteins through its binding domain or, affecting cell hydrophobicity, it improves cell aggregation and biofilm formation, as reported by Habimana et al. (25) for the cell wall protease PrtP in Lactococcus lactis.
Biofilm formation ability provides an ecological advantage for the survival and persistence of this species, which is strongly adapted to dairy environments. Interestingly, proteolytic activity and biofilm formation are associated with the prtS gene, which is located on a mobile genetic element that has been demonstrated to be a horizontally acquired trait (6–8). The mechanism of prtS island excision involves two copies of the ISSth1 insertion sequence flanking the prtS gene and is similar to that already described by Delorme et al. (8) in different S. thermophilus strains, and to that observed by Dandoy et al. (6), inserting by natural transformation the island in PrtS− strains. Unlike other S. thermophilus prtS islands (8), in UC8547, there is a single copy of IS1167.
The comparison of the whole-genome sequence of UC8547 and its PrtS− spontaneous mutant demonstrated the natural excision event in an isogenic model and confirmed the involvement of ISSth1 in the recombination, supporting the observation of IS-dependent recombination that leads to population heterogeneity in S. thermophilus (7). These authors, using the engineered endogenous clustered regularly interspaced short palindromic repeat 3 (CRISPR3) type II system of strain LMD-9 targeted to the lacZ genomic island, showed that IS elements flanking this expendable genomic island are responsible for large chromosomal deletions. Moreover, in the same study, using the engineered CRISPR3 repeat-spacer arrays targeted to a 30-bp internal region of the prtS gene, the excision of the prtS island was induced (7). We observed that the excision event involving the prtS island occurred at a relatively high frequency when the culture was grown in medium containing hydrolyzed proteins, such as in LM17, and the selective pressure of milk was absent. This phenomenon results in a heterogeneous population of fast and slow acidifiers in milk.
Additionally, we have noticed that PrtS influences the cell hydrophobicity and the aggregation phenotype when S. thermophilus is grown in broth. Since this phenotype has been associated with high-frequency plasmid conjugation in other lactic acid bacteria, such as Lactococcus, Lactobacillus, and Enterococcus (26–28), we assessed the ability to acquire the conjugative plasmid pAMβ1 of S. thermophilus UC8547 and its prtS-null mutants. The frequency of pAMβ1 transfer was substantially higher in the wild-type strain than in its mutants. A relationship between biofilm formation and high frequency of pAMβ1 conjugal transfer was observed in Lactococcus lactis expressing the CluA protein, a binding substance that induces cell aggregation, high conjugal transfer, and biofilm formation (29).
The recognition of the role of IS in the instability of mobile genetic elements coding for technologically important traits, such as the Prt+ and biofilm phenotypes, provides clearer insight into the genome plasticity of S. thermophilus and its adaptation to environmental conditions: in the presence of milk this bacterium, which is highly adapted to the dairy environment, maintains the ability to persist in the habitat and to retrieve amino acids from milk proteins. Thus, it can be hypothesized that the biofilm forming on milk residues provides an ecological advantage for the persistence of S. thermophilus in the dairy environment, such as in pipelines and vats, and the proteolytic activity allows a rapid restoration of growth when milk is present. The identification of the molecular mechanisms behind biofilm formation in the milk environment represents an important finding in terms of both ecological advantage for persistence in this habitat and practical consequences in the dairy industry.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. thermophilus strains were cultivated in M-17 medium (Merck KGaA, Darmstadt, Germany) containing 1% lactose (LM17) or 1% glucose (GM17) at 42°C. A total of 25 strains were tested in the study; the strains were used as industrial starter cultures or isolated from raw milk cheeses. The proteinase phenotype was determined on bacterial colonies grown on fast-slow differential agar (FSDA) medium after 48 h, as previously described (6, 30). Protease-positive strains form large yellow opaque colonies surrounded by a yellow area different from the small translucent colonies of the slow-acidifying phenotype. The acidification rate was assessed by growing S. thermophilus strains in 10% (wt/vol) reconstituted skim milk (RSM) (Oxoid, Thermo Fisher Scientific, Inc.) treated at 110°C for 10 min as follows: an overnight culture grown in LM17 was inoculated (1%) in RSM, incubated at 42°C, and the pH was measured over 16 h. In the studies with PrtS-negative mutant, 5g/liter hydrolyzed casein (tryptone; Sigma-Aldrich) was added to RSM before heat treatment. A high rate of acidification was considered when the pH was 4.8 or lower after 6 h of incubation.
Lactococcus lactis was cultivated in GM17 at 30°C. E. coli was grown in Luria-Bertani medium at 37°C. When needed, erythromycin (4 μg/ml for S. thermophilus and 200 μg/ml for E. coli) was added to broth and agar media.
Plasmid and DNA/RNA manipulations.
The plasmid pRV300 harboring erythromycin resistance (31) was utilized as a vector to perform a Campbell-like integration. A 595-bp internal fragment of the prtS gene was amplified by PCR with oligonucleotides prtSf (5′-GTGAGGCTTTGGCAGCTAAC-3′) and prtSr (5′-TCGCGATATAGACCGGATTC-3′), digested with EcoRI-ClaI (Sigma-Aldrich, St. Louis, MO), and ligated with pRV300 digested with the same enzymes. The obtained construct was electroporated in Escherichia coli TB1 and named pPC7020. This plasmid was electrotransformed in S. thermophilus UC8547 as previously described (32), and mutants were selected on LM17 with 4 μg/ml erythromycin. The Campbell-like integration was confirmed by PCR, and a mutant, named UC8547Δ16, was selected for the study.
Total RNA was extracted from log-phase S. thermophilus cultures grown in LM17 using the RNeasy minikit (Qiagen, Inc., Hilden, Germany), with an additional step of treatment with RNase-free DNase (Qiagen). The concentration of RNA was determined by measuring the absorbance at 260 nm using an Ultrospec 2100 Pro UV-visible spectrophotometer (GE Healthcare Life Sciences, Buckinghamshire, UK). Reverse transcription reactions were performed on total RNA using the Reverse-iTTM one-step RT-PCR kit (ABgene, Epsom, UK), according to the supplier's recommendations, in a final volume of 25 μl containing 30 ng of total RNA. Reaction mixtures were incubated at 47°C for 30 min, followed by denaturation at 94°C for 2 min. cDNA products were amplified as described above (33). PCR products were analyzed on 0.8% agarose gels. To control for the residual presence of DNA, RT-PCR was also performed in the absence of reverse transcriptase. The experiments were performed in triplicate.
Biofilm formation.
The biofilm formation capacity by S. thermophilus was assessed under static conditions over stainless steel (SS) by crystal violet staining. Five sterile spheres of SS (0.5 mm diameter) were introduced in a 5-ml tube containing 3.3% (wt/vol) diluted reconstituted skim milk (dRSM). The tube was inoculated with 50 μl of an overnight S. thermophilus culture and incubated at 42°C for 18 h. The spheres were removed from milk, washed three times with saline solution to remove unattached cells, and then resuspended in a 0.3% crystal violet solution for 15 min. The crystal violet solution was removed and the spheres rinsed three times with deionized water to remove the residual dye. Then, 2 ml of 95% (vol/vol) ethanol was added and the tube vortexed for 1 min. The ethanol solution was transferred to a 1-cm-path cuvette and the absorbance at 590 nm determined. Three independent replicates were performed.
Biofilm formation under dynamic conditions was studied using a flow chamber system composed of 5 glass tubes (10 cm long; internal diameter, 0.4 mm) in parallel, into which a stainless wire (0.2-mm diameter) was introduced. The tubes were connected by a Teflon pipeline to a solution dispenser, and the flow rate (1 ml/min) was maintained using a peristaltic pump. The apparatus was chemically disinfected with a succession of disinfection solutions (0.5 M NaOH and 50% ethanol) and washed with sterile saline solution. The apparatus was maintained at 42°C. An overnight culture of the S. thermophilus strain grown in RSM was introduced into the system at a 1 ml/min flow rate, and when the pipeline was full, the flow was stopped and the whole system was incubated at 42°C for 1 h. After this step, the nutrient solution, dRSM, dRSM–0.5% hydrolyzed casein (tryptone; Sigma-Aldrich), or LM17 was introduced inside the apparatus at a flow rate of 1 ml/min. At defined time points (0, 3, 6, 18, 24, and 30 h), the flow was stopped and aliquots of the wire (1 cm) and of the eluate were taken. Planktonic cell numbers were obtained by serially diluting the collected milk samples. The count of sessile cells was achieved by analyzing a fragment of the wire, collected under aseptic conditions and transferred to a sterile tube. To remove nonadhered cells, two washing steps with sterile saline solution were performed and then 1 ml of phosphate buffer (pH 7) was added to the tube. Detachment of sessile cells was performed by vortexing for 3 min. Counts were obtained by plating onto LM17 agar and incubating at 42°C. All the biofilm formation experiments were performed in triplicate.
MATS and aggregation assays.
MATS was performed to determine the cell hydrophobicity, as previously described (34), by measuring the affinity of cells for hexadecane. Aggregation experiments were performed, as already described (26), in LM17 and GM17 media.
SEM analysis.
Samples for SEM were prepared as follows. A 1-cm portion of the stainless steel wire was dehydrated stepwise in 75%, 85%, 95%, and 100% ethanol for 1 h each at room temperature. Critical point drying was performed in a Bal-Tec CPD030 critical point dryer. Samples were gold coated, as described by Palumbo et al. (35), and observed with a Philips XL30 environmental scanning electron microscope (ESEM).
Isolation of biofilm formation-negative mutants.
The selection of nonaggregative mutants was achieved by subculturing S. thermophilus UC8547 in LM17 for 30 days. The inoculation of a new tube was performed with 10 μl from the top of an overnight culture. After 30 days, a culture lacking the aggregation phenotype was streaked on LM17 agar plates, and the resulting colonies were tested for aggregative properties.
Conjugation experiments.
Mating experiments were performed as follows. Lactococcus lactis SH4174 carrying the erythromycin resistance broad-host-range conjugative plasmid pAMβ1 was used as a donor strain. The S. thermophilus strains UC8547, UC8547Δ3, and UC8547Δ16 were used as recipients. Donor and recipient log-phase cultures (0.2 ml) grown in GM17 were mixed in a 1:1 ratio, plated on a GM17 plate, and incubated for 24 h at 30°C. Transconjugants were selected on LM17 agar with 10 μg/ml erythromycin at 45°C, a limiting temperature for the donor strain.
Whole-genome sequencing and bioinformatic analyses.
Shotgun sequencing of S. thermophilus strains UC8547 and UC8547Δ3 was performed. The genomes were sequenced using an Illumina HiSeq 1000 platform. Quality-filtered reads were assembled using the Velvet software (version 1.1.04) (36), and contig sequences were annotated in the RAST server (37) and the NCBI Prokaryotic Genome Annotation Pipeline. To fill the gap between contigs 20 and 61 of the S. thermophilus UC8547 genome, containing a region flanking the prtS island, primers were designed immediately upstream of the gap and used for amplification and sequencing. Multiple alignments were performed using Geneious 9.0.5 software. Basic bioinformatics analysis was performed with the SnapGene Viewer 3.0.1 software. The OrthoMCL software was used for the identification of orthologous genes within the genomes of the two S. thermophilus strains. The protein sequences obtained from the annotation of the two S. thermophilus genomes were subjected to a BLAST all-versus-all search using a cutoff E value of 10−5, and the results were submitted to OrthoMCL to generate groups of orthologous genes (OGs).
Accession number(s).
The S. thermophilus genome sequences have been deposited in GenBank with accession numbers NZ_MCHW00000000.1 for UC8547 and NZ_MCHV00000000.1 for UC8547Δ3.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the MicroEmiRo project, cofunded by the Emilia-Romagna Region through the POR FESR 2014–2020 funds (European Regional Development Fund).
We thank Sacco System S.r.l. for providing strain UC8547.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02840-16.
REFERENCES
- 1.Hols P, Hancy F, Fontaine L, Grossiord B, Prozzi D, Leblond-Bourget N, Decaris B, Bolotin A, Delorme C, Ehrlich SD, Guédon E, Monnet V, Renault P, Kleerebezem M. 2005. New insights in the molecular biology and physiology of Streptococcus thermophilus revealed by comparative genomics. FEMS Microbiol Rev 29:435–463. doi: 10.1016/j.femsre.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 2.Bolotin A, Quinquis B, Renault P, Sorokin A, Ehrlich SD, Kulakauskas S, Lapidus A, Goltsman E, Mazur M, Pusch GD, Fonstein M, Overbeek R, Kyprides N, Purnelle B, Prozzi D, Ngui K, Masuy D, Hancy F, Burteau S, Boutry M, Delcour J, Goffeau A, Hols P. 2004. Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nat Biotechnol 22:1554–1558. doi: 10.1038/nbt1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Makarova KS, Koonin EV. 2007. Evolutionary genomics of lactic acid bacteria. J Bacteriol 189:1199–1208. doi: 10.1128/JB.01351-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rasmussen TB, Danielsen M, Valina O, Garrigues C, Johansen E, Pedersen MB. 2008. Streptococcus thermophilus core genome: comparative genome hybridization study of 47 strains. Appl Environ Microbiol 74:4703–4710. doi: 10.1128/AEM.00132-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fernandez-Espla MD, Garault P, Monnet V, Rul F. 2000. Streptococcus thermophilus cell wall-anchored proteinase: release, purification, and biochemical and genetic characterization. Appl Environ Microbiol 66:4772–4778. doi: 10.1128/AEM.66.11.4772-4778.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dandoy D, Fremaux C, de Frahan MH, Horvath P, Boyaval P, Hols P, Fontaine L. 2011. The fast milk acidifying phenotype of Streptococcus thermophilus can be acquired by natural transformation of the genomic island encoding the cell-envelope proteinase PrtS. Microb Cell Fact 10:S21. doi: 10.1186/1475-2859-10-S1-S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Selle K, Klaenhammer TR, Barrangou R. 2015. CRISPR-based screening of genomic island excision events in bacteria. Proc Natl Acad Sci U S A 112:8076–8081. doi: 10.1073/pnas.1508525112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delorme C, Bartholini C, Bolotine A, Dusko Ehrlich S, Renault P. 2010. Emergence of a cell wall protease in the Streptococcus thermophilus population. Appl Environ Microbiol 76:451–460. doi: 10.1128/AEM.01018-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Couvigny B, Thérial C, Gautier C, Renault P, Briandet R, Guédon E. 2015. Streptococcus thermophilus biofilm formation: a remnant trait of ancestral commensal life? PLoS One 10:e0128099. doi: 10.1371/journal.pone.0128099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Jong P, te Giffel MC, Kiezebrink EA. 2002. Prediction of the adherence, growth and release of microorganisms in production chains. Int J Food Microbiol 74:13–25. doi: 10.1016/S0168-1605(01)00713-9. [DOI] [PubMed] [Google Scholar]
- 11.Knight GC, Nicol RS, McMeekin TA. 2004. Temperature step changes: a novel approach to control biofilms of Streptococcus thermophilus in a pilot plant-scale cheese-milk pasteurisation plant. Int J Food Microbiol 93:305–318. doi: 10.1016/j.ijfoodmicro.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 12.Hup G, Bangma A, Stadhouders J, Bouman S. 1980. Growth of thermoresistant streptococci in cheese-milk pasteurisers: I. Some observations in cheese factories. North Eur Dairy J 46:245–251. [Google Scholar]
- 13.Hood SK, Zottola ZE. 1995. Biofilms in food processing. Food Control 6:9–18. doi: 10.1016/0956-7135(95)91449-U. [DOI] [Google Scholar]
- 14.Palmer J, Flint S, Brooks J. 2007. Bacterial cell attachment, the beginning of a biofilm. J Ind Microbiol Biotechnol 34:577–588. doi: 10.1007/s10295-007-0234-4. [DOI] [PubMed] [Google Scholar]
- 15.Lortal S, Di Blasi A, Madec MN, Pediliggieri C, Tuminello L, Tanguy G, Fauquant J, Lecuona Y, Campo P, Carpino S, Licitra G. 2009. Tina wooden vat biofilm: a safe and highly efficient lactic acid bacteria delivering system in PDO Ragusano cheese making. Int J Food Microbiol 132:1–8. doi: 10.1016/j.ijfoodmicro.2009.02.026. [DOI] [PubMed] [Google Scholar]
- 16.Scatassa ML, Gaglio R, Macaluso G, Francesca N, Randazzo W, Cardamone C, Grigoli Di A, Moschetti G, Settanni L. 2015. Transfer, composition and technological characterization of the lactic acid bacterial populations of the wooden vats used to produce traditional stretched cheeses. Food Microbiol 52:31–41. [DOI] [PubMed] [Google Scholar]
- 17.Vescovo M, Morelli L, Bottazzi V, Gasson MJ. 1983. Conjugal transfer of broad-host-range plasmid pam31 into enteric species of lactic acid bacteria. Appl Environ Microbiol 46:753–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clewell DB, Yagi Y, Dunny GM, Schultz SK. 1974. Characterization of three plasmid deoxyribonucleic acid molecules in a strain of Streptococcus faecalis: identification of a plasmid determining erythromycin resistance. J Bacteriol 117:283–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rabe M, Verdes D, Seeger S. 2011. Understanding protein adsorption phenomena at solid surfaces. Adv Colloid Interface Sci 162:87–106. doi: 10.1016/j.cis.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 20.Hedberg Y, Karlsson ME, Blomberg E, Odnevall Wallinder I, Hedberg J. 2014. Correlation between surface physicochemical properties and the release of iron from stainless steel AISI 304 in biological media. Colloids Surf B Biointerfaces 122:216–222. doi: 10.1016/j.colsurfb.2014.06.066. [DOI] [PubMed] [Google Scholar]
- 21.Kim JC, Lund DB. 1998. Milk protein/stainless steel interaction relevant to the initial stage of fouling in thermal processing. J Food Process Eng 21:369–386. doi: 10.1111/j.1745-4530.1998.tb00459.x. [DOI] [Google Scholar]
- 22.Barnes L-M, Lo MF, Adams MR, Chamberlain AHL. 1999. Effect of milk proteins on adhesion of bacteria to stainless steel surfaces. Appl Environ Microbiol 65:4543–4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al-Makhlafi H, Nasir A, Mcguire J, Daeschel M. 1995. Adhesion of Listeria monocytogenes to silica surfaces after sequential and competitive adsorption of bovine serum albumin and beta-lactoglobulin. Appl Environ Microbiol 61:2013–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stingele F, Neeser JR, Mollet B. 1996. Identification and characterization of the eps (exopolysaccharide) gene cluster from Streptococcus thermophilus Sfi6. J Bacteriol 178:1680–1690. doi: 10.1128/jb.178.6.1680-1690.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Habimana O, Le Goff C, Juillard V, Bellon-Fontaine MN, Buist G, Kulakauskas S, Briandet R. 2007. Positive role of cell wall anchored proteinase PrtP in adhesion of lactococci. BMC Microbiol 7:36. doi: 10.1186/1471-2180-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reniero R, Cocconcelli P, Bottazzi V, Morelli L. 1992. High frequency of conjugation in Lactobacillus mediated by an aggregation-promoting factor. J Gen Microbiol 138:763–768. doi: 10.1099/00221287-138-4-763. [DOI] [Google Scholar]
- 27.van der Lelie D, Chavarri F, Venema G, Gasson MJ. 1991. Identification of a new genetic determinant for cell aggregation associated with lactose plasmid transfer in Lactococcus lactis. Appl Environ Microbiol 57:201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dunny GM, Leonard BAB, Hedberg PJ. 1995. Pheromone-inducible conjugation in Enterococcus faecalis: interbacterial and host-parasite chemical communication. J Bacteriol 177:871–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo H, Wan K, Wang HH. 2005. High-frequency conjugation system facilitates biofilm formation and pAMbeta1 transmission by Lactococcus lactis. Appl Environ Microbiol 71:2970–2978. doi: 10.1128/AEM.71.6.2970-2978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huggins AR, Sandine WE. 1984. Differentiation of fast and slow milk-coagulating isolates in strains of lactic streptococci. J Dairy Sci 67:1674–1679. doi: 10.3168/jds.S0022-0302(84)81491-5. [DOI] [Google Scholar]
- 31.Leloup L, Ehrlich SD, Zagorec M, Morel-Deville F. 1997. Single-crossover integration in the Lactobacillus sakei chromosome and insertional inactivation of the ptsI and lacL genes. Appl Environ Microbiol 63:2117–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marciset O, Mollet B. 1994. Multifactorial experimental design for optimizing transformation: electroporation of Streptococcus thermophilus. Biotechnol Bioeng 43:490–496. doi: 10.1002/bit.260430609. [DOI] [PubMed] [Google Scholar]
- 33.Vandecasteele SJ, Peetermans WE, Merckx R, Van Ranst M, Van Eldere J. 2002. Use of gDNA as internal standard for gene expression in staphylococci in vitro and in vivo. Biochem Biophys Res Commun 291:528–534. doi: 10.1006/bbrc.2002.6465. [DOI] [PubMed] [Google Scholar]
- 34.Rosenberg M, Gutnick D, Rosenberg E. 1980. Adherence of bacteria to hydrocarbons: a simple method for measuring cell-surface hydrophobicity. FEMS Microbiol Lett 9:29–33. doi: 10.1111/j.1574-6968.1980.tb05599.x. [DOI] [Google Scholar]
- 35.Palumbo E, Favier CF, Deghorain M, Cocconcelli PS, Grangette C, Mercenier A, Vaughan EE, Hols P. 2004. Knockout of the alanine racemase gene in Lactobacillus plantarum results in septation defects and cell wall perforation. FEMS Microbiol Lett 233:131–138. doi: 10.1016/j.femsle.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 36.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.