ABSTRACT
Detection of human wastewater contamination in recreational waters is of critical importance to regulators due to the risks posed to public health. To identify such risks, human wastewater-associated microbial source tracking (MST) markers have been developed. At present, however, a greater understanding of the suitability of these markers for the detection of diluted human wastewater in environmental waters is necessary to predict risk. Here, we compared the process limit of detection (PLOD) and process limit of quantification (PLOQ) of six human wastewater-associated MST markers (Bacteroides HF183 [HF183], Escherichia coli H8 [EC H8], Methanobrevibacter smithii nifH, human adenovirus [HAdV], human polyomavirus [HPyV], and pepper mild mottle virus [PMMoV]) in relation to a fecal indicator bacterium (FIB), Enterococcus sp. 23S rRNA (ENT 23S), and three enteric viruses (human adenovirus serotypes 40/41 [HAdV 40/41], human norovirus [HNoV], and human enterovirus [EV]) in beach water samples seeded with raw and secondary-treated wastewater. Among the six MST markers tested, HF183 was the most sensitive measure of human fecal pollution and was quantifiable up to dilutions of 10−6 and 10−4 for beach water samples seeded with raw and secondary-treated wastewater, respectively. Other markers and enteric viruses were detected at various dilutions (10−1 to 10−5). These MST markers, FIB, and enteric viruses were then quantified in beach water (n = 12) and sand samples (n = 12) from South East Queensland (SEQ), Australia, to estimate the levels of human fecal pollution. Of the 12 sites examined, beach water and sand samples from several sites had quantifiable concentrations of HF183 and PMMoV markers. Overall, our results indicate that while HF183 is the most sensitive measure of human fecal pollution, it should be used in conjunction with a conferring viral marker to avoid overestimating the risk of gastrointestinal illness.
IMPORTANCE MST is an effective tool to help utilities and regulators improve recreational water quality around the globe. Human fecal pollution poses significant public health risks compared to animal fecal pollution. Several human wastewater-associated markers have been developed and used for MST field studies. However, a head-to-head comparison in terms of their performance to detect diluted human fecal pollution in recreational water is lacking. In this study, we cross-compared the performance of six human wastewater-associated markers in relation to FIB and enteric viruses in beach water samples seeded with raw and secondary-treated wastewater. The results of this study will provide guidance to regulators and utilities on the appropriate application of MST markers for tracking the sources of human fecal pollution in environmental waters and confer human health risks.
KEYWORDS: microbial source tracking, human wastewater, fecal indicator bacteria, beach water, enteric viruses
INTRODUCTION
Recreational water bodies polluted with raw and inadequately treated wastewater can pose a significant risk to human health due to the presence of enteric pathogens (1–3). Accidental ingestion or inhalation of such polluted waters can cause a variety of enteric and nonenteric illnesses in swimmers and other recreational water users (4). Raw and treated wastewater can be released into the waterways from defective wastewater treatment plants (WWTPs) (5), broken sewage pipes, malfunctioning septic systems (6), stormwater runoff (7), and during extreme weather events, such as flooding (8). Outbreaks of gastroenteritis in Australia from recreational water exposure are far more common than those attributed to drinking water. In Australia, 42 reported gastroenteritis outbreaks linked to recreational water use were reported between 2001 and 2007, compared to 10 outbreaks that were linked to drinking water consumption (9). Therefore, the microbiological quality of recreational waters is of concern to water quality regulators and health departments.
Direct monitoring of pathogens in recreational waters can provide valuable information on the potential health risks. However, pathogen distribution and abundance can be highly varied due to source and receiving waters, making the direct monitoring approach impractical and uneconomical. Escherichia coli and Enterococcus spp. have previously been used as fecal indicator bacteria (FIB), and based on their concentrations, guideline values have been developed by the water quality regulators to assess the human health risk. However, a shortcoming of this form of monitoring is that concentrations of FIB above the guideline values do not always indicate the presence of pathogens (10, 11). Another significant shortcoming of FIB monitoring is that their presence in a waterbody does not provide information about whether they originated from animal or human feces, thus greatly hindering remediation efforts (12).
The advent of microbial source tracking (MST) tools has led to the development of more efficient water quality monitoring programs (1, 3, 7, 11). MST tools have the ability to rapidly quantify the host-associated genes (known as molecular markers) found in bacteria, protozoa, and viruses from the feces of various animal species, including humans. Commonly used human wastewater-associated markers include the Bacteroides HF183 (HF183) (12), the nifH marker from Methanobrevibacter smithii (13), and the recently developed E. coli H8 (EC H8) markers (14, 15). Viral markers have also received significant attention due to their high host association. These viral markers include human adenovirus (HAdV), human polyomavirus (HPyV), and a plant virus, the pepper mild mottle virus (PMMoV) (11, 16, 17). Host prevalence and host specificity are often considered the two most important performance characteristics of MST markers. Nonspecific (found in nontarget hosts) and nonprevalent (rare) markers tend to yield false-positive or -negative results in field studies (18). Several studies have determined the host prevalence and specificity of the HF183 (19), EC H8 (14), nifH (20), HAdV (21), HPyV (22), and PMMoV (17) markers by analyzing fecal and wastewater samples from various animals. The high host prevalence and specificity values of the aforementioned markers support their potential for human fecal pollution tracking. Limited information, however, is available on the assay process limit of detection (PLOD) or process limit of quantification (PLOQ) for these markers to detect human wastewater pollution in recreational waters. The PLOD and PLOQ determine the smallest volume of wastewater that can be reliably detected and quantified, respectively, after the sample is subjected to filtration, DNA/RNA extraction, and quantitative PCR (qPCR) analysis. The PLOD assessment of the HF183 (3) and PMMoV markers (23) has been undertaken in the United States. However, a comparative study investigating the PLOD and PLOQ of the multiple markers simultaneously in relation to FIB and enteric viruses has not yet been reported.
In this study, we cross-compared the PLOD and PLOQ of six human wastewater-associated MST markers (EC H8, HF183, nifH, HAdV, HPyV, and PMMoV) in beach water samples seeded with raw and secondary-treated wastewater using qPCR assays. In addition, the PLOD and PLOQ values of FIB (Enterococcus sp. 23S rRNA [ENT 23S]) and enteric viruses (HAdV 40/41, human norovirus [HNoV)] genotype II [GII], and enterovirus [EV]) were also determined in parallel to MST markers. This was done to identify the most suitable marker(s) of human fecal pollution and their occurrence in relation to FIB and enteric viruses in a scenario where beach water samples are amended with various amount of fresh raw and secondary-treated wastewater. We also determined to what extent these FIB, MST markers, and enteric viruses are present in ambient recreational beach water and sand samples from South East Queensland (SEQ), Australia, to understand the health risks associated with human fecal pollution.
RESULTS
qPCR performance characteristics.
The qPCR standards had a linear range of quantification from 3 ×106 to 3 copies per 3 μl of DNA extracts. The amplification efficiencies, correlation coefficients, and slope were determined by analyzing the standards. The amplification efficiencies ranged from 81.2 to 119%. The correlation coefficient (r2) ranged from 0.921 to 1.00. The slope ranged from −2.931 to −3.872. The qPCR performance characteristic ranges were within the prescribed Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines (24). The qPCR performance characteristics for the individual assays are shown in Table S2.
Concentrations of DNA/RNA targets in raw and secondary-treated wastewater.
The concentrations of FIB, MST markers, and enteric viruses in raw and secondary-treated wastewater samples are shown in Fig. 1a and b. In raw wastewater, the mean concentration of ENT 23S was 1.31 × 106 copies per ml. Among the bacterial markers, HF183 had the highest concentration (6.15 × 106 copies per ml), followed by EC H8 (4.75 × 106 copies per ml). The concentration of the nifH marker (2.60 × 10 copies per ml) was two orders of magnitude lower than those of the HF183 and EC H8 markers. Among the viral MST markers, HPyV had the highest mean concentration (2.56 × 105 copies per ml), followed by PMMoV (5.72 × 104 copies per ml) and HAdV (1.37 × 104 copies per ml). The concentrations of enteric viruses were two to three orders of magnitude lower than those of the FIB and MST markers. Among the three enteric viruses tested, HAdV 40/41 (2.99 × 103 copies per ml) had the highest concentration, followed by EV (1.79 × 103 copies per ml) and HNoV (9.66 × 102 copies per ml).
FIG 1.
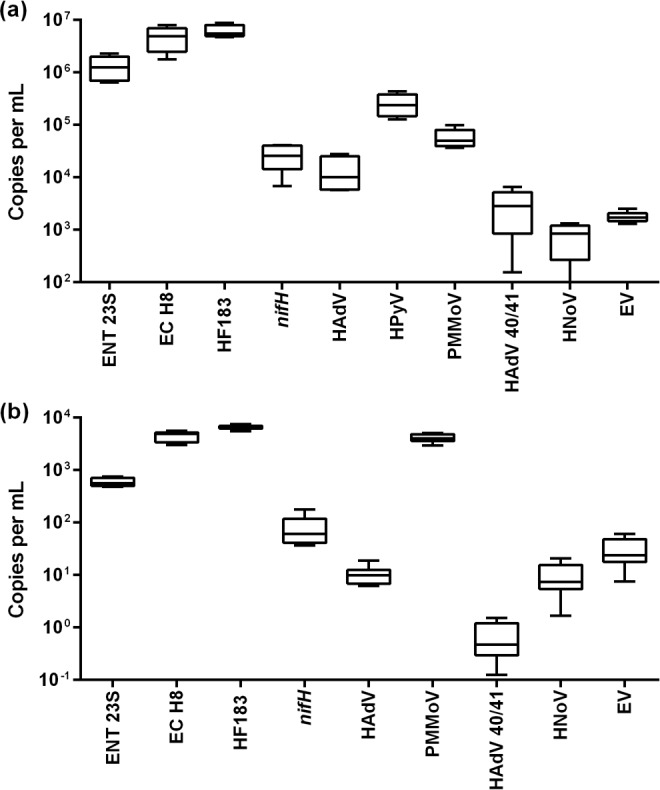
Box-and-whisker plots of the concentrations (copies per milliliter) of fecal indicator bacteria (FIB), bacterial and viral microbial source tracking (MST) markers, and enteric viruses in raw (a) and secondary-treated (b) wastewater collected from a wastewater treatment plant. The lower and upper boxes denote the 25th and 75th percentiles. The lower and upper bars represent the 5th and 95th percentiles. Shown on a vertical logarithmic scale.
The ENT 23S concentration was 5.90 × 102 copies per ml in secondary-treated wastewater. Similarly to raw wastewater, the HF183 had the highest mean concentration (6.53 × 103 copies per ml), followed by EC H8 (4.32 × 103 copies per ml). The nifH marker concentration in secondary-treated wastewater (7.89 × 101 copies per ml) was again two orders of magnitude lower than those of the HF183 and EC H8 markers. In contrast to raw wastewater, PMMoV had the highest mean concentration (4.11 × 103 copies per ml), followed by HAdV (1.04 × 101 copies per ml) in the secondary-treated wastewater. Notably, the HPyV marker was below the PLOD in secondary-treated wastewater and therefore is not shown in Fig. 1b. Dissimilar to the concentration levels of enteric viruses in raw wastewater, HAdV 40/41 (0.70 copies per ml) had the lowest concentration, followed by HNoV (9.62 copies per ml) and EV (2.95 × 101 copies per ml).
The Kruskal-Wallis one-way analysis of variance (ANOVA) was undertaken to determine if there are any significant differences in the target concentrations in raw and secondary-treated wastewater samples. Dunn's multiple comparisons posttest indicated that the concentrations of ENT 23S in raw wastewater were significantly different from those of EC H8 and HF183 (P < 0.05). The concentrations of the EC H8 and the HF183 markers were significantly different (both P < 0.05) from those of other markers and enteric viruses. The concentrations of ENT 23S in secondary-treated wastewater were significantly different from those of the EC H8, HF183, and PMMoV markers. EC H8 concentrations were significantly different from all other targets except PMMoV. The HF183 concentration in secondary-treated wastewater was significantly different from those of the other targets. The concentrations of PMMoV were significantly different from the concentrations of nifH, HAdV, HAdV 40/41, HNoV, and EV.
Correlations among DNA/RNA targets in raw and secondary-treated wastewater.
In raw wastewater, there was a highly strong correlation (r > 0.7; P < 0.05) among ENT 23S, EC H8, HF183, nifH, HAdV, HPyV, PMMoV, and HAdV 40/41. However, HPyV (r = 0.68; P < 0.05) and PMMoV (r = 0.60; P < 0.05) moderately correlated with the bacterial MST marker nifH. EV moderately or poorly correlated with other targets, while HNoV showed mostly negative correlations with all other DNA and RNA targets tested in this study, except EC H8 (Fig. 2a). In secondary-treated wastewater, the abundance of FIB, MST markers, and enteric viruses did not show strong correlations like those with raw wastewater (Fig. 2b). Only five positive strong correlations were observed between ENT 23S and nifH (r = 0.93; P < 0.05), ENT 23S and HNoV (r = 0.71; P < 0.05), EC H8 and PMMoV (r = 0.87; P < 0.05), HF183 and HNoV (r = 0.94; P < 0.05), and nifH and HNoV (r = 0.89). Moderate correlations were observed between ENT 23S and HAdV (r = 0.52; P < 0.05), HF183 and nifH (r = 0.43; P < 0.05), HAdV and nifH (r = 0.67; P < 0.05), and HAdV 40/41 and EV (r = 0.45; P < 0.05). The remaining pairwise correlations among DNA and RNA targets were either poor or negative (Fig. 2b). Since the concentration of the HPyV marker was below the PLOD in secondary-treated wastewater, this marker was excluded from the correlation analysis.
FIG 2.
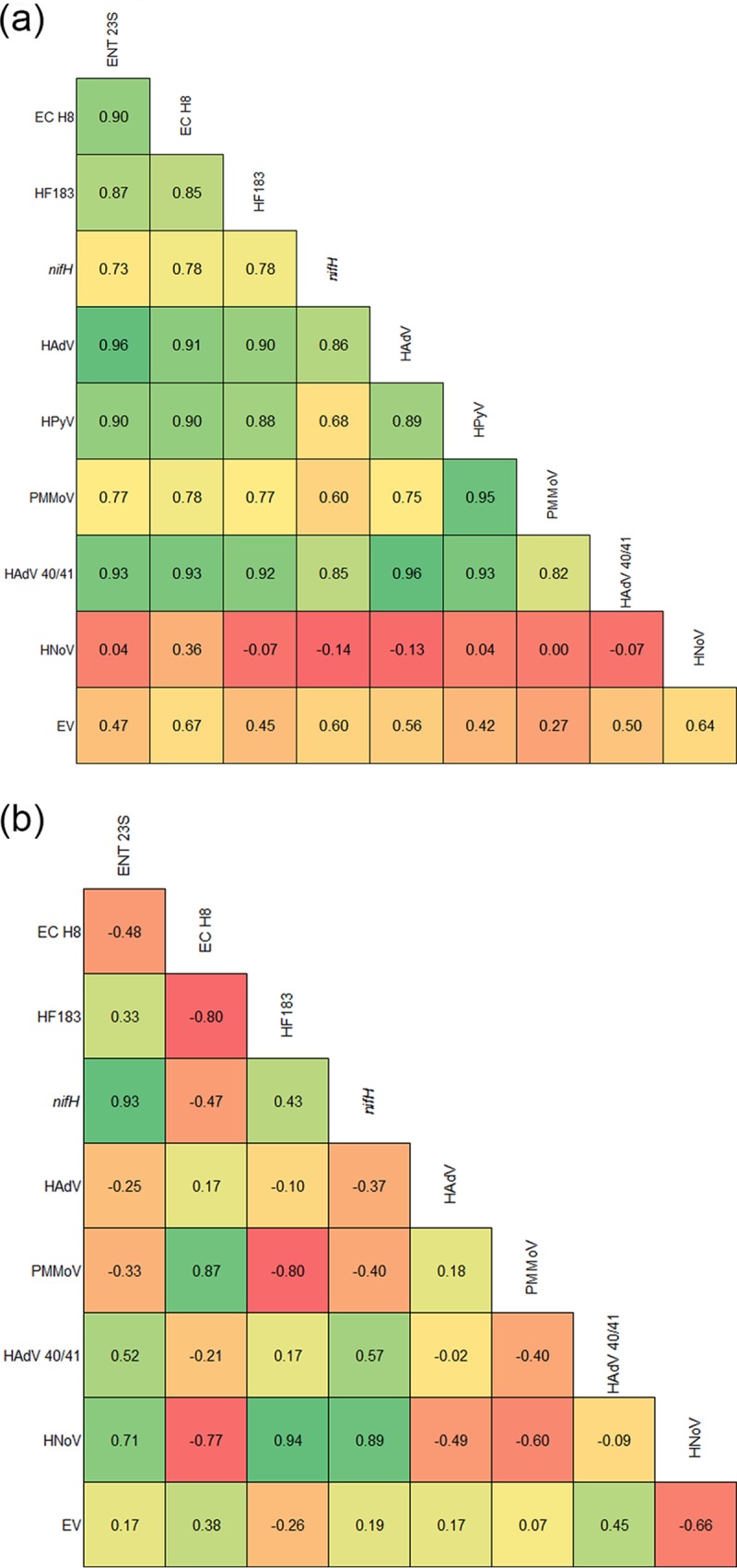
Correlations (Spearman's rank correlation) among fecal indicator bacteria, microbial source tracking (MST) markers, and enteric viruses in raw wastewater (a) and secondary-treated wastewater (b). The Spearman's r values are presented in the box. Green boxes represent positive and red boxes represent negative correlations.
PLOD and PLOQ of DNA/RNA targets in serially diluted samples of filtered beach water seeded with raw wastewater.
Thirty percent of the beach water samples seeded with raw wastewater had PCR inhibitors. A 10-fold serial dilution relieved PCR inhibition. The PLOD, PLOQ, and concentrations of all DNA and RNA targets in serially diluted filtered beach water samples seeded with raw wastewater are shown in Table 1. The concentration of ENT 23S was 3.2 × 101 copies per 3 μl of DNA at the dilution level of 10−5. Among the MST markers, HF183 was quantifiable (5.2 × 101 copies per 3 μl of DNA) at the dilution level of 10−6. EC H8 was the second most sensitive marker and was quantifiable (1.0 × 102 copies per 3 μl of DNA) at the dilution level of 10−4. HPyV and PMMoV were quantifiable at the dilution level of 10−3, while the HAdV and nifH markers were quantifiable at the dilution 10−2. HAdV 40/41 and EV were quantifiable up to a dilution level of 10−2. HNoV were detected, but the level was below the PLOQ when 3 ml of raw wastewater was seeded into the beach water sample and could not be detected at the dilution level of 10−1.
TABLE 1.
Concentrations of fecal indicator bacteria, bacterial and viral microbial source tracking markers, and enteric viruses per 3 μl of DNA/RNA extracted from beach water samples seeded with raw wastewater
| Dilution and corresponding amt of raw wastewater into beach water samples | Concn (mean ± SD) (per 3 μl of DNA/RNA) ina: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ENT 23S (FIB) | Bacterial or viral MST marker |
Enteric virus |
||||||||
| EC H8 | HF183 | nifH | HAdV | HPyV | PMMoV | HAdV 40/41 | HNoV | EV | ||
| 3 ml | 3.3 × 105 ± 1.3 × 105 | 4.2 × 105 ± 5.2 × 104 | 1.0 × 106 ±1.1 × 105 | 2.7 × 103 ± 8.3 × 102 | 2.0 × 103 ± 3.7 × 102 | 4.4 × 104 ± 5.8 × 103 | 2.6 × 103 ± 2.8 × 102 | 3.1 × 102 ± 1.7 × 102 | <PLOQ | 5.7 ×102 ± 2.6 ×102 |
| 10−1 (300 μl) | 6.1 × 104 ± 1.3 × 104 | 4.3 × 104 ± 9.8 × 103 | 2.2 × 105 ± 6.0 × 104 | 3.5 × 102 ± 6.0 × 101 | 2.3 × 102 ± 1.5 × 102 | 7.5 × 103 ± 6.8 × 103 | 6.0 × 102 ± 1.6 × 102 | 9.4 × 101 ± 4.1 × 101 | <PLOD | 7.0 ×101 ± 3.1 ×101 |
| 10−2 (30 μl) | 2.8 × 103 ± 2.3 × 103 | 1.6 × 103 ± 1.1 × 103 | 1.1 × 104 ± 7.4 × 103 | 1.3 × 101 ± 1.2 × 101 | 1.1 × 101 ± 9.9 × 100 | 8.0 × 102 ± 3.2 × 101 | 1.4 × 102 ± 5.2 × 101 | <PLOD | <PLOD | <PLOD |
| 10−3 (3 μl) | 5.7 × 102 ± 3.7 × 102 | 3.8 × 102 ± 9.3 × 101 | 2.9 × 103 ± 8.1 × 102 | <PLOD | <PLOD | 9.1 × 101 ± 1.4 × 101 | 4.9 × 101 ± 2.4 × 101 | <PLOD | <PLOD | <PLOD |
| 10−4 (300 nl) | 1.3 × 102 ± 4.4 × 101 | 1.0 × 102 ± 4.3 × 101 | 6.7 × 102 ± 3.3 × 102 | <PLOD | <PLOD | <PLOD | <PLOD | <PLOD | <PLOD | <PLOD |
| 10−5 (30 nl) | 3.2 × 101 ± 1.5 × 101 | <PLOD | 2.9 × 102 ± 2.1 × 102 | <PLOD | <PLOD | <PLOD | <PLOD | <PLOD | <PLOD | <PLOD |
| 10−6 (3 nl) | <PLOD | <PLOD | 5.2 × 101 ± 7.9 × 101 | <PLOD | <PLOD | <PLOD | <PLOD | <PLOD | <PLOD | <PLOD |
FIB, fecal indicator bacteria; MST, microbial source tracking; PLOQ, process limit of quantification; PLOD, process limit of detection.
PLOD and PLOQ of DNA/RNA targets in serially diluted samples of filtered beach water seeded with secondary-treated wastewater.
In all, 37.5% of the beach water samples seeded with secondary-treated wastewater had PCR inhibitors. A 10-fold serial dilution relieved PCR inhibition. The PLOD, PLOQ, and concentrations of all DNA and RNA targets in serially diluted filtered beach water samples seeded with secondary-treated wastewater are shown in Table 2. ENT 23S was quantifiable (8.5 × 101 copies per 3 μl of DNA) up to a dilution level of 10−4. Among the MST markers, both EC H8 (1.5 × 101 copies per 3 μl of DNA) and HF183 (8.6 × 101 copies per 3 μl of DNA) were quantifiable up to a dilution level of 10−4. PMMoV was quantifiable (1.6 × 101 copies per 3 μl of DNA) to dilution level of 10−3. The HAdV and nifH markers were quantifiable up to a dilution level of 10−1. Human enteric virus and HAdV 40/41 were detectable but nonquantifiable in 150 ml of seeded secondary-treated wastewater. HNoV was below the PLOD and EV was quantifiable in a 150-ml seeded secondary-treated wastewater sample. However, at dilution 10−1, it fell below the PLOQ, and at dilution 10−2, it was below the PLOD.
TABLE 2.
Concentrations of fecal indicator bacteria, bacterial and viral microbial source tracking markers, and enteric viruses per 3 μl of DNA/RNA extracted from beach water samples seeded with secondary-treated wastewater
| Dilution and corresponding amt of secondary-treated wastewater into beach water samples | Concn (mean ± SD) (per 3 μl of DNA/RNA) ina: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| FIB (ENT 23S) | Bacterial or viral MST marker |
Enteric virus |
|||||||
| EC H8 | HF183 | nifH | HAdV | PMMoV | HAdV 40/41 | HNoV | EV | ||
| 150 ml | 2.4 × 103 ± 7.5 × 101 | 5.8 × 103 ± 3.8 × 102 | 2.0 × 104 ± 7.6 × 102 | 2.2 × 102 ± 7.0 × 101 | 2.1 × 101 ± 8.7 × 100 | 3.6 × 103 ± 3.9 × 102 | <PLOQ | <PLOD | 2.9 ×101 ± 1.1 × 101 |
| 10−1 (15 ml) | 1.6 × 103 ± 4.9 × 102 | 1.3 × 103 ± 2.3 × 102 | 4.7 × 103 ± 9.4 × 102 | 3.5 × 101 ± 9.3 × 100 | 3.7 × 100 ± 3.0 × 100 | 4.8 × 102 ± 2.4 × 101 | <PLOD | <PLOD | <PLOQ |
| 10−2 (1.5 ml) | 8.7 × 102 ± 7.4 × 102 | 2.7 × 102 ± 2.2 × 102 | 6.3 × 102 ± 4.0 × 102 | <PLOD | <PLOD | 1.8 × 102 ± 3.8 × 101 | <PLOD | <PLOD | <PLOD |
| 10−3 (150 μl) | 9.4 × 102 ± 9.6 × 102 | 1.8 × 102 ± 2.0 × 102 | 3.3 × 102 ± 3.5 × 102 | <PLOD | <PLOD | 1.6 × 101 ± 1.5 × 101 | <PLOD | <PLOD | <PLOD |
| 10−4 (15 μl) | 8.5 × 101 ± 5.0 × 101 | 1.5 × 101 ± 1.7 × 101 | 8.6 × 101 ± 1.4 × 102 | <PLOD | <PLOD | <PLOD | <PLOD | <PLOD | <PLOD |
| 10−5 (1.5 μl) | <PLODa | <PLOD | <PLOD | <PLOD | <PLOD | <PLOD | <PLOD | <PLOD | <PLOD |
| 10−6 (150 nl) | <PLOD | <PLOD | <PLOD | <PLOD | <PLOD | <PLOD | <PLOD | <PLOD | <PLOD |
FIB, fecal indicator bacteria; MST, microbial source tracking; PLOQ, process limit of quantification; PLOD, process limit of detection.
Concentrations of culturable FIB in recreational beach water samples in SEQ.
The E. coli and Enterococcus species concentrations in recreational beach water samples are shown in Fig. 3. At each site except Paradise Point and Southport, the Enterococcus species concentrations exceeded the Australian and New Zealand Environment and Conservation Council (ANZECC) guidelines (<35 CFU per 100 ml for primary recreational water). The highest concentration (165 CFU per 100 ml) of Enterococcus spp. was found at Deception Bay. For E. coli, each site except Jabiru Island (151 CFU per 100 ml) had a concentration below the recommended ANZECC guideline (<150 CFU per 100 ml). Using Spearman's rank correlation, it was determined that the E. coli and Enterococcus species concentrations were significantly correlated with each other in these sites (P < 0.05).
FIG 3.
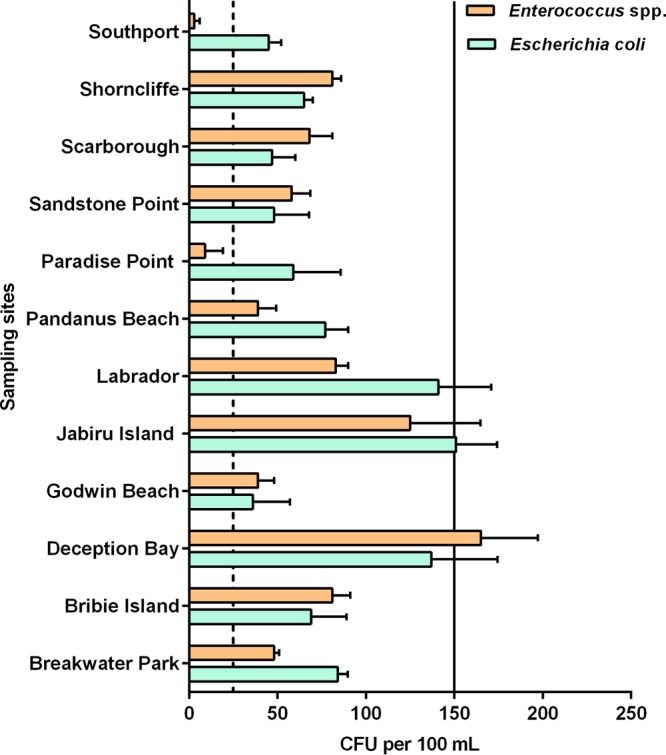
Concentrations of Escherichia coli and Enterococcus spp. in water samples collected from 12 recreational beaches in South East Queensland, Australia. The dashed line represents guideline value (35 CFU per 100 ml) for Enterococcus spp. and the solid line represents the guideline value (150 CFU per 100 ml) for E. coli.
MST markers and enteric viruses in beach water and sand samples in SEQ.
In all, 41.6% of the 24 beach water (n = 12) and sand (n = 12) samples had PCR inhibition. A 10-fold serial dilution was made for these samples to relieve PCR inhibition. All uninhibited and serially diluted samples were used in analysis. The prevalence and concentrations of FIB, MST markers, and enteric viruses in the 12 recreational beach sites from SEQ are shown in Table 3. ENT 23S was detected in 91.6% (11 of 12) of the recreational water samples. The highest concentration of ENT 23S, at 3.8 × 103 per 100 ml of water, was found at Deception Bay. Fifty percent of the beach water samples had quantifiable concentrations of the HF183 marker, and the concentration ranged from 1.8 × 103 to 1.3 × 102 copies per 100 ml of water. PMMoV was quantifiable in 33.3% of the water samples, and the concentrations ranged from 3.6 × 103 to 8.6 × 103 copies per 100 ml of water. HAdV was detected (1 of 3 qPCR replicates was positive) in the water sample from Pandanus Beach. However, the level of HAdV in this sample was below the PLOQ. EV was detected at one site (Southport), but it was below the PLOQ. The MST markers EC H8, nifH, HPyV, and enteric viruses HAdV 40/41 and HNoV could not be detected in any of the recreational water samples. For the sand samples, ENT 23S was detected and quantifiable in 83.3% of the sites. HF183 was detected and quantifiable at 33 and 25% of the sand samples, respectively. HAdV was not detected in sand samples. PMMoV and EV were not tested for in the sand samples.
TABLE 3.
Prevalence and concentrations of ENT 23S, HF183, and enteric viruses in environmental water and sand samples collected from 12 recreational beaches in South East Queensland, Australia
| Sampling site | Matrix | No. of positive samples/no. of samples tested (concn per 100 ml of water or per g of sand [wet wt])a |
||||
|---|---|---|---|---|---|---|
| FIB (ENT 23S) | Bacterial or viral MST marker |
Enteric virus (EV) | ||||
| HF183 | HAdV | PMMoV | ||||
| Breakwater Park | Water | 3/3 (1.2 × 103 ± 6.5 × 102) | <PLOD | <PLOD | 1/3 (<PLOQ) | <PLOD |
| Sand | 3/3 (7.5 × 103 ± 2.2 × 102) | 2/3 (3.6 × 102) | <PLOD | NT | NT | |
| Bribie Island | Water | 2/3 (8.7 × 101) | 2/3 (2.8 × 102) | <PLOD | <PLOD | <PLOD |
| Sand | 3/3 (1.2 × 102 ± 1.8 × 102) | 1/3 (2.9 × 103) | <PLOD | NT | NT | |
| Deception Bay | Water | 3/3 (3.8 × 103 ± 7.3 × 102) | <PLOD | <PLOD | 1/3 (3.6 × 103) | <PLOD |
| Sand | 3/3 (1.1 × 102 ± 3.2 × 102) | 3/3 (<PLOQ) | <PLOD | NT | NT | |
| Godwin Beach | Water | 1/3 (1.4 × 102) | <PLOD | <PLOD | 1/3 (5.6 × 103) | <PLOD |
| Sand | 3/3 (8.5 × 103 ± 6.3 × 102) | <PLOD | <PLOD | NT | NT | |
| Jabiru Island | Water | 3/3 (4.7 × 102 ± 3.2 × 102) | 3/3 (2.5 × 102 ± 1.9 ×102) | <PLOD | <PLOD | <PLOD |
| Sand | 3/3 (3.7 × 103 ± 4.5 × 102) | <PLOD | <PLOD | NT | NT | |
| Labrador | Water | 1/3 (4.1 × 101) | 1/3 (1.8 ×103) | <PLOD | <PLOD | <PLOD |
| Sand | 3/3 (4.4 × 103 ± 2.4 × 102) | <PLOD | <PLOD | NT | NT | |
| Pandanus Beach | Water | 2/3 (2.2 × 102) | <PLOD | 1/3 (<PLOQ) | <PLOD | <PLOD |
| Sand | 3/3 (2.1 × 104 ± 1.0 × 104) | <PLOD | <PLOD | NT | NT | |
| Paradise Point | Water | 2/3 (1.8 × 102) | 2/3 (1.7 × 103) | <PLOD | <PLOD | <PLOD |
| Sand | <PLODa | <PLOD | <PLOD | NT | NT | |
| Sandstone Point | Water | 3/3 (3.3 × 102 ± 1.8 × 102) | <PLOD | <PLOD | <PLOD | <PLOD |
| Sand | 2/3 (2.5 × 102) | 1/3 (4.1 × 103) | <PLOD | NT | NT | |
| Scarborough | Water | 3/3 (3.3 × 102 ± 9.5 × 101) | 2/3 (2.5 × 102) | <PLOD | 1/3 (8.6 × 103) | <PLOD |
| Sand | 3/3 (6.3 × 102 ± 2.4 × 102) | <PLOD | <PLOD | NT | NT | |
| Shorncliffe | Water | 2/3 (2.3 × 102) | <PLOD | <PLOD | 2/3 (5.7 × 103) | <PLOD |
| Sand | 3/3 (2.7 × 103 ± 3.9 × 102) | <PLOD | <PLOD | NT | NT | |
| Southport | Water | <PLOD | 1/3 (1.3 × 102) | <PLOD | <PLOD | 1/3 (<PLOQ) |
| Sand | 3/3 (8.1 × 103 ± 2.0 × 103) | <PLOD | <PLOD | NT | NT | |
FIB, fecal indicator bacteria; MST, microbial source tracking; PLOD, process limit of detection; PLOQ, process limit of quantification; NT, not tested.
DISCUSSION
Human wastewater is known to contain >100 pathogenic viruses, some with the ability to cause disease and illness in humans at extremely low dosages (25). Thus, the pollution of recreational water with human wastewater poses a more significant risk to humans than animal fecal wastewater (26). This has led to the development of MST tools which utilize molecular markers to inform targeted solutions to minimize fecal pollution in water. Only a few studies compared or determined the PLOD (defined as the smallest volume of wastewater that could be subjected to the complete sample preparation process, including dilution, filtration, and DNA/RNA extraction, and still be reliably detected in qPCR) of molecular markers to detect human wastewater in environmental water samples. For example, Ahmed et al. (27) compared the PLOD values of the HF183, Enterococcus faecium, HAdV esp, and HPyV in fresh and seawater samples in Australia. While in Florida, USA, Staley et al. (3) determined the PLOD values for HF183 and HPyV in various types of surface water samples. Despite experimental differences, both studies found HF183 to be the most sensitive marker of human fecal pollution. The potential of PMMoV as a human wastewater marker has also been reported in a recent study, when its PLOD in coastal waters was investigated by Symonds et al. (23). The limited data highlight the knowledge gap that this study aimed to address by undertaking a large-scale cross comparison of the PLOD/PLOQ values for six human-wastewater-associated MST markers in relation to FIB and enteric viruses in beach water samples seeded with fresh raw and secondary-treated wastewater.
Before the seeding experiment, the concentrations of all DNA and RNA targets were measured in raw and secondary wastewater samples using qPCR assays. The HF183 concentrations were the highest among all targets in both raw and secondary-treated wastewater. The mean concentrations of the HF183 obtained in this study were similar to the mean concentrations of HF183 found in human wastewater around the globe (12). Limited information is available on the concentration of the EC H8 marker in human wastewater. However, in a previous study, we reported that approximately 50% of E. coli isolates in human wastewater carried the EC H8 marker (14). Despite a lack of comparative information, the results from this study indicated that the concentrations of EC H8 in raw wastewater could be quite high, being only slightly less than the concentration of the HF183. These high levels indicated that EC H8 may be a suitable marker for tracking human wastewater pollution in environmental waters.
The concentrations of HAdV and HPyV in raw and secondary-treated wastewater were similar to those reported in other published studies (28–31). Some researchers reported that PMMoV has the potential to be an overly sensitive marker, due to its high concentrations in human wastewater (17, 32, 33). While this presents an advantage for the PMMoV as a sensitive marker, its presence may also overestimate risk (23). A study by Rosario et al. (17) determined the average concentrations of PMMoV at a Florida Keys WWTP over a 2-week period and found that levels ranged from 8.04 × 105 to 1.9 × 106 copies per ml in raw wastewater and 2.02 × 104 to 1.01 × 106 copies per ml in treated wastewater. The levels of PMMoV found in this study were 1 to 2 orders of magnitude lower in raw wastewater and 1 to 3 orders of magnitude lower in secondary-treated wastewater than those reported by Rosario et al. (17), thus indicating that PMMoV may not be overly sensitive, as others have previously reported. HNoV can be excreted in high quantities from infected individuals and has been detected at concentrations of 200 copies per ml in raw wastewater (34). The result from this study (9.62 × 102 copies per ml of raw wastewater) was similar to that of the Hewitt and colleagues study in New Zealand (34). The concentrations of HAdV 40/41 and EV in raw and secondary-treated wastewater were found to be similar to those previously reported (35–37).
The FIB, MST markers, and the enteric virus HAdV 40/41 had strong positive correlations in raw wastewater but largely weak correlations in secondary-treated wastewater samples. Similar correlations between MST markers in raw wastewater have been reported among ENT 23S, HF183, HAdV, and HPyV (38). The lack of correlation with secondary-treated wastewater is likely due to low and varied target concentrations as a result of the treatment process.
In the raw wastewater seeding experiment, the PLOD of the HF183 marker was remarkably low and was quantifiable to a dilution level of 10−6. Staley et al. (3) reported similar results, with the quantification of HF183 to a dilution level of 10−6 in 500-ml ambient water samples seeded with 5 ml of raw wastewater. EC H8 was the second most sensitive marker in terms of PLOD and was quantifiable at a dilution level of 10−5. Among the viral markers, PMMoV had the lowest PLOD and was quantifiable at a dilution level of 10−3. PMMoV was three orders of magnitude less sensitive than the HF183 marker. A similar level of PMMoV detection (10−2) was reported in a recent study in Florida, USA (23).
Due to the decreased concentration of targets in secondary-treated wastewater, the detection and quantification of FIB and MST markers were not seen in the higher dilution levels, and the enteric viruses were not detectable in any diluted samples. Similarly to the raw wastewater, the HF183, EC H8, and PMMoV markers were detected at dilution levels of 10−5, 10−5, and 10−3, respectively. The detection of FIB, MST markers, and enteric viruses in these samples to such a high dilution level confirms that pollution of recreational waters with secondary-treated wastewater may still represent a risk to humans.
Overall, HF183, EC H8, and PMMoV were detected at high dilution levels (10−3 to 10−6), but their presence at these dilutions was not directly indicative of human health risks because enteric viruses were not observed at a dilution level of <10−2. From these results, it can be assumed that while high concentrations of bacterial markers, such as HF183 and EC H8, in environmental waters can be indicative of human wastewater pollution, their presence alone cannot be assumed to confer risk. When these markers are used alone, quantitative data must be reported to identify the magnitude of the pollution. We recommend that the HF183 or EC H8 marker be used for initial wastewater detection, followed by further testing with more specific viral MST markers, such as PMMoV, HAdV, or HPyV, in decreasing order of preference. In this study, we determined the PLOD/PLOQ of targets in serially diluted beach water samples seeded with raw and secondary-treated wastewater. This may not be representative of a true environmental sample that contains various background microflora. Therefore, the results presented in this study should be interpreted with care, as it is possible that the PLOD/PLOQ of targets may be less sensitive for true environmental samples.
The application of enteric viruses for wastewater pollution monitoring could greatly improve the efficiency of monitoring procedures because risks could be directly estimated based on viral concentrations. However, due to the vast abundance of human enteric viruses, using only a few enteric viruses alone to confer risk is inadvisable. Therefore, the use of multiple MST markers for wastewater detection, if possible, is still the preferred method for application to a real-world setting.
In this study, environmental water and sand samples from 12 recreational beaches were collected and analyzed for the presence of the 10 DNA and RNA targets. Six of the 12 sites had quantifiable concentrations of the HF183 marker, most likely due to their proximity to stormwater drains. Four of the 12 sites also had quantifiable concentrations of PMMoV, highlighting the fact that stormwater drains may indeed be the primary sources of human fecal pollution in these sites. The Scarborough site was positive for two MST markers (HF183 and PMMoV), strongly suggesting the presence of human wastewater pollution.
The three enteric viruses tested in this study (HAdV 40/41, HNoV, and EV) were not detected above the PLOD at any of the sites. However, the presence of other enteric viruses (not tested in this study) cannot be ruled out (25). Therefore, it is suggested that further water quality monitoring of these sites and an exploratory quantitative microbial risk assessment (QMRA) should be performed to assess the risks associated with the use of these recreational waters (3, 23). To expand on previous studies that have also detected MST markers in sands and sediments in the United States (39, 40), 12 sand samples were analyzed for the presence of ENT 23S and five DNA markers. ENT 23S was quantified in 83% of the sand samples, and HF183 was quantified at three sites. These results indicated that SEQ recreational beach sands could harbor MST markers and be potential reservoirs for enteric pathogens. Based on the results, we recommend that the microbiological quality of beach sand also needs to be monitored in SEQ. This would allow for the establishment of guidelines to assess public health risks and would be useful for implementing beach sanitation programs for beachgoers.
Several points uncovered in this study require further investigation. First, the majority of wastewater in Australia undergoes complete treatment, including chlorination, before being released into the environment. As a result, the concentrations of MST markers and enteric viruses are likely to be low, except in scenarios where treatment plants fail due to extreme weather events (flooding), broken pipes, or illegal discharges. However, the concentration of MST markers in fully treated wastewater should be investigated to ensure that pathogen levels at wastewater outfalls do not present human health risks. Second, sand samples from SEQ should be analyzed for the presence of enteric viruses to better understand the associated risks. Last, this study was conducted using fresh wastewater, which means the PLOD/PLOQ analysis of the MST markers and enteric viruses is based on the target concentrations that would likely to be present immediately following a recent contamination event. However, as water quality assessments are not always undertaken immediately following pollution events, the concentrations of MST markers in aged wastewater samples should also be investigated. In addition, comparative inactivation studies of FIB, MST markers, and enteric viruses in the water column and beach sands should be undertaken.
MATERIALS AND METHODS
Wastewater sampling and analysis.
Composite fresh raw and secondary-treated wastewater samples were collected from the primary influent and secondary settling tank of a wastewater treatment plant (WWTP), respectively. The WWTP serves a population of approximately 250,000 people. The treatment process consists of primary treatment, a secondary treatment (activated sludge process), UV disinfection, and chlorination prior to discharge in the Brisbane River. The secondary-treated wastewater samples used in this study were collected before UV disinfection and chlorination. The wastewater samples were collected at 8:00 a.m. and transported to the laboratory on ice. Upon arrival, the samples were stored at 4°C and processed within 3 h of collection.
To quantify the background concentrations of DNA targets (ENT 23S, HF183, EC H8, nifH, HAdV, HAdV 40/41, and HPyV), triplicate 10-ml raw wastewater and 25-ml secondary-treated wastewater samples were filtered through negatively charged 47 mm, 0.45-μm-pore-size HA membranes (HAWP04700; Merck Millipore, Tokyo, Japan). For RNA targets (PMMoV, HNoV, and EV), triplicate 100-ml raw wastewater and 250-ml secondary-treated wastewater samples were also filtered through negatively charged 47-mm, 0.45-μm-pore-size HA membranes.
Beach water sampling for seeding experiments.
For seeding experiments, beach water samples were collected from a recreational beach located in Redcliffe in SEQ. To the best of our knowledge, based on a sanitary survey, these sites were not exposed to point or nonpoint sources of fecal pollution. Triplicate water samples from three sites (not shown in Fig. 4) were collected at a water depth of approximately 25 cm. Water samples were collected in sterile 10-liter containers and transported to the laboratory. The samples were filtered through negatively charged 90-mm, 0.45-μm-pore-size HA membranes (HAWP09000; Merck Millipore, Tokyo, Japan). Following the filtration step, 100 ml of each water sample was filtered through 0.45-μm nitrocellulose membranes (Advantec, Tokyo, Japan), placed on Chromocult coliform agar and enterococci agar (Merck, KGaA, Germany), and incubated at 37°C for 24 h. No colonies were observed on the agar plates, indicating that the beach water samples collected for seeding experiment did not contain any FIB after filtration. To confirm the absence of background MST markers that may affect the PLOD and PLOQ assessments, the samples (1 liter) were screened for HF183 and HAdV. HF183 and HAdV were concentrated using a previously described method (11). First, the pH of each filtered beach water sample was lowered to 3.5 using 2.0 N HCl and then filtered through negatively charged 90 mm, 0.45-μm-pore-size HA membranes. The HF183 and HAdV qPCR assays were performed in a 20-μl reaction volume using 10 μl of SsoFast EvaGreen supermix (Bio-Rad Laboratories, CA, USA) (HF183) or 10 μl of SsoAdvanced universal probes supermix (Bio-Rad) (HAdV), 300 nM each primer (HF183), 200 nM each primer and probe (HAdV), and 3 μl of template DNA. The samples were negative for HF183 and HAdV.
FIG 4.
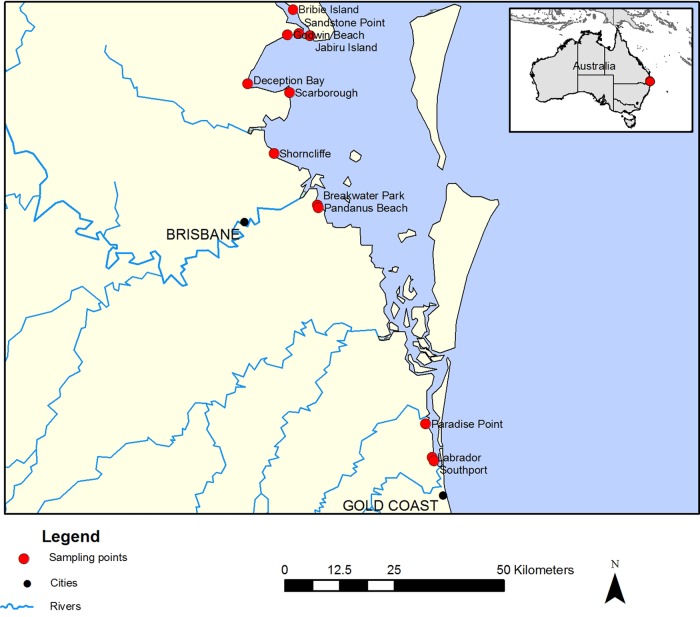
Map showing the beach water and sand sampling locations in South East Queensland, Australia.
Wastewater seeding experiment.
For DNA targets, 3 ml of raw wastewater was seeded into 297 ml of the filtered beach water samples collected from sites A to C. Similarly, 150 ml of secondary-treated wastewater was seeded into 150 ml of the filtered beach water samples collected from sites A to C. A higher volume of secondary-treated wastewater was added to compensate for low target concentrations in secondary-treated wastewater due to activated sludge process. Tenfold serial dilutions (10−1 to 10−6) were then made for all seeded samples. The pH of each sample was adjusted to 3.5 with 2.0 N HCl before being filtered through negatively charged 47-mm, 0.45-μm-pore-size HA membranes.
For RNA targets, 3 ml of raw wastewater was seeded into 297 ml of the filtered beach water samples collected from site A only. Similarly, 150 ml of secondary-treated wastewater was seeded into 150 ml of the filtered beach water samples collected from site A only. Tenfold serial dilutions (10−1 to 10−6) were then made, followed by adjustment of the pH of each seeded sample to 3.5 with 2.0 N HCl. The samples were filtered through negatively charged 47-mm, 0.45-μm-pore-size HA membranes (HAWP04700; Merck Millipore, Tokyo, Japan).
qPCR standards.
Standard curves for the nifH, HPyV, HAdV 40/41, PMMoV, HNoV, and EV qPCR assays were constructed using synthesized plasmid DNA (pIDTSMART with ampicillin resistance [Integrated DNA Technologies, Coralville, IA, USA]). qPCR standards for ENT 23S and EC H8 were prepared from the genomic DNA of Enterococcus faecalis ATCC 19433 and E. coli ATCC 23226. qPCR standards for HF183 and HAdV were prepared from the plasmid DNA. The purified genomic or plasmid DNA was serially diluted to create a standard ranging from 1 × 106 to 1 copy per μl of DNA. A 3-μl template from each serial dilution was used to prepare a standard curve for each qPCR assay. For each standard, the genomic copies were plotted against the cycle number at which the fluorescence signal increased above the quantification cycle (Cq) value. The amplification efficiency (E) was determined by analysis of the standards and was estimated from the slope of the standard curve to be E = 10−1/slope.
qPCR analysis.
The primers and probes are shown in Table S1. Each qPCR amplification was performed separately for each target. EC H8 and HF183 qPCR amplifications were performed in a 20-μl reaction mixture using 10 μl of SsoFast EvaGreen supermix (Bio-Rad Laboratories, CA, USA), 400 nM each primer (EC H8 assay), 300 nM each primer (HF183 assay), and 3 μl of template DNA. To separate the specific products from the nonspecific products, including primer dimers, a melting-curve analysis was performed for EC H8 and HF183 assays. During melting-curve analysis, the temperature was increased from 65 to 95°C at 0.5°C increments. Melting-curve analysis showed a distinct peak at a temperature of 78.5°C ± 0.5°C for HF183 and 92.0 ± 0.5°C for EC H8, indicating positive and correct amplifications.
ENT 23S, nifH, HAdV, HPyV, and HAdV 40/41 qPCR assays were performed in 20-μl reaction mixtures using 10 μl of SsoAdvanced universal probes supermix (Bio-Rad Laboratories, CA, USA), 500 nM each primer and 400 nM probe (ENT 23S), 800 nM each primer and 240 nM probe (nifH), 200 nM each primer and 200 nM probe (HAdV), 240 nM each primer and 160 nM probe (HPyV), 400 nM each primer and 100 nM probe (HAdV 40/41), and 3 μl of template DNA (Table S1).
For RNA targets (PMMoV, HNoV, and EV), cDNA was synthesized using the SuperScript III first-strand synthesis reverse transcriptase kit (Invitrogen, Carlsbad, CA, USA), according to the manufacturer's instructions. Six microliters of extracted RNA was used to generate cDNA. PMMoV, HNoV, and EV qPCR assays were performed in 20-μl reaction mixtures using 10 μl of SsoAdvanced universal probes supermix (Bio-Rad Laboratories, CA, USA), 200 nM each primer and 80 nM probe (PMMoV), 250 nM each primer and 100 nM probe (HNoV), 300 nM forward primer, 900 nM reverse primer, and 125 nM probe (EV) and 3 μl of template cDNA (Table S1).
Limit of detection and quantification.
To determine whether a sample was positive, negative, or quantifiable, defined criteria were established. The qPCR assay limit of detection (ALOD) was defined as the number of copies that could be detected in 2 out of 3 qPCR assays, while the assay limit of quantification (ALOQ) was the number of copies that could be quantified in 2 out of 3 qPCR assays. The process limit of detection (PLOD) was defined as the smallest volume of wastewater that could be subjected to the complete sample preparation process, including dilution, filtration, and DNA/RNA extraction and still be reliably detected/quantified (PLOQ) in 2 out of 3 qPCR reactions (23).
Recreational beach water and sand sampling.
A field study was undertaken to determine the extent of human wastewater pollution in 12 recreational beaches along the SEQ coast (Fig. 4). The northernmost site was located on Bribie Island, while the southernmost site was located in Southport on the Gold Coast. All sites were public beach areas known to be popular with bathers and recreational water users. Stormwater pipes were identified as a major source of human fecal pollution in these sites (Table S3). At each recreational beach, a 10-liter water sample was collected in a sterile container at an approximate water depth of 25 cm. In addition, beach sand cores were collected by manually inserting a clear plastic tube (internal diameter, 2 cm) below the water surface, 10 cm into the sand. All water and sand samples were immediately transported to the laboratory and processed within 4 to 6 h after collection.
The concentrations of culturable E. coli and Enterococcus spp. in the water samples were determined using culture-based methods, as described earlier. For the quantification of DNA and RNA targets, the pH of the recreational water sample was lowered to 3.5 using 2.0 N HCl. For DNA targets, 1.8 liters of each sample was then filtered through negatively charged 90-mm, 0.45-μm-pore-size HA membranes. For the RNA targets, 800 ml of each water sample was filtered through negatively charged 90-mm, 0.45-μm-pore-size HA membranes. Sand cores were only tested for the DNA targets. Five grams of sand from the top layer of each sand core (approximately 1 to 5 cm depth) was used to extract DNA.
DNA extraction and PCR inhibition analysis.
DNA samples were extracted from the membranes using the Mo Bio PowerWater DNA isolation kit (Mo Bio Laboratories, Carlsbad, CA, USA). RNA samples were extracted using the Mo Bio PowerWater RNA isolation kit. All DNA and RNA samples were stored at −80°C. An experiment was conducted to determine the effect of potential PCR inhibitory substances on the quantitative detection of DNA and RNA targets in (i) wastewater, (ii) wastewater-seeded beach water samples, and (iii) beach water and sand samples using a Sketa22 real-time assay (41). Samples with a 2-Cq delay were considered to have PCR inhibitors.
Quality control.
All equipment used for collection, filtration, and sample storage was sterilized by bleaching and autoclaving prior to use. During water sampling, container blanks carrying sterile distilled water were taken to each site and left open during the collection procedures. These samples were processed in an identical manner to all other samples to confirm the absence of contamination. During the DNA and RNA extraction activities, reagent blanks were run and analyzed for each extraction procedure. During qPCR analysis, all DNA and RNA samples were run in triplicate with three negative controls (sterile water) on 96-well plates using the CFX 96 thermocycler (Bio-Rad Laboratories, CA, USA).
Statistical analysis.
The concentrations of DNA and RNA targets in raw and secondary-treated wastewater samples were not normally distributed (as determined by a Kolmogorov-Smirnov test). Therefore, the nonparametric Kruskal-Wallis one-way analysis of variance (ANOVA) with Dunn's posttest was performed to determine if there were any significant differences in FIB, MST marker, and enteric virus concentrations in raw and secondary-treated wastewater. The nonparametric Spearman's rank correlation with a two-tailed test was used to establish the relationship among targets in raw and secondary-treated wastewater samples. In general, when r was >0.7, the targets were considered to have a strong positive correlation, an r of ≥0.4 but <0.7 was a moderate correlation, and an r of >0.2 but <0.4 was a weak correlation. GraphPad Prism 6 was used for statistical analysis (GraphPad Software, Inc.). Spearman's rank correlation was also performed to determine the correlation between culturable E. coli and Enterococcus species concentrations in recreational water samples. All statistical analyses were evaluated at an α value of 0.05.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge funding from CSIRO and the School of Agriculture and Food Sciences, The University of Queensland.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00028-17.
REFERENCES
- 1.Chase E, Hunting J, Staley C, Harwood V. 2012. Microbial source tracking to identify human and ruminant sources of faecal pollution in an ephemeral Florida river. J Appl Microbiol 113:1396–1406. doi: 10.1111/jam.12007. [DOI] [PubMed] [Google Scholar]
- 2.Graczyk TK, Sunderland D, Tamang L, Lucy FE, Breysse PN. 2007. Bather density and levels of Cryptosporidium, Giardia, and pathogenic microsporidian spores in recreational bathing water. Parasitol Res 101:1729–1731. doi: 10.1007/s00436-007-0734-1. [DOI] [PubMed] [Google Scholar]
- 3.Staley C, Gordon K, Schoen M, Harwood V. 2012. Performance of two quantitative PCR methods for microbial source tracking of human sewage and implications for microbial risk assessment in recreational waters. Appl Environ Microbiol 78:7317–7326. doi: 10.1128/AEM.01430-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petri WA Jr, Miller M, Binder HJ, Levine MM, Dillingham R, Guerrant RL. 2008. Enteric infections, diarrhea, and their impact on function and development. J Clin Invest 118:1277–1290. doi: 10.1172/JCI34005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wolf L, Held I, Eiswirth M, Hötzl H. 2004. Impact of leaky sewers on groundwater quality. Clean 32:361–373. doi: 10.1002/aheh.200400538. [DOI] [Google Scholar]
- 6.Sowah RA, Habteselassie MY, Radcliffe DE, Bauske E, Risse M. 2017. Isolating the impact of septic systems on fecal pollution in streams of suburban watersheds in Georgia, United States. Water Res 108:330–338. doi: 10.1016/j.watres.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 7.Sidhu JPS, Ahmed W, Gernjak W, Aryal R, McCarthy D, Palmer A, Kolotelo P, Toze S. 2013. Sewage pollution in urban stormwater runoff as evident from the widespread presence of multiple microbial and chemical source tracking markers. Sci Total Environ 463:488–496. doi: 10.1016/j.scitotenv.2013.06.020. [DOI] [PubMed] [Google Scholar]
- 8.Cann KF, Thomas DR, Salmon RL, Wyn-Jones AP, Kay D. 2012. Extreme water-related weather events and waterborne disease. Epidemiol Infect 141:671–686. doi: 10.1017/S0950268812001653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dale K, Kirk M, Sinclair M, Hall R, Leder K. 2010. Reported waterborne outbreaks of gastrointestinal disease in Australia are predominantly associated with recreational exposure. Aust N Z Public Health 34:527–530. doi: 10.1111/j.1753-6405.2010.00602.x. [DOI] [PubMed] [Google Scholar]
- 10.Harwood V, Levine A, Scott T, Chivukula V, Lukasik J, Farrah SR, Rose JB. 2005. Validity of the indicator organism paradigm for pathogen reduction in reclaimed water and public health protection. Appl Environ Microbiol 71:3163–3170. doi: 10.1128/AEM.71.6.3163-3170.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McQuaig S, Griffith J, Harwood VJ. 2012. Association of fecal indicator bacteria with human viruses and microbial source tracking markers at coastal beaches impacted by nonpoint source pollution. Appl Environ Microbiol 78:6423–6432. doi: 10.1128/AEM.00024-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahmed W, Hughes B, Harwood V. 2016. Current status of marker genes of Bacteroides and related taxa for identifying sewage pollution in environmental waters. Water 8:231. doi: 10.3390/w8060231. [DOI] [Google Scholar]
- 13.Ufnar JA, Wang SY, Christiansen J, Yampara-Iquise H, Carson C, Ellender RD. 2006. Detection of the nifH gene of Methanobrevibacter smithii: a potential tool to identify sewage pollution in recreational waters. J Appl Microbiol 101:44–52. doi: 10.1111/j.1365-2672.2006.02989.x. [DOI] [PubMed] [Google Scholar]
- 14.Ahmed W, Triplett C, Gomi R, Gyawali P, Hodgers L, Toze S. 2015. Assessment of genetic markers for tracking the sources of human wastewater associated Escherichia coli in environmental waters. Environ Sci Technol 49:9341–9346. doi: 10.1021/acs.est.5b02163. [DOI] [PubMed] [Google Scholar]
- 15.Gomi R, Matsuda T, Matsui Y, Yoneda M. 2014. Fecal source tracking in water by next-generation sequencing technologies using host-specific Escherichia coli genetic markers. Environ Sci Technol 48:9616–9623. doi: 10.1021/es501944c. [DOI] [PubMed] [Google Scholar]
- 16.Rusiñol M, Fernandez-Cassi X, Hundesa A, Vieira C, Kern A, Eriksson I, Ziros P, Kay D, Miagostovich M, vargha M, Allard A, Vantarakis A, Wyn-Jones P, Bofill-Mas S, Girones R. 2014. Application of human and animal viral microbial source tracking tools in fresh and marine waters from five different geographical areas. Water Res 59:119–129. doi: 10.1016/j.watres.2014.04.013. [DOI] [PubMed] [Google Scholar]
- 17.Rosario K, Symonds E, Sinigalliano C, Stewart J, Breitbart M. 2009. Pepper mild mottle virus as an indicator of fecal pollution. Appl Environ Microbiol 75:7261–7267. doi: 10.1128/AEM.00410-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stoeckel DM, Harwood VJ. 2007. Performance, design, and analysis in microbial source tracking studies. Appl Environ Microbiol 73:2405–2415. doi: 10.1128/AEM.02473-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Odagiri M, Schriewer A, Hanley K, Wuertz S, Misra P, Panigrahi P, Jenkins MW. 2015. Validation of Bacteroidales quantitative PCR assays targeting human and animal fecal contamination in the public and domestic domains in India. Sci Total Environ 502:462–470. doi: 10.1016/j.scitotenv.2014.09.040. [DOI] [PubMed] [Google Scholar]
- 20.Ahmed W, Sidhu J, Toze S. 2012. Evaluation of the nifH gene marker of Methanobrevibacter smithii for the detection of sewage pollution in environmental waters in Southeast Queensland, Australia. Environ Sci Technol 46:543–550. doi: 10.1021/es203372u. [DOI] [PubMed] [Google Scholar]
- 21.Hundesa A, Maluquer de Motes C, Albinana-Gimenez N, Rodriguez-Manzano J, Bofill-Mas S, Suñen E, Rosina-Girones R. 2009. Development of a qPCR assay for the quantification of porcine adenoviruses as an MST tool for swine fecal contamination in the environment. J Virol Methods 158:130–135. doi: 10.1016/j.jviromet.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 22.Kirs M, Caffaro-Filho RA, Wong M, Harwood VJ, Moravcik P, Fujioka RS. 2016. Human-associated Bacteroides spp. and human polyomaviruses as microbial source tracking markers in Hawaii. Appl Environ Microbiol 82:6757–6767. doi: 10.1128/AEM.01959-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Symonds E, Sinigalliano C, Gidley M, Ahmed W, McQuaig-Ulrich S, Breitbart M. 2016. Faecal pollution along the southeastern coast of Florida and insight into the use of pepper mild mottle virus as an indicator. J Appl Microbiol 121:1469–1481. doi: 10.1111/jam.13252. [DOI] [PubMed] [Google Scholar]
- 24.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 25.Fong TT, Lipp EK. 2005. Enteric viruses of humans and animals in aquatic environments: health risks, detection, and potential water quality assessment tools. Microbiol Mol Biol Rev 69:357–371. doi: 10.1128/MMBR.69.2.357-371.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soller JA, Bartrand T, Ashbolt NJ, Ravenscroft J, Wade TJ. 2010. Estimating the primary etiologic agents in recreational freshwaters impacted by human sources of faecal contamination. Water Res 44:4736–4747. doi: 10.1016/j.watres.2010.07.064. [DOI] [PubMed] [Google Scholar]
- 27.Ahmed W, Goonetilleke A, Powell D, Chauhan K, Gardner T. 2009. Comparison of molecular markers to detect fresh sewage in environmental waters. Water Res 40:4908–4917. doi: 10.1016/j.watres.2009.09.047. [DOI] [PubMed] [Google Scholar]
- 28.Albinana-Gimenez N, Clemente-Casares P, Bofill-Mas S, Hundesa A, Ribas F, Girones R. 2006. Distribution of human polyomaviruses, adenoviruses, and hepatitis E virus in the environment and in a drinking-water treatment plant. Environ Sci Technol 40:7416–7422. doi: 10.1021/es060343i. [DOI] [PubMed] [Google Scholar]
- 29.Fong T, Phanikumar M, Xagoraraki I, Rose J. 2009. Quantitative detection of human adenoviruses in wastewater and combined sewer overflows influencing a Michigan river. Appl Environ Microbiol 76:715–723. doi: 10.1128/AEM.01316-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haramoto E, Kitajima M, Katayama H, Ohgaki S. 2010. Real-time PCR detection of adenoviruses, polyomaviruses, and torque teno viruses in river water in Japan. Water Res 44:1747–1752. doi: 10.1016/j.watres.2009.11.043. [DOI] [PubMed] [Google Scholar]
- 31.McQuaig S, Scott T, Lukasik J, Paul J, Harwood V. 2009. Quantification of human polyomaviruses JC virus and BK virus by TaqMan quantitative PCR and comparison to other water quality indicators in water and fecal samples. Appl Environ Microbiol 75:3379–3388. doi: 10.1128/AEM.02302-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamza IA, Jurzik L, Überla K, Wilhelm M. 2011. Evaluation of pepper mild mottle virus, human picobirnavirus and torque teno virus as indicators of fecal contamination in river water. Water Res 45:1358–1368. doi: 10.1016/j.watres.2010.10.021. [DOI] [PubMed] [Google Scholar]
- 33.Kuroda K, Nakada N, Hanamoto S, Inaba M, Katayama H, Do AT, Nga TTV, Oguma K, Hayashi T, Takizawa S. 2015. Pepper mild mottle virus as an indicator and a tracer of fecal pollution in water environments: comparative evaluation with wastewater-tracer pharmaceuticals in Hanoi, Vietnam. Sci Total Environ 506–507:287–298. doi: 10.1016/j.scitotenv.2014.11.021. [DOI] [PubMed] [Google Scholar]
- 34.Hewitt J, Greening GE, Leonard M, Lewis GD. 2013. Evaluation of human adenovirus and human polyomavirus as indicators of human sewage contamination in the aquatic environment. Water Res 47:6750–6761. doi: 10.1016/j.watres.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 35.Haramoto E, Katayama H, Oguma K, Ohgaki S. 2007. Quantitative analysis of human enteric adenoviruses in aquatic environments. J Appl Microbiol 103:2153–2159. doi: 10.1111/j.1365-2672.2007.03453.x. [DOI] [PubMed] [Google Scholar]
- 36.Prado T, Silva DM, Guilayn WC, Rose TL, Gaspar AM, Miagostovich MP. 2011. Quantification and molecular characterization of enteric viruses detected in effluents from two hospital wastewater treatment plants. Water Res 45:1287–1297. doi: 10.1016/j.watres.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 37.Belguith K, Hassen A, Aouni M. 2006. Comparative study of four extraction methods for enterovirus recovery from wastewater and sewage sludge. Bioresour Technol 97:414–419. doi: 10.1016/j.biortech.2005.03.022. [DOI] [PubMed] [Google Scholar]
- 38.Ahmed W, Sidhu J, Smith K, Beale D, Gyawali P, Toze S. 2015. Distributions of fecal markers in wastewater from different climatic zones for human fecal pollution tracking in Australian surface waters. Appl Environ Microbiol 82:1316–1323. doi: 10.1128/AEM.03765-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eichmiller JJ, Hicks RE, Sadowsky MJ. 2013. Distribution of genetic markers of fecal pollution on a freshwater sandy shoreline in proximity to wastewater effluent. Environ Sci Technol 47:3395–3402. doi: 10.1021/es305116c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shah AH, Abdelzaher AM, Phillips M, Hernandez R, Solo-Gabriele HM, Kish J, Scorzetti G, Fell JW, Diaz MR, Scott TM, Lukasik J, Harwood VJ, McQuaig S, Sinigalliano CD, Gidley ML, Wanless D, Ager A, Lui J, Stewart JR, Plano LRW, Fleming LE. 2011. Indicator microbes correlate with pathogenic bacteria, yeasts and helminthes in sand at a subtropical recreational beach site. J Appl Microbiol 110:1571–1583. doi: 10.1111/j.1365-2672.2011.05013.x. [DOI] [PubMed] [Google Scholar]
- 41.Haugland RA, Siefring SC, Wymer LJ, Brenner KP, Dufour AP. 2005. Comparison of Enterococcus measurements in freshwater at two recreational beaches by quantitative polymerase chain reaction and membrane filter culture analysis. Water Res 39:559–568. doi: 10.1016/j.watres.2004.11.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.