ABSTRACT
Cellulosomes are considered to be one of the most efficient systems for the degradation of plant cell wall polysaccharides. The central cellulosome component comprises a large, noncatalytic protein subunit called scaffoldin. Multiple saccharolytic enzymes are incorporated into the scaffoldins via specific high-affinity cohesin-dockerin interactions. Recently, the regulation of genes encoding certain cellulosomal components by multiple RNA polymerase alternative σI factors has been demonstrated in Clostridium (Ruminiclostridium) thermocellum. In the present report, we provide experimental evidence demonstrating that the C. thermocellum cipA gene, which encodes the primary cellulosomal scaffoldin, is regulated by several alternative σI factors and by the vegetative σA factor. Furthermore, we show that previously suggested transcriptional start sites (TSSs) of C. thermocellum cipA are actually posttranscriptional processed sites. By using comparative bioinformatic analysis, we have also identified highly conserved σI- and σA-dependent promoters upstream of the primary scaffoldin-encoding genes of other clostridia, namely, Clostridium straminisolvens, Clostridium clariflavum, Acetivibrio cellulolyticus, and Clostridium sp. strain Bc-iso-3. Interestingly, a previously identified TSS of the primary scaffoldin CbpA gene of Clostridium cellulovorans matches the predicted σI-dependent promoter identified in the present work rather than the previously proposed σA promoter. With the exception of C. cellulovorans, both σI and σA promoters of primary scaffoldin genes are located more than 600 nucleotides upstream of the start codon, yielding long 5′-untranslated regions (5′-UTRs). Furthermore, these 5′-UTRs have highly conserved stem-loop structures located near the start codon. We propose that these large 5′-UTRs may be involved in the regulation of both the primary scaffoldin and other cellulosomal components.
IMPORTANCE Cellulosome-producing bacteria are among the most effective cellulolytic microorganisms known. This group of bacteria has biotechnological potential for the production of second-generation biofuels and other biocommodities from cellulosic wastes. The efficiency of cellulose hydrolysis is due to their cellulosomes, which arrange enzymes in close proximity on the cellulosic substrate, thereby increasing synergism among the catalytic domains. The backbone of these multienzyme nanomachines is the scaffoldin subunit, which has been the subject of study for many years. However, its genetic regulation is poorly understood. Hence, from basic and applied points of view, it is imperative to unravel the regulatory mechanisms of the scaffoldin genes. The understanding of these regulatory mechanisms can help to improve the performance of the industrially relevant strains of C. thermocellum and related cellulosome-producing bacteria en route to the consolidated bioprocessing of biomass.
KEYWORDS: 5′-UTR, Clostridium thermocellum, cellulosome, cipA, gene regulation, promoters, scaffoldin, sigma factors
INTRODUCTION
Certain Gram-positive anaerobic bacteria, epitomized by Clostridium thermocellum, produce extracellular multienzyme systems known as cellulosomes to efficiently degrade plant cell wall polysaccharides (1). The backbone of cell-associated cellulosomes is a scaffolding protein that incorporates various carbohydrate-active enzymes (CAZymes) into the complex (1). The scaffolding proteins, termed scaffoldins, contain multiple repetitions of cohesin modules that tightly bind CAZymes and/or other cellulosomal proteins by their associated dockerin modules (1, 2). The entire enzymatic complex can be displayed on the bacterial surface, with a dominant role for the primary scaffold, or can be released as cell-free cellulosomal systems based on secondary scaffoldins such as ScaE (Cthe_0736) (2). In C. thermocellum, the attachment of the cellulosome to the cell surface is carried out by the anchoring proteins OlpB, Orf2p, and SdbA (1, 2). Additionally, cellulosomes are attached to the cellulosic substrate by carbohydrate-binding modules (CBMs) present in the primary scaffoldins and/or in the CAZymes (1).
The importance of the primary scaffoldin proteins in cellulosome-producing bacteria has been demonstrated in C. thermocellum by using mutant strains deficient in the scaffoldin CipA (2–6). However, the regulation of the primary scaffoldin genes in cellulosome-producing bacteria is poorly understood. An early study performed by Dror and colleagues (7) found that the expression of the C. thermocellum cipA gene is growth dependent. Under conditions of carbon limitation, the expression of cipA was shown to be higher at low growth rates than at high growth rates (7). Additionally, using primer extension analysis, Dror and colleagues identified two transcriptional start sites (TSSs) upstream of cipA (7). According to the known promoter consensus sequences at that time, and using Bacillus subtilis as reference, Dror and colleagues suggested RNA polymerase (RNAP) sigma factors σL (σ54) and σA (RpoD) as responsible for the expression of cipA (7).
Interestingly, the C. thermocellum genome encodes eight alternative RNAP sigma σI factors (σI1 to σI8), of which six, σI1 to σI6, were shown to be involved in regulating cellulosomal components (8–11). The cognate membrane-associated anti-σI factors (RsgI1 to RsgI6) contain C-terminal CBMs that are displayed on the cell surface, suggesting that they serve as putative sensors for extracellular polysaccharide substrates (8–11). Recently, using a heterologous B. subtilis host system, we showed that the C. thermocellum alternative RNAP factors σI3 and σI6 recognized a predicted σI-dependent promoter of the primary scaffoldin gene cipA, located approximately 850 bp from the start codon (10). This suggests that C. thermocellum cipA is likely to be regulated by these two alternative σI factors.
In the present study, we provide evidence that the putative promoters suggested to be responsible for the expression of the C. thermocellum cipA gene (7) were incorrectly proposed. Recent technological advancements have now allowed us to differentiate between the authentic TSSs and posttranscriptional processed sites (PSs). Here, we used a combination of computational methods and laboratory experimentation to demonstrate that in addition to C. thermocellum, other cellulosome-producing bacteria, such as C. straminisolvens JCM 21531 (12), Clostridium sp. strain Bc-iso-3 (13), C. clariflavum DSM 19732 (14), Acetivibrio cellulolyticus CD2 (ATCC 33288) (15), and C. cellulovorans 743B (16), also exhibit a highly conserved σI-dependent promoter upstream of the primary scaffoldin gene. Additionally, with the exception of C. cellulovorans, a highly conserved σA promoter was also identified in the above-mentioned bacteria. Both σI- and σA-dependent promoters are distant from the start codon of the scaffoldin genes. This suggests that the transcription from both σI- and σA-dependent promoters may produce mRNA with long 5′-untranslated regions (5′-UTRs) that are probably involved in posttranscriptional regulation of the cellulosomal primary scaffoldin genes.
RESULTS AND DISCUSSION
Reanalyzing the TSSs of the C. thermocellum cipA gene.
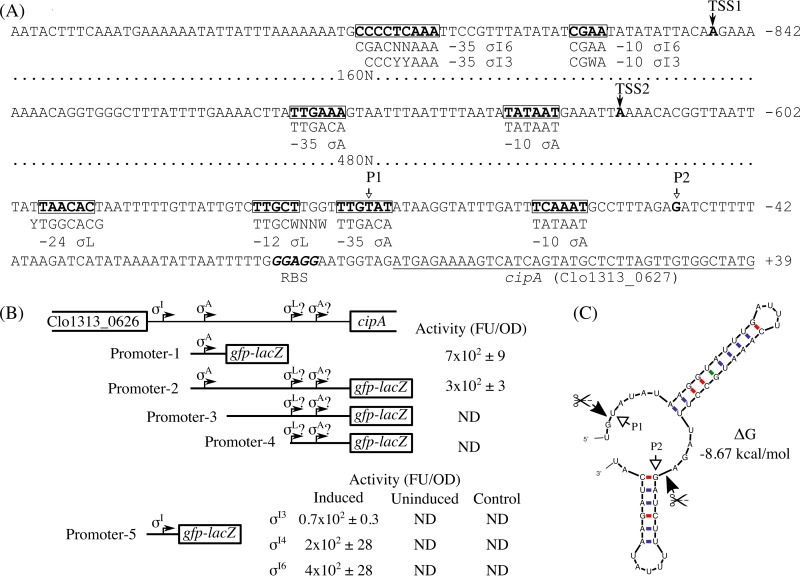
In order to verify our previously predicted σI-dependent promoter upstream of C. thermocellum cipA (10), we mapped the TSSs using the 5′ rapid amplification of cDNA ends (RACE) technique. During our analysis, in order to upregulate the expression of the multiple alternative σI factors, C. thermocellum was grown under the conditions reported by Nataf and colleagues, in a medium containing cellulose and xylan (9). Additionally, during the mapping of the TSSs, a sample of total RNA extracted from C. thermocellum was treated with Terminator 5′-phosphate-dependent exonuclease (TEX) (see Fig. S1A in the supplemental material) in order to discriminate between TSSs and PSs. This enzyme is a 5′→3′ exonuclease that digests processed RNA (5′ monophosphated), allowing an enrichment of primary transcripts (5′ triphosphate) (Fig. S1A). As shown in Fig. 1A, we identified in the present work TSS1 and TSS2 in the TEX-treated RNA sample, which are located 846 and 616 bp upstream of cipA, respectively. TSS1 corresponds to the previously predicted σI-dependent promoter (10), and TSS2 corresponds to a putative promoter of the vegetative σA factor (Fig. 1A). Interestingly, in this TEX-treated sample, we were not able to detect the two previously described 5′-mRNA ends P1 and P2 that were predicted to be TSSs associated with σL and σA, respectively (7) (Fig. 1A). As can be observed in Fig. S1A, PCR analysis of the 5′-RACE-Ready cDNA obtained from TEX-treated RNA gave two main PCR products, of 1,114 bp and 884 bp, which were analyzed by Sanger sequencing and correspond to TSS1 and TSS2, respectively. Interestingly, PCR analysis of the 5′-RACE-Ready cDNA prepared from RNA not treated with TEX gave only one PCR product, of approximately 300 bp (Fig. S1B). This result shows that TSS1 and TSS2 are being processed; hence, Dror and colleagues (7) could not detect them.
FIG 1.
Analysis of the TSSs of the C. thermocellum cipA gene. (A) Localization of putative promoters within the C. thermocellum cipA upstream region. TSS1 and TSS2, identified in the present work, are indicated by filled arrows. The putative TSSs identified by Dror and colleagues (P1 and P2 [7]) are indicated by empty arrows. The proposed promoter elements are framed, and promoter consensus sequences are shown beneath them. In TSS1, the σI6 and σI3 promoter consensus sequences shown beneath were identified in a previous report (10). The ribosomal binding site (RBS) is italicized, and the cipA coding sequence is underlined. The number shown at the right of each sequence specifies the position of the last 3′-terminal nucleotide of each sequence in relation to the first codon of cipA (“−” upstream and “+” downstream). Segments of 160 and 480 nucleotides, immediately downstream of −842 and −602, respectively, are not shown. The nucleotide code Y represents C or T, W represents A or T, and N represents any nucleotide. (B) Analysis of the C. thermocellum cipA promoters based on deletions of promoter regions. Promoter activity was measured by quantifying the fluorescence in a microplate reader. The promoter activity shown is the average from three independent experiments. During assays of the alternative σI factors, Promoter-3 was used as a negative control and the cells were induced. FU, fluorescence units; OD, optical density; ND, not detected. (C) Putative mRNA secondary structures identified near the C. thermocellum cipA coding sequence. The analysis was performed with the program “mfold” (17) by using the default parameters but with an elevated temperature (60°C) consistent for the thermophile. P1 and P2 are the 5′-RNA ends mapped by Dror and colleagues (7). The scissors indicate putative processing sites.
In order to analyze whether the σA-dependent promoter proposed by Dror and colleagues (7) is functional or not, we fused different fragments of the upstream region of cipA to a gfp-lacZ reporter operon (Fig. 1B). Additionally, we included in the analysis the proposed σA promoter identified in the present work and the σI-dependent promoter previously identified by us (10) (Fig. 1B). The promoter activities were studied in a heterologous B. subtilis host system that was developed previously (10), due to the limited set of genetic tools available to work directly in C. thermocellum. Recognition of the σI-dependent promoter upstream of cipA by the C. thermocellum alternative σI3 and σI6 factors was already shown (10), and here we extended the analysis to the other C. thermocellum alternative σI factors. As shown in Fig. 1B, we observed promoter activity only with fragments of the upstream region of cipA that contain the σI promoter previously identified and the σA promoter identified in the present work. Moreover, in addition to the C. thermocellum alternative σI3 and σI6 factors, in the present work we showed that the σI promoter was also recognized by the alternative σI4 factor (Fig. 1B). C. thermocellum σI1, σI2, σI5, σI7, and σI8 were unable to recognize the cipA σI promoter.
During the TSS analysis performed by Dror and colleagues, the major 5′-end of the cipA mRNA detected was from a putative σL promoter in cells at exponential growth rate using cellulose or cellobiose as a carbon source (7). Furthermore, only the mRNA 5′-end associated with the putative σL promoter was detected at exponential growth rate when cellobiose was used as a carbon source (7). We therefore decided to perform a search for a σL-encoding gene in C. thermocellum. The bioinformatic analysis was performed by using the B. subtilis σL protein sequence as query. Surprisingly, however, no σL-encoding gene was detected in C. thermocellum. Analysis of the mRNA sequence located in the vicinity of the mRNA 5′-ends mapped by Dror and colleagues (7) by using the program “mfold” (17) showed that two putative stem-loop structures are predicted to form with a folding free energy (ΔG) of −8.67 kcal/mol (Fig. 1C). Interestingly, the PSs mapped by Dror and colleagues (7) are found next to these putative stem-loop structures (Fig. 1C). These structures, located close to the first codon of cipA, are likely involved in the stability of the coding region by protecting the mRNA from the action of exoribonucleases, supporting the observed mRNA 5′-end obtained from total RNA not treated with TEX in contrast to the TEX-treated RNA (Fig. S1).
Primary cellulosomal scaffoldin genes are regulated by alternative σI factors in several cellulosome-producing bacteria.
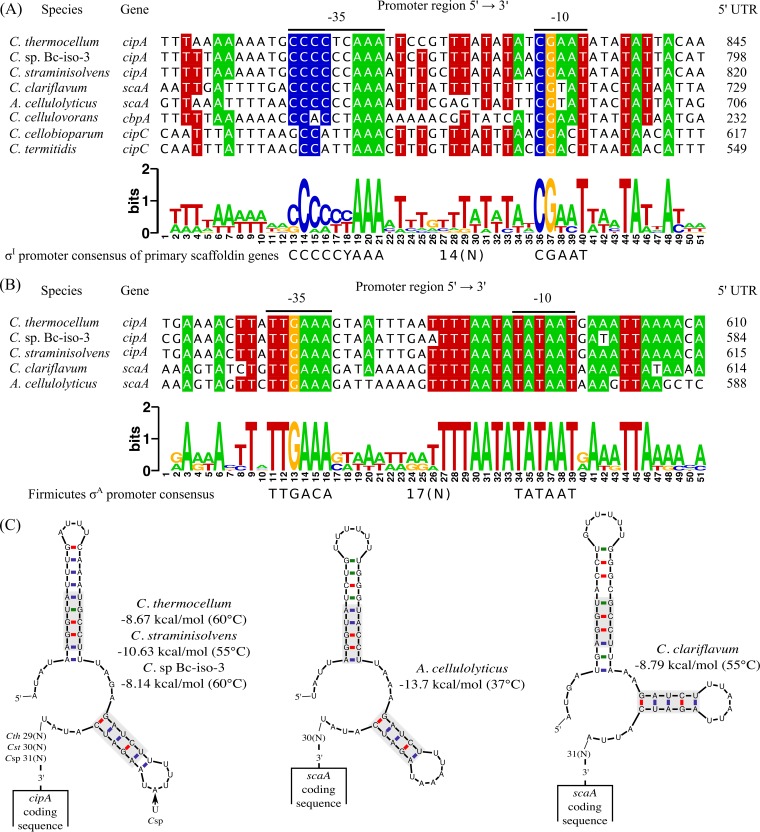
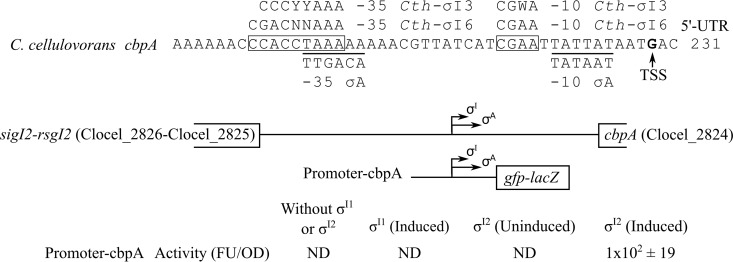
Previous results showed that in addition to C. thermocellum, other cellulosome-producing bacteria also have multiple alternative σI factors that may be involved in the regulation of cellulosomal components (10). Consequently, we performed bioinformatic analysis in order to predict putative σI-dependent promoters upstream of the primary scaffoldin gene of cellulosome-producing bacteria. By using the program “Pattern Locator” (18), we were able to identify a highly conserved σI-dependent promoter upstream of the primary scaffoldin gene in the following bacteria: Clostridium sp. Bc-iso-3 (cipA-like, ODM27873.1), C. straminisolvens JCM 21531 (cipA-like, GAE91018.1), C. clariflavum 4-2a (scaA, Clocl_3306), A. cellulolyticus CD2 (scaA, AAF06064.1), C. termitidis (cipC-like, EMS74252.1), C. cellobioparum (cipC-like, NZ_KK211293.1), and C. cellulovorans 743B (cbpA, Clocel_2824) (Fig. 2A). These highly conserved σI-dependent promoters of primary scaffoldin genes have the consensus sequence CCCCCYAAA(14N)CGAAT (where Y is C or T, and N is any nucleotide) (Fig. 2A). Interestingly, the previously identified TSS of the C. cellulovorans cbpA gene, which was proposed to be under the control of σA (19), also matches the predicted σI-dependent promoter identified in the present work (Fig. 3). Moreover, further bioinformatic analysis revealed that upstream of the C. cellulovorans cbpA gene (Clocel_2824) there is an operon composed of a σI gene (sigI2, Clocel_2826) and its cognate anti-σI gene (rsgI2, Clocel_2825) (Fig. 3). These observations suggest that the previously identified TSS of C. cellulovorans cbpA is also probably related to the σI-dependent promoter identified in the present work (Fig. 2A).
FIG 2.
Identification of highly conserved regulatory elements upstream of the primary scaffoldin genes. (A) Alignment of the experimentally validated σI promoter of C. thermocellum cipA and orthologous sequences from other cellulosome-producing bacteria. Predicted −35 and −10 promoter elements are indicated by a line above the alignment. Distances between the promoter region sequences used for the alignment and the first codon of corresponding scaffoldin genes are shown below “5′-UTR.” The WebLogo was generated with the sequences shown in the alignment. The nucleotide code Y represents C or T, and N is any nucleotide. (B) Alignment of the experimentally validated σA promoter of C. thermocellum cipA and orthologous sequences from other cellulosome-producing bacteria. For details, see the legend for panel A. (C) Putative 5′-terminal mRNA secondary structures identified near the primary scaffoldin-coding gene sequence of C. thermocellum (Cth), Clostridium sp. Bc-iso-3 (Csp), C. straminisolvens (Cst), C. clariflavum, and A. cellulolyticus. The secondary structures were predicted using the temperature of the optimal growth conditions of each bacterium as shown in the figure. The conserved nucleotides in the stem-loops are indicated in gray boxes. Nucleotide pairing is shown in red for C-G, in blue for A-U, and in green for G-U. The structure on the left has one nucleotide replacement (U instead of A) in Csp, which is shown with an arrow. Distances between the 3′-nucleotide of the stem-loop structures and the first codon of corresponding scaffoldin genes are numbered, such as in Cth 29(N), Cst 30(N), and Csp 31(N), where N represents any nucleotide.
FIG 3.
Analysis of C. cellulovorans cbpA promoters. The TSS identified by Han and colleagues (19) is indicated by the vertical arrow. The σI promoter elements proposed in the present work are framed, and the σA promoter elements proposed by Han and colleagues (19) are underlined. The promoter consensus sequences are aligned with the proposed promoter elements. The C. thermocellum σI6 and σI3 promoter consensus sequences (Cth-σI6 and Cth-σI3, respectively) shown above were identified in a previous report (10). The nucleotide code Y represents C or T, W represents A or T, and N represents any nucleotide. Promoter activity was measured by quantifying the fluorescence in a microplate reader. The promoter activity shown is the average from three independent experiments. FU, fluorescence units; OD, optical density; ND, not detected.
In order to confirm that the predicted σI-dependent promoter of the C. cellulovorans cbpA can be recognized by the C. cellulovorans σI2 factor, we fused the predicted σI-dependent promoter including the previously identified σA-dependent promoter to a gfp-lacZ reporter operon (Fig. 3). Recognition of the predicted σI-dependent promoter of cbpA by the C. cellulovorans σI2 factor was studied in a heterologous B. subtilis host system (10). The paralogous C. cellulovorans σI1 (sigI1, Clocel_0130) factor was also included in the analysis. As shown in Fig. 3, only the C. cellulovorans σI2 factor was able to recognize the predicted σI-dependent promoter of cbpA. Furthermore, when the promoters fused to the gfp-lacZ reporter operon were introduced into the B. subtilis host system without any σI factor, neither LacZ activity (data not shown) nor green fluorescence protein (GFP) was detected (Fig. 3). Taking these results together, it appears that C. cellulovorans cbpA is regulated only by the C. cellulovorans σI2 factor rather than σA and σI1.
Additional bioinformatic analysis showed that in addition to the newly identified σA promoter upstream of C. thermocellum cipA, the upstream regions of the primary scaffoldin genes of Clostridium sp. Bc-iso-3, C. straminisolvens, C. clariflavum, and A. cellulolyticus also contain a putative σA-dependent promoter (Fig. 2B). Alignment of these putative σA promoters reveals the highly conserved homology of their sequences (Fig. 2B), suggesting that the primary scaffoldin genes of this group of cellulosome-producing bacteria are regulated by σA as in the C. thermocellum cipA gene. Interestingly, with the exception of C. cellulovorans, the σI and σA promoters in this group of bacteria are located more than 600 nucleotides from the start codon (Fig. 2A and B). The localization of promoters at such a distance from the translational start site of the primary scaffoldin gene has also been reported in Clostridium cellulolyticum (20, 21). The TSS of the primary scaffoldin gene cipC of C. cellulolyticum was thus identified at 637 or 638 nucleotides upstream of the start codon (20, 21).
Given the conservation in both promoter sequences and distance to the coding region (Fig. 2A and B), the long 5′-UTRs of the primary scaffoldin genes are likely to have a regulatory role. This suggestion is supported by a report by Olson and colleagues (6) in which a cipA knockout mutant of C. thermocellum including 819 nucleotides upstream of the start codon was prepared. The latter stretch contained the σA promoter identified in the present work and which was deleted. After complementation of this mutant with a plasmid carrying a copy of cipA including the 819 nucleotides upstream of the start codon, the mutant recovered its capacity to solubilize Avicel (crystalline cellulose) (6), although the complemented mutant had a lag phase of 100 h before commencing rapid cellulose hydrolysis. More strikingly, in the complemented mutant, expression of the anchoring protein gene olpB, positioned immediately downstream of cipA and reported to be transcribed independently of cipA (7), was drastically affected, reducing its protein abundance 11-fold (6). Moreover, the complementation of the mutant with the above-mentioned plasmid caused a pleiotropic effect, changing drastically the gene expression of other cellulosomal components and even the expression of genes encoding CAZymes not associated with the cellulosome (6). Hence, the long 5′-UTR of C. thermocellum cipA might have other regulatory elements, including possible yet-unidentified binding sites of transcriptional factors. It is important to mention that, although the complemented mutant recovered its capacity to solubilize Avicel, the rate of Avicel solubilization was one-third as high as that for the empty-vector control strain (6). This result could be the consequence of not including in the plasmid used for complementing cipA the σI promoter identified in the present work.
Scaffoldin genes that are regulated by alternative σI factors have conserved predicted 5′-terminal stem-loop structures near the translational start sites.
Secondary structures, like simple stem-loops in the 5′-UTR of mRNA, can improve the stability of the messenger (22). This phenomenon was observed by Maamar and colleagues (20) during mapping of the C. cellulolyticum cip-cel operon, which encodes the primary cellulosomal scaffoldin CipC and 11 cellulosomal components (mainly cellulases). Putative mRNA secondary structures were predicted near the first codon of the two most expressed cellulase genes, cel48F and cel9E, of the cip-cel operon. These authors demonstrated that the predicted secondary structures may have a role in increasing the stability of the mRNA of cel48F and cel9E. Recently, Xu and colleagues analyzed the relationship between stoichiometry of different processed mRNAs of the C. cellulolyticum cip-cel operon and the secondary structures identified between different coding sequences of the cip-cel operon (23). An important observation in the study was that the structures of the loops identified in the C. cellulolyticum cip-cel operon are conserved among orthologous operons from different mesophilic cellulolytic clostridial species (23).
In order to analyze whether the stem-loop structure identified close to the first codon of the C. thermocellum cipA gene is conserved in bacteria that use σI factors to regulate the primary scaffoldin gene, we analyzed the upstream region of the primary scaffoldin genes of this group of bacteria using the “mfold” program (17). As shown in Fig. 2C, C. straminisolvens, Clostridium sp. Bc-iso-3, C. clariflavum, and A. cellulolyticus have predicted 5′-terminal stem-loop structures near the translational start site of the scaffoldin genes, resembling that of C. thermocellum. The stem-loop structures in these 5′-terminal mRNAs have folding free energies (ΔG) of −8.14 kcal/mol or less (Fig. 2C). Furthermore, a high conservation in shape, distance to the start codon (between 29 and 31 nucleotides), and sequence (as indicated by the gray boxes in Fig. 2C) is seen this group of bacteria (Fig. 2C). Hence, these 5′-terminal stem-loops near the translational start sites of the primary scaffoldin genes are likely involved in the stability of the mRNA and in the stoichiometry of the scaffoldin gene, relative to the downstream genes.
Cellulolytic clostridia that produce complex cellulosomes harbor multiple σI factors and share similar regulatory features of the primary scaffoldin gene.
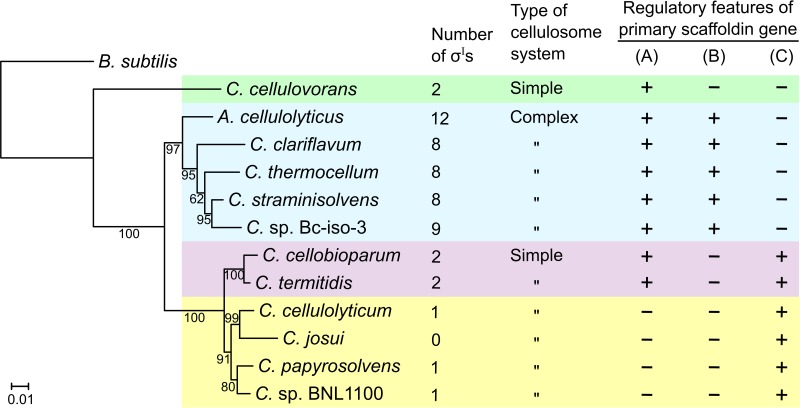
A summary of the regulatory features of the primary scaffoldin genes from different cellulosome-producing species identified in the present work is given in Fig. 4. This figure includes a phylogenetic analysis of the 16S-rRNA gene sequences, where C. thermocellum, C. straminisolvens, Clostridium sp. Bc-iso-3, C. clariflavum, and A. cellulolyticus cluster together. Interestingly, these cellulolytic clostridia produce highly elaborate cellulosomal systems that can be termed complex cellulosomes (1) (light blue-highlighted clade in Fig. 4). The above-mentioned group of clostridia shares three regulatory features of the primary scaffoldin gene: a highly conserved σI-dependent promoter, a stem-loop structure near the start codon, and a highly conserved σA-dependent promoter (Fig. 4).
FIG 4.
Phylogeny of 16S rRNA gene sequences derived from cellulolytic clostridia producing different types of cellulosome and regulatory features of their primary scaffoldin gene. The phylogenetic tree was inferred from multiple 16S rRNA gene sequence alignments using the neighbor-joining method implemented in MEGA 7 (35). The B. subtilis 16S rRNA gene sequence was used as an outgroup. Bootstrap percentages retrieved from 2,000 replications are shown at the nodes. The scale bar (0.01) indicates the number of nucleotide substitutions per site. The type of regulatory feature(s) in the given species (A to C, described below) is marked with “+” if identified and “−” if not: A, presence of σI-like promoter (as shown in Fig. 2A); B, presence of conserved σA-like promoter and conserved putative stem-loop structure resembling those of C. thermocellum (as shown in Fig. 2B and C, respectively); C, presence of conserved σA-like promoter resembling that of C. cellulolyticum (as shown in Fig. S3 in the supplemental material).
Additionally, Fig. 4 shows that as the cellulosome becomes more elaborate, the bacterium harbors more σI factors. For example, C. cellulovorans, C. cellobioparum, and C. termitidis (simple-cellulosome producers) harbor two σIs, C. thermocellum (complex-cellulosome producer) harbors 8 σIs, and A. cellulolyticus (exceptionally complex-cellulosome producer) harbors 12 σIs (1, 8, 15, 16, 24). In the cases of clostridia that produce simple cellulosomal systems, such as those of Clostridium cellulolyticum (25) or Clostridium papyrosolvens (26), their genomes encode only one σI factor, and in the case of Clostridium josui, σI was not detected (Fig. 4). We assume that in cellulolytic clostridia harboring only one σI factor, its function is probably to regulate genes involved in the maintenance of cell envelope integrity and homeostasis, as in B. subtilis (27).
It is important to mention that Clostridium sp. Bc-iso-3 is a newly isolated cellulolytic bacterium from an industrial anaerobic digester (13), whose closest relatives are C. straminisolvens and C. thermocellum (97.6% and 95.7% identity, respectively, based on comparison of 16S-rRNA gene sequences [Fig. 4]). Our analysis revealed that the Clostridium sp. Bc-iso-3 genome encodes a large set of cellulosomal proteins, including a CipA-like primary scaffoldin (ODM27873.1) and an OlpB-like anchoring scaffoldin (ODM27874.1), suggesting that this species produces complex cellulosomes. Additionally, its genome contains nine σI genes (sigI1, ODM26107.1; sigI2, ODM25887.1; sigI3, ODM25838.1; sigI4, ODM25738.1; sigI5, ODM24951.1; sigI6, ODM26859.1; sigI7, ODM27255.1; sigI8, ODM25355.1; sigI9, ODM27746.1). Taken together, these observations suggest that in clostridia that produce complex cellulosomes, the regulatory features of the primary scaffoldin gene and the number of σI factors have evolved together with the complexity of the cellulosome (Fig. 4).
Apart from C. cellulovorans, simple-cellulosome-producing bacteria are clustered in the phylogenetic tree shown in Fig. 4 (clades highlighted in pink and yellow). Further analysis revealed that this bacterial cluster shares a highly conserved σA-like promoter (see Fig. S3 in the supplemental material). An alignment of the above-mentioned promoters (Fig. S3) showed that they share a consensus sequence different from that of the σA-dependent promoter of the primary scaffoldin gene of complex-cellulosome producers (Fig. 2B). For example, while the −10 region of the σA-dependent promoter of the primary scaffoldin gene of complex-cellulosome producers has the classic TATAAT sequence (Fig. 2B), the −10 region of the σA-dependent promoter of simple-cellulosome producers is less conserved but contains the −10 extended element, TG (Fig. S3).
Interestingly, C. termitidis and C. cellobioparum (pink-highlighted clade in Fig. 4) are intermediate in their type of cellulosome production, between complex-cellulosome producers (light blue-highlighted clade in Fig. 4) and simple-cellulosome producers (yellow-highlighted clade in Fig. 4) in relation to the regulatory features of the primary scaffoldin gene (Fig. 4). The primary scaffoldin gene of C. cellobioparum and C. termitidis has a putative σI-dependent promoter resembling that of C. thermocellum (Fig. 2A) and a σA-dependent promoter resembling that of C. cellulolyticum (20, 21) (Fig. S3). In the case of C. cellulovorans, however, we could not predict a σA-dependent promoter of cbpA, while its σI-dependent promoter is located relatively closer to the start codon (234 nucleotides) (Fig. 3) than are the other σI-dependent promoters of primary scaffoldin genes (Fig. 2A). These observations can be explained by the distant phylogenetic relationship between C. cellulovorans and other cellulosome-producing clostridia (Fig. 4).
Conclusions.
In the present work, we identified three highly conserved regulatory elements upstream of the C. thermocellum cipA gene: σI- and σA-dependent promoters and stem-loop structures near the first codon (Fig. 2). Both promoter sequences are far away from the translational start site of C. thermocellum cipA, creating a large 5′-UTR. The presence of a large 5′-UTR in the scaffoldin gene is common in all of the cellulosome-producing bacteria described in this communication. This implies that such large 5′-UTRs might have an important role in the regulation of the scaffoldin gene and, probably, of some other cellulosomal components; however, their precise function remains to be elucidated.
MATERIALS AND METHODS
Bacterial strains, growth media, and culture conditions.
C. thermocellum DSM 1313 (LQ8) was obtained from the DSMZ (German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany). C. thermocellum was grown in batch culture at 60°C and 250 rpm in 100-ml serum bottles (Wheaton, Millville, NJ, USA) in a medium described elsewhere (9). The medium included microcrystalline cellulose (10-g/liter final concentration; 50-μm particle size) as the carbon source. Additionally, in order to ensure upregulation of all C. thermocellum σI-factor genes, the medium was supplemented with birchwood xylan (final concentration, 1 g/liter) as reported previously (9). The B. subtilis strains constructed in this work are isogenic derivatives of B. subtilis CO02 (10). B. subtilis was grown routinely at 37°C in liquid (at 250 rpm) or on solid LB broth (Lennox, Difco, BD Diagnostics, Maryland, USA). When appropriate, antibiotics were included at the following final concentrations: 5 μg/ml chloramphenicol (Cam) or 3 μg/ml erythromycin (Erm). The induction of genes under the PxylA promoter was carried out with d-xylose (10 g/liter final). Antibiotics and sugars were purchased from Sigma-Aldrich (Missouri, USA).
DNA manipulation techniques.
The oligonucleotide primers and plasmids used in the present study are shown in Tables 1 and 2, respectively. Standard procedures were employed for DNA isolation, PCR, restriction enzyme digestion, transformations, and gel electrophoresis as described elsewhere (28). Plasmids were built using a combination of standard molecular cloning techniques (28) and ligase-independent cloning using the In-Fusion HD Cloning kit (Clontech Laboratories, Inc., California, USA). DNA sequences from C. thermocellum and C. cellulovorans 743B were PCR amplified using genomic DNA as the template. C. cellulovorans 743B (DSM 3052) genomic DNA was obtained from the DSMZ. DNA for cloning was PCR amplified using DreamTaq Green PCR master mix (Fermentas, Thermo Fisher Scientific Inc., Massachusetts, USA). Colony PCR was performed using Hy-Taq Ready Mix (Hy Laboratories Ltd., Rehovot, Israel). PCR primers were purchased from Hy·Labs (Hy Laboratories Ltd.). Restriction enzymes and ligase were purchased from Fermentas. PCR and agarose gel products were isolated and purified using the Hy·Labs Gel/PCR Extraction kit (Hy Laboratories Ltd.). Agarose gel products with DNA fragments higher than 10 kb were purified using the Qiaex II gel extraction kit (Qiagen, Hilden, Germany). Purification of plasmids was carried out using the Presto Mini Plasmid kit (Geneaid Biotech Ltd., Shijr, Taiwan). DNA manipulations using kits were conducted according to supplier protocols. All clones were verified by PCR and sequencing in the Instrumentation and Service Center of the Life Sciences Faculty at Tel Aviv University.
TABLE 1.
Primers used in the present work
| Designation | Name | Sequencea (5′→3′) | Usage or gene amplified |
|---|---|---|---|
| P0 | SMARTer II A Oligonucleotide | AAGCAGTGGTATCAACGCAGAGTACXXXXX | 5′-RACE analysis |
| P1 | Universal Primer Long | CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT | |
| P2 | Universal Primer Short | CTAATACGACTCACTATAGGGC | |
| P3 | cipA.gsp | GATTACGCCAAGCTTTGCGCTATCAAAGCTCTTGCTAGGATCCG | |
| P4 | cipA.ngsp | GATTACGCCAAGCTTCCCAATACGAAGTCGCAATTGGCCATTCC | |
| P5 | Fw.CtsigI1.IF | GGGGGAAATGGGATCATGGAAGTCCGGAAAATTAATACCC | C. thermocellum sigI1 |
| P6 | Rv.CtsigI1.IF | CGCGGGAGCTCGGATTCACTTTCCTCCGTCCATAG | |
| P7 | Fw.CtsigI2.IF | GGGGGAAATGGGATCATGATTGATTTGTTTTCCCCTAAG | C. thermocellum sigI2 |
| P8 | Rv.CtsigI2.IF | CGCGGGAGCTCGGATTTAATGTGACATATTATTTTGTGCTCCG | |
| P9 | Fw.CtsigI4.IF | GGGGGAAATGGGATCATGCTAAACGTCCAGCTG | C. thermocellum sigI4 |
| P10 | Rv.CtsigI4.IF | CGCGGGAGCTCGGATTTATATTATTTTTCCCTCCTTTCCTCAG | |
| P11 | Fw.CtsigI5.IF | GGGGGAAATGGGATCATGCTATTTGTTTCTGC | C. thermocellum sigI5 |
| P12 | Rv.CtsigI5.IF | CGCGGGAGCTCGGATCCGTTGCACATATTGAATATCTC | |
| P13 | Fw.CtsigI7.IF | GGGGGAAATGGGATCATGTATTCTGTTACTATAAACCAAAGAG | C. thermocellum sigI7 |
| P14 | Rv.CtsigI7.IF | CGCGGGAGCTCGGATTTACATTATTCCACCTCCCAGTTTAC | |
| P15 | Fw.CtsigI8.IF | GGGGGAAATGGGATCATGATAAATTTAGGCTCATACAACTTAC | C. thermocellum sigI8 |
| P16 | Rv.CtsigI8.IF | CGCGGGAGCTCGGATTCACCGTCCTAATGTATCATATATG | |
| P17 | Fw.CcvsigI1.IF | GGGGGAAATGGGATCATGGATGATACACCAATAACTG | C. thermocellum sigI1 |
| P18 | Rv.CcvsigI1.IF | CGCGGGAGCTCGGATTTAACTAACTAACATTCTTCACTATCC | |
| P19 | Fw.CcvsigI2.IF | GGGGGAAATGGGATCATGGAGGAGAGGTTAGATTTG | C. cellulovorans sigI2 |
| P20 | Rv.CcvsigI2.IF | CGCGGGAGCTCGGATTTCACTCCTCCTCAATATTTTTGACT | |
| P21 | Fw.GFP.IF | GCCGCTGCAGGGATCCGCTGATTAACTAATAAGGAGGAC | gfp |
| P22 | Rv.GFP.IF | TATGTACTGTGGATCTTATTTATACAATTCATCCATACCATG | |
| P23 | Fw.cipA-PsigA.Eco | GTGAATTCGTACATAGAGTTTGAAAACAGGTG | cipA Promoter-1 |
| P24 | Rv.cipA-PsigA.Bam | GTGGATCCTCTCTAACAATGAATCTG | |
| P25 | Rv.cipA-UTR.Bam | GTGGATCCCTACCATTCCTCCCAAAAATTAATATTTTATATG | cipA Promoter-2 (with P23) |
| P26 | Fw.cipA-UTR.Eco | GCGAATTCCAGATTCATTGTTAGAGAGAGAATTG | cipA Promoter-3 (with P25) |
| P27 | Fw.cipA-UTR2.Eco | GAGAATTCACCACTCCCCAATATTTATCG | cipA Promoter-4 (with P25) |
| P28 | Fw.cipA-PsigI.Eco | TGGAATTCGGGGGAGAAAAGTAATTTTG | cipA Promoter-5 |
| P29 | Rv.cipA-PsigI.Bam | TCGGATCCTTATCTGGGCATCTTCTTTC | |
| P30 | Fw.PcbpA.Eco | GCGAATTCTGGTGACACAATATTACATTGGAGG | Promoter-cbpA |
| P31 | Rv.PcbpA.Bam | CGGGATCCATGGTTCTTTCTTTAGCTACTAC | |
| P32 | GanQ-Cnf | ATATACATTGCCCGTCGGTC | Confirmation integration at B. subtilis lacA locus |
| P33 | Erm-Cnf | GCAATGAAACACGCCAAAG | |
| P34 | GanB-Cnf | CAATGGCAGCGGCATATCC | |
| P35 | XylR-Cnf | GGAGCGGTTTCTATCGTTATTGATTC | |
| P36 | Ycg-Cnf | GGAAGCGTTCACAGTTTCG | Confirmation integration at B. subtilis amyE locus |
| P37 | LacZ-Cnf | TCCTGGAGCCCGTCAGTATC | |
| P38 | Ldh-Cnf | CAATGACCACAAGCTCATCTG | |
| P39 | Cat-Cnf | CTATTCAGGAATTGTCAGATAGGC |
X, undisclosed base in the proprietary SMARTer oligonucleotide sequence.
TABLE 2.
Plasmids used in the present work
| Plasmid use and name | Relevant genotypea | Brief description |
|---|---|---|
| Analysis of alternative σI factor in B. subtilis | ||
| pAX01b | bla lacA3′ xylR PxylA erm lacA5′ | Integration vector |
| pAX01-SigI1 | bla lacA3′ xylR PxylA-sigI1Ct erm lacA5′ | pAX01-derived plasmids for the expression of C. thermocellum σI1, σI2, σI4, σI5, σI7, and σI8 |
| pAX01-SigI2 | bla lacA3′ xylR PxylA-sigI2Ct erm lacA5′ | |
| pAX01-SigI4 | bla lacA3′ xylR PxylA-sigI4Ct erm lacA5′ | |
| pAX01-SigI5 | bla lacA3′ xylR PxylA-sigI5Ct erm lacA5′ | |
| pAX01-SigI7 | bla lacA3′ xylR PxylA-sigI7Ct erm lacA5′ | |
| pAX01-SigI8 | bla lacA3′ xylR PxylA-sigI8Ct erm lacA5′ | |
| pAX01-SigI1-Ccel | bla lacA3′ xylR PxylA-sigI1Cc erm lacA5′ | pAX01-derived plasmids for the expression of C. cellulovorans σI1 and σI2 |
| pAX01-SigI2-Ccel | bla lacA3′ xylR PxylA-sigI2Cc erm lacA5′ | |
| pAX01-SigI3c | bla lacA3′ xylR PxylA-sigI3Ct erm lacA5′ | pAX01-derived plasmids for the expression of C. thermocellum σI3 and σI6 |
| pAX01-SigI6c | bla lacA3′ xylR PxylA-sigI6Ct erm lacA5′ | |
| Construction of the GFP-LacZ reporter system | ||
| pDR111_GFP(Sp)b | bla amyE3′ spec Phyperspank-gfp(Sp) lacI amyE5′ | Template for the amplification of gfp |
| pBS1CLacZb | bla amyE5′ cat mcs rfp mcs lacZ amyE3′ | Integration vector with a LacZ reporter system |
| pBS1C-GFP-LacZ | bla amyE5′ cat mcs rfp mcs gfp-lacZ amyE3′ | pBS1CLacZ-derived plasmid with a GFP-LacZ reporter system |
| Analysis of C. thermocellum cipA and C. cellulovorans cbpA promoters in B. subtilis | ||
| pPromoter-1 | bla amy5′ cat Promoter-1Ct-gfp-lacZ amyE3′ | pBS1C-GFP-LacZ-derived plasmids which have Promoter-1, -2, -3, -4, and -5 of C. thermocellum cipA and Promoter-cbpA of C. cellulovorans fused to the gfp-lacZ reporter system |
| pPromoter-2 | bla amy5′ cat Promoter-2Ct-gfp-lacZ amyE3′ | |
| pPromoter-3 | bla amy5′ cat Promoter-3Ct-gfp-lacZ amyE3′ | |
| pPromoter-4 | bla amy5′ cat Promoter-4Ct-gfp-lacZ amyE3′ | |
| pPromoter-5 | bla amy5′ cat Promoter-5Ct-gfp-lacZ amyE3′ | |
| pPromoter-cbpA | bla amy5′ cat Promoter-cbpACc-gfp-lacZ amyE3′ |
mcs, multiple cloning site; rfp, red fluorescence protein gene; gfp, green fluorescence protein gene.
Plasmid obtained from the Bacillus Genetic Stock Center.
Plasmid constructed by us in a previous work (10).
Mapping of the TSSs.
Mid-log culture cells (10 ml) of C. thermocellum were treated with RNAprotect Bacteria Reagent (Qiagen, Hilden, Germany). RNA was isolated with the RNeasy minikit (Qiagen), and the DNA was digested using DNase I Digestion Set (Sigma-Aldrich). In order to deplete processed RNA (5′ monophosphated) and map primary transcripts (5′ triphosphated), RNA (10 μg) was treated with Terminator 5′-phosphate-dependent exonuclease (TEX; Epicentre, Wisconsin, USA). A final cleaning was performed using RNA Clean & Concentrator-5 (Zymo Research, USA).
Mapping of 5′-ends was performed by using the rapid amplification of cDNA ends (5′-RACE) technique with the SMARTer RACE 5′/3′ kit (Clontech Laboratories, TaKaRa, California, USA) as per the manufacturer's instructions. In order to map primary transcripts and processed RNAs, 1 μg of total RNA treated or untreated with TEX was subjected to reverse transcription (RT)-PCR using the SMARTScribe Reverse transcriptase, random primers, and the SMARTer II A oligonucleotide (enzyme and primers were provided in the kit). Subsequently, 20 μl of 5′-RACE-Ready cDNA was brought to a final volume of 70 μl with Tricine-EDTA buffer (provided in the kit). This solution (2.5 μl) was submitted to a PCR amplification using primer P3 (Table 1) and Universal Primer A Mix, which is a combination of Universal Primer Long and Universal Primer Short (provided in the kit) (Table 1). The RACE PCR touchdown cycling parameters were as suggested by the manufacturer's instructions. Then, this PCR product was diluted 1:50 in Tricine-EDTA buffer (provided in the kit). Next, 5 μl of this solution was subjected to a second PCR touchdown cycle with primer P4 (Table 1) and Universal Primer Short (provided in the kit). Finally, the PCR products were gel purified, cloned into pRACE using the In-Fusion HD Cloning kit (provided in the kit), and sequenced as per the manufacturer's instructions.
Construction of plasmids.
To express the C. thermocellum and C. cellulovorans σI factors in B. subtilis, we used the pAX01 integration vector (29). pAX01 integrates at the B. subtilis lacA chromosomal locus, carries an erm resistance cassette as a selectable marker, and has the xylose-inducible promoter PxylA (29). First, pAX01 was linearized with the restriction enzyme BamHI. Subsequently, the DNA sequences carrying the C. thermocellum genes sigI1, sigI2, sigI4, sigI5, sigI7, and sigI8 and the C. cellulovorans genes sigI1 and sigI2 were PCR amplified using the primer pairs P5-P6, P7-P8, P9-P10, P11-P12, P13-P14, P15-P16, P17-P18, and P19-P20, respectively (Table 1). Finally, each PCR product was cloned into the linearized pAX01 vector using the In-Fusion HD Cloning kit, thereby obtaining the pAX01-derived plasmids pAX01-SigI1, pAX01-SigI2, pAX01-SigI4, pAX01-SigI5, pAX01-SigI7, pAX01-SigI8, pAX01-SigI1-Ccel, and pAX01-SigI2-Ccel, respectively (Table 2). In order to express the C. thermocellum σI3 and σI6 factors in B. subtilis, we used the pAX01-derived plasmids pAX01-SigI3 and pAX01-SigI6 (Table 2) that were constructed in our previous work (10).
To study the promoter recognition by the alternative σI factors or the vegetative σA factor, we built the pBS1C-GFP-LacZ integration vector that contains a promoterless operon consisting of gfp and lacZ reporter genes (see Fig. S2 in the supplemental material). This vector integrates at the B. subtilis amyE locus and carries a cam resistance cassette as a selectable marker. The pBS1C-GFP-LacZ plasmid was built using the pBS1ClacZ plasmid as backbone (30). First, the pBS1ClacZ plasmid, which carries a lacZ reporter gene, was linearized with the restriction enzyme BamHI. Subsequently, the DNA sequence encoding the GFP was amplified with the primers P21-P22 (Table 1) using the pDR111_gfp(sp) (31) plasmid as the template. Finally, the PCR product was cloned using the In-Fusion HD Cloning kit into the linearized pBS1ClacZ vector, obtaining the pBS1C-GFP-LacZ plasmid (Table 2; Fig. S2).
The analysis of the C. thermocellum cipA promoter was performed by cloning different fragments of the upstream intergenic region into the pBS1C-GFP-LacZ plasmid as indicated in Fig. 1B. The analysis of the C. cellulovorans cbpA promoter was performed by including both the previously proposed σA promoter (19) and the σI promoter identified in the present work, as indicated in Fig. 3. Promoters 1, 2, 3, 4, and 5 from C. thermocellum cipA (Fig. 1B) were amplified with the primer pairs P23-P24, P23-P25, P26-P25, P27-P25, and P28-P29, respectively (Table 1). Promoter-cbpA from C. cellulovorans was amplified with the primer pair P30-P31 (Table 1). Subsequently, each PCR product was digested with restriction enzymes EcoRI and BamHI. Finally, each digested PCR product was cloned into pBS1C-GFP-LacZ, which had been digested previously with the same restriction enzymes, thereby obtaining the pBS1C-GFP-LacZ-derived plasmids pPromoter-1, pPromoter-2, pPromoter-3, pPromoter-4, pPromoter-5, and pPromoter-cbpA, respectively (Table 2).
Construction of B. subtilis strains and measurement of promoter activity.
The B. subtilis strains constructed in the present work are listed in Table 3. To avoid the interference of the native B. subtilis σI during the analysis of C. thermocellum and C. cellulovorans σI factors, we used the B. subtilis CO02 strain, which is devoid of its sigI-rsgI operon (10). B. subtilis was transformed by using the natural competence method (32). First, the pAX01-derived plasmids, carrying C. thermocellum or C. cellulovorans σI genes, were transformed into B. subtilis CO02, thereby yielding B. subtilis strains SigI1Ct, SigI2Ct, SigI3Ct, SigI4Ct, SigI5Ct, SigI6Ct, SigI7Ct, SigI8Ct, SigI1Cc, and SigI2Cc (Table 3). To analyze the activation of the cipA σI-dependent promoter, the pPromoter-5 plasmid was transformed into the pAX01-derived B. subtilis strains that harbor C. thermocellum σI factors, thus generating B. subtilis strains SigI1Ct-Promoter5, SigI2Ct-Promoter5, SigI3Ct-Promoter5, SigI4Ct-Promoter5, SigI5Ct-Promoter5, SigI6Ct-Promoter5, SigI7Ct-Promoter5, and SigI8Ct-Promoter5 (Table 3). As a negative control, pAX01-derived B. subtilis strains that harbor C. thermocellum σI factors were transformed with the pPromoter-3 plasmid, thereby yielding the B. subtilis strains SigI1Ct-Promoter3, SigI2Ct-Promoter3, SigI3Ct-Promoter3, SigI4Ct-Promoter3, SigI5Ct-Promoter3, SigI6Ct-Promoter3, SigI7Ct-Promoter3, and SigI8Ct-Promoter3 (Table 3).
TABLE 3.
Bacillus subtilis strainsa constructed in the present work
| Strain derivation and name | Relevant genotype | Factors or promoters harbored |
|---|---|---|
| From pAX01 | ||
| SigI1Ct | CO02 lacA::(Pxyl-sigI1Ct erm) | C. thermocellum σI factors |
| SigI2Ct | CO02 lacA::(Pxyl-sigI2Ct erm) | |
| SigI3Ctb | CO02 lacA::(Pxyl-sigI3Ct erm) | |
| SigI4Ct | CO02 lacA::(Pxyl-sigI4Ct erm) | |
| SigI5Ct | CO02 lacA::(Pxyl-sigI5Ct erm) | |
| SigI6Ctb | CO02 lacA::(Pxyl-sigI6Ct erm) | |
| SigI7Ct | CO02 lacA::(Pxyl-sigI7Ct erm) | |
| SigI8Ct | CO02 lacA::(Pxyl-sigI8Ct erm) | |
| SigI1Cc | CO02 lacA::(Pxyl-sigI1Cc erm) | C. cellulovorans σI factors |
| SigI2Cc | CO02 lacA::(Pxyl-sigI2Cc erm) | |
| From pBS1C-GFP-LacZ | ||
| Promoter-1 | CO02 amyE::(Promoter-1Ct-gfp-lacZ cat) | Different fragments of the upstream region of C. thermocellum cipA used to analyze their activation by σA |
| Promoter-2 | CO02 amyE::(Promoter-2Ct-gfp-lacZ cat) | |
| Promoter-3 | CO02 amyE::(Promoter-3Ct-gfp-lacZ cat) | |
| Promoter-4 | CO02 amyE::(Promoter-4Ct-gfp-lacZ cat) | |
| Promoter-cbpA | CO02 amyE::(Promoter-cbpACc-gfp-lacZ cat) | C. cellulovorans cbpA promoter used to analyze its activation by σA |
| From pAX01 and pBS1C-GFP-LacZ | ||
| SigI1Ct-Promoter-5 | CO02 lacA::(Pxyl-sigI1Ct erm) amyE::(Promoter-5Ct-gfp-lacZ cat) | σI-dependent promoter of C. thermocellum cipA used to analyze its activation by the different C. thermocellum σI factors |
| SigI2Ct-Promoter-5 | CO02 lacA::(Pxyl-sigI2Ct erm) amyE::(Promoter-5Ct-gfp-lacZ cat) | |
| SigI3Ct-Promoter-5 | CO02 lacA::(Pxyl-sigI3Ct erm) amyE::(Promoter-5Ct-gfp-lacZ cat) | |
| SigI4Ct-Promoter-5 | CO02 lacA::(Pxyl-sigI4Ct erm) amyE::(Promoter-5Ct-gfp-lacZ cat) | |
| SigI5Ct-Promoter-5 | CO02 lacA::(Pxyl-sigI5Ct erm) amyE::(Promoter-5Ct-gfp-lacZ cat) | |
| SigI6Ct-Promoter-5 | CO02 lacA::(Pxyl-sigI6Ct erm) amyE::(Promoter-5Ct-gfp-lacZ cat) | |
| SigI7Ct-Promoter-5 | CO02 lacA::(Pxyl-sigI7Ct erm) amyE::(Promoter-5Ct-gfp-lacZ cat) | |
| SigI8Ct-Promoter-5 | CO02 lacA::(Pxyl-sigI8Ct erm) amyE::(Promoter-5Ct-gfp-lacZ cat) | |
| SigI1Ct-Promoter-3 | CO02 lacA::(Pxyl-sigI1Ct erm) amyE::(Promoter-3Ct-gfp-lacZ cat) | Promoter-3 of C. thermocellum cipA as negative control during the analysis of the different C. thermocellum σI factors |
| SigI2Ct-Promoter-3 | CO02 lacA::(Pxyl-sigI2Ct erm) amyE::(Promoter-3Ct-gfp-lacZ cat) | |
| SigI3Ct-Promoter-3 | CO02 lacA::(Pxyl-sigI3Ct erm) amyE::(Promoter-3Ct-gfp-lacZ cat) | |
| SigI4Ct-Promoter-3 | CO02 lacA::(Pxyl-sigI4Ct erm) amyE::(Promoter-3Ct-gfp-lacZ cat) | |
| SigI5Ct-Promoter-3 | CO02 lacA::(Pxyl-sigI5Ct erm) amyE::(Promoter-3Ct-gfp-lacZ cat) | |
| SigI6Ct-Promoter-3 | CO02 lacA::(Pxyl-sigI6Ct erm) amyE::(Promoter-3Ct-gfp-lacZ cat) | |
| SigI7Ct-Promoter-3 | CO02 lacA::(Pxyl-sigI7Ct erm) amyE::(Promoter-3Ct-gfp-lacZ cat) | |
| SigI8Ct-Promoter-3 | CO02 lacA::(Pxyl-sigI8Ct erm) amyE::(Promoter-3Ct-gfp-lacZ cat) | |
| SigI1Cc-Promoter-cbpA | CO02 lacA::(Pxyl-sigI1Cc erm) amyE::(Promoter-cbpACc-gfp-lacZ cat) | σI-dependent promoter of C. cellulovorans cbpA used to analyze its activation by the different C. cellulovorans σI factors |
| SigI2Cc-Promoter-cbpA | CO02 lacA::(Pxyl-sigI2Cc erm) amyE::(Promoter-cbpACc-gfp-lacZ cat) |
In order to analyze the activation of the different fragments of the upstream region of C. thermocellum cipA by σA, B. subtilis CO02 was transformed with pPromoter-1, pPromoter-2, pPromoter-3, and pPromoter-4, thus yielding B. subtilis strains Promoter-1, Promoter-2, Promoter-3, and Promoter-4 (Table 3). To analyze the activation of the C. cellulovorans cbpA promoter by σA, the pPromoter-cbpA plasmid was transformed into B. subtilis CO02, thus generating the B. subtilis strain Promoter-cbpA (Table 3). Finally, to analyze the activation of the C. cellulovorans cbpA promoter by σI factors, the pAX01-derived strains harboring either C. cellulovorans σI1 or σI2 were transformed with pPromoter-cbpA, thereby obtaining the B. subtilis strains SigI1Cc-Promoter-cbpA and SigI1Cc-Promoter-cbpA, respectively (Table 3).
Chromosomal integration of plasmids by a double-crossover event was confirmed by colony PCR. Chromosomal integration at the B. subtilis lacA locus was confirmed with primer pairs P32-P33 and P34-P35 (Table 1). Chromosomal integration at the B. subtilis amyE locus was confirmed with primer pairs P36-P37 and P38-P39 (Table 1). The different B. subtilis strains obtained were stored at −80°C in 20% (vol/vol) glycerol. To measure the fluorescence associated with the GFP reporter system, B. subtilis strain samples were taken from the −80°C glycerol stock and inoculated in 5 ml of LB broth with Cam. Subsequently, the cells were grown overnight at 37°C with shaking (250 rpm). The next day, 2 μl of the cells were inoculated in a 96-well plate with 200 μl of LB broth supplemented with xylose. The 96-well plate was incubated at 37°C with medium agitation in a microplate reader (Biotek Synergy HT; Biotek, Vermont, USA). The fluorescence was measured using the excitation filter 485/20 and the emission filter 528/20, and the relative fluorescence levels of the cultures were calculated as reported elsewhere (31) when the cells reached an optical density at 600 nm (OD600) of 1.
Computational analysis.
Primary DNA sequence analyses were performed with the Clone Manager 9 Professional Edition software (Scientific & Educational Software, Durham, NC). DNA promoter motif searches were performed with the Pattern Locator program (18) (http://www.cmbl.uga.edu/software/patloc.html) and analyzed with the Jalview software (33) (http://www.jalview.org/). The search was carried out using the general motifs for σI-dependent promoter of cellulosome-producing bacteria, which are AAA in the −35 region and CGWA in the −10 region (where W is A or T), with a spacer of 13 or 14 nucleotides (10). DNA sequence logos were generated with the program WebLogo (34) (http://weblogo.berkeley.edu/logo.cgi). RNA secondary structures were predicted with the “mfold” program (17) with the default parameters (http://unafold.rna.albany.edu/?q=mfold). The phylogenetic tree of cellulosome-producing bacteria was inferred from multiple 16S rRNA gene sequence alignments using the neighbor-joining method implemented in MEGA7 software (35).
Supplementary Material
ACKNOWLEDGMENTS
We appreciate the technical support provided by Milana Voronov-Goldman (Tel Aviv University).
Y.S. holds the Erwin and Rosl Pollak Chair in Biotechnology at the Technion-Israel Institute of Technology. E.A.B. is the incumbent of The Maynard I. and Elaine Wishner Chair of Bio-organic Chemistry at the Weizmann Institute of Science.
I.M.-G. and L.O.D.O. were supported by the Mexican Council of Science and Technology with a postdoctoral fellowship and a Ph.D. scholarship, respectively (CONACyT proposal number 238334 and CONACyT scholarship number 440354, respectively). This research was supported by the Israel Science Foundation (grant no. 24/11 to R.L. and I.B., 177/14 to Y.S., and 1349/13 to E.A.B.) and by the US-Israel Binational Science Foundation (BSF; grant no. 2011049 to Y.S.). Additional support was issued to R.L. and I.B. by The Sidney E. Frank Foundation through the ISF.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.03088-16.
REFERENCES
- 1.Artzi L, Bayer EA, Moraïs S. 2017. Cellulosomes: bacterial nanomachines for dismantling plant polysaccharides. Nat Rev Microbiol 15:83–95. doi: 10.1038/nrmicro.2016.164. [DOI] [PubMed] [Google Scholar]
- 2.Xu Q, Resch MG, Podkaminer K, Yang S, Baker JO, Donohoe BS, Wilson C, Klingeman DM, Olson DG, Decker SR, Giannone RJ, Hettich RL, Brown SD, Lynd LR, Bayer EA, Himmel ME, Bomble YJ. 2016. Dramatic performance of Clostridium thermocellum explained by its wide range of cellulase modalities. Sci Adv 2:e1501254. doi: 10.1126/sciadv.1501254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bayer EA, Kenig R, Lamed R. 1983. Adherence of Clostridium thermocellum to cellulose. J Bacteriol 156:818–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lamed R, Naimark J, Morgenstern E, Bayer EA. 1987. Specialized cell surface structures in cellulolytic bacteria. J Bacteriol 169:3792–3800. doi: 10.1128/jb.169.8.3792-3800.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zverlov VV, Klupp M, Krauss J, Schwarz WH. 2008. Mutations in the scaffoldin gene, cipA, of Clostridium thermocellum with impaired cellulosome formation and cellulose hydrolysis: insertions of a new transposable element, IS1447, and implications for cellulase synergism on crystalline cellulose. J Bacteriol 190:4321–4327. doi: 10.1128/JB.00097-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olson DG, Giannone RJ, Hettich RL, Lynd LR. 2013. Role of the CipA scaffoldin protein in cellulose solubilization, as determined by targeted gene deletion and complementation in Clostridium thermocellum. J Bacteriol 195:733–739. doi: 10.1128/JB.02014-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dror TW, Rolider A, Bayer EA, Lamed R, Shoham Y. 2003. Regulation of expression of scaffoldin-related genes in Clostridium thermocellum. J Bacteriol 185:5109–5116. doi: 10.1128/JB.185.17.5109-5116.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kahel-Raifer H, Jindou S, Bahari L, Nataf Y, Shoham Y, Bayer EA, Borovok I, Lamed R. 2010. The unique set of putative membrane-associated anti-sigma factors in Clostridium thermocellum suggests a novel extracellular carbohydrate-sensing mechanism involved in gene regulation. FEMS Microbiol Lett 308:84–93. doi: 10.1111/j.1574-6968.2010.01997.x. [DOI] [PubMed] [Google Scholar]
- 9.Nataf Y, Bahari L, Kahel-Raifer H, Borovok I, Lamed R, Bayer EA, Sonenshein AL, Shoham Y. 2010. Clostridium thermocellum cellulosomal genes are regulated by extracytoplasmic polysaccharides via alternative sigma factors. Proc Natl Acad Sci U S A 107:18646–18651. doi: 10.1073/pnas.1012175107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muñoz-Gutiérrez I, Ortiz de Ora L, Rozman Grinberg I, Garty Y, Bayer EA, Shoham Y, Lamed R, Borovok I. 2016. Decoding biomass-sensing regulons of Clostridium thermocellum alternative sigma-I factors in a heterologous Bacillus subtilis host system. PLoS One 11:e0146316. doi: 10.1371/journal.pone.0146316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sand A, Holwerda EK, Ruppertsberger NM, Maloney M, Olson DG, Nataf Y, Borovok I, Sonenshein AL, Bayer EA, Lamed R, Lynd LR, Shoham Y. 2015. Three cellulosomal xylanase genes in Clostridium thermocellum are regulated by both vegetative SigA (σA) and alternative SigI6 (σI6) factors. FEBS Lett 589:3133–3140. doi: 10.1016/j.febslet.2015.08.026. [DOI] [PubMed] [Google Scholar]
- 12.Kato S, Haruta S, Cui ZJ, Ishii M, Yokota A, Igarashi Y. 2004. Clostridium straminisolvens sp. nov., a moderately thermophilic, aerotolerant and cellulolytic bacterium isolated from a cellulose-degrading bacterial community. Int J Syst Evol Microbiol 54:2043–2047. doi: 10.1099/ijs.0.63148-0. [DOI] [PubMed] [Google Scholar]
- 13.Sun L, Schürer A. 2016. Draft genome sequence of the cellulolytic strain Clostridium sp. Bc-iso-3 isolated from an industrial-scale anaerobic digester. Genome Announc 4:e01188-16. doi: 10.1128/genomeA.01188-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Izquierdo JA, Goodwin L, Davenport KW, Teshima H, David B, Detter C, Tapia R, Han S, Land M, Hauser L, Jeffries CD, Han J, Pitluck S, Nolan M, Chen A, Huntemann M, Mavromatis K, Mikhailova N, Liolios K, Wokye T, Lynd RL. 2012. Complete genome sequence of Clostridium clariflavum DSM 19732. Stand Genomic Sci 6:104–115. doi: 10.4056/sigs.2535732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dassa B, Borovok I, Lamed R, Henrissat B, Coutinho P, Hemme CL, Huang Y, Zhou J, Bayer EA. 2012. Genome-wide analysis of Acetivibrio cellulolyticus provides a blueprint of an elaborate cellulosome system. BMC Genomics 13:210. doi: 10.1186/1471-2164-13-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamaru Y, Miyabe H, Kuroda K, Nakanishi A, Matsushima C, Doi RH, Ueda M. 2011. Comparison of the mesophilic cellulosome-producing Clostridium cellulovorans genome with other cellulosome-related clostridial genomes. Microb Biotechnol 4:64–73. doi: 10.1111/j.1751-7915.2010.00210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mrázek J, Xie S. 2006. Pattern locator: a new tool for finding local sequence patterns in genomic DNA sequences. Bioinformatics 22:3099–3100. doi: 10.1093/bioinformatics/btl551. [DOI] [PubMed] [Google Scholar]
- 19.Han SO, Yukawa H, Inui M, Doi RH. 2003. Transcription of Clostridium cellulovorans cellulosomal cellulase and hemicellulase genes. J Bacteriol 185:2520–2527. doi: 10.1128/JB.185.8.2520-2527.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maamar H, Abdou L, Boileau C, Valette O, Tardif C. 2006. Transcriptional analysis of the cip-cel gene cluster from Clostridium cellulolyticum. J Bacteriol 188:2614–2624. doi: 10.1128/JB.188.7.2614-2624.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abdou L, Boileau C, De Philip P, Pagès S, Fiérobe HP, Tardif C. 2008. Transcriptional regulation of the Clostridium cellulolyticum cip-cel operon: a complex mechanism involving a catabolite-responsive element. J Bacteriol 190:1499–1506. doi: 10.1128/JB.01160-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emory SA, Bouvet P, Belasco JG. 1992. A 5′-terminal stem-loop structure can stabilize mRNA in Escherichia coli. Genes Dev 6:135–148. doi: 10.1101/gad.6.1.135. [DOI] [PubMed] [Google Scholar]
- 23.Xu C, Huang R, Teng L, Jing X, Hu J, Cui G, Wang Y, Cui Q, Xu J. 2015. Cellulosome stoichiometry in Clostridium cellulolyticum is regulated by selective RNA processing and stabilization. Nat Commun 6:6900. doi: 10.1038/ncomms7900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munir RI, Schellenberg J, Henrissat B, Verbeke TJ, Sparling R, Levin DB. 2014. Comparative analysis of carbohydrate active enzymes in Clostridium termitidis CT1112 reveals complex carbohydrate degradation ability. PLoS One 9:e1042060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Desvaux M. 2005. The cellulosome of Clostridium cellulolyticum. Enzyme Microb Technol 37:373–385. doi: 10.1016/j.enzmictec.2004.04.025. [DOI] [Google Scholar]
- 26.Zepeda V, Dassa B, Borovok I, Lamed R, Bayer EA, Cate JH. 2013. Draft genome sequence of the cellulolytic bacterium Clostridium papyrosolvens C7 (ATCC 700395). Genome Announc 1:e00698-13. doi: 10.1128/genomeA.00698-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tseng C-L, Shaw G-C. 2008. Generic evidence for the actin homolog gene mreBH and the bacitracin resistance gene bcrC as targets of the alternative sigma factor SigI of Bacillus subtilis. J Bacteriol 190:1561–1567. doi: 10.1128/JB.01497-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook J, Russell D. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 29.Härtl B, Wehrl W, Wiegert T, Homuth G, Schumann W. 2001. Development of a new integration site within the Bacillus subtilis chromosome and construction of compatible expression cassettes. J Bacteriol 183:2696–2699. doi: 10.1128/JB.183.8.2696-2699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Radeck J, Kraft K, Bartels J, Cikovic T, Dürr F, Emenegger J, Kelterborn S, Sauer C, Fritz G, Gebhard S, Mascher T. 2013. The Bacillus BioBrick Box: generation and evaluation of essential genetic building blocks for standardized work with Bacillus subtilis. J Biol Eng 7:29. doi: 10.1186/1754-1611-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Overkamp W, Beilharz K, Detert Oude Weme R, Solopova A, Karsens H, Kovacs AT, Kok J, Kuipers OP, Veening J-W. 2013. Benchmarking various green fluorescent protein variants in Bacillus subtilis, Streptococcus pneumoniae, and Lactococcus lactis for live cell imaging. Appl Environ Microbiol 79:6481–6490. doi: 10.1128/AEM.02033-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cutting SM, Vander Horn PB. 1990. Genetic analysis, p 27–74. In Harwood CR, Cutting SM (ed), Molecular biological methods for Bacillus. John Wiley & Sons Ltd., Chichester, England. [Google Scholar]
- 33.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. 2009. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crooks GE, Hon G, Chandonia J-M, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.