Abstract
O-Linked N-acetylglucosamine transferase (OGT) catalyzes O-GlcNAcylation of target proteins and regulates numerous biological processes. OGT is encoded by a single gene that yields nucleocytosolic and mitochondrial isoforms. To date, the role of the mitochondrial isoform of OGT (mOGT) remains largely unknown. Using high throughput proteomics, we identified 84 candidate mitochondrial glycoproteins, of which 44 are novel. Notably, two of the candidate glycoproteins identified (cytochrome oxidase 2 (COX2) and NADH:ubiquinone oxidoreductase core subunit 4 (MT-ND4)) are encoded by mitochondrial DNA. Using siRNA in HeLa cells, we found that reducing endogenous mOGT expression leads to alterations in mitochondrial structure and function, including Drp1-dependent mitochondrial fragmentation, reduction in mitochondrial membrane potential, and a significant loss of mitochondrial content in the absence of mitochondrial ROS. These defects are associated with a compensatory increase in oxidative phosphorylation per mitochondrion. mOGT is also critical for cell survival; siRNA-mediated knockdown of endogenous mOGT protected cells against toxicity mediated by rotenone, a complex I inhibitor. Conversely, reduced expression of both nucleocytoplasmic (ncOGT) and mitochondrial (mOGT) OGT isoforms is associated with increased mitochondrial respiration and elevated glycolysis, suggesting that ncOGT is a negative regulator of cellular bioenergetics. Last, we determined that mOGT is probably involved in the glycosylation of a restricted set of mitochondrial targets. We identified four proteins implicated in mitochondrial biogenesis and metabolism regulation as candidate substrates of mOGT, including leucine-rich PPR-containing protein and mitochondrial aconitate hydratase. Our findings suggest that mOGT is catalytically active in vivo and supports mitochondrial structure, health, and survival, whereas ncOGT predominantly regulates cellular bioenergetics.
Keywords: energy metabolism, glycosylation, mitochondria, mitochondrial metabolism, O-GlcNAcylation
Introduction
Protein O-linked N-acetylglucosamine glycosylation (O-GlcNAcylation)5 is emerging as an important mechanism for the regulation of numerous physiological processes in eukaryotic cells (1). Using UDP-GlcNAc as a donor, O-GlcNAc transferase (OGT) transfers GlcNAc to specific serine and/or threonine residues of protein targets. UDP-GlcNAc is derived from the nutrient-sensing hexosamine biosynthetic pathway, which utilizes 2–4% of the total glucose transported into the cell (2). O-GlcNAcylation is a dynamic process regulated by the interplay between OGT and O-GlcNAcase (OGA), the enzyme that catalyzes the removal of O-GlcNAc from protein targets. Deregulation of O-GlcNAcylation is associated with multiple diseases, including diabetes (3), cancer (4), cardiovascular diseases (5), and Alzheimer's disease (6). Furthermore, the interplay of O-GlcNAcylation with phosphorylation plays a significant role in regulation of multiple biological processes and diseases (7–11).
Although OGT is encoded by a single gene in mammals, multiple OGT splice variants have been detected (12). Furthermore, the identification of a mitochondrially targeted isoform of OGT (mOGT) that is catalytically active in vitro (13) suggested that O-GlcNAcylation may also occur in the mitochondria. Over the past 6 years, we and other groups have identified glycosylated isoforms of multiple proteins known to regulate different aspects of mitochondrial function (14–21). Most of these proteins are nuclear DNA-encoded proteins. However, the identification of two O-GlcNAcylated mitochondrial DNA-encoded proteins (cytochrome oxidase 1 (MTCO1) (21) and NADH:ubiquinone oxidoreductase core subunit 4 (MT-ND4) (20)) raises the possibility that the mOGT isoform regulates O-GlcNAcylation of proteins within the mitochondria. The recent identification of a mitochondrially localized OGA and a UDP-GlcNAc transporter support the notion that mitochondria have the machinery required for regulating functional O-GlcNAc on/off cycling (22).
Cellular O-GlcNAcylation is tightly dependent on the flux through the hexosamine biosynthetic pathway and therefore on the metabolic state of the cell. Solid evidence supports a detrimental role of chronically elevated O-GlcNAc levels in conditions of cellular damage, such as in hyperglycemia in diabetes models (23). Furthermore, O-GlcNAcylation of mitochondrial proteins is emerging as a potential mechanism for regulating cell energetics, redox signaling, and cell survival pathways. For example, O-GlcNAc cycling modulates the recruitment and activity of the mitochondrial fission protein dynamin-related protein 1 (Drp1) (16), and O-GlcNAcylation of the mitochondrial adaptor protein Milton regulates the anterograde movement of mitochondria in axons of neurons (24). Moreover, O-GlcNAcylation of respiratory chain complex proteins (complex I and complex III subunits) is associated with mitochondrial dysfunction in cardiac myocytes under hyperglycemic conditions (5). Last, overexpression of the nucleocytoplasmic OGT isoform (ncOGT) decreases mitochondrial respiration and leads to abnormal formation of mitochondrial cristae (25). These contrasting roles underscore the critical need to close the gap in our knowledge regarding the molecular mechanisms that govern O-GlcNAcylation of mitochondrial proteins.
Although a myriad of studies have associated the physiological functions of global cellular O-GlcNAcylation with ncOGT activity, comparatively little is known regarding the role of the mOGT isoform in regulating mitochondrial function. Overexpression of mOGT triggers apoptosis, a phenotype that is dependent on the mitochondrial localization of mOGT and its catalytic activity (26). This observation suggests that deregulated mitochondrial O-GlcNAcylation plays a role in programmed cell death signaling. However, the role of endogenous mOGT in cell physiology remains largely unknown.
In this study, we report 84 candidate mitochondrial glycoproteins in different tissues, of which 44 represent novel, putative O-GlcNAc-modified proteins. Two of these glycoproteins are mtDNA-encoded proteins, suggesting the involvement of a mitochondrially localized glycosyltransferase, such as mOGT. By using siRNA-mediated approaches, we show that mOGT plays a crucial role in maintaining mitochondrial structure and membrane integrity as well as in modulating cell survival in response to toxic insults. Finally, we identified four candidate glycoproteins that may exhibit reduced glycosylation when endogenous mOGT levels are decreased by siRNA, suggesting that mOGT is catalytically active in vivo and that glycosylation of these proteins may be part of the mechanism by which mOGT regulates mitochondrial physiology and function.
Results
Large Scale Screening Identifies 84 Mitochondrial Glycoprotein Candidates
Our previous candidate-driven studies of mitochondrial protein glycosylation (15) and recent studies (20) suggest that the glycosylation of mitochondrial proteins might not be rare and instead might only be rarely reported. Thus, to investigate the glycosylation of mitochondrial proteins in a more comprehensive manner, we performed a high throughput proteomics study to identify candidate glycoproteins. Specifically, we used three independent but complementary approaches for glycoprotein enrichment and detection in mitochondrial fractions from various tissues. These were (i) lectin affinity chromatography to purify glycoproteins and glycopeptides (in separate experiments) from mitochondria enriched from bovine heart tissue, (ii) enzymatic labeling of bovine heart mitochondria with UDP-azido-GalNAc via the mutant galactosyltransferase GalT1(Y289L), and (iii) azido-GalNAc metabolic labeling coupled with click chemistry and streptavidin enrichment to capture glycoproteins from mitochondria enriched from rat neuroblastoma B103 cells.
Collectively, these studies yielded 84 glycoprotein candidates with known mitochondrial function (supplemental Table 1). We note that LC-MS/MS provided only protein identifications of mitochondrial proteins that were enriched via one of the aforementioned glycosylation-enrichment approaches. Therefore, their status as O-GlcNAcylation targets will only be certain once the modification sites are unequivocally determined by using alternate approaches that are better suited to maintain the labile O-GlcNAc modification. Of these 84 candidate proteins, 44 represent novel reports of candidate mitochondrial glycoproteins, whereas the others were previously reported (17–19, 21, 27), and notably, 35 were recently reported as O-GlcNAcylated by Ma et al. (20) (supplemental Table 1). Among these glycoprotein candidates, we observed two mitochondrially encoded proteins: cytochrome c oxidase subunit 2, which is a novel report, and NADH-ubiquinone oxidoreductase chain 4, which was recently identified by Ma et al. (20) as O-GlcNAcylated. Only six of the 84 glycoprotein candidates (7%) were identified in both heart and neuroblastoma tissues. This observation probably reflects that the list of O-GlcNAcylated proteins is incomplete, and the amount of overlap may increase as the lists become more comprehensive.
When these 84 glycoprotein candidates are divided into categories according to their function, 45.2% are proteins involved in regulation of oxidative phosphorylation. Other well represented categories are lipid metabolism (7.1%), tricarboxylic acid cycle (8.3%), amino acid metabolism (4.8%), mitochondrial structure (4.8%), transporters (9.3%), and protein turnover (2.5%). These findings support the notion that glycosylation may play an important role in regulating mitochondrial metabolic pathways and consequently cellular metabolism.
mOGT Can Be Specifically Knocked Down by siRNA
Based on our glycoproteomics observations, we surmised that at least some mitochondrial proteins might become glycosylated inside the mitochondria by a mitochondrially localized glycosyltransferase, such as mOGT. To address the role of endogenous mOGT in mitochondrial protein glycosylation and function, we designed several siRNAs directed against the unique sequences of the mOGT or ncOGT splice variants (Table 1, regions underlined in black in Fig. 1A). The efficiency of siRNAs targeting OGT was tested in HeLa cells, a cell line that has been previously used to characterize the structure and function of mOGT (12, 26). Using a validated qPCR assay, we verified that transfection with two siRNAs designed against sequences that are unique to the mOGT isoform (mOGT siRNAs 1 and 2) significantly reduced mOGT mRNA levels by 45–65% when compared with the non-targeting (NT) siRNA as control (Fig. 1B). mOGT siRNAs 1 and 2 do not significantly reduce the mRNA levels of ncOGT (Fig. 1B), suggesting that they are specific for mOGT. In contrast, several siRNAs directed at sequences unique to ncOGT reduced the levels of both ncOGT and mOGT mRNAs (shown for one ncOGT siRNA; Fig. 1B), whereas the short isoform (75 kDa) was spared (data not shown).
TABLE 1.
Sequences of siRNAs used in this study
| siRNA | Sequence |
|---|---|
| Non-targeting siRNA (sense) | 5′-CAACCUGCAAUGGAUAGGUGAGUCA |
| mOGT siRNA 1 (sense) | 5′-CCCUUUCCCUUUACCUCCUUUCCCU |
| mOGT siRNA 2 (sense) | 5′-CCCUUUACCUCCUUUCCCUCCCATC |
| mOGT siRNA 3 (sense) | 5′-CAAGCGAGCCUAUGCUGCAGGGUCA |
| ncOGT siRNA 2 (sense) | 5′-CAGCACAGAACCAACGAAACGUAUG |
| ncOGT siRNA 3 (sense) | 5′-CUACUACCUCCAAAUACGUUCUUGC |
| ncOGT siRNA 4 (sense) | 5′-GAGACAAGAGCCAGACAAUACUGGT |
FIGURE 1.
Validation of OGT-isoform specific siRNAs knockdown in HeLa cells. A, schematic of human ncOGT and mOGT protein sequences. Black bars, unique sequences in each isoform of OGT targeted by siRNAs. MTS, mitochondrial targeting sequence; TPR, tetricopeptide domain; CDI, catalytic domain I; CDII, catalytic domain II. B, RT-qPCR quantification of mOGT and ncOGT mRNA levels in cells transfected with the indicated siRNAs. Scatter plots show individual data points (percentage of expression relative to tubulin, normalized to that of NT siRNA) ± S.E. (error bars) compiled from at least three experiments (***, p < 0.0001 versus NT siRNA; NS, not significant versus NT siRNA; #, p < 0.05 versus mOGT siRNAs (1 and 2); n = 3 experiments, one-way ANOVA, Bonferroni-corrected Tukey's test). C, Western blotting analyses of mitochondrial and cytosolic fractions derived from HeLa cells immunoblotted with an anti-OGT antibody, showing enrichment of a 103-kDa OGT isoform (mOGT) in the mitochondrial fraction and a 117-kDa OGT immunoreactive band in cytosolic fractions. Cytoplasm lanes were loaded with 15 and 20 μg of protein (shown in the left and right lanes, respectively) from the same sample. D and E, Western blotting analyses of ncOGT levels (D) or mOGT levels (E) in cells transfected with the indicated siRNAs for 3 days. Details on quantification and normalization of OGT expression levels are described under “Densitometric Analyses.” Scatter plots show individual data points ± S.E. compiled from at least three experiments (**, p < 0.05 versus NT siRNA; NS, not significant versus NT siRNA; ***, p < 0.001 versus NT siRNA; #, p < 0.05 versus mOGT siRNAs (siRNAs 1 and 2); data points ± S.E., n = 7–9 Western blots, one-way ANOVA, Bonferroni-corrected Tukey's test). F, 3D reconstruction image showing partial co-localization of OGT (green) with mitochondria (red). Scale bar, 10 μm. G, representative Western blots of cells transfected with the indicated siRNAs and immunoblotted for OGT (top) and loading control (β-tubulin or β-actin). Western blotting images were cropped to remove irrelevant lanes for visual clarity. H, immunofluorescence analyses of mean OGT immunofluorescence in nucleus in paraformaldehyde-fixed HeLa cells transfected with the indicated siRNAs for 3 days. Scatter plots show mean OGT fluorescence per cell with S.E. *, p < 0.05 versus NT siRNA; #, p < 0.05 versus mOGT siRNAs (siRNAs 1 and 2), n = 8–27 cells/condition from two experiments, one-way ANOVA, Bonferroni-corrected Tukey's test). I, immunofluorescence analyses of the percentage of endogenous OGT puncta that colocalize with mitochondria in paraformaldehyde-fixed HeLa cells transfected with the indicated siRNAs and immunostained with anti-OGT antibody and mito-p60 antibody. Scatter plots show mean percentage of OGT that colocalizes with mitochondria per cell with S.E. indicated (*, p < 0.05 versus NT siRNA, n = 15–30 cells/condition, one-way ANOVA, Bonferroni-corrected Tukey's test).
Next, we tested the ability of siRNAs to reduce the protein levels of the OGT isoforms by Western blotting (Fig. 1, C–E and G) and to affect OGT levels and localization by immunofluorescence (Fig. 1, H and I). Using a commercial anti-OGT antibody, we observed enrichment of a ∼103 kDa immunoreactive band in the crude mitochondrial fraction (Fig. 1C), as reported previously for mOGT (12). mOGT siRNA 1 or 2 significantly reduced the protein levels of the 103-kDa mOGT isoform (Fig. 1, E and G) as well as the number of OGT puncta co-localizing with a mitochondrial marker (Figs. 1 (F and I) and 2I). mOGT siRNAs did not significantly reduce ncOGT protein levels (Fig. 1D) or nucleus-localized OGT (Fig. 1H). Stringent statistical analysis of the compiled qPCR, Western blotting, and immunofluorescence data indicate that mOGT siRNAs 1 and 2 consistently and significantly decrease the levels of the 103-kDa isoform and mitochondrially localized OGT but do not decrease the levels of the 117-kDa isoform or nucleus-localized OGT (one-way ANOVA, Bonferroni-corrected Tukey's test, 95% confidence interval, α = 0.05, 12 experiments included).
FIGURE 2.
mOGT regulates mitochondrial morphology. A–D, epifluorescence micrographs of paraformaldehyde-fixed cells immunolabeled for OGT (green), TOM20 (red), counterstained with DAPI (blue), showing co-localization of OGT with mitochondria in control (NT) (A), pan-OGT (B), mOGT1 (C), and mOGT2 (D) siRNA-transfected cells. Scale bar, 10 μm. Each channel is shown separately for visual clarity. E–H, representative epifluorescence micrographs of paraformaldehyde-fixed cells transfected with NT siRNA (E), pan-OGT siRNA (F), mOGT1 (G), and mOGT2 (H) siRNA and immunostained for the outer mitochondrial membrane-localized protein TOM20 to visualize mitochondria. Shown are compiled quantification of the number of mitochondrially colocalized OGT puncta per cell (I), mitochondrial content, as determined by the percentage of cytosol occupied by mitochondria stained with MitoTracker Green (J), and mitochondrial interconnectivity in cells transfected with the indicated siRNAs (K). The scatter plot shown in I was compiled from two independent experiments with S.E. (error bars) shown (n = 9–20 cells). For bar graphs shown in J and K, data were compiled from three experiments and expressed as means ± S.E. (n = 100–150 cells/condition), one-way ANOVA, Bonferroni correction for multiple comparisons. *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus NT siRNA.
In contrast, ncOGT siRNAs significantly reduced the protein levels of both the ncOGT and mOGT bands (shown for one representative ncOGT siRNA in Fig. 1 (D, E, and G)). In addition, ncOGT siRNAs significantly reduced both nucleus-localized OGT levels and colocalization of OGT puncta with mitochondrial markers (Fig. 1, H and I). Statistical comparisons between mOGT siRNA 1 or 2 and pan-OGT siRNA show no significant differences for their effect on mOGT levels but significant differences for their effect on ncOGT levels.
Together, these observations show that mOGT siRNAs 1 and 2 are specific for mOGT, whereas ncOGT siRNAs behave as pan-OGT siRNAs. Therefore, we used these pan-OGT siRNAs to assess the functional consequences of reducing the expression of both mitochondrial and nucleocytoplasmic OGT isoforms in subsequent experiments. It remains unclear why the ncOGT siRNAs affect the expression of both splice variants of OGT. However, it is worth noting that the temporal splicing events that give rise to mOGT and ncOGT mRNAs have not been elucidated. One plausible explanation is that ncOGT mRNA can be generated as an independent mRNA derived from more than one precursor RNA, whereas mOGT mRNA may be derived via downstream alternative splicing of a single parental transcript that contains exon 5 (which contains the sequence uniquely found in the mOGT transcript) and other shared elements of ncOGT, giving rise to the two splice variants.
mOGT Regulates Mitochondrial Content and Morphology
Mitochondria are dynamic organelles that elongate, fragment (fission), or form highly interconnected networks. Given that ncOGT regulates mitochondrial morphology and content by glycosylating the mitochondrial fission modulator dynamin-related protein 1 (Drp1) (16), we investigated whether endogenous mOGT plays a role in mitochondrial dynamics and morphology. We analyzed mitochondrial morphology and mitochondrial content in cells transfected with the validated mOGT siRNAs 1 and 2 (Fig. 1) by using well characterized parameters of mitochondrial morphology (28, 29) as described under “Experimental Procedures.” In control cells transfected with NT siRNA, mitochondria predominantly showed a tubular and elongated morphology, some of which colocalized with OGT (Fig. 2A). siRNA-mediated knockdown of both OGT isoforms (pan-OGT siRNA) efficiently reduced OGT staining throughout the cell (Fig. 2B). In contrast, mOGT siRNA-transfected cells showed a specific reduction of OGT immunofluorescence co-localizing with a mitochondrial marker (Fig. 2, C, D, and I). These data further validate the specificity of the mOGT siRNAs by immunofluorescence. mOGT siRNAs 1 and 2 also promoted significant mitochondrial fragmentation (Fig. 2, G, H, and K). Because mitochondrial fragmentation is an obligatory step in mitochondrial autophagy (turnover) and alters mitochondrial content (30), we investigated whether mitochondrial fragmentation induced by mOGT siRNA affects mitochondrial content. We found that siRNA-mediated reduction of endogenous mOGT leads to significant mitochondrial content loss (Fig. 2J). Surprisingly, the reduction of both isoforms by pan-OGT siRNA does not affect mitochondrial content and interconnectivity (Fig. 2, F, J, and K), although there is a robust decrease in OGT staining (Fig. 2B). This finding suggests a specific role for the mOGT isoform in maintaining normal mitochondrial morphology. Reducing the levels of endogenous mOGT with a third mOGT siRNA (siRNA 3) also causes mitochondrial fragmentation and a decrease in mitochondrial content (supplemental Fig. 1). Overall, the congruence in phenotypes among cells transfected with three different siRNAs designed to target the mOGT unique sequences further supports the specificity of these siRNAs for mOGT.
In some pathological conditions, mitochondrial ROS contributes to mitochondrial fragmentation, reduced mitochondrial health, and a loss of mitochondrial levels (31). To determine whether increased mitochondrial fragmentation is associated with increased mitochondrial ROS, we stained HeLa cells transfected with NT or mOGT siRNAs with the red fluorescent, cell-permeable superoxide indicator MitoSOX. Whereas rotenone treatment robustly increased the levels of mitochondrial superoxide, transfection with mOGT siRNAs did not lead to a significant increase in mitochondrial ROS levels (supplemental Fig. 2). This observation suggests that mitochondrial ROS does not contribute to mitochondrial fragmentation or mitochondrial loss in mOGT siRNA-transfected HeLa cells.
Effects of Reduction of Endogenous mOGT on Mitochondrial Respiration and Glycolysis
HeLa cells are routinely grown in high glucose medium and are adapted to rely on a high rate of glycolysis to survive, which consequently suppresses their maximal respiratory capacity (32) through the Crabtree/Warburg effect (33). Hence, we investigated the effects of reducing the levels of endogenous mOGT by siRNA on the bioenergetic status of HeLa cells, in conditions favorable for either glycolysis or mitochondrial oxidative phosphorylation (32). Mitochondrial respiration was measured in live cells transfected with mOGT or pan-OGT siRNAs by employing an XF24e Metabolic Analyzer (Seahorse Biosciences, North Billerica, MA). The complete oxygen consumption rate (OCR) profile of NT, mOGT1, mOGT2, and pan-OGT siRNA-transfected cells is depicted in Fig. 3A for a representative experiment. In brief, HeLa cells transfected with pan-OGT siRNA in high glucose conditions (25 mm glucose) showed enhanced mitochondrial respiration as evidenced by a significant increase in basal OCR (Fig. 3B), maximal OCR (Fig. 3C), and spare respiratory capacity (Fig. 3D). Under these same conditions, mitochondrial respiration was not significantly affected in cells transfected with mOGT siRNAs when OCRs were normalized to the number of cells analyzed (protein concentration of cell lysates (Fig. 3, B–D).
FIGURE 3.
Mitochondrial respiration is altered in mOGT siRNA transfected cells grown in glucose. A, representative oxygraph showing OCRs over time in cells transfected with the indicated siRNAs. Shown are compiled analyses of basal OCR (B), maximum OCR (C), and spare respiratory capacity (reserve capacity) (D) normalized to either the number of cells (B–D) or to mitochondrial content index (percentage of cytosol occupied by mitochondria as per Fig. 2J). E–G, scatter plots from one representative experiment are shown (from three independent experiments) and expressed as means ± S.E. (error bars), one-way ANOVA, n = 12 wells/condition, Bonferroni correction for multiple comparisons. *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus NT siRNA.
However, our image-based analysis indicated that cells with reduced mOGT grown in high glucose contained significantly less mitochondria than control cells (Fig. 2J). Because OCR values essentially reflect readouts of mitochondrial function, we considered it pertinent to take into account the reduction of mitochondrial content when analyzing OCR values. When OCRs were normalized to mitochondrial content (OCRs divided by the mitochondrial content index determined in Fig. 2J), cells transfected with mOGT siRNAs displayed a significant increase in maximum OCRs but not significant changes in spare respiratory capacity (reserve capacity) compared with NT siRNA-transfected cells (Fig. 3, F and 3G). These observations suggest that a reduction of endogenous mOGT is associated with an increase in mitochondrial respiration per mitochondrion as a possible compensatory mechanism to overcome a deficiency in mitochondrial content.
To assess the role of endogenous mOGT in conditions favorable for mitochondrial oxidative phosphorylation, OCRs were then measured in cells transfected with mOGT siRNAs and cultured in medium lacking glucose but supplemented with galactose (Fig. 4A). These cells exhibited a modest, non-significant reduction in baseline OCR, maximal OCR, and spare respiratory capacity (reserve capacity) when normalized to total cellular protein levels in mOGT siRNA-transfected cells compared with NT siRNA-transfected cells (Fig. 4, B–D).
FIGURE 4.
Mitochondrial respiration is impaired in mOGT siRNA-transfected cells grown in galactose. A, representative oxygraph showing OCRs over time in cells transfected with the indicated siRNAs. Shown are compiled analyses of basal OCR (B), maximum OCR (C), and spare respiratory capacity (D) in cells grown in glucose-free medium containing galactose normalized to either the number of cells (B–D) or to mitochondrial content index (percentage of cytosol occupied by mitochondria based on MitoTracker Green staining of cells as per Fig. 2J). E–G, scatter plots from one representative experiment are shown (from three independent experiments) and expressed as means ± S.E. (error bars), n = 12 wells/condition, one-way ANOVA, Bonferroni correction for multiple comparisons. *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus NT siRNA.
A significant decrease in mitochondrial content was also observed when cells grown in the absence of glucose (galactose-containing medium), were treated with mOGT siRNA 1, mOGT siRNA 2, and pan-OGT siRNA as compared with cells treated with NT siRNA. The mean and S.D. of percentages of cytosol occupied by mitochondria in galactose-fed cells (calculated after staining with Mitotracker Green FM) were 33.9 ± 2.1% for NT siRNA, 17.9 ± 2.0% for mOGT siRNA 1, 19.2 ± 2.7% for mOGT siRNA 2, and 27.0 ± 2.1% for pan-OGT siRNA (both mOGT siRNAs p < 0.0001 versus NT siRNA; pan-OGT siRNA p < 0.024 versus NT siRNA). When OCRs were normalized to mitochondrial content, we did observe non-significant increases in baseline OCRs and in the spare respiratory capacity (reserve capacity) of cells transfected with mOGT siRNAs (Fig. 4, E–G). Similar to what was observed in high glucose conditions, pan-OGT siRNA-transfected cells showed an increase in maximum OCRs (Fig. 4, C and F) and spare respiratory capacity (Fig. 4, D and G) when normalized to either total cellular protein or mitochondrial levels in the absence of glucose. It is worth noting that NT siRNA-transfected cells exhibited the expected increase in basal OCRs when grown in the absence of glucose compared with glycolytic conditions, consistent with the Crabtree/Warburg effects (34) (9 pmol/min/μg of protein in glucose medium (Fig. 3B) versus 20 pmol/min/μg of protein in galactose medium (Fig. 4B). However, cells transfected with pan-OGT siRNA exhibited similar basal OCRs in both glucose-free medium supplemented with galactose and high glucose conditions (Figs. 4B and 5B, 15 pmol/min/μg of protein in both glucose and galactose media). These data suggest that mitochondria from pan-OGT siRNA-transfected cells are respiring at their maximal capacity regardless of glucose concentration present in the medium.
FIGURE 5.
Glycolysis is affected in cells transfected with pan-OGT siRNA. Shown are compiled analyses of non-glycolytic ECARs (A), basal glycolysis (ECARs measured following the addition of glucose) (B), glycolytic capacity (maximal ECARs after oligomycin treatment) (C), and glycolytic reserve in cells transfected with NT, pan-OGT, and mOGT siRNAs (D). Scatter plots from one representative experiment are shown (from three independent experiments) and expressed as means ± S.E. (error bars), one-way ANOVA, Bonferroni correction for multiple comparisons. *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus NT siRNA).
To determine the effect of reduced mOGT levels on glycolysis, siRNA-transfected cells were analyzed with the glycolysis stress test. In cells habituated to high glucose conditions and transfected with mOGT or pan-OGT siRNAs versus NT siRNA, no significant differences were detected in non-glycolytic acidification (Fig. 5A) or in baseline glycolysis (Fig. 5B). Maximal glycolysis can be experimentally induced by the addition of the ATP synthase inhibitor oligomycin through a phenomenon that is akin to the Crabtree effect (34). Interestingly, although pan-OGT siRNA-transfected cells showed a robust increase in maximal glycolytic capacity and glycolytic reserve compared with NT siRNA-transfected cells, cells transfected with mOGT siRNAs showed only a modest, non-significant increase (Fig. 5, C and D). These observations are consistent with a role for the global pool of OGT in the regulation of glycolysis in HeLa cells, and a mitochondria-specific role in the regulation of oxidative phosphorylation in HeLa cells for endogenous mOGT.
siRNA-mediated Reduction of Endogenous mOGT Decreases Mitochondrial Membrane Potential
Fragmented mitochondria typically exhibit reduced mitochondrial membrane potential (28), which is a sign of mitochondrial damage, and are more susceptible to lysosome-mediated degradation (30). Because siRNA-mediated reduction of endogenous mOGT levels remodels mitochondria and increases mitochondrial respiration per mitochondrion (Figs. 2 and 3), we investigated the effects of reducing endogenous levels of mOGT on the mitochondrial transmembrane potential, which is an index of mitochondrial health (28, 35). The transmembrane potential was assessed by staining cells with TMRM, a red fluorescent potentiometric dye that accumulates in respiring mitochondria. Cells transfected with mOGT siRNAs 1 and 2 showed significantly reduced mean TMRM fluorescence in mitochondria compared with control (NT siRNA-transfected) cells (Fig. 6). As an additional control for potential off-target effects of the siRNA constructs, the mitochondrial transmembrane potential was also evaluated for cells transfected with a third mOGT siRNA. As with mOGT siRNAs 1 and 2, reducing the levels of endogenous mOGT with mOGT siRNA 3 also caused a decrease in mitochondrial transmembrane potential (supplemental Fig. 1). In contrast, cells transfected with pan-OGT siRNA showed significantly increased TMRM staining compared with NT siRNA-transfected cells, suggesting that global knockdown of OGT increased mitochondrial health and mitochondrial coupling (Fig. 6D).
FIGURE 6.
siRNA-mediated reduction of mOGT alters mitochondrial transmembrane potential. Shown are representative epifluorescence micrographs of HeLa cells transfected with non-targeting siRNA (A), pan-OGT siRNA (B), or mOGT1 siRNA (C) and stained with TMRM to visualize mitochondrial membrane potential by confocal microscopy. D, compiled quantification of the mean fluorescence intensity of TMRM in HeLa cells transfected with the indicated siRNAs. Data were compiled from three independent experiments and expressed as means ± S.E. (error bars), n = 120–150 cells/condition, one-way ANOVA, Bonferroni correction for multiple comparisons. ****, p < 0.0001 versus NT siRNA.
Reduction of mOGT Decreases Mitochondrial Content and Elicits Fragmentation through Drp1-dependent Fission
Drp1 is a GTPase that translocates from the cytosol to mitochondria to facilitate fragmentation of mitochondria (fission) (36). To test whether the mitochondrial fragmentation observed in mOGT siRNA-transfected cells is facilitated by endogenous Drp1, we employed a Drp1 mutant with a K38A substitution (Drp1-DN). Drp1-DN is impaired for GTPase activity, and it inhibits the fission activity of endogenous Drp1 in a dominant negative fashion (31). HeLa cells were transfected with NT or mOGT2 siRNAs for 1 day, followed by retransfection with GFP as a control or with GFP-tagged Drp1-DN (Drp1-DN-GFP) to elicit mitochondrial fusion by inhibiting endogenous Drp1. As expected, transfection with Drp1-DN-GFP increased mitochondrial interconnectivity in NT siRNA HeLa cells (Fig. 7E). Furthermore, transient expression of GFP-Drp1-DN reversed mitochondrial fragmentation in mOGT2 siRNA-transfected cells. This data suggests that classic Drp1-dependent fission is required for the mitochondrial fragmentation observed in cells with reduced endogenous mOGT (Fig. 7, A–C and E). In addition, co-transfecting cells with Drp1-DN-GFP significantly reversed the loss of mitochondrial content and transmembrane potential seen in cells transfected with mOGT2 siRNA alone (Fig. 7, C, D, and F). These data indicate that the Drp1-mediated mitochondrial fission machinery mediates the mitochondrial fragmentation, loss of mitochondrial content, and reduced transmembrane potential induced by reduction of endogenous mOGT, which are mitochondrial phenotypes of mOGT siRNA transfection that are not associated with increased mitochondrial ROS (supplemental Fig. 2).
FIGURE 7.
Mitochondrial pathology in cells with reduced levels of endogenous mOGT depends on intact Drp1 activity. A–C, representative epifluorescence micrographs of HeLa cells transiently co-expressing the indicated siRNAs/plasmids and stained with TMRM to visualize mitochondria. Whereas cells transfected with mOGT siRNA (B) exhibit reduced TMRM staining, fission, and mitochondrial content, transient co-expression of Drp1-DN reverses mitochondrial fragmentation, content, and decreased staining with TMRM in mOGT siRNA-transfected cells (C). D–F, compiled quantifications of mean mitochondrial transmembrane potential (mean pixel intensity of TMRM fluorescence) (D), mean mitochondrial interconnectivity (average area/perimeter ratio) (E), and mitochondrial content (percentage of cytosol occupied by mitochondria) (F) in cells transfected with the indicated conditions; means ± S.E. (error bars), one-way ANOVA, Bonferroni correction for multiple comparisons, n = 69–273 cells/condition pooled from at least two experiments; **, p < 0.05; ***, p < 0.001; ****, p < 0.0001.
mOGT Is Essential for Cell Survival in Response to Toxic Insults
Because our data show that decreasing endogenous levels of mOGT lead to changes in mitochondrial structure, content, and function (Figs. 2 and 6), we next investigated the effects of siRNA-mediated reduction of endogenous mOGT on cell survival. In brief, cells transfected with OGT siRNAs for 3 days were exposed to increasing concentrations of rotenone, a well characterized mitochondria-specific toxin that promotes necrosis by inhibiting complex I and robustly eliciting mitochondrial ROS (supplemental Fig. 2). Cell survival was measured with the lactate dehydrogenase (LDH) release assay, which is an active measure of cell death. Whereas cell viability was not significantly affected by transfection with mOGT siRNAs in the absence of rotenone (Fig. 8A), cells transfected with mOGT siRNAs were significantly more resistant to rotenone treatment than NT siRNA-transfected cells (Fig. 8B). In addition, it is worth noting that image analyses of DAPI-stained cells suggest that mOGT siRNA-expressing cells do not show significant differences in cell numbers relative to NT siRNA-transfected cells (percentage change in number of DAPI-stained nuclei/field relative to NT siRNA as follows: 119.4 ± 15.3% for mOGT siRNA 1, 97.0 ± 9.8% for mOGT siRNA 2, and 94.6 ± 17.4% for pan-OGT siRNA). This result suggests that mOGT does not modulate cell number or proliferation. Although the loss of endogenous mOGT by itself does not increase oxidative stress (supplemental Fig. 2), our cell survival data suggest that mOGT regulates cell survival in response to oxidative stress.
FIGURE 8.

siRNA-mediated reduction of mOGT impacts cell survival. A, compiled analyses of cell survival as assessed by the LDH release assay (percentage of LDH release) in cells transfected with the indicated siRNAs and treated with either vehicle control (A) or increasing concentrations of rotenone (B). Data from one representative experiment are shown (from three independent experiments) and expressed as means ± S.E. (error bars), one-way ANOVA, n = 12 wells/treatment, Bonferroni correction for multiple comparisons; **, p < 0.01 versus NT siRNA; ***, p < 0.001; ****, p < 0.0001 versus NT siRNA at the indicated toxin concentration.
mOGT Glycosylates a Restricted Set of Mitochondrial Proteins
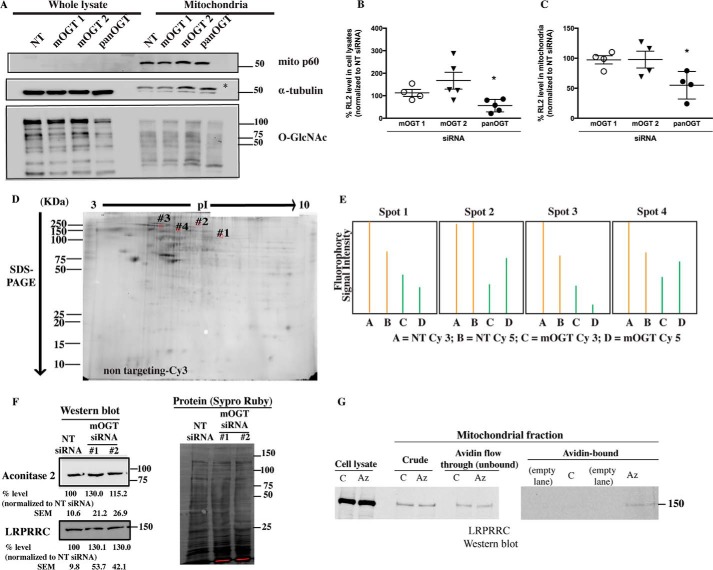
Because our overall data indicated that reducing the endogenous levels of mOGT significantly affects mitochondrial structure and some aspects of mitochondrial function, we then investigated whether alterations in mitochondrial structure and function caused by a reduction of endogenous levels of mOGT are associated with altered mOGT-mediated O-GlcNAcylation of mitochondrial proteins. By employing a well characterized anti-O-GlcNAc antibody (RL2), we assessed the O-GlcNAcylation levels of proteins in mitochondria-enriched fractions and total lysates derived from cells transfected with NT, pan-OGT, or mOGT siRNAs. Cells transfected with three different mOGT siRNAs (siRNAs 1, 2, and 3) did not exhibit a significant decrease in O-GlcNAcylation of proteins in mitochondrial fractions or in whole cell lysates (Fig. 9 (A–C) and supplemental Fig. 1A). In contrast, siRNA-mediated reduction of both ncOGT and mOGT by pan-OGT siRNA caused a robust decrease in the total levels of O-GlcNAcylated proteins in mitochondrial fractions and in whole lysates (Fig. 9, A–C). This result suggests that the ncOGT isoform contributes to the majority of mitochondrial protein O-GlcNAcylation, presumably by glycosylation of nucleus-encoded mitochondrial proteins before their import into the mitochondria (Fig. 9A). However, these data do not rule out the possibility that mOGT is catalytically active in live cells, where it may glycosylate a small number of proteins that are not detectable by SDS-PAGE.
FIGURE 9.
Identification of novel candidate substrates of mOGT by proteomics approaches. A, a representative Western blot of whole lysates and mitochondria fractions from HeLa cells transfected with NT, mOGT 1, mOGT 2 and pan-OGT siRNAs probed with RL2 (anti-O-GlcNAc), tubulin, and mito-p60 antibodies shows that mOGT knockdown does not visibly alter the O-GlcNAcylation of proteins; B and C, compiled analysis of mean RL2 immunoreactivity in cell lysates (B) and mitochondrial fractions (C) of HeLa cells transfected with the indicated siRNAs for 3 days. The scatter plots show individual data points of mean RL2 intensity relative to NT siRNA. Transfection with pan-OGT siRNA, but not with the indicated mOGT siRNAs, causes a significant reduction in O-GlcNAcylation in cell lysates and mitochondrial fractions from HeLa cells (*, p < 0.05 versus NT siRNA, n = 3–5 experiments, one-way ANOVA, Tukey's test). D, 2D Click-DIGE of proteins from mitochondrial fractions of cells transfected with NT siRNA and labeled with Cy3, shown as a reference to indicate the positions of the four spots that showed changes in fluorescence intensity in three independent experiments. E, fluorescence signal intensity of spots that were found to be down-regulated (>2-fold) in mOGT siRNA-transfected cells (green peaks indicated by A and B) compared with NT siRNA-transfected cells (orange peaks indicated by C and D). F, protein levels of aconitase 2 and leucine-rich PPR motif containing protein are not significantly decreased by mOGT siRNA-mediated knockdown. Cell lysates from cells transfected with NT, mOGT 1, and mOGT 2 siRNAs were subjected to SDS-PAGE and probed with anti-aconitase 2 and anti-LRPPRC antibodies; band intensities were quantified as described under “Densitometric Analyses.” Representative Western blots are shown. The protein level of aconitase 2 or LRPPRC relative to loading protein is expressed as a percentage relative to NT siRNA from three experiments. G, a small fraction of leucine-rich PPR motif-containing protein is glycosylated. HeLa cells were incubated with 50 μm tetraacetylated azido-GalNAc (Az) or 50 μm GalNAc (C) for 48 h. Equal amounts of mitochondria isolated from GalNAc and azido-GalNAc cell lysates were reacted with biotin-alkyne, and biotinylated proteins were captured with streptavidin beads. Error bars, S.E.
To address this hypothesis, we analyzed glycosylation differences in mitochondria-enriched fractions by Click-DIGE, a method that is more sensitive than SDS-PAGE for detecting subtle protein glycosylation differences in complex biological samples (37) (see “Experimental Procedures”). In agreement with the RL2 Western blots, we observed that reducing endogenous mOGT expression by siRNA does not visibly alter the glycosylation of the majority of proteins in enriched mitochondrial fractions. To identify minor alterations in fluorescence between control (NT siRNA-transfected) and mOGT siRNA-transfected cells, we applied a stringent computer-automated image analysis for three independent Click-DIGE experiments, each of them consisting of reciprocally labeled samples (a total of six 2D gels). We observed that transfection of HeLa cells with mOGT siRNA resulted in the consistent reduction (>2-fold decrease) of the fluorescent signals of four protein spots as compared with NT siRNA transfected cells (Fig. 9, D and E). These spots were excised, and proteins were identified by LC-MS/MS as described in the supplemental Methods, leading to the identification of four candidate glycoproteins. The identified proteins were as follows: spot 1, glycerol 3-phosphate dehydrogenase, methylcrotonyl-CoA carboxylase subunit α, mitochondrial aconitate hydratase; spot 2, leucine-rich PPR motif-containing protein (Table 2). Western blots show that the protein levels of aconitate hydratase and leucine-rich PPR motif-containing protein do not decrease in cells transfected with mOGT siRNAs (Fig. 9F). Hence, these data rule out the possibility that differences in the fluorescence intensity of spots could be attributed to differences in protein expression levels. It is worth noting that aconitate hydratase was also identified as a wheat germ agglutinin (WGA)-binding glycoprotein in our high throughput proteomics screen (supplemental Table 1). Moreover, aconitate hydratase was recently identified as O-GlcNAcylated at multiple sites (Ser-35, Ser-420, Ser-471, and Ser-562) in rat cardiac mitochondria (20). Similarly, LRPPRC was also identified as a glycoprotein candidate by glycoprotein metabolic labeling and subsequent enrichment. HeLa cells were metabolically labeled with tetraacetylated N-azidoacetylgalactosamine (azido-GalNAc) followed by a click chemistry reaction with biotin-alkyne. Anti-LRPPRC Western blotting showed enrichment of LRPPRC in the mitochondrial protein fraction captured by avidin from cells incubated with azido-GalNAc but no enrichment of LRPPRC in avidin-captured mitochondrial proteins from cells incubated with the control sugar (GalNAc) and otherwise subjected to the same protocol (Fig. 9G). We note that detection of glycosylated LRPPRC in the avidin-captured fraction required metabolic labeling of an excessive amount of biological material (∼30 10-cm dishes of cells in a single experiment), supporting our previous finding that glycosylated isoforms of some mitochondrial proteins may constitute a minute fraction of the total protein (15). Additional mitochondrial proteins were identified from spots 3 and 4 from the Click-DIGE experiments, but these were excluded from further analysis because they did not meet molecular weight and isoelectric point criteria (see “Experimental Procedures”). In aggregate, our results support a model in which mOGT probably targets a restricted set of mitochondrial proteins. The identification of four candidate protein targets of mOGT warrants further studies to identify the candidates' putative O-GlcNAcylation sites and assess whether they are genuine targets of mOGT.
TABLE 2.
Four glycoprotein candidates identified in spots with reduced glycosylation in mOGT siRNA-transfected cells
Raw mass spectrometry data were deposited in the MassIVE database, data set IDs: MSV000080316 and MSV000080317. The protein threshold was 95%, the minimum number of peptides was 2, and the peptide threshold was 90%.
| Spot | Protein name | Accession number (UNIPROTKB) | Unique peptides | Coverage |
|---|---|---|---|---|
| % | ||||
| 1 | Glycerol-3-phosphate dehydrogenase, mitochondrial | P43304 | 4 | 5.6 |
| 1 | Methycrotonyl-CoA carboxylase subunit α, mitochondrial | Q96RQ3 | 4 | 8.7 |
| 1 | Aconitate hydratase, mitochondrial | Q99798 | 2 | 3.3 |
| 2 | Leucine-rich PPR motif-containing protein, mitochondrial | P42704 | 2 | 1.9 |
Discussion
OGT Isoforms Regulate Mitochondrial Structure and Functions
Recent studies have shown that mitochondrial morphology and bioenergetic function are affected by alterations in O-GlcNAc cycling. However, these studies were performed via ncOGT and ncOGA overexpression (21, 25) or by using OGT and OGA inhibitors that are likely to affect all isoforms of OGT (20, 22). A role of mOGT in regulating mitochondrial metabolism and function has been proposed (38), but the function of mOGT was investigated in the context of overexpression (26).
Accordingly, the role of endogenous mOGT in vivo remains largely unknown, due in part to the lack of isoform-specific knock-out models. Thus, we designed and validated siRNAs that specifically and reliably reduce mOGT expression. Our study is the first to report physiological functions of endogenous mOGT on mitochondrial function and structure. In particular, our data show that siRNA-mediated reduction of endogenous mOGT elicits (i) Drp1-mediated mitochondrial fission, (ii) significant mitochondrial content loss and fragmentation, (iii) a significant decline in transmembrane potential, and (iv) a compensatory increase in mitochondrial respiration, possibly in response to reduced mitochondrial levels. Notably, we consistently detected mitochondrial phenotypes (mitochondrial fragmentation, loss of mitochondrial content, and reduced transmembrane potential) in the absence of pathological levels of mitochondrial ROS when the endogenous mOGT expression was only partially reduced by siRNA (∼45–60% knockdown in all experiments shown). Coupled with prior data indicating that overexpression of mOGT induces apoptosis (26), these observations suggest that the level of endogenous mOGT is critical for the maintenance of mitochondrial structure and function, and they suggest a role for the mOGT isoform in regulating mitochondrial content and organelle health.
OGT Isoforms Regulate Mitochondrial Structure and Function
Although the loss of mitochondrial content requires Drp1-mediated fission, the molecular mechanisms leading to a decline in mitochondrial content in mOGT-deficient cells remain to be elucidated. Interestingly, we observed that the reduction of both nucleocytoplasmic and mitochondrial isoforms of OGT (pan-OGT) elevates basal and maximal mitochondrial respiration, consistent with a previous study that showed that overexpression of ncOGT suppresses basal and maximal mitochondrial respiration in conditions of O-GlcNAc cycling disruption (25).
In contrast, the reduction of mOGT alone does not affect basal or maximal OCR or spare respiratory capacity per cell but significantly increases these parameters when normalized per mitochondrial content. These findings suggest that a reduction of endogenous mOGT up-regulates mitochondrial respiration, possibly in response to decreased mitochondrial levels. The observation that cells with reduced mOGT levels failed to increase basal or maximal OCR to similar levels as NT siRNA-transfected cells when grown in the absence of glucose suggests that mOGT modulates mitochondrial respiration in response to glucose levels via the Crabtree/Warburg effect.
In addition, our data suggest that mOGT regulates mitochondrial number and structure. The finding that siRNA-mediated reduction of mOGT decreases mitochondrial content may indicate enhanced mitochondrial turnover of damaged mitochondria (39) or impaired mitochondrial biogenesis. Given that ROS is a trigger for mitophagy, the observation that mOGT siRNA-transfected cells do not show increased mitochondrial ROS suggests that mitophagy may not be activated in mOGT siRNA-transfected cells. Therefore, the reduced mitochondrial content observed in mOGT siRNA-transfected cells is probably a consequence of reduced mitochondrial biogenesis in the absence of induced mitochondrial autophagy (40).
Our data also suggest that mOGT plays a dual role in regulating cell survival. Indeed, a reduction in mOGT levels significantly protects against treatment with rotenone, a toxin that promotes necrotic cell death. The effect of mOGT on cell survival in the presence of oxidative stress may be explained by the altered metabolic and mitochondrial morphology status of the cell. Rotenone is a complex I inhibitor that elicits a significant increase in mitochondrial superoxide levels and preferentially targets highly metabolically active cells. Hence, it is likely that the mOGT siRNA-induced decrease in transmembrane potential could favor reduced complex I activity, which induces cells to become less metabolically active (more quiescent) and consequently less sensitive to rotenone toxicity.
Collectively, these data suggest that ncOGT is a glucose-sensing protein that negatively modulates mitochondrial respiration and glycolysis, whereas mOGT is a regulator of mitochondrial structural integrity, cell death pathways that converge at the mitochondrion, and probably up-regulates mitochondrial respiration to compensate for mitochondrial loss. These isoform-specific effects of OGT on mitochondrial function are probably regulated by a combination of glucose availability, mitochondrial content, and oxidative stress. This work shows that reduction of endogenous mOGT expression elicits distinct bioenergetic and mitochondrial related morphological phenotypes supporting a major role of endogenous mOGT in integrating mitochondrial function with cellular metabolism and survival.
Identification of Candidate mOGT-dependent Glycoproteins
The underlying mechanism by which mOGT regulates mitochondrial structure and function remains unknown. The in vitro O-GlcNAc transferase activity of mOGT (41), taken together with the recent detection of O-GlcNAcase within the mitochondria and identification of a mitochondrial transporter that imports UDP-GlcNAc (22), is consistent with a model in which O-GlcNAc cycling occurs inside the mitochondria. Notably, two mitochondrial DNA-encoded proteins, COX1 and MT-ND4, have been previously reported to be O-GlcNAcylated (20, 21). In the present study, we identified cytochrome c oxidase subunit 2 (COX2), an additional mtDNA-encoded protein, as a candidate glycoprotein. Accordingly, we tested the hypothesis that endogenous mOGT mediates the glycosylation of mitochondrial proteins. We found that reduction of endogenous levels of mOGT causes a subtle decrease in O-GlcNAcylation of mitochondrial proteins, suggesting that endogenous mOGT mediates O-GlcNAcylation of select mitochondrial proteins in vivo. Therefore, the observed mitochondria-specific phenotypes associated with reduced expression of mOGT may correlate with differences in O-GlcNAcylation of a small set of mitochondrially localized proteins.
Four candidate glycoproteins were identified by LC-MS/MS from regions of the 2D gels where O-GlcNAc modification was reduced by mOGT siRNA. These findings are subject to the following caveats. First, although we provide evidence that a minute fraction of LRPPRC is glycosylated in vivo, only one of the candidates (aconitate hydratase 2, Aco2) has been unequivocally identified as O-GlcNAcylated in a previous study via identification of the O-GlcNAcylation sites (20). Second, a lack of good commercially available antibodies and insufficient sensitivity with conventional immunoprecipitation and Western blotting approaches (data not shown) have thus far precluded the detection of these presumably low abundance glycosylated candidate proteins to further corroborate whether mOGT differentially glycosylates these four proteins. Finally, we cannot rule out that additional candidate targets of mOGT would be affected if mOGT expression were further reduced or completely abolished. However, those analyses will depend on the development of cell culture or animal models using other techniques (e.g. CrispR-Cas9 gene editing).
Interestingly, all four of the candidate mOGT-dependent glycoproteins identified are nucleus-encoded proteins, which suggests that glycosylation of these proteins may occur following their import to mitochondria. Indeed, the observation that reduction of endogenous levels of mOGT causes a decrease in O-GlcNAcylation of mitochondrial proteins suggests that endogenous mOGT may glycosylate select mitochondrial proteins in vivo to regulate mitochondrial structural integrity, mitochondrial function, and cell survival (Fig. 10).
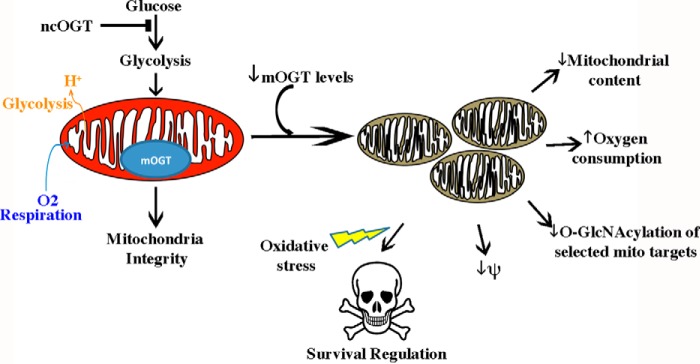
FIGURE 10.
Conceptual model on the regulation of mitochondrial function/structure by endogenous mOGT. Conceptual schematic summarizing the results of this study. Under physiological conditions, steady state levels of O-GlcNAcylation in the mitochondria are essential for normal mitochondrial structure, oxidative phosphorylation, and glycolysis. The transient loss of nucleo-/cytoplasmic OGT leads to a global decrease in mitochondrial glycosylation, which is associated with overt mitochondrial respiration and glycolysis alterations without affecting mitochondrial structure. In contrast, transient reduction of mOGT leads to a decrease in glycosylation of a restricted set of mitochondrial targets. These posttranslational events in the mitochondrion up-regulate Drp1-dependent mitochondrial fission, which is causally linked to loss of mitochondrial content, a decline in transmembrane potential, and alterations in cell survival signaling pathways. Arrows indicate progression of the pathway, whereas anti-parallel lines depict a halt in a specific pathway. Yellow lightning bolt, toxic insult (e.g. rotenone).
The candidate mOGT-dependent proteins identified in this study include (i) mitochondrial glycerol-3-phosphate dehydrogenase, which is a pivotal enzyme in many energetic pathways (42); (ii) aconitate hydratase, which is an essential enzyme for tricarboxylic acid metabolism and oxidative phosphorylation (43); (iii) methylcrotonyl-CoA carboxylase (MCCC) subunit α, which is an enzyme involved in leucine and isovaleric acid catabolism and is associated with a recessive genetic disorder termed MCCC-deficient syndrome (44, 45); and (iv) leucine-rich PPR-containing protein (LRPPRC), a protein involved in RNA translation and stability of mitochondrially encoded COX subunits, which are critical steps in the regulation of mitochondrial biogenesis (46, 47). Because loss of endogenous LRPPRC is associated with aberrations in mitochondrial morphology and dysfunction in vivo (47), it is conceivable that mOGT-mediated O-GlcNAcylation of LRPPRC may be part of the mechanism by which mOGT siRNA transfection generates the observed abnormal phenotypes in mitochondrial content and morphology. In addition, up-regulation of mitochondrial respiration may occur via glycosylation differences of metabolic enzymes (e.g. aconitate hydratase, glycerol phosphate dehydrogenase). Overall, this report warrants future studies to verify that these candidates are indeed mOGT targets and to assess the functional involvements of mOGT-mediated O-GlcNAcylation of each substrate.
Role of mOGT in Pathogenesis of Human Diseases
Dysregulation of O-GlcNAcylation is associated with several pathological conditions, including diabetes (3), cancer (4), cardiovascular diseases (5), and Alzheimer's disease (6). Mitochondrial dysfunction has been linked to development of insulin resistance and may underlie various aspects of cellular dysfunction in diabetic cardiomyopathy (3). Indeed, exposure of cardiac myocytes to high glucose increases mitochondrial protein O-GlcNAcylation; impairs the activity of complex I, III, and IV; and lowers cellular ATP and mitochondrial calcium content (21). High glucose also increases mitochondrial fragmentation and decreases mitochondrial membrane potential (48). Furthermore, a recent study revealed that the levels of OGT and OGA are dramatically altered in mitochondria from diabetic mice hearts with significant mislocalization of OGT in the mitochondrial matrix and reduced interaction of OGT with complex IV (22). In addition, deregulation of O-GlcNAcylation has also been implicated in the pathology of neurodegenerative diseases, in particular in Alzheimer's disease (49), and underlies the pathology of both diabetes mellitus and Alzheimer's disease (50). In line with this concept, our data suggest that endogenous pools of OGT isoforms are critical for regulating essential cell functions and that alterations in the expression levels of OGT, whether induced by oxidative stress or disease, can alter the bioenergetics status and cell survival under stress conditions. Specifically, our data support a critical role of mOGT in maintaining mitochondrial structure and function under normal conditions and indicate that a partial reduction of mOGT levels elicits Drp1-dependent mitochondrial pathology. However, it remains to be elucidated whether increased endogenous levels of OGT elicited by certain disease states (e.g. diabetes) represents a compensatory but beneficial response in the context of disease. Our data suggest that endogenous mOGT may play a beneficial role by up-regulating compensatory responses to maintain mitochondrial integrity and function during oxidative stress, whereas ncOGT may play a negative role in mitochondrial respiration and glycolysis. Hence, this report warrants future studies that interrogate the physiological roles of each isoform of OGT in vivo in the context of health and disease.
Experimental Procedures
Glycoprotein Enrichment and Identification from Mitochondrial Fractions
Detailed experimental methods for glycoprotein enrichment and LC-MS/MS analysis are provided in the supplemental Methods. Briefly, our high throughput screening to identify mitochondrial glycoprotein candidates was composed of three independent approaches: (i) lectin affinity purification with WGA of proteins and peptides from enriched bovine heart mitochondria, (ii) enzymatic labeling with GalT1 (Y289L) and UDP-azido-GalNAc of glycoproteins from enriched bovine heart mitochondria, and (iii) metabolic labeling of rat neuroblastoma B103 cells with tetraacetylated azido-GalNAc and subsequent analysis of the azido-labeled glycoproteins within the enriched mitochondrial fraction. Metabolically and enzymatically azido-labeled glycoproteins were subjected to copper-catalyzed azide-alkyne cycloaddition (click chemistry reaction) to covalently tag azido-glycoproteins with a biotin tag, before stringent purification over streptavidin-Sepharose. Protein identifications for glycoproteins that were enriched with all three methods were determined by LC-MS/MS analysis. Only peptide identifications with a >95% probability as determined by the PeptideProphet algorithm (51) were accepted as valid. Only protein identifications with >99% probability (as determined by the Protein Prophet algorithm (52)) and at least two unmodified and unique identified peptides were accepted as valid. Proteins with similar peptides that failed to be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. The decoy false discovery rate was calculated as described (53) and was 0% for all experiments.
Experimental Design and Statistical Rationale
The sample size for this study is 4; the study includes one enzymatic labeling experiment, one metabolic labeling experiment, and two lectin affinity purification experiments (one with proteins and one with peptides). Negative controls for each experiment were performed and analyzed by LC-MS/MS in parallel. For WGA purification, the negative control was the last PBS wash of the WGA-agarose immediately before specific elution of the captured glycoproteins (or glycopeptides) with PBS supplemented with the competitive sugar, GlcNAc. For enzymatic labeling, proteins were mock-labeled with buffer in place of GalT1 (Y289L). For metabolic labeling, cells were incubated with GalNAc in place of azido-GalNAc. Enzymatic and metabolic labeling negative controls were subjected to the same click chemistry reaction and streptavidin enrichment steps as the experimental samples. In place of technical or biological replicates, we chose to use three independent yet complementary approaches to glycoprotein enrichment (e.g. lectin affinity purification, metabolic labeling, enzymatic labeling). All mitochondrial protein identifications meeting the above criteria were reported; no additional statistical analysis was used to exclude data points or analyze the data.
mOGT siRNA Design, Sequences, and Validation of siRNA
HeLa cells were maintained in DMEM containing 10% FBS at 37 °C and 5% CO2. To analyze the effects of mOGT versus global OGT knockdown on various aspects of cell metabolism and survival, HeLa cells seeded in 6-well plates were transfected with 120 pmol of NT siRNA or siRNAs targeting mOGT (mOGT 1 and 2) or ncOGT with Lipofectamine 2000 or Oligofectamine (Life Technologies, Inc.) for 72 h. Knockdown efficiency was evaluated by real-time quantitative PCR. In brief, RNA was extracted using the RNeasy minikit (Qiagen), cDNA synthesis was performed with the iScript synthesis kit (Bio-Rad), and qPCR with SYBR as a probe was done according to the manufacturer's instructions. The following sense and antisense primers were employed to measure the efficacy of siRNA-mediated reduction of mOGT and ncOGT by qPCR: mOGT, 5′-GAACTCCAGCATAGAGCCTTATAG-3′ and 3′-GTGCAAACCACCATGTTCAG-5′; ncOGT, 5′-CTGGACAGATCTGCTCACTTTAG-3′ and 3′-CAACTGCCCTCTTTCCTTGTA-5′. All cell culture reagents and siRNA were obtained from Invitrogen unless otherwise indicated. Given the high sequence conservation between ncOGT and mOGT mRNA isoforms, the design of siRNA sequences was based on unique regions of each isoform (Fig. 2A). A total of four siRNAs designed to target mOGT and three siRNAs designed to target ncOGT were tested in this study, the respective sequences are listed in Table 1. The following two mOGT siRNAs showed a robust decrease in mOGT expression levels and were used in this study: mOGT siRNA 1, 5′-CCCUUUCCCUUUACCUCCUUUCCCU-3′; mOGT siRNA 2, 5′-CCUUUACCUCCUUUCCCUCCCAUC-3′; mOGT siRNA 3, 5′-CAAGCGAGCCUAUGCUGCAGGGUCA-3′. The following siRNA showed knockdown against both nucleocytoplasmic OGT and mitochondrial OGT (pan-OGT): 5′-CAGCACAGAACCAACGAAACGUAUG-3′. A non-targeting siRNA hairpin was used as a control (NT, 5′-CAACCUGCAAUGGAUAGGUGAGUCA-3′).
Mitochondrial Enrichment and Western Blotting
Mitochondria enriched fractions derived from HeLa cells transfected with NT, mOGT1, mOGT2, or ncOGT siRNAs were isolated using a mitochondrial isolation kit (Sigma) per the manufacturer's recommendation and as previously published (28, 31). The purity of the mitochondrial and cytosolic fractions was assessed by Western blotting. Briefly, 30 μg of protein, as assessed by the Bradford assay, was separated by SDS-PAGE, transferred to PVDF, and blocked with 2% BSA in Tris-buffered saline containing Triton X-100 (TBST) for 1 h. The membrane was incubated with various primary antibodies (OGT (Cell Signaling Technologies), catalog no. 5368; α-tubulin (Sigma-Aldrich), T9026; mito-p60 (Biogenex), MU213-UC; RL2 (Thermo Scientific), MA1-072; aconitate hydratase (Abcam), ab129105; GPD-2 (Thermo Fisher), PA5-30049; LRPPRC (Abcam), ab97505; MCCC1 (Abcam), ab197282) at 1:1000 dilution overnight followed by secondary HRP probing (1:5000 dilution) (GE Healthcare). The immunoreactive bands were detected by chemiluminescence. For each experiment, protein loading was verified by staining the membranes with the total protein stain, Sypro Ruby (Life Technologies), following protein transfer and by reblotting the PVDF membranes for β-tubulin or β-actin.
Densitometric Analyses
Densitometric analyses of OGT isoforms in Western blots were performed to assess the efficiency of reduction of endogenous isoforms of OGT (mOGT or ncOGT) by siRNAs. The integrated density (mean and area of pixels within immunoreactive bands) for each OGT isoform (117 kDa for ncOGT and 103 kDa for mOGT) was measured by selecting each band using the rectangular selection tool of ImageJ (version 1.42; National Institutes of Health, Bethesda, MD) and was adjusted by subtracting the integrated density of the nonspecific background (i.e. subtracting the integrated density of a rectangle of equal size from elsewhere on the blot). The adjusted integrated density of each OGT isoform was then divided by the specific integrated density of the respective loading control used for each Western blot (β-tubulin, ATP synthase α-subunit, or β-actin). The mOGT/loading control or ncOGT/loading control ratios for each experimental condition (mOGT- or pan-OGT siRNA-transfected cells) were then normalized to NT siRNA-transfected cells and expressed in the figures as a percentage of NT siRNA control. For Fig. 9 (B and C), the same method of quantification was applied for quantifying levels of O-GlcNAcylation in protein lysates and mitochondrial fractions (RL2 levels), as shown in Fig. 9 (B and C) with a minor modification. In brief, the specific integrated density of the RL2 bands (background-adjusted) was measured for the entire lane, divided by the specific integrated density of β-tubulin, and then normalized to NT siRNA control (percentage of NT siRNA control).
Densitometric analysis of aconitase 2 and LRPPRC were performed by dividing the mean integrated density of the aconitase 2 or LRPPRC immunoreactive bands by the mean integrated density of the entire lane of proteins stained by Sypro Ruby. Each protein of interest was adjusted for background as indicated above. The resulting data were then normalized to NT siRNA and expressed as percentage of NT siRNA control from three experiments. Notably, none of the siRNAs (mOGT or ncOGT siRNAs) significantly alter the expression levels of the loading controls (β-actin (Santa Cruz Biotechnology, Inc.), sc-81178; ATP synthase α-subunit or β-tubulin (Abcam), ab6046) used to normalize OGT levels (data not shown).
Mitochondrial Morphology, Colocalization of OGT with Mitochondria, and Mitochondrial Content Assays
We used image-based fluorescence assays to study the effects of reducing isoforms of OGT on mitochondrial membrane structure and content. In brief, HeLa cells transfected with NT, ncOGT, mOGT1, or mOGT2 siRNAs were plated in chambered coverglasses (Nunc) at 35,000 cells/well for 72 h. Following each transfection, cells were fixed in 4% paraformaldehyde, washed in PBS, permeabilized in PBS containing 0.1% Triton X-100 for 10 min, and blocked in 2% BSA. Cells were immunostained for OGT (Abcam) at a 1:1000 dilution and for the outer mitochondrial membrane-localized protein TOM20 as a mitochondrial marker (Santa Cruz Biotechnologies) at a 1:2000 dilution overnight at 4 °C. Fixed cells were then incubated with the appropriate Alexa 488-conjugated donkey anti-rabbit IgG or Alexa 545-conjugated donkey anti-mouse secondary IgG antibodies (1:400; Molecular Probes, Inc., Eugene, CA). Cells were then washed with PBS and counterstained with DAPI (1.25 μg/ml; Molecular Probes) to visualize nuclei. Immunolabeled fixed cells were imaged at 25 °C at a magnification of ×100 by using an EVOS-FL Cell Imaging System equipped with EVOS Light cubes specific for GFP (excitation/emission of 470/510 nm), RFP (excitation/emission of 531/593 nm), and Cy5 (excitation/emission of 628/692 nm) and at a magnification of ×20 (numeric aperture 0.45) or ×40 (numeric aperture 0.0.60).
To analyze mitochondrial morphology, the indices of mitochondrial interconnectivity (area/perimeter ratio per mitochondrion) were quantified by using well validated ImageJ macros (Mitochondrial and Mitophagy Morphology macros, available at the ImageJ Information and Documentation Portal). These macros allow for semi-automated image analysis of individual mitochondrial particles and mitochondrial content (28, 54). To analyze the effects of reducing endogenous mOGT on cell number, the number of DAPI-stained nuclei per epifluorescence field in NT siRNA- or mOGT siRNA-transfected HeLa cells was manually counted by an observer who was blind to the transfection status of the cells, and the data were expressed as number of DAPI-stained cells/epifluorescence field.
Image-based analyses were used to quantify the extent by which OGT siRNAs (mOGT siRNA or pan-OGT siRNA) decrease the levels of OGT isoforms in high resolution RGB epifluorescence images (TIFF) of paraformaldehyde-fixed cells immunolabeled with anti-OGT and stained with DAPI. Using ImageJ, the mean OGT fluorescence signal within DAPI-stained nuclei was measured, fluorescence background was subtracted (region of interest that was of equal pixel area yet devoid of cells), and the mean background-adjusted OGT signal/nuclei was then calculated. The mean fluorescence levels of nuclear OGT were quantified for ≥30 cells/transfection condition from two experiments (mOGT siRNA- or pan-OGT siRNA). To measure the extent to which OGT siRNAs affect the colocalization of OGT with mitochondria by immunocytochemistry, the number and percentage of OGT puncta that colocalize with mitochondria per cell was quantified in cells immunostained for OGT and mitochondria using ImageJ and similar image-based methodology as published (55). All image-based quantifications (counting the number of OGT puncta/cell or assessing the percentage of OGT colocalizing with mitochondria) were performed manually by an observer who was blind to the transfection status.
Bioenergetic Analysis; Mitochondrial Oxygen Consumption and Glycolysis
The effects of siRNA-mediated knockdown of mOGT on cellular bioenergetics were evaluated by measuring the OCRs and extracellular acidification rates (ECARs) by using an XF24e extracellular flux analyzer (Seahorse Biosciences). Whereas OCR is used as an index of mitochondria-specific metabolic activity, ECAR is a proxy for glycolysis. Cells transfected with NT, mOGT1, mOGT2, or ncOGT siRNAs were trypsinized 2 days post-transfection and reseeded at 40,000 cells/well in a XF24e cell culture microplate in quadruplicate wells under various media conditions as described below. Cells seeded in either high glucose (25 mm) or galactose (10 mm) were supplemented with 2 mm glutamine and 1 mm sodium pyruvate and grown at 37 °C and 5% CO2 for 24 h before the assay. OCRs in transfected HeLa cells were analyzed at baseline and following the sequential addition of 1 μm oligomycin, 300 nm FCCP, and 1 μm rotenone/antimycin A to determine the ATP-linked OCRs, maximal OCRs, and mitochondrially derived OCRs, respectively, as published previously (28). Spare respiratory capacity was calculated as described previously (56, 57). In brief, the basal OCRs were subtracted from the maximal OCRs induced by FCCP by applying the formula, spare respiratory capacity = FCCP-induced maximal OCR − basal OCR. To directly assess for glycolysis, the glycolysis stress assay was performed in transfected HeLa cells per the manufacturer's recommendations (Seahorse Biosciences). In brief, cells were incubated in glucose-free medium for 1 h before treatment with the following compounds: 25 mm glucose, 1 μm oligomycin, and 2 mm deoxyglucose to measure basal glycolysis, glycolytic capacity, and non-glycolytic acidification rates, respectively. ECARs were recorded as milli-pH (pH × 10−3) (mpH/min). The glycolytic reserve was then obtained by subtracting ECARs associated with baseline glycolysis from ECARs associated with the glycolytic capacity. At the end of each assay, OCRs and ECARs were normalized to protein concentration (μg/ml) for its respective well. In addition, OCRs were normalized to a mitochondrial marker to analyze the effects of knocking down OGT isoforms on oxidative phosphorylation per mitochondrial content.
Transmembrane Potential Assays
To study the effects of reducing endogenous mOGT or both pools of OGT (mOGT and ncOGT) on mitochondrial membrane potential, HeLa cells transfected with NT, pan-OGT, mOGT1, or mOGT2 siRNAs in DMEM containing 20 mm glucose for 72 h were stained with the red fluorescent, cell-permeable potentiometric dye tetramethylrhodamine methyl ester (TMRM) at a concentration of 100 nm and incubated at 37 °C for 45 min before washing once with warm medium. The mean TMRM-specific fluorescence intensity in mitochondria was normalized to the background fluorescence intensity measured within the nucleus as described previously (28, 35).
Drp1 Functional Assays
Mitochondrially targeted GFP (cytochrome oxidase VIII import sequence, mito-GFP), and GFP-tagged wild-type and dominant negative K38A Drp1 in the EGFP-N1 backbone were generously provided by Dr. Stefan Strack (University of Iowa). To assess the effects of inhibiting endogenous Drp1 activity on transmembrane potential, mitochondrial content, and morphology in cells with reduced expression of mOGT, cells seeded in 4-well-chambered slides were first transfected with mOGT or NT siRNAs at 20 pmol/well for 24 h before retransfecting with 0.5 μg of mitochondria-targeted GFP as a control plasmid or GFP-tagged Drp1-DN to inhibit endogenous Drp1 at a Lipofectamine concentration of 0.20%. Cells were maintained in 37 °C and 5% CO2 for an additional 48 h (72-h total transfection time) before processing cells for image-based quantifications of transmembrane potential, mitochondrial content, and morphology as further described above.
Cell Survival Assessment
To study the effects of reducing mOGT on cell survival, NT, mOGT 1 and 2 siRNA-transfected cells were plated at 2500 cells/well in black-sided 96-well plates and incubated with DMSO as vehicle control or with increasing concentrations of the mitochondrial complex I inhibitor rotenone (at 250, 500, or 1000 nm) for 24 h at 37 °C and 5% CO2. Cell survival was assessed using the LDH assay per the manufacturer's instructions (Thermo Fisher).
Click-DIGE
To identify candidate proteins of mOGT-mediated O-GlcNAcylation, Click-DIGE was performed as described previously (37) on mitochondria-enriched fractions obtained from HeLa cells transfected with either NT or mOGT1 siRNAs. Briefly, HeLa cells transfected with NT or mOGT1 siRNA were metabolically labeled with 25 mm tetraacetylated N-azidoacetylgalactosamine (C33365, Thermo Fisher) for 48 h. Mitochondria-enriched fractions of metabolically labeled NT and mOGT siRNA samples (50 μg of protein/reaction) were subjected to separate click chemistry reactions with size-matched alkyne-Cy3 or alkyne-Cy5 (Lumiprobe) in the presence of 5 mm TBTA (Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine), 5 mm ascorbic acid, 5 mm CuSO4, 47% DMSO for 1 h at room temperature in the dark. Reacted proteins were precipitated by chloroform/methanol (4:1), resulting in the following samples: NT-alkyne-Cy3, mOGT1-alkyne-Cy3, NT-alkyne-Cy5, and mOGT1-alkyne-Cy5.
Equal amounts of protein samples derived from lysates of NT and mOGT siRNA-transfected cells were reacted with different fluorophores and then combined in pairs, resulting in two sets of samples labeled with reciprocal fluorophores (NT-Cy3 + mOGT-Cy5 and NT-Cy5 + mOGT-Cy3). Combined samples were resuspended in Destreak Rehydration Solution (GE Healthcare) and ampholytes (Biolyte 3-10, Bio-Rad). Samples were run in separate 2D electrophoresis gels in the following conditions: strip 3–7, 11 cm, immobilized pH gradient (Bio-Rad) total 26,000 V-h for 1D electrophoresis and 8–16% Tris-HCl gel (Bio-Rad), run at 200 V for 1 h for 2D electrophoresis. Gels were imaged with a Typhoon laser imager immediately following electrophoresis (Cy3, excitation 550 nm and emission 570 nm; Cy5, excitation 650 nm and emission 670 nm BP30). Gels were fixed in methanol/acetic acid, stained with Sypro Ruby (Bio-Rad) overnight for total protein staining, and imaged with a Typhoon laser scanner.
Cy3 and Cy5 images were aligned, and spot analysis was performed using PDQuest software and by applying the following corrections and exclusion criteria to the images: correction for streaking, ignoring false positive spots, excluding spots that do not follow the same expression pattern regardless of the fluorophore (e.g. correcting for intrinsic differences in fluorophore intensity), and excluding spots that do not show up in both reciprocal gels. The inclusion criteria applied to select spots of interest were as follows: ≥2-fold decrease in mOGT siRNA versus NT siRNA-transfected cells in both reciprocally labeled samples and in three independent Click-DIGE experiments. Spots were cut with the ProPik2 (PerkinElmer Life Sciences) instrument and sent for identification of protein candidates by LC-MS/MS. Spot processing and LC-MS/MS details are described in the supplemental Methods. Only peptide identifications with a >90% probability as determined by the PeptideProphet algorithm (51) were accepted as valid. Only protein identifications with >95% probability (as determined by the Protein Prophet algorithm (52)) and at least two unmodified and unique identified peptides were accepted as valid. The mass spectrometry data were filtered by hits that contained the correct molecular weight and isoelectric point (pI) in the 2D gel.
Experimental Design and Statistical Rationale
The sample size for this study is 3; the study includes three biological replicates of the same experiment. All mitochondrial protein identifications meeting the above criteria were reported; no additional statistical analysis was used to exclude data points or analyze the data.
Statistical Analyses
Unless indicated otherwise, all experiments were performed at least three times, and the results are presented as means ± S.E. Statistical analyses with an α of 0.05 were performed by applying one-way ANOVAs followed by a Bonferroni-corrected test for multiple group comparisons (>3) or by performing t tests for comparing two groups.
Author Contributions
A. B. M. conceived the central hypothesis of isoform-specific defects, R. K. D. and P. M. B. jointly conceived the study, J. L. S. contributed to Figs. 1–6, 8, 9, Table 1, and supplemental Figs. 1 and 2; R. Y. D. contributed to Figs. 1, 2, and 6–8 and supplemental Figs. 1 and 2; A. B. M. contributed to supplemental Table 1; J. L. S., A. B. M., R. K. D., R. Y. D., and P. M. B. designed the experiments; J. L. S., R. Y. D., and A. B. M. performed the experiments; and J. L. S., P. M. B., R. K. D., and A. B. M. wrote and edited the manuscript.
Supplementary Material
Acknowledgments
We acknowledge Dr. Dave Quilici and Rebekah Wolsey at the Nevada Proteomics Center for assistance with LC-MS/MS-mediated identification and analyses of candidate targets of mOGT.
This project was funded by National Institutes of Health Grants P20 GM103554 (a COBRE grant in “Cell Biology of Signaling Across Membranes”) and R21 AG047623 (Role of Protein Glycosylation on Mitochondrial Function). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Table 1, Figs. 1 and 2, and Methods.
- O-GlcNAcylation
- O-linked N-acetylglucosamine glycosylation
- OGT
- O-GlcNAc transferase
- mOGT
- mitochondrial OGT
- ncOGT
- nucleocytoplasmic OGT
- NT
- non-targeting
- OGA
- O-GlcNAcase
- qPCR
- quantitative PCR
- ANOVA
- analysis of variance
- DIGE
- differential in-gel electrophoresis
- OCR
- oxygen consumption rate
- WGA
- wheat germ agglutinin
- FCCP
- carbonyl cyanide p-trifluoromethoxyphenylhydrazone
- TMRM
- tetramethylrhodamine, methylester, perchlorate.
References
- 1. Hart G. W. (2014) Minireview series on the thirtieth anniversary of research on O-GlcNAcylation of nuclear and cytoplasmic proteins: nutrient regulation of cellular metabolism and physiology by O-GlcNAcylation. J. Biol. Chem. 289, 34422–34423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Marshall S., Bacote V., and Traxinger R. R. (1991) Discovery of a metabolic pathway mediating glucose-induced desensitization of the glucose transport system: role of hexosamine biosynthesis in the induction of insulin resistance. J. Biol. Chem. 266, 4706–4712 [PubMed] [Google Scholar]
- 3. Vaidyanathan K., and Wells L. (2014) Multiple tissue-specific roles for the O-GlcNAc post-translational modification in the induction of and complications arising from type II diabetes. J. Biol. Chem. 289, 34466–34471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ma Z., and Vosseller K. (2014) Cancer metabolism and elevated O-GlcNAc in oncogenic signaling. J. Biol. Chem. 289, 34457–34465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Marsh S. A., Collins H. E., and Chatham J. C. (2014) Protein O-GlcNAcylation and cardiovascular (patho)physiology. J. Biol. Chem. 289, 34449–34456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhu Y., Shan X., Yuzwa S. A., and Vocadlo D. J. (2014) The emerging link between O-GlcNAc and Alzheimer disease. J. Biol. Chem. 289, 34472–34481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Butkinaree C., Park K., and Hart G. W. (2010) O-Linked β-N-acetylglucosamine (O-GlcNAc): extensive crosstalk with phosphorylation to regulate signaling and transcription in response to nutrients and stress. Biochim. Biophys. Acta 1800, 96–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gu Y., Ande S. R., and Mishra S. (2011) Altered O-GlcNAc modification and phosphorylation of mitochondrial proteins in myoblast cells exposed to high glucose. Arch. Biochem. Biophys. 505, 98–104 [DOI] [PubMed] [Google Scholar]
- 9. Hart G. W., Slawson C., Ramirez-Correa G., and Lagerlof O. (2011) Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 80, 825–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang Z., Gucek M., and Hart G. W. (2008) Cross-talk between GlcNAcylation and phosphorylation: site-specific phosphorylation dynamics in response to globally elevated O-GlcNAc. Proc. Natl. Acad. Sci. U.S.A. 105, 13793–13798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhong J., Martinez M., Sengupta S., Lee A., Wu X., Chaerkady R., Chatterjee A., O'Meally R. N., Cole R. N., Pandey A., and Zachara N. E. (2015) Quantitative phosphoproteomics reveals crosstalk between phosphorylation and O-GlcNAc in the DNA damage response pathway. Proteomics 15, 591–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Love D. C., Kochran J., Cathey R. L., Shin S. H., and Hanover J. A. (2003) Mitochondrial and nucleocytoplasmic targeting of O-linked GlcNAc transferase. J. Cell Sci. 116, 647–654 [DOI] [PubMed] [Google Scholar]
- 13. Hanover J. A., Yu S., Lubas W. B., Shin S. H., Ragano-Caracciola M., Kochran J., and Love D. C. (2003) Mitochondrial and nucleocytoplasmic isoforms of O-linked GlcNAc transferase encoded by a single mammalian gene. Arch. Biochem. Biophys. 409, 287–297 [DOI] [PubMed] [Google Scholar]
- 14. Burnham-Marusich A. R., Snodgrass C. J., Johnson A. M., Kiyoshi C. M., Buzby S. E., Gruner M. R., and Berninsone P. M. (2012) Metabolic labeling of Caenorhabditis elegans primary embryonic cells with azido-sugars as a tool for glycoprotein discovery. PLoS One 7, e49020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Burnham-Marusich A. R., and Berninsone P. M. (2012) Multiple proteins with essential mitochondrial functions have glycosylated isoforms. Mitochondrion 12, 423–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gawlowski T., Suarez J., Scott B., Torres-Gonzalez M., Wang H., Schwappacher R., Han X., Yates J. R 3rd, Hoshijima M., and Dillmann W. (2012) Modulation of dynamin-related protein 1 (DRP1) function by increased O-linked-β-N-acetylglucosamine modification (O-GlcNAc) in cardiac myocytes. J. Biol. Chem. 287, 30024–30034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Clark P. M., Dweck J. F., Mason D. E., Hart C. R., Buck S. B., Peters E. C., Agnew B. J., and Hsieh-Wilson L. C. (2008) Direct in-gel fluorescence detection and cellular imaging of O-GlcNAc-modified proteins. J. Am. Chem. Soc. 130, 11576–11577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kung L. A., Tao S. C., Qian J., Smith M. G., Snyder M., and Zhu H. (2009) Global analysis of the glycoproteome in Saccharomyces cerevisiae reveals new roles for protein glycosylation in eukaryotes. Mol. Syst. Biol. 5, 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nandi A., Sprung R., Barma D. K., Zhao Y., Kim S. C., Falck J. R., and Zhao Y. (2006) Global identification of O-GlcNAc-modified proteins. Anal. Chem. 78, 452–458 [DOI] [PubMed] [Google Scholar]
- 20. Ma J., Liu T., Wei A. C., Banerjee P., O'Rourke B., and Hart G. W. (2015) O-GlcNAcomic profiling identifies widespread O-linked β-N-acetylglucosamine modification (O-GlcNAcylation) in oxidative phosphorylation system regulating cardiac mitochondrial function. J. Biol. Chem. 290, 29141–29153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hu Y., Suarez J., Fricovsky E., Wang H., Scott B. T., Trauger S. A., Han W., Hu Y., Oyeleye M. O., and Dillmann W. H. (2009) Increased enzymatic O-GlcNAcylation of mitochondrial proteins impairs mitochondrial function in cardiac myocytes exposed to high glucose. J. Biol. Chem. 284, 547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Banerjee P. S., Ma J., and Hart G. W. (2015) Diabetes-associated dysregulation of O-GlcNAcylation in rat cardiac mitochondria. Proc. Natl. Acad. Sci. U.S.A. 112, 6050–6055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wells L., Vosseller K., and Hart G. W. (2003) A role for N-acetylglucosamine as a nutrient sensor and mediator of insulin resistance. Cell Mol. Life Sci. 60, 222–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pekkurnaz G., Trinidad J. C., Wang X., Kong D., and Schwarz T. L. (2014) Glucose regulates mitochondrial motility via Milton modification by O-GlcNAc transferase. Cell 158, 54–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tan E. P., Villar M. T., E L., Lu J., Selfridge J. E., Artigues A., Swerdlow R. H., and Slawson C. (2014) Altering O-linked β-N-acetylglucosamine cycling disrupts mitochondrial function. J. Biol. Chem. 289, 14719–14730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shin S. H., Love D. C., and Hanover J. A. (2011) Elevated O-GlcNAc-dependent signaling through inducible mOGT expression selectively triggers apoptosis. Amino Acids 40, 885–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Trinidad J. C., Barkan D. T., Gulledge B. F., Thalhammer A., Sali A., Schoepfer R., and Burlingame A. L. (2012) Global identification and characterization of both O-GlcNAcylation and phosphorylation at the murine synapse. Mol. Cell. Proteomics 11, 215–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dagda R. K., Gusdon A. M., Pien I., Strack S., Green S., Li C., Van Houten B., Cherra S. J. 3rd, Chu C. T. (2011) Mitochondrially localized PKA reverses mitochondrial pathology and dysfunction in a cellular model of Parkinson's disease. Cell Death Differ. 18, 1914–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dagda R. K., Pien I., Wang R., Zhu J., Wang K. Z., Callio J., Banerjee T. D., Dagda R. Y., and Chu C. T. (2014) Beyond the mitochondrion: cytosolic PINK1 remodels dendrites through protein kinase A. J. Neurochem. 128, 864–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Twig G., Elorza A., Molina A. J., Mohamed H., Wikstrom J. D., Walzer G., Stiles L., Haigh S. E., Katz S., Las G., Alroy J., Wu M., Py B. F., Yuan J., Deeney J. T., et al. (2008) Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 27, 433–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dagda R. K., Cherra S. J. 3rd, Kulich S. M., Tandon A., Park D., and Chu C. T. (2009) Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J. Biol. Chem. 284, 13843–13855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gusdon A. M., and Chu C. T. (2011) To eat or not to eat: neuronal metabolism, mitophagy, and Parkinson's disease. Antioxid. Redox Signal. 14, 1979–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Suchorolski M. T., Paulson T. G., Sanchez C. A., Hockenbery D., and Reid B. J. (2013) Warburg and Crabtree effects in premalignant Barrett's esophagus cell lines with active mitochondria. PLoS One 8, e56884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Diaz-Ruiz R., Rigoulet M., and Devin A. (2011) The Warburg and Crabtree effects: on the origin of cancer cell energy metabolism and of yeast glucose repression. Biochim. Biophys. Acta 1807, 568–576 [DOI] [PubMed] [Google Scholar]
- 35. Dagda R. K., Barwacz C. A., Cribbs J. T., and Strack S. (2005) Unfolding-resistant translocase targeting: a novel mechanism for outer mitochondrial membrane localization exemplified by the Bβ2 regulatory subunit of protein phosphatase 2A. J. Biol. Chem. 280, 27375–27382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Smirnova E., Griparic L., Shurland D. L., and van der Bliek A. M. (2001) Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell 12, 2245–2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Burnham-Marusich A. R., Plechaty A. M., and Berninsone P. M. (2014) Size-matched alkyne-conjugated cyanine fluorophores to identify differences in protein glycosylation. Electrophoresis 35, 2621–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bond M. R., and Hanover J. A. (2015) A little sugar goes a long way: the cell biology of O-GlcNAc. J. Cell Biol. 208, 869–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Klionsky D. J., and Emr S. D. (2000) Autophagy as a regulated pathway of cellular degradation. Science 290, 1717–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mitchell T., Chacko B., Ballinger S. W., Bailey S. M., Zhang J., and Darley-Usmar V. (2013) Convergent mechanisms for dysregulation of mitochondrial quality control in metabolic disease: implications for mitochondrial therapeutics. Biochem. Soc. Trans. 41, 127–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lazarus B. D., Love D. C., and Hanover J. A. (2006) Recombinant O-GlcNAc transferase isoforms: identification of O-GlcNAcase, yes tyrosine kinase, and tau as isoform-specific substrates. Glycobiology 16, 415–421 [DOI] [PubMed] [Google Scholar]
- 42. Mráček T., Drahota Z., and Houštěk J. (2013) The function and the role of the mitochondrial glycerol-3-phosphate dehydrogenase in mammalian tissues. Biochim. Biophys. Acta 1827, 401–410 [DOI] [PubMed] [Google Scholar]
- 43. Beinert H., and Kennedy M. C. (1993) Aconitase, a two-faced protein: enzyme and iron regulatory factor. FASEB J. 7, 1442–1449 [DOI] [PubMed] [Google Scholar]
- 44. Chu C. H., and Cheng D. (2007) Expression, purification, characterization of human 3-methylcrotonyl-CoA carboxylase (MCCC). Protein Expr. Purif. 53, 421–427 [DOI] [PubMed] [Google Scholar]
- 45. Holzinger A., Röschinger W., Lagler F., Mayerhofer P. U., Lichtner P., Kattenfeld T., Thuy L. P., Nyhan W. L., Koch H. G., Muntau A. C., and Roscher A. A. (2001) Cloning of the human MCCA and MCCB genes and mutations therein reveal the molecular cause of 3-methylcrotonyl-CoA: carboxylase deficiency. Hum. Mol. Genet. 10, 1299–1306 [DOI] [PubMed] [Google Scholar]
- 46. Ruzzenente B., Metodiev M. D., Wredenberg A., Bratic A., Park C. B., Cámara Y., Milenkovic D., Zickermann V., Wibom R., Hultenby K., Erdjument-Bromage H., Tempst P., Brandt U., Stewart J. B., Gustafsson C. M., and Larsson N. G. (2012) LRPPRC is necessary for polyadenylation and coordination of translation of mitochondrial mRNAs. EMBO J. 31, 443–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mourier A., Ruzzenente B., Brandt T., Kühlbrandt W., and Larsson N. G. (2014) Loss of LRPPRC causes ATP synthase deficiency. Hum. Mol. Genet. 23, 2580–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Makino A., Suarez J., Gawlowski T., Han W., Wang H., Scott B. T., and Dillmann W. H. (2011) Regulation of mitochondrial morphology and function by O-GlcNAcylation in neonatal cardiac myocytes. Am. J. Physiol. Regul. Integr. Comp. Physiol. 300, R1296–R1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lefebvre T., Dehennaut V., Guinez C., Olivier S., Drougat L., Mir A. M., Mortuaire M., Vercoutter-Edouart A. S., and Michalski J. C. (2010) Dysregulation of the nutrient/stress sensor O-GlcNAcylation is involved in the etiology of cardiovascular disorders, type-2 diabetes and Alzheimer's disease. Biochim. Biophys. Acta 1800, 67–79 [DOI] [PubMed] [Google Scholar]
- 50. Lozano L., Lara-Lemus R., Zenteno E., and Alvarado-Vásquez N. (2014) The mitochondrial O-linked N-acetylglucosamine transferase (mOGT) in the diabetic patient could be the initial trigger to develop Alzheimer disease. Exp. Gerontol. 58, 198–202 [DOI] [PubMed] [Google Scholar]
- 51. Keller A., Nesvizhskii A. I., Kolker E., and Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 52. Nesvizhskii A. I., Keller A., Kolker E., and Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 53. Käll L., Storey J. D., MacCoss M. J., and Noble W. S. (2008) Assigning significance to peptides identified by tandem mass spectrometry using decoy databases. J. Proteome Res. 7, 29–34 [DOI] [PubMed] [Google Scholar]
- 54. Dagda R. K., Merrill R. A., Cribbs J. T., Chen Y., Hell J. W., Usachev Y. M., and Strack S. (2008) The spinocerebellar ataxia 12 gene product and protein phosphatase 2A regulatory subunit Bβ2 antagonizes neuronal survival by promoting mitochondrial fission. J. Biol. Chem. 283, 36241–36248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dagda R. K., Zhu J., Kulich S. M., and Chu C. T. (2008) Mitochondrially localized ERK2 regulates mitophagy and autophagic cell stress: implications for Parkinson's disease. Autophagy 4, 770–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Keuper M., Jastroch M., Yi C. X., Fischer-Posovszky P., Wabitsch M., Tschöp M. H., and Hofmann S. M. (2014) Spare mitochondrial respiratory capacity permits human adipocytes to maintain ATP homeostasis under hypoglycemic conditions. FASEB J. 28, 761–770 [DOI] [PubMed] [Google Scholar]
- 57. Pace C., Banerjee T. D., Welch B., Khalili R., Dagda R. K., and Angermann J. (2016) Monomethylarsonous acid, but not inorganic arsenic, is a mitochondria-specific toxicant in vascular smooth muscle cells. Toxicol. in Vitro 35, 188–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.