ABSTRACT
In order to elucidate interactions between sulfate reduction and dechlorination, we systematically evaluated the effects of different concentrations of sulfate and sulfide on reductive dechlorination by isolates, constructed consortia, and enrichments containing Dehalococcoides sp. Sulfate (up to 5 mM) did not inhibit the growth or metabolism of pure cultures of the dechlorinator Dehalococcoides mccartyi 195, the sulfate reducer Desulfovibrio vulgaris Hildenborough, or the syntroph Syntrophomonas wolfei. In contrast, sulfide at 5 mM exhibited inhibitory effects on growth of the sulfate reducer and the syntroph, as well as on both dechlorination and growth rates of D. mccartyi. Transcriptomic analysis of D. mccartyi 195 revealed that genes encoding ATP synthase, biosynthesis, and Hym hydrogenase were downregulated during sulfide inhibition, whereas genes encoding metal-containing enzymes involved in energy metabolism were upregulated even though the activity of those enzymes (hydrogenases) was inhibited. When the electron acceptor (trichloroethene) was limiting and an electron donor (lactate) was provided in excess to cocultures and enrichments, high sulfate concentrations (5 mM) inhibited reductive dechlorination due to the toxicity of generated sulfide. The initial cell ratio of sulfate reducers to D. mccartyi (1:3, 1:1, or 3:1) did not affect the dechlorination performance in the presence of sulfate (2 and 5 mM). In contrast, under electron donor limitation, dechlorination was not affected by sulfate amendments due to low sulfide production, demonstrating that D. mccartyi can function effectively in anaerobic microbial communities containing moderate sulfate concentrations (5 mM), likely due to its ability to outcompete other hydrogen-consuming bacteria and archaea.
IMPORTANCE Sulfate is common in subsurface environments and has been reported as a cocontaminant with chlorinated solvents at various concentrations. Inconsistent results for the effects of sulfate inhibition on the performance of dechlorination enrichment cultures have been reported in the literature. These inconsistent findings make it difficult to understand potential mechanisms of sulfate inhibition and complicate the interpretation of bioremediation field data. In order to elucidate interactions between sulfate reduction and reductive dechlorination, this study systematically evaluated the effects of different concentrations of sulfate and sulfide on reductive dechlorination by isolates, constructed consortia, and enrichments containing Dehalococcoides sp. This study provides a more fundamental understanding of the competition mechanisms between reductive dechlorination by Dehalococcoides mccartyi and sulfate reduction during the bioremediation process. It also provides insights on the significance of sulfate concentrations on reductive dechlorination under electron donor/acceptor-limiting conditions during in situ bioremediation applications. For example, at a trichloroethene-contaminated site with a high sulfate concentration, proper slow-releasing electron donors can be selected to generate an electron donor-limiting environment that favors reductive dechlorination and minimizes the sulfide inhibition effect.
KEYWORDS: reductive dechlorination, sulfate reduction, sulfide generation, inhibition, competition
INTRODUCTION
Perchloroethene (PCE), trichloroethene (TCE), and their daughter products dichloroethene (DCE) and vinyl chloride are common soil and groundwater contaminants with established toxicity and mutagenicity toward many organisms (1–3). In situ bioremediation processes that stimulate the growth of anaerobic microbial communities capable of reductively dechlorinating these contaminants to harmless ethene are of great interest (4). Among reported dechlorinating species, Dehalococcoides mccartyi is the only known bacterium that can reductively dechlorinate PCE and TCE all the way to ethene (1). D. mccartyi requires H2 as its exclusive electron donor, acetate and CO2 as carbon sources and vitamin B12 as a cofactor (5–7). Although reductive dechlorination can occur under a variety of redox conditions (8), dechlorination commonly only accounts for a small fraction of electron flow in microbial communities during bioremediation (9–11). Other terminal-electron-accepting processes, such as sulfate reduction, iron reduction, nitrate reduction, methanogenesis, homoacetogenesis, and volatile fatty acid formation, typically account for a large fraction of the electron flow in these systems (see Table S1 in the supplemental material).
Sulfate is common in subsurface environments and is often reported as a cocontaminant with chlorinated solvents at various concentrations (0.2 to 30 mM) (12–17). The effects of sulfate and its reduction product sulfide on other terminal electron accepting processes have been explored in anaerobic digestion, sulfate reduction, and nitrification, as well as reductive dechlorination processes (17–21). Hydrogen sulfide (H2S) has been shown to inhibit the growth of sulfate-reducing bacteria at 16 mM due to both its intrinsic toxicity and indirect toxicity by precipitation with iron as ferric sulfide (19).
The dechlorination of solvents under sulfate-reducing conditions is complicated and less well studied. Although successful organic-stimulated bioremediation of solvents has been observed in aquifers containing sulfate, the typical approaches involve injecting an excess of electron donor in order to deplete sulfate and to avoid competition for hydrogen between dechlorination and sulfate reduction (22, 23). This approach was shown to be successful at some field sites; however, it has proven to be unsuccessful at sites with high sulfate concentrations or complex geochemical conditions (15, 24–26).
There are a limited number of laboratory studies with detailed information on the effects of sulfate on dechlorination (21, 26–28). In addition, some conflicting results due to sulfate addition, ranging from enhanced dechlorination (27, 29, 30) to inhibited or incomplete dechlorination (15, 27, 29, 31, 32), as well as no observed effect on dechlorination (16, 25), have been reported over the past decade. A review of published field data from TCE-contaminated sites with sulfate concentrations ranging from 39 to 4,800 mg liter−1 reported the overall trend that as sulfate concentrations increased, dechlorination reactions became incomplete or delayed (26). In addition, among these previous studies, there have only been a few that used microbial communities with the confirmed presence of D. mccartyi (15, 28, 30, 32, 33) and cellular quantification has been lacking. Further work is needed to clarify the significance of sulfate concentrations on reductive dechlorination under electron donor/acceptor-limiting conditions. In addition, the effects of sulfide, the sulfate reduction product, on dechlorination need to be systematically evaluated.
In this study, we hypothesize two main mechanisms for the observed failure of complete dechlorination during bioremediation in sulfate-containing environments: (i) the inhibition of enzymes involved in dechlorination by the sulfate reduction product sulfide and (ii) the predominance and faster growth kinetics of sulfate-reducing bacteria, compared to D. mccartyi at high H2 concentrations (electron acceptor limitation). In order to test these hypotheses, we investigated the inhibitory effect of sulfate and sulfide on (i) pure D. mccartyi strain and supporting microorganisms, (ii) constructed syntrophic consortia at different cell ratios and electron donor/acceptor-limited conditions, and (iii) a methanogenic dechlorinating enrichment culture with high and low sulfate amendments. Transcriptomic analysis of D. mccartyi was used to investigate gene expression patterns during sulfide inhibition in order to better understand the mechanism of inhibition. This study provides a fundamental understanding of the effects of sulfate reduction on reductive dechlorination by D. mccartyi.
RESULTS
Sulfate and sulfide effects on axenic cultures.
An environmentally high sulfate concentration (5 mM) did not affect cell growth or dechlorination rates of strain 195 (data not shown), a bacterium unable to reduce sulfate to sulfide. We also tested the effect of sulfide (the reduction product of sulfate) on the cell growth of strain 195 and found that with 5% inoculation, it took 6, 10, and 14 days to dechlorinate 75 μmol of TCE in the presence of 0, 2, and 5 mM sulfide, respectively. The cell yield of strain 195 decreased about 65% as sulfide concentrations increased from 0 to 5 mM (see Fig. S1A in the supplemental material). For S. wolfei (another bacterium incapable of sulfate reduction) grown with crotonate as electron donor, cell growth was not inhibited by 5 mM sulfate addition, while 5 mM sulfide decreased cell yields by 40% compared to the control group (see Fig. S1C in the supplemental material). For the Desulfovibrio vulgaris Hildenborough (DvH) isolate, which is capable of sulfate reduction, when sulfide concentrations were >10 mM, the cell growth was inhibited (see Fig. S1B in the supplemental material).
Effect of sulfate reduction on dechlorination under electron acceptor limitation.
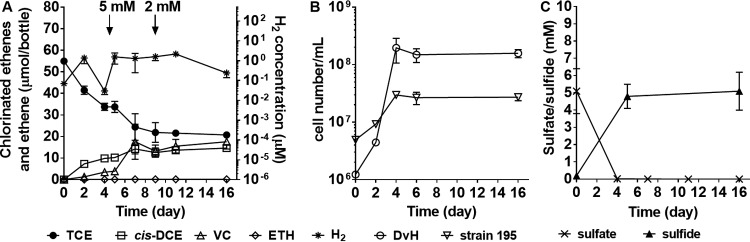
Syntrophic coculture DvH/195 grows sustainably on lactate and TCE with DvH fermenting lactate to acetate and H2 that are used by strain 195 as a carbon source and an electron donor for the reductive dechlorination of TCE, respectively (34). DvH can reduce sulfate and hence produce sulfide. In this study, 12 mM lactate was amended to the coculture initially as an electron donor, whereas 5 mM sulfate and 0.55 mM TCE were added as electron acceptors (Fig. 1B). Based on stoichiometry, 11.1 mM lactate would be required to reduce both electron acceptors: 10 mM lactate for sulfate reduction to sulfide and 1.1 mM lactate for TCE reduction to ethene, creating electron acceptor limitation. Aqueous H2 concentrations increased to 1.4 ± 0.6 μM on day 2 in the sulfate-fed coculture (Fig. 2A) compared to 43.1 ± 3.7 μM in the control group without sulfate amendment (lactate fermentation only, data not shown). When H2 in the coculture dropped below 0.1 μM on day 4, another 5 mM lactate was amended to the culture, and H2 slightly increased to >1.0 μM on day 5, indicating that lactate fermentation was proceeding. However, TCE dechlorination rates decreased by 62% from day 4 to day 9, and no cell growth was observed. On day 9, another 2 mM lactate was added to the coculture and the H2 concentration slightly increased to 2.0 μM, but both dechlorination and cell growth stalled from day 9 to day 16 (Fig. 2B). The 5 mM sulfate was depleted within 4 days. On day 5 no sulfate was detected, whereas the sulfide concentration was measured to be 4.8 ± 0.7 mM (Fig. 2C). At the end of the experiment (day 16), the cell number ratio of strain 195 to DvH was about 1:6 in contrast to the no-sulfate control (coculture grown on lactate and TCE), where the ratio was 4.3:1, similar to previously reported ratios (34). Sulfide can precipitate metals that are necessary nutrients and hence make them inaccessible to the cells (35). In order to demonstrate that the lack of dechlorination observed in this study was due to sulfide inhibition instead of trace metal insufficiency caused by sulfide precipitation, at the end of the experiment (day 16) the headspaces of the experimental bottles were flushed for 40 min with sterilized nitrogen gas to remove sulfide, and then the bottles were re-amended with 0.5 mM TCE and 1 mM lactate. Complete TCE dechlorination was observed after 5 days (data not shown). In addition, at the end of the experiment (day 16), trace metal concentrations in the liquid medium were observed to be at the same micromolar levels as in the positive controls (no sulfate amendment).
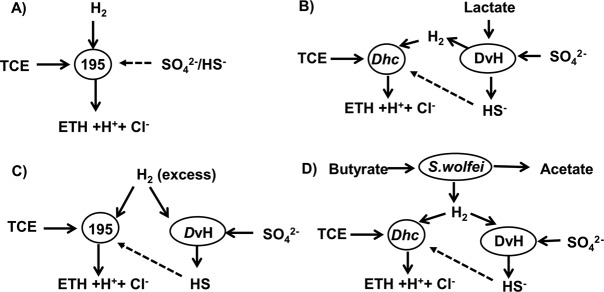
FIG 1.
Ecological interactions between strain 195, DvH, and S. wolfei in constructed consortia and the potential inhibitory effects of sulfate/sulfide. (A) Potential inhibitory effects of sulfate/sulfide on strain 195; (B) DvH and 195 in a syntrophic coculture with lactate as electron donor and sulfate addition; (C) DvH and 195 with H2 fed in excess as electron donor and sulfate addition; (D) DvH, 195, and S. wolfei triculture (S. wolfei/DvH/195) with butyrate as an electron donor and TCE and sulfate as electron acceptors. Acetate is the carbon source for the growth of 195. Dashed lines indicate a potential inhibitory effect.
FIG 2.
Coculture (DvH/195) electron acceptor limitation experiment. (A) TCE dechlorination activity and H2 production with arrows showing lactate amendments of 5 and 2 mM; (B) cell numbers; (C) sulfate and sulfide concentrations. Symbols represent means of biological triplicates, and error bars indicate standard deviations. The absence of error bars indicates the error was smaller than the symbol.
To further investigate the effect of sulfate reduction on reductive dechlorination under electron acceptor limitation, the activity of coculture DvH/195 was quantified at different inoculum cell ratios (see Table S2 in the supplemental material) with excess H2 as electron donor (H2/CO2 headspace, 90:10 [vol/vol]; Fig. 1C). When 2 mM sulfate was amended to the coculture, no negative effects on dechlorination or cell growth were observed compared to the positive control (no sulfate amendment) among different cell ratios, whereas sulfate was fully reduced to sulfide (see Fig. S2 in the supplemental material). However, when 5 mM sulfate was amended to the coculture, all sulfate was reduced by the end of the experiment at all cell ratios with reduced product sulfide (see Fig. S3 in the supplemental material) and TCE degradation stalled after day 4 (Fig. 3A; see also Fig. S3). Both the sulfate reduction rates and growth rates of DvH were higher when inhibition of dechlorination occurred, and >99.5% of consumed electron equivalents (i.e., H2) went to sulfate reduction (Table 1) rather than to dechlorination.
FIG 3.
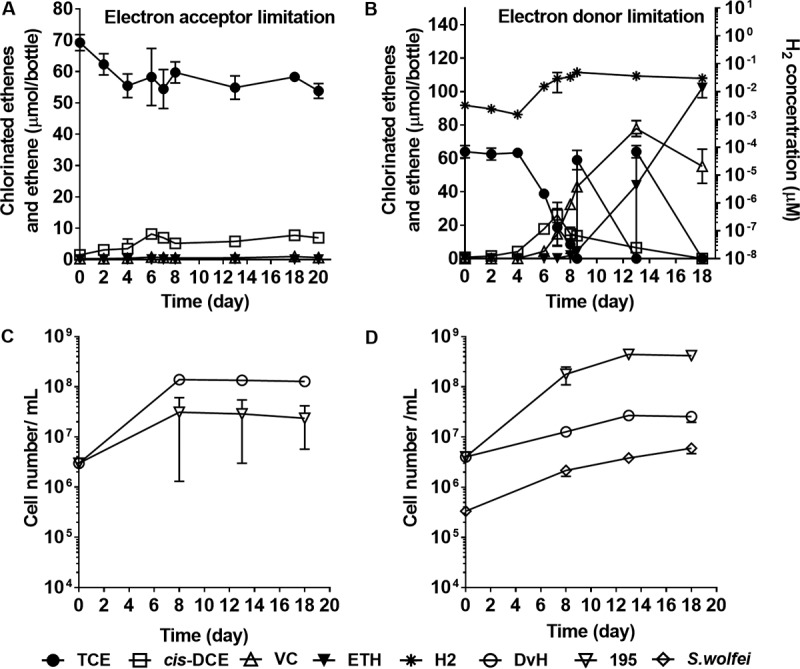
Constructed consortia amended with 5 mM sulfate, along with TCE and H2. (A and C) Coculture DvH/195 (inoculum ratio, 1:1) with H2/CO2 headspace; (B and D) triculture S. wolfei/DvH/195 (inoculum ratio, 0.08:1:1) with 6 mM butyrate. Symbols represent the means of biological triplicates, and error bars indicate standard deviations. The absence of error bars indicates the error was smaller than the symbol.
TABLE 1.
Substrate utilization rates and electron equivalent consumption by constructed consortia
| Electron donor, consortium members | DvH/195 or DvH/195/S. wolfei ratioa | Limiting substrate | SO42− concn (mM) | Mean ± SEM |
Consumed electron equivalent ratio of sulfate reduction to dechlorinationc | Inhibition effect | |||
|---|---|---|---|---|---|---|---|---|---|
| Cl− release rateb (μmol Cl− day−1) | Sulfate reduction rate (μmol of sulfate day−1) | Specific dechlorination rate (×10−10 μmol cell−1 day−1) | Specific sulfate reduction rate (×10−9 μmol cell−1 day−1) | ||||||
| H2, DvH/195 | 3:1 | Acceptor | 2 | 20.2 ± 0.3 | 30.0 ± 1.5 | 6.2 ± 1.2 | 4.2 ± 0.6 | 1.8:1 | No |
| 1:1 | Acceptor | 2 | 22.1 ± 0.4 | 28.4 ± 1.7 | 6.5 ± 1.1 | 4.4 ± 0.3 | 1.7:1 | No | |
| 1:3 | Acceptor | 2 | 22.6 ± 0.3 | 27.9 ± 1.1 | 5.5 ± 0.7 | 3.8 ± 0.2 | 1.6:1 | No | |
| 3:1 | Acceptor | 5 | 1.1 ± 0.1 | 40.4 ± 0.5 | 5.5 ± 0.5 | 2.1 ± 0.3 | 346:1 | Yes | |
| 1:1 | Acceptor | 5 | 1.4 ± 0.3 | 41.3 ± 0.8 | 5.7 ± 0.1 | 3.2 ± 0.7 | 282:1 | Yes | |
| 1:3 | Acceptor | 5 | 1.6 ± 0.4 | 37.6 ± 2.1 | 5.1 ± 0.3 | 2.2 ± 0.5 | 199:1 | Yes | |
| Butyrate, S. wolfei/DvH/195 | 3:1:0.08 | Donor | 2 | 29.8 ± 0.9 | 15.0 ± 1.1 | 4.6 ± 0.7 | 5.0 ± 1.3 | 1.6:1 | No |
| 1:1:0.08 | Donor | 2 | 25.7 ± 0.6 | 14.3 ± 0.3 | 4.6 ± 0.3 | 7.1 ± 1.8 | 1.8:1 | No | |
| 1:3:0.24 | Donor | 2 | 25.1 ± 0.1 | 13.8 ± 0.7 | 4.2 ± 0.3 | 6.3 ± 1.1 | 1.6:1 | No | |
| 3:1:0.08 | Donor | 5 | 23.2 ± 0.7 | 13.1 ± 1.1 | 8.0 ± 2.1 | 6.6 ± 1.2 | 1.8:1 | No | |
| 1:1:0.08 | Donor | 5 | 23.1 ± 0.3 | 10.7 ± 2.1 | 5.3 ± 1.1 | 5.3 ± 0.7 | 1.8:1 | No | |
| 1:3:0.24 | Donor | 5 | 23.7 ± 0.1 | 11.9 ± 0.8 | 5.4 ± 0.7 | 4.0 ± 0.3 | 1.6:1 | No | |
The initial cell ratios are from Table S2 in the supplemental material.
Cl− release rates were calculated for 14-day experimental periods in each experiment (see Fig. S2 to S5 in the supplemental material). The sulfate reduction rate was calculated for the same experimental period.
Consumed electron equivalents were calculated based on the half-reactions listed in Table S1 in the supplemental material, with 2 electrons per Cl− released and 8 electrons per sulfate reduced.
Effect of sulfate reduction on dechlorination under electron donor limitation.
In order to study the competition for H2 by dechlorination and sulfate reduction under electron donor limitation, we maintained a triculture of the S. wolfei, DvH, and 195 strains (S. wolfei/DvH/195) on 5 mM butyrate, 0.7 mM TCE, and 2 mM sulfate (Fig. 1D) with different initial cell ratios of DvH to strain 195 (see Table S2 in the supplemental material). In these cultures, S. wolfei ferments butyrate to acetate (used for biosynthesis) and H2, which competes with 195 for dechlorination and with DvH for sulfate reduction, thus maintaining the requisite low H2 concentrations to sustain energetically unfavorable butyrate degradation by S. wolfei. DvH does not use butyrate as an electron donor for sulfate reduction. Based on stoichiometry (see equations in Table S1 in the supplemental material), 5.4 mM butyrate would be required to fully reduce each of the electron acceptors: 2 mM sulfate to hydrogen sulfide and 0.7 mM TCE to ethene. We first fed 5 mM butyrate to the triculture to generate electron donor-limiting conditions. An additional 0.7 mM TCE and 1 mM butyrate were subsequently amended to the culture when the previous dose of TCE was depleted. During the experimental period, H2 remained between 0.03 and 0.13 μM for all cell ratios (see Fig. S4 in the supplemental material), which is above the threshold for either dechlorination or sulfate reduction (36, 37) and was comparable to that maintained in the control group (without sulfate addition). TCE dechlorination rates were not considerably affected by the sulfate additions (2 or 5 mM) for all initial cell ratios (Fig. 3B; see also Fig. S4 and S5 in the supplemental material). With equal starting cells of DvH and strain 195, the sulfate reduction rates (14.3 ± 0.3 μmol day−1) decreased to about half of those in the electron acceptor-limited condition (28.4 ± 1.7 μmol day−1) at 2 mM sulfate and to about one-quarter with 5 mM sulfate at electron acceptor-limited condition, whereas only 26.8 to 28.0% of sulfate was reduced in all three cell ratios (see Fig. S5 in the supplemental material).
In order to study the continuous competition of sulfate reduction and dechlorination under an electron donor-limiting condition, we constructed the triculture S. wolfei/DvH/195 with initial cell ratios of 0.08:1:1 (see Table S2 in the supplemental material). First, 5 mM butyrate, 2 mM sulfate, and 0.7 mM TCE were amended, and the triculture was routinely subcultured into fresh medium (10% [vol/vol]) every 14 days after TCE was fully reduced to ethene (data not shown). After three subculture events, we monitored TCE dechlorination performance and cell growth in the triculture (see Fig. S6 in the supplemental material). Strain 195 increased to 1.9 × 108 ± 0.2 × 108 ml−1, which was similar to the control group with no sulfate amendment (1.8 × 108 ± 0.2 × 108 ml−1), whereas S. wolfei (1.2 × 107 ± 0.3 × 107 ml−1) increased to 50% higher than the control (0.8 × 107 ± 0.1 × 107 ml−1). DvH cell numbers increased to 1.4 × 107 ± 0.2 × 107 ml−1 on day 10 and then decreased to 0.8 × 107 ± 0.1 × 107 ml−1 by the end of the experiment (62% lower than that in the initially constructed triculture 2.1 × 107 ± 0.1 × 107 ml−1). The cell ratio (S. wolfei/DvH/195) was stably maintained at 1:1:16, and the dechlorination rate was not affected by sulfate addition (2 mM) after the three subcultures. Interestingly, cell aggregates were observed in the late exponential phase of each subculture event (see Fig. S7 in the supplemental material).
Effects of sulfate on dechlorination in a groundwater enrichment.
A methanogenic reductive-dechlorinating enrichment culture (ANAS) was used to test sulfate effects on dechlorination under different electron-limiting conditions (Fig. 4). Two sulfate concentrations (2 and 5 mM) were amended to ANAS with 20 mM lactate and 0.2 mM TCE for electron acceptor limitation. When electron acceptor was limiting, H2 was produced within 2 days and achieved mM levels. TCE dechlorination stalled on day 6 at both sulfate concentrations, and H2 levels dropped to <2 nM (data not shown). In order to avoid electron donor limitation, another 20 mM lactate was re-amended to the bottles on day 6, and TCE reduction resumed within 2 days in the 2 mM sulfate cultures, whereas H2 levels remained >20 nM. However, no further TCE reduction was observed in the 5 mM sulfate cultures, although the H2 levels (6.0 nM) were above the threshold of dechlorination by day 8.
FIG 4.
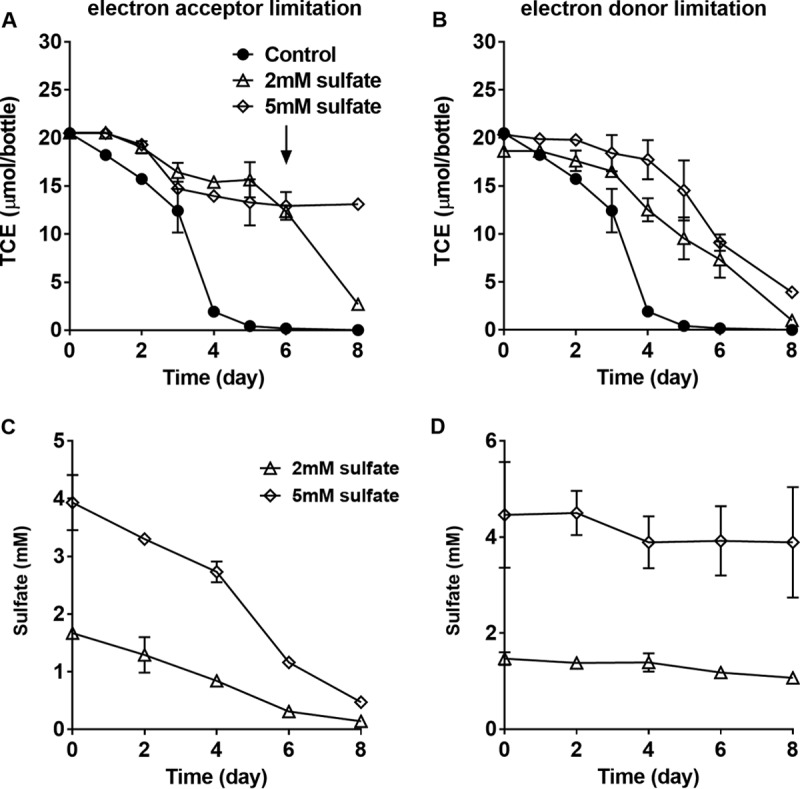
(A and B) TCE dechlorination profile of enrichment culture ANAS with different initial sulfate concentrations: with 20 mM lactate amendment (A) and with H2 amendment (B) as an electron donor. (C and D) Sulfate consumption during the experiment: with 20 mM lactate amendment (C) and with H2 amendment (D) as an electron donor. Error bars indicate standard deviations of biological triplicates. The arrow indicates 20 mM lactate amendment to the culture on day 6.
Under electron donor limitation, micromolar levels of H2 (∼0.15 μM) were intermittently added to maintain the low H2 concentrations expected. Dechlorination rates were the same with sulfate amendments as in the control (Fig. 4B) with cis-DCE (81.7% ± 3.3% with 5 mM sulfate, 90.8% ± 1.5% with 2 mM sulfate) as the main reduction product on day 8 (data not shown). Sulfate reduction rates decreased significantly to 3.7 ± 0.2 μmol day−1 (at 2 mM sulfate) and 7.0 ± 1.3 μmol day−1 (at 5 mM sulfate) compared to the electron acceptor limitation at 15.3 ± 1.1 μmol day−1 (with 2 mM sulfate) and 43.3 ± 0.8 μmol day−1 (with 5 mM sulfate). Methane production occurred in the control (data not shown) but was not observed in the enrichment with sulfate amendments within the experimental period (8 days) due to the low aqueous H2 concentrations (<100 nM), an observation consistent with previous research (36).
Transcriptomic study of strain 195 with sulfide inhibition.
The effects of sulfide addition on D. mccartyi 195 gene expression were studied in order to better understand the inhibition mechanism. Sulfide (10 mM) was amended to strain 195 at the mid-log phase of growth on day 4. Cell samples were collected after 48 h of additional incubation (day 6), when control bottles (without sulfide amendment) reached the late exponential growth phase. TCE dechlorination rates were lower in the sulfide-amended bottles than in the controls (0.3 versus 1.3 mmol/liters/day). Transcriptomic analysis showed that 115 genes were significantly downregulated, whereas 207 genes were significantly upregulated (a ≥2-fold change) in the presence of sulfide.
The short-term exposure to 10 mM sulfide did not change the expression pattern of genes encoding dehalogenases (see Table S3 in the supplemental material). However, the downregulated genes include Hym [Fe]-hydrogenase (DET0146 to DET0148), ATP synthase (DET0558 to DET0565), and genes related to biosynthesis (see Table S3 in the supplemental material). Upregulated gene expressions were observed in a subset of genes encoding ferrous iron transport protein (DET0095 to DET0097) and phosphate ABC transporters (DET0138 to DET0142) and genes related to nitrogen regulation and transport (DET1124 to DET1125).
DISCUSSION
Inconsistent results for the effects of sulfate inhibition on the performance of dechlorination enrichment cultures have been reported in the literature. El Mamouni et al. (31) reported that 10 mM sulfate addition to soil had no significant effect on TCE dechlorination by indigenous microorganisms, whereas higher sulfate concentrations (15 and 20 mM) yielded slower dechlorination. Heimann et al. (30) reported that 2.5 mM sulfate inhibited dechlorination by a mixed anaerobic culture by reducing the H2 supply to low nanomolar H2. Conversely, sulfate did not affect dechlorination when rapid fermentation of lactate resulted in accumulation of hydrogen to levels >100 nM. Aulenta et al. (27) reported that 3.7 mM sulfate adversely affected the rate of reductive dechlorination of an enriched dechlorinating community. These inconsistent findings make it difficult to understand potential mechanisms of sulfate inhibition and complicate the interpretation of bioremediation field data.
Inhibition mechanism.
This study demonstrates that sulfide rather than sulfate exhibits inhibitory effects on the dechlorination and growth of D. mccartyi, the fermenting bacterium S. wolfei, and the sulfate-reducing bacterium DvH. The cell yield of strain 195 decreased significantly at high sulfide concentrations (5 mM), whereas TCE dechlorination slowed, indicating that D. mccartyi decoupled growth from dechlorination when sulfide was introduced to the system at moderate to high concentrations. This result agrees with a previous study showing that sulfate did not inhibit D. mccartyi FL2 at high concentrations (10 mM) (21). Sulfide exerts inhibitory effects on a variety of cultures with different thresholds (21, 38–42). A previous study reported that insoluble metal sulfide formation from mg/liter concentrations of heavy metals deactivated sulfate-reducing bacteria by acting as a physical barrier to the cells (35). At remediation sites, sulfate reduction can overlap with iron reduction, which can lead to precipitated iron sulfide. The biogenic iron sulfide may reduce part of the sulfide toxicity and also perform abiotic chloroethene degradation when electron donor is in excess (43, 44). The overall TCE remediation may benefit from the resulting iron sulfide formation. However, this abiotic chloroethene degradation process was not investigated in this study. In our experimental setup, D. mccartyi was grown as planktonic cells, and trace metals were supplied at micromolar levels, below those needed to generate significant insoluble precipitation. In addition, our results demonstrated that sulfide inhibition is reversible, similar to the study conducted by Samhan-Arias et al. (45), indicating that inhibition was not due to a trace metal deficiency. In the pH range used in these experiments (ca. 7.0 to 7.3), H2S and HS− each count for half of the sulfide present in the culture (38). In some organisms, the toxicity of H2S has been attributed to its ability to inhibit cytochrome c-oxidase in a similar manner to hydrogen cyanide that prevents cellular respiration and inhibits the activity of a number of metal-containing enzymes by forming complex bonds with metals (46). Although D. mccartyi genomes do not encode cytochromes, they do encode many metal-containing enzymes, including the critical reductive dehalogenases. The downregulated genes for ATP synthase, biosynthesis, and Hym hydrogenase during sulfide inhibition agree with the physiological observation of lower cell yields and reduced dechlorination rates in strain 195. A similar gene expression pattern of membrane-bound electron transferring complexes was observed in a previous transcriptomic study of DvH during inhibition by nitrate-reducing bacteria (47). In contrast, sulfide inhibition of metal-containing enzymes resulted in the upregulated expression of genes encoding metal-containing enzymes involved in energy metabolism. The added counter ion sodium (in the form of sodium sulfide) was unlikely be inhibitory to the growth of strain 195, because in the sulfate inhibition experiment, we tested up to 10 mM sodium sulfate and did not observe any inhibitory effect on the growth or the dechlorination performance of strain 195.
Sulfate effects on dechlorination during electron acceptor limitation.
Faster growth kinetics of sulfate-reducing bacteria under electron acceptor (TCE) limitation caused sulfide accumulation that inhibits the growth of D. mccartyi at high initial sulfate concentrations (5 mM). The results from the DvH/195 coculture with lactate and sulfate addition showed that 5 mM initial sulfate inhibited both dechlorination and the growth of strain 195. The same inhibition effect on dechlorination was also observed when DvH/195 was supplied with excess electron donor and in the ANAS enrichment culture supplied with lactate. This inhibition is consistent with previous observations that 5 mM sulfide inhibited dehalogenation in soil-free microcosms (29) and a recent field-scale enhanced reductive dechlorination study that also showed that reductive dechlorination was negatively impacted by a sulfate concentration >5 mM when ethanol was supplied in excess as an electron donor (15). Another recent field-scale study showed that sulfate at <2 mM did not inhibit reductive dechlorination in hyporheic zones (16), an observation that agrees with our observation that a low sulfate concentration (2 mM) did not inhibit DvH/195 or ANAS enrichment. No methane production was observed in sulfate-supplied ANAS under electron acceptor limitation due to the low H2 production (∼10 nM) compared to the control (∼100 nM). Sulfate-reducing bacteria outcompete methanogens for H2, even at 2 mM initial sulfate.
Sulfate effect on dechlorination under electron donor limitation.
The dechlorination rate and cell yield of strain 195 were little affected at the tested sulfate concentrations (ca. 2 to 5 mM) in the triculture under electron donor limitation (Fig. 1D). The sulfate reduction rate by DvH was slower than that for electron acceptor limitation (Fig. 1C and Table 1) due to the competition with strain 195 for H2, which agrees with a previous report using an enrichment culture growing at electron donor-limiting condition (25). Reductive dechlorination accounted for ca. 36 to 38% of consumed electrons, whereas sulfate reduction accounted for ca. 62 to 64% of consumed electrons. Similar observations were made using sediment slurries, in which the presence of PCE accounted for approximately 50% of the reducing equivalents with the remainder directed to sulfate reduction (48). In contrast, in cases where dehalogenation was inhibited (Table 1), hydrogen was provided in excess to the system to avoid hydrogen competition between sulfate reduction and dehalogenation. Therefore, accumulated sulfide was the reason for the inhibited dehalogenation.
The long-term maintenance of triculture S. wolfei/DvH/195 consortia under electron donor-limiting conditions showed that with the same initial cell inoculation of DvH and 195, 195 became dominant after several subculturing events, demonstrating D. mccartyi outcompetes DvH for available H2 even in sulfate-rich (5 mM) environments. This finding is consistent with a recent study using a butyrate-fed dechlorinating enrichment culture in which D. mccartyi became the predominant species regardless of the sulfate concentrations (0.6 to 11.2 mM) (32). Further, the H2 utilization half-velocity coefficient for sulfate reduction (KS-H2, sul) was reported to be 0.2 to 2.4 μM (49–51), whereas for reductive dechlorination the reported value (KS-H2, dechlorination) is in the range of 2 to 7 nM (50, 51), indicating that reductive dechlorination has a much higher H2 affinity and can outcompete sulfate reduction at lower hydrogen concentrations. Scanning electron microscopy photos of the triculture (see Fig. S7 in the supplemental material) growing on butyrate (electron donor limitation) show cell aggregate formation between the fermenting syntrophs and the H2-consuming bacteria, demonstrating that the cells tend to form a physical proximity under the syntrophic condition under electron donor limitation. This result is similar to a previous study showing that cell aggregates formed between S. wolfei and strain 195 during syntrophic growth (52).
The observed dechlorination rate of the sulfate-supplied ANAS enrichment culture under electron donor limitation was similar to that for the control group (no sulfate added). This agrees with the observations made using constructed consortia that reductive dechlorination rates are not affected by sulfate amendment (2 and 5 mM) under electron donor limitation. Also, the higher abundance of D. mccartyi (>30%) in the enrichment culture compared to the co- and tricultures may be another reason that dechlorination outcompetes sulfate reduction (53).
Cell ratio effect.
Few studies have examined the effect of cell ratios of sulfate reducers to dechlorinators on dechlorination performance (54). In sulfate-rich environments, sulfate reducers may be the dominant species compared to D. mccartyi, resulting in competition for H2. We showed here that different initial cell ratios of sulfate-reducing bacteria to D. mccartyi (from 0.3 to 3.0) resulted in no significant differences in dechlorination profiles or cell growth (Table 1), demonstrating that initial cell ratios are not the most critical factor for controlling the inhibition of reductive dechlorination in sulfate-reducing environments.
In conclusion, sulfide instead of sulfate is responsible for the inhibitory effects on dechlorination and growth by D. mccartyi. Under electron acceptor-limited conditions, sulfate concentrations are the key factor that determines the extent of dechlorination, with high sulfate concentrations exhibiting inhibition due to the toxicity of the sulfate reduction product sulfide. Under electron donor-limited conditions, D. mccartyi can successfully dechlorinate in anaerobic microbial communities regardless of sulfate concentrations, demonstrating the ability of D. mccartyi to effectively compete against other hydrogen-consuming bacteria. The inhibitory concentrations of sulfide on Dehalococcoides strains could be incorporated to current kinetic modeling in order to better predict the reductive dechlorination process in a sulfate-reducing environment during bioremediation practice.
MATERIALS AND METHODS
Bacterial cultures and growth conditions.
Dehalococcoides mccartyi strain 195 (strain 195) was grown in defined medium with an H2/CO2 (90:10) headspace, 0.6 mM TCE as an electron acceptor, and 2 mM acetate as a carbon source (6) (Fig. 1A). Desulfovibrio vulgaris Hildenborough (DvH) was grown in the same defined medium with an N2/CO2 headspace, 10 mM lactate as an electron donor, and 5 mM sulfate as an electron acceptor. Syntrophomonas wolfei was grown on crotonate in 160-ml serum bottles as described previously (55). Bacterial cocultures of S. wolfei and strain 195 (S. wolfei and strain 195 were each inoculated at 5% of the total liquid volume) were sustainably maintained on 5 mM butyric acid (5% [vol/vol] inoculation) with 0.6 mM TCE as described previously (52). Bacterial cocultures of DvH and strain 195 (DvH/195, 5% [vol/vol] inoculation) were sustainably maintained on 5 mM lactic acid (5% [vol/vol] inoculation) with 0.6 mM TCE as described previously (34). The methanogenic dechlorinating community ANAS was previously enriched from contaminated soil obtained from the Alameda Naval Air Station (CA). The culture has been maintained in the laboratory for more than 15 years in a continuously stirred semibatch fed reactor, and its community structure and dechlorination performance have been previously described (53, 56, 57).
In order to study the competition between reductive dechlorination and sulfate reduction under electron acceptor limitation (electron donor in excess), strain 195 and DvH were grown in defined medium with an H2/CO2 headspace (Fig. 1C) with 0.7 mM TCE and 2 or 5 mM sulfate (see Table S2 in the supplemental material). For electron donor limitation experiments, tricultures containing S. wolfei/DvH/195 were constructed in defined medium (see Table S2 in the supplemental material) with 7.0 mM butyric acid, 0.7 mM TCE and 2 mM (or 5 mM) sulfate and N2/CO2 (80:20, vol/vol) headspace (Fig. 1D). For both electron donor- and acceptor-limiting conditions, TCE (0.7 mM per dose) was amended to the cultures when the previous dose was depleted. All experiments were performed in triplicate. After three subculturing events (5% [vol/vol] inoculation), S. wolfei/DvH/195 triculture cells were harvested during late exponential phase (day 6) and analyzed by scanning electron microscopy as described previously (52).
Chemical analysis.
Chloroethenes and ethene were measured by using an FID-gas chromatograph with 100-μl headspace samples, and hydrogen and carbon monoxide were measured by RGD-gas chromatography with 300-μl headspace sample as described previously (56, 58). The mass of each compound was calculated based on gas-liquid equilibrium by using Henry's law constants at 34°C. Organic acids, including butyrate and acetate, were analyzed by high-performance liquid chromatography as described previously (56). Sulfate concentration was measured by suppressed ion chromatography (Dionex ICS 1100) on a Dionex IonPac AERS500 column (4 mm) with 4.5 mM Na2CO3 and 0.8 mM NaHCO3 as the eluent. The sulfide concentration was measured at the end of the experiments by the methylene blue method (59). Trace metal concentrations were analyzed on an Agilent Technologies 7700 series ICP-MS (60).
DNA extraction and cell number quantification.
Liquid samples (1.5 ml) were collected during the incubation for cell density measurements, and the cells were harvested by centrifugation (21,000 × g, 10 min at 4°C). Genomic DNA was extracted from the cell pellets by using a Qiagen DNeasy blood and tissue kit according to the manufacturer's instructions for Gram-positive bacteria. Quantitative PCR using SYBR green-based detection reagents was used to quantify the gene copy numbers for each bacterium with S. wolfei 16S rRNA gene primers (forward, 5′-GTATCGACCCCTTCTGTGCC-3′; reverse, 5′-CCCCAGGCGGGATACTTATT-3′) (61), DvH 16S rRNA gene primers (forward, 5′-AATCGGAATCACTGGGCGTA-3′; reverse, 5′-CCCTGACTTACCAAGCAGCC-3′) (34), and D. mccartyi tceA gene primers (forward, 5′-ATCCAGATTATGACCCTGGTGAA-3′; reverse, 5′-GCGGCATATATTAGGGCATCTT-3′), as previously described (62). Cell number calculation was normalized based on the target gene copy numbers in each genome of the bacterium.
RNA preparation and transcriptome analysis.
A 100 mM stock solution of sulfide-S (the sum of all speciations of H2S-S, HS−-S, and S2−-S) was prepared from Na2S·9H2O in the defined culture medium. Sulfide-S at 10 mM was amended to strain 195 cultures on day 4 during the mid-log growth phase when 50% of TCE was degraded. Cultures were sampled on day 6 when control bottles exhibited late exponential growth (around 75% of 78 μmol of TCE was dechlorinated). In order to collect sufficient material for transcriptomic microarray analysis, 60 bottles of sulfide-S-amended strain 195 cultures and 18 bottles of control (strain 195, no sulfide-S addition) were inoculated and grown from triplicate bottles of the isolate. For each biological triplicate, the cells from 20 bottles were collected by vacuum filtration on day 6 for the experimental group and the control (300-ml culture per filter, 0.2-μm-pore-size autoclaved GVWP filter [Durapore membrane; Millipore, Billerica, MA]). Each filter was placed in a 2-ml orange-cap microcentrifuge tube, frozen with liquid nitrogen, and stored at −80°C until further processing was performed. RNA extraction and preparation were described previously (52).
Transcriptomic microarray analysis.
The Affymetrix GeneChip microarray used in this study has been described previously (63). Briefly, the chip contains 4,744 probe sets that represent more than 98% of the open reading frames from four published Dehalococcoides genomes (strain 195, VS, BAV1, and CBDB1). cDNA was synthesized from 9 μg of RNA, and then each cDNA sample was fragmented, labeled, and hybridized to each array. All procedures were performed with minimal modifications to the protocols in section 3 of the GeneChip Expression Analysis Technical Manual (Affymetrix, Santa Clara, CA). The microarray data analysis methods were as described previously (34, 57).
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by NIEHS (P42-ES04705-14) and NSF (CBET-1336709) research grants.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.03384-16.
REFERENCES
- 1.Bhatt P, Kumar MS, Mudliar S, Chakrabarti T. 2007. Biodegradation of chlorinated compounds: a review. Crit Rev Environ Sci Technol 37:165–198. doi: 10.1080/10643380600776130. [DOI] [Google Scholar]
- 2.U.S. Environmental Protection Agency. 2011. IRIS toxicological review of trichloroethylene. U.S. Environmental Protection Agency, Washington, DC: https://cfpub.epa.gov/ncea/iris_drafts/recordisplay.cfm?deid=237625. [Google Scholar]
- 3.Moran MJ, Zogorski JS, Squillace PJ. 2007. Chlorinated solvents in groundwater of the United States. Environ Sci Technol 41:74–81. doi: 10.1021/es061553y. [DOI] [PubMed] [Google Scholar]
- 4.Pandey J, Chauhan A, Jain RK. 2009. Integrative approaches for assessing the ecological sustainability of in situ bioremediation. FEMS Microbiol Rev 33:324–375. doi: 10.1111/j.1574-6976.2008.00133.x. [DOI] [PubMed] [Google Scholar]
- 5.Maymó-Gatell X, Anguish T, Zinder SH. 1999. Reductive dechlorination of chlorinated ethenes and 1,2-dichloroethane by “Dehalococcoides ethenogenes” 195. Appl Environ Microbiol 65:3108–3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He J, Holmes VF, Lee PK, Alvarez-Cohen L. 2007. Influence of vitamin B-12 and cocultures on the growth of Dehalococcoides isolates in defined medium. Appl Environ Microbiol 73:2847–2853. doi: 10.1128/AEM.02574-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yi S, Seth EC, Men YJ, Stabler SP, Allen RH, Alvarez-Cohen L, Taga ME. 2012. Versatility in corrinoid salvaging and remodeling pathways supports corrinoid-dependent metabolism in Dehalococcoides mccartyi. Appl Environ Microbiol 78:7745–7752. doi: 10.1128/AEM.02150-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradley PM, Chapelle FH. 2010. Biodegradation of chlorinated ethenes, p 39–67. In Stroo HF, Ward CH (ed), In situ remediation of chlorinated solvent plumes. SERDP/ESTCP Environmental Remediation Technology. Springer, Heidelberg, Germany. [Google Scholar]
- 9.Yu SH, Semprini L. 2002. Comparison of trichloroethylene reductive dehalogenation by microbial communities stimulated on silicon-based organic compounds as slow-release anaerobic substrates. Water Res 36:4985–4996. doi: 10.1016/S0043-1354(02)00222-1. [DOI] [PubMed] [Google Scholar]
- 10.Ma X, Novak PJ, Clapp LW, Semmens MJ, Hozalski RM. 2003. Evaluation of polyethylene hollow-fiber membranes for hydrogen delivery to support reductive dechlorination in a soil column. Water Res 37:2905–2918. doi: 10.1016/S0043-1354(03)00111-8. [DOI] [PubMed] [Google Scholar]
- 11.Lee IS, Bae JH, Yang Y, McCarty PL. 2004. Simulated and experimental evaluation of factors affecting the rate and extent of reductive dehalogenation of chloroethenes with glucose. J Contam Hydrol 74:313–331. doi: 10.1016/j.jconhyd.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 12.Bagley DM, Gossett JM. 1990. Tetrachloroethene transformation to trichloroethene and cis-1,2-dichloroethene by sulfate-reducing enrichment cultures. Appl Environ Microbiol 56:2511–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee MD. 1998. New perspectives on microbial dehalogenation of chlorinated solvents: insights from the field. Annu Rev Microbiol 52:423–452. doi: 10.1146/annurev.micro.52.1.423. [DOI] [PubMed] [Google Scholar]
- 14.Miao Z, Brusseau ML, Carroll KC, Carreón-Diazconti C, Johnson B. 2012. Sulfate reduction in groundwater: characterization and applications for remediation. Environ Geochem Health 34:539–550. doi: 10.1007/s10653-011-9423-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pérez-de-Mora A, Zila A, McMaster ML, Edwards EA. 2014. Bioremediation of chlorinated ethenes in fractured bedrock and associated changes in dechlorinating and nondechlorinating microbial populations. Environ Sci Technol 48:5770–5779. doi: 10.1021/es404122y. [DOI] [PubMed] [Google Scholar]
- 16.Freitas JG, Rivett MO, Roche RS, Durrant M, Walker C, Tellam JH. 2015. Heterogeneous hyporheic zone dechlorination of a TCE groundwater plume discharging to an urban river reach. Sci Total Environ 505:236–252. doi: 10.1016/j.scitotenv.2014.09.083. [DOI] [PubMed] [Google Scholar]
- 17.Zanaroli G, Negroni A, Häggblom MM, Fava F. 2015. Microbial dehalogenation of organohalides in marine and estuarine environments. Curr Opin Biotechnol 33:287–295. doi: 10.1016/j.copbio.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 18.Chen Y, Cheng JJ, Creamer KS. 2008. Inhibition of anaerobic digestion process: a review. Bioresour Technol 99:4044–4064. doi: 10.1016/j.biortech.2007.01.057. [DOI] [PubMed] [Google Scholar]
- 19.Reis MAM, Almeida JS, Lemos PC, Carrondo MJT. 1992. Effect of hydrogen sulfide on growth of sulfate reducing bacteria. Biotechnol Bioeng 40:593–600. doi: 10.1002/bit.260400506. [DOI] [PubMed] [Google Scholar]
- 20.Joye SB, Hollibaugh JT. 1995. Influence of sulfide inhibition of nitrification on nitrogen regeneration in sediments. Science 270:623–625. doi: 10.1126/science.270.5236.623. [DOI] [Google Scholar]
- 21.He J, Sung Y, Krajmalnik-Brown R, Ritalahti KM, Löffler FE. 2005. Isolation and characterization of Dehalococcoides sp. strain FL2, a trichloroethene (TCE)- and 1,2-dichloroethene-respiring anaerobe. Environ Microbiol 7:1442–1450. doi: 10.1111/j.1462-2920.2005.00830.x. [DOI] [PubMed] [Google Scholar]
- 22.Morse JJ, Alleman BC, Gossett JM, Zinder SH, Fennell DE, Sewell GW. 1997. A treatability test for evaluating the potential applicability of the Reductive Anaerobic Biological In Situ Treatment Technology (RABITT) to remediate chloroethenes: a draft technical protocol developed for the Environmental Security Technology Certification Program, Department of Defense. U.S. Government Printing Office, Washington, DC. [Google Scholar]
- 23.Aulenta F, Majone M, Tandoi V. 2006. Enhanced anaerobic bioremediation of chlorinated solvents: environmental factors influencing microbial activity and their relevance under field conditions. J Chem Technol Biotechnol 81:1463–1474. doi: 10.1002/jctb.1567. [DOI] [Google Scholar]
- 24.Fennell DE, Gossett JM. 2003. Microcosms for site-specific evaluation of enhanced biological reductive dehalogenation, p 385–420. In Häggblom MM, Bossert ID (ed), Dehalogenation: microbial processes and environmental applications. Kluwer Academic, Boston, MA. [Google Scholar]
- 25.Aulenta F, Pera A, Rossetti S, Papini MP, Majone M. 2007. Relevance of side reactions in anaerobic reductive dechlorination microcosms amended with different electron donors. Water Res 41:27–38. doi: 10.1016/j.watres.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 26.Pantazidou M, Panagiotakis I, Mamais D, Zikidi V. 2012. Chloroethene biotransformation in the presence of different sulfate concentrations. Ground Water Monit Remediation 32:106–119. doi: 10.1111/j.1745-6592.2011.01374.x. [DOI] [Google Scholar]
- 27.Aulenta F, Beccari M, Majone M, Papini MP, Tandoi V. 2008. Competition for H2 between sulfate reduction and dechlorination in butyrate-fed anaerobic cultures. Process Biochem 43:161–168. doi: 10.1016/j.procbio.2007.11.006. [DOI] [Google Scholar]
- 28.Berggren DRV, Marshall IPG, Azizian MF, Spormann AM, Semprini L. 2013. Effects of sulfate reduction on the bacterial community and kinetic parameters of a dechlorinating culture under chemostat growth conditions. Environ Sci Technol 47:1879–1886. doi: 10.1021/es304244z. [DOI] [PubMed] [Google Scholar]
- 29.Hoelen TP, Reinhard M. 2004. Complete biological dehalogenation of chlorinated ethylenes in sulfate containing groundwater. Biodegradation 15:395–403. doi: 10.1023/B:BIOD.0000044592.33729.d6. [DOI] [PubMed] [Google Scholar]
- 30.Heimann AC, Friis AK, Jakobsen R. 2005. Effects of sulfate on anaerobic chloroethene degradation by an enriched culture under transient and steady-state hydrogen supply. Water Res 39:3579–3586. doi: 10.1016/j.watres.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 31.El Mamouni R, Jacquet R, Gerin P, Agathos SN. 2002. Influence of electron donors and acceptors on the bioremediation of soil contaminated with trichloroethene and nickel: laboratory- and pilot-scale study. Water Sci Technol 45:49–54. [PubMed] [Google Scholar]
- 32.Panagiotakis I, Mamais D, Pantazidou M, Rossetti S, Aulenta F, Tandor V. 2014. Predominance of Dehalococcoides in the presence of different sulfate concentrations. Water Air Soil Pollut 225:1785–1799. doi: 10.1007/s11270-013-1785-9. [DOI] [Google Scholar]
- 33.Atashgahi S, Lu Y, Zheng Y, Saccenti E, Suarez-Diez M, Ramiro-Garcia J, Eisenmann H, Elsner M, Stams JMA, Springael D, Dejonghe W, Smidt H. 6 October 2016. Geochemical and microbial community determinants of reductive dechlorination at a site biostimulated with glycerol. Environ Microbiol doi: 10.1111/1462-2920.13531. [DOI] [PubMed] [Google Scholar]
- 34.Men Y, Feil H, VerBerkmoes NC, Shah MB, Johnson DR, Lee PK, West KA, Zinder SH, Andersen GL, Alvarez-Cohen L. 2011. Sustainable syntrophic growth of Dehalococcoides ethenogenes strain 195 with Desulfovibrio vulgaris Hildenborough and Methanobacterium congolense: global transcriptomic and proteomic analyses. ISME J 6:410–421. doi: 10.1038/ismej.2011.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Utgikar VP, Harmon SM, Chaudhary N, Tabak HH, Govind R, Haines JR. 2002. Inhibition of sulfate-reducing bacteria by metal sulfide formation in bioremediation of acid mine drainage. Environ Toxicol 17:40–48. doi: 10.1002/tox.10031. [DOI] [PubMed] [Google Scholar]
- 36.Yang YR, McCarty PL. 1998. Competition for hydrogen within a chlorinated solvent dehalogenating anaerobic mixed culture. Environ Sci Technol 32:3591–3597. doi: 10.1021/es980363n. [DOI] [Google Scholar]
- 37.Lovley DR, Goodwin S. 1988. Hydrogen concentrations as an indicator of the predominant terminal electron-accepting reactions in aquatic sediments. Geochim Cosmochim Acta 52:2993–3003. doi: 10.1016/0016-7037(88)90163-9. [DOI] [Google Scholar]
- 38.Karhadkar PP, Audic J-M, Faup GM, Khanna P. 1987. Sulfide and sulfate inhibition of methanogenesis. Water Res 21:1061–1066. doi: 10.1016/0043-1354(87)90027-3. [DOI] [Google Scholar]
- 39.Okabe S, Nielsen PH, Jones WL, Characklis WG. 1995. Sulfide product inhibition of Desulfovibrio desulfuricans in batch and continuous cultures. Water Res 29:571–578. doi: 10.1016/0043-1354(94)00177-9. [DOI] [Google Scholar]
- 40.Magnuson JK, Stern RV, Gossett JM, Zinder SH, Burris DR. 1998. Reductive dechlorination of tetrachloroethene to ethene by a two-component enzyme pathway. Appl Environ Microbiol 64:1270–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin RC, Yang GF, Zhang QQ, Ma C, Yu JJ, Xing BS. 2013. The effect of sulfide inhibition on the anammox process. Water Res 47:1459–1469. doi: 10.1016/j.watres.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 42.Zhou Z, Xing C, An Y, Hu D, Qiao W, Wang L. 2014. Inhibitory effects of sulfide on nitrifying biomass in the anaerobic-anoxic-aerobic wastewater treatment process. J Chem Technol Biotechnol 89:214–219. doi: 10.1002/jctb.4104. [DOI] [Google Scholar]
- 43.He YT, Wilson JT, Su C, Wilkin RT. 2015. Review of abiotic degradation of chlorinated solvents by reactive iron minerals in aquifers. Ground Water Monit Remediation 35:57–75. [Google Scholar]
- 44.Hyun SP, Hayes KF. 2015. Abiotic reductive dechlorination of cis-DCE by ferrous monosulfide mackinawite. Environ Sci Pollut Res 22:16463–16474. doi: 10.1007/s11356-015-5033-2. [DOI] [PubMed] [Google Scholar]
- 45.Samhan-Arias AK, Garcia-Bereguain MA, Gutierrez-Merino C. 2009. Hydrogen sulfide is a reversible inhibitor of the NADH oxidase activity of synaptic plasma membranes. Biochem Biophys Res Commun 388:718–722. doi: 10.1016/j.bbrc.2009.08.076. [DOI] [PubMed] [Google Scholar]
- 46.Truong DH, Eghbal MA, Hindmarsh W, Roth SH, O'Brien PJ. 2006. Molecular mechanisms of hydrogen sulfide toxicity. Drug Metab Rev 38:733–744. doi: 10.1080/03602530600959607. [DOI] [PubMed] [Google Scholar]
- 47.Haveman SA, Greene EA, Voordouw G. 2005. Gene expression analysis of the mechanism of inhibition of Desulfovibrio vulgaris Hildenborough by nitrate-reducing, sulfide-oxidizing bacteria. Environ Microbiol 7:1461–1465. doi: 10.1111/j.1462-2920.2005.00834.x. [DOI] [PubMed] [Google Scholar]
- 48.Mazure CS, Jones WJ. 2001. Hydrogen concentrations in sulfate-reducing estuarine sediments during PCE dehalogenation. Environ Sci Technol 35:4783–4788. doi: 10.1021/es0110372. [DOI] [PubMed] [Google Scholar]
- 49.Sonne-Hansen J, Westermann P, Ahring BK. 1999. Kinetics of sulfate and hydrogen uptake by the thermophilic sulfate-reducing bacteria Thermodesulfobacterium sp. strain JSP and Thermodesulfovibrio sp. strain R1Ha3. Appl Environ Microbiol 65:1304–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kouznetsova I, Mao X, Robinson C, Barry DA, Gerhard JI, McCarty PL. 2010. Biological reduction of chlorinated solvents: batch-scale geochemical modeling. Adv Water Res 33:969–986. doi: 10.1016/j.advwatres.2010.04.017. [DOI] [Google Scholar]
- 51.Cupples AM, Spormann AM, McCarty PL. 2003. Growth of a Dehalococcoides-like microorganism on vinyl chloride and cis-dichloroethene as electron acceptors as determined by competitive PCR. Appl Environ Microbiol 69:953–959. doi: 10.1128/AEM.69.2.953-959.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mao X, Stenuit B, Polasko A, Alvarez-Cohen L. 2015. Efficient metabolic exchange and electron transfer within a syntrophic trichloroethene-degrading coculture of Dehalococcoides mccartyi 195 and Syntrophomonas wolfei. Appl Environ Microbiol 81:2015–2024. doi: 10.1128/AEM.03464-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brisson VL, West KA, Lee PK, Tringe SG, Brodie EL, Alvarez-Cohen L. 2012. Metagenomic analysis of a stable trichloroethene-degrading microbial community. ISME J 6:1702–1714. doi: 10.1038/ismej.2012.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Drzyzga O, Gerritse J, Dijk JA, Elissen H, Gottschal JC. 2001. Coexistence of a sulphate-reducing Desulfovibrio species and the dehalorespiring Desulfitobacterium frappieri TCE1 in defined chemostat cultures grown with various combinations of sulphate and tetrachloroethene. Environ Microbiol 3:92–99. doi: 10.1046/j.1462-2920.2001.00157.x. [DOI] [PubMed] [Google Scholar]
- 55.Beaty PS, McInerney MJ. 1987. Growth of Syntrophomonas wolfei in pure culture on crotonate. Arch Microbiol 147:389–393. doi: 10.1007/BF00406138. [DOI] [Google Scholar]
- 56.Lee PKH, Johnson DR, Holmes VF, He J, Alvarez-Cohen L. 2006. Reductive dehalogenase gene expression as a biomarker for physiological activity of Dehalococcoides spp. Appl Environ Microbiol 72:6161–6168. doi: 10.1128/AEM.01070-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.West KA, Lee PKH, Johnson DR, Zinder SH, Alvarez-Cohen L. 2013. Global gene expression of Dehalococcoides within a robust dynamic TCE-dechlorinating community under conditions of periodic substrate supply. Biotechnol Bioeng 110:1333–1341. doi: 10.1002/bit.24819. [DOI] [PubMed] [Google Scholar]
- 58.Freeborn RA, West KA, Bhupathiraju VK, Chauhan S, Rahm BG, Richardson RE, Alvarez-Cohen L. 2005. Phylogenetic analysis of TCE-dechlorinating consortia enriched on a variety of electron donors. Environ Sci Technol 39:8358–8368. doi: 10.1021/es048003p. [DOI] [PubMed] [Google Scholar]
- 59.Cline JD. 1969. Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol Oceanogr 14:454–458. doi: 10.4319/lo.1969.14.3.0454. [DOI] [Google Scholar]
- 60.Brisson VL, Zhuang WQ, Alvarez-Cohen L. 2016. Bioleaching of rare earth elements from monazite sand. Biotechnol Bioeng 113:339–348. doi: 10.1002/bit.25823. [DOI] [PubMed] [Google Scholar]
- 61.Sieber JR, Sims DR, Han C, Kim E, Lykidis A, Lapidus AL, McDonnald E, Rohlin L, Culley DE, Gunsalus R, McInerney MJ. 2010. The genome of Syntrophomonas wolfei: new insights into syntrophic metabolism and biohydrogen production. Environ Microbiol 12:2289–2301. doi: 10.1111/j.1462-2920.2010.02237.x. [DOI] [PubMed] [Google Scholar]
- 62.Johnson DR, Lee PKH, Holmes VF, Alvarez-Cohen L. 2005. An internal reference technique for accurately quantifying specific mRNAs by real-time PCR with application to the tceA reductive dehalogenase gene. Appl Environ Microbiol 71:3866–3871. doi: 10.1128/AEM.71.7.3866-3871.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee PKH, Cheng D, Hu P, West KA, Dick GJ, Brodie EL, Andersen GL, Zinder SH, He J, Alvarez-Cohen L. 2011. Comparative genomics of two newly isolated Dehalococcoides strains and an enrichment using a genus microarray. ISME J 5:1014–1024. doi: 10.1038/ismej.2010.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.