Abstract
UHRF2 has been implicated as a novel regulator for both DNA methylation (5mC) and hydroxymethylation (5hmC), but its physiological function and role in DNA methylation/hydroxymethylation are unknown. Here we show that in mice, UHRF2 is more abundantly expressed in the brain and a few other tissues. Uhrf2 knock-out mice are viable and fertile and exhibit no gross defect. Although there is no significant change of DNA methylation, the Uhrf2 null mice exhibit a reduction of 5hmC in the brain, including the cortex and hippocampus. Furthermore, the Uhrf2 null mice exhibit a partial impairment in spatial memory acquisition and retention. Consistent with the phenotype, gene expression profiling uncovers a role for UHRF2 in regulating neuron-related gene expression. Finally, we provide evidence that UHRF2 binds 5hmC in cells but does not appear to affect the TET1 enzymatic activity. Together, our study supports UHRF2 as a bona fide 5hmC reader and further demonstrates a role for 5hmC in neuronal function.
Keywords: dioxygenase, DNA demethylation, DNA methylation, epigenetics, mouse genetics, neurobiology, protein-DNA interaction
Introduction
As an evolutionarily conserved epigenetic modification, DNA methylation at the C5 position of cytosine (5mC)4 in mammalian cells mainly occurs at the CG dinucleotides and has been shown to play critical roles in development and regulation of gene expression and genome stability (1–3). Recent studies have demonstrated that 5mC can be oxidized by the TET (ten eleven translocation) family of dioxygenases (4–6). TET proteins oxidize 5mC consecutively to generate 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC), all of which have been implicated as intermediates of active DNA demethylation (5–7).
Among the oxidative derivatives of 5mC, 5hmC is much more abundant than 5-formylcytosine and 5-carboxylcytosine and appears to be stable (8–10). Thus, besides as an intermediate of active demethylation, 5hmC, also termed DNA hydroxymethylation, has been postulated to function as a distinct epigenetic modification. This concept has inspired efforts to search for 5hmC-specific binding or effector proteins that may at least in part transmit the function of 5hmC in epigenetic regulation. So far, a few proteins have been shown to preferentially bind 5hmC or both 5mC and 5hmC (11, 12), including UHRF2 (12).
UHRF2, also known as NIRF, was originally identified as a novel RING finger protein implicated in cell cycle regulation (13). At the levels of amino acid sequence and domain organization, UHRF2 is highly related to UHRF1, a protein that has emerged as an evolutionarily conserved epigenetic regulator essential for DNMT1-mediated DNA maintenance methylation in mammals (14, 15). Both UHRF1 and UHRF2 contain a SET and RING-associated domain that specifically recognizes hemimethylated CpG and a tandem Tudor domain and a plant homeodomain that bind cooperatively the H3 tails with H3K9me2/3, and a C-terminal RING finger domain that confers a ubiquitin E3 ligase activity (16–18). Despite their sequence and biochemical similarity, we and others showed previously that UHRF2 cannot substitute for UHRF1 in supporting DNA maintenance methylation (16, 17). It is noteworthy that UHRF2 was not only identified as a specific reader of 5hmC but also shown to promote TET1 enzymatic activity (12). The preferential binding of 5hmC by UHRF2 but not UHRF1 was further confirmed by our structural study (19). However, as a sole protein implicated in reading of three epigenetic markers, H3K9 methylation, 5mC, and 5hmC, the physiological functions of UHRF2 in development and DNA methylation and/or demethylation are unknown.
Recent studies have begun to reveal a critical role for epigenetic modification of DNA, including 5mC and 5hmC, and TET1 enzyme in neurogenesis, learning, memory consolidation, and extinction (20–24). In fact, 5hmC was initially demonstrated to be highly enriched in the brain (25). In this study, we have generated a Uhrf2 knock-out mouse model to investigate the physiological function of UHRF2. Although the Uhrf2 null mice are viable, fertile, and grossly normal, they display reduced levels of 5hmC but normal 5mC in the brain and other tissues. Furthermore, the Uhrf2 null mice display a partial impairment in spatial memory. We also provide evidence that UHRF2 binds 5hmC in vivo but does not appear to affect TET1 enzymatic activity.
Results
The UHRF2 Expression Pattern in Mice
To investigate the physiological function of UHRF2 in mice, we first analyzed the pattern of UHRF2 expression. We prepared protein extracts from various mouse tissues and examined the levels of UHRF2 proteins by Western blotting analysis. Using the GAPDH proteins as a loading control, we detected the presence of UHRF2 proteins in various tissues. A representative result in Fig. 1A showed that the UHRF2 protein was mostly detected in several tissues, including the thymus, spleen, lung, adrenal gland, and ovary. In addition, UHRF2 was also detected in several tissues in the brain (cerebellum, hippocampus, and cerebral cortex). The Western blotting analysis thus indicates that UHRF2 expresses in a tissue-specific manner in mice.
FIGURE 1.
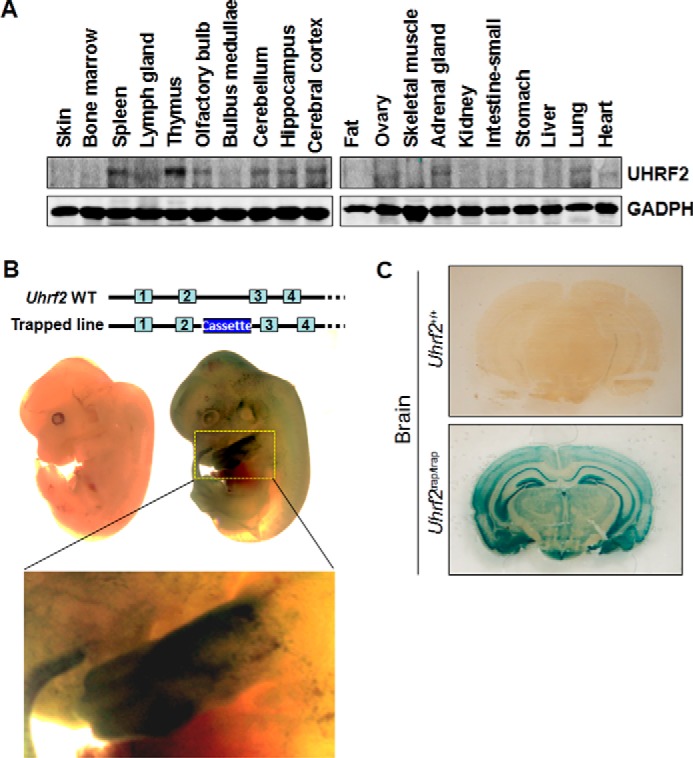
UHRF2 expression in mice. A, analysis of UHRF2 expression in various mouse tissues by Western blotting analysis. The protein extracts were prepared from various tissues of 8-week-old mice. Western blotting analysis was performed with a homemade anti-human UHRF2 antibody. GAPDH served as a loading control. B, analysis of UHRF2 expression in Uhrf2 gene-trapping embryos by whole-mount staining. The littermates of the wild-type and Uhrf2/Uhrf2β-geo heterozygous embryos were processed for β-geo staining. Note that no staining was observed for the wild-type embryo. Also shown is an enlarged picture for fore limb bud. C, analysis of the UHRF2 expression in the brain. Shown are 8-week Uhrf2/Uhrf2β-geo mice by β-geo staining.
To further define the UHRF2 expression pattern in mice, we obtained a Uhrf2 gene trapping ES cell line (AD0406) from the Mutant Mouse Regional Resource Center (MMRRC). Within this ES cell line, a gene trap vector containing a splice acceptor sequence upstream of a reporter gene, β-geo, was inserted into the second intron of Uhrf2 genomic DNA (Fig. 1B, top). This insertional mutation created a fusion transcript containing exon 1 and exon 2 of Uhrf2 joined to the β-geo marker, consequently disrupting the rest of Uhrf2 transcripts. The Uhrf2 trapping mice were successfully derived from this ES cell line. We found that both heterozygous and homozygous Uhrf2 trapping mice were viable and phenotypically normal (data not shown). Making use of the β-geo marker, which is under the control of native Uhrf2 promoter, we performed β-geo whole-mount staining of embryonic day 12.5 Uhrf2/Uhrf2β-geo heterozygous embryos. The representative results are shown in Fig. 1B. Whereas no obvious staining was observed for the control wild-type embryos, a general weak β-geo staining was observed for the Uhrf2/Uhrf2β-geo littermates, except the forelimbs, which displayed a strong staining.
Recent studies have begun to reveal a critical role for DNA methylation, 5hmC, and TET1 enzyme in neurogenesis, learning, and memory (20–24). Because UHRF2 has been identified as a reader for both 5mC and 5hmC (12, 16, 17, 19), we next wished to determine in more detail the UHRF2 expression pattern in the brain. Again using β-geo staining, we examined the UHRF2 expression in the brain derived from WT and Uhrf2/Uhrf2β-geo heterozygous mice at the age of 10 weeks. The results in Fig. 1C showed a relatively strong staining in the hippocampus and cortex, indicating that the UHRF2 is highly expressed in the hippocampus and cortex regions.
Uhrf2 Knock-out Mice Are Viable, Fertile, and Grossly Normal
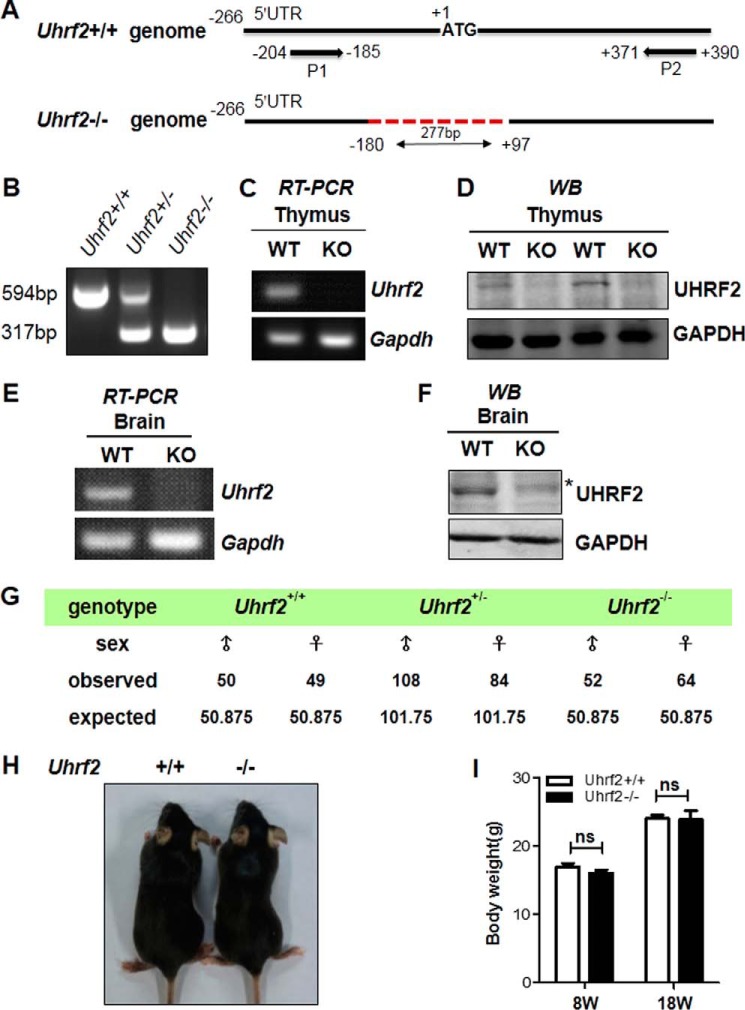
We extensively characterized the Uhrf2β-geo heterozygous and homozygous mice and observed no obvious phenotypes in embryonic development, growth, fertility, and health (data not shown). Although Western blotting analysis detected no UHRF2 proteins in the thymus of Uhrf2β-geo homozygous mice (data not shown), we could not exclude the possibility that the Uhrf2 exon 2 might not be 100% spliced to the acceptor upstream of the β-geo, thus producing a low level of normal Uhrf2 mRNA and consequently a low level of UHRF2 protein that is below the detection of Western blotting. To ascertain the physiological function of UHRF2, we thus generated a Uhrf2 knock-out mouse model via CRISPR-CAS9 technology (26). DNA sequencing analysis revealed a 277-bp deletion corresponding to bp −180 to +97 of the Uhrf2 coding region and thus expected to completely inactivate the Uhrf2 gene (Fig. 2A). The Uhrf2 wild-type and deletion allele could be distinguished by PCR-based genome typing using P1 and P2 primers as illustrated in the top panel of Fig. 2A and by the representative results in Fig. 2B. The lack of Uhrf2 transcripts in the Uhrf2−/− mice was confirmed by qPCR analysis of total RNAs prepared from the thymus (Fig. 2C) and brain (Fig. 2E), respectively, of wild-type and Uhrf2−/− mice using a pair of Uhrf2-specific primers. By Western blotting analysis, we further confirmed the lack of UHRF2 proteins in the protein extracts prepared from the thymus tissues (Fig. 2D) and brain (Fig. 2F) of wild-type and Uhrf2−/− mice.
FIGURE 2.
The Uhrf2 null mice are viable, fertile, and grossly normal. A, diagram illustrating 277-bp deletion of Uhrf2 genomic DNA generated by CRISPR-Cas9 and genotyping strategy by PCR. The position is relative to the first ATG of Uhrf2 mRNA. P1, upstream PCR primer; P2, downstream PCR primer. B, representative result of PCR-based genotyping of wild-type, heterozygous, and homozygous Uhrf2 deletion mutant mice. The wild-type genomic DNA gave rise to a 594-bp PCR product, whereas the deletion mutant resulted in a 317-bp PCR product. C, RT-PCR analysis of the levels of Uhrf2 mRNA in the thymus from wild-type and Uhrf2−/− mice. As a positive control, the levels of Gapdh mRNA were also analyzed by RT-PCR. D, Western blotting analysis of UHRF2 proteins in the thymus from wild-type and Uhrf2−/− mice. Thymus tissues were prepared from two pairs of wild-type and Uhrf2−/− littermates and subjected to Western blotting analysis. GAPDH served as a loading control. E, RT-PCR analysis of the levels of Uhrf2 mRNA in the brain from wild-type and Uhrf2−/− mice. F, Western blotting analysis of UHRF2 proteins in the brain from wild-type and Uhrf2−/− mice. G, summary of genotyping results of breeding between Uhrf2+/− mice. H, no gross morphological difference between the wild-type and Uhrf2 null mice. I, no difference in body weight between the wild-type and Uhrf2 null mice. The representative body weights were measured at 8 and 18 weeks, respectively. ns, not significant. Error bars, S.E.
Through extensive breeding between the heterozygous mice, we obtained a Mendelian ratio of the wild-type, heterozygous (Uhrf2+/−) and homozygous (Uhrf2−/−) mice (Fig. 2E), indicating that Uhrf2 is not required for embryonic development. Under the regular diet, we also did not observe any significant difference between the wild-type and Uhrf2−/− mice in growth, size (Fig. 2F), and body weight at ages of 8 and 18 weeks (Fig. 2G). In addition, both Uhrf2−/− male and female mice were fertile. We thus conclude that Uhrf2 is not required for mouse embryonic development or for fertility and general health.
Reduced 5hmC in Genomic DNA of Mutant Mouse Brain
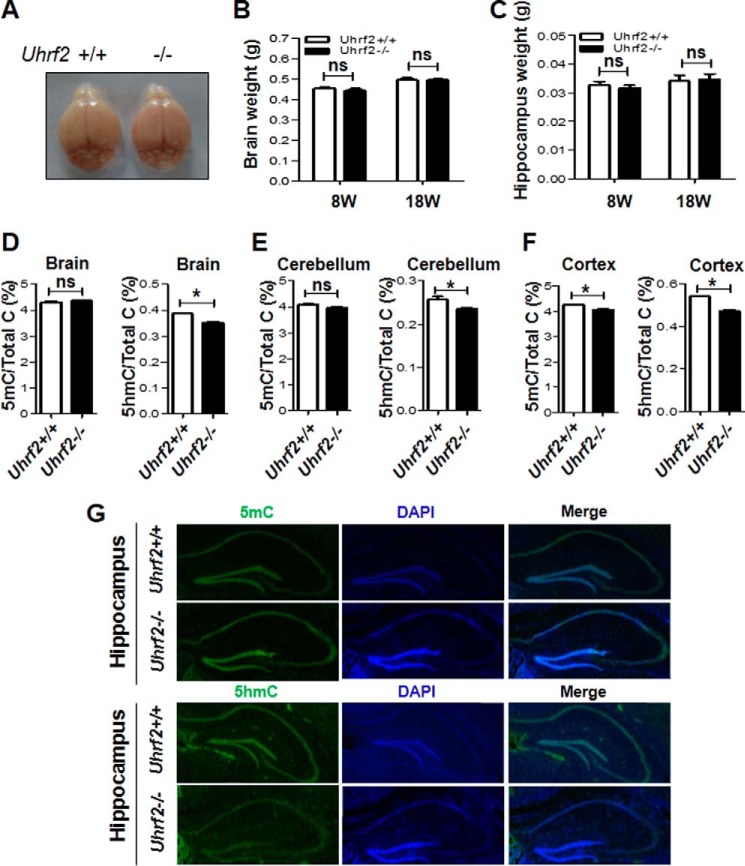
Recent studies indicate that TET1 is not required for general health of mice but has a role in control of 5hmC in the hippocampus, neurogenesis, and learning and memory (22, 23). Because UHRF2 was identified as a 5hmC reader and shown to enhance TET1 activity, we next examined whether Uhrf2 knock-out affected the levels of 5mC and 5hmC in the brain. By comparing brains from seven pairs of wild-type and Uhrf2−/− littermates at ages of 8 and 18 weeks, respectively, we found that there were no detectable difference in the anatomy (Fig. 3A), brain weight (Fig. 3B), and hippocampus weight (Fig. 3C). We then prepared genomic DNA from entire brains, cerebellums, and cortexes derived from three pairs of wild-type and Uhrf2−/− littermates. The resulting genomic DNA was subjected to quantitative analysis for the levels of 5mC and 5hmC by liquid chromatography. As shown in Fig. 3D, on average, ∼4.3 and 4.4% of cytosines were methylated in the wild-type and Uhrf2−/− brains, respectively, indicating that Uhrf2 knock-out had no significant influence on DNA methylation. However, Uhrf2 knock-out resulted in a moderate but statistically significant reduction of the level of 5hmC in the brain (from 0.386% in the wild-type to 0.352% in the Uhrf2−/−). Similarly, we found that Uhrf2 knock-out resulted in a moderate reduction of 5hmC in both cerebellum and cortex (Fig. 3, E and F). Interestingly, although Uhrf2 knock-out did not significantly affect the levels of 5mC in the cerebellum (Fig. 3E), a small but statistically significant reduction of 5mC was again observed in the cortex (Fig. 3F). Altogether, Uhrf2 knock-out appears to affect the levels of 5hmC more than 5mC.
FIGURE 3.
Loss of UHRF2 results in reduced levels of 5hmC in the brain. A, no gross morphological difference between the brains of wild-type and Uhrf2 null mice. The data were from three pairs of Uhrf2+/+ and Uhrf2−/− littermate mice. B, no significant difference in the brain weight between the wild-type and Uhrf2 null mice. The data were from three pairs of Uhrf2+/+ and Uhrf2−/− littermate mice. C, no significant difference in the hippocampus weight between the wild-type and Uhrf2 null mice. The data were from seven pairs of Uhrf2+/+ and Uhrf2−/− littermate mice. D–F, Uhrf2 null mice have reduced levels of 5hmC in the brain (D), cerebellum (E), and cortex (F). The genomic DNA was prepared from the entire brain, cerebellum, and cortex of three pairs of Uhrf2+/+ and Uhrf2−/− littermate mice and subjected to measurement of 5mC and 5hmC by HPLC. G, immunohistochemistry showing reduced levels of 5hmC in the hippocampus of Uhrf2−/− mice. The brain tissue sections from Uhrf2+/+ and Uhrf2−/− mice were analyzed for the levels of 5mC and 5hmC by immunohistochemistry using 5mC- or 5hmC-specific antibody as indicated. The tissue sections were also stained by DAPI to reveal DNA. *, p < 0.05; ns, not significant. Error bars, S.E.
Because our aforementioned data indicated that UHRF2 is highly expressed in the hippocampus, we performed immunohistochemistry (IHC) analysis of the hippocampus using antibodies against 5mC and 5hmC. As shown in Fig. 3G, we found no difference in the 5mC staining of the hippocampus derived from either the wild-type or Uhrf2−/− mice. Interestingly, in comparison with the wild-type hippocampus, a reduced level of 5hmC was consistently observed for the Uhrf2−/− hippocampus. Although the IHC results were not quantitative, it nevertheless suggests that the loss of UHRF2 leads to a more severe reduction of 5hmC in the hippocampus region, where UHRF2 is highly expressed. Taken together, these data suggest that UHRF2 regulates the levels of 5hmC in the brain and especially in the hippocampus.
Uhrf2 Null Mice Show Normal Locomotor Activity and Anxiety Level
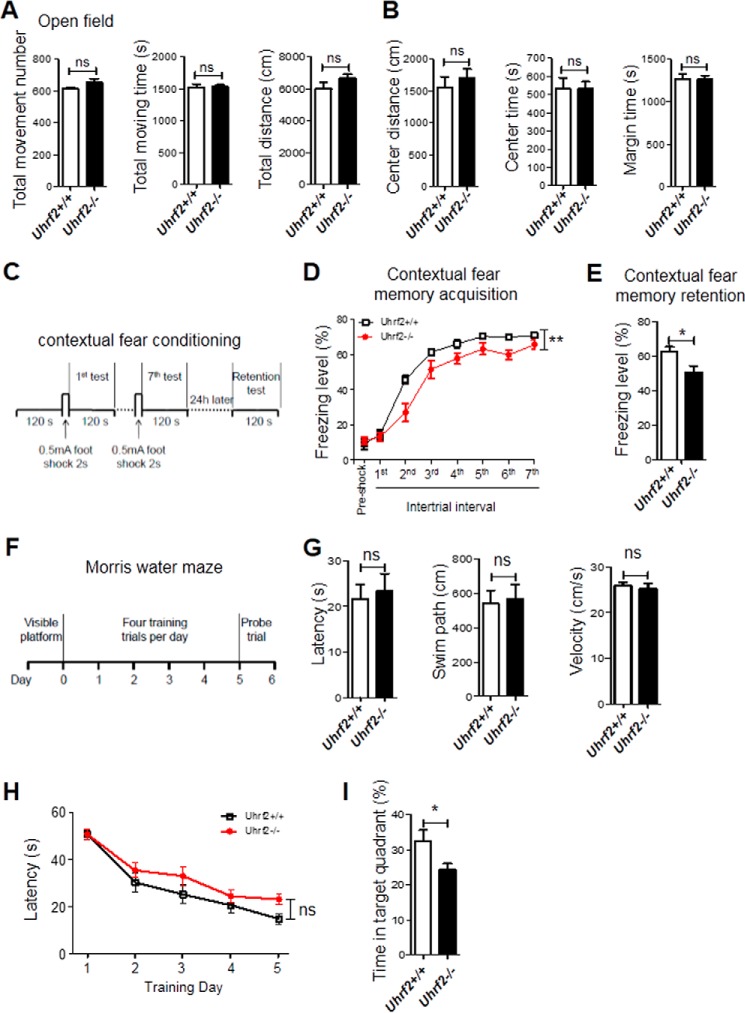
We next examined whether loss of UHRF2 has potential impact upon basal behavioral performance of adult mice. The open field test was utilized to examine locomotor activity and exploratory and anxious behaviors. We found no significant difference between Uhrf2+/+ and Uhrf2−/− mice in the locomotor activity (Fig. 4A; Student's t test, p > 0.05). Furthermore, compared with Uhrf2+/+ mice, Uhrf2−/− mice had a comparable anxiety level in an open field test (Fig. 4B; Student's t test, p > 0.05). Taken together, these results indicate that loss of UHRF2 may not influence locomotor activity and anxiety level.
FIGURE 4.
Uhrf2 null mice show normal locomotor activity and anxiety level but impaired learning and memory. A and B, no significant difference in locomotor activity and anxiety level between Uhrf2+/+ and Uhrf2−/− mice. A, total movement number, total moving time, and total distance in the open field. B, distance of movement in the center, moving time in the center, and margin in the open field (30-min observation; n = 10 + 10). C-E, Uhrf2 KO mice show impaired contextual fear memory acquisition and retention in a contextual fear conditioning test. C, schematic diagram of the contextual fear conditioning test design. D, freezing level of mice in contextual fear conditioning during training. E, freezing level of mice in contextual fear conditioning memory retention (n = 10 + 9). F–I, Uhrf2 KO mice show impaired spatial reference memory in a Morris water maze test. F, schematic diagram of the Morris water maze test. G, similar escape latency, length of swim path, and swim velocity in the visible platform test, indicating that Uhrf2 KO mice have normal visual acuity and mobility. H, escape latency of mice reaching the platform during the training phase of the Morris water maze test. I, percentage of time mice spent in the target quadrant during the probe trial test (n = 9 + 9; ns, not significant; *, p < 0.05; error bars, S.E.).
Uhrf2 Null Mice Show Impaired Spatial Memory Acquisition and Retention
DNA methylation and hydroxymethylation are important for neurobehavioral phenomena (20–24). Because UHRF2 is highly expressed in hippocampus and Uhrf2 knock-out results in reduced 5hmC in hippocampus, we surmised that Uhrf2 knock-out might affect hippocampus-dependent learning and memory. To verify our hypothesis, mice were individually conditioned with seven unconditioned stimulus/conditioned stimulus (US/CS; foot shock/shock chamber) pairings for 120 s, as illustrated in Fig. 4C, and the contextual fear memory acquisition was examined by recording the percentage of time spent in freezing response during a 120-s intertrial interval without foot shock. Although both Uhrf2+/+ and Uhrf2 null mice displayed a progressive increase in freezing response following the US/CS shock pairs (Fig. 4D; two-way repeated measures ANOVA, significant main effect of US/CS pairs, F(1, 17) = 130.42, p < 0.001), there was a significant difference between the Uhrf2+/+ and Uhrf2 null mice (Fig. 4D; two-way repeated-measures ANOVA, significant main effect of genotype, F(1, 17) = 16.361, p < 0.01), indicating that Uhrf2 null mice had impaired contextual fear memory acquisition. Moreover, the post hoc analysis revealed a significant difference at the second (p < 0.01), fourth (p < 0.05), and sixth (p < 0.01) inter-trial interval. In addition, two groups of mice displayed similar freezing behavior before the conditioning (Fig. 4D; Student's t test, p > 0.05), suggesting that the Uhrf2 null mice were not more prone to freezing behavior when introduced to a novel environment. Moreover, compared with the Uhrf2+/+ mice, Uhrf2 null mice also displayed significantly lower freezing scores on the retention of contextual fear memory (Fig. 4E; Student's t test, p < 0.05). Together, these results show that hippocampus-dependent contextual fear memory acquisition and retention are impaired in Uhrf2 null mice.
We also subjected Uhrf2+/+ mice and Uhrf2 null littermate mice to a Morris water maze test according to a protocol in Fig. 4F. The measurement indexes of the visible platform test showed that Uhrf2 null mice had normal vision and motivation (Fig. 4G; Student's t test, p > 0.05, respectively). During training phase, there is no significant difference in learning curve between two groups of mice (Fig. 4H; two-way repeated measures ANOVA, significant main effect of genotype, F(1, 16) = 3.119, p > 0.05). However, during the probe trial, Uhrf2 null mice spent a significantly less time in the target quadrant than Uhrf2+/+ mice (Fig. 4I; Student's t test, p < 0.05), suggesting that Uhrf2 null mice exhibited the deficits in hippocampus-dependent spatial reference memory. Taken all together, these results suggest that loss of UHRF2 could impair hippocampus-dependent spatial memory.
Uhrf2 Knock-out Causes Deregulated Expression of Neuron-related Genes
To examine whether Uhrf2 knock-out affected gene expression in the hippocampus, we prepared total RNAs from the hippocampus of the Uhrf2+/+ and Uhrf2−/− mice and analyzed the gene expression profiling by RNA-seq. By using 1.5-fold change as a cut-off, we found that Uhrf2 knock-out led to up-regulation of 151 genes and down-regulation of 99 genes, indicating a limited effect of loss of UHRF2 on gene expression. Interestingly, gene ontology analysis showed that the affected genes are enriched for function in neuroactive ligand-receptor interaction, including adrenergic receptor a2b, dopamine receptor D1 (Drd1a), and dopamine receptor D2 (Drd2) and Fos. To validate the RNA-seq data, we carried out quantitative RT-PCR analysis to measure the levels of Adora2a, Fos, Drd1a, Drd2, and Gabra2a transcripts from hippocampus tissues of the wild-type and Uhrf2−/− mice. As shown in Fig. 5C, we confirmed the increased expression for these genes in the Uhrf2−/− mice. From our RNA-seq data, we found that the expression levels of Tet1, Tet2, and Tet3 were not affected by the loss of UHRF2 (Fig. 5D). Thus, the reduced levels of 5hmC in the brain and hippocampus of the Uhrf2−/− mice are unlikely to be due to alteration of gene expression of the Tet1, Tet2, and Tet3 genes. Taken together, we conclude that loss of UHRF2 affects the expression of a limited number of neuron-related genes, which may in part account for the impaired memory formation in the Uhrf2−/− mice.
FIGURE 5.
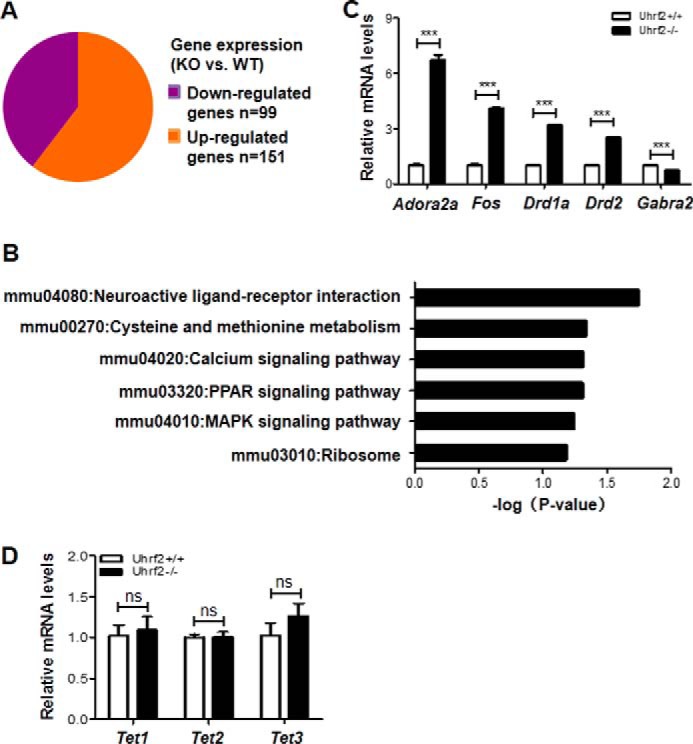
UHRF2 regulates neuron-related gene expression. A, up- and down-regulated genes in the brains of Uhrf2−/− mice revealed by RNA-seq analysis. RNA-seq was performed with mixed total RNAs prepared from the brains of three Uhrf2+/+ and Uhrf2−/− mice. A 1.5-fold difference was used as the cut-off for differentially expressed genes between Uhrf2−/− and Uhrf2+/+. B, gene ontology enrichment analysis of differentially expressed genes. C, verification of differentially expressed genes by quantitative RT-PCR analysis. The level of mRNA in the Uhrf2+/+ mice for each testing gene was set as 1. ***, p < 0.001. D, quantitative RT-PCR analysis showing the relative expression levels of Tet1, Tet2, and Tet3 in the brains of three Uhrf2+/+ and Uhrf2−/− mice. ns, not significant. Error bars, S.E.
UHRF2 Is Unlikely to Be Required for TET1 Dioxygenase Activity
UHRF2 has been shown previously to enhance TET1 (TET1cat) activity in a cell-based assay and function as a 5hmC-binding protein (12). Having established that Uhrf2 knock-out led to reduced 5hmC in mice, we wished to test whether the reduced 5hmC in the Uhrf2−/− mice was due to the role of UHRF2 in promoting TET1 activity. To this end, we compared the activity for TET1 catalytic domain (TET1cat) to catalyze 5hmC in the wild-type and Uhrf2−/− mouse embryonic fibroblast (MEF) cells. We found that expression of TET1cat in the wild-type MEFs by transient transfection lentiviral infection led to substantially increased 5hmC as detected by immunofluorescent staining using an anti-5hmC antibody (Fig. 6A). Similarly, we found that expression of TET1cat in the Uhrf2−/− MEF cells also resulted in substantially increased 5hmC (Fig. 6A). From multiple experiments and a large number of transfected cells, we did not observe statistically any significant difference in catalyzing 5hmC by TET1cat in the wild-type and Uhrf2−/− MEFs. These results indicate that UHRF2 in MEFs is not required for TET1cat activity. To test this further, we examined the ability for co-expressed FLAG-tagged UHRF2 to enhance TET1cat activity in HeLa cells. We found that, on the basis of a large number of transfected cells, co-expression of FLAG-UHRF2 with TET1cat, like co-expression of FLAG-UHRF1 with TET1cat, did not enhance the levels of 5hmC over that generated by TET1cat alone (Fig. 6B). Thus, neither is UHRF2 in MEFs required for the TET1 activity nor is UHRF2 in HeLa able to significantly promote TET1 activity, suggesting that the reduced 5hmC in the Uhrf2−/− mice is unlikely to be due to a role for UHRF2 in promoting TET1 activity.
FIGURE 6.
UHRF2 binds 5hmC in cells but does not appear to affect TET1 enzymatic activity. A, expression of Myc-TET1cat resulted in similarly elevated levels of 5hmC in the wild-type and Uhrf2−/− MEFs. The wild-type and Uhrf2−/− MEF cells were infected with lentiviral Myc-TET1cat, and 2 days after viral transduction, the cells were analyzed for expression of Myc-TET1cat and 5hmC by double immunostaining. B, neither UHRF2 nor UHRF1 enhanced TET1cat activity. The FLAG-UHRF2 (F-UHRF2) or FLAG-UHRF1 (F-UHRF1) was co-transfected with Myc-TET1cat into HeLa cells. The cells were subsequently double-immunostained with anti-FLAG antibody to detect the expression of FLAG-UHRF2 or FLAG-UHRF1 and anti-5hmC antibody to detect the levels of 5hmC. Note that the 5hmC-positive cells should be positive for expression of Myc-TET1cat. C, targeting TET1cat to lac operon loci resulted in recruitment (co-localization) of UHRF2 but not UHRF1. GFP-Lac-TET1cat was co-expressed with either FLAG-UHRF2 or FLAG-UHRF1 in CHO DG44 cells, and GFP fluorescence and FLAG immunostaining were performed to examine the co-localization of GFP-tagged TET1cat and FLAG-tagged UHRF2 or UHRF1. Note that co-localization of CFP-Lac-TET1cat was observed for FLAG-UHRF2 but not UHRF1. Also note that no co-localization of CFP-Lac and FLAG-UHRF2 was observed. D, targeting TET1cat to lac operon loci resulted in localized 5hmC. GFP-Lac-TET1cat or control GFP-Lac was transiently expressed in CHO DG44 cells, and immunostaining for 5hmC was performed. No cell with a strong 5hmC focus was observed for the cells transfected with GFP-Lac, whereas ∼15% cells were positive for a strong 5hmC focus for cells transfected with GFP-Lac-TET1cat. Note that due to the denatured condition, no GFP signal was observed upon immunostaining of 5hmC. The ∼15% 5hmC focus-positive cells were close to the transfection efficiency of CHO DG44 cells. E, no protein-protein interaction between TET1cat and UHRF2 or UHRF1. UHRF2 or UHRF1 was expressed as a GFP fusion protein, and TET1cat was expressed as a Myc-tagged protein in NIH3T3 cells. Both GFP-UHRF1 and GFP-UHRF2 bound to pericentromeric heterochromatin regions, illustrated as DAPI foci. Note that Myc-TET1cat was not co-localized with either GFP-UHRF2 or GFP-UHRF1.
UHRF2 Binds 5hmC in Cells
Although UHRF2 was identified as a specific reader of 5hmC by affinity purification and this activity was subsequently confirmed by structural study (12, 19), it has yet to be shown whether UHRF2 binds specifically 5hmC in cells. To test whether UHRF2 binds 5hmC in cells, we made use of a DG44 CHO cell line, which contains large numbers of Lac operator sequences stably integrated in a single chromosomal site (27). As a representative result, expression of either a CFP-Lac or a fusion protein of CFP-Lac-TET1cat in DG44 cells generated a bright CFP focus due to the binding of CFP-Lac or CFP-Lac-TET1cat fusion proteins to the locus with large numbers of Lac binding sites (Fig. 6C, left, white arrow). By immunofluorescent staining using anti-5hmC antibody, we demonstrated that targeting TET1cat to the Lac locus resulted in a strong localized accumulation of 5hmC, as revealed by a bright 5hmC focus in cells transfected with CFP-Lac-TET1cat but not the control CFP-Lac (Fig. 6D). Due to the denaturing conditions used for 5hmC staining, no CFP fluorescence could be observed in this experiment (data not shown). Thus, targeting TET1cat to the Lac locus is sufficient to generate localized 5hmC.
We next tested whether the localized 5hmC generated by CFP-Lac-TET1cat could recruit UHRF2. As a control, we also tested whether CFP-Lac-TET1cat could recruit UHRF1, which is not known for binding of 5hmC. As shown in Fig. 6C, we found that co-expression of CFP-Lac-TET1cat and FLAG-UHRF2 resulted in a nice co-localization of CFP-Lac-TET1cat and FLAG-UHRF2 at one big focus (marked by a white arrow). In contrast, no co-localization between CFP-Lac-TET1cat and FLAG-UHRF1 was observed under the same experimental conditions. Furthermore, no co-localization was observed between CFP-Lac and FLAG-UHRF2, indicating that the co-localization is specific to TET1cat. Importantly, the co-localization of CFP-Lac-TET1cat and FLAG-UHRF2 in this assay is not due to a direct interaction between these two proteins, because we found that there was no co-localization of GFP-UHRF2 and Myc-TET1cat when they were co-expressed in NIH3T3 cells (Fig. 6E). Note that in NIH3T3 cells, both GFP-UHRF1 and GFP-UHRF2 displayed a focal distribution pattern that overlapped with the densely stained DAPI foci, in agreement with the reported heterochromatin localization for both UHRF1 and UHRF2 in NIH3T3 cells (17, 28). Taken together, we concluded that the observed co-localization of CFP-Lac-TET1cat and FLAG-UHRF2 in DG44 cells is a result of UHRF2 binding of 5hmC generated by Lac locus-associated CFP-Lac-TET1cat.
Discussion
In this study, we have taken the gene ablation approach to investigate the physiological role of UHRF2, a novel protein that has been shown to bind three epigenetic markers (H3K9me2/3, 5mC, and 5hmC) and has been implicated in epigenetic and cell cycle regulation in mice. Although our study reveals that UHRF2 is not required for mouse embryonic development, growth, and general health, we show that it is highly expressed in the brain, especially in hippocampus, and influences the level of 5hmC and mouse memory acquisition and retention.
In this study, we show that, in contrast to the early embryonic lethality of Uhrf1 knock-out mice (15), Uhrf2 knock-out mice are viable and fertile and exhibit no gross defect. This phenotype is consistent with the previous conclusion that UHRF2 is not required for DNA maintenance methylation (16, 17). The lack of obvious phenotype for Uhrf2 knock-out mice may also be explained by its low expression in mice and/or potential functional redundancy with Uhrf1. For instance, we have shown recently that both UHRF1 and UHRF2 proteins can negatively regulate de novo DNA methylation by targeting DNMT3A for ubiquitination and subsequent proteasome-dependent degradation (29). Thus, the role of UHRF2 in regulation of DNMT3A in mice could be compensated by the presence of UHRF1. This functional redundancy may also explain the insignificant effect of Uhrf2 knock-out on DNA methylation in the brain and other tissues, such as liver, that we have tested. The lack of obvious defect in development and growth also indicates that Uhrf2 knock-out has no significant effect on cell cycle regulation, suggesting that the effect of UHRF2 on cell cycle might be linked to its overexpression in cancer cells (30).
By Western blotting analysis and β-geo staining of Uhrf2 trapping mice, we found that UHRF2 is relatively highly expressed in the brain, especially in the hippocampus. Furthermore, by quantitative HPLC analysis (Fig. 3, D and F) and/or semiquantitative IHC analysis (Fig. 3G), we found that Uhrf2 knock-out results in reduced levels of 5hmC in the entire brain, cerebellum, cortex, and hippocampus. In this regard, it is noteworthy that TET1 also displays an enriched expression in the hippocampus and that Tet1 knock-out results in reduced 5hmC in the hippocampus (22, 23). Tet1 knock-out mice are also grossly normal but exhibit a defect in neurogenesis and learning and memory in one study and a defect in memory extinction in another study (22, 23). Thus, the phenotype of Uhrf2 knock-out mice is somewhat similar to that of Tet1 knock-out, displaying reduced 5hmC in the brain and hippocampus and a defect in memory acquisition and retention. Despite a similar reduction of 5hmC in Tet1 and Uhrf2 knock-out mice, the effect on gene expression appears to be different, because the affected genes identified in our study did not overlap significantly with those identified in a previous study (23). In this regard, it is also noteworthy that a study by Kaas et al. (31) demonstrated that TET1cat controls neuronal gene expression and memory formation in a dioxygenase activity-independent manner. This is consistent with the findings that TET proteins also interact with O-linked β-GlcNAc (O-GlcNAc) transferase (OGT) and play roles in regulating and/or targeting OGT to chromatin (32–36). Thus, UHRF2 may overlap with TET1 in 5hmC-dependent gene expression, but it has no role in OGT-related neuronal gene expression.
Although Uhrf2 knock-out results in reduced levels of 5hmC in multiple tissues that we have tested, this is unlikely to be due to a role for UHRF2 in enhancing TET1 activity for the following reasons. First, we found that ectopically expressed TET1cat was equally active in catalyzing 5hmC in the wild-type and Uhrf2−/− MEFs (Fig. 6A). Second, we observed that co-expression of UHRF2 with TET1cat in HeLa cells did not promote the generation of 5hmC by TET1cat (Fig. 6B). On the basis of the above observation, we disfavor the idea that the reduced levels of 5hmC in the Uhrf2−/− mice are due to a direct effect of UHRF2 on TET1 activity.
We provide in vivo evidence that UHRF2 is a bona fide 5hmC reader protein. We demonstrated that UHRF2 binds 5hmC in cells (Fig. 6C). Under the same conditions, we found that UHRF1 did not bind 5hmC in cells (Fig. 6C). Thus, in agreement with the previous in vitro studies (12, 19), we demonstrate for the first time that UHRF2 binds 5hmC in cells and thus is a bona fide 5hmC reader. Because Uhrf2 knock-out does not appear to affect the expression of TET1/2/3 (Fig. 5D) or the activity of TET1 (Fig. 6, A and B), we postulate that UHRF2 may regulate the cellular levels of 5hmC by its binding of 5hmC. One possibility is that the binding of UHRF2 may inhibit the further oxidation of 5hmC by TET family proteins and therefore stabilize the levels of 5hmC. Future work is required to determine whether UHRF2 regulates the cellular levels of 5hmC and affects neuronal function indeed through its ability to bind 5hmC. Together, our study demonstrates that UHRF2 regulates the levels of 5hmC in the brain, especially in the hippocampus, and reveals a role for UHRF2 in regulating neuronal function in mice. We also provide evidence supporting UHRF2 as a bona fide reader for 5hmC. On the basis of the phenotype similarity between Tet1 and Uhrf2 knock-out mice, it is tempting to suggest a working model in which UHRF2 is probably a major 5hmC reader protein, especially in the hippocampus. Our study further supports a role for DNA hydroxymethylation in neuronal function.
Experimental Procedures
Plasmids and Antibodies
The pEGFP-UHRF1, pEGFP-UHRF2, FLAG-UHRF1, FLAG-UHRF2, and FLAG-TET1cat were as described (17, 36, 37). CFP-Lac-TET1cat was generated by cloning the catalytic domain of TET1 into pECFP-C1 vector. All plasmids were verified by DNA sequencing. The antibodies used were as follows: Myc (AbMART), FLAG (Sigma-Aldrich), GAPDH (AbMART), UHRF2 (homemade), and 5mC (AnaSpec). The anti-5hmC antibody was a kind gift from Dr. Degui Chen (SIBCB, CAS).
Cell Culture, Transient Transfection, and Lentiviral Transduction
HeLa, NIH3T3, and MEF cells were routinely maintained with regular Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (Gibco). Transient transfection of HeLa and DG44-CHO cells with plasmids was carried out using Lipofectamine 2000 (Invitrogen) essentially according to the manufacturer's instructions. Lentiviral particle and cell transduction were performed as described previously (38). The Uhrf2 gene trapping ES cell line (AD0406) was obtained from the Mutant Mouse Regional Resource Center.
Western Blotting Analysis of Mouse Tissues
Tissues were dissected from 2-month-old wild-type mice and broken up by a tissue grinding apparatus. The cells were then lysed with 1 ml of radioimmune precipitation assay buffer at 4 °C for 2 h. After centrifugation at 12,000 rpm for 10 min at 4 °C, the clean extracts were obtained and subjected to Western blotting analysis with 8% SDS-PAGE. The immunoreactive proteins were detected by the Odyssey laser digital imaging system.
Whole-mount Staining of Brain Sections
The whole brains from 2-month-old wild-type and Uhrf2−/− mice were coated with optimal cutting temperature compound in a microtome cryostat and pinned in a 25-mm diameter bracket. The sections were fixed in 4% PFA in PBS for 15 min at 4 °C, rinsed three times with detergent washing solution (2 mm MaCl2, 0.01% sodium deoxycholate, 0.02% Nonidet P-40), and then incubated with staining solution (7.2 mm NaCl, 5 mm K3Fe(CN)6, 5 mm K4Fe(CN)6 in detergent washing solution) in the dark for 20 h at 37 °C. Images were acquired with an upright microscope (DM750, Lycra).
Genotyping of the Wild-type and Uhrf2 Null Mice
Tail clips were subjected to a standard DNA extraction procedure. 1 μl of DNA solution was used to provide 50–200 ng of genomic DNA for the PCR template. 2× power TaqPCR Master Mix was used for amplifying the Uhrf2 target region with the primer pairs 5′-GGTTTCCTTCCACCGAGGAG-3′ (forward) and 5′-GACCCTCGGAGGCTATGTCC-3′ (reverse). The PCR procedure was as follows: 98 °C for 5 min, followed by 35 cycles of 98 °C for 30 s, 60 °C for 45 s, and 72 °C for 45 s. The expected size of the PCR product was 594 bp for the wild-type allele and 317 bp for the Uhrf2 deletion allele.
Measurement of 5mC and 5hmC by HPLC
The measurement of 5mC and 5hmC of genomic DNAs from various tissues were performed as described (22).
Immunohistochemistry Analysis of 5mC and 5hmC
The brain sections were prepared as described previously for whole-mount staining. Samples were fixed in 4% PFA in PBS for 20 min at room temperature and then incubated with HCl (2 m) for 30 min at 37 °C and neutralized with Tris·HCl (pH 9.0) for 10 min at room temperature. After washing, sections were incubated in blocking solution (3% BSA in PBS) for 12 h at 4 °C. The primary antibodies (5mC, 1:1000; 5hmC, 1:1000) were then added and incubated for 12 h at 4 °C, followed by the secondary antibodies for 1 h at room temperature. Last, the sections were stained with 4,6-diamidino-2-phenylindole (DAPI) for 15 min at 37 °C. Images were acquired by an upright microscope (DM750, Lycra).
Immunostaining of 5hmC in Cell Culture
For immunostaining of 5hmC, HeLa or MEF cells were washed with iced 1× PBS (137 mm NaCl, 2.7 mm KCl, 2 mm KH2PO4, 10 mm Na2HPO4) before fixation in 4% fresh paraformaldehyde in PBS for 15 min. Subsequently, the cells were treated with 2 n HCl at room temperature for 30 min, followed by neutralizing with 100 mm Tris-HCl (pH 8.0) at room temperature for 30 min and blocking with 5% BSA in PBS for 1 h in 37 °C incubator. The incubation with 5hmC primary antibody was carried out at 4 °C overnight, and incubation with the secondary antibody was performed at room temperature for 1 h. DNA was then stained with DAPI. Images were acquired using the Leica SP5 system.
Open Field
Each mouse was placed in the center of an opaque Plexiglas cage (27 × 27 × 38 cm) (Coulbourn Instruments, Holliston, MA) equipped with photo beam sensor rings to monitor the locomotor activity of the mouse. The mouse was allowed to explore the environment for 30 min in the opaque Plexiglas cage. Total distance and time traveled by an individual at the margin and center of the cage were measured using a Tru-scan DigBahv-locomotion activity analysis system (Coulbourn Instruments).
Contextual Fear Conditioning
The procedure for fear conditioning was similar to the protocol described previously (39). The freezing behavior was monitored by the FreezeFrame system (Coulbourn Instruments). During the training phase, mice were individually placed in the conditioning chamber (CS) and were individually allowed to explore freely the environment for 120 s, and then the US (0.50 mA, 2 s) was delivered to the mouse's foot. After seven CS/US pairings with a 120-s inter-trial interval, mice were allowed to stay in the chamber for 120 s and then returned to their home cages. Contextual fear memory acquisition was measured as the amount of time spent in freezing response during each 120-s inter-trial interval without shock stimulus, and contextual fear memory retention was examined by placing mice back in the same conditioning chamber without shock at 24 h after seven CS/US pairings. Data are presented as the mean ± S.E. Two-way repeated measures ANOVA was used for statistical analysis.
Morris Water Maze Test
The spatial learning and memory test was assessed in the Morris water maze. In brief, during visible platform training, a visible cue was put on the platform, which was submerged. The mouse was trained to swim to the platform from each quadrant (randomly), and the mouse was kept on it for 15 s. Then a training session was conducted 24 h later and lasted for 5 days, the cue was removed from the submerged platform, and the submerged platform was moved to another quadrant. The platform was kept in the same (target) quadrant during the entire training course of the experiment. In the training session, the mice were gently released into the water, always facing the tank wall. The mice were trained to find the hidden platform using distal cues available on the curtain. Mice were trained with 4 trials/day, and each trial had a different starting position. Once they found the platform, the mice were permitted to remain on it for 15 s. If the mice did not find the platform within 60 s, they were guided to the platform and also allowed to stay on it for 15 s. Then they were taken out, dried, and placed back in the home cage. During each training trial, the time used to reach the hidden platform (escape latency) was recorded. Twenty-four hours after the last training day, a probe test was performed to assess memory. During the probe test, the platform was removed from the tank, and the mice were allowed to swim freely. The time rats spent in each quadrant and the swim path were recorded.
Co-localization Assay in DG44-CHO Cells
To examine co-localization in DG44-CHO cells, CFP-Lac-TET1cat was co- transfected with FLAG-UHRF1 or FLAG-UHRF2 into DG44-CHO cells. 36 h after transfection, cells were processed for immunostaining using anti-FLAG. The images were acquired and examined for co-localization of CFP-Lac-TET1 with the FLAG-UHRF1 or FLAG-UHRF2.
RNA-seq and Gene Ontology Analysis
Total RNAs were prepared from the hippocampus of three pairs of wild-type and Uhrf2−/− littermates and mixed, respectively. RNA-seq sequencing was performed by Berry Genomics Co., Ltd. (Beijing, China). Gene ontology enrichment analysis was performed as described.
Statistics of Behavioral Data
Student's t test was used for the open field, contextual fear conditioning retention test, visible platform test, and spatial probe test. Two-way repeated measures ANOVA was used for contextual fear conditioning training and water maze training. The statistical analysis was performed using GraphPad Prism software.
Author Contributions
R. C., Q. Z., and X. D. conducted both animal and DNA methylation and hydroxylation analyses. P. K. performed whole-mount in situ hybridization for wild-type and Uhrf2 gene-trapping mice. G. D. C. performed quantitative HPLC analysis of 5hmC. P. Y. and M. X. helped R. C. and X. D. to carry out mice behavior study. H. Z. and Q. W. carried out RNA-seq data analysis. P. C. provided essential reagents and performed 5mC measurement. D. L., J. S., G. X., P. Z., and X. C. supervised the experiments and conceived the ideas. J. W. conceived the ideas and wrote the paper with X. C. and J. L.
Acknowledgments
We thank members of the Wong laboratory for valuable discussion. We are also grateful to Dr. Degui Chen for providing anti-5hmC antibody.
This work was supported by National Science and Technology Major Project “Key New Drug Creation and Manufacturing Program” of China Grant 2014ZX09507002-002, Ministry of Science and Technology of China Grant 2015CB910402, National Natural Science Foundation of China Grant 91419303, and Science and Technology Commission of Shanghai Municipality Grants 14XD1401700 and 11DZ2260300 (to J. W.) and National Natural Science Foundation of China Grants 91219202 and 31525013 and Ministry of Science and Technology of China Grant 2015CB856200 (to G. L.). The authors declare that they have no conflicts of interest with the contents of this article.
- 5mC
- DNA methylation at the C5 position of cytosine
- 5hmC
- 5-hydroxymethylcytosine
- US
- unconditioned stimulus
- CS
- conditioned stimulus
- H3K9
- histone H3 Lys-9
- ES cell
- embryonic stem cell
- IHC
- immunohistochemistry
- ANOVA
- analysis of variance
- RNA-seq
- RNA sequencing
- MEF
- mouse embryo fibroblast
- OGT
- O-GlcNAc transferase
- PFA
- paraformaldehyde.
References
- 1. Jones P. A. (2012) Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 13, 484–492 [DOI] [PubMed] [Google Scholar]
- 2. Li E., and Zhang Y. (2014) DNA methylation in mammals. Cold Spring Harb. Perspect. Biol. 6, a019133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Klose R. J., and Bird A. P. (2006) Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 31, 89–97 [DOI] [PubMed] [Google Scholar]
- 4. Tahiliani M., Koh K. P., Shen Y., Pastor W. A., Bandukwala H., Brudno Y., Agarwal S., Iyer L. M., Liu D. R., Aravind L., and Rao A. (2009) Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. He Y. F., Li B. Z., Li Z., Liu P., Wang Y., Tang Q., Ding J., Jia Y., Chen Z., Li L., Sun Y., Li X., Dai Q., Song C. X., Zhang K., et al. (2011) Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ito S., Shen L., Dai Q., Wu S. C., Collins L. B., Swenberg J. A., He C., and Zhang Y. (2011) Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shen L., Song C. X., He C., and Zhang Y. (2014) Mechanism and function of oxidative reversal of DNA and RNA methylation. Annu. Rev. Biochem. 83, 585–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bachman M., Uribe-Lewis S., Yang X., Williams M., Murrell A., and Balasubramanian S. (2014) 5-Hydroxymethylcytosine is a predominantly stable DNA modification. Nat. Chem. 6, 1049–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yu M., Hon G. C., Szulwach K. E., Song C. X., Zhang L., Kim A., Li X., Dai Q., Shen Y., Park B., Min J. H., Jin P., Ren B., and He C. (2012) Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 149, 1368–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Booth M. J., Branco M. R., Ficz G., Oxley D., Krueger F., Reik W., and Balasubramanian S. (2012) Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science 336, 934–937 [DOI] [PubMed] [Google Scholar]
- 11. Mellén M., Ayata P., Dewell S., Kriaucionis S., and Heintz N. (2012) MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell 151, 1417–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Spruijt C. G., Gnerlich F., Smits A. H., Pfaffeneder T., Jansen P. W., Bauer C., Münzel M., Wagner M., Müller M., Khan F., Eberl H. C., Mensinga A., Brinkman A. B., Lephikov K., Müller U., et al. (2013) Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell 152, 1146–1159 [DOI] [PubMed] [Google Scholar]
- 13. Mori T., Li Y., Hata H., Ono K., and Kochi H. (2002) NIRF, a novel RING finger protein, is involved in cell-cycle regulation. Biochem. Biophys. Res. Commun. 296, 530–536 [DOI] [PubMed] [Google Scholar]
- 14. Bostick M., Kim J. K., Estève P. O., Clark A., Pradhan S., and Jacobsen S. E. (2007) UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317, 1760–1764 [DOI] [PubMed] [Google Scholar]
- 15. Sharif J., Muto M., Takebayashi S., Suetake I., Iwamatsu A., Endo T. A., Shinga J., Mizutani-Koseki Y., Toyoda T., Okamura K., Tajima S., Mitsuya K., Okano M., and Koseki H. (2007) The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450, 908–912 [DOI] [PubMed] [Google Scholar]
- 16. Pichler G., Wolf P., Schmidt C. S., Meilinger D., Schneider K., Frauer C., Fellinger K., Rottach A., and Leonhardt H. (2011) Cooperative DNA and histone binding by Uhrf2 links the two major repressive epigenetic pathways. J. Cell. Biochem. 112, 2585–2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang J., Gao Q., Li P., Liu X., Jia Y., Wu W., Li J., Dong S., Koseki H., and Wong J. (2011) S phase-dependent interaction with DNMT1 dictates the role of UHRF1 but not UHRF2 in DNA methylation maintenance. Cell Res. 21, 1723–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mori T., Li Y., Hata H., and Kochi H. (2004) NIRF is a ubiquitin ligase that is capable of ubiquitinating PCNP, a PEST-containing nuclear protein. FEBS Lett. 557, 209–214 [DOI] [PubMed] [Google Scholar]
- 19. Zhou T., Xiong J., Wang M., Yang N., Wong J., Zhu B., and Xu R. M. (2014) Structural basis for hydroxymethylcytosine recognition by the SRA domain of UHRF2. Mol. Cell 54, 879–886 [DOI] [PubMed] [Google Scholar]
- 20. Feng J., Zhou Y., Campbell S. L., Le T., Li E., Sweatt J. D., Silva A. J., and Fan G. (2010) Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat. Neurosci. 13, 423–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Day J. J., and Sweatt J. D. (2010) DNA methylation and memory formation. Nat. Neurosci. 13, 1319–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang R. R., Cui Q. Y., Murai K., Lim Y. C., Smith Z. D., Jin S., Ye P., Rosa L., Lee Y. K., Wu H. P., Liu W., Xu Z. M., Yang L., Ding Y. Q., Tang F., et al. (2013) Tet1 regulates adult hippocampal neurogenesis and cognition. Cell Stem Cell 13, 237–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rudenko A., Dawlaty M. M., Seo J., Cheng A. W., Meng J., Le T., Faull K. F., Jaenisch R., and Tsai L. H. (2013) Tet1 is critical for neuronal activity-regulated gene expression and memory extinction. Neuron 79, 1109–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lister R., Mukamel E. A., Nery J. R., Urich M., Puddifoot C. A., Johnson N. D., Lucero J., Huang Y., Dwork A. J., Schultz M. D., Yu M., Tonti-Filippini J., Heyn H., Hu S., Wu J. C., et al. (2013) Global epigenomic reconfiguration during mammalian brain development. Science 341, 1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kriaucionis S., and Heintz N. (2009) The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324, 929–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li D., Qiu Z., Shao Y., Chen Y., Guan Y., Liu M., Li Y., Gao N., Wang L., Lu X., Zhao Y., and Liu M. (2013) Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat. Biotechnol. 31, 681–683 [DOI] [PubMed] [Google Scholar]
- 27. Nye A. C., Rajendran R. R., Stenoien D. L., Mancini M. A., Katzenellenbogen B. S., and Belmont A. S. (2002) Alteration of large-scale chromatin structure by estrogen receptor. Mol. Cell. Biol. 22, 3437–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Papait R., Pistore C., Grazini U., Babbio F., Cogliati S., Pecoraro D., Brino L., Morand A. L., Dechampesme A. M., Spada F., Leonhardt H., McBlane F., Oudet P., and Bonapace I. M. (2008) The PHD domain of Np95 (mUHRF1) is involved in large-scale reorganization of pericentromeric heterochromatin. Mol. Biol. Cell 19, 3554–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jia Y., Li P., Fang L., Zhu H., Xu L., Cheng H., Zhang J., Li F., Feng Y., Li Y., Li J., Wang R., Du J. X., Li J., Chen T., et al. (2016) Negative regulation of DNMT3A de novo DNA methylation by frequently overexpressed UHRF family proteins as a mechanism for widespread DNA hypomethylation in cancer. Cell Discov. 2, 16007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mori T., Ikeda D. D., Yamaguchi Y., Unoki M., and NIRF Project (2012) NIRF/UHRF2 occupies a central position in the cell cycle network and allows coupling with the epigenetic landscape. FEBS Lett. 586, 1570–1583 [DOI] [PubMed] [Google Scholar]
- 31. Kaas G. A., Zhong C., Eason D. E., Ross D. L., Vachhani R. V., Ming G. L., King J. R., Song H., and Sweatt J. D. (2013) TET1 controls CNS 5-methylcytosine hydroxylation, active DNA demethylation, gene transcription, and memory formation. Neuron 79, 1086–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen Q., Chen Y., Bian C., Fujiki R., and Yu X. (2013) TET2 promotes histone O-GlcNAcylation during gene transcription. Nature 493, 561–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Deplus R., Delatte B., Schwinn M. K., Defrance M., Méndez J., Murphy N., Dawson M. A., Volkmar M., Putmans P., Calonne E., Shih A. H., Levine R. L., Bernard O., Mercher T., Solary E., et al. (2013) TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 32, 645–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shi F. T., Kim H., Lu W., He Q., Liu D., Goodell M. A., Wan M., and Songyang Z. (2013) Ten-eleven translocation 1 (Tet1) is regulated by O-linked N-acetylglucosamine transferase (Ogt) for target gene repression in mouse embryonic stem cells. J. Biol. Chem. 288, 20776–20784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vella P., Scelfo A., Jammula S., Chiacchiera F., Williams K., Cuomo A., Roberto A., Christensen J., Bonaldi T., Helin K., and Pasini D. (2013) Tet proteins connect the O-linked N-acetylglucosamine transferase Ogt to chromatin in embryonic stem cells. Mol. Cell 49, 645–656 [DOI] [PubMed] [Google Scholar]
- 36. Zhang Q., Liu X., Gao W., Li P., Hou J., Li J., and Wong J. (2014) Differential regulation of the ten-eleven translocation (TET) family of dioxygenases by O-linked β-N-acetylglucosamine transferase (OGT). J. Biol. Chem. 289, 5986–5996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu X., Gao Q., Li P., Zhao Q., Zhang J., Li J., Koseki H., and Wong J. (2013) UHRF1 targets DNMT1 for DNA methylation through cooperative binding of hemi-methylated DNA and methylated H3K9. Nat. Commun. 4, 1563. [DOI] [PubMed] [Google Scholar]
- 38. Moore C. B., Guthrie E. H., Huang M. T., and Taxman D. J. (2010) Short hairpin RNA (shRNA): design, delivery, and assessment of gene knockdown. Methods Mol. Biol. 629, 141–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Walker D. L., and Davis M. (2008) Amygdala infusions of an NR2B-selective or an NR2A-preferring NMDA receptor antagonist differentially influence fear conditioning and expression in the fear-potentiated startle test. Learn. Mem. 15, 67–74 [DOI] [PMC free article] [PubMed] [Google Scholar]