Abstract
The study represents the genetic overview of non-polio enteroviruses (NPEV) isolated from acute flaccid paralysis (AFP) cases in Yunnan Province from 2006 to 2010. Molecular typing based on VP1 nucleotide sequence was carried out on 98 NPEV isolates, and 33 serotypes were identified. EV-B was detected most frequently with an overall prevalence of 71.4%, followed by EV-A (18.4%) and EV-C (10.2%). No EV-D was identified. NPEV positive rate was higher in children <3 years of age and in summer and autumn months. Clinically, 68.4% patients presented with fever, and 16 cases (16.3%) were classified as Guillain-Barré syndrome, followed by myositis (13.3%). The phylogenetic analysis on the VP1 and 3D regions of prevalent serotypes provided evidence for recombination events among them. EV-A71, an important pathogen previously demonstrated to be associated with paralysis, had also been detected (n = 8) in this study and they all belonged to genotype C4. Great genetic divergence between Yunnan isolates and strains from other regions of the world was revealed. The findings of the study are of great importance for further research on molecular evolution of EV under the circumstance of no specialized EV surveillance system in China.
The genus Enterovirus within the family Picornaviridae consists of positive sense single stranded RNA viruses, including polioviruses (PV), coxsackieviruses A (CVA) and B (CVB), echoviruses (E), and the numbered enteroviruses (EVs)1,2. Based on phylogenetic relationships in multiple genome regions, the serotypes of human enteroviruses are classified into 7 species, EV-A to D and human rhinovirus (HRV)-A to C3,4. Each species comprises a large number of different serotypes, whose number is constantly increasing due to continual discovery of new serotypes5,6,7. EVs are transmitted through the fecal–oral route and infect millions of people worldwide each year, particularly children. Although usually associated with asymptomatic infections, EVs can cause severe diseases including acute flaccid paralysis (AFP), meningitis, encephalitis, and hand, foot and mouth disease (HFMD)8,9,10,11. Though PV is considered as the main cause of AFP, many non-polio AFP cases have been usually reported12,13,14.
Since the early 1980s, the polio incidence in China has dropped significantly due to widespread use of the oral polio vaccine (OPV). The last case of laboratory-confirmed wild-type poliovirus (WPV) occurred in Yunnan Province in 1993. In connection with the Global Eradication Programme of Poliomyelitis led by the World Health Organisation (WHO), there is ongoing surveillance of AFP in China, and the monitoring system has been fully developed in Yunnan since 1994. In addition to PV, non-polio enteroviruses (NPEV) have been frequently isolated through AFP surveillance. Previous study has described the serotypes and molecular epidemiology of NPEV in Yunnan in 1997–2000 and 200415. In this study, to understand the possible alteration in serotype spectrums in following years, especially under the circumstance of national HFMD epidemic since 2007, we investigate the NPEV isolates from AFP cases and present different epidemiological and clinical features associated with enterovirus infections in Yunnan Province from 2006 to 2010. Besides, we describe the phylogenetic relationships of the EV-B predominant serotypes based on VP1 and 3D regions, and analyze the recombination among these serotypes and other reference strains.
Results
Isolation
During the 5-year study from 2006 to 2010, PVs and NPEVs were isolated from 51 and 98 AFP cases, respectively. Of 51 PV positive cases, PVs could be isolated from the double specimens of 38 cases and from the single specimen of 13 cases. All PV positive samples produced cytopathic effect (CPE) both in human rhabdosarcoma (RD) and human poliovirus receptor-CD155 expressing recombinant murine (L20B) cells. These isolates were identified as PV by neutralization test, and 42 strains were identified as single-type PV (PV1 = 8, PV2 = 23, PV3 = 11) and 9 strains were identified as mixed-type PV [PV(1 + 2) = 5, PV(1 + 3) = 2, PV(2 + 3) = 2]. PV isolates were forwarded to the Regional Reference Polio Laboratory. Fifty strains were confirmed to be Sabin-like strains, and another one (sample NO. 10-YN-2010AFP) was confirmed to be vaccine-derived poliovirus (VDPV) with 9 VP1 nucleotide substitutions. The comparison of the nucleotide/amino acid mutations between local VDPV strain (10-YN-2010AFP) and Sabin 2 has been provided in Supplementary Table S1. The VDPV case was a 1 year old boy with no inoculation history of OPV. On 60-day follow up after the onset of paralysis, the VDPV cases still presented residual paralysis. Further investigation revealed no secondary cases of VDPV from the close contacts and other healthy children in local area.
Of 98 NPEV positive cases, NPEVs could be isolated from the double specimens of 86 cases and from the single specimen of 12 cases. All samples from these 98 cases produced CPE only in RD cells while not in L20B cells. The isolation rate of NPEV was 8.9% (98/1097) during the 5 years. VP1 RT-PCR and sequencing were performed and molecular typing was conducted. The sequences of 98 strains were all successfully amplified by using 6 pairs of primer designed for EV-A to C. So the primers directed towards EV-D have not been used. The results revealed that the 98 NPEVs belong to 33 serotypes (Table 1): 18 isolates (18/98, 18.4%) were classified into 7 serotypes of EV-A (including CVA: 8/98, 8.2%; numbered EVs: 10/98, 10.2%), 70 isolates (70/98, 71.4%) into 22 serotypes of EV-B (including E: 62/98, 63.3%; CVB: 4/98, 4.1%; and CVA9: 4/98, 4.1%), and 10 isolates (10/98, 10.2%) into 4 serotypes of EV-C (including CVA: 6/98, 6.1%; numbered EVs: 4/98, 4.1%). EV-B was considered the predominant species in Yunnan Province. No isolates belonged to EV-D species. E13 (9.2%), E6 (8.2%), E25 (8.2%), EV-A71 (7.6%) and E30 (6.2%) were the most frequently isolated serotypes. Twelve serotypes (CVA8, EV-A76, EV-A90, E2, E11, E12, E20, E24, E32, CVB2, CVA11 and CVA17) were detected only once.
Table 1. Serotypes and annual isolation of NPEVs in Yunnan Province, 2006–2010.
| Numbers of isolates | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Serotypes | 2006 | 2007 | 2008 | 2009 | 2010 | N | % | nt (aa) divergence with prototype strain (%) | nt (aa) divergence among Yunnan isolates (%) |
| EV-A | |||||||||
| EV-A71 | 0 | 2 | 3 | 0 | 3 | 8 | 8.2 | 16.8–18.2(8.4–9.8) | 0.1–5.7(0–2.8) |
| CVA2 | 2 | 0 | 1 | 0 | 0 | 3 | 3.1 | 18.2–19.6(7.7–9.4) | 8.4–19.5(5.6–10.3) |
| CVA5 | 0 | 2 | 0 | 0 | 0 | 2 | 2.0 | 18.5–18.7(3.8–4.6) | 2.3(2.3) |
| CVA8 | 1 | 0 | 0 | 0 | 0 | 1 | 1.0 | 15.2(1.6) | — |
| CVA10 | 1 | 1 | 0 | 0 | 0 | 2 | 2.0 | 21.0–23.3(4.5–5.2) | 4.5(0.7) |
| EV-A76 | 0 | 0 | 1 | 0 | 0 | 1 | 1.0 | 13.2(2.5) | — |
| EV-A90 | 1 | 0 | 0 | 0 | 0 | 1 | 1.0 | 9.1(1.1) | — |
| EV-B | |||||||||
| E13 | 2 | 2 | 1 | 2 | 2 | 9 | 9.2 | 23.1–24.0 (7.5–9.4) | 0–15(0–1.9) |
| E6 | 1 | 2 | 2 | 2 | 1 | 8 | 8.2 | 20.3–22.7(2.9–3.7) | 0–21.5(0–2.9) |
| E25 | 3 | 2 | 3 | 0 | 0 | 8 | 8.2 | 17.3–18.1(1.7–3.4) | 0–8.8(0–1.7) |
| E30 | 5 | 1 | 0 | 0 | 0 | 6 | 6.1 | 15.8–16.6(1.5–3.2) | 0–3.9(0–1.2) |
| E14 | 1 | 1 | 1 | 2 | 0 | 5 | 5.1 | 19.9–22.8(3.4–4.3) | 6.8–17.7(0–2.6) |
| E3 | 0 | 2 | 1 | 1 | 0 | 4 | 4.1 | 15.9–16.5(1.3–2.6) | 0.2–5.4(0.6–1.3) |
| CVA9 | 2 | 0 | 2 | 0 | 0 | 4 | 4.1 | 19.3–20.1(3.2) | 2.9–14.4(0–2.4) |
| E4 | 0 | 2 | 1 | 0 | 0 | 3 | 3.1 | 17.4–18.5(5.5–6.3) | 0–4.4(0–0.8) |
| E19 | 3 | 0 | 0 | 0 | 0 | 3 | 3.1 | 21.2 (6.4) | 0(0) |
| CVB3 | 1 | 0 | 1 | 1 | 0 | 3 | 3.1 | 20.1–20.7(2.7–3.1) | 5.8–7.7(0–0.4) |
| E7 | 1 | 0 | 0 | 1 | 0 | 2 | 2.0 | 18.6–20.4(4.1–5.3) | 5.3(2.0) |
| E15 | 0 | 2 | 0 | 0 | 0 | 2 | 2.0 | 22.3–24(9.5–13.1) | 17.2(4.4) |
| E16 | 0 | 2 | 0 | 0 | 0 | 2 | 2.0 | 18.1–18.3(3.5) | 4.7(0) |
| E21 | 2 | 0 | 0 | 0 | 0 | 2 | 2.0 | 17.6(2.1) | 0.1(0) |
| E29 | 2 | 0 | 0 | 0 | 0 | 2 | 2.0 | 21.2(4.7–5.5) | 1.8(2.4) |
| E2 | 1 | 0 | 0 | 0 | 0 | 1 | 1.0 | 23.2(8.2) | — |
| E11 | 0 | 0 | 1 | 0 | 0 | 1 | 1.0 | 20.7(8.8) | — |
| E12 | 0 | 0 | 1 | 0 | 0 | 1 | 1.0 | 19.1(3.1) | — |
| E20 | 1 | 0 | 0 | 0 | 0 | 1 | 1.0 | 20.1(2.8) | — |
| E24 | 0 | 0 | 0 | 0 | 1 | 1 | 1.0 | 18.4(3.6) | — |
| E32 | 1 | 0 | 0 | 0 | 0 | 1 | 1.0 | 19.8(8.9) | — |
| CVB2 | 1 | 0 | 0 | 0 | 0 | 1 | 1.0 | 17.1(2.8) | — |
| EV-C | |||||||||
| CVA24 | 2 | 0 | 2 | 0 | 0 | 4 | 4.1 | 18.9–21.5(4.3–10.8) | 13.4–20.0(3.4–10.1) |
| EV-C96 | 2 | 0 | 1 | 1 | 0 | 4 | 4.1 | 20.4–21.9(10.2–12.2) | 8.6–12.9(4.9–8.3) |
| CVA11 | 1 | 0 | 0 | 0 | 0 | 1 | 1.0 | 23.4(5.8) | — |
| CVA17 | 0 | 0 | 0 | 1 | 0 | 1 | 1.0 | 19.8(4.4) | — |
| Total | 37 | 21 | 22 | 11 | 7 | 98 | 100 | — | — |
Homologous comparison
Among EV-A members, two CVA10 strains isolated in 2006–2007 exhibited great nucleotide divergence from prototype strain in the range of 21.0–23.3%. Kowalik (AY421767) as the CVA10 prototype strain was first isolated from a patient suffered from meningitis in New York, 195316. Similarly, the three CVA2 isolates had 8.4–19.5% nucleotide divergence with each other. However, the EV-A71 isolates in this study had relatively less nucleotide divergence (range, 0.1–5.7%). Two new enterovirus serotypes in this species were found, namely, EV-A76 and EV-A90.
E13, E6 and E25 were the three most common serotypes, and they all belonged to EV-B species. E13 and E6 were detected every year during the surveillance period. VP1 nucleotide sequence alignment showed that the nine isolates of E13 had high nucleotide divergence (23.1–24.0%) with the prototype strain, Del Carmen (AY302539) which was isolated from 4-month-old female in Philippine Islands, 195317. E30 had 15.8–16.6% nucleotide divergence with the prototype strain Bastianni (AF162711) which was isolated from a patient suffered from aseptic meningitis in New York, 195818. E14 could be detected with 1–2 strains every year during 2006–2009, but the 5 strains exhibited high nucleotide divergence ranging from 6.8–17.7%. Two strains of E15 isolated in the same year demonstrated a relatively high nucleotide divergence of 17.2%, which indicated that they originated from different chains of transmission.
Among the 4 EV-C serotypes identified in this study, CVA11 was not detected until 2006, and only one CVA17 was isolated, suggesting that these two serotypes circulated in Yunnan at a low prevalence. Four isolates of EV-C96 had 20.4–21.9% nucleotide divergence from the prototype strain BAN00-10488 (EF015886) isolated in Bangladesh, 20007. This serotype is frequently isolated from healthy persons or AFP patients in China15,19.
Epidemiological and clinical data
Of total 98 isolates, 46 (46.9%) were isolated from males and 52 (53.1%) from females with a sex ratio of 1∶1.1. NPEV infections were found to be significantly higher in pre-school children aged <3 years (64/98, 65.3%) than in school children aged 4–6 years (17/98, 17.4%), 7–11 years (12/98, 12.3%), and 12–15 years (5/98, 5.1%).
The clinical data of all 98 NPEV positive patients were analyzed. Sixty-seven (68.4%) patients were found to have fever at the onset of paralysis. Other clinical symptoms included diarrhea (9 cases, 9.2%), neck stiffness (6 cases, 6.1%), muscle pain (33 cases, 33.7%) and upper respiratory tract infection (7 cases, 7.1%). All 98 AFP cases presented paralysis during onset. Single lower limb paralysis (43 cases, 43.9%) and lower limbs paralysis (32 cases, 32.7%) accounted for most cases, and followed by unilateral paralysis (13 cases, 13.3%), limbs paralysis (8 cases, 8.2%) and single upper limb paralysis (2 cases, 2.0%). The most common diagnosis for NPEV positive AFP cases was Guillain-Barré Syndrome (16 cases, 16.3%), followed by myositis (13 cases, 13.3%) and viral encephalitis (10 cases, 10.2%). On 60-day follow up after the onset of paralysis, residual paralysis was still present in 20 patients (20.4%), including 13 cases (13.3%) in the age of 0–5 years and 7 cases (7.1%) in the age of 6–14 years. The numbers of cases with residual paralysis on single lower limb, lower limbs, upper limbs, unilateral limbs and single upper limb were 8, 4, 4, 3 and 1, respectively. Twelve NPEV serotypes were found in association with the 20 residual paralysis cases which included E13 and EV-A71 (3 each), E6, E19, E21 and E30 (2 each), CVA2, CVA24, CVA9, E11, E25 and EV-A76 (1 each). There were 21 children died within 60 days of the onset of paralysis owing to the severity of disease, but no enterovirus were isolated in RD or L20B cells from the specimens of the 21 patients.
Monthly distribution of NPEVs
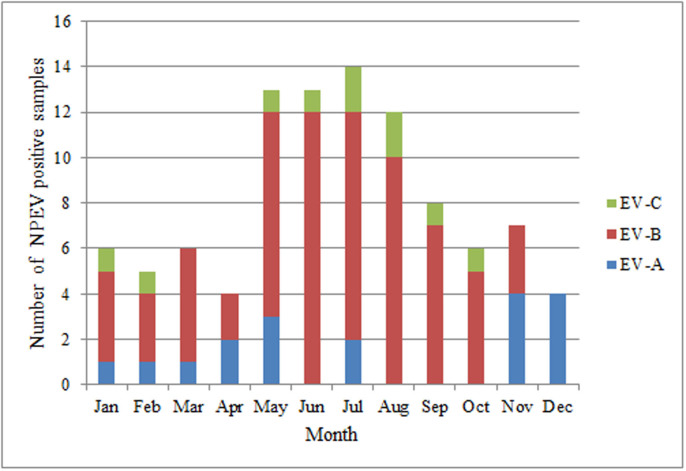
NPEV positive AFP cases were detected throughout the year. The distribution of 33 serotypes in 12 months was shown in Fig. 1. Obviously, most NPEV positive samples were isolated in summer and autumn, namely April–June (30.6%), July–September (34.7%) than January–March (17.3%) and October–December (17.3%), which was consistent with the typical pattern of enteroviral infections20,21. No distinct aggregation of a single serotype in every month was observed during the 5-year study.
Figure 1. Monthly distribution of EV species in AFP cases in Yunnan Province from 2006 to 2010.
(PV was excluded from HEV-C).
Isolation and phylogeny of EV-A71
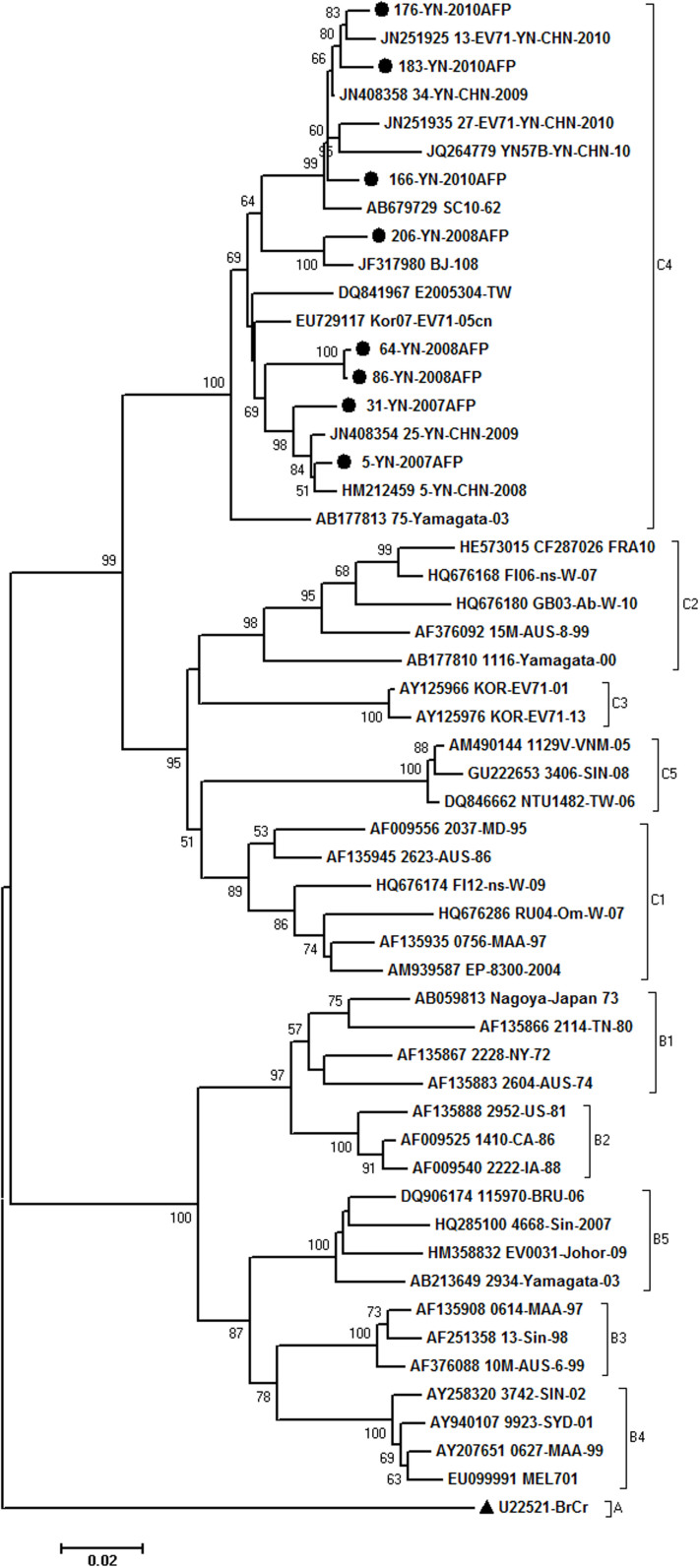
Eight strains of EV-A71 were detected during the 5 years. The phylogenetic relationship with 55 reference isolates from other regions of the world was analyzed based on complete VP1 sequences (Fig. 2). Yunnan EV-A71 strains grouped with the isolates from Japan, Korea, Taiwan and the mainland China, and have been classified into subgenotype C4. Seven EV-A71 was closely related with the Yunnan EV-A71 isolated from HFMD patients, and the other one (strain 206-08) was clustered close to the Beijing strain, with 100% bootstrap value, which revealed the possibility of long-term transmission of the virus. These EV-A71 showed 93.3–98.9% identity with other reference strains of C4 and >10% divergence with the strains of other subgenotypes. Nucleotide identities within Yunnan strains were 94.3–99.0%.
Figure 2. Phylogenetic tree depicting the relationships among the VP1 sequences of the EV-A71 isolates.
The local Yunnan strains are indicated by blank round, and the prototype strain of EV-A71 is indicated by black triangle.
Phylogeny of prevalent serotypes and recombination analysis
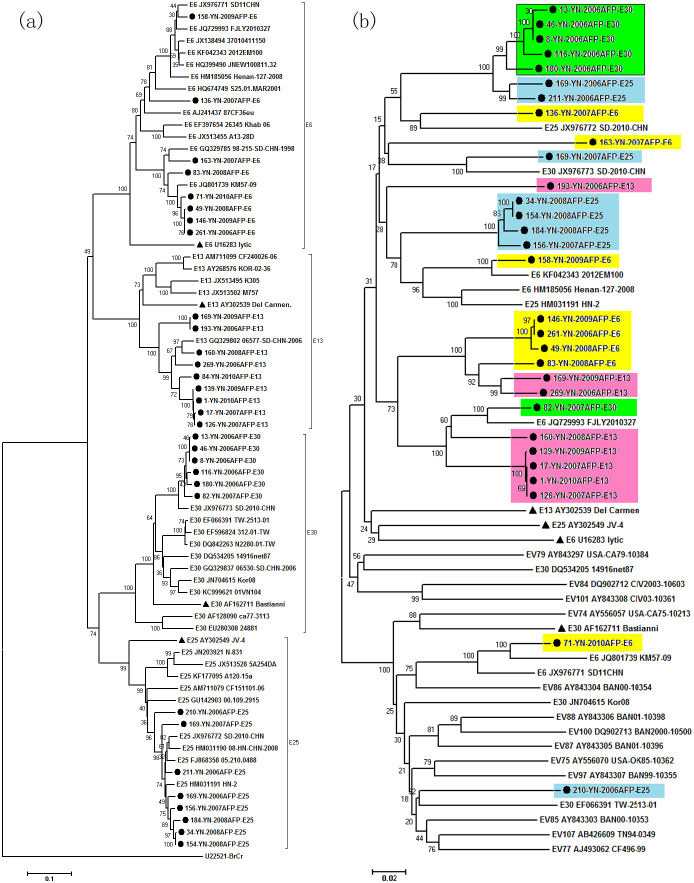
Of the 33 different types, E13, E6, E25 and E30 were predominant EV-B serotypes during the 5 years. To examine the extent of intra- or inter-serotype recombination, VP1 and 3D regions of the 31 strains of these 4 serotypes were sequenced. Phylogenetic trees (Fig. 3) were constructed on the VP1 and 3D sequences in comparison with the prototype strains and reference strains. In the VP1 tree, all isolates formed monophyletic clusters with their prototype strains and corresponding reference strains according to their serotype, as is expected since VP1 is the bases of serotype designation.
Figure 3. Phylogenetic trees on VP1 (a) and 3D (b) regions of 4 prevalent serotypes in Yunnan Province and reference strains.
The local Yunnan strains are indicated by blank round, and prototype strains of the 4 serotypes (E30: Bastianni, E6: Lytic, E25: JV-4 and E13: Del Camen) are indicated by black triangle.
In comparison on the phylogeny between VP1 and 3D trees, some strains (E13: 160-08, 139-09, 17-07, 1-10, 126-07; E6: 146-09, 261-06, 49-08, 83-08; E25: 156-07, 184-08, 34-08, 154-08; E30: 13-06, 46-06, 8-06, 116-06, 180-06) displayed consistent grouping (Fig. 3). The Pairwise p-distance showed the nucleotide divergence among each serotype showed little genetically in the two region: E30: 0–2.5%, E25: 0.1–3.0%, E6: 0–6.7%, E13: 0.4–11.3% in VP1 region respectively, and E6: 0–7.6%, E13: 0–8.7%, E25: 0–2.2%, E25: 0–4.2% in 3D region. There is thus no evidence for recombination among above strains.
In contrast, among the local strains, five E30 strains were clustered with two E25 strains with 100% bootstrap value. Similarly, four E6 strains were clustered with two E13, and five E13 strains were clustered with one E30, all with 100% bootstrap values. Hence, it could be inferred that inter-recombination events occured among these serotypes. Some strains displayed incongruent tree topologies and inconsistent inter-serotype clustering in VP1 and 3D regions. For example, strains 169-07-E25, 136-07-E6 and 82-07-E30 were closely related to reference strains of different serotypes (respectively, JX976773-E30, JX976772-E25 and JQ729993-E6) in 3D region with high bootstrap values. Other strains (163-07-E6, 193-06-E13, 210-06-E25 and 71-10-E6) were no longer clustered with their prototype strains and corresponding reference strains in the 3D tree. So, the results presented here suggest intra- and inter-recombination events among these prevalent strains. The clinical data of patients whom those strains were isolated from was provided in Supplementary Table S2, but from which we still could not come to conclusion by analyzing the relevance between the clinical features of these patients and recombination strains based on these limited information.
Discussion
During 2006–2010, in the 51 AFP cases associated with PVs, Sabin-like strains were detected from 50 cases, and another type 2 VDPV strain was isolated from a 1-year-old patient. This was the first detecting of VDPV in Yunnan Province. VDPV defined as Sabin vaccine viruses that show greater than 1% drift within the VP1 region of the genome, represents a major challenge to the eradication of polio. The first outbreak of poliomyelitis associated with VDPVs was reported in the Dominican Republic and Haiti from 2000 to 200122. Subsequent VDPV outbreaks occurred in the Philippines, China, Indonesia, Cambodia, Madagascar and more recently in Myanmar and Nigeria, the Philippines, China, Indonesia, Cambodia and Madagascar23,24,25,26. Thus, VDPVs will become increasingly important as the prevalence of wild polioviruses decreases, and the outbreaks also highlight the importance of maintaining sensitive poliovirus laboratory surveillance.
After more than 40 years of use and many billions of doses administrated worldwide, OPV has been associated with very few adverse events. The most commonly recognized adverse event is vaccine associated paralytic poliomyelitis (VAPP). Nevertheless, the incidence of VAPP is low and similar in most countries, such as Latin America, England, and so on27,28,29. In the United States, the risk of VAPP in first-dose OPV recipients is about 1 case per 750,000 children immunized30. Similarly, in this study, 6 cases (<2 years old) were diagnosed as VAPP of the 51 cases associated with PVs during the 5-year surveillance. Among 6 VAPP cases, 3 had ever received one dose of OPV, and the other had none. Of the 6 VAPP cases, 5 were caused by type 2 Sabin-Like strains, another one by type 3. During 2006–2010, about 30,463,390 doses of OPV were used in routine immunization and supplementary immunization in Yunnan Province, which indicated that the risk of VAPP was very low indeed.
In addition to PVs, many NPEVs have been isolated from AFP surveillance. In terms of distribution of NPEV types in AFP cases, there were slight changes on the composition ratio of NPEV comparing to that of a few years ago. The proportion of coxsackieviruses, echoviruses and the numbered enteroviruses were 24.21%, 65.26% and 10.53%, respectively in previous 5 years (1997–2000, and 2004) in Yunnan Province15, and 22.45%, 63.26% and 14.28%, respectively in this study, indicating that more new EV types have been identified in recent years. EV-B is the most common and diversified EV species. During 10 years of AFP surveillance in Yunnan Province, EV-B species account for 73.3% of total isolation, which is in accordance with the study from Taiwan31. The predominant serotypes have changed partly during the 10 years. In addition to the E13 and E6 which were still the main serotypes, other predominant serotypes E14 (5.6%), E12 (5.6%) and CVA24 (5.6%) had been replaced by E25 (8.2%), EV-A71 (8.2%) and E30 (6.1%). E13 and E6 are frequently associated with acute infections in children and are recognized as important causes of outbreaks of viral meningitis worldwide32,33,34,35. But to the best of our knowledge, no documented outbreaks of aseptic meningitis caused by these serotypes had been reported yet in Yunnan Province.
AFP has multiple etiologies, including GBS, transverse myelitis and transient or occasionally permanent paralyses associated with NPEV infections36. In this study, of total 1097 cases, the major condition of illness was GBS (226 cases, 20.6%) which is consistent with the findings in other countries37,38,39. The other clinical symptoms were myelitis, myositis, viral encephalitis, traumatic Neuritis, meningitis, myopathy, cerebellitis and acute infantile hemiplegia. During the 5-year AFP surveillance, there were 202 cases presenting residual paralysis on 60-day follow-up, in which poliovirus and NPEV were isolated from 28 and 20 patients, respectively. Besides EV infection, AFP can be resulted from many other causes, and EVs just account for a small proportion. Therefore, further studies are needed to explore the spectrum of the pathogens. There were 21 children died within 60 days of the onset of paralysis owing to the severity of disease. However, for the patient with death, no association could be found out between the age and NPEV/PV infection, and the death may be attributed to other causes rather than infection of EVs.
EV-A71 is divided into three genogroups: A, B, and C. BrCr, the prototype strain, is the only member of genogroup A. Most EV-A71 isolates belong to either genogroups B or C, which are each further divided into five genotypes or subgenogroups, B1–B5 and C1–C540. EV-A71 infection has become a major public health problem since 1997 in developed and developing countries in the Asia-Pacific region and it has a potential to spread further. In China, large scale outbreak of HFMD associated with EV-A71 emerged in Shandong Province in 200741. The nationwide epidemics of EV-A71 started in 2008, beginning as an outbreak of unknown viral infection in Anhui Province, and spreading into other provinces quickly42,43,44. Since 2009, HFMD associated with EV-A71 has been popular throughout Yunnan Province. Evidence from molecular epidemiology confirmed that these outbreak or pandemic in the mainland China were caused by subgenotype C4. Interestingly, in recent years, the number of EV-A71 isolates from AFP cases also increased. Eight EV-A71 strains—all belonging to C4 subgenotype—were identified in the study, as is different with the results from previous study with only 1 EV-A71 isolates in Yunnan in 1997–2000 and 200415. It is well known that EV-A71 often causes epidemics of severe neurological diseases especially in children <5 years of age. For the residual paralysis cases of this study, although EV-A71 was detected in 3 cases at the onset of paralysis, it was still to be noted that EV-A71 was found in the recovered cases, and it was reported that EV-A71 can also be detected from healthy children20. Hence, the severity of infection (sever or asymptomatic) may be explained by other factors such as differences in the pre-existing immunity of pediatric population, differences in host genetic susceptibility, and so on.
Recombination has been widely reported between enteroviruses. Enterovirus species B has been shown undergo much more frequent recombination events than found for species A45,46,47. Genomic recombinations are well known to contribute to genetic variations and evolution of enteroviruses48. Isolates sharing similar VP1 genes but differing in other parts of the genome (3C or 3D) may display different epidemiological or clinical properties49. Since recombination events usually occur in non-structural coding regions, and in order to investigate the genetic relationship between our predominant serotypes (all belonged to EV-B) and reference strains, phylogenetic trees based on VP1 and 3D regions were constructed. The analysis suggested that closely related VP1 regions can be associated with divergent 3D regions, and the intra- and inter-recombination events occurred during the evolution of these viruses. Thus more epidemiological and clinical investigation of NPEV infections is needed to learn about the different pathogenic properties of the virus which experienced recombination.
In conclusion, this study presented a genetic overview on the NPEVs isolates from AFP cases in Yunnan Province during 2006–2010, and explored the possible link between NPEV infections and AFP cases.
Methods
Ethics statement
Ethical approval was given by the Ethics Review Committee of the Yunnan Center for Disease Control and Prevention, and the study was conducted in compliance with the principles of the Declaration of Helsinki. Written informed consent for the use of the clinical samples was obtained from the legal guardian of the patient.
Patients and sample collection
Yunnan Province is located in southwest China with an area of 390,000 square kilometers and a population of 45.966 million (2010 census data). A total of 1097 AFP cases aged <15 years were reported from 16 prefectures under the AFP surveillance system in Yunnan Province between 2006 and 2010. All cases had at least one stool specimen (double specimens collected in1085 cases, single specimen collected in 12 cases) with an interval of 24–48 hr between collections and within 14 days from the date of onset of paralysis. Specimens were transported to the provincial polio laboratory under cold chain conditions for detection. Patient information was recorded on a standard questionnaire including demographic information, clinical presentation, and other epidemiological data. Finally, a 60-day follow-up for AFP cases was carried.
Virus isolation, neutralization test
All stool specimens collected from AFP patients were processed according to the WHO protocols accepted for the program50. RD and L20B cell lines were used to isolate the viruses. A total of 200 μl of treated specimen was inoculated into culture tubes and the inoculated cells were examined every day. The stool specimens producing CPE only in RD cells and not in L20B cells were considered to contain NPEV51. Within 14 days, when CPE was obtained, the viruses were harvested and stored under frozen conditions (−20°C). The micro-neutralization assay was carried out in 96-well tissue culture plates using poliovirus type-specific rabbit polyclonal antisera [National Institute for Public Health and the Environment (RIVM), Bilthoven, The Netherlands].
Amplification, sequencing and typing
Viral RNA extraction and RT-PCR were performed using QIAamp viral RNA mini kit (Qiagen, Valencia, CA, USA) and Access Quick™ RT-PCR system (Promega, USA), respectively according to the manufacturers' instructions. Primers pairs16,41,52 using for VP1 RT-PCR and nucleotide sequencing were listed in Table 2, and primer pairs were designed to amplify 3D region of EV-B viruses. RT-PCR were carried out as followed: 5 μl viral RNA was added to RT-PCR mixtures (total volume, 40 μl) containing Access Quick Master Mix 2×, RNA Template, and 1 μM each of the primers. The mixtures were incubated at 48°C for 45 min and then at 94°C for 2 min, followed by 35 cycles of amplification (94°C for 10 s, 50°C for 10 s and 65°C for 1 min) and a final extension at 65°C for 5 min. The PCR products were electrophoresed on a 2.0% agarose gel, and purified using a QIAquick Gel Extraction Kit (Qiagen). Purified products were then labeled directly by cycle sequencing reaction with BigDye Terminator v.1.1 (Applied Biosystems, USA), and were analyzed by ABI PRISM 3100 Genetic Analyzer (Applied Biosystems, USA). The VP1 sequences obtained were compared with sequences available in the GenBank database by using the Basic Local Alignment Search Tool (BLAST) from the National Center for Biotechnology Information. Virus isolates showing >75% nucleotide sequence identity with a known EV type were designated as the relative type53.
Table 2. Primers used for VP1 PCR amplification and sequencing.
| Primer | Sequence (5′–3′) | Gene | Location (nt) |
|---|---|---|---|
| 48652 | TGGTAICARACIAAITWYGTIGTNCC | VP3 | 2297–2322 |
| 48752 | ATGTWYGYICCICCIGGIGCNCC | VP1 | 2894–2916 |
| 48852 | GTIGGRTAICCITCITARAACCAYTG | VP1 | 3063–3038 |
| 48952 | AYIGCICCISWITGYTGNCC | 2A | 3348–3329 |
| 18753 | ACIGCIGYIGARACIGGNCA | VP1 | 2612–2631 |
| 01153 | GCICCIGAYTGITGICCRAA | 2A | 3408–3389 |
| 00816 | GCRTGCAATGAYTTCTCWGT | VP3 | 2411–2430 |
| 01316 | GGIGCRTTICCYTCIGTCCA | VP1 | 3051–3032 |
| 49452 | GAYGAYWSITTIACIGAIGGNGG | VP3 | 2306–2328 |
| 49652 | CCRTCITARAARTGISIRTANGC | VP1 | 3089–3111 |
| 04016 | ATGTAYRTICCIMCIGGIGC | VP1 | 2951–2970 |
| EV-A71-S41 | GCAGCCCAAAAGAACTTCAC | VP3 | 2372–2392 |
| EV-A71-A41 | AAGTCGCGAGAGCTGTCTTC | 2A | 3434–3454 |
Nucleotide sequence analysis
The sequences of viruses were aligned with prototype strains and other reference strains obtained from GenBank database using the BioEdit software (version 7.0.5.3)54. The phylogenetic trees were constructed by MEGA 4.055 using the neighbor-joining method with a Kimura two-parameter model. Bootstrapping was performed with 1000 duplicates and bootstrap values greater than 80% were considered statistically significant for grouping.
Nucleotide sequence accession numbers
Sequences reported in this study were deposited in the GenBank sequence database under accession numbers JQ886640–JQ886662, JQ968944–JQ969016, AB740160–AB740168 and KJ754035–KJ754095.
Supplementary Material
Supplementary Table S1 and S2
Acknowledgments
This study was supported by the National Natural Science Foundation of China (project no. 81160198) (http://www.nsfc.gov.cn).
Footnotes
The authors declare no competing financial interests.
Author Contributions J.T., H.Y. and Z.D. conceived the study and drafted the paper, J.T., H.Y. and Z.T. gathered and analyzed the data, and J.Z., B.T., Z.Z. and L.Z. helped to interpret results and contributed to the writing.
References
- Brown B., Oberste M. S., Maher K. & Pallansch M. A. Complete genomic sequencing shows that polioviruses and members of human enterovirus species C are closely related in the noncapsid coding region. J. Virol. 77, 8973–8984 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coxsackievirus, echovirus, and other enteroviruses. Infectious Diseases. (Lippincott Williams & Wilkins, 2003). [Google Scholar]
- Picornaviridae. Virus Taxonomy: Eighth Report of the International Committee on Taxonomy of Viruses. (Elsevier, Amsterdam, 2005). [Google Scholar]
- Picornaviridae. Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses. (Elsevier, San Diego, 2011). [Google Scholar]
- Lau S. K. et al. Clinical features and complete genome characterization of a distinct human rhinovirus (HRV) genetic cluster, probably representing a previously undetected HRV species, HRV-C, associated with acute respiratory illness in children. J. Clin. Microbiol. 45, 3655–3664 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberste M. S. et al. Molecular identification of 13 new enterovirus types, EV79-88, EV97, and EV100-101, members of the species Human Enterovirus B. Virus Res. 128, 34–42 (2007). [DOI] [PubMed] [Google Scholar]
- Brown B. A. et al. Resolving ambiguities in genetic typing of human enterovirus species C clinical isolates and identification of enterovirus 96, 99 and 102. J. Gen.Virol. 90, 1713–1723 (2009). [DOI] [PubMed] [Google Scholar]
- Enteroviruses: Polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses. Fields Virology, 5th ed. (Lippincott Williams & Wilkins, Philadelphia, 2007). [Google Scholar]
- Julian K. G. et al. Aseptic meningitis epidemic during a West Nile virus avian epizootic. Emerg. Infect. Dis. 9, 1082–1088 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios G. & Oberste M. S. Enteroviruses as agents of emerging infectious diseases. J. Neurovirol. 11, 424–433 (2005). [DOI] [PubMed] [Google Scholar]
- Ang L. W. et al. Epidemiology and control of hand, foot and mouth disease in Singapore, 2001–2007. Ann. Acad. Med. Singapore. 38, 106–112 (2009). [PubMed] [Google Scholar]
- Dhole T. N. et al. Non-polio enteroviruses in acute flaccid paralysis children of India: vital assessment before polio eradication. J. Paediatr. Child. Health. 45, 409–413 (2009). [DOI] [PubMed] [Google Scholar]
- World Health Organization. Tracking progress towards global polio eradication, 2010–2011. Wkly. Epidemiol. Rec. 87, 153–160 (2012).22536628 [Google Scholar]
- Chen C. Y. et al. Acute flaccid paralysis in infants and young children with enterovirus 71 infection: MR imaging findings and clinical correlates. AJNR Am. J. Neuroradiol. 22, 200–205 (2001). [PMC free article] [PubMed] [Google Scholar]
- Bingjun T. et al. Molecular typing and epidemiology of non-polio enteroviruses isolated from Yunnan province, the People's Republic of China. J. Med.Virol. 80, 670–679 (2008). [DOI] [PubMed] [Google Scholar]
- Oberste M. S., Maher K., Kilptrick D. R. & Pallansch M. A. Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification. J. Virol. 73, 1941–1948 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammon W. M., Yohn D. S., Pavia R. A. & Sather G. ECHO virus type 13. I. Isolation and characteristics. Proc. Soc. Exp. Biol. Med. 100, 425–429 (1959). [DOI] [PubMed] [Google Scholar]
- Plager H. & Decher W. A. newly-recognized enterovirus isolated from cases of aseptic meningitis. Am. J. Hyg. 77, 26–28 (1963). [DOI] [PubMed] [Google Scholar]
- Xu A. et al. The complete genome analysis of two enterovirus 96 strains isolated in China in 2005–2009. Virus Genes 42, 323–330 (2011). [DOI] [PubMed] [Google Scholar]
- Wu W. et al. Molecular identification and analysis of human enteroviruses isolated from healthy children in Shenzhen, China from 2010 to 2011. PLoS ONE 8, e64889 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxmivandana R., Yergolkar P., Gopalkrishna V. & Chitambar S. D. Characterization of the non-polio enterovirus infections associated with acute flaccid pralysis in south-western India. PLoS ONE 8, e61650 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kew O. et al. Outbreak of poliomyelitis in Hispaniola associated with circulating type 1 vaccine-derived poliovirus. Science 296, 356–359 (2002). [DOI] [PubMed] [Google Scholar]
- Rousset D. et al. Recombinant vaccine–derived poliovirus in Madagascar. Emerg. Infect. Dis. 9, 885–887 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X. et al. An outbreak of poliomyelitis caused by type 1 vaccine-derived poliovirus in China. J. Infect. Dis. 194, 545–551 (2006). [DOI] [PubMed] [Google Scholar]
- World Health Organization. Global update on vaccine-derived polioviruses, January 2006-August 2007. Wkly. Epidemiol. Rec. 82, 337–344 (2007). [PubMed] [Google Scholar]
- Yang C. F. et al. Circulation of endemic type 2 vaccine-derived poliovirus in Egypt from 1983 to 1993. J. Virol. 77, 8366–8377 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrus J. K., Strebel P. M., de Quadros C. A. & Olivé J. M. Risk of vaccine-associated paralytic poliomyelitis in Latin America, 1989–91. Bull World Health Organ 73, 33–40 (1995). [PMC free article] [PubMed] [Google Scholar]
- Joce R., Wood D., Brown D. & Begg N. Paralytic poliomyelitis in England and Wales, 1985–91. Br. Med. J. 305, 79–82 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler K. A., Banerjee K., Hlady G. W., Andrus J. K. & Sutter R. W. Vaccine-associated paralytic poliomyelitis in India during 1999: decreased risk despite massive use of oral polio vaccine. Bull World Health Organ. 80, 210–216 (2002). [PMC free article] [PubMed] [Google Scholar]
- Center for Disease Control and Prevention. Poliomyelitis prevention in the United States: introduction of a sequential vaccination schedule of inactivated poliovirus vaccine followed by oral poliovirus vaccine. Recommendations of the Advisory Committee on Immunization Practices (ACIP). MMWR Recomm. Rep. 46, 1–25 (1997). [PubMed] [Google Scholar]
- Lo C. W. et al. Application of a molecular method for the classification of human enteroviruses and its correlation with clinical manifestations. J. Microbiol. Immunol. Infect. 43, 354–359 (2010). [DOI] [PubMed] [Google Scholar]
- Avellon A. et al. Molecular analysis of echovirus 13 isolates and aseptic meningitis, Spain. Emerg. Infect. Dis. 9, 934–941 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheon D. S. et al. Isolation and molecular identification of echovirus 13 isolated from patients of aseptic meningitis in Korea, 2002. J. Med.Virol. 73, 439–442 (2004). [DOI] [PubMed] [Google Scholar]
- Papa A. et al. Molecular epidemiology of echovirus 6 in Greece. Eur. J. Clin. Microbiol. Infect. Dis. 28, 682–687 (2009). [DOI] [PubMed] [Google Scholar]
- Mao N. et al. An aseptic meningitis outbreak caused by echovirus 6 in Anhui Province, China. J. Med. Virol. 82, 441–445 (2010). [DOI] [PubMed] [Google Scholar]
- Andrus J. K., de Quadros C., Olivé J. M. & Hull H. F. Screening of cases of acute flaccid paralysis for poliomyelitis eradication: ways to improve specificity. Bull World Health Organ. 70, 591–596 (1992). [PMC free article] [PubMed] [Google Scholar]
- Whitfield K. & Kelly H. Using the two-source capture–recapture method to estimate the incidence of acute flaccid paralysis in Victoria, Australia. Bull World Health Organ. 80, 846–851 (2002). [PMC free article] [PubMed] [Google Scholar]
- Saeed M. et al. Epidemiology and clinical findings associated with enteroviral acute flaccid paralysis in Pakistan. BMC Infect. Dis. 7, 6 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poorolajal J. et al. Evaluation of acute flaccid paralysis in Hamadan, Iran from 2002 to 2009. Epidemiol. Health 33, e2011011 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown B. A., Oberste M. S., Alexander J. P. Jr, Kennett M. L. & Pallansch M. A. Molecular epidemiology and evolution of enterovirus 71 strains isolated from 1970 to 1998. J. Virol. 73, 9969–9975 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. et al. An outbreak of hand, foot, and mouth disease associated with subgenotype C4 of human enterovirus 71 in Shandong, China. J. Clin. Virol. 44, 262–267 (2009). [DOI] [PubMed] [Google Scholar]
- Yang F. et al. Enterovirus 71 outbreak in the People's Republic of China in 2008. J. Clin. Microbiol. 47, 2351–2352 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. et al. An emerging recombinant human enterovirus 71 responsible for the 2008 outbreak of hand foot and mouth disease in Fuyang city of China. Virol. J. 7, 94 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing W. et al. Hand, foot, and mouth disease in China, 2008-12: an epidemiological study. Lancet Infect. Dis. 14, 308–318 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberste M. S., Maher K. & Pallansch M. A. Evidence for frequent recombination within species human enterovirus B based on complete genomic sequences of all thirty-seven serotypes. J. Virol. 78, 855–867 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg A. M., Andersson P., Savolainen C., Mulders M. N. &Hovi T. Evolution of the genome of Human enterovirus B: incongruence between phylogenies of the VP1 and 3CD regions indicates frequent recombination within the species. J. Gen. Virol. 84, 1223–1235 (2003). [DOI] [PubMed] [Google Scholar]
- Tapparel C. et al. New respiratory enterovirus and recombinant rhinoviruses among circulating picornaviruses. Emerg. Infect. Dis. 15, 719–726 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. et al. Complete genome analysis of the C4 subgenotype strains of enterovirus 71: predominant recombination C4 viruses persistently circulating in China for 14 years. PLoS ONE 8, e56341 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C. Y. et al. A retrospective overview of enterovirus infection diagnosis and molecular epidemiology in the public hospitals of Marseille, France (1985–2005). PLoS ONE 6, e18022 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isolation and identification of polioviruses. WHO Polio Laboratory Manual, 4th edn. (World Health Organization, Geneva, 2004). [Google Scholar]
- The department of immunization, vaccines and biological. Supplement to the WHO Polio Laboratory Manual. (World Health Organization, Geneva, 2007). [Google Scholar]
- Oberste M. S. et al. Species-specific RT-PCR amplification of human enteroviruses: a tool for rapid species identification of uncharacterized enteroviruses. J. Gen. Virol. 87, 119–128 (2006). [DOI] [PubMed] [Google Scholar]
- Oberste M. S. et al. Comparison of classic and molecular approaches for the identification of untypeable enteroviruses. J. Clin. Microbiol. 38, 1170–1174 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T. A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser. 41, 95–98 (1999). [Google Scholar]
- Tamura K., Dudley J., Ne M. & Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1 and S2