Abstract
Glycosylation and deglycosylation are impressive mechanisms that allow plants to regulate the biological activity of an array of secondary metabolites. Although glycosylation improves solubility and renders the metabolites suitable for transport and sequestration, deglycosylation activates them to carry out biological functions. Herein, we report the functional characterization of CsBGlu12, a β-glucosidase from Crocus sativus. CsBGlu12 has a characteristic glucoside hydrolase 1 family (α/β)8 triose-phosphate isomerase (TIM) barrel structure with a highly conserved active site. In vitro enzyme activity revealed that CsBGlu12 catalyzes the hydrolysis of flavonol β-glucosides and cello-oligosaccharides. Site-directed mutagenesis of any of the two conserved catalytic glutamic acid residues (Glu200 and Glu414) of the active site completely abolishes the β-glucosidase activity. Transcript analysis revealed that Csbglu12 is highly induced in response to UV-B, dehydration, NaCl, methyl jasmonate, and abscisic acid treatments indicating its possible role in plant stress response. Transient overexpression of CsBGlu12 leads to the accumulation of antioxidant flavonols in Nicotiana benthamiana and confers tolerance to abiotic stresses. Antioxidant assays indicated that accumulation of flavonols alleviated the accretion of reactive oxygen species during abiotic stress conditions. β-Glucosidases are known to play a role in abiotic stresses, particularly dehydration through abscisic acid; however, their role through accumulation of reactive oxygen species (ROS) scavenging flavonols has not been established. Furthermore, only one β-glucosidase 12 homolog has been characterized so far. Therefore, this work presents an important report on characterization of CsBGlu12 and its role in abiotic stress through ROS scavenging.
Keywords: enzyme kinetics, flavonoid, plant, reactive oxygen species (ROS), stress
Introduction
Plants are sessile organisms that cannot escape their predators and the harsh environmental conditions. However, during the course of evolution, plants have learned to defend themselves and adapt to different types of biotic and abiotic stresses by synthesizing a diverse assortment of secondary metabolites. Many of these secondary metabolites are stored in inactive glycosylated forms. The glycosylation chemically stabilizes and enhances the solubility of the metabolites and renders them fit for storage in the vacuole and more so to protect the plant from the noxious effects of its own defense system (1). These glyconjugates are activated by the hydrolysis of the β-glucosidic bond by a class of enzymes called β-glucosidases. β-Glucosidases (EC 3.2.1.2.1) belong to family 1 of glycoside hydrolases and perform diverse and vital functions in plants, which include, but are not limited to, the activation of lignin precursors (2), release of glucose from oligosaccharides (3), release of phytohormones from inactive glycosides (4), and activation of several defense compounds (5–9). This process of glycosylation and deglycosylation offers an efficient mechanism that regulates the homeostasis of secondary metabolites and is not limited to plants only but extends to other domains of life like archaea, eubacteria, and animals. Furthermore, the subcellular localization, substrate specificity, and conditions under which β-glucosidases come into contact with their physiological substrates determine their biological role (10) and remain an area of intensive study. It is, therefore, important to deepen our knowledge on how plants regulate the individual members of β-glucosidases in response to developmental and environmental cues and how the released metabolites modulate the plant's physiological response.
Crocus sativus L. (Iridaceae) is a sterile triploid and vegetatively propagating plant. The dried stigmas of C. sativus form the commercial saffron. Saffron is considered as an important source of apocarotenoids and several flavonol glucosides like quercetin and kaempferol, which are considered important for its organoleptic properties (11, 12). The flavonol glucosides may serve as a reservoir for the discharge of free aglycones, which play vital roles in defense against biotic and abiotic stresses (13–16). Although glucosyltransferases involved in the glucosylation of flavonol β-glucosides in Crocus and across the plant kingdom have been characterized, the β-glucosidases involved in their hydrolysis have not been explored.
To date, only one close homolog of Crocus β-glucosidase from rice (Os4Bglu12) (17) has been characterized from plant glucoside hydrolase 1 family, and only a few other have been characterized for their possible function. Although a few of these characterized β-glucosidases are reported to play role in abiotic stress through accumulation of abscisic acid, we for the first time report the role of β-glucosidase, CsBGlu12, from C. sativus in abiotic stress tolerance through accumulation of ROS3-scavenging flavonols. CsBGlu12 has an (α/β)8 TIM barrel conformation typical of glycoside hydrolase 1 family of β-glucosidases. The gene exhibits higher expression in floral tissues and is regulated in a development-specific manner. The purified recombinant enzyme catalyzes the hydrolysis of cello-oligosaccharide and flavonol β-glucoside in vitro. However, mutation of any of the two catalytic glutamic acid residues completely abolishes the CsBGlu12 activity. The transcription of Csbglu12 is highly induced in response to several abiotic stress stimuli. Moreover, transient overexpression of CsBGlu12 in Nicotiana benthamiana leads to the accumulation of antioxidant flavonols that confer tolerance to UV-B, salinity, and dehydration stresses through scavenging of ROS.
Results
Identification, Cloning, and Phylogenetic Analysis of CsBGlu12
Previously, our laboratory developed a transcriptome database from Crocus stigma and flower tissues (18). Fifteen sequences from this database exhibited homology with plant β-glucosidases. Among these gene sequences, one sequence (comp32077_c0_seq1) showed higher expression in stigma, which is the main site for the synthesis of flavonoids in C. sativus. Based on its expression, the full-length gene was cloned, which was 1524 bp long, coding for 507 amino acids. The predicted molecular mass of the protein was 57.31 kDa. Theoretical isoelectric point (pI) and extinction coefficient of the deduced protein were 7.6 and 108,430 m−1 cm−1 respectively.
The phylogenetic analysis of CsBGlu12 was performed to gain insights about its evolutionary relation with β-glucosidase orthologs from other plants. CsBGlu12 exhibited high sequence similarity with β-glucosidases from Brachypodium distachyon, Oryza brachyantha, and Oryza sativus. Furthermore, CsBGlu12 and other β-glucosidases used in phylogenetic analysis diverged in accordance to the classification of monocots and dicots (supplemental Fig. S1).
CsBGlu12 Has (β/α)8 TIM Barrel Conformation Typical of GH1 Family
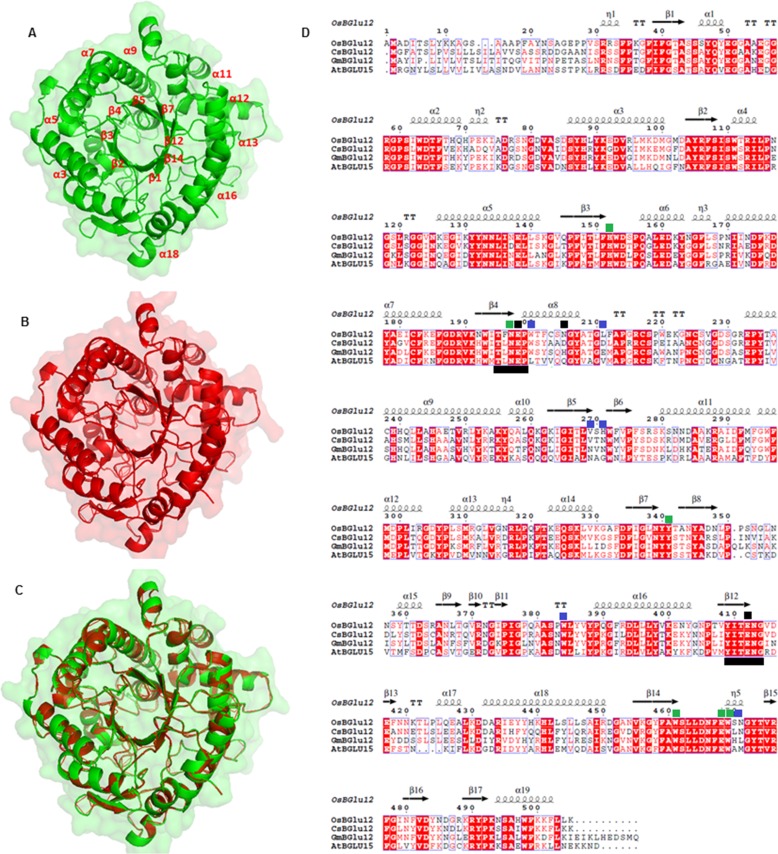
To gain insights about the structure of CsBGlu12, a three-dimensional homology model was developed by SWISS-MODEL software with rice Os4Bglu12 as a template. The results indicated that CsBGlu12 has a characteristic (β/α)8 TIM barrel conformation. Moreover, the active site of CsBGlu12 consists of evolutionarily conserved amino acids residues (His154, Asn199, Tyr343, Trp463, Glu470, Trp202, and Asp207) indicative of an active β-glucosidase and non-conserved aglycone-binding residues (Trp202, Asp207, Asp213, Val271, Ser281, Trp386, and Leu472) responsible for substrate specificity of plant β-glucosidases. The catalytic residues include two highly conserved glutamates Glu200 and Glu414 located in the characteristic and conserved peptide motifs Thr-(Phe/Leu)-Asn-Glu-Pro (T(F/L)NEP) and Tyr-Ile-Thr-Glu-Asn-Gly (YITENG) of CsBGlu12 (Fig. 1). These features are characteristic of many other members of the GH1 family of β-glucosidases (19), and the residues are considered critical for the β-glucosidase activity (20, 21).
FIGURE 1.
Structural analysis of CsBGlu12. A, homology model of CsBGlu12 carried out by SWISS-MODEL software depicting characteristic (β/α)8 TIM barrel- shaped structure; B, rice Os4BGlu12 (3ptk) used as template; and C, superimposed structure of CsBGlu12 and Os4BGlu12. D, multiple sequence alignment carried out by ClustalW and analyzed by ESPript 3 software. The alignment shows secondary structure elements of the (α/β)8 barrel structure, containing the two catalytic glutamates (black squares), highly conserved residues involved in glucose binding (green squares) and residues involved in aglycone binding (blue squares). Conserved peptide motifs Thr-(Phe/Leu)-Asn-Glu-Pro (T(F/L)NEP) and Tyr-Ile-Thr-Glu-Asn-Gly (YITENG) are underlined in black. Red boxes denote the sites of perfect sequence identity. The protein sequences used in this study include C. sativus CsBGlu12 (KX790358), A. thaliana AtBGlu15 (O64879.1), O. sativa Os4BGlu12 (Q7XKV4.2), and Glycine max GmBGlu12 (XP_006590951).
Docking studies carried on a panel of substrates revealed that CsBGlu12 exhibits high specificity for cello-oligosaccharides and flavonol 3-O-β-glucosides, moderate affinity for phenolic β-glucosides, and low affinity for coumarin and other glucosides. As representative substrates, the free energy change (ΔG) of the best pose of the enzyme-ligand complex was −8.71 and −7.62 kcal/mol for cellobiose and kaempferol 3-O-β glucoside, whereas the intermolecular energy was −7.01 and −6.20 kcal/mol, respectively. The free energy change and intermolecular energy for 1-O-sinopyl-β-d-glucose was −3.11 and −2.7 kcal/mol, respectively. However, the free energy changes inCsBGlu12 for coumarin and other glucosides analyzed were comparatively lower. The interacting residues of CsBGlu12 included His154, Trp155, Asp156, Asn199, Glu200, Trp202, Tyr204, Ala215, Asp207, Asp213, Val2671, Asn273, Ser279, Met295, Tyr343, Trp386, Leu387, Thr413, Glu414, Leu472, Glu470, and Trp471 (supplemental Fig. S2). The list of substrates used for docking analysis and their respective free energy changes are given in supplemental Table S1.
CsBGlu12 Exhibits Higher Affinity for Flavonol β-Glucosides and Cello-oligosaccharides
For heterologous expression, the full-length cDNA of Csbglu12 was cloned into pGEX-4T-1 vector and expressed in Escherichia coli as a fusion protein with the N-terminal GST. The protein expression was determined at different time intervals from 0 to 8 h after induction with 1 mm IPTG at 30 °C. The maximum expression of the protein was observed at 1 mm IPTG induction for 6 h at 30 °C. The recombinant enzyme exhibited an apparent molecular mass of ∼57 kDa.
To confirm the specificity, β-glucosidase activity of recombinant CsBGlu12 enzyme was determined against the substrates selected on the basis of docking analysis. Relative activities of CsBGlu12 were determined considering the activity of the enzyme against cellobiose as 100% (Fig. 2). CsBGlu12 exhibited activity against the cello-oligosaccharides, flavonoid and phenolic β-glucosides; however, no visible CsBGlu12 activity was observed for coumarin and other glucosides tested.
FIGURE 2.
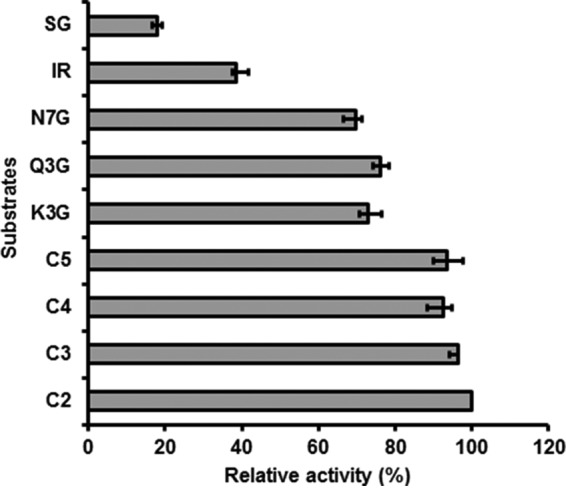
Relative activity (%) of CsBGlu12 against a panel of substrates. Highest activity of cellobiose was taken as 100%, and activity of other compounds was calculated relative to the activity of cellobiose. C2, C3, C4, C5, Q3G, K3G, N7G, IR, and SG represent cellobiose, cellotriose, cellotetraose, cellopentose, quercetin 3-O-β-glucoside, kaempferol 3-O-β-glucoside, naringenin 7-O-β-glucoside, Iridin, and 1-O-sinopyl-β-glucose. All the values represent means of the three independent replicates ± S.D.
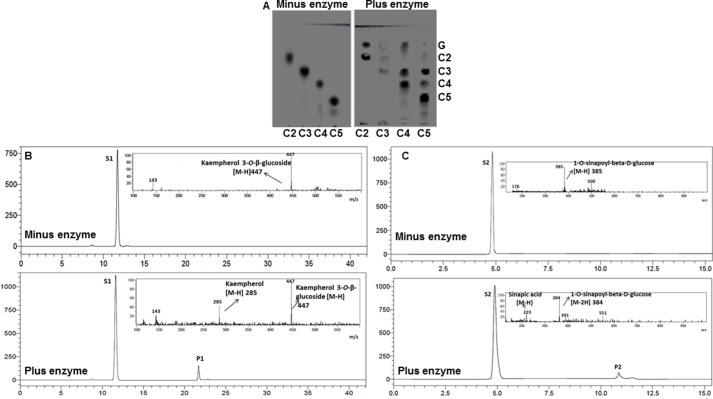
The reaction products were analyzed by thin layer chromatography (TLC) in the case of cello-oligosaccharides and by LC-MS in case of other glucosides. The TLC showed hydrolysis of cello-oligosaccharides as evident from the detection of shorter cello-oligosaccharides and glucose (Fig. 3A). Similarly, LC-MS analysis showed product peaks P1 (m/z 285) and P2 (m/z 223) corresponding to kaempferol and sinapic acid formed by hydrolysis of substrates S1, kaempferol 3 O-β-glucoside (m/z 447), and S2, 1-O-sinopyl-β-d-glucose (m/z 385), respectively (Fig. 3, B and C).
FIGURE 3.
In vitro hydrolysis of representative substrates by CsBGlu12. A, TLC of cello-oligosaccharides in the absence and presence of enzyme CsBGlu12. G, C2, C3, C4, and C5 depict glucose, cellobiose, cellotriose, cellotetraose, and cellopentose. LC-MS chromatograms of Kaempferol 3-O-β-glucoside in absence and in presence of enzyme (B) and 1-O-sinapoyl-β-d-glucose in absence and in presence of the enzyme CsBGlu12 (C) are shown. Substrates S1, kaempferol 3-O-β-glucoside (m/z 447), and S2, 1-O-sinapoyl-β-d-glucose (m/z 385), were converted to products P1 (m/z 285) and P2 (m/z 223), respectively.
Furthermore, kinetic parameters were determined for substrates hydrolyzed by CsBGlu12 (Table 1 and supplemental Fig. S3). The enzyme showed higher affinity for cello-oligosaccharides and flavonol β-glucosides followed by phenolic β-glucosides. Among the cello-oligosaccharides, CsBGlu12 showed more specificity for cellobiose as depicted by its lower Km value (46.44 μm) and higher catalytic efficiency (17.90 mm−1 s−1). Furthermore, among the flavonoid substrates tested, CsBGlu12 showed higher affinity for kaempferol 3-O-β-glucoside (Km: 39.22 μm) and quercetin 3-O-β-glucoside (Km: 54.96 μm). However, higher Km values were observed for phenolic glucosides suggesting comparatively lower affinity of the recombinant enzyme for these substrates.
TABLE 1.
Substrate saturation kinetic parameters of CsBGlu12
The parameters were studied in reaction mixture containing different concentrations of the substrates (1.95–500 μm). Kcat was calculated assuming the molecular mass of the recombinant CsBGlu12 subunit was 57 kDa. All values are the means ± S.D. of three separate preparations of the recombinant enzyme. No detectable activity was observed for coumarin, phenyethanoid, and apocarotenoid glucosides.
| Substrate | Km | Kcat | Kcat/Km |
|---|---|---|---|
| μm | s−1 | s−1mm−1 | |
| Cellobiose | 46.44 ± 4.60 | 0.83 ± 0.02 | 17.90 ± 0.3 |
| Cellotriose | 61.42 ± 3.30 | 0.80 ± 0.03 | 13.02 ± 0.3 |
| Cellotetraose | 62.25 ± 1.78 | 0.78 ± 0.02 | 12.53 ± 0.2 |
| Cellopentose | 74.81 ± 3.81 | 0.78 ± 0.03 | 10.42 ± 0.3 |
| Kaempferol 3-O-β-glucoside | 39.22 ± 3.27 | 0.71 ± 0.02 | 18.10 ± 0.2 |
| Quercetin 3-O-β-glucoside | 54.16 ± 4.21 | 0.92 ± 0.02 | 17.00 ± 0.2 |
| Naringenin 7-O-β-glucoside | 60.21 ± 3.26 | 1.05 ± 0.06 | 17.40 ± 1.6 |
| Iridin | 72.80 ± 4.97 | 1.29 ± 0.05 | 7.70 ± 0.3 |
| 1-O-Sinopyl-β-d-glucose | 132.4 ± 3.60 | 1.46 ± 0.04 | 2.56 ± 0.1 |
CsBGlu12 Activity Is Affected by pH, Metal Ions, and Chaotropic Agents
A comparison of hydrolytic activity across a wide pH range showed that recombinant CsBGlu12 had optimal activity at pH 5.5 (supplemental Fig. S4). The pH optimum is characteristic of other plant β-glucosidases (22). The effect of various additives, which included several metal ions and chaotropic agents on in vitro enzyme activity of recombinant CsBGlu12, was also evaluated. Most of the metal ions reduced the activity of the enzyme to the extent of 10–98% (Table 2). The highest inhibition was caused by Hg2+ (10 mm) followed by Cu2+ (1 mm), whereas Ca2+ had minimum inhibitory effect on the in vitro enzyme activity. In comparison with the metal ions, the recombinant enzyme was comparatively less affected by chaotropic agents. In the presence of SDS (0.05%) and urea (4 mm), the enzyme activity was inhibited by 50 and 60%, respectively. However, EDTA (1 mm), the nonionic detergent Triton X-100 (1%), and reducing agent 2-mercaptoethanol (1%) had little or no effect on the enzyme activity. These observations are also in agreement with the studies carried out previously (23).
TABLE 2.
Effects of metal cations, SDS, Triton X-100, urea, EDTA, and 2-mercaptoethanol on the enzyme activity of recombinant CsGlu12 and cellobiose used as substrate
| Additive | Concentration | Relative activity (%) |
|---|---|---|
| Control | 100 | |
| Ca2+ | 1 mm | 90 |
| Mg2+ | 1 mm | 77 |
| Cu2+ | 1 mm | 46 |
| Hg2+ | 10 mm | 02 |
| K+ | 1 mm | 73 |
| Zn2+ | 1 mm | 62 |
| SDS | 0.05% | 50 |
| Urea | 4 mm | 60 |
| EDTA | 1 mm | 103 |
| Triton X-100 | 1% | 92 |
| 2-Mercaptoethanol | 1% | 111 |
Evolutionarily Conserved Glu200 and Glu414 Residues Are Critical for CsBGlu12 Activity
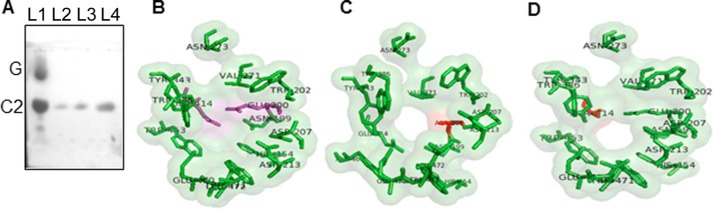
The catalytic residues Glu200 and Glu414 are highly conserved among plant β-glucosidases and have been considered critical for their β-glucosidase activity. To confirm their significance in CsBGlu12, site-directed mutagenesis of these two residues was carried out. Two single substitution mutations were developed by replacing Glu200 and Glu414 with Ala200 and Ala414, respectively. The two mutant proteins (M1 and M2) were expressed and purified, and their activities were evaluated against cellobiose and kaempferol 3-O-β-glucoside. No detectable β-glucosidase activity was observed for either of the two mutant proteins (Fig. 4A). These results were consistent with the bioinformatic analysis of the active site of the two mutant CsBGlu12 proteins, which showed that the mutations lead to visible change in the structure of the active site of CsBGlu12 (Fig. 4, B–D).
FIGURE 4.
In vitro activity of mutant CsBGlu12 proteins. Two mutant proteins designated as M1 (Glu200–Ala200) and M2 (Glu414–Ala414) were generated, and their activity was determined against cellobiose. A, TLC of wild type (WT) and mutant CsBGlu12. Lane L1, WT protein plus cellobiose; lane L2, cellobiose only; lane L3, M1 plus cellobiose; and lane L4, M2 plus cellobiose. G and C2 depict glucose and cellobiose, respectively. B, homology models of WT active site. Pink color, residues depict two catalytic Glu residues, M1 active site (C) and M2 active site (D). Red color residues depict mutated residues.
CsBGlu12 Localizes to Vacuole
Subcellular localization of enzymes provides key insights about their biological functions. Prediction of subcellular localization of CsBGlu12 with PSORT indicated vacuolar and/or chloroplast location. To confirm the localization experimentally, Csbglu12 was cloned in PAM-PAT-35S-YFP vector. The fusion gene Csbglu12-YFP was introduced into onion (Allium cepa) epidermal cells by particle bombardment. Although the control YFP accumulated throughout the cell, CsBGlu12-YFP was localized in the vacuole (Fig. 5).
FIGURE 5.
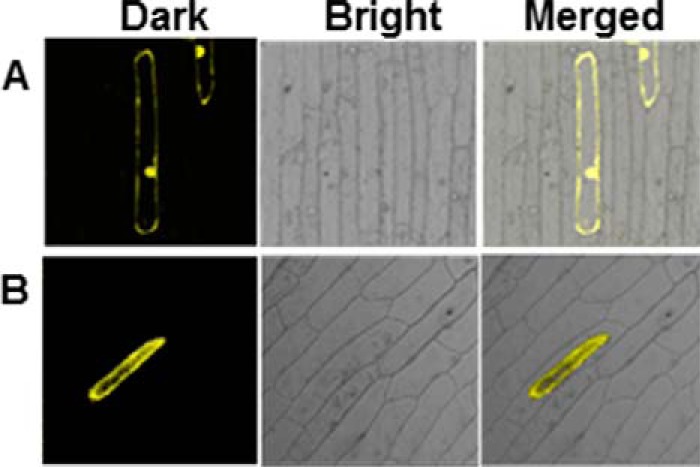
Subcellular localization of CsBGlu12. Dark field, bright field, and merged images of YFP (A) vacuolar localized CsBGlu12-YFP (B) in onion peel.
Expression Pattern of Csbglu12 Correlated with the Accumulation of Flavonols
To gain an understanding about the biological function of Csbglu12, its expression profile was investigated in various tissues, at different developmental stages, and in response to stresses and phytohormones using qRT-PCR. Two genes (18S and GAPDH) were used independently as endogenous control to normalize the data. In both the cases, the expression pattern followed the same trend. The results obtained with 18S are presented in Fig. 6, and those with GAPDH are given in supplemental Fig. S5.
FIGURE 6.
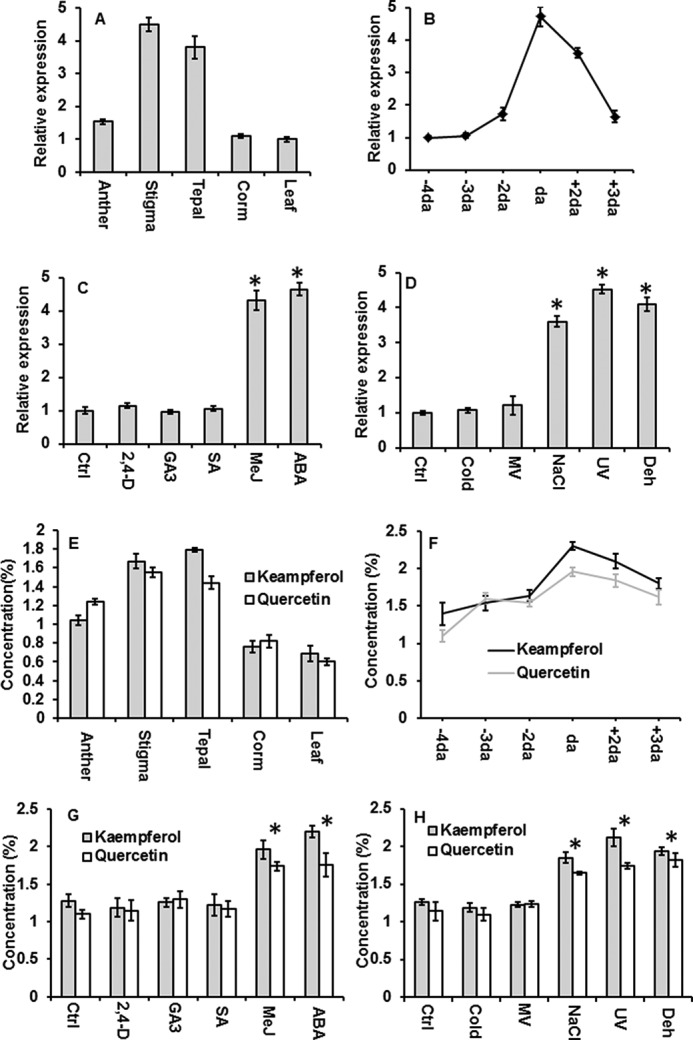
Expression of Csbglu12 and accumulation of unconjugated flavonols. Expression of Csbglu12 was determined using quantitative real time PCR of different tissues (A) and different developmental stages of flower (B) as follows: 4 days before anthesis (−4da; 3 days before anthesis (−3da); 2 days before anthesis (−2da); day of anthesis (da); 2 days after anthesis (+2da); and 3 days after anthesis. C, hormonal treatments as follows: control (Ctrl); MeJ, 2,4-dichlorophenoxyacetic acid (2,4-D); ABA, salicylic acid (SA), and gibberellic acid (GA3). D, stress treatments as follows: sodium chloride (NaCl); ultraviolet-B (UV); methyl viologen (MV); dehydration (Dehyd); and 4 °C (cold). 18S was used as endogenous control. Concentration of flavonols (Kaempferol and Quercetin) was determined in different tissues (E), different developmental stages (F), different hormonal treatments (G), and stress treatments (H). All experiments were carried out in triplicate and expressed as mean ± S.D. Differences between the control and treatments were analyzed by Student's t test and considered statistically significant at *, p < 0.05.
We observed that Csbglu12 shows relatively higher expression in stigma followed by tepal and anther; however, low transcript levels were observed in corm and leaf (Fig. 6A).
Because transcript levels of Csbglu12 were higher in floral tissues, the expression of Csbglu12 was examined for 4, 3, and 2 days before anthesis, on the day of anthesis, and 2 and 3 days after anthesis. The expression of Csbglu12 was observed to increase from 4 days before anthesis until the day of anthesis and then exhibited a sharp decline during post-anthesis stage of the flower development (Fig. 6B). In the case of stress treatments, Csbglu12 was significantly induced in response to dehydration, NaCl, and UV-B treatments. It also showed enhanced expression in response to methyl jasmonate (MeJ) and ABA treatments (Fig. 6, C and D).
As CsBGlu12 exhibited high affinity for flavonol β-glucosides, the concentrations of free (unconjugated) flavonols (quercetin and kaempferol) in different tissues and developmental stages of flower and in response to various stresses and phytohormones were estimated. The accumulation of flavonols followed a similar trend as that of Csbglu12 expression (Fig. 6, E and F). Thus, the expression of Csbglu12 was consistent with the deglycosylation of flavonol β-glucosides.
Transient Expression of CsBGlu12 in N. benthamiana Leads to Accumulation of Flavonols
To validate the role CsBGlu12 in stress, the open reading frame of Csbglu12 and mutant Csbglu12 versions (M1 and M2) were cloned in plant overexpression vector pBI121 under 35S promoter. The gene constructs pB1121-Csbglu12, pB1121-M1, and pB1121-M2 were transiently overexpressed in N. benthamiana leaves. The leaves of overexpression lines and uninoculated plants were collected after 72 h, and the expression of the transgene was estimated by semiquantitative RT-PCR. The results revealed high expression of Csbglu12 in N. benthamiana CsBGlu12 overexpression line, although no transcripts were detected in uninoculated plants (supplemental Fig. S6). Furthermore, CsBGlu12 activity was determined with kaempferol 3-O-β-glucoside and cellobiose as the substrates. The β-glucosidase activity of the leaf extracts of overexpression lines was 4.7- and 5.2-fold higher than that of the uninoculated plants confirming the overexpression of CsBGlu12 in N. benthamiana. However, no significant differences were observed in β-glucosidase activity of mutant CsBGlu12 overexpression lines (M1 and M1) and uninoculated plants.
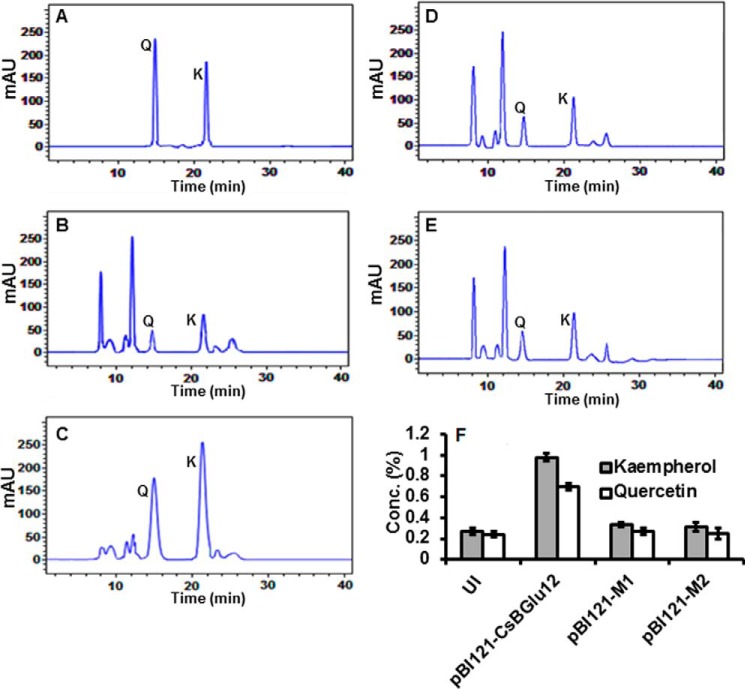
As a result of higher β-glucosidase activity in the CsBGlu12 overexpression line, we speculated that it may exhibit accumulation of comparatively higher concentrations of free flavonols (quercetin and kaempferol) due to hydrolysis of corresponding flavonol β-glucosides. Therefore, we quantified the kaempferol and quercetin contents in both uninoculated and overexpression lines (pBI121-Csbglu12, pBI121-M1, and pBI121-M2) of N. benthamiana by HPLC. Interestingly, we observed that the concentrations of unconjugated kaempferol and quercetin contents were comparatively higher (3.4- and 2.9-fold, respectively) in plants overexpressing normal CsBGlu12 as compared with those overexpressing mutant forms of protein and the uninoculated plants (Fig. 7, A–F).
FIGURE 7.
Quantification of flavonols (kaempferol and quercetin) by HPLC in uninoculated and transiently overexpressing N. benthamiana plants (pBI121-CsBGlu12, pBI121-M1, and pB121-M2). Chromatogram of reference compounds (A), uninoculated N. benthamiana (B), transiently overexpressing pBI121-CsBGlu12 plants (C), transiently overexpressing pBI121-M1 plants (D), transiently pBI121-M2 overexpressing plants (E), and quantification of flavonols in uninoculated (UI), pBI121-CsBGlu12, pBI121-M1 and pB121-M2 overexpressing plants (F). Q and K represent quercetin and kaempferol, respectively. All experiments were carried out in triplicate and expressed as mean ± S.D. mAU, milliabsorption units.
Accumulation of Flavonols Confers Abiotic Stress Tolerance through ROS Scavenging
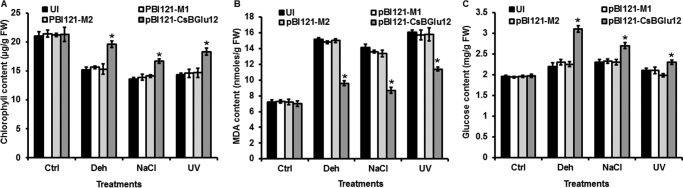
To evaluate the uninoculated plants and the plants transiently overexpressing normal CsBGlu12 and its two mutant forms (M1 and M2) for stress tolerance, their leaves were cut into small circular discs of equal diameter and subjected to different stress treatments in Petri plates. UV-B, dehydration, and NaCl treatments were given for 24 h. The leaf discs were collected, and total chlorophyll and MDA contents were determined (Fig. 8). The chlorophyll content of leaf discs of uninoculated plants and all three overexpression lines was similar under control conditions; however, under all the stresses, there was a significant decrease in chlorophyll content of uninoculated plants and the plants overexpressing mutant (M1 and M2) proteins. The plants overexpressing normal Csbglu12 gene had significantly higher chlorophyll content (Fig. 8A). MDA content of CsBGlu12 overexpression lines was lower than the uninoculated plants, and the plants overexpressing mutant proteins under stress conditions (Fig. 8B). As the soluble sugar is indicative of stress response in plants, its content in uninoculated and all the overexpression lines was determined. Under stress conditions the total soluble sugar content was higher in plants overexpressing CsBGlu12 than those of uninoculated plants and the plants overexpressing mutant proteins (Fig. 8C). Taken together, these results suggest that CsBGlu12 enhances tolerance to various abiotic stresses.
FIGURE 8.
Assessment of effect of abiotic stress on physiology (A), total chlorophyll (B), MDA and soluble sugar content of uninoculated (UI) and overexpressing lines (pBI121-CsBGlu12, pBI121-M1, and pB121-M2) (C). Differences between the uninoculated (UI) and transiently overexpressing N. benthamiana plants were analyzed using Student's t test and considered statistically significant at *, p < 0.05. Differences and standard deviations were calculated from three biological replicates. Ctrl, control; Deh, dehydration.
Stress conditions may also induce expression of endogenous Csbglu12 homologs in N. benthamiana that may result in enhanced production of quercetin and kaempferol and subsequent tolerance to stress. To confirm the role of CsBGlu12 in stress tolerance, we determined quercetin and kaempferol content in uninoculated plants and CsBGlu12, M1, and M2 overexpression lines. Results indicated a significant increase in quercetin and kaempferol concentration in plants overexpressing normal CsBGlu12 only providing a strong clue thatCsBGlu12 is involved in flavonol accumulation under these conditions (supplemental Fig. S7).
To gain insights into the underlying mechanism for abiotic stress tolerance in CsBGlu12-overexpressing lines, the radical scavenging activity of the crude leaf extract from each plant was assayed using 2,2-diphenyl-1-picrylhydrazyl (DPPH) (24). Results revealed that the radical scavenging activity of crude leaf extracts of plants overexpressing normal CsBGlu12 protein was comparatively higher than the uninoculated plants and plants overexpressing mutant (M1 and M2) forms of proteins (Fig. 9A). To examine the flavonols in overexpression lines that mitigate ROS in vivo, 3,3′-diaminobenzidine (DAB) staining, a marker of H2O2 accumulation (25) was carried out on the leaf discs of N. benthamiana plants subjected to dehydration, salt, and UV-B stress. The patterns of DAB staining revealed that higher flavonol accumulation in plants overexpressing CsBGlu12 mitigated H2O2 accumulation under abiotic stress conditions (Fig. 9B).
FIGURE 9.
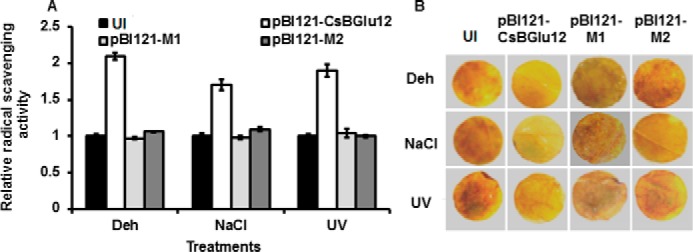
Evaluation of antioxidant activity of uninoculated and overexpression lines. A, radical scavenging activity of crude leaf extracts of uninoculated and overexpression lines (pBI121-CsBGlu12, pBI121-M1, and pB121-M2) determined by DPPH assay. The values represent mean of three biological replicates ± S.D. Deh, dehydration. B, H2O2 generation in uninoculated (UI) and overexpression lines (pBI121-CsBGlu12, pBI121-M1, and pB121-M2) detected by 3,3′-diaminobenzidine polymerization. The representative results were obtained from three biological replicates.
Discussion
With recent advancements in the study of plant secondary metabolism, it has become quite obvious that biosynthesis, localization, and biological activity of secondary metabolites is not only regulated at a transcriptional and post-transcriptional level but also encompasses the reversible glycosylation of these metabolites by regioselective glycosyltransferases and β-glucosidases. Currently, one of the major challenges in this field is to gain better understanding of how plants regulate the function of individual members of glycosyltransferase/β-glucosidase families in distinct tissues, developmental stages, and in response to biotic and abiotic stress stimuli (26). Plants contain a myriad of β-glucosidases, and it is not always clear which one is accountable for a specific function (27, 28). Moreover, β-glucosidases and their substrates are compartmentalized separately and come into contact only under certain conditions. To assess their biological function, it is imperative to know about the physiological substrates of a particular β-glucosidase and the conditions under which they come into contact. Against this backdrop, 15 β-glucosidases were identified from C. sativus transcriptome (18). Among these, Csbglu12 exhibited higher expression in stigma of the flower, the main site of biosynthesis and accumulation of many secondary metabolites in C. sativus, which include, but are not limited to, flavonols and apocarotenoids (29, 30). Therefore, Csbglu12 was selected for further study. Crocus metabolites, particularly flavonols, like many other plant secondary metabolites are stored as glucosylated forms. Under certain conditions, β-glucosidases come into play and release active aglycones from their respective inactive glucosides. So far, none of the β-glucosidases have been isolated and characterized from C. sativus thus making it necessary to identify these enzymes, their substrate preferences, and the conditions under which they perform their roles. This will enable a better understanding of the biological function of secondary metabolites of plants in general and Crocus in particular. Moreover, phylogenetic analysis of CsBGlu12 clustered it with a yet largely uncharacterized β-glucosidases (supplemental Fig. S1), making its study more important. Among very close homologs of CsBlu12, only one β-glucosidase (Os4BGlu12) has been characterized, but its role in planta is yet to be established (17, 31).
CsBglu12 possesses (β/α)8 TIM barrel conformation characteristic of the GH1 family of glycoside hydrolases (Fig. 1). The active site consists of a glycone-binding pocket with highly conserved amino acid residues and non-conserved aglycone-binding pocket (Fig. 1D). The presence of non-conserved aglycone-binding amino acid residues is considered important for the substrate specificity of β-glucosidases (32). Consistent with this, many related β-glucosidases are often unable to catalyze the hydrolysis of closely related β-glucosides (33, 34). Phylogenetic and docking analysis revealed that CsBGlu12 exhibits higher affinity for flavonoid β-glucosides and cello-oligosaccharides and low affinity for phenolic glucoside and other glucosides as evident from free energy change (ΔG) for the best pose of the CsBGlu12-ligand complex (supplemental Fig. S2 and supplemental Table S1).
Consistent with the docking analysis, recombinant CsBGlu12 exhibits higher in vitro activity against cello-oligosaccharides and flavonoid glucosides followed by phenolic glucosides. However, it was incapable of catalyzing the hydrolysis of coumarin and other glucosides (Figs. 2 and 3). The Kcat/Km values for cello-oligosaccharides range from 10.42 ± 0.3 to 17.90 ± 0.3 mm−1 s−1. The steady decrease in Kcat/Km values from cellobiose to cellopentose indicates that CsBGlu12 has a higher efficiency for shorter cello-oligosaccharides, which may be explained by the presence of a short binding cleft in CsBGlu12. Similar observations were made on rice Os3BGlu6, which catalyzes hydrolysis of β(1→3)- and β(1→2)-linked disaccharides more efficiently than longer oligosaccharides (35). Furthermore, among the natural glucosides tested, the flavonol glucosides like kaempferol 3-O-β-glucoside (Kcat/Km, 18.10 mm−1 s−1), quercetin 3-O-β-glucoside (Kcat/Km, 17.0 mm−1 s−1), and naringenin 7-O-β-glucoside (Kcat/Km, 17.40 mm−1 s−1) are hydrolyzed more rapidly than iridin (Kcat/Km, 7.70 mm−1 s−1) and the phenolic glucoside 1-O-sinopyl-β-d-glucose (Kcat/Km, 2.56 mm−1 s−1) (Table 1). This suggests that kaempferol 3-O-β-glucoside, quercetin 3-O-β-glucoside, and naringenin 7-O-β-glucoside may serve as hydrophobic substrates in planta. The predicted (as suggested by PSORT) and experimentally validated (Fig. 5) vacuolar localization and lower optimal pH (5.5) (supplemental Fig. S4) of CsBGlu12 are in conformity with its substrate specificity for flavonol β-glucosides that are found in this compartment.
Besides the glycone- and aglycone-binding pockets, the active site of CsBGlu12 harbors two highly conserved glutamic acid residues (Glu200 and Glu414) located in the characteristic and conserved peptides Thr-(Phe/Leu)-Asn-Glu-Pro and Tyr-Ile-Thr-Glu-Asn-Gly. These two glutamic acid residues are considered critical for the catalytic activity of several plant β-glucosidases (36). However, the role of these residues as catalytic nucleophiles has not been validated in any of the CsBGlu12 orthologs from other plants. To validate the significance of these two Glu residues in the catalytic activity of CsBGlu12, two mutant proteins, M1(E200A) and M2(E414A), were generated. It was observed that both of the mutants exhibit complete loss of activity against the tested substrates (Fig. 4A). Our results are in agreement with studies carried out on other plant β-glucosidases. In cassava, cyanogenic β-glucosidase (36, 37), and in Rauvolfia, strictosidine β-glucosidase (38), mutation of the catalytic Glu residues leads to complete abolishment of β-glucosidase activity. Homology modeling of the active site of mutant forms (M1 and M2) with rice Os4BGlu12 as a template indicates that the mutation of any of the two key Glu residues results in visible change in the architecture of the active site of CsBGlu12 which, in part, may be responsible for the abolishment of the β-glucosidase activity (Fig. 4, B–D). However, further investigations are required for confirmation.
Analysis of gene expression has often been useful to gain insights about the probable function of β-glucosidases (28). Expression analysis revealed that the transcription of Csbglu12 is regulated in a tissue- and development-specific manner. The transcript levels of Csbglu12 were higher in the floral tissues as compared to corm and leaf (Fig. 6A). Furthermore, the expression of Csbglu12 increases upto anthesis and then exhibits a sharp decline during the post-anthesis stage of floral development (Fig. 6B). The expression of Csbglu12 was in agreement with the accumulation pattern of unconjugated flavonols (Fig. 6, E and F), providing proof for the involvement of this enzyme in the deglycosylation process of flavonol glucosides. Furthermore, the accumulation of free and biologically active flavonols at these specific stages of floral development may be explained by the fact that flavonols are synthesized and accumulated in the floral tissues until anthesis to prevent the newly opened reproductive tissues from several biotic and abiotic stresses (39–42). Consequently, the expression of Csbglu12 during the anthesis stage is higher to release the active flavonols from their corresponding glucosides. Furthermore, in C. sativus, flower senescence sets in immediately after anthesis, and secondary metabolites are remobilized from the senescent flower to other plant parts especially the underground corm. During this stage, the transcript levels of Csbglu12 might be lower so as to enhance flux toward glucosylated forms of metabolites to increase their solubility and to remobilize them to other plant parts for storage (30, 43). The expression of Csbglu12 is also induced in response to dehydration, NaCl and UV-B treatments, as well as in response to hormonal treatments like MeJ and ABA (Fig. 6, C and D). This expression of Csbglu12 was accompanied by the concomitant increase in the accumulation of flavonols (Fig. 6, G and H), providing a clue toward the role of this enzyme in abiotic stress. Several other β-glucosidases have also been found to be induced in response to stress stimuli (44, 45).
Transient expression of CsBGlu12 leads to the accumulation of antioxidant flavonols (kaempferol and quercetin) in N. benthamiana indicating that CsBGlu12 catalyzes the hydrolysis of kaempferol- and quercetin 3-O-β-glucoside in planta. However, no significant accumulation of flavonols was observed in N. benthamiana expressing catalytically inactive M1 and M2 proteins (Fig. 7). Moreover, disc assays revealed that transiently overexpressing CsBGlu12 plants are more tolerant to stresses than uninoculated ones and the catalytically inactive M1 and M2 overexpression lines as was evident from higher total chlorophyll and lower MDA contents (Fig. 8, A and B). Abiotic stresses affect photosynthetic apparatus in plants and cause reduction in chlorophyll content, which is often used as a measure of extent of stress (29). Also, ROS generated during stress leads to membrane damage due to lipid peroxidation, and MDA is released as a by-product. Therefore, higher chlorophyll and lower MDA levels are indicators of higher stress tolerance. Soluble sugar content of transient overexpression lines was also comparatively higher under stress (Fig. 8C). It has been reported that soluble sugars maintain high antioxidant protection, and it is one of the mechanisms in plants to cope with abiotic stress (46, 47). Although there are a few reports about the role of β-glucosidases in abiotic stress, most of them suggest synthesis and accumulation of ABA as possible mechanisms. To the best of our knowledge, this work represents the first study that reports the role of β-glucosidase in stress through accumulation of antioxidant flavonols. Radical scavenging activity by DPPH and DAB staining revealed that crude leaf extracts of transient overexpression lines of CsBGlu12 had higher antioxidant activity than the uninoculated plants and catalytically inactive M1 and M2 overexpression lines of N. benthamiana (Fig. 9). Thus, our results suggest that the stress tolerance of CsBGlu12 overexpression lines may be due to the accumulation of flavonols (kaempferol and quercetin), which are known to play key roles in abiotic stress (48). There are many instances where kaempferol and quercetin levels were shown to increase in response to UV-B irradiation (49). It has also been reported that a brief exposure of plants to UV-B provides a regulatory signal, eliciting the accumulation of flavonols to confer protection to UV-induced damage due to their absorption in this region (50, 51). This is further supported by Arabidopsis thaliana tt5 and tt6 mutants that are highly sensitive to UV-B radiation due to reduced levels of UV-absorbing flavonoids (52). Similarly, some recently carried out experiments on A. thaliana revealed that dehydration stress induced alternation in the concentration of glycosides of kaempferol and quercetin (53). Abiotic stresses such as UV-B, salt, and dehydration cause the production of substantial amounts of ROS in plants leading to cell damage (48, 54). Because flavonols, like quercetin and kaempferol, are good scavengers of ROS, accumulation of these compounds may explain the UV-B, dehydration, and salt stress tolerance of the transiently overexpressing CsBGlu12 N. benthamiana plants. Taken together, we conclude that CsBGlu12 deglycosylates flavonol β-glucosides in planta leading to the accumulation of active flavonols that confer tolerance to various abiotic stresses, one of the mechanisms of which is ROS scavenging. Moreover, during hydrolysis of these flavonol β-glucosides, glucose is also released, which may serve as an alternative source of energy during insufficient photosynthesis under stress.
Experimental Procedures
Plant Material and Elicitor Treatment
C. sativus L. plants were collected from an experimental farm at the Indian Institute of Integrative Medicine, Srinagar, India (longitude, 34°5′24″N; latitude, 74°47′24″ and altitude 1585 m above sea level) as reported previously (11, 55). The representative voucher specimen was submitted at the Janaki Ammal Herbarium, CSIR-IIIM, Jammu, India, under accession number 22893. For tissue and developmental stage-specific expression, samples were collected from different tissues (stigma, anther, tepal, corm, and leaf) and developmental stages. To assess the effect of stress stimuli on gene expression, C. sativus corms were grown in pots in greenhouse conditions at 26 ± 2 °C. Stress induction was carried out as described previously (55). Briefly, 200 mm NaCl, 5 μm methyl viologen, and 200 mm mannitol solutions were added to Crocus plants growing in separate pots after intermittent intervals for induction of salinity, oxidative and dehydration stresses, respectively. For UV stress, plants were irradiated with UV-B light of 1500 μJ/m2. Cold stress was induced by shifting the plants to 4 °C. All the treatments were given for 24 h, and the plants grown under normal conditions were kept as control. For treatment with phytohormones, the plants were sprayed with 0.1 mm salicylic acid, 50 μm 2,4-dichlorophenoxyacetic acid, 0.1 mm MeJ, and 0.1 mm gibberellic acid. For all these treatments, flower tissue was harvested after 24 h. All the tissue samples were frozen in liquid nitrogen and stored at −80 °C for further experimental use.
RNA Isolation and cDNA Synthesis
For total RNA isolation, RNeasy isolation kit (Qiagen) was used, and the procedure was carried out according to the manufacturer's instructions. The quality of RNA samples was determined by gel electrophoresis and A260/A280 ratio using a spectrophotometer. The genomic DNA contamination was removed by DNase I (Fermentas) treatment. The cDNA synthesis was done using RevertAid cDNA synthesis kit (Fermentas), and the procedure was carried out in accordance with the manufacturer's instructions.
Identification and Cloning of Csbglu12
Previously our laboratory had carried out the whole transcriptome sequencing of C. sativus (18). From this transcriptome, we identified 15 β-glucosidase cDNA sequences. Of these β-glucosidases, Csbglu12 was selected for further study based on it higher expression in the stigma part of the flower. The full-length amplification of Csbglu12 was performed using the primers listed in supplemental Table S2 following cycling conditions: one cycle of 95 °C for 3 min, 35 cycles of 94 °C for 40 s, 57 °C for 1 min, and 72 °C for 2 min followed by a final extension of 72 °C for 10 min in a thermal cycler (Applied Biosystems). The amplicon was visualized by gel electrophoresis and then purified by gel extraction kit (Qiagen). The purified amplicon was cloned in pTZ57R/T vector (Fermentas), and then transformed into E. coli DH5α host strain. The plasmid containing the gene was sequenced by automated DNA sequencer (Applied Biosystems). The sequence obtained was finally confirmed by NCBI Blast.
In Silico Analysis
The complete nucleotide sequence of Csbglu122 was translated using translation tool from ExPASy, and the properties of deduced amino acid sequence were predicted using ProtParam and Phobius programs. Subcellular localization was predicted by PSORT server. Secondary structure analysis was carried out by ESPript 3 software (56). For phylogenetic analysis, sequences were recovered from the GenBankTM by BLASTp algorithm at the National Centre for Biotechnology Information (NCBI) using CsBGlu12 sequence as query. Sequences were aligned by the ClustalW program using default parameters, and the phylogenetic tree was generated using the Neighbor-Joining method by MEGA 6 software (57). The three-dimensional structure of CsBGlu12 was created using SWISS-MODEL workspace with crystal structure of O. sativa Os4BGlu12 (Protein Data Bank code 3ptk) as template. The stereochemical analysis of the modeled protein was done using the Ramachandran plot obtained from RAMPAGE. Docking analysis was carried out on CsBGlu12 protein using DockingServer as described previously (55, 58, 59).
Heterologous Expression and Purification of CsBGlu12
For heterologous expression of CsBGlu12, the gene was cloned in bacterial expression vector pGEX4T-1 at SalI and NotI restriction sites, and the construct was transformed into the E. coli strain BL21 (DE3). The heterologous expression was carried out as described previously (12). Briefly, the pGEX4T-1-Csbglu122 construct was transformed into the E. coli strain BL21 (DE3) following the supplier's recommendations (Novagen, Madison, WI), and positive colonies were selected on LB media containing ampicillin (100 mg/ml). Individual positive colonies were grown overnight in 5 ml of LB, and 1% of overnight cultures was used to inoculate fresh LB, and cells were grown at 30 °C until the A600 of 0.6–0.8 was reached. 1 mm IPTG was added to the exponentially growing cells to induce the gene expression, and 1 ml of induced cultures was harvested after every 2-h interval from 0 to 8 h. The harvested 1-ml culture samples were centrifuged at 13,000 × g for 5 min at 4 °C. The cells were resuspended in 1 ml of PBS buffer and disrupted by sonication on ice for a 15- to 20-s burst time period. The lysate was centrifuged at 13,000 × g for 20 min at 4 °C, and the supernatant was loaded on 10% SDS-polyacrylamide gel after heating the sample with 2× SDS loading dye at 99 °C for 10 min. For purification of the recombinant protein, transformed E. coli cells were cultured at 37 °C until absorbance at 600 nm (A600) was 0.5. Expression was induced by addition of 1 mm IPTG followed by incubation at 30 °C for 6 h. The culture was then centrifuged at 13,000 × g for 5 min at 4 °C, and the cell pellet was resuspended in 1 ml of PBS buffer and lysed by 20 mm DTT and 0.2 mg/ml lysozyme followed by sonication on ice for 20–25 s. The soluble fraction was recovered by centrifugation at 13,000 × g for 20 min at 4 °C and incubated overnight with glutathione-Sepharose beads (1 ml liter−1 of culture) (GE Healthcare) at 4 °C. The beads were washed five times with 10 bed volumes of 1× phosphate-buffered saline (PBS). The GST tag was removed by incubating with thrombin protease (<10 cleavage units/ml) at 24 °C for 12 h. This was followed by centrifugation at 3500 rpm. Afterward, supernatant was collected and incubated with benzamidine-Sepharose (10 μl/unit of thrombin protease) for 30 min at 24 °C to remove thrombin. The cleaved protein was run on the 10% SDS-polyacrylamide gel, and concentration was determined using Bradford assay.
CsBGlu12 pH Optimum, in Vitro Enzyme Assays, and Enzyme Kinetics
The optimum pH for CsBGlu12 was determined as described previously (35). Briefly, 1 μg of enzyme was incubated in a reaction volume of 140 μl containing 100 mm universal buffer (citric acid disodium hydrogen phosphate). The pH was varied from 3 to 8.5 with 0.5 pH unit increments, and 500 μm cellobiose was used as substrate. The reaction was terminated by the addition of 100 μl of 2 m sodium carbonate or by boiling for 5 min.
CsBGlu12 activity was determined in 100 mm sodium acetate (pH 5.), as described previously (35). The reactions were stopped after an overnight incubation (30 °C) by boiling for 5 min. The products from the reactions were spotted onto silica gel F254 plates (Merck) and run in ethyl acetate/acetic acid/water (2:1:1, v/v) in the case of oligosaccharides and ethyl acetate/acetic acid/methanol/water (15:2:1:2, v/v) in the case of other glycosides. The thin layer chromatograms (TLC) were visualized under UV light at 254 nm and developed by dipping in 10% (v/v) sulfuric acid in methanol followed by heating at 120 °C until the appearance of dark spots. Flavonol and phenolic glucosides were also analyzed by LC-MS.
Kinetic parameters were determined for substrates of interest, based on the initial relative activity assays using standard assay conditions with varied substrate concentrations (1.95–500 μm). Glucose release was measured by the glucose oxidase assay to monitor hydrolysis of glucosides, as described previously (35). The kinetic constants Km and Vmax values were calculated with non-linear regression analysis using GraphPad Prism 6 software. Effect of additives on enzyme activity was evaluated using a standard protocol (23).
LC-ESI-MS Analysis
LC-MS analysis was carried as described previously (22, 60) with slight modifications using Nexera UHPLC (130 megapascals) equipped with MS-8030 (Shimadzu). Enable RP-C18 column (250 × 4.6 mm, 5 μm) was used for the analysis. The injection volume was 5 μl and flow rate 0.3 ml/min. Peaks of the substrates and their products were detected at 360 (flavonols) and 330 nm (phenolics). Ion trap mass spectrometer interfaced with electrospray ionization was used to detect the masses of the eluted compounds and therefore identification of the peaks. Analysis was carried out in the negative ion mode. The drying gas flow was kept at 1 liter min−1, and the nebulizer pressure was set at 35 p.s.i. Drying gas temperature was kept at 300 °C and capillary voltages at 4 kV. The capillary exit of the source was tuned to −140 V, and the end plate was offset by −500 V. Eluted masses were scanned in the range 100–1000 m/z and a maximum acquired time of 200,000 μs.
Site-directed Mutagenesis
Substitutions at Glu200 and Glu414 with Ala200 and Ala414 were generated following the QuikChange protocol (Stratagene) using the Csbglu12 construct as a template. The primers used for site directed mutagenesis are given in supplemental Table 2.
Subcellular Localization
The subcellular localization of CsBGlu12 was studied by performing transient expression assay in onion epidermal cells as described previously (11). For this, Csbglu12 was cloned in PAM-PAT-35S to produce Csbglu12-YFP fusion protein. The fusion construct of Csbglu12-YFP was bombarded on to the onion peels using biolistic gene delivery device PDS-1000/He (Bio-Rad). The onion peels were then incubated for 24 h and then visualized under a confocal microscope.
Expression Profiling of Csbglu12
The expression profile of Csbglu12 was studied using quantitative RT-PCR. The qRT-PCR was performed in triplicate in ABI StepOne real time (Applied Biosystems) using SYBR Green Master Mix (Fermentas) and gene-specific primers. The relative quantification (ΔΔ-CT) method was used to evaluate quantitative variation between the samples examined. The reaction was performed in a total volume of 20 μl, which included 10 μl of 2× SYBR Green Master Mix, 0.2 μm gene-specific primers, and 100 ng of template cDNA. The cycling conditions were 95 °C for 20 s, followed by 40 cycles of 95 °C for 15 s, and 58 °C for 1 min. Two genes (18S and GAPDH) were used independently as endogenous controls to normalize all data.
Transient Expression in N. benthamiana
N. benthamiana plants were grown in a growth chamber set at 23–25 °C under a 16-h light/8-h dark cycle. Agrobacterium tumefaciens suspension preparation and infiltrations were done as described previously (61). A. tumefaciens strain LBA4404 containing pBI121 Csbglu12, pBI121-M1, or pBI121-M2 were cultured separately at 28 °C to stationary phase in LB medium in the presence of both kanamycin (50 mg liter−1) and rifampicin (50 mg liter−1). The culture was centrifuged at 2000 × g followed by resuspension of the pellet in MMA buffer to an absorbance at 600 nm (A600) of 1.2 and kept at room temperature for 2–4 h. The MMA buffer consisted of 10 mm MES (pH 5.6), 10 mm MgCl2, and 100 mm acetosyringone. The suspensions were pressure infiltrated into 4-week-old N. benthamiana leaves using a syringe. The tissue was harvested 3 days after infiltration for further analysis.
For enzyme activity of N. benthamiana leaf tissues transiently overexpressing CsBGlu12, about 1 g of leaf tissue was chilled in 3 volumes of ice-chilled extraction buffer. The extraction buffer consisted of 100 mm sodium acetate (pH 5.5) containing 1 mm EDTA, 12 mm β-mercaptoethanol, and 10 mm ascorbate. The homogenate was filtered and centrifuged for 10 min at 10,000 × g at 4 °C. The supernatant was desalted on a PD10 column (GE Healthcare) equilibrated with the same buffer and used for CsBGlu12 assays.
Disc assay was carried out as described previously (62). Briefly, leaf discs of equal diameter were excised from healthy and fully expanded tobacco leaves of uninoculated, M1-, M2-, and CsBGlu12-overexpressing plants using a cork borer. For inducing stress, the leaf discs were subjected to UV-B (1500 μJ/m2), salt stress (200 mm), and dehydration stress (200 mm mannitol) for 24 h.
Estimation of Total Chlorophyll, MDA, and Soluble Sugar Content
Chlorophyll content of uninoculated and transient overexpression lines of N. benthamiana was determined according to the procedure described previously (55). Briefly, 500 mg of leaf tissue from the plants was homogenized in 2 ml of 80% (w/v) cold acetone. The homogenate was centrifuged at 3500 × g for 5 min. The supernatant was collected, and its absorbance was recorded at 663 and 645 nm with only 80% (w/v) cold acetone as blank using a Nano-Drop spectrophotometer (Thermo Fisher Scientific). The absorbance of the blank was subtracted from the absorbance of the sample, and total chlorophyll content was calculated by Equation 1,
| (Eq. 1) |
where A663 and A645 are the absorbances at 663 and 645 respectively.
MDA content of the samples was determined as described previously (55). Briefly, 500 mg of leaves were homogenized in 0.5 ml of 0.1% (w/v) trichloroacetate (TCA) and centrifuged at 10,000 × g for 10 min. Afterward, 0.5 ml of the supernatant was mixed with 0.5% (w/v) thiobarbituric acid. After incubating at 95 °C for 30 min, the reaction was quickly cooled on ice and centrifuged at 10,000 × g for 5 min. The absorbance of the supernatant was recorded at 532 nm and corrected for nonspecific turbidity by subtracting the absorbance at 600 nm. MDA content was calculated as shown in Equation 2,
| (Eq. 2) |
where V = volume of extract and W = fresh weight of sample. Total sugar content in leaf samples was estimated as described previously (63).
Extraction and Quantification of Quercetin and Kaempferol
The tissues samples (stigma, anther, tepal, leaf, corm, and flowers of different developmental stages) of C. sativus and leaf tissue of both uninoculated and transiently overexpressing N. benthamiana lines were air dried at room temperature and crushed to fine powder. The powdered samples were serially extracted with dichloromethane/MeOH in the ratio of 1:1 (v/v). The procedure was carried out three times and each time with a fresh solvent. The filtrates of all the three extractions were combined and passed through Whatman No. 1 paper filter. The solvents were removed at 45 °C under reduced pressure using a rotary evaporator (Sigma). The stock solutions (1 mg/ml) of quercetin and kaempferol along with extracts were freshly dissolved in methanol and filter sterilized with 0.25-μm membrane filters (Millipore). The HPLC (Shimadzu CLASS-VP Version 6.14 SPI model) equipped with RP-18e column (E-Merck, 5 μm, 4.6 × 250 nm), a photodiode array detector (SPD-M10A VP model), and a pump (LC-10AT VP model) was used for the analysis of flavonols (quercetin and kaempferol). Determination of flavonol content was carried out as described previously (64). The solvent system was 97.8% (v/v H2O), 2% CH3CN, 0.2% H3PO4 (A), and 97.8% (v/v CH3CN), 2% H2O, 0.2% H3PO4 (B) with a gradient elution of 0–30 min, 20% B; 30–35 min, 45% B; 35–38 min, 55% B; 38–40 min, 55% B; and 40–45 min, 20% B; at a flow rate of 0.5 ml/min. Injection volume of the sample was 10 μl, and the column temperature was kept at 30 °C. The identification and quantification of the two flavonols were done on the basis of retention time of reference compounds. Relative contents of the two flavonols (quercetin and kaempferol) were determined and expressed as percentage peak area.
Radical Scavenging Activity Assay and DAB Staining
Radical scavenging activity was determined by DPPH method as described previously (65, 66). Briefly, leaves of transient overexpression lines and uninoculated plants were extracted with 5 μl of solvent (MeOH/H2O/CH3COOH = 9:10:1) per mg of fresh weight. Afterward, 50 μl of the extract was added to 450 μl of DPPH and incubated at room temperature for 5 min. Finally, DPPH absorbance was measured at 517 nm. DAB staining of leaf discs was carried as described previously (25).
Statistical Analysis
All the experiments were carried out and analyzed with three biological replicates. The values of chlorophyll, MDA, and flavonol contents were expressed as mean ± S.D. Differences between the uninoculated plants and transiently overexpressing lines were analyzed using Student's t test, and statistical significance was considered at p < 0.05.
Author Contributions
N. A. and R. A. V. conceived and designed the experiments. S. A. B. performed the bulk of the experiments as a part of his Ph.D. program. Experiments and bioinformatics analyses were supported by N. A. Data were analyzed by S. A. B. and N. A. R. A. V. and N. A. contributed reagents/materials/analysis tools. S. A. B. and N. A. wrote the paper.
Supplementary Material
Acknowledgments
We thank Dr. Gulzar Ahmad, Natural Product Chemistry Division, Indian Institute of Integrative Medicine, Sanat Nagar, Srinagar, India, for kindly providing some of the substrates. We thank Murtaza Gani, Instrumentation Division, Indian Institute of Integrative Medicine, Sanat Nagar, Srinagar, India, for technical help with LC/MS analysis. We also acknowledge help from Dr. Prashant Mishra, Plant Biotechnology Division, IIIM Jammu, India, for providing Nicotiana plants.
This work was supported by Grant SIMPLE (BSC-0109) from the Council of Scientific and Industrial Research and MLP 3012 from Indian Institute of Integrative Medicine. This paper has institutional manuscript number IIIM/1993/2017. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Tables S1 and S2 and Figs. S1–S7.
- ROS
- reactive oxygen species
- MDA
- malondialdehyde
- DPPH
- 2,2-diphenyl-1-picrylhydrazyl
- DAB
- 3,3′-diaminobenzidine
- IPTG
- or isopropyl β-d-thiogalactopyranoside
- MeJ
- methyl jasmonate
- ABA
- abscisic acid
- qRT
- quantitative RT
- TIM
- triose-phosphate isomerase.
References
- 1. Jones P., and Vogt T. (2001) Glycosyltransferases in secondary plant metabolism: tranquilizers and stimulant controllers. Planta 213, 164–174 [DOI] [PubMed] [Google Scholar]
- 2. Dharmawardhana D. P., Ellis B. E., and Carlson J. E. (1995) A β-glucosidase from lodgepole pine xylem specific for the lignin precursor coniferin. Plant Physiol. 107, 331–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Leah R., Kigel J., Svendsen I., and Mundy J. (1995) Biochemical and molecular characterization of a barley glucosidase. J. Biol. Chem. 270, 15789–15797 [DOI] [PubMed] [Google Scholar]
- 4. Kleczkowski K., and Schell J. (1995) Phytohormone conjugates: nature and function. Crit. Rev. Plant Sci. 14, 283–298 [Google Scholar]
- 5. Nisius A. (1988) The stroma center in avena plastids–an aggregation of β-glucosidase responsible for the activation of oat leaf saponins. Planta 173, 474–481 [DOI] [PubMed] [Google Scholar]
- 6. Poulton J. E. (1990) Cyanogenesis in plants. Plant Physiol. 94, 401–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jones P. R., Andersen M. D., Nielsen J. S., Høj P. B., and Møller B. L. (2000) in Evolution of Metabolic Pathways (Romeo J. T., Ibrahim R., Varin L., and De Luca V., eds) pp. 191–247, Elsevier Science Ltd., New York [Google Scholar]
- 8. Halkier B. A., and Gershenzon J. (2006) Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 57, 303–333 [DOI] [PubMed] [Google Scholar]
- 9. Suzuki H., Takahashi S., Watanabe R., Fukushima Y., Fujita N., Noguchi A., Yokoyama R., Nishitani K., Nishino T., and Nakayama T. (2006) An isoflavone conjugate-hydrolyzing β-glucosidase from the roots of soybean (Glycine max) seedlings–purification, gene cloning, phylogenetics, and cellular localization. J. Biol. Chem. 281, 30251–30259 [DOI] [PubMed] [Google Scholar]
- 10. Rouyi C., Baiya S., Lee S. K., Mahong B., Jeon J. S., Ketudat-Cairns J. R., and Ketudat-Cairns M. (2014) Recombinant expression and characterization of the cytoplasmic rice β-glucosidase Os1BGlu4. PLoS ONE 9, e96712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ashraf N., Jain D., and Vishwakarma R. A. (2015) Identification, cloning and characterization of an ultrapetala transcription factor CsULT1 from Crocus: a novel regulator of apocarotenoid biosynthesis. BMC Plant Biol. 15, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Moraga A. R., Mozos A. T., Ahrazem O., and Gómez-Gómez L. (2009) Cloning and characterization of a glucosyltransferase from Crocus sativus stigmas involved in flavonoid glucosylation. BMC Plant Biol. 9, 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Graham T. L., Kim J. E., and Graham M. Y. (1990) Role of constitutive isoflavone conjugates in the accumulation of glyceollin in soybean infected with Phytophthora megasperma. Mol. Plant Microbe Interact. 3, 157–166 [Google Scholar]
- 14. Kessmann H., Edwards R., Geno P. W., and Dixon R. A. (1990) Stress responses in alfalfa (Medicago sativa L.) V. Constitutive and elicitor-induced accumulation of isoflavonoid conjugates in cell suspension cultures. Plant Physiol. 94, 227–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mackenbrock U., and Barz W. (1991) Elicitor-induced formation of pterocarpan phytoalexins in chickpea (Cicer arietinum L.) cell suspension cultures from constitutive isoflavone conjugates upon inhibition of phenylalanine ammonia lyase. Z. Naturforsch. C. 46, 43–50 [Google Scholar]
- 16. Dakora F. D., and Phillips D. A. (1996) Diverse functions of isoflavonoids in legumes transcend antimicrobial definitions of phytoalexins. Physiol. Mol. Plant Pathol. 49, 1–20 [Google Scholar]
- 17. Opassiri R., Maneesan J., Akiyama T., Pomthong B., Jin S., Kimura A., and Cairns J. R. (2010) Rice Os4BGlu12 is a wound-induced β-glucosidase that hydrolyzes cell wall-β-glucan-derived oligosaccharides and glycosides. Plant Sci. 179, 273–280 [Google Scholar]
- 18. Baba S. A., Mohiuddin T., Basu S., Swarnkar M. K., Malik A. H., Wani Z. A., Abbas N., Singh A. K., and Ashraf N. (2015) Comprehensive transcriptome analysis of Crocus sativus for discovery and expression of genes involved in apocarotenoid biosynthesis. BMC Genomics 16, 698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Czjzek M., Cicek M., Zamboni V., Bevan D. R., Henrissat B., and Esen A. (2000) The mechanism of substrate (aglycone) specificity in β-glucosidases is revealed by crystal structures of mutant maize β-glucosidase-DIMBOA, -DIMBOAGlc, and -dhurrin complexes. Proc. Natl. Acad. Sci. U.S.A. 97, 13555–13560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Davies G. J., and Henrissat B. (2002) Plant glyco-related genomics. Structural enzymology of carbohydrate-active enzymes: implications for the post-genomic era. Biochem. Soc. Trans. 30, 291–297 [DOI] [PubMed] [Google Scholar]
- 21. Withers G. S., Warren R. A., Street I. P., Rupitz K., Kempton J. B., and Aebersold R. (1990) Unequivocal demonstration of the involvement of a glutamate residue as a nucleophile in the mechanism of a retaining glycosidase. J. Am. Chem. Soc. 112, 5887–5889 [Google Scholar]
- 22. Roepke J., and Bozzo G. G. (2015) Arabidopsis thaliana β-glucosidase BGLU15 attacks flavonol 3-O-β-glucoside-7-O-α-rhamnosides. Phytochemistry 109, 14–24 [DOI] [PubMed] [Google Scholar]
- 23. Nomura T., Quesada A. L., and Kutchan T. M. (2008) The new β-d-glucosidase in terpenoid-isoquinoline alkaloid biosynthesis in Psychotria ipecacuanha. J. Biol. Chem. 283, 34650–34659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Seyoum A., Asres K., and El-Fiky F. K. (2006) Structure-radical scavenging activity relationships of flavonoids. Phytochemistry 67, 2058–2070 [DOI] [PubMed] [Google Scholar]
- 25. Thordal-Christensen H., Zhang Z. G., Wei Y. D., and Collinge D. B. (1997) Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 11, 1187–1194 [Google Scholar]
- 26. Le Roy J., Huss B., Creach A., Hawkins S., and Neutelings G. (2016) Glycosylation is a major regulator of phenylpropanoid availability and biological activity in plants. Front. Plant Sci. 7, 735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ketudat Cairns J. R., and Esen A. (2010) β-Glucosidases. Cell. Mol. Life Sci. 67, 3389–3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ketudat Cairns J. R., Mahong B., Baiya S., and Jeonc J.-S. (2015) β-Glucosidases: multitasking, moonlighting or simply misunderstood? Plant Sci. 241, 246–259 [DOI] [PubMed] [Google Scholar]
- 29. Baba S. A., Malik A. H., Wani Z. A., Mohiuddin T., Abbas N., Shah Z., and Ashraf N. (2015) Phytochemical analysis and antioxidant activity of different tissue types of Crocus sativus and oxidative stress alleviating potential of saffron extract in plants, bacteria, and yeast. S. Afr. J. Bot. 99, 80–87 [Google Scholar]
- 30. Trapero A., Ahrazem O., Rubio-Moraga A., Jimeno M. L., Gómez M. D., and Gómez-Gómez L. (2012) Characterization of a glucosyltransferase enzyme involved in the formation of kaempferol and quercetin sophorosides in Crocus sativus. Plant Physiol. 159, 1335–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sansenya S., Opassiri R., Kuaprasert B., Chen C. J., and Cairns J. R. (2011) The crystal structure of rice (Oryza sativa L.) Os4BGlu12, an oligosaccharide and tuberonic acid glucoside-hydrolyzing β-glucosidase with significant thioglucohydrolase activity. Arch. Biochem. Biophys. 510, 62–72 [DOI] [PubMed] [Google Scholar]
- 32. Koudounas K., Banilas G., Michaelidis C., Demoliou C., Rigas S., and Hatzopoulos P. (2015) A defence-related Olea europaea β-glucosidase hydrolyses and activates oleuropein into a potent protein cross-linking agent. J. Exp. Bot. 66, 2093–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hemscheidt T., and Zenk M. H. (1980) Glucosidases involved in indole alkaloid biosynthesis of Catharanthus cell cultures. FEBS Lett. 110, 187–191 [DOI] [PubMed] [Google Scholar]
- 34. Schübel H., Stöckigt J., Feicht R., and Simon H. (1986) Partial purification and characterization of raucaffricine β-d-glucosidase from plant cell suspension cultures of Rauwolfia serpentina BENTH. Helv. Chim. Acta 69, 538–547 [Google Scholar]
- 35. Seshadri S., Akiyama T., Opassiri R., Kuaprasert B., and Cairns J. K. (2009) Structural and enzymatic characterization of Os3BGlu6, a rice β-glucosidase hydrolyzing hydrophobic glycosides and (1→3)-and (1→2)-linked disaccharides. Plant Physiol. 151, 47–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Keresztessy Z., Brown K., Dunn M. A., and Hughes M. A. (2001) Identification of essential active-site residues in the cyanogenic β-glucosidase (linamarase) from cassava (Manihot esculenta Crantz) by site-directed mutagenesis. Biochem. J. 353, 199–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Keresztessy Z., Kiss L., and Hughes M. A. (1994) Investigation of the active site of the cyanogenic β-d-glucosidase (linamarase) from Manihot esculenta Crantz (cassava). II. Identification of Glu-198 as an active site carboxylate group with acid catalytic function. Arch. Biochem. Biophys. 315, 323–330 [DOI] [PubMed] [Google Scholar]
- 38. Barleben L., Panjikar S., Ruppert M., Koepke J., and Stöckigt J. (2007) Molecular architecture of strictosidine glucosidase: the gateway to the biosynthesis of the monoterpenoid indole alkaloid family. Plant Cell 19, 2886–2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tringali C. (ed) (2003) Bioactive Compounds from Natural Sources: Isolation, Characterization and Biological Properties, CRC Press, Inc., Boca Raton, FL [Google Scholar]
- 40. Harborne J. B., and Williams C. A. (2000) Advances in flavonoid research since 1992. Phytochemistry 55, 481–504 [DOI] [PubMed] [Google Scholar]
- 41. Middleton E. Jr., and Kandaswami C. (1994) in The Impact of Plant Flavonoids on Mammalian Biology: Implications for Immunity, Inflammation and Cancer (Harborne J. B., ed) pp. 619–652, Chapman and Hall Ltd., London [Google Scholar]
- 42. Cushnie T. P., and Lamb A. J. (2005) Antimicrobial activity of flavonoids. Int. J. Antimicrob. Agents 26, 343–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rubio-Moraga A., Trapero A., Ahrazem O., and Gómez-Gómez L. (2010) Crocins transport in Crocus sativus: the long road from a senescent stigma to a newborn corm. Phytochemistry 71, 1506–1513 [DOI] [PubMed] [Google Scholar]
- 44. Yoo D., Hara T., Fujita N., Waki T., Noguchi A., Takahashi S., and Nakayama T. (2013) Transcription analyses of GmICHG, a gene coding for a β-glucosidase that catalyzes the specific hydrolysis of isoflavone conjugates in Glycine max (L.) Merr. Plant Sci. 208, 10–19 [DOI] [PubMed] [Google Scholar]
- 45. Wang P., Liu H., Hua H., Wang L., and Song C. P. (2011) A vacuole localized β-glucosidase contributes to drought tolerance in Arabidopsis. Chinese Sci. Bull. 56, 3538–3546 [Google Scholar]
- 46. Djilianov D., Ivanov S., Moyankova D., Miteva L., Kirova E., Alexieva V., Joudi M., Peshev D., and Van den Ende W. (2011) Sugar ratios, glutathione redox status and phenols in the resurrection species Haberlea rhodopensis and the closely related non-resurrection species Chirita eberhardtii. Plant Biol. 13, 767–776 [DOI] [PubMed] [Google Scholar]
- 47. Scarpeci T. E., and Valle E. M. (2008) Rearrangement of carbon metabolism in Arabidopsis thaliana subjected to oxidative stress condition: an emergency survival strategy. Plant Growth Regulations 54, 133–142 [Google Scholar]
- 48. Agati G., Brunetti C., Di Ferdinando M., Ferrini F., Pollastri S., and Tattini M. (2013) Functional roles of flavonoids in photoprotection: new evidence, lessons from the past. Plant Physiol. Biochem. 72, 35–45 [DOI] [PubMed] [Google Scholar]
- 49. Warren J. M., Bassman J. H., Fellman J. K., Mattinson D. S., and Eigenbrode S. (2003) Ultraviolet-B radiation alters phenolic salicylate and flavonoid composition of Populus trichocarpa leaves. Tree Physiol. 23, 527–535 [DOI] [PubMed] [Google Scholar]
- 50. Favory J. J., Stec A., Gruber H., Rizzini L., Oravecz A., Funk M., Albert A., Cloix C., Jenkins G. I., Oakeley E. J., Seidlitz H. K., Nagy F., and Ulm R. (2009) Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J. 28, 591–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Heijde M., and Ulm R. (2012) UV-B photoreceptor-mediated signalling in plants. Trends Plant Sci. 17, 230–237 [DOI] [PubMed] [Google Scholar]
- 52. Li J., Ou-Lee T. M., Raba R., Amundson R. G., and Last R. L. (1993) Arabidopsis flavonoid mutants are hypersensitive to UV-B irradiation. Plant Cell 5, 171–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nakabayashi R., Mori T., and Saito K. (2014) Alternation of flavonoid accumulation under drought stress in Arabidopsis thaliana. Plant Signal. Behav. 9, e29518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nakabayashi R., Yonekura-Sakakibara K., Urano K., Suzuki M., Yamada Y., Nishizawa T., Matsuda F., Kojima M., Sakakibara H., Shinozaki K., Michael A. J., Tohge T., Yamazaki M., and Saito K. (2014) Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 77, 367–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Baba S. A., Jain D., Abbas N., and Ashraf N. (2015) Overexpression of Crocus carotenoid cleavage dioxygenase, CsCCD4b, in Arabidopsis imparts tolerance to dehydration, salt and oxidative stresses by modulating ROS machinery. J. Plant Physiol. 189, 114–125 [DOI] [PubMed] [Google Scholar]
- 56. Robert X., and Gouet P. (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tamura K., Stecher G., Peterson D., Filipski A., and Kumar S. (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Morris G. M., Goodsell D. S., Halliday R. S., Huey R., Hart W. E., Belew R. K., and Olson A. J. (1998) Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 19, 1639–1662 [Google Scholar]
- 59. Solis F. J., and Wets R. J. (1981) Minimization by random search techniques. Mathamat. Operat. Res. 6, 19–30 [Google Scholar]
- 60. Roepke J., and Bozzo G. G. (2013) Biocatalytic synthesis of quercetin 3-O-glucoside-7-O-rhamnoside by metabolic engineering of Escherichia coli. Chembiochem. 14, 2418–2422 [DOI] [PubMed] [Google Scholar]
- 61. Sainsbury F., Thuenemann E. C., and Lomonossoff G. P. (2009) pEAQ: Versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol. J. 7, 682–693 [DOI] [PubMed] [Google Scholar]
- 62. Jami S. K., Clark G. B., Turlapati S. A., Handley C., Roux S. J., and Kirti P. B. (2008) Ectopic expression of an annexin from Brassica juncea confers tolerance to abiotic and biotic stress treatments in transgenic tobacco. Plant Physiol. Biochem. 46, 1019–1030 [DOI] [PubMed] [Google Scholar]
- 63. Scott T. A., and Melvin E. H. (1953) Determination of dextran with anthrone. Anal. Chem. 25, 1656–1661 [Google Scholar]
- 64. Samappito S., Page J., Schmidt J., De-Eknamkul W., and Kutchan T. M. (2002) Molecular characterization of root-specific chalcone synthases from Cassia alata. Planta 216, 64–71 [DOI] [PubMed] [Google Scholar]
- 65. Nakajima J., Tanaka I., Seo S., Yamazaki M., and Saito K. (2004) LC/PDA/ ESI-MS profiling and radical scavenging activity of anthocyanins in various berries. J. Biomed. Biotechnol. 2004, 241–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tohge T., Matsui K., Ohme-Takagi M., Yamazaki M., and Saito K. (2005) Enhanced radical scavenging activity of genetically modified Arabidopsis seeds. Biotechnol. Lett. 27, 297–303 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.