Abstract
Members of the adhesion G protein-coupled receptor (aGPCR) family carry an agonistic sequence within their large ectodomains. Peptides derived from this region, called the Stachel sequence, can activate the respective receptor. As the conserved core region of the Stachel sequence is highly similar between aGPCRs, the agonist specificity of Stachel sequence-derived peptides was tested between family members using cell culture-based second messenger assays. Stachel peptides derived from aGPCRs of subfamily VI (GPR110/ADGRF1, GPR116/ADGRF5) and subfamily VIII (GPR64/ADGRG2, GPR126/ADGRG6) are able to activate more than one member of the respective subfamily supporting their evolutionary relationship and defining them as pharmacological receptor subtypes. Extended functional analyses of the Stachel sequences and derived peptides revealed agonist promiscuity, not only within, but also between aGPCR subfamilies. For example, the Stachel-derived peptide of GPR110 (subfamily VI) can activate GPR64 and GPR126 (both subfamily VIII). Our results indicate that key residues in the Stachel sequence are very similar between aGPCRs allowing for agonist promiscuity of several Stachel-derived peptides. Therefore, aGPCRs appear to be pharmacologically more closely related than previously thought. Our findings have direct implications for many aGPCR studies, as potential functional overlap has to be considered for in vitro and in vivo studies. However, it also offers the possibility of a broader use of more potent peptides when the original Stachel sequence is less effective.
Keywords: adhesion, cyclic AMP (cAMP), G protein, G protein-coupled receptor (GPCR), inositol phosphate, NFAT transcription factor, peptides, signal transduction
Introduction
Adhesion G protein-coupled receptors (aGPCRs)4 are a family of GPCRs with 33 members in humans (1). Recent advances in elucidation of their signal transduction pathways and dissecting their physiological relevance (reviewed in Refs. 2 and 3) render these receptors one of the fastest emerging scientific fields among the GPCR superfamily. Their importance is also reflected by the fact that genetic variants and mutations of several aGPCRs have implications in severe diseases (2), stressing the huge pharmacological relevance of this receptor family. Although it is widely accepted that these receptors couple to G proteins (reviewed in Refs. 2 and 4), their mode of activation remained enigmatic until recently.
It has been demonstrated that several aGPCRs can be activated through a tethered peptide agonist (Stachel) (5–8) located within the GPCR autoproteolysis-inducing (GAIN) domain (9). Many aGPCRs seem to be autocatalytically cleaved at a motif within the GAIN domain, termed GPCR proteolysis site (GPS), yielding an N-terminal fragment (NTF) and a C-terminal fragment (CTF) (10) (Fig. 1A). As the tethered peptide agonist comprises the sequence C-terminal of this designated cleavage site, it has been proposed that the cryptic tethered agonist is liberated through removal of the NTF (11) by this cleavage. A similar mechanism of proteolytic activation has been previously described for protease-activated receptors (12), where an internal agonistic peptide sequence within the N terminus is exposed following protease-mediated cleavage.
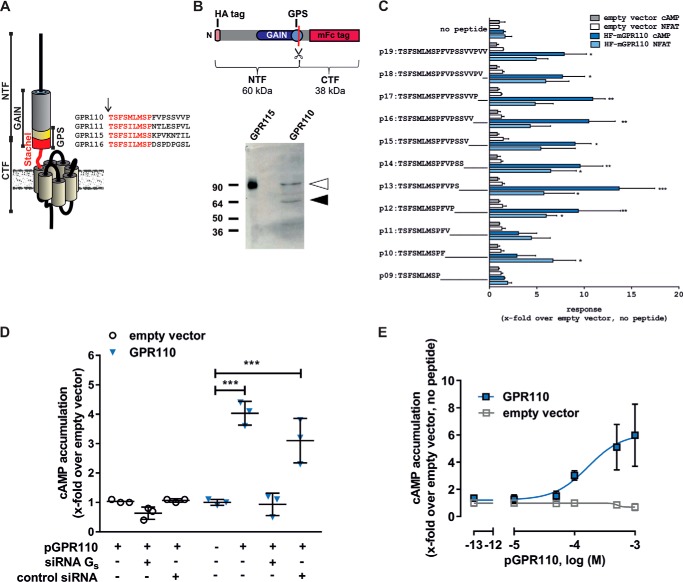
FIGURE 1.
Signal transduction and activation of GPR110. A, schematic presentation of a prototypical aGPCR structure. Adhesion GPCR share a common domain called the GAIN domain. At the very C-terminal part of this GAIN domain lies the GPS at which many full-length aGPCRs are autoproteolytically cleaved into an N-terminal fragment (NTF) and a C-terminal fragment (CTF). The recently discovered tethered agonist sequence (Stachel, red cylinder and line) is located between the cleavage site of the GPS and the first transmembrane helix. The sequence of each estimated Stachel sequence is given with the core activating region marked in red letters. B, GPR110 is cleaved at the GPS. A construct containing an N-terminal HA tag and a C-terminal mFc tag was used for cleavage analysis. Upon cleavage at the GPS two fragments, NTF (60 kDa) and CTF (38 kDa), occur (top panel). GPR110 is autoproteolytically processed as the NTF can be detected using an anti-HA antibody (bottom panel, black arrowhead). Note that cleavage appears to be incomplete as a second band can be detected (white arrowhead), which is only slightly smaller than a potential full-length construct (98 kDa). A similar construct of GPR115, which contains a non-cleaving GPS, served as control, yielding one band representing the full-length receptor (90 kDa). C, screening for agonistic peptides derived from GPR110 Stachel sequence. A Stachel-derived peptide library of GPR110 was tested in cAMP and NFAT reporter gene assay as described under ”Methods.“ Data are given as x-fold over empty vector (basal pcDps: for cAMP, 5.9 ± 5.1 nm; for luciferase activity, 322.5 ± 95.7 counts). D, peptide-induced activation of GPR110 in cAMP assay is induced by coupling to the Gs protein. Data are given as x-fold over empty vector, which serves as a negative control (empty vector: cAMP: 2.0 ± 0.1 nm). E, different pGPR110 concentrations were tested on empty vector- and GPR110-transfected cells in cAMP assay. Basal empty vector levels were 100.9 ± 84.2 nm. All assay data are given as mean ± S.D. of three independent experiments, each performed in triplicate. Statistics applied compare basal activity of a given construct to peptide-induced activation (see ”Methods“ for further detail).
However, fully efficient cleavage is not a requirement for activation of all aGPCRs as in vitro and in vivo studies suggested that were using cleavage-impairing receptor variants (13, 14). The mutations usually introduced address two positions, HisP−2 and Ser/ThrP+1, yet only the latter should truly inhibit cleavage, whereas the first was shown to lead to a drastic reduction in cleavage efficiency (10) and could thereby still contribute to receptor function in long term experiments. However, there are even aGPCRs with a naturally disrupted cleavage motif such as GPR111/ADGRF2 and GPR115/ADGRF4 (15), whereas GPR123/ADGRA1 lacks the GPS. The aGPCR GPR114/ADGRG5 harbors a cleavage motif, yet it does not release its NTF under in vitro conditions (16). For GPR114/ADGRG5 we postulated that there is a prebound state of the agonist, which isomerizes upon changes occurring at the NTF, e.g. ligand binding or mechanical force (16, 17). Isomerization of a prebound ligand (retinal) is the classical activation scenario in rhodopsin.
Even though peptides derived from GPR126 and GPR133 specifically activate the respective receptor (5), the core sequence of the Stachel is highly conserved among aGPCRs. It is therefore conceivable that Stachel-derived peptides of closely related members have the ability to activate several aGPCRs.
To test this hypothesis we investigated the potential Stachel sequence of the aGPCR subfamily VI (2). This subfamily comprises five receptor genes, Gpr110, Gpr111, Gpr113/ADGRF3, Gpr115, and Gpr116/ADGRF5, of which Gpr110, Gpr111, Gpr115, and Gpr116 cluster in mammalian genomes and have very likely emerged from one common ancestor during vertebrate evolution (15). Because of their high conservation and the availability of tissues from mice deficient for Gpr110, Gpr111, and Gpr115, this group is highly suitable for in-depth analyses of tethered agonist specificity.
Here, we show that some peptides derived from the Stachel sequences of members of subfamily VI can activate not only the receptor of origin, but also others in in vitro expression systems and in tissues ex vivo. The observed agonist promiscuity seems to be common among aGPCRs because we identified additional examples in and between different aGPCR subfamilies. This knowledge is important for designing in vitro and in vivo studies dealing with the activation of aGPCRs.
Our data suggest revising the current grouping into nine subfamilies because several aGPCRs should rather be referred to as pharmacological receptor subtypes than members of different subfamilies despite distinct differences in the 7-TM domain.
Results
GPR110 and GPR116 Harbor a Receptor-activating Stachel Sequence
Adhesion GPCR subfamily VI, consisting of the closely related aGPCRs GPR110, GPR111, GPR113/ADGRF3, GPR115, and GPR116, is an ideal assembly of receptors to study agonist specificity. The receptors GPR110, GPR111, GPR115, and GPR116 display high sequence similarities in the Stachel regions (Fig. 1A). Yet, GPR111 and GPR115 have a disrupted cleavage site preventing autoproteolysis (15), whereas GPR110 and GPR116 harbor a classical cleavage motif. That cleavage indeed occurs as this motif has previously been shown for GPR116 (18). Western blot analyses of GPR110 constructs heterologously expressed in HEK293T cells revealed that this aGPCR is also cleaved at the GPS (Fig. 1B).
We generated N and C terminally epitope-tagged versions of GPR110, GPR111, GPR115, and GPR116 for signal transduction assays. However, for GPR111 no cell surface expression was detected with or without the addition of an expression-enhancing Rho epitope (13) (Table 1). Therefore, GPR111 was omitted from further experiments of this study as GPR115 could also represent the features of the non-cleaved receptors of subfamily VI.
TABLE 1.
Expression analysis of GPR110, GPR111, GPR115, and GPR116
COS-7 cells were transfected with the given construct. All aGPCR constructs contained an N-terminal HA- and C-terminal FLAG tag. To increase cell surface expression, a rhodopsin N terminus (Rho) was inserted directly downstream from the HA tag in the indicated constructs. For expression studies, cell surface and sandwich ELISA were used to measure cell surface and total cellular expression levels, respectively. Specific optical density (OD) readings (OD value of double HA/FLAG-tagged ADP receptor P2Y12 (31) (positive control, pC) constructs minus OD value of mock-transfected cells) are given as percentage of double HA/FLAG-tagged P2Y12. For the cell surface ELISA, the nonspecific OD value (pcDps) was 0.025 ± 0.026 (set 0%) and the OD value of wt P2Y12 was 1.304 ± 0.236 (set 100%). OD readings of 0.080 ± 0.039 (set 0%) and 2.217 ± 0.730 (set 100%) were found in sandwich ELISA (total expression) for the negative control vector (pcDps) and the wt P2Y12, respectively.
| receptor | N-terminal tag | Cell surface ELISA | Whole cell ELISA |
|---|---|---|---|
| % of pC | |||
| Mouse GPR110 | HA | 89.7 ± 18.1 | 98.2 ± 1.9 |
| Mouse GPR111 | HA | 1.6 ± 0.5 | 39.0 ± 19.1 |
| HA-Rho | 4.2 ± 2.4 | 72.9 ± 17.6 | |
| Mouse GPR115 | HA | 75.7 ± 13.0 | 81.6 ± 3.9 |
| Mouse GPR116 | HA | 11.8 ± 11.4 | 81.5 ± 22.7 |
| HA-Rho | 99.1 ± 17.8 | 89.8 ± 7.2 | |
Previously published data and our own studies suggest GPR110 (6, 19) and GPR116 (20) couple Gq/11 proteins and GPR110 also couples to the Gs protein (see below). Therefore, we used a cAMP formation assay (Gs), NFAT (the activation of the nuclear factor of activated T cells), and inositol 1-phosphate (IP1) assay (both assays monitor Gq activation) as read-outs for testing of Stachel-derived peptide libraries of GPR110, GPR116 and GPR115 (Figs. 1C and 2, A and C).
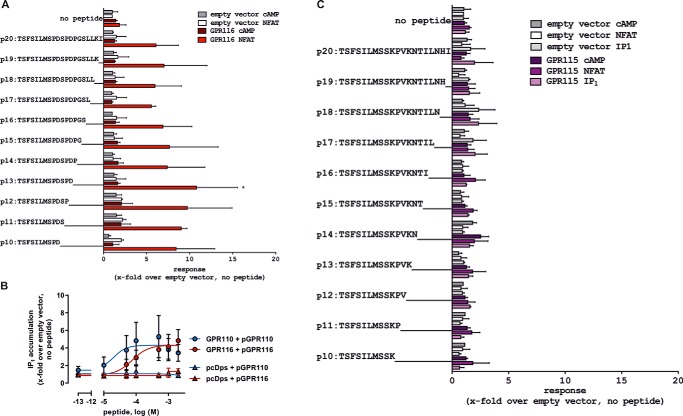
FIGURE 2.
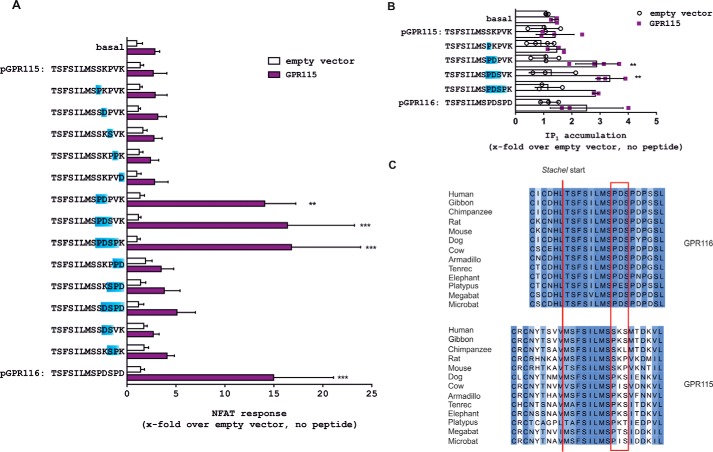
Signal transduction and activation of aGPCRs GPR115 and GPR116. A, screening for agonistic peptides derived from the GPR116 Stachel sequence. A Stachel-derived peptide library of GPR116 was tested in cAMP and NFAT reporter gene assay. Data are given as x-fold over empty vector (basal pcDps: for cAMP, 5.9 ± 5.1 nm; for luciferase activity, 322.5 ± 95.7 counts). B, concentration-response analysis of Stachel-derived peptides pGPR110 and pGPR116 on their receptor of origin and empty vector control in IP1 accumulation assay. Data are given as x-fold over empty vector, which serves as a negative control (IP1 level, 67.5 ± 15.4 nm). C, screening for agonistic peptides derived from GPR115 Stachel sequence. A Stachel-derived peptide library of GPR115 was tested in cAMP, NFAT reporter gene, and IP1 accumulation assays. Data are given as x-fold over empty vector, which serves as a negative control (basal pcDps: for cAMP, 5.9 ± 5.1 nm; for luciferase activity, 322.5 ± 95.7 counts; for IP1 level, 94.4 ± 41.7 nm). All assay data are given as mean ± S.D. of three independent experiments, each performed in triplicate. Statistics applied compare basal activity of a given construct to peptide-induced activation (see ”Methods“ for further detail).
Based on previous experiences with Stachel-derived peptides we used a standard concentration of 1 mm for all peptides of the libraries. GPR110 elicited a significant signal in cAMP accumulation and NFAT luciferase assays with Stachel sequence-derived peptides of 12 or more amino acids (aa). The best activation was found for a 13-AA long peptide (pGPR110) (Fig. 1C), which is in accordance with the previously identified agonist sequence by Stoveken et al. (6). In contrast to this previous publication, we found that peptide-induced activation of GPR110 is transduced through both Gs and Gq proteins. To exclude indirect cAMP formation by Gαs-independent mechanisms, Gs protein-mediated cAMP formation in GPR110-transfected and peptide-stimulated cells was abolished after Gαs-specific siRNA-mediated knockdown (Fig. 1D). The EC50 value for pGPR110 in cAMP assays was 167.2 μm (Fig. 1E). Recently, Gs-protein coupling of GPR110 has been demonstrated by another group through the newly identified endogenous agonist called synaptamide (21).
For GPR116, a 13-AA long peptide (pGPR116) displayed the highest efficiency in NFAT, whereas no activity was seen for any of the peptides in the cAMP assay (Fig. 2A). This indicates a more specific coupling of GPR116 to Gq/11 proteins. For more direct comparison of the downstream second messengers, concentration-response curves of pGPR110 and pGPR116 were performed in an IP1 accumulation assay using homogeneous time resolved fluorescence technology instead of the convenient but very indirect NFAT receptor assay. Here, EC50 values of 19.61 and 75.8 μm in GPR110- and GPR116-transfected cells were found, respectively (Fig. 2B). GPR115 activation was neither detectable in cAMP, NFAT, nor IP1 assay by any of the GPR115-derived peptides of the chosen library (Fig. 2C), even though its cell surface expression was comparable with GPR110 and GPR116 (Table 1).
To check whether insufficient solubility of the peptides could be the cause of the observed differences in peptide efficacy or rather lack of function we performed dynamic light scattering measurements (see supplemental Fig. S1). We found that pGPR115 and pGPR116 were well dissolved, whereas pGPR110 showed more aggregates associated with a reduced solubility. Therefore, the observed differences in agonistic peptide function do not correlate with their solubility.
Function of Agonistic Peptides in ex Vivo Studies
Previous expression studies (15) and qPCR analyses (Fig. 3A) revealed that Gpr110 is most prominently expressed in renal papillae, whereas GPR115 and GPR116 are more widely distributed, with GPR116 being present in nearly every tissue tested. Yet, signals mediated by GPR116 and GPR110 are distinguishable from each other as GPR116 activation mainly triggers a cascade via a Gq protein, whereas GPR110 additionally activates Gs. We chose lung tissue and cells isolated from renal papilla to study GPR116 and GPR110 activation, respectively. Seminal vesicle served as negative control tissue.
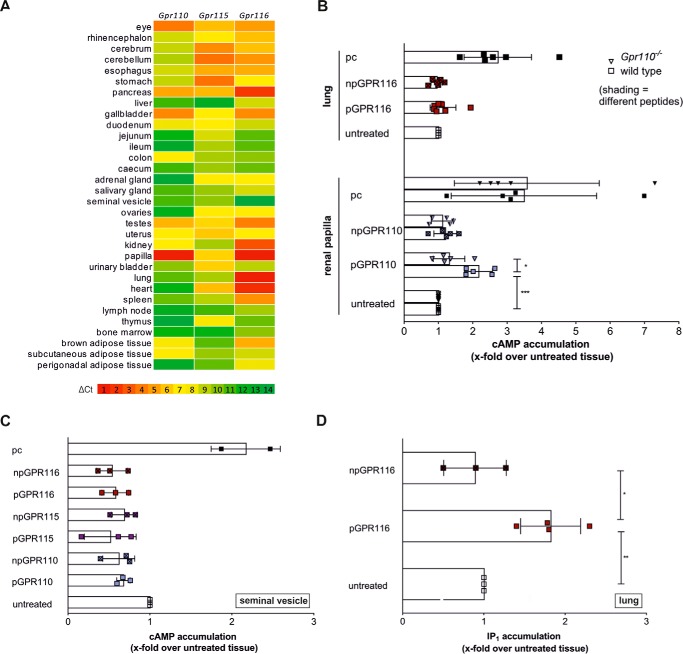
FIGURE 3.
Ex vivo analysis of aGPCR activation by Stachel-derived peptides. A, expression of Gpr110, Gpr115, and Gpr116 determined by quantitative RT-PCR in tissues of wt mice. Gpr110 is expressed strongly in renal papilla. Gpr115 is abundant and Gpr116 is detected ubiquitously. Only very low levels of all receptor transcripts are present in seminal vesicles. B, peptide-stimulated cAMP response in renal papilla and lung. Tissues were isolated from wt and Gpr110 knock-out mice and stimulated as described under ”Methods.“ npGPR110 and npGPR116 serves as a control peptides with no agonistic properties, forskolin served as positive control (pc). Basal cAMP levels (untreated) were 2.4 ± 0.6 nm (wt renal papilla), 2.3 ± 0.4 nm (Gpr110−/− renal papilla), and 17.1 ± 3.6 nm (lung). Statistics compare raw activities (see ”Methods“ for further detail). C, no cAMP response of pGPR110, pGPR115, and pGPR116 at seminal vesicles. Basal cAMP levels (untreated) were 9.0 ± 4.6 nm, forskolin served as positive control (pc). D, pGPR116 activates its receptor GPR116 ex vivo in lung tissue. IP1 accumulation was measured by employing an ELISA-based IP1 accumulation assay. The mean IP1 level of untreated tissue was 729 ± 220 nm. All data are given as mean x-fold over untreated tissue control ± S.D. of at least three independent experiments, each performed in triplicates. Statistics compare raw activities (see ”Methods“ for further detail).
We first investigated signaling of GPR110 upon stimulation with the agonistic peptide using the cAMP accumulation assay. The expected increase in cyclic AMP levels was measurable ex vivo in renal papilla (Fig. 3B) and upon forskolin application, which served as positive control in seminal vesicle (Fig. 3C). Treatment with pGPR110 (5 mm) led to a 2.2-fold increase of cAMP concentration in renal papillae ex vivo compared with tissue treated with same concentration of DMSO without the peptide (marked as “untreated”). Renal papillae isolated from Gpr110-deficient mice (15) (Fig. 3B) did not show any response upon stimulation with pGPR110, indicating that the cAMP signal detected in wild-type (wt) papillae is specific for GPR110 activation. To exclude any nonspecific effects of the peptide, a control peptide of 10 AA in length (npGPR110) with no activation capacity in in vitro assays (Fig. 1C) was applied. This peptide did not induce any significant changes in cAMP levels (Fig. 3B).
Ex vivo activation of GPR116 in lung tissue did not show changes in cAMP concentration upon treatment with pGPR116 (Fig. 3B), whereas a 1.8-fold increase in IP1 concentration was detected compared with untreated control tissue (Fig. 3D). A negative control peptide of 20 AA length (npGPR116, Fig. 2A) did not induce any signal (Fig. 3D).
Peptides Derived from the Stachel Sequences of GPR110 and GPR116 Activate Other Members of Subfamily VI
We have recently postulated that the tethered peptide agonists of aGPCRs have a core region spanning the first 6–8 highly conserved AA, which is essential for agonistic activity (5). As the core sequences of GPR110, GPR115, and GPR116 are almost identical (Fig. 1A) it is reasonable to assume that Stachel-derived peptides are promiscuous for members of subfamily VI. We therefore tested the active peptides of GPR110 and GPR116 and an analogous 13-AA peptide derived from GPR115 in cAMP and IP1 assays. In cAMP assays only activation of GPR110 through its own derived peptide was observed (Fig. 4A). However, IP1 assays showed promiscuous activation of both GPR110 and GPR116, through pGPR110 and pGPR116 (Fig. 4B) plus an activation of GPR110 through the respective peptide derived from the GPR115 Stachel region. Also, GPR115 raised IP1 levels upon stimulation with pGPR116 showing that the receptor itself is capable of signaling. These data confirm that subfamily VI-derived Stachel peptides are promiscuous between individual members (summarized in Fig. 4C).
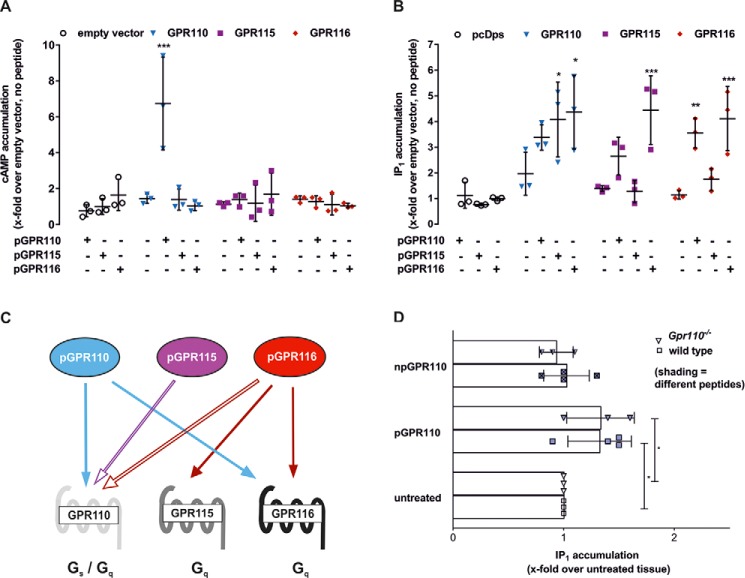
FIGURE 4.
Specific Stachel sequence-derived peptides of subfamily VI aGPCRs are able to activate several receptors within the group in vitro and ex vivo. A and B, pharmacological characterization of activation capabilities of various Stachel sequence-derived peptides on different aGPCRs. A, cAMP assays, and B, IP1 assays were performed on empty vector (pcDps)-, GPR110-, GPR115-, and GPR116-transfected cells. pGPR110 and pGPR116 can stimulate GPR110 and GPR116. It is noteworthy that GPR116 activation is only detectable in IP1 assays. In the same assay, GPR115 can be activated through pGPR116 and pGPR115 activates GPR110. Each transfection was stimulated with all peptides. Data are given as x-fold over empty vector without stimulation, which serves as a negative control (pcDps: for cAMP, 53.6 ± 28.9 nm, for IP1 activity, 67.5 ± 15.4 nm). All assay data are given as mean ± S.D. of three independent experiments, each performed in triplicate. Statistics applied compare basal activity of given construct to peptide-induced activation (see ”Methods“ for further detail). C, schematic depiction of the activating abilities among the peptides of subfamily VI. Open arrow indicates partial activation of GPR110 through pGPR116 and pGPR115. D, pGPR110 activates GPR116 in lung tissue. Wild-type and Gpr110 knock-out mouse tissue were stimulated with respective peptide or negative control peptide npGPR110. IP1 levels of untreated tissue were 729 ± 220 (wt) and 809 ± 202 nm (Gpr110−/−). Data are given as mean ± S.D. of at least three independent experiments, each performed in triplicates. Statistics compare raw activities (see ”Methods“ for further detail).
Ex vivo assays with lung tissue supported the in vitro results showing activation of GPR116 through pGPR110 in IP1 measurements (Fig. 4D). Verification of overlapping activation of peptides derived from GPR115 and GPR116 ex vivo in cAMP or IP1 assays is unfortunately hampered by the overlapping expression and preference in signal transduction of all three receptors (Fig. 3A).
Mutations Rescue Activity of pGPR115 at GPR115
As GPR115 displayed signal transduction upon stimulation with pGPR116, and is therefore per se functional, we speculated that distinct differences in the GPR115-derived peptide sequence to pGPR116 are responsible for lack-of-function of pGPR115 at GPR115. Systematic site-directed mutagenesis of AA positions that differ between pGPR115 and pGPR116 revealed two mutations, S9P and K10D, enabling pGPR115 activity at GPR115 in NFAT and IP1 assay (Fig. 5, A and B), even though AA positions 9 and 10 are not part of the previously described core activating region of the Stachel sequence (16). It is noteworthy that only the combination of the two mutants was capable of rescuing the agonistic property of pGPR115, yet the proline at position 9 (Fig. 1A) is the most prominent difference between GPR115 and the other members of this aGPCR subfamily. Comparing the cleavage motif and Stachel sequences of different orthologs of GPR116 and GPR115 may provide information whether the Stachel sequence of GPR115 has displayed an AA sequence that renders it inactive. As shown in Fig. 5C, GPR115 contains a proline at position 9 of the Stachel sequence in most mammalian orders at the beginning of its evolutionary appearance. This proline at position 9 is substituted by serine in rat, mouse, chimpanzee, and humans. Analysis of over 100 mammalian orthologs revealed that only proline and serine at position 9 are tolerated. Therefore, activity levels of the tethered agonist should not be hampered by either of the amino acids. This assumption is supported by the fact that the rescue of pGPR115 activity depended on the combination of S9P and K10D.
FIGURE 5.
Residues in the Stachel sequence essential for pGPR115 activity. A, functional rescue of pGPR115 by substitution of two amino acids. Subsequent mutagenesis was performed to gradually change pGPR115 into pGPR116. Peptides were analyzed in an NFAT reporter assay toward their potential to activate GPR115. Decisive positions to restore activity are +9 and +10 (S9P, K10D). Data are given as x-fold over empty vector without stimulation, which serves as a negative control (pcDps: 110.9 ± 57.7 counts). B, evaluation of selected peptide mutants in IP1 assay. Data are given as x-fold over empty vector without stimulation, which serves as a negative control (pcDps: 56.0 ± 27.7 nm). All data represent mean ± S.D. of three independent experiments each performed in triplicate. Statistics applied compare basal activity of given construct to peptide-induced activation (see ”Methods“ for further detail). C, alignment of the cleavage motifs and Stachel sequences of GPR116 and GPR115 in various species. GPR115 displays a distorted cleavage motif throughout evolution. Residues in the Stachel sequence identified as being essential for tethered agonist activity (PDS in GPR116, red box) are subject to alterations in GPR115 in different species.
The lysine in GPR115 is highly conserved among mammals and an asparagine is not found in any recent species sequenced yet at this position. It remains unknown whether the tethered agonist sequence has ever fit to the GPR115 binding pocket or whether the arrangement of serine and lysine within the peptide sequence leads to secondary effects of the peptide structure that impairs functional interaction with the receptor, whereas the Stachel sequence itself is active within the receptor structure. What we know is that GPR115 is a receptor capable of eliciting second messenger signals (Figs. 4B and 5, A and B) and pGPR115 is an active peptide at GPR110, where it causes changes in IP1 levels (Fig. 4B).
Cross-activation of aGPCRs by Stachel-derived Peptide Agonists Occurs between Members of Different aGPCR Groups
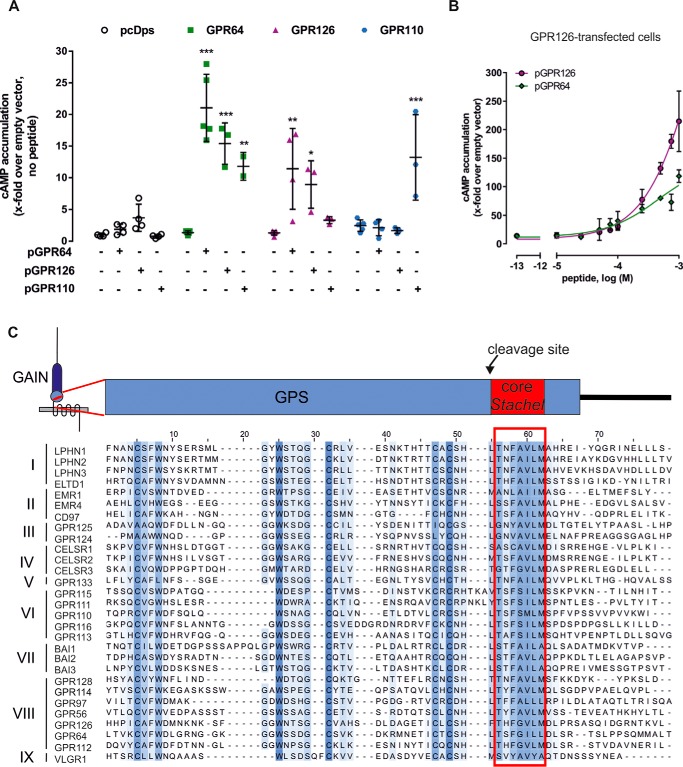
Due to their common ancestry and AA sequence homology receptor cross-activation by Stachel-derived peptides of subfamily VI is reasonable. We next asked whether cross-activation can also occur between receptors within other subfamilies and across subfamilies. We have previously shown that peptides derived from GPR126 cannot activate GPR133 and vice versa (5). Also, the peptide agonist of GPR64 cannot activate GPR110 and GPR133 (7). However, reflecting on the results from this study on subfamily VI, a cross-activation between GPR126 and GPR64 is likely, as they are closely related according to sequence homology (2). Indeed, peptides derived from GPR64 (pGPR64) and GPR126 (pGPR126) cross-activated the respective receptor in cAMP accumulation assays (Fig. 6A), again supporting their close evolutionary relationship as receptor subtypes. Concentration-response curves of the cross-activating peptides pGPR126 and pGPR64 at GPR126-transfected cells revealed that pGPR64 showed a shifted potency toward higher peptide concentrations compared with pGPR126 (Fig. 6B). As both peptides did not induce saturation in cAMP levels EC50 values cannot be determined.
FIGURE 6.
Activation between different aGPCR subfamilies. A, COS-7 cells were transfected with the given aGPCR and stimulated with 500 μm pGPR64, pGPR110, and pGPR126. Cyclic AMP accumulation is given as x-fold over empty vector, which serves as a negative control (pcDps: 41.1 ± 3.7 nm). B, comparing concentration-response curves between pGPR126 and pGPR64 on GPR126 showed higher potency and efficacy of pGPR126 on its originating receptor. Increasing concentrations of the given peptide were used to stimulate cAMP accumulation. Cyclic AMP accumulation is given as x-fold over empty vector, which serves as a negative control (pcDps: 0.27 ± 0.13 nm). All data represent mean ± S.D. of three independent experiments each performed in triplicate. Statistics applied compare basal activity of given construct to peptide-induced activation (see ”Methods“ for further detail). C, evolutionary conservation of the Stachel sequences C-terminal of the GPS cleavage site in all mouse aGPCRs. Sequences were retrieved from NCBI/GenBankTM and aligned using Jalview (32).
Cross-activation was also tested between subfamilies. As shown in Fig. 6A, pGPR110 elicited a significant signal on GPR64-transfected cells, indicating that cross-activation can occur between subfamilies VIII and VI. Comparing the Stachel sequences of all murine aGPCR paralogs striking similarities between members of different families are obvious that could be the basis for more cross-activating peptides (Fig. 6C).
Discussion
In the present study we analyzed the activation capabilities of Stachel peptides derived from aGPCRs of subfamily VI. Because no proper expression of GPR111 could be achieved we focused on GPR110, GPR115, and GPR116 as they are all similarly well expressed (Table 1) and they comprise a conserved evolutionary cluster. We found that several Stachel sequence-derived peptides are able to activate more than one member of this subfamily. Interestingly, promiscuous activation of GPR110 through peptides from other receptors only occurs in the Gq protein pathway but not in the Gs protein-mediated signaling pathway. Furthermore, our data indicate that peptides of some receptors can also activate receptors from other subfamilies.
We identified and characterized agonistic peptides derived from the Stachel sequences of GPR110 and GPR116. GPR110 displays dual coupling to Gs and Gq protein-mediated pathways. In contrast, GPR116 can only signal via the Gq/11 protein-mediated pathway. We showed that the Stachel sequence-derived peptides of GPR110 and GPR116 can also efficiently activate the other receptor in vitro and ex vivo (Fig. 4).
GPR115 could not be activated by any peptide derived from the putative GPR115 Stachel sequence. Based on these data it is tempting to speculate that GPR115 is a non-functional aGPCR (pseudogene) exposed to neutral drift. This assumption could be supported by the observation that GPR115 exhibits a disrupted cleavage site and does not undergo autoproteolysis (15). Thus, it can further be speculated that by not liberating the NTF through cleavage, exposure of the tethered agonist is hampered, rendering the whole receptor non-functional.
However, there are several points arguing against this assumption. 1) We demonstrate here that pGPR116 is capable of activating GPR115 (Fig. 4B), which indicates that the receptor has not lost its ability to be activated by a peptide agonist. 2) Several studies revealed that activation of an aGPCR is independent of the cleavability of the receptor in vitro (13, 5) and in vivo (14). This indicates that cleavage is not generally essential for tethered agonist exposure and receptor activation. 3) We show in this study that the core peptide region for each Stachel-derived peptide begins C terminally of the cleavage motif mutated in GPR115 and is almost identical between GPR110, GPR115, and GPR116 (Fig. 1). 4) It has been shown that aGPCRs can signal via G protein independent pathways and potentially without the Stachel sequence (14, 22, 23).
Although it is certain that the 7-TM domain of GPR115 is able to activate G proteins, it is not proven that its tethered agonist is functional in the context of the full-length receptor. We showed that replacement of two positions with the respective AA of pGPR116 is required to restore function of pGPR115 (Fig. 5, A and B), whereas the wt pGPR115 elicited a signal on GPR110. It remains elusive how GPR115 is activated under physiological conditions.
Our functional data using Stachel sequence peptides demonstrate that aGPCRs appear to be pharmacologically more closely related than predicted through previous phylogenetic analysis (2). Although aGPCRs are highly different in their ectodomains, our analyses show that the internal agonist and its binding pocket seem to bear structural similarities. However, we believe that cross-activation via the tethered agonist mechanism by direct interactions of different aGPCR members in vivo is only likely in the case where the affinity of a heterologous Stachel sequence is significantly higher than that for the own Stachel sequence. Reflecting on the current pharmacological definition of receptor subtypes by shared endogenous agonists, one has to consider several aGPCRs as receptor subtypes rather than grouping them into different phylogenetically defined subfamilies (2). For example, ADP/ATP receptor subtypes and histamine receptor subtypes are phylogenetically as distantly related as many aGPCRs when only the 7-TM domain core of aGPCR is considered for analysis (Fig. 7).
FIGURE 7.
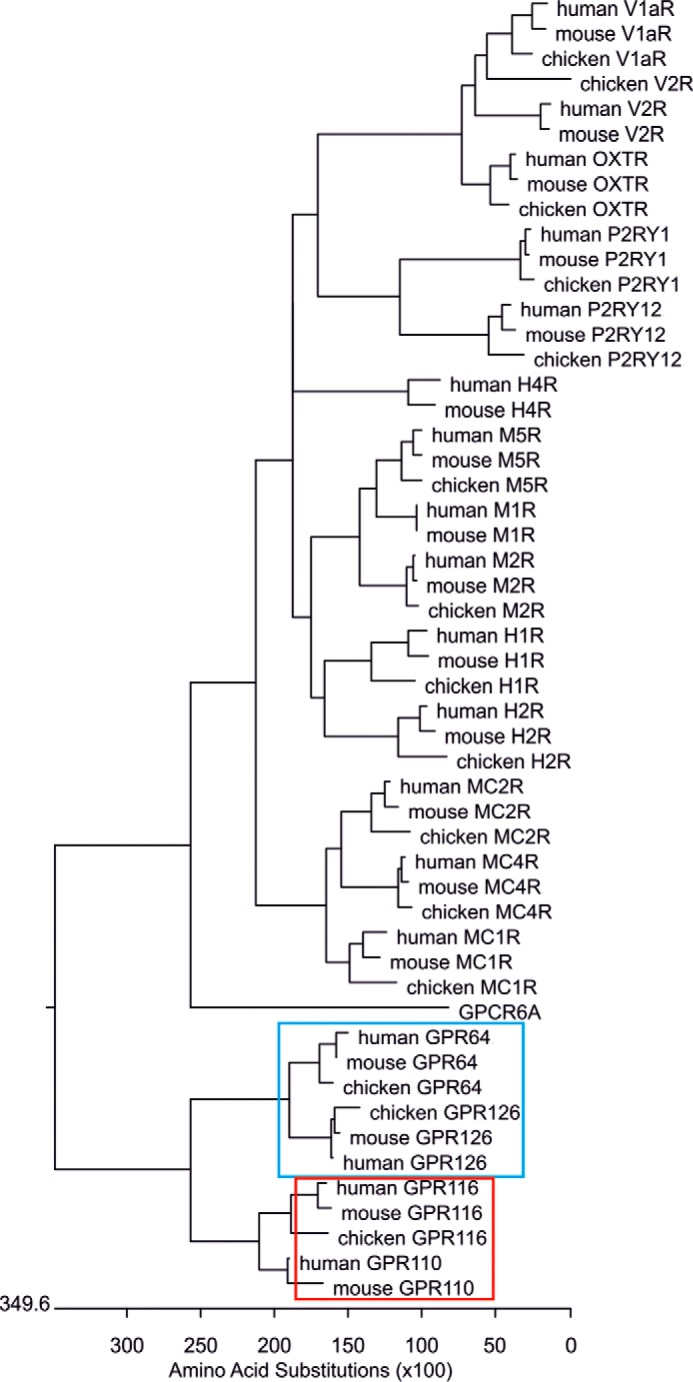
Phylogenetic distances between GPCR subtypes. Pharmacological receptor subtypes are considered when individual receptors share the same endogenous agonist. For example, acetylcholine activates muscarinic acetylcholine receptor subtypes 1–5 (M1R, M2R, and M5R shown in this tree). In peptide GPCR field, subtypes are also considered when several closely related peptides activate the same individual receptors. For example, different melanocortins and ACTH activate the five melanocortin receptor subtypes 1–5 (MC1R, MC2R, and MC4R shown in this tree). Furthermore, the vasopressin receptor subtypes V1aR and V2R are activated by vasopressin, but also by high concentrations of oxytocin including the oxytocin receptor (OXTR) as subtype into the vasopressin/oxytocin receptor group. To determine the structural divergence of the individual receptors and receptor subtypes a phylogenetic tree was constructed on the basis of the amino acid identity of the respective human, mouse, and chicken receptor orthologues (to assure the same phylogenetic time) using the identity matrix and ClustalW implemented in DNAStar. The branch lengths of the individual receptors and receptor groups in the phylogenetic tree are a measure of their phylogenetic distances. GPCR6A (member of the glutamate receptor family) was used as outgroup. The phylogenetic distances of GPR110 and GPR116 (subfamily VI, red box) as well as of GPR126 and GPR64 (subfamily VIII, blue box) are comparable with those found, e.g. in muscarinic acetylcholine receptors (M1R, M2R, M5R). The phylogenetic distances of the nucleotide receptor subtypes P2RY1 and P2RY12, both sharing ADP as agonist, are as long as the distances between the aGPCR subfamilies VI and VIII. Histamine receptor subtypes do not cluster (compare H1R, H2R, with H4R) but are still considered as receptor subtypes. This indicates that phylogenetic distances are only a weak measure to predict and group receptor subtypes in respect to pharmacological properties. As shown in this study, Stachel sequence-derived peptides can activate members across previously defined subfamilies of aGPCR (2). Their phylogenetic distances of several aGPCR subfamilies are within the range of distances of well established GPCR subtypes. Consequently, current grouping of aGPCR into 8 subfamilies needs to be reconsidered on the basis of functional data.
Another main conclusion of our analyses is that the use of Stachel sequence-derived peptides for in vivo studies, and investigations with cell lines endogenously expressing aGPCRs, have to be properly controlled due to potential cross-activation issues. On the other hand, within well controlled settings the peptide promiscuity demonstrated in this study can be helpful whenever the original peptide sequence is experimentally challenging to handle. For instance, this can be the case when a peptide is highly hydrophobic, rendering it unfeasible to work with. An alternate, more hydrophilic peptide derived from another aGPCR could be used instead. In that same line, our study shows that synthetic peptides might not always mimic the natural Stachel sequence. It is possible that the structure of the isolated peptide does not allow for binding of the receptor, whereas the natural conformation within the protein structure would facilitate this interaction. Therefore, alternate peptides may be invaluable tools for aGPCR activation until other agonistic compounds, e.g. small molecule agonists, are established.
Experimental Procedures
Materials
If not stated otherwise, all standard substances were purchased from Sigma-Aldrich (Taufkirchen, Germany), Merck (Darmstadt, Germany), and C. Roth GmbH (Karlsruhe, Germany). Cell culture material and primers were obtained from Life Technologies (Darmstadt, Germany).
Peptide Synthesis
Solid phase peptide synthesis of the peptides was performed on an automated peptide synthesizer MultiPep from Intavis AG (Köln, Germany) using standard Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry. The final side chain deprotection and cleavage from the solid support employed a mixture of TFA, water, and thioanisole (95:2.5:2.5, vol %) for the peptides. The peptides were purified to >95% purity using preparative RP-HPLC (Shimadzu LC-8, Duisburg, Germany) equipped with a PLRP-S column (300 × 25 mm, Agilent, Waldbronn, Germany). For both analytical and preparative use, the mobile phases were water (A) and acetonitrile (B), respectively, each containing 0.1% TFA. Samples were eluted with a linear gradient from 5% B to 90% B in 30 min for analytical runs and in 90 min for preparative runs. Finally, all peptides were characterized by analytical HPLC (Agilent 1100) and MALDI-MS (Bruker Microflex LT, Bremen, Germany), which gave the expected [M+H]+ mass peaks.
Generation of Construct for GPR110 Cleavage Analysis
Amino acids 18–585 of Gpr110 (accession number AY140952) were amplified from murine kidney cDNA with the following primers: 5′-GCGGCCCAGCCGGCCAGGCGCGCCGTACGAAGCTTGACCGATGGCTTCCTGCAGCAGAAA-3′ and 5′-TAGTAGCGCGGCCGCGATCCATTTTACCACAGGAACAAC-3′.
The forward primer contained a hemagglutinin (HA) tag introduced downstream of the signal peptide sequence in the cDNA and a HindIII site 5′ of the tag. The reverse primer introduced a NotI site. The PCR product was cut with the respective enzymes, purified, and ligated into a modified vector pSecTag2A containing an mFc tag with a stop codon upstream of the myc epitope (kind gift from Dr. M. Stacey, University of Leeds), which was digested with the same restriction nucleases as the PCR fragment. The construct for Gpr115 has been described previously (15).
Transfection for GPR110 Cleavage Analysis
24 h prior to transfection, 2.5 × 106 HEK293T cells were seeded in 6-cm dishes containing 4 ml of DMEM. Cells were transfected using calcium phosphate precipitation: 500 μl of a 25 mm CaCl2 solution containing 15 mg of DNA were mixed with 500 μl of 2× HBS (50 mm HEPES, pH 7.05, 10 mm KCl, 12 mm dextrose, 280 mm NaCl, 1.5 mm Na2HPO4). Cells were incubated in this solution for 8 h. Medium was discarded and 4 ml of CD293 medium were added.
Protein Preparation and Western Blot Analysis
Conditioned medium was collected from cells 86 h post-transfection, pelleted by centrifugation, and filtered through a 0.45-mm filter. Protein was concentrated by methanol/chloroform precipitation (24) and subject to electrophoresis using a 12.5% SDS-PAGE gel as described previously (25). Subsequently, proteins were transferred to a polyvinylidene difluoride membrane using a tank blotting approach. Blots were then probed with rat anti-HA high affinity antibody (Roche Applied Science), 1:10,000, overnight at 4 °C. After washing, the membrane was incubated for 1 h at room temperature with a horseradish peroxidase-conjugated goat anti-rat antibody (Jackson ImmunoResearch, Suffolk, UK) (1:10,000). Western blots were developed using enhanced chemiluminescence (ECL) detection (Amersham Biosciences, Freiburg, Germany).
Generation of Wild-type and Mutant Adhesion GPCR Constructs
Full-length human and mouse aGPCR sequences were amplified from human monocyte cDNA library or mouse organ cDNA libraries, respectively, and directly cloned into the mammalian expression vector pcDps (26). A description of the aGPCRs cloned and a list of the primers used are given in supplemental Table S1. For detection purposes and to increase cell surface expression an HA epitope and, when required, a sequence encoding the N-terminal 20 amino acids of bovine rhodopsin N terminus (as described in Refs. 13 and 27), were inserted directly downstream of the predicted signal peptide (SignalP 4.1 server) of aGPCRs by a PCR-based site-directed mutagenesis and fragment replacement strategy. Furthermore, a FLAG epitope was introduced at the very C terminus of all aGPCRs.
Point mutations were inserted in the wt receptors by site-directed mutagenesis PCR. The sequences of all wt aGPCRs and derived constructs were verified by sequencing.
In Vitro Functional Assays
Adhesion GPCRs were heterologously expressed in COS-7 or HEK293T cells grown in Dulbecco's minimum essential medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 units/ml of penicillin, and 100 μg/ml of streptomycin at 37 °C and 5% CO2 in a humidified atmosphere. For assays cells were split into 96-well plates (1.5 × 104 cells/well for COS-7 and 3.0 × 104 cells/well for HEK293T cells) and transfected with LipofectamineTM 2000 (Life Technologies) according to the manufacturer's protocol. 50 ng of plasmid DNA/well were used. To perform concentration-response curves in cAMP assay 7.5 × 105 COS-7 cells were seeded into small flasks of 25 cm2 surface.
Purified peptides were dissolved in 100% DMSO achieving a 100 mm solution, which was further diluted into 10 mm stocks (10% DMSO) using a 50 mm, pH 8, Tris buffer, and finally pH controlled. Peptide concentrations used in assays are usually 1 mm concentration, if not stated otherwise in the figure legend. The 1 mm peptide solution contains 1% DMSO and 10% of the Tris buffer used for dilution. Empty vector-transfected cells were treated with assay medium containing the same concentration of DMSO and buffer.
For cAMP measurements, 48 h after transfection, COS-7 cells were first incubated with 3-isobutylmethylxanthine (1 mm)-containing medium for 5 min and then stimulated with isobutylmethylxanthine containing medium including the given agonist for 30 min at 37 °C. After stimulation cells were lysed in LI buffer (PerkinElmer Life Sciences, Rodgau, Germany) and kept frozen at −20 °C until measurement. To measure cAMP concentration, the Alpha Screen cAMP assay kit (PerkinElmer Life Sciences) was used according to the manufacturer's protocol. The accumulated cAMP was measured in 384-well white OptiPlate microplates (PerkinElmer Life Sciences) with the Fusion AlphaScreen multilabel reader (PerkinElmer Life Sciences). For Gαs knock-down experiments 5 pmol/well of Gαs-specific siRNA (sc-29328; Santa Cruz Biotechnology, Heidelberg, Germany) and control siRNA (sc-37007; Santa Cruz Biotechnology) were used.
For direct quantitative measurement of IP1, COS-7 cells were grown in DMEM as previously described. To perform the assay, Cisbio's IP-One Tb kit (Cisbio, Codolet, France) was used. 48 h after transfection, cells were stimulated 30 min at 37 °C with 35 μl of 1× IP1 stimulation buffer (Cisbio) containing the respective reagents. Cells were lysed in 30 μl of lysis buffer (Cisbio) per well and kept frozen at −20 °C until measurement. The accumulated IP1 was measured according to the manufacturer's protocol in ProxiPlate-384 Plus microplates (PerkinElmer Life Sciences) with the EnVision Multilabel Reader (PerkinElmer Life Sciences).
For the luciferase reporter gene assay (NFAT) HEK293T cells were used. 24 h after seeding, cells were co-transfected with the given expression plasmid and the activation of the NFAT luciferase reporter plasmid (50 ng/well; PathDetect trans-reporting System). After transfection, cells were maintained in medium with 10% serum throughout the experiments. One day after transfection, cells were treated with the respective reagents for 5 h at 37 °C. The assay was terminated by washing the cells twice with PBS and addition of 100 μl of luciferase assay reagent (SteadyLite, PerkinElmer Life Sciences) and fluorescence was measured with a Victor 2 1420 Multilabel counter (PerkinElmer Life Sciences). Second messenger and reporter gene assay data were analyzed using GraphPad Prism version 6.0 for Windows (GraphPad Software Inc., San Diego, CA).
To estimate cell surface expression of receptors carrying an N-terminal HA tag, an indirect cellular ELISA was used (28). To assess the amounts of full-length HA/FLAG double-tagged constructs in the cell a sandwich ELISA was performed as previously described (29).
Animals
Mice heterozygous for a disruption of the Gpr110 locus on a 129S6/SvEv-background (15) were bred to obtain homozygous Gpr110 knock-out (129S-Gpr110 tm1(LacZ)Tcam) and wt mice. Mice were bred under specific pathogen-free conditions, a 12:12 h light/dark cycle, 21 °C, and 55% humidity. Mice had free access to food and water. Breeding and sacrificing for tissue removal were conducted in accordance with European Directive 2010/63/EU on the protection of animals used for scientific purposes and were performed with permission from the Animal Care and Use Committee (ACUC #T24/16) and the Government of the State of Saxony, Germany.
Quantitative PCR
Tissues were removed from mice sacrificed using CO2, ground, and incubated in TRIzol®. Total RNA was extracted following the manufacturer's protocol. Total RNA was purified using SV Total RNA Isolation System (Promega, Mannheim, Germany). cDNA was obtained with the reverse transcriptase Omniscript RT kit (Qiagen, Hilden, Germany) and random hexamer primers. qPCRs were performed using GoTaq® qPCR Master Mix (Promega GmbH, Mannheim, Germany) in triplicates according to the manufacturer's protocol on an Mx3000PTM (Stratagene, Santa Clara, CA). β2-Microglobulin was used as internal normalization control. For primer sequences see supplemental Table S2.
Ex Vivo Functional Assays
Organs were taken from mice sacrificed using CO2. Renal papilla was dissected immediately following kidney removal. Organs were cut with a scalpel and incubated for 45 min at 37 °C and 200 rpm with tissue-specific enzyme solution. Renal papillae solution was 520 units/ml of collagenase (type II), 3,000 units/ml of hyaluronidase (type V from sheep testes), 4,000 units/ml of DNase I in phenol red-free DMEM for cAMP accumulation assay or phosphate-free DMEM for IP-One ELISA analyses, respectively. Lung, heart and seminal vesicle solution was 78 units/ml of collagenase and 4,000 units/ml of DNase I in phenol red-free DMEM for cAMP accumulation assay or phosphate-free DMEM for IP-One ELISA analyses, respectively. Subsequently, cells were separated using a canula (0.9 × 70 mm) and pelleted. Cells of renal papilla were resuspended in 100 μl, cells of all other organs in 200 μl of phenol red-free DMEM containing 1 mm isobutylmethylxanthine (cAMP accumulation assay) or stimulation buffer (IP-One ELISA, Cisbio), respectively. 15 μl of cell suspensions were transferred into a 96-well microtiter plate and stimulated with 5 mm peptide solution, 10 μm forskolin, or 5% DMSO for 30 min, 37 °C, 5% CO2 in a humidified atmosphere. For measuring cAMP accumulation, cells were lysed in LI buffer (PerkinElmer Life Sciences) and cAMP concentrations were measured using the ALPHAScreenTM cAMP assay kit (PerkinElmer Life Sciences) according to the manufacturer's protocol. IP levels were determined by using the IP-One ELISA kit (Cisbo) following the manufacturer's instructions for 96-well plates, with the exception that cells were lysed for 1 h. Color reaction was quantified using the ELISA reader SunriseTM (Tecan, Männedorf, Switzerland).
Phylogenetic Analyses
The phylogenetic tree was constructed on the basis of amino acid identities of human, mouse, and chicken aGPCR and rhodopsin-like GPCR orthologs (to assure the same phylogenetic time) using the identity matrix (default parameters) and Clustal W implemented in the software package of DNAStar®. The accession numbers of the analyzed protein sequences are given in supplemental Table S3. Only the amino acid sequences of the aGPCR CTFs were included in the alignment. GPCR6A (member of the glutamate receptor family) was used as outgroup. Branch lengths of the individual receptors and receptor groups in the phylogenetic tree (neighbor-joining method (30)) are a measure of their phylogenetic distances.
Data Analysis
Measurements were performed at least three times in triplicates. Data are given as mean ± S.D. Graphical and statistical analyses were performed using Prism version 6.0 (GraphPad Software Inc., San Diego, CA). Statistics were performed using a one-way analysis of variance with Bonferroni as post-hoc test: *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Author Contributions
I. L., S. P., and T. S. designed experiments; L. M. D., J. W., C. W., K. U. S., J. S., S. P., and I. L. performed experiments; L. M. D., J. W., C. W., S. P., and I. L. analyzed results; I. L., S. P., and T. .S wrote the paper; all authors edited and approved of the manuscript.
Supplementary Material
Acknowledgments
We thank Samantha Hughes for proofreading of the manuscript, Tina Steinhardt for excellent technical assistance, Norbert Sträter, Renato Weiße, and Björn Kieslich for dynamic light scattering measurements of dissolved peptides.
This work was supported by Deutsche Forschungsgemeinschaft Grant FOR2149, BMBF (IFB AdipositasDiseases Leipzig) Grant ADI-K767, the European Social Fund, and the Free State of Saxony. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Fig. S1 and Tables S1–S3.
- aGPCR
- adhesion G protein-coupled receptor
- 7-TM
- seven transmembrane
- CTF
- C-terminal fragment
- GAIN
- GPCR autoproteolysis-inducing
- GPS
- GPCR autoproteolysis site
- NFAT
- nuclear factor of activated T cells
- NTF
- N-terminal fragment
- IP1
- inositol 1-phosphate
- pc
- positive control
- AA
- amino acid
- qPCR
- quantitative PCR.
References
- 1. Fredriksson R., Lagerström M. C., Lundin L.-G., and Schiöth H. B. (2003) The G-protein-coupled receptors in the human genome form five main families: phylogenetic analysis, paralogon groups, and fingerprints. Mol. Pharmacol. 63, 1256–1272 [DOI] [PubMed] [Google Scholar]
- 2. Hamann J., Aust G., Araç D., Engel F. B., Formstone C., Fredriksson R., Hall R. A., Harty B. L., Kirchhoff C., Knapp B., Krishnan A., Liebscher I., Lin H.-H., Martinelli D. C., Monk K. R., et al. (2015) International Union of Basic and Clinical Pharmacology: XCIV. adhesion G protein-coupled receptors. Pharmacol. Rev. 67, 338–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liebscher I., Schöneberg T., and Prömel S. (2013) Progress in demystification of adhesion G protein-coupled receptors. Biol. Chem. 394, 937–950 [DOI] [PubMed] [Google Scholar]
- 4. Paavola K. J., and Hall R. A. (2012) Adhesion G protein-coupled receptors: signaling, pharmacology, and mechanisms of activation. Mol. Pharmacol. 82, 777–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liebscher I., Schön J., Petersen S. C., Fischer L., Auerbach N., Demberg L. M., Mogha A., Cöster M., Simon K. U., Rothemund S., Monk K. R., and Schöneberg T. (2014) A tethered agonist within the ectodomain activates the adhesion G protein-coupled receptors GPR126 and GPR133. Cell Rep. 9, 2018–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stoveken H. M., Hajduczok A. G., Xu L., and Tall G. G. (2015) Adhesion G protein-coupled receptors are activated by exposure of a cryptic tethered agonist. Proc. Natl. Acad. Sci. U.S.A. 112, 6194–6199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Demberg L. M., Rothemund S., Schöneberg T., and Liebscher I. (2015) Identification of the tethered peptide agonist of the adhesion G protein-coupled receptor GPR64/ADGRG2. Biochem. Biophys. Res. Commun. 464, 743–747 [DOI] [PubMed] [Google Scholar]
- 8. Müller A., Winkler J., Fiedler F., Sastradihardja T., Binder C., Schnabel R., Kungel J., Rothemund S., Hennig C., Schöneberg T., and Prömel S. (2015) Oriented cell division in the C. elegans embryo is coordinated by G-protein signaling dependent on the adhesion GPCR LAT-1. PLoS Genet. 11, e1005624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Araç D., Boucard A. A., Bolliger M. F., Nguyen J., Soltis S. M., Südhof T. C., and Brunger A. T. (2012) A novel evolutionarily conserved domain of cell-adhesion GPCRs mediates autoproteolysis. EMBO J. 31, 1364–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lin H.-H., Chang G.-W., Davies J. Q., Stacey M., Harris J., and Gordon S. (2004) Autocatalytic cleavage of the EMR2 receptor occurs at a conserved G protein-coupled receptor proteolytic site motif. J. Biol. Chem. 279, 31823–31832 [DOI] [PubMed] [Google Scholar]
- 11. Petersen S. C., Luo R., Liebscher I., Giera S., Jeong S.-J., Mogha A., Ghidinelli M., Feltri M. L., Schöneberg T., Piao X., and Monk K. R. (2015) The adhesion GPCR GPR126 has distinct, domain-dependent functions in Schwann cell development mediated by interaction with laminin-211. Neuron 85, 755–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vu T. K., Hung D. T., Wheaton V. I., and Coughlin S. R. (1991) Molecular cloning of a functional thrombin receptor reveals a novel proteolytic mechanism of receptor activation. Cell 64, 1057–1068 [DOI] [PubMed] [Google Scholar]
- 13. Bohnekamp J., and Schöneberg T. (2011) Cell adhesion receptor GPR133 couples to Gs protein. J. Biol. Chem. 286, 41912–41916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Prömel S., Frickenhaus M., Hughes S., Mestek L., Staunton D., Woollard A., Vakonakis I., Schöneberg T., Schnabel R., Russ A. P., and Langenhan T. (2012) The GPS motif is a molecular switch for bimodal activities of adhesion class G protein-coupled receptors. Cell Rep. 2, 321–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Prömel S., Waller-Evans H., Dixon J., Zahn D., Colledge W. H., Doran J., Carlton M. B., Grosse J., Schöneberg T., Russ A. P., and Langenhan T. (2012) Characterization and functional study of a cluster of four highly conserved orphan adhesion-GPCR in mouse. Dev. Dyn. 241, 1591–1602 [DOI] [PubMed] [Google Scholar]
- 16. Wilde C., Fischer L., Lede V., Kirchberger J., Rothemund S., Schöneberg T., and Liebscher I. (2016) The constitutive activity of the adhesion GPCR GPR114/ADGRG5 is mediated by its tethered agonist. FASEB J. 30, 666–673 [DOI] [PubMed] [Google Scholar]
- 17. Liebscher I., and Schöneberg T. (2016) Tethered agonism: a common activation mechanism of adhesion GPCRs. Handb. Exp. Pharmacol. 234, 111–125 [DOI] [PubMed] [Google Scholar]
- 18. Abe J., Fukuzawa T., and Hirose S. (2002) Cleavage of Ig-Hepta at a “SEA” module and at a conserved G protein-coupled receptor proteolytic site. J. Biol. Chem. 277, 23391–23398 [DOI] [PubMed] [Google Scholar]
- 19. Gupte J., Swaminath G., Danao J., Tian H., Li Y., and Wu X. (2012) Signaling property study of adhesion G-protein-coupled receptors. FEBS Lett. 586, 1214–1219 [DOI] [PubMed] [Google Scholar]
- 20. Tang X., Jin R., Qu G., Wang X., Li Z., Yuan Z., Zhao C., Siwko S., Shi T., Wang P., Xiao J., Liu M., and Luo J. (2013) GPR116, an adhesion G-protein-coupled receptor, promotes breast cancer metastasis via the Gαq-p63RhoGEF-Rho GTPase pathway. Cancer Res. 73, 6206–6218 [DOI] [PubMed] [Google Scholar]
- 21. Lee J.-W., Huang B. X., Kwon H., Rashid M. A., Kharebava G., Desai A., Patnaik S., Marugan J., and Kim H.-Y. (2016) Orphan GPR110 (ADGRF1) targeted by N-docosahexaenoylethanolamine in development of neurons and cognitive function. Nat. Commun. 7, 13123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kishore A., Purcell R. H., Nassiri-Toosi Z., and Hall R. A. (2016) Stalk-dependent and Stalk-independent signaling by the adhesion G protein-coupled receptors GPR56 (ADGRG1) and BAI1 (ADGRB1). J. Biol. Chem. 291, 3385–3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Patra C., van Amerongen M. J., Ghosh S., Ricciardi F., Sajjad A., Novoyatleva T., Mogha A., Monk K. R., Mühlfeld C., and Engel F. B. (2013) Organ-specific function of adhesion G protein-coupled receptor GPR126 is domain-dependent. Proc. Natl. Acad. Sci. U.S.A. 110, 16898–16903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wessel D., and Flügge U. I. (1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138, 141–143 [DOI] [PubMed] [Google Scholar]
- 25. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 26. Okayama H., and Berg P. (1992) A cDNA cloning vector that permits expression of cDNA inserts in mammalian cells 1983. Biotechnology 24, 270–279 [PubMed] [Google Scholar]
- 27. Liberles S. D., and Buck L. B. (2006) A second class of chemosensory receptors in the olfactory epithelium. Nature 442, 645–650 [DOI] [PubMed] [Google Scholar]
- 28. Schöneberg T., Schulz A., Biebermann H., Grüters A., Grimm T., Hübschmann K., Filler G., Gudermann T., and Schultz G. (1998) V2 vasopressin receptor dysfunction in nephrogenic diabetes insipidus caused by different molecular mechanisms. Human Mutat. 12, 196–205 [DOI] [PubMed] [Google Scholar]
- 29. Römpler H., Yu H.-T., Arnold A., Orth A., and Schöneberg T. (2006) Functional consequences of naturally occurring DRY motif variants in the mammalian chemoattractant receptor GPR33. Genomics 87, 724–732 [DOI] [PubMed] [Google Scholar]
- 30. Saitou N., and Nei M. (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 [DOI] [PubMed] [Google Scholar]
- 31. Cöster M., Wittkopf D., Kreuchwig A., Kleinau G., Thor D., Krause G., and Schöneberg T. (2012) Using ortholog sequence data to predict the functional relevance of mutations in G-protein-coupled receptors. FASEB J. 26, 3273–3281 [DOI] [PubMed] [Google Scholar]
- 32. Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., and Barton G. J. (2009) Jalview Version 2: a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.