Abstract
The β1-adrenergic receptor (β1AR) is a G protein-coupled receptor (GPCR) and the predominant adrenergic receptor subtype in the heart, where it mediates cardiac contractility and the force of contraction. Although it is the most important target for β-adrenergic antagonists, such as β-blockers, relatively little is yet known about its regulation. We have shown previously that β1AR undergoes constitutive and regulated N-terminal cleavage participating in receptor down-regulation and, moreover, that the receptor is modified by O-glycosylation. Here we demonstrate that the polypeptide GalNAc-transferase 2 (GalNAc-T2) specifically O-glycosylates β1AR at five residues in the extracellular N terminus, including the Ser-49 residue at the location of the common S49G single-nucleotide polymorphism. Using in vitro O-glycosylation and proteolytic cleavage assays, a cell line deficient in O-glycosylation, GalNAc-T-edited cell line model systems, and a GalNAc-T2 knock-out rat model, we show that GalNAc-T2 co-regulates the metalloproteinase-mediated limited proteolysis of β1AR. Furthermore, we demonstrate that impaired O-glycosylation and enhanced proteolysis lead to attenuated receptor signaling, because the maximal response elicited by the βAR agonist isoproterenol and its potency in a cAMP accumulation assay were decreased in HEK293 cells lacking GalNAc-T2. Our findings reveal, for the first time, a GPCR as a target for co-regulatory functions of site-specific O-glycosylation mediated by a unique GalNAc-T isoform. The results provide a new level of β1AR regulation that may open up possibilities for new therapeutic strategies for cardiovascular diseases.
Keywords: ADAM, adrenergic receptor, G protein-coupled receptor (GPCR), glycoprotein, glycosyltransferase, matrix metalloproteinase (MMP), protein processing, receptor structure-function
Introduction
β-Adrenergic receptors (βARs)4 are G protein-coupled receptors (GPCRs) that activate intracellular signaling pathways mainly via the stimulatory Gs protein after binding of agonists such as adrenaline and noradrenaline (1, 2). The βARs exist as three subtypes, β1, β2, and β3, and the former two are important in the regulation of the excitation-contraction coupling of the myocardium. The β1AR is the predominant βAR subtype expressed in the heart and the main mediator of the endogenous catecholamine-stimulated positive chronotropy and inotropy (1, 2). Thus, it is the most important target receptor for β-adrenergic antagonists, also called β-blockers, which are widely used in the treatment of cardiac diseases, such as chronic heart failure, coronary artery disease, arrhythmias, and hypertension. However, these therapeutic agents have limited effectiveness in some patients and also exert adverse effects. Consequently, there is a growing need to better understand the underlying mechanisms in cardiac function and disease to develop alternative and more individualized treatment options that can improve clinical outcomes.
During chronic heart failure, a persistent compensatory increase of catecholamines causes β1AR desensitization and down-regulation. The density of β1ARs at the plasma membrane is reduced by 50%, whereas that of β2ARs does not seem to change (3). The resulting partial loss of β1AR function is believed to be an adaptive mechanism to counteract the cardiotoxicity of chronic adrenergic signaling (4). We have shown previously that β1AR undergoes limited N-terminal cleavage by metalloproteinases in heterologous expression systems as well as in vivo in rat neonatal cardiomyocytes (5, 6), indicating that limited proteolysis has a regulatory function in receptor down-regulation. We identified two cleavage sites for the human receptor at positions 31R↓L32 and 52P↓L53 in the extracellular N terminus and demonstrated that the cleavage at the major site 31R↓L32 is augmented by agonist-mediated activation of the receptor (5, 6). We also found that β1AR undergoes GalNAc-type O-glycosylation (hereafter simply referred to as O-glycosylation) (5), and the NetOGlyc4.0 prediction algorithm predicts the presence of five O-glycan sites in the receptor N-terminal extracellular sequence (7).
O-Glycosylation is found on a majority of proteins passing through the secretory pathway (7). It is a unique type of protein glycosylation because it is differentially regulated by a large family of up to 20 distinct polypeptide GalNAc-transferase isoenzymes (GalNAc-Ts) that catalyze the addition of the first GalNAc residue to Ser and Thr residues (8). These GalNAc-T isoforms have distinct, albeit partly overlapping, acceptor substrate specificities, and they are differentially expressed in cells and tissues, providing a unique scenario for differential and dynamic regulation of site-specific O-glycosylation (8). Whereas tightly spaced O-glycans found in unstructured regions of proteins that serve as linkers between folded domains or as stem regions of membrane proteins may serve to protect these regions from general proteolysis (9–12), it is becoming clear that individual O-glycosites may serve more specific and co-regulatory roles of limited proteolytic processing events with important biological functions. The first example with clear medical relevance was discovered studying the rare disease familial tumoral calcinosis caused by a deficiency in GALNT3, encoding one of the many GalNAc-Ts (13). The loss of GalNAc-T3 was found to eliminate a single O-glycosylation site in fibroblast growth factor 23 adjacent to a proprotein convertase (PC) processing site that inactivates this protein, a major regulator of phosphate homeostasis leading to hyperphosphatemia (14). More recently, it has become apparent that site-specific O-glycosylation close to processing sites (±3 residues) plays a more general role in the co-regulation of PC processing, and thus the substrate specificities of individual members of the large GalNAc-T family provide a huge potential for differential co-regulation (15). In line with this, we have also found that site-specific O-glycosylation appears to fine-tune ectodomain shedding regulated by a disintegrin and metalloproteinases (ADAMs) (16, 17), increasing the potential for even wider roles of O-glycosylation in co-regulating protein functions.
A limited number of GPCRs are known to be O-glycosylated, including the V2 vasopressin (18), δ opioid (19–21), κ opioid (22), bradykinin B2 receptors (23), and the C-C chemokine receptor type 5 (24). The functional significance of the modification is largely unexplored, but in one example, O-glycosylation in the receptor N terminus has been shown to affect ligand binding (24). Recent progress in the proteome-wide identification and prediction of O-glycosylation sites (7) points to a characteristic presence of O-glycans in the N-terminal region of GPCRs with more than 300 members having identified or predicted O-glycosylation (7, 25). Here, we focused on β1AR, a rhodopsin-type GPCR, that has predicted O-glycosites adjacent to previously described cleavage sites in the extracellular N terminus (5). We hypothesized that site-specific O-glycosylation by one (or more) GalNAc-T isoforms co-regulates the receptor N-terminal cleavage. First, we demonstrate that the GalNAc-T2 isoform selectively glycosylates the β1AR N-terminal ectodomain and that glycosylation affects the metalloproteinase-mediated cleavage in vitro. Then, using cells deficient in O-glycosylation, a panel of genetically engineered cell lines with different repertoires of GalNAc-Ts, and a Galnt2−/− rat model, we confirm that the GalNAc-T2-mediated O-glycosylation affects β1AR processing in vivo. Finally, using a cell line without GalNAc-T2, we show that O-glycosylation modulates receptor signaling. Our study provides evidence that GalNAc-T2 co-regulates the processing of β1AR and modulates its downstream signaling. The results warrant further studies assessing the consequent biomedical implications of these findings for heart failure and other diseases related to β1AR signaling.
Results
GalNAc-T Isoform-specific O-Glycosylation of the β1AR N Terminus
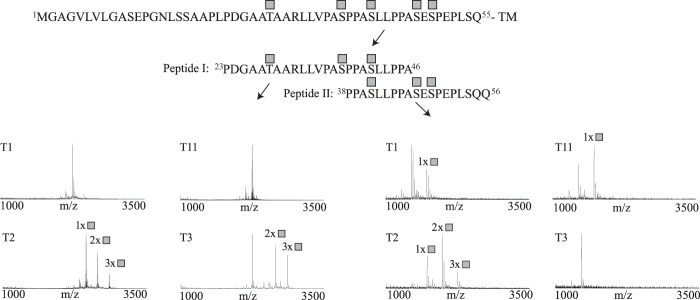
Using enzymatic deglycosylation assays, we have previously demonstrated that the human β1AR (hβ1AR) is O-glycosylated (5). To map the specific sites and to determine potential GalNAc-T isoform-specific activity, we designed two short peptide sequences (peptide I, 23PDGAATAARLLVPASPPASLLPPA46; peptide II, 38PPASLLPPASESPEPLSQQ56) covering the central part of the receptor N terminus flanking two previously identified cleavage sites at 31R↓L32 and 52P↓L53 (Fig. 1; see also Fig. 3A). The peptides were used for in vitro O-glycosylation assays with a panel of 10 different recombinantly expressed GalNAc-Ts (T1, T2, T3, T4, T5, T11, T12, T13, T14, and T16). This in vitro analysis has been shown previously to be quite predictive of O-glycosylation in vivo (7, 17, 26). Glycosylated peptides were characterized by electrospray ionization-linear ion trap-Fourier transform-MS for site identification. We found that GalNAc-T2 glycosylated both peptides at three positions corresponding to a total of five residues, Thr-28, Ser-37, Ser-41, Ser-47, and Ser-49, in the β1AR N terminus (Fig. 1). GalNAc-T3 glycosylated the peptides at three positions (Thr-28, Ser-37, and Ser-41) (Fig. 1) and GalNAc-T1 and GalNAc-T11 at one position each (not determined). However, these latter reactions were slow and never reached completion in an overnight experiment. Thus, the results suggest that GalNAc-T2 is the major isoform controlling O-glycosylation of β1AR and initiates O-glycosylation at five Ser/Thr residues in the N-terminal ectodomain of the receptor.
FIGURE 1.
In vitro analysis of GalNAc-T isoform specificity and a summary of detected glycosylation sites of hβ1AR. Shown is in vitro O-glycosylation analysis with GalNAc-T1, T2, T3, T4, T5, T11, T12, T13, T14, and T16 of two overlapping peptides (peptides I and II) covering the β1AR N terminus. Reactions were followed by MALDI-TOF-MS, and spectra are shown for a 24-h incubation time point for T1, T2, T3, and T11. GalNAc-T2 was found to be the most efficient at glycosylating the two peptides, with a total of five sites corresponding to Thr-28, Ser-37, Ser-41, Ser-47, and Ser-49 in the full-length protein. GalNAc-T3 modified three sites at Thr-28, Ser-37, and Ser-41; GalNAc-T1 and GalNAc-T11 modified one position each (not determined). The spectra show relative intensity. The complete hβ1AR N-terminal sequence and peptide sequences are shown above the spectra. The locations of the attached GalNAcs are shown as gray squares. The results are representative of three independent experiments. TM, transmembrane domain.
FIGURE 3.
N-terminal cleavage of hβ1AR is enhanced in CHO-ldlD cells deficient in GalNAc-type O-glycosylation. A, receptor model highlighting the extracellular N terminus with five O-glycosylation sites (gray squares) and the two cleavage sites and one N-glycosylation site identified previously (5). The two peptides (PI and PII) used in the in vitro assays and the N- and C-terminal epitope tags are also indicated. B–E, G, and H, the WT and the R31H/L32A cleavage site mutant Myc-hβ1AR-FLAG and the Myc-hδOR-FLAG were transiently expressed in CHO-K1, CHO-ldlD, or HEK293 cells for 24 h. For ldlD cells, the low serum culture medium (2% FBS (E and G) and 0.5% FBS (H)) was supplemented or not with Gal (20 μm) and/or GalNAc (400 μm). The CHO-K1 cells were cultured at 5% FBS. Expressed receptors were immunoprecipitated from solubilized membranes (B–D) or cellular lysates (E, G, and H) and analyzed by Western blotting. For C and D, the purified receptors were first digested or not with Endo H or PNGase F. The ratios of full-length (black bars) and cleaved (white bars) β1AR species in ldlD cells shown in E are depicted in F. The repeated measures two-way analysis of variance followed by Tukey's post hoc test was used to compare the ratios in cells supplemented with both Gal and GalNAc with those cultured in the presence of no sugars or only with GalNAc (a). The ratios observed in the two latter culture conditions were also compared (b). ***, p < 0.001, n = 5. Open circles, full-length hβ1ARs; open triangles, C-terminal fragments cleaved at 31R↓L32. The hδOR mature and precursor forms are indicated with a closed square and circle, respectively. Ab, antibody; IP, immunoprecipitation; WB, Western blotting. Error bars, S.E. The results are representative of two (B), five (E), four (G), and three (H) independent experiments. Deglycosylation (C and D) was performed once.
Site-specific O-Glycosylation by GalNAc-T2 Modulates the in Vitro Proteolytic Cleavage of β1AR N-terminal Peptides
To systematically evaluate the effect of GalNAc O-glycosylation on the metalloproteinase-mediated β1AR cleavage, we performed in vitro cleavage assays of peptides and GalNAc glycopeptides using four ADAMs (ADAM8, -10, -12, and -17) and nine matrix metalloproteinases (MMP1, -2, -3, -7, -8, -9, -12, -13, and -14). Of the four ADAMS analyzed, we found that ADAM17 cleaved peptide II at 52P↓L53 and that this was inhibited by GalNAc-T2 glycosylation (Fig. 2A). A comparable effect of glycosylation was observed at 31R↓L32 of peptide I (Fig. 2A). Furthermore, we found that ADAM10 cleaved peptide I at 30A↓R31; however, glycosylation did not appear to affect the processing. ADAM8 and -12 did not cleave either of the two peptides (data not shown). Of the panel of nine MMPs tested, several members of this metalloproteinase family cleaved the peptides at 31R↓L32 and 52P↓L53 as observed with ADAM17. MMP2 and -3 cleaved peptide I at 31R↓L32, (Fig. 2C) (data not shown), and MMP3, -7, -8, -12, -13, and -14 cleaved peptide II at 52P↓L53 (Fig. 2C) (data not shown). Unexpectedly, we found that MMP2, -7, -8, -9, -12, and -13 cleaved both peptides at a previously uncharacterized cleavage site at 41S↓L42 (Fig. 2C) (data not shown), and this cleavage was also inhibited by O-glycosylation, as exemplified by the MMP9 cleavage (Fig. 2B). Here we analyzed the effect of a simple GalNAc O-glycan; however, it is likely that extended O-glycans with sialic acids have more pronounced effects on the adjacent proteolysis (14). Taken together, these results indicate that several metalloproteinases in the ADAM and MMP families are able to cleave the β1AR N-terminal peptides at multiple sites.
FIGURE 2.
Site-specific O-glycosylation modulates the in vitro cleavage of hβ1AR N-terminal peptides. A, in vitro cleavage analysis of β1AR peptide I and II and respective glycopeptides by ADAM17 monitored by MALDI-TOF. Products formed after 30-min, 1-h, and 4-h incubations are shown. ADAM17 was able to cleave both the naked peptide and the O-glycopeptide at the 31R↓L32 and 52P↓L53 cleavage sites. However, the cleavage of the glycopeptides was less efficient. B, in vitro cleavage analysis of β1AR peptide I and O-glycopeptide by MMP9. Cleavage was observed at 41S↓L42, and again, the glycopeptide was less efficiently cleaved. C, schematic overview of the total observed cleavage sites by ADAM10, ADAM17, and MMPs, and their relation to the observed glycosylation sites. The boldface font indicates that a protection from cleavage was observed. The spectra show relative intensity. The locations of the attached GalNAcs are shown as gray squares. The results shown are representative of three (A) and two (B) independent experiments.
O-Glycans Affect β1AR N-terminal Cleavage in Intact Cells
To further investigate the role of β1AR O-glycosylation, we used the CHO-ldlD cell model system, in which a deficient UDP-Gal/UDP-GalNAc C4-epimerase allows to control O-glycosylation by the addition of exogenous Gal and GalNAc sugars to the cell culture medium (11, 27). Without sugars added to the medium, CHO-ldlD cells do not perform O-glycosylation and galactosylation of N-glycans, whereas with the addition of GalNAc, O-glycosylation is limited to simple GalNAc O-glycans, and by adding both GalNAc and Gal, O- and N-glycosylation are almost fully restored to the level of the parental cell line CHO-K1.
We first expressed β1AR in CHO-K1 cells to confirm that the expressed receptor forms are comparable with those shown previously for HEK293 cells (5, 6). Western blotting using the FLAG M2 antibody that recognizes the C-terminal epitope tag revealed two receptor forms (69 and 54 kDa) in both CHO-K1 and HEK293 cells (Fig. 3B), whereas only the larger 69-kDa form was detected with the c-Myc antibody directed against the N-terminal epitope tag (Fig. 3B). As expected, the higher molecular weight species was digested with peptide-N-glycosidase F (PNGase F) but not with endo-N-acetylglucosaminidase H (Endo H) (Fig. 3C), indicating that it represents the full-length receptor carrying fully processed N-glycans. In contrast, the smaller 54-kDa species was resistant to both enzymes, in line with the expectation that it corresponds to the C-terminal fragment cleaved at the main cleavage site at 31R↓L32 and not to the similar sized receptor precursor (5, 6). In the same conditions, Endo H readily digested the human δ-opioid receptor (hδOR) precursor (Fig. 3D) (19, 28). A 47-kDa β1AR C-terminal fragment cleaved at 52P↓L53 (5) was seen occasionally and only in HEK293 cells (see Fig. 4A).
FIGURE 4.
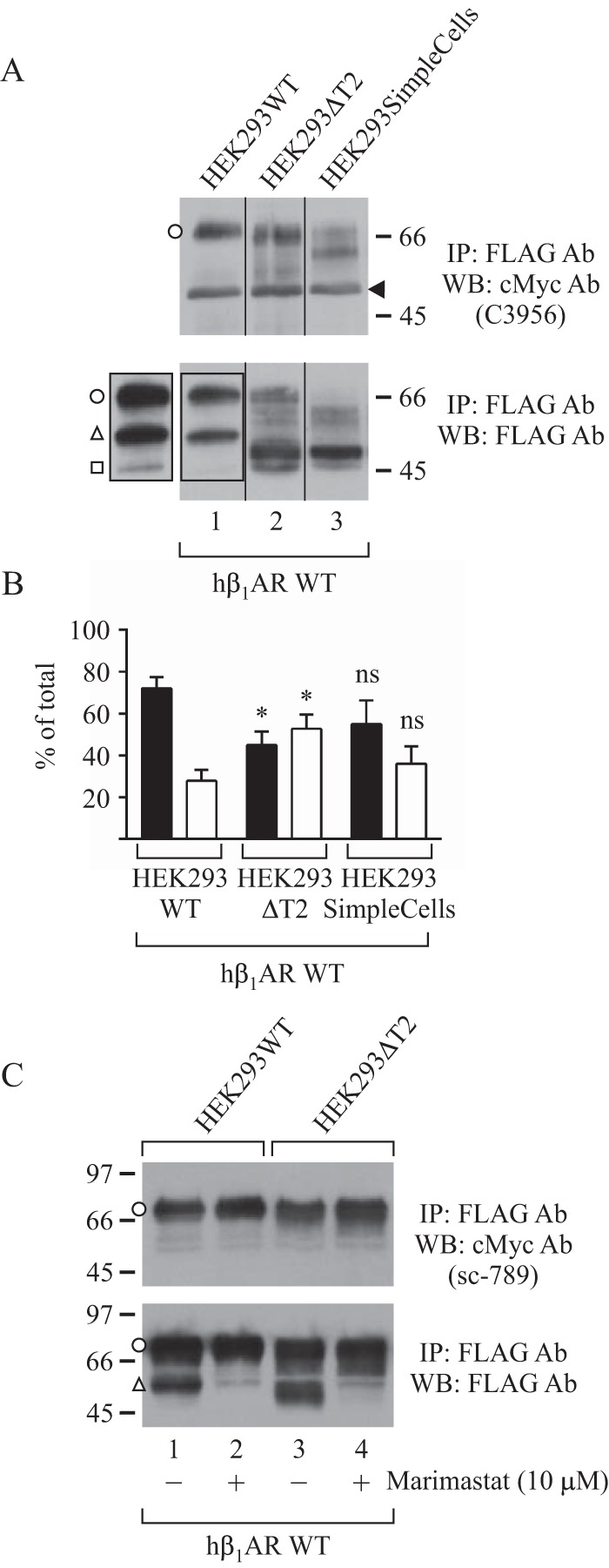
N-terminal cleavage and O-glycosylation of hβ1AR in glycoengineered HEK293 cell lines. The WT Myc- and FLAG-tagged hβ1AR was transiently expressed in the indicated cell lines for 24 h. Receptors were immunoprecipitated from solubilized membranes and analyzed by SDS-PAGE and Western blotting. In A, a longer exposure is shown for the outlined area of lane 1. The ratios of full-length (black bars) and cleaved receptor forms (white bars) detected with the FLAG M2 antibody are shown in B. The two-way analysis of variance followed by Tukey's post hoc test was used to compare the ratios in HEK293ΔT2 cells and SimpleCells with the corresponding values in the HEK293WT cells. *, p < 0.05; ns, not significant; n = 4. For C, the metalloproteinase inhibitor marimastat was added or not to the culture medium 4 h after starting the transfection and was maintained thereafter. The results shown are representative of four (A) and two (C) independent experiments. The C-terminal receptor fragment cleaved at 52P↓L53 is indicated with an open square, and a nonspecific protein detected with the Myc antibody (A, top) is shown with an arrowhead. Open circles, full-length hβ1ARs; open triangles, C-terminal fragments cleaved at 31R↓L32; open squares, C-terminal fragments cleaved at 52P↓L53. IP, immunoprecipitation; WB, Western blotting. Error bars, S.E.
When expressed in the CHO-ldlD cells without Gal/GalNAc addition, β1AR showed a significant increase in cleavage compared with CHO-K1 cells. The full-length receptor in the ldlD cells migrated at about 57 kDa, and its relative amount was decreased compared with the full-length 69-kDa receptor form seen in CHO-K1 cells (Fig. 3E, lanes 6 and 10, respectively). Concomitantly, there was a significant increase in the relative amount of the corresponding cleaved receptor forms (Fig. 3, E (lanes 1 and 5) and F). Upon the addition of Gal, no clear changes in the relative amount of cleaved receptor forms were detected compared with the corresponding control cells (Fig. 3E, lanes 1 and 2). However, as expected, the migration of the full-length receptor species was slowed down consistently with the expected restoration of receptor N-glycosylation (Fig. 3E, lanes 2 and 7). Importantly, the addition of GalNAc resulted in a significant decrease of receptor cleavage (Fig. 3, E (lane 3) and F) and in the appearance of a heterogeneous smear of receptor forms (Fig. 3E, lanes 3 and 8), probably reflecting differentially O-glycosylated receptors. Finally, when both Gal and GalNAc were added, the cleavage was restored to the same level seen in CHO-K1 cells, (Fig. 3, E (lanes 4 and 5) and F). Taken together, these results are fully in line with the in vitro data and show that β1AR O-glycosylation has a protective effect on the proteolysis of the receptor N terminus. Furthermore, the results indicate that the addition of GalNAc alone provides detectable, albeit partial, protection from proteolytic cleavage.
We have shown previously that a mutation of 31RL32 to 31HA32, flanking the major β1AR cleavage site, inhibits cleavage at this site almost completely when the mutant receptor is expressed in HEK293 cells (5). Thus, we tested the effect of the lack of O-glycosylation on the cleavage of the mutant receptor. As shown in Fig. 3G, the R31H/L32A mutant was cleaved almost to the same extent as the WT receptor when expressed in CHO-ldlD cells without Gal and GalNAc (compare lanes 1 in Fig. 3, G and E). In comparison, GalNAc addition into the culture medium blocked the cleavage almost completely, to the same extent as the full restoration of O-glycosylation by adding both Gal and GalNAc (Fig. 3G, lanes 2 and 3, respectively). These results provide further evidence that the accessibility of metalloproteinases to the receptor cleavage site(s) is significantly modulated by the adjacent O-glycans. Interestingly, for the R31H/L32A mutant (but not for the WT), a smear of smaller molecular mass receptor forms below the 69-kDa full-length receptor was detected when the cells were cultured at lower serum conditions (0.5% FBS instead of 2% FBS used routinely) (Fig. 3H). This is probably because of up-regulated metalloproteinases in the more rigorous culture conditions. This finding could be related to our observation that the tested peptides could be cleaved at alternative sites in the in vitro cleavage assays (Fig. 2). Thus, taking this evidence together, we conclude that the intact β1AR N terminus is a substrate for several metalloproteinases and/or is cleaved at several adjacent sites.
The β1AR N-terminal Cleavage Is Co-regulated by GalNAc-T2 in Human Cell Lines HEK293 and HepG2
The in vitro results pointed to GalNAc-T2 as the major enzyme modifying β1AR but also showed that GalNAc-T3 and to a lesser extent GalNAc-T1 and -T11 were potentially able to glycosylate the β1AR N-terminal peptides. Importantly, although widely expressed, GalNAc-T3 is not found in the heart (8). We therefore used two different isogenic cell line model systems genetically engineered to lack specific GalNAc-Ts to probe isoform specificity in cells with different GalNAc-T expression profiles. HEK293 cells are known to express GalNAc-T1, T2, T3, and T11 (7) and have been used extensively to study the N-terminal cleavage and function of β1AR (5, 6, 29). HepG2 cells express GalNAc-T2, T1, and T11, but not GalNAc-T3 (30), making the GalNAc-T expression profile of HepG2 cells comparable with that described for the heart (8). Thus, we used HEK293 and HepG2 cells with a knock-out of GALN-T2 (ΔT2), HepG2 cells with a knock-out of GALN-T1 (ΔT1) or T11 (ΔT11), and a HepG2 rescue cell line (ΔT2+T2) with site-directed re-insertion of the GalNAc-T2 coding cDNA into the adeno-associated virus integration site 1 (AAVS1) (30). Furthermore, to confirm the protective effect of truncated O-glycans observed in the CHO-ldlD cells, we used a HEK293 COSMC knock-out cell model, designated HEK293SimpleCells (31). Cosmc is a chaperone for the C1GalT1 core 1 enzyme. Thus, O-glycans are not elongated in SimpleCells, which consequently produce truncated GalNAc O-glycans similar to those produced in CHO-ldlD cells cultured in the presence of GalNAc alone.
When expressed in HEK293ΔT2 cells, β1AR migrated slightly faster on SDS-PAGE compared with the HEK293WT cells and presented a heterogeneous smear below the major receptor form (Fig. 4A, lanes 2 and 1, respectively, first panel). This suggests a loss of one or more O-glycans in the former cell line. Importantly, there was a significant increase in the relative amount of the two N-terminally cleaved β1AR forms detected with the FLAG M2 antibody (Fig. 4, A (lane 2, second panel) and B). The smaller of these cleaved forms represents a receptor fragment cleaved at 52P↓L53, because it co-migrated with the 47-kDa form observed in the HEK293WT cells (Fig. 4A, lanes 1 and 2, second panel). The larger form probably corresponds to a C-terminal fragment with fewer O-glycans, cleaved at the main cleavage site at 31R↓L32. The smallest receptor form was not seen in all experiments.
We next expressed β1AR in HEK293SimpleCells. The observed clear decrease in the apparent molecular mass of the full-length receptor (Fig. 4A, lane 3, first panel) is in line with the expectation that O-glycans are not elongated in these cells (7). In some experiments, the receptor was more extensively cleaved in these cells compared with the HEK293WT cells (Fig. 4A, lanes 3 and 1, respectively, second panel). However, this difference was not consistent and did not reach statistical significance (Fig. 4B). Thus, these results suggest that in HEK293 cells, truncated O-glycans are able to protect the receptor from cleavage, at least partially, in a similar manner as in CHO-ldlD cells.
To confirm that the enhanced receptor processing observed in the HEK293ΔT2 cells was mediated by metalloproteinases, we treated the cells with a broad range metalloproteinase inhibitor, marimastat, during transfection. This decreased the amount of the smaller N-terminally cleaved receptor forms with a concomitant increase in the amount of full-length receptors (Fig. 4C, lanes 3 and 4). Similar results were obtained for the HEK293WT cells (Fig. 4C, lanes 1 and 2). This verifies that the smallest receptor forms detected in these cells are the result of metalloproteinase-mediated cleavage.
To investigate the role of GalNAc-T2-mediated O-glycosylation in a cell line without GalNAc-T3, we next expressed β1AR in a panel of isogenic HepG2 cells with knock-out or knock-in of specific GalNAc-Ts. We observed a significant increase in receptor cleavage in HepG2ΔT2 cells that was reversed in the rescue HepG2ΔT2+T2 cell line (Fig. 5, A (lanes 1–3) and B). The cleavage of β1AR was consistently more pronounced in the WT HepG2 cells compared with the corresponding HEK293 cells (compare Fig. 4A with Fig. 5A and Fig. 4B with Fig. 5B), which is probably due to partial compensatory glycosylation in HEK293 cells or other cell-dependent differences in O-glycosylation capacities and/or protease activities in the two cell lines.
FIGURE 5.

N-terminal cleavage and O-glycosylation of hβ1AR in glycoengineered HepG2 cell lines. The WT Myc- and FLAG-tagged hβ1AR and the N-terminally truncated (Δ2–31 and Δ2–52) mutants with a C-terminal FLAG tag were transiently expressed in the indicated cell lines for 48 h. Receptors were immunoprecipitated from cellular lysates and analyzed by SDS-PAGE and Western blotting. In A, a shorter exposure is shown for the outlined area of lane 2. The ratios of the WT full-length (black bars) and cleaved receptor forms (white bars) shown in A are depicted in B. Two-way analysis of variance followed by Tukey's post hoc test was used to compare the ratios in HepG2ΔT2 and HepG2ΔT2+T2 cells with the corresponding values in the HepG2WT cells (a). The values obtained for the ΔT2 and ΔT2+T2 cells were also compared (b). ***, p < 0.001; ns, not significant; n = 3–5. For D, immunoprecipitated receptors were subjected or not to deglycosylation with neuraminidase and O-glycosidase before SDS-PAGE. The results shown are representative of five (A, lanes 1–3), three (A, lanes 4–6), four (C), and one (D) independent experiment. Open circles, full-length hβ1ARs; open triangles, C-terminal fragments cleaved at 31R↓L32. IP, immunoprecipitation; WB, Western blotting. Error bars, S.E.
The cleaved receptor fragments seen in the HepG2ΔT2 cells migrated as a doublet (Fig. 5A, lane 2), most likely representing differentially O-glycosylated and non-modified receptor fragments cleaved at the major cleavage site at 31R↓L32. This was supported by the observation that these forms co-migrated with receptor species truncated at 31R↓L32 (Fig. 5A, compare lanes 2 and 5) and was further verified by enzymatic de-O-glycosylation (Fig. 5D). No clear changes in the pattern or intensity of receptor forms were observed in HepG2ΔT1 or HepG2ΔT11 cell lines (Fig. 5C) (data not shown), although some variability between experiments was found, especially in the case of HepG2ΔT1 cells. Taken together, these results indicate that in the two tested cell lines, HEK293 and HepG2, which express a diverse set of GalNAc-Ts, the major isoform modifying β1AR is GalNAc-T2. Other transferases, most likely GalNAc-T3, can, however, partially compensate for GalNAc-T2, possibly involving specific Ser residues in the receptor N terminus.
Impaired O-Glycosylation Attenuates β1AR Signaling
To investigate whether impaired O-glycosylation and the consequent enhanced N-terminal cleavage of β1AR might alter its functional activity, we analyzed the ability of isoproterenol, a βAR agonist, to induce cAMP production in transfected HEK293ΔT2/T3 and corresponding WT cells in dose-response experiments. As shown in Fig. 6, the EC50 for isoproterenol was higher in ΔT2/T3 KO cells compared with WT cells (26.1 ± 2.7 and 16.8 ± 3.9 nm, respectively; p = 0.04), and the maximum stimulation was significantly decreased by 40.4 ± 5.7% (p = 0.009). In comparison, no change in cAMP accumulation was detected in the two cell lines when the endogenously expressed gastric inhibitor peptide receptor (32) was stimulated by an increasing concentration of gastric inhibitor peptide (supplemental Fig. 1). Thus, these data indicate that β1AR O-glycans have a modulatory role in receptor function, having an impact on receptor number as well as on receptor signaling. This occurs most likely via O-glycan-mediated co-regulation of receptor proteolytic processing.
FIGURE 6.
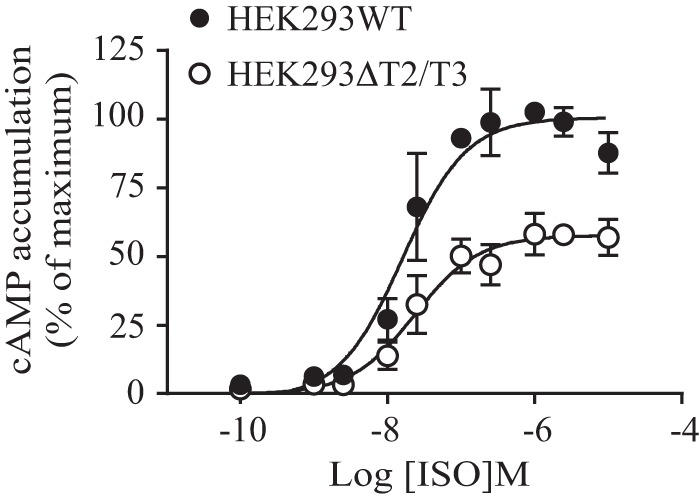
Isoproterenol-induced cAMP accumulation is attenuated in HEK293ΔT2/T3 cells. The WT Myc- and FLAG-tagged hβ1AR was transiently transfected into HEK293ΔT2/T3 or HEK293WT cells for 48 h. Cells were stimulated with increasing concentrations of isoproterenol for 60 min, and cAMP was measured using the Cisbio Bioassays cAMP assay. The data represent the mean ± S.E. (error bars) of three independent experiments performed in duplicate. The values were normalized to the maximum signal obtained for the HEK293WT cells, which was set to 100%.
β1AR Proteolytic Processing Is Co-regulated by GalNAc-T2 in Vivo in Rat Hearts
To further investigate the potential co-regulatory function of GalNAc-T2 in the β1AR N-terminal processing and to confirm the in vitro and cell line findings, we compared the cleavage pattern of β1AR in WT and Galnt2−/− rat heart ventricles. Because the N-terminal sequence of the rat β1AR differs slightly from the human sequence, we first confirmed the GalNAc-T2 isoform-specific glycosylation of peptides corresponding to the rat sequence by in vitro analysis (Fig. 7C). We then analyzed the endogenous β1AR in the rat heart by SDS-PAGE and Western blotting with an antibody directed against the receptor C-terminal domain. This antibody was shown previously to detect both full-length and N-terminally cleaved forms of the rat receptor (6). As shown in Fig. 7, the Galnt2−/− rat hearts exhibited a clear increase in the relative amount of cleaved β1ARs. This was observed when receptors solubilized from isolated membranes were analyzed directly or after immunoprecipitation (Fig. 7, A and B, respectively). The lack of GalNAc-T3 expression in the heart (8), probably explains the extensive receptor processing in the heart muscle comparable with the observations in the HepG2 cell line model system.
FIGURE 7.
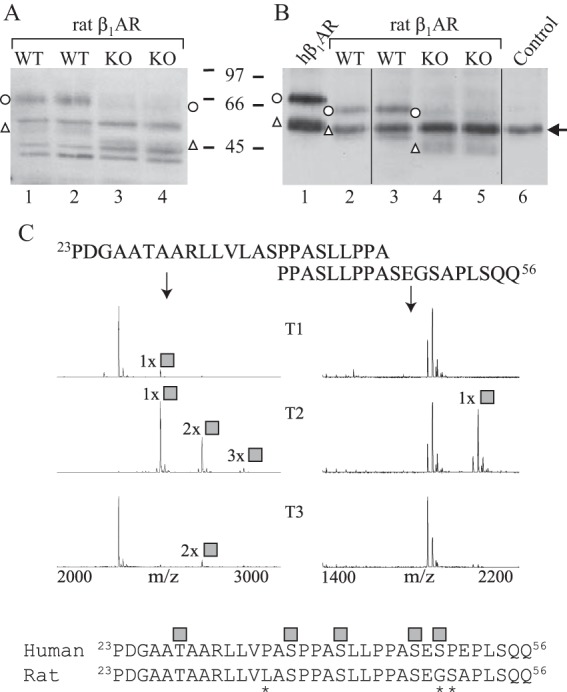
β1AR is cleaved extensively in Galnt2−/− rat hearts. A and B, cellular membranes were isolated from heart ventricles of Galnt2−/− and corresponding WT rats, and receptors were analyzed by SDS-PAGE and Western blotting with an antibody directed against the receptor C-terminal domain (sc-568). The analysis was performed either directly after solubilization (A) or after immunoprecipitation (B). In B, hβ1AR stably expressed in HEK293i cells was used as a control (lane 1). No sample was included for immunoprecipitation for lane 6. Arrow, antibody heavy chain. Open circles, full-length hβ1ARs; open triangles, C-terminal fragments cleaved at 31R↓L32. The experiment was replicated twice using a total of three Galnt2−/− and three WT rat hearts. C, overlapping peptides covering amino acid positions 23–46 of the rat β1AR N terminus were tested for glycosylation by 10 recombinant GalNAc-Ts and analyzed by MALDI-TOF. The 24-h incubation time points are shown. GalNAc-T2 was found to be the most efficient in glycosylating the peptides, whereas GalNAc-T1 and GalNAc-T3 were found to glycosylate the peptide 23PDGAATAARLLVLASPPASLLPPA46 to a minor extent. Shown below is the alignment of the human and rat β1AR N termini; asterisks denote differences in sequence. The identified O-glycosylation sites of the human receptor are shown as gray squares. The spectra show relative intensity. The results shown are representative of two independent experiments.
Discussion
The complex regulation of site-specific O-glycosylation involving up to 20 polypeptide GalNAc-T isoforms offers a compelling system to fine-tune and enhance differential regulation of important proteolytic processing events (15). This has been demonstrated for PC processing regulated by PCs as well as more recently for ectodomain shedding regulated by metalloproteinases of the ADAM and MMP families (15, 17). Here, we expanded the targets for co-regulatory functions of site-specific O-glycosylation to include the cell surface-exposed N-terminal domains of GPCRs using β1AR as an example. We have shown previously that β1AR is O-glycosylated and undergoes limited and regulated processing by metalloproteinases affecting receptor turnover (5, 6) and now demonstrate that O-glycosylation of the receptor is specifically regulated by a single GalNAc-T isoform at specific sites close to the metalloproteinase cleavage sites. Furthermore, we confirm in vitro and in vivo that the metalloproteinase-mediated processing of β1AR is co-regulated by GalNAc-T2 site-specific O-glycosylation and demonstrate that this phenomenon has an effect on receptor function. This is the first example of site-specific O-glycan fine-tuning of GPCR cleavage, and we predict that over 300 GPCRs (30%) could be O-glycosylated and have potential for similar co-regulation by O-glycosylation.
GPCRs are a large group of integral membrane proteins with seven transmembrane segments and a highly variable cell surface-exposed N-terminal sequence. They undergo a variety of post-translational modifications, including N- and O-glycosylation. However, still only a very limited number of GPCRs are known to be O-glycosylated and their actual glycosites determined (7, 18, 19, 22–24, 33). With the introduction of the SimpleCell O-glycoproteomics strategy, our knowledge of O-glycosites has greatly advanced (7), although initially only few glycosites in GPCRs were detected (7, 25). One obstacle is the challenge in detecting juxtamembrane regions, such as GPCR N termini in the bottom-up mass spectrometry that depends on appropriate peptide digestions (34). With the advancing analysis of SimpleCells derived from different organs and improvements in mass spectrometry workflow and instrumentation, we have now identified more than 50 GPCR N termini with one or more O-glycosites,5 supporting the NetOGly4.0 prediction of >300 GPCRs being O-glycosylated (7, 25).
The N-terminal proteolytic cleavage of GPCRs has been demonstrated to participate in various molecular mechanisms. Adhesion GPCRs have a highly conserved GPCR proteolysis site (GPS), which is cleaved by an intramolecular autocatalytic reaction (35). The proteinase-activated receptors (PARs) are a subfamily of GPCRs activated by serine proteases (36), although a metalloproteinase is also involved in the regulated proteolysis of PAR1 (35). For both adhesion receptors and PARs, the proteolytic processing of the receptor N terminus leads to an exposure of a cryptic tethered peptide sequence that acts as an agonist and activates the cognate receptors (35). A growing number of other GPCRs are also reported to undergo N-terminal cleavage mediated by metalloproteinases, including the parathyroid hormone receptor (37), thyrotropin receptor (38), endothelin B receptor (39), C5a receptor (40), GPR37 (41), and GPR37L1 (42) in addition to β1AR (5). The functional roles of the N-terminal cleavage of these GPCRs are not fully understood, although several are cleaved in an activation-dependent manner. Interestingly, all of these receptors are predicted to be O-glycosylated in their N termini, and in several cases, the O-glycosites have been determined experimentally.5 Thus, there is mounting evidence to suggest that limited N-terminal proteolysis and O-glycosylation may overlap topologically, thus opening the possibility for a broader functional interplay for GPCRs.
The global discovery of an interplay between O-glycosylation and proteolytic processing has been hampered by limited insight into the location of O-glycosylation and proteolytic processing within the protein sequence or domains. A major challenge is the lack of a clear consensus motif for general GalNAc O-glycosylation or even GalNAc-T isoform-specific glycosylation, although prediction algorithms can provide some guidance (7, 43). Metalloproteinases also lack clear consensus motifs, and a substantial degree of redundancy exists between different isoforms (44, 45). Consequently, peptide libraries and substrate degradomes of knock-out cell lines and animals have been applied to provide information of the preference for sequence features and topology in substrate recognition and used to identify specific substrates (46–50). Due to these challenges, we used here in vitro analyses with peptide substrates derived from the N-terminal sequence of β1AR to probe the existence and interplay between site-specific O-glycosylation and metalloproteinase-mediated processing. We previously used this strategy and found a good correlation with in vivo functions (17, 26), and here we could unambiguously validate our findings in cell and animal model systems in vivo. It is important to note that our studies only relate to the direct interplay at the substrate level (i.e. the N terminus of β1AR), and we found previously that such interplay of processes occurs within ±3–5 residues (51). Clearly, both players, being the proteolytic processing and O-glycosylation events, have additional layers of complex regulation.
We demonstrated in the in vitro assays that O-glycosylation blocks metalloproteinase-mediated cleavage at 52P↓L53 almost completely and inhibits cleavage at 31R↓L32 of the β1AR N-terminal peptides. We also identified an additional cleavage site at 41S↓L42 utilized by a number of MMPs, which was also blocked by O-glycosylation. Thus, our in vitro studies show the potential for multiple cleavage sites in the β1AR N terminus regulated by different enzymes. This observation was supported indirectly by results obtained using the cell line models, where mutating the main cleavage site of the receptor at 31R↓L32 led to heterogeneity in the expressed receptor forms detected in CHO-ldlD cells. A similar observation was made previously when the mutant receptor was expressed in HEK293 cells (5). In our previous studies, we have demonstrated that the N-terminal cleavage of β1AR at 31R↓L32 is modulated by agonist-mediated receptor activation (5). Thus, further studies are needed to decipher regulation of β1AR processing sites and the ADAMs and MMPs involved in specific situations. A wide range of molecular mechanisms and physiological stimuli are known to regulate ADAM and MMP activities (52), and currently our understanding of the complex regulatory mechanisms is limited.
Similarly, our understanding of the regulation of site-specific O-glycosylation is still incomplete. Although O-glycosylation is widely found on the majority of proteins trafficking the secretory pathway, we predict that only a subset of these are regulated in cells (25). Thus, most O-glycosites are covered by redundancies among the many polypeptide GalNAc-Ts, and a complete loss of a single GalNAc-T isoform does not affect the majority of O-glycosites. However, a subset of glycosites that are often found in isolated sites in proteins are specifically controlled by individual GalNAc-T isoforms and hence can be differentially regulated (25). Here, we found that the N-terminal region of β1AR is selectively controlled by the GalNAc-T2 isoform with only minor contributions from a few other isoforms. Furthermore, a complete loss of GalNAc-T2 reduced glycosylation and enhanced the metalloproteinase-mediated processing of the receptor in vivo. It is therefore likely that the site-specific O-glycosylation of β1AR is controlled through regulation of GalNAc-T2. The GALNT2 gene has been associated with dyslipidemia, and a specific regulatory role of this isoform in O-glycosylation of the lipase inhibitor angiopoietin-like 3 has been proposed (25, 30). Importantly, it appears that a liver-specific enhancer element in the intron 1 of the GALNT2 gene selectively regulates this function (54). Furthermore, the murine Galnt2 gene has been shown to be up-regulated in adrenergic-deficient mouse hearts during development, supporting a regulatory link between GalNAc-T2 and β-adrenergic signaling in the heart (55).
Another important aspect, at the substrate level, of the interplay between proteolytic processing and site-specific O-glycosylation is protein sequence variations. Sequence variations in regions of processing and/or O-glycosylation with clinical consequences are often found, but due to the inherent difficulties in predicting processing and O-glycosylation, experimental analysis is required for the evaluation of effects. Here, we mapped the exact locations of O-glycosites in β1AR and identified Ser-49 as an amino acid modified by O-glycosylation. Interestingly, a common single nucleotide polymorphism (S49G) with functional relevance has been identified at this site (56, 57). The less common Gly-49 variant exhibits a higher degree of desensitization and more profound agonist-promoted down-regulation compared with the Ser-49 variant when expressed in cell lines (29, 58). Furthermore, several clinical associations have been suggested for the polymorphism, although the results have been inconsistent (59). The S49G replacement clearly eliminates the O-glycosite at this position, but it remains to be determined to what degree this amino acid replacement affects glycosylation of other sites in the region and ultimately proteolytic processing. However, it is compelling to hypothesize that the differences between the variants observed in cells, and the potential clinical associations, are due to the loss of O-glycosylation and to dysregulation of processing. We are currently exploring this possibility.
In summary, our study demonstrates the existence of a regulatory interplay between site-specific O-glycosylation and proteolytic processing in the extracellular N terminus of β1AR. We developed isogenic cell systems and a knock-out rat model, which will be used in future studies to address the physiological consequence of this interplay. Finally, based on the prevalence of O-glycosylation identified and predicted in GPCR N termini, we expect that the phenomenon described here is more widely present in this large family of important drug targets.
Experimental Procedures
Glycosyltransferase Assays
Recombinant glycosyltransferases were expressed as soluble secreted truncated proteins in insect cells (60). In vitro activity assays for GalNAc-T glycosylation of peptides (Schafer-N, NeoBioSci) were performed as described before and monitored with MALDI-TOF (17, 26). Samples were purified for subsequent site determination and cleavage experiments by HPLC using a multistep gradient on a Dionex Ultimate 3000 LC system (Thermo Scientific) with a Kinetex 2.6u C18 100A 100 × 4.60-mm column (Phenomenex).
Characterization of O-Glycosites by Electron Transfer Dissociation-MS2
Samples were dissolved in methanol/water (1:1) containing 1% formic acid and introduced by direct infusion via a TriVersa NanoMate ESI-Chip interface (Advion BioSystems) at a flow rate of 100 nl/min and 1.4 kV spray voltage. Mass spectra were acquired in positive ion Fourier transform mode using parameters similar to previous studies (17, 61), except at a nominal resolving power of 30,000 or 60,000. Electron transfer dissociation-MS2 spectra were analyzed by comparison with theoretical c and z• fragment m/z values calculated for all positional combinations of one HexNAc residue distributed on all potential Ser and Thr glycosylation sites in the sequence. Calculations were performed using the web-based Protein Prospector MS-Product software routine. Furthermore, samples were analyzed on a setup composed of an EASY-nanoLC 1000 (Thermo Scientific) connected via a nanoSpray Flex ion source to an LTQ-OrbitrapVelos Pro hybrid mass spectrometer. Glycopeptides were dissolved in 0.1% formic acid and separated on an in-house packed reverse phase column (1.9-μm Reprosil-Pure-AQ C18 particles, Dr. Maisch GmbH). The MS1 precursor ion scan was performed in the Orbitrap using a nominal resolution of 30,000 followed by two MS2 scan events (15,000 resolving power at m/z 400) utilizing HCD and ETD fragmentation modes. Glycopeptide identification and glycosite assignments were accomplished by Proteome Discoverer version 1.4 with final validation through manual inspection of the assigned peaks.
ADAM and MMP Cleavage of Peptides and Glycopeptides
In vitro metalloproteinase cleavage activity was assayed by adding 150–600 nm ADAM8, -10, -12, or -17 (Enzo Life Sciences) or MMP1, -2, -3, -7, -8, -9, -12, -13, or -14 (produced as described previously (46)), using 10 μg of peptide or glycopeptide substrate in a total volume of 25 μl. Reactions were performed in 25 mm Tris-HCl, pH 9 (ADAM17); 25 mm Tris-HCl, pH 9, 2 mm CaCl2, 0.0005% Brij-35 (ADAM10); 20 mm Tris-HCl, pH 8, 0.0005% Brij-35 (ADAM12); or 20 mm Tris-HCl, pH 8, 25 mm CaCl2, and 0.0005% Brij-35 (ADAM8). Reactions were incubated at 37 °C (ADAM17, -8, -10, and -12) or 30 °C (ADAM8). MMP cleavage assays were performed at 37 °C in 50 mm Tris-HCl, pH 7.5, 200 mm NaCl, 5 mm CaCl2, and 0.05% Brij-35. MMP1, -2, -3, -7, -8, -9, and -13 were activated before the assay by incubation with 10 mm α-amino-3-hydroxy-5-methylisoxazole-propionic acid. Product development was evaluated after 0.5, 1, and 4 h by MALDI-TOF-MS.
DNA Constructs and Cell Lines
DNA constructs for the WT hβ1AR, the R31H/L32A cleavage site mutant, and the Δ2–31 and Δ2–52 truncation mutants have been described previously (5). The construct for the hδOR Cys-27 variant has been described (28).
Cell Lines
The stably transfected HEK293i cell lines expressing the Myc- and FLAG epitope-tagged hβ1AR or hδOR in an inducible manner have been described previously (5, 28). HEK293SimpleCells with a knock-out of COSMC and HEK293 and HepG2 cell lines with a knock-out and knock-in of specific GalNAc-Ts (HEK293ΔT2, HepG2ΔT2, HepG2ΔT2+T2, and HepG2ΔT1) have been described previously (17, 24). A HepG2 cell line with a knock-out of GalNAc-T11 (HepG2ΔT11) was produced by ZFN gene-mediated targeting of GALNT11. The ZFN targeting construct for GALNT11 was custom-produced (Sigma-Aldrich) with the following binding sites with the cutting site indicated in parenthesis: 5′-GACCGCTTGGGCTAC(CACAGA)GATGTGCCAGACACAAGG-3′. In short, the HepG2 cells (a kind gift from Novo Nordisk) were transfected with one vial of mRNA (Sigma-Aldrich) using nucleofection on an Amaxa Nucleofector (Lonza). The clones were confirmed to have GALNT11 mutations by PCR and sequencing using the oligonucleotide 5′-TCGCTGACTAACTTCACTCTTTTG-3′ and its reverse complement (62). CHO-K1 cells and the CHO-ldlD cell line were obtained from the ATCC cell culture collection. A HEK293 cell line with a knock-out of both GalNAc-T2 and -T3 was prepared by gene targeting using GFP-tagged CRISPR/Cas9. The 20-nucleotide guide sequences targeting human GALNT2 (5′-GTGAAACGTGATCACCACGC-3′) and GALNT3 (5′-TATGGAAGTAACCATAACCG-3′) were designed using the online tool. The single-guide RNAs were co-transfected with the Cas9-PBKS plasmid using Lipofectamine 3000 according to the manufacturer's instructions (Thermo Fisher Scientific). At 24 h after transfection, GFP-positive cells were enriched by FACS and cultured for 1–2 weeks. The sorted cells were then single-sorted again for GFP-negative cells into 96-well plates. Knock-out clones with frameshift mutations were identified by IDAA (indel detection by amplicon analysis) with the following primers: GALNT2, 5′-CCATCCCAGTTGGTCAGTCT-3′ (forward) and 5′-CTGTGCTGAGCAGTCAGGAG-3′ (reverse); GALNT3, 5′-TCCCTCCAGGTGAGTGTTTC-3′ (forward) and 5′-AAAGCAAACAGTGTGTACATATTCAA-3′ (reverse). The mutated sequences of the selected knock-out clone were confirmed by sanger sequencing for each gene, and the loss of the enzyme was characterized by immunocytochemistry with in house monoclonal antibodies to GalNAc-T1 (4D8), GalNAc-T2 (4C4), and GalNAc-T3 (2D10).
All cell lines were cultured in a humidified atmosphere at 37 °C with 5% CO2. HEK293 and HepG2 cells were cultured in DMEM containing 10% FBS, and CHO-K1 and CHO-ldlD cells were cultured in F-12 Ham nutrient mixture with Kaighn's modification containing 5% FBS. The media were supplemented with 100 units/ml penicillin and 0.1 mg/ml streptomycin and with selection antibiotics in the case of the stably transfected HEK293i cells (5, 28). The cell culture reagents were obtained from Sigma-Aldrich, Thermo Fisher Scientific, or Invivogen. For experiments, cells were seeded onto 6- or 10-cm plates and cultured for 1–3 days to 60–80% confluence. For CHO-K1 and CHO-ldlD cells, FBS concentration was lowered to 2 or 0.5%. Cells were transfected with 1–2 μg (WT, R31H/L32A)/3 μg (Δ2–31, Δ2–52) of receptor constructs for 24–48 h using Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer's instructions or, alternatively, using linear 25-kDa polyethyleneimine (Polysciences) (41). Both reagents were used at a 1:3 DNA/reagent ratio. The metalloproteinase inhibitor marimastat (Tocris Bioscience; Fig. 4C), and the monosaccharides Gal (20 μm) and GalNAc (400 μm) (Sigma-Aldrich; Fig. 3, E, G, and H) were added to the culture medium 4 h after starting the transfection. For some experiments (Fig. 5), cycloheximide (20 μg/ml; Sigma-Aldrich) was added 4 h before cells were harvested to deplete receptor precursors that co-migrate with the main C-terminal fragment on SDS-PAGE (5). Receptor expression in the stably transfected HEK293i cells was induced with 0.5 μg/ml tetracycline for 6 h (Fig. 7B) or 24 h (Fig. 3D). Cells were lifted from plates using warm 2 mm EDTA/PBS and harvested in ice-cold PBS, quick frozen in liquid nitrogen, and stored at −70 °C.
Galnt2−/− Rats
The GalNAc-T2 knock-out rats were prepared by the SAGE Laboratories as described elsewhere (53). Hearts were collected from 12-week-old male homozygote Galnt2−/− and corresponding WT rats, and ventricular samples were quick frozen in liquid nitrogen.
cAMP Accumulation Assay
The assay was performed by applying homogeneous time-resolved FRET using the cAMP cell-based assay kit from Cisbio Bioassays according to recommendations. Briefly, cells were detached from culture plates 48 h post-transfection, resuspended in DMEM with 1 mm 3-isobutyl-1-methylxanthine (Sigma-Aldrich), counted, and transferred in duplicates to white 384 microplates (Greiner Bio-One) (2,500 cells/well in a total volume of 5 μl). Appropriate concentrations of isoproterenol (Sigma-Aldrich) were then added in a total volume of 5 μl, and the cells were incubated for 60 min at 37 °C. The reaction was terminated by adding 10 μl of the kit's lysis buffer containing cAMP-d2 (acceptor) and anti-cAMP-Eu3+ Cryptate (donor). The signal was measured after 60 min using the EnSpire multilabel reader (PerkinElmer Life Sciences). The cAMP generated was interpolated from a cAMP standard curve generated in parallel for each experiment.
Preparation and Solubilization of Membranes and Whole Cell Extracts
Cellular membranes from transfected cells or from rat heart ventricles were prepared and solubilized, and total cellular lysates were prepared as described previously (6, 28). The detergent n-dodecyl-β-d-maltoside was replaced with Triton X-100 (Sigma-Aldrich).
Immunoprecipitation of Solubilized Receptors
Solubilized receptors from transfected cells and rat heart ventricles were purified by immunoprecipitation using the immobilized mouse monoclonal FLAG M2 antibody (Sigma-Aldrich, A2220) and the polyclonal rabbit V-19 antibody (Santa Cruz Biotechnology, Inc., sc-568), respectively. The latter antibody, which is directed against the receptor C terminus, was cross-linked to magnetic beads (Pierce Protein A/G magnetic beads, Thermo Scientific) according to the manufacturer's instructions. The one-step immunoprecipitation with the FLAG M2 antibody was performed as described previously (5), and purified receptors were eluted with SDS-sample buffer. Immunoprecipitation with the V-19 antibody was performed using the Pierce Crosslink Magnetic IP/CO-IP kit. The buffers were supplemented with 0.1% Triton X-100, 2 mm EDTA, 0.5 mm PMSF, 2 mm 1,10-phenanthroline 5 μg/ml leupeptin, 5 μg/ml soybean trypsin inhibitor, and 10 μg/ml benzamidine.
Deglycosylation of Immunoprecipitated Receptors
For deglycosylation, samples were eluted from the FLAG M2 antibody affinity resin with 1% SDS in 50 mm sodium phosphate, pH 5.5, and diluted eluates were digested with Endo H (50 milliunits/ml), PNGase F (50 units/ml), neuraminidase (50 milliunits/ml), and O-glycosidase (50 milliunits/ml) obtained from Roche Applied Science, as described (5).
SDS-PAGE and Western Blotting
The analysis of purified receptors was performed by SDS-PAGE and Western blotting as described (6). The blots were probed with FLAG M2 (0.1 μg/ml; Sigma-Aldrich, F3165), FLAG M2-HRP (1:50,000–80,0000; Sigma-Aldrich, A8592), c-Myc A14 (1:1,000–3,000; Santa Cruz Biotechnology, sc-789), c-Myc (1:1,000; Sigma-Aldrich, C3956), or V-19 (1:1,000; Santa Cruz Biotechnology) antibodies and, when appropriate, followed by HRP-conjugated donkey anti-mouse F(ab′)2/anti-rabbit F(ab′)2 antibodies (1:10,000–40,000; GE Healthcare, NA9310 and NA9340, respectively). Pierce ECL or ECL Plus detection reagents (Thermo Scientific) or the Luminata Classico or Crescendo Western HRP substrate (Merck Millipore) were used to reveal the receptor bands. The relative intensities of bands on ECL films were analyzed by densitometric scanning with the Umax PowerLook 1120 color scanner (GE Healthcare) and Image Master 2D Platinum version 6.0 software (GE Healthcare), and quantified using ImageJ version 1.45s, subtracting the local background from each lane.
Data Analysis
Data were analyzed using GraphPad Prism version 7.02 (GraphPad Software, La Jolla, CA). Statistical analyses were performed using the paired t test, the one-sample t test, or the two-way analysis of variance followed by Tukey's multiple-comparison post hoc test. The limit of significance was set at p < 0.05. The data are presented as mean ± S.E.
Author Contributions
C. K. G. performed and analyzed the experiments in Figs. 1, 2, 6, and 7C and supplemental Fig. 1 and wrote the first draft of the manuscript. H. E. T. performed and analyzed the experiments in Figs. 3 (B–E, G, and H) and 4C, and H. K. performed and analyzed the experiments in Figs. 4A and 5. J. J. L. constructed the receptor model in Fig. 3A and participated in the analysis of results of the cell models. S. W. made the HepG2ΔT11 cell line, Y. N. made the HEK293ΔT2/T3 cell line, and L. H. H. assisted in performing and analyzing the experiments in Fig. 6 and supplemental Fig. 1. C. M. O. provided the panel of recombinant MMPs. K. T. S. coordinated the preparation of the Galnt2−/− rats, and U. E. P.-R. performed and analyzed the experiment in Fig. 7 (A and B) and analyzed the data in Figs. 3F, 4B, 5B, and 6 and supplemental Fig. 1. K. T. S., H. C., and U. E. P.-R., conceived and coordinated the study and wrote the final version of the paper. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgment
We thank Miia Vierimaa for expert technical assistance.
This work was supported by the Danish Research Councils (including a Sapere Aude Research Talent Grant to K. T. S.), a program of excellence grant from the University of Copenhagen, the Novo Nordisk Foundation, Danish National Research Foundation Grant DNRF107 (to H. C.), the Neye Foundation (to C. G.), and the MRC Oulu and Magnus Ehrnrooth Foundation (to U. E. P.-R.).

This article contains supplemental Fig. 1.
C. K. Goth and H. Clausen, unpublished data.
- βAR
- β-adrenergic receptor
- hβ1AR
- human β1-adrenergic receptor
- ADAM
- a disintegrin and metalloproteinase
- GalNAc-T
- polypeptide GalNAc-transferase
- GPCR
- G protein-coupled receptor
- MMP
- matrix metalloproteinase
- PAR
- proteinase-activated receptors
- PC
- proprotein convertase
- PNGase F
- peptide-N-glycosidase F
- Endo H
- endo-N-acetylglucosaminidase H
- hδOR
- human δ-opioid receptor.
References
- 1. Rockman H. A., Koch W. J., and Lefkowitz R. J. (2002) Seven-transmembrane-spanning receptors and heart function. Nature 415, 206–212 [DOI] [PubMed] [Google Scholar]
- 2. Dorn G. W. 2nd, and Liggett S. B. (2009) Mechanisms of pharmacogenomic effects of genetic variation within the cardiac adrenergic network in heart failure. Mol. Pharmacol. 76, 466–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bristow M. R., Hershberger R. E., Port J. D., Minobe W., and Rasmussen R. (1989) β1- and β2-adrenergic receptor-mediated adenylate cyclase stimulation in nonfailing and failing human ventricular myocardium. Mol. Pharmacol. 35, 295–303 [PubMed] [Google Scholar]
- 4. Bristow M. R. (2000) β-Adrenergic receptor blockade in chronic heart failure. Circulation 101, 558–569 [DOI] [PubMed] [Google Scholar]
- 5. Hakalahti A. E., Vierimaa M. M., Lilja M. K., Kumpula E. P., Tuusa J. T., and Petäjä-Repo U. E. (2010) Human β1-adrenergic receptor is subject to constitutive and regulated N-terminal cleavage. J. Biol. Chem. 285, 28850–28861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hakalahti A. E., Khan H., Vierimaa M. M., Pekkala E. H., Lackman J. J., Ulvila J., Kerkelä R., and Petäjä-Repo U. E. (2013) β-adrenergic agonists mediate enhancement of β1-adrenergic receptor N-terminal cleavage and stabilization in vivo and in vitro. Mol. Pharmacol. 83, 129–141 [DOI] [PubMed] [Google Scholar]
- 7. Steentoft C., Vakhrushev S. Y., Joshi H. J., Kong Y., Vester-Christensen M. B., Schjoldager K. T., Lavrsen K., Dabelsteen S., Pedersen N. B., Marcos-Silva L., Gupta R., Bennett E. P., Mandel U., Brunak S., Wandall H. H., et al. (2013) Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 32, 1478–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bennett E. P., Mandel U., Clausen H., Gerken T. A., Fritz T. A., and Tabak L. A. (2012) Control of mucin-type O-glycosylation: a classification of the polypeptide GalNAc-transferase gene family. Glycobiology 22, 736–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Magrané J., Casaroli-Marano R. P., Reina M., Gåfvels M., and Vilaró S. (1999) The role of O-linked sugars in determining the very low density lipoprotein receptor stability or release from the cell. FEBS Lett. 451, 56–62 [DOI] [PubMed] [Google Scholar]
- 10. May P., Bock H. H., Nimpf J., and Herz J. (2003) Differential glycosylation regulates processing of lipoprotein receptors by γ-secretase. J. Biol. Chem. 278, 37386–37392 [DOI] [PubMed] [Google Scholar]
- 11. Kingsley D. M., Kozarsky K. F., Segal M., and Krieger M. (1986) Three types of low density lipoprotein receptor-deficient mutant have pleiotropic defects in the synthesis of N-linked, O-linked, and lipid-linked carbohydrate chains. J. Cell Biol. 102, 1576–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kozarsky K., Kingsley D., and Krieger M. (1988) Use of a mutant cell line to study the kinetics and function of O-linked glycosylation of low density lipoprotein receptors. Proc. Natl. Acad. Sci. U.S.A. 85, 4335–4339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Topaz O., Shurman D. L., Bergman R., Indelman M., Ratajczak P., Mizrachi M., Khamaysi Z., Behar D., Petronius D., Friedman V., Zelikovic I., Raimer S., Metzker A., Richard G., and Sprecher E. (2004) Mutations in GALNT3, encoding a protein involved in O-linked glycosylation, cause familial tumoral calcinosis. Nat. Genet. 36, 579–581 [DOI] [PubMed] [Google Scholar]
- 14. Kato K., Jeanneau C., Tarp M. A., Benet-Pagès A., Lorenz-Depiereux B., Bennett E. P., Mandel U., Strom T. M., and Clausen H. (2006) Polypeptide GalNAc-transferase T3 and familial tumoral calcinosis: secretion of fibroblast growth factor 23 requires O-glycosylation. J. Biol. Chem. 281, 18370–18377 [DOI] [PubMed] [Google Scholar]
- 15. Schjoldager K. T., and Clausen H. (2012) Site-specific protein O-glycosylation modulates proprotein processing: deciphering specific functions of the large polypeptide GalNAc-transferase gene family. Biochim. Biophys. Acta 1820, 2079–2094 [DOI] [PubMed] [Google Scholar]
- 16. Boskovski M. T., Yuan S., Pedersen N. B., Goth C. K., Makova S., Clausen H., Brueckner M., and Khokha M. K. (2013) The heterotaxy gene GALNT11 glycosylates Notch to orchestrate cilia type and laterality. Nature 504, 456–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goth C. K., Halim A., Khetarpal S. A., Rader D. J., Clausen H., and Schjoldager K. T. (2015) A systematic study of modulation of ADAM-mediated ectodomain shedding by site-specific O-glycosylation. Proc. Natl. Acad. Sci. U.S.A. 112, 14623–14628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sadeghi H., and Birnbaumer M. (1999) O-Glycosylation of the V2 vasopressin receptor. Glycobiology 9, 731–737 [DOI] [PubMed] [Google Scholar]
- 19. Petaja-Repo U. E., Hogue M., Laperriere A., Walker P., and Bouvier M. (2000) Export from the endoplasmic reticulum represents the limiting step in the maturation and cell surface expression of the human δ opioid receptor. J. Biol. Chem. 275, 13727–13736 [DOI] [PubMed] [Google Scholar]
- 20. Markkanen P. M. H., and Petäjä-Repo U. E. (2008) N-Glycan-mediated quality control in the endoplasmic reticulum is required for the expression of correctly folded δ-opioid receptors at the cell surface. J. Biol. Chem. 283, 29086–29098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lackman J. J., Markkanen P. M. H., Hogue M., Bouvier M., and Petäjä-Repo U. E. (2014) N-Glycan-dependent and -independent quality control of human δ opioid receptor N-terminal variants. J. Biol. Chem. 289, 17830–17842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li J. G., Chen C., and Liu-Chen L. Y. (2007) N-Glycosylation of the human κ opioid receptor enhances its stability but slows its trafficking along the biosynthesis pathway. Biochemistry 46, 10960–10970 [DOI] [PubMed] [Google Scholar]
- 23. Michineau S., Alhenc-Gelas F., and Rajerison R. M. (2006) Human bradykinin B2 receptor sialylation and N-glycosylation participate with disulfide bonding in surface receptor dimerization. Biochemistry 45, 2699–2707 [DOI] [PubMed] [Google Scholar]
- 24. Bannert N., Craig S., Farzan M., Sogah D., Santo N. V., Choe H., and Sodroski J. (2001) Sialylated O-glycans and sulfated tyrosines in the NH2-terminal domain of CC chemokine receptor 5 contribute to high affinity binding of chemokines. J. Exp. Med. 194, 1661–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schjoldager K. T., Joshi H. J., Kong Y., Goth C. K., King S. L., Wandall H. H., Bennett E. P., Vakhrushev S. Y., and Clausen H. (2015) Deconstruction of O-glycosylation-GalNAc-T isoforms direct distinct subsets of the O-glycoproteome. EMBO Rep. 16, 1713–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kong Y., Joshi H. J., Schjoldager K. T., Madsen T. D., Gerken T. A., Vester-Christensen M. B., Wandall H. H., Bennett E. P., Levery S. B., Vakhrushev S. Y., and Clausen H. (2015) Probing polypeptide GalNAc-transferase isoform substrate specificities by in vitro analysis. Glycobiology 25, 55–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Krieger M., Reddy P., Kozarsky K., Kingsley D., Hobbie L., and Penman M. (1989) Analysis of the synthesis, intracellular sorting, and function of glycoproteins using a mammalian cell mutant with reversible glycosylation defects. Methods Cell Biol. 32, 57–84 [DOI] [PubMed] [Google Scholar]
- 28. Leskelä T. T., Markkanen P. M. H., Pietilä E. M., Tuusa J. T., and Petäjä-Repo U. E. (2007) Opioid receptor pharmacological chaperones act by binding and stabilizing newly synthesized receptors in the endoplasmic reticulum. J. Biol. Chem. 282, 23171–23183 [DOI] [PubMed] [Google Scholar]
- 29. Rathz D. A., Brown K. M., Kramer L. A., and Liggett S. B. (2002) Amino acid 49 polymorphisms of the human beta1-adrenergic receptor affect agonist-promoted trafficking. J. Cardiovasc. Pharmacol. 39, 155–160 [DOI] [PubMed] [Google Scholar]
- 30. Schjoldager K. T., Vakhrushev S. Y., Kong Y., Steentoft C., Nudelman A. S., Pedersen N. B., Wandall H. H., Mandel U., Bennett E. P., Levery S. B., and Clausen H. (2012) Probing isoform-specific functions of polypeptide GalNAc-transferases using zinc finger nuclease glycoengineered SimpleCells. Proc. Natl. Acad. Sci. U.S.A. 109, 9893–9898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Steentoft C., Vakhrushev S. Y., Vester-Christensen M. B., Schjoldager K. T., Kong Y., Bennett E. P., Mandel U., Wandall H., Levery S. B., and Clausen H. (2011) Mining the O-glycoproteome using zinc-finger nuclease-glycoengineered SimpleCell lines. Nat. Methods 8, 977–982 [DOI] [PubMed] [Google Scholar]
- 32. Atwood B. K., Lopez J., Wager-Miller J., Mackie K., and Straiker A. (2011) Expression of G protein-coupled receptors and related proteins in HEK293, AtT20, BV2, and N18 cell lines as revealed by microarray analysis. BMC Genomics 12, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Halim A., Rüetschi U., Larson G., and Nilsson J. (2013) LC-MS/MS characterization of O-glycosylation sites and glycan structures of human cerebrospinal fluid glycoproteins. J. Proteome Res. 12, 573–584 [DOI] [PubMed] [Google Scholar]
- 34. Levery S. B., Steentoft C., Halim A., Narimatsu Y., Clausen H., and Vakhrushev S. Y. (2015) Advances in mass spectrometry driven O-glycoproteomics. Biochim. Biophys. Acta 1850, 33–42 [DOI] [PubMed] [Google Scholar]
- 35. Monk K. R., Hamann J., Langenhan T., Nijmeijer S., Schöneberg T., and Liebscher I. (2015) Adhesion G protein-coupled receptors: from in vitro pharmacology to in vivo mechanisms. Mol. Pharmacol. 88, 617–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Macfarlane S. R., Seatter M. J., Kanke T., Hunter G. D., and Plevin R. (2001) Proteinase-activated receptors. Pharmacol. Rev. 53, 245–282 [PubMed] [Google Scholar]
- 37. Klenk C., Schulz S., Calebiro D., and Lohse M. J. (2010) Agonist-regulated cleavage of the extracellular domain of parathyroid hormone receptor type 1. J. Biol. Chem. 285, 8665–8674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Couet J., Sar S., Jolivet A., Hai M. T., Milgrom E., and Misrahi M. (1996) Shedding of human thyrotropin receptor ectodomain: involvement of a matrix metalloprotease. J. Biol. Chem. 271, 4545–4552 [DOI] [PubMed] [Google Scholar]
- 39. Grantcharova E., Furkert J., Reusch H. P., Krell H. W., Papsdorf G., Beyermann M., Schulein R., Rosenthal W., and Oksche A. (2002) The extracellular N terminus of the endothelin B (ETB) receptor is cleaved by a metalloprotease in an agonist-dependent process. J. Biol. Chem. 277, 43933–43941 [DOI] [PubMed] [Google Scholar]
- 40. van den Berg C. W., Gonçalves-de-Andrade R. M., Okamoto C. K., and Tambourgi D. V. (2012) C5a receptor is cleaved by metalloproteases induced by sphingomyelinase D from Loxosceles spider venom. Immunobiology 217, 935–941 [DOI] [PubMed] [Google Scholar]
- 41. Mattila S. O., Tuusa J. T., and Petäjä-Repo U. E. (2016) The Parkinson's-disease-associated receptor GPR37 undergoes metalloproteinase-mediated N-terminal cleavage and ectodomain shedding. J. Cell Sci. 129, 1366–1377 [DOI] [PubMed] [Google Scholar]
- 42. Coleman J. L., Ngo T., Schmidt J., Mrad N., Liew C. K., Jones N. M., Graham R. M., and Smith N. J. (2016) Metalloprotease cleavage of the N terminus of the orphan G protein-coupled receptor GPR37L1 reduces its constitutive activity. Sci. Signal. 9, ra36. [DOI] [PubMed] [Google Scholar]
- 43. Gerken T. A., Raman J., Fritz T. A., and Jamison O. (2006) Identification of common and unique peptide substrate preferences for the UDP-GalNAc:polypeptide α-N-acetylgalactosaminyltransferases T1 and T2 derived from oriented random peptide substrates. J. Biol. Chem. 281, 32403–32416 [DOI] [PubMed] [Google Scholar]
- 44. Weber S., and Saftig P. (2012) Ectodomain shedding and ADAMs in development. Development 139, 3693–3709 [DOI] [PubMed] [Google Scholar]
- 45. Page-McCaw A., Ewald A. J., and Werb Z. (2007) Matrix metalloproteinases and the regulation of tissue remodelling. Nat. Rev. Mol. Cell Biol. 8, 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Eckhard U., Huesgen P. F., Schilling O., Bellac C. L., Butler G. S., Cox J. H., Dufour A., Goebeler V., Kappelhoff R., Keller U. A., Klein T., Lange P. F., Marino G., Morrison C. J., Prudova A., et al. (2016) Active site specificity profiling of the matrix metalloproteinase family: proteomic identification of 4300 cleavage sites by nine MMPs explored with structural and synthetic peptide cleavage analyses. Matrix Biol. 49, 37–60 [DOI] [PubMed] [Google Scholar]
- 47. Kleifeld O., Doucet A., auf dem Keller U., Prudova A., Schilling O., Kainthan R. K., Starr A. E., Foster L. J., Kizhakkedathu J. N., and Overall C. M. (2010) Isotopic labeling of terminal amines in complex samples identifies protein N-termini and protease cleavage products. Nat. Biotechnol. 28, 281–288 [DOI] [PubMed] [Google Scholar]
- 48. Kuhn P. H., Colombo A. V., Schusser B., Dreymueller D., Wetzel S., Schepers U., Herber J., Ludwig A., Kremmer E., Montag D., Muller U., Schweizer M., Saftig P., Brase S., and Lichtenthaler S. F. (2016) Systematic substrate identification indicates a central role for the metalloprotease ADAM10 in axon targeting and synapse function. Elife 10.7554/eLife.12748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Minond D., Lauer-Fields J. L., Cudic M., Overall C. M., Pei D., Brew K., Visse R., Nagase H., and Fields G. B. (2006) The roles of substrate thermal stability and P2 and P1′ subsite identity on matrix metalloproteinase triple-helical peptidase activity and collagen specificity. J. Biol. Chem. 281, 38302–38313 [DOI] [PubMed] [Google Scholar]
- 50. Schlage P., and auf dem Keller U. (2015) Proteomic approaches to uncover MMP function. Matrix Biol. 44, 232–238 [DOI] [PubMed] [Google Scholar]
- 51. Schjoldager K. T., Vester-Christensen M. B., Goth C. K., Petersen T. N., Brunak S., Bennett E. P., Levery S. B., and Clausen H. (2011) A systematic study of site-specific GalNAc-type O-glycosylation modulating proprotein convertase processing. J. Biol. Chem. 286, 40122–40132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Khokha R., Murthy A., and Weiss A. (2013) Metalloproteinases and their natural inhibitors in inflammation and immunity. Nat. Rev. Immunol. 13, 649–665 [DOI] [PubMed] [Google Scholar]
- 53. Khetarpal S. A., Schjoldager K. T., Christoffersen C., Raghavan A., Edmondson A. C., Reutter H. M., Ahmed B., Ouazzani R., Peloso G. M., Vitali C., Zhao W., Somasundara A. V., Millar J. S., Park Y., Fernando G., et al. (2016) Loss of function of GALNT2 lowers high-density lipoproteins in humans, nonhuman primates, and rodents. Cell Metab. 24, 234–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cavalli M., Pan G., Nord H., and Wadelius C. (2016) Looking beyond GWAS: allele-specific transcription factor binding drives the association of GALNT2 to HDL-C plasma levels. Lipids Health Dis. 15, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Osuala K., Baker C. N., Nguyen H. L., Martinez C., Weinshenker D., and Ebert S. N. (2012) Physiological and genomic consequences of adrenergic deficiency during embryonic/fetal development in mice: impact on retinoic acid metabolism. Physiol. Genomics 44, 934–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Maqbool A., Hall A. S., Ball S. G., and Balmforth A. J. (1999) Common polymorphisms of β1-adrenoceptor: identification and rapid screening assay. Lancet 353, 897. [DOI] [PubMed] [Google Scholar]
- 57. Börjesson M., Magnusson Y., Hjalmarson A., and Andersson B. (2000) A novel polymorphism in the gene coding for the β1-adrenergic receptor associated with survival in patients with heart failure. Eur. Heart J. 21, 1853–1858 [DOI] [PubMed] [Google Scholar]
- 58. Levin M. C., Marullo S., Muntaner O., Andersson B., and Magnusson Y. (2002) The myocardium-protective Gly-49 variant of the β1-adrenergic receptor exhibits constitutive activity and increased desensitization and down-regulation. J. Biol. Chem. 277, 30429–30435 [DOI] [PubMed] [Google Scholar]
- 59. Ahles A., and Engelhardt S. (2014) Polymorphic variants of adrenoceptors: pharmacology, physiology, and role in disease. Pharmacol. Rev. 66, 598–637 [DOI] [PubMed] [Google Scholar]
- 60. Wandall H. H., Hassan H., Mirgorodskaya E., Kristensen A. K., Roepstorff P., Bennett E. P., Nielsen P. A., Hollingsworth M. A., Burchell J., Taylor-Papadimitriou J., and Clausen H. (1997) Substrate specificities of three members of the human UDP-N-acetyl-α-d-galactosamine:Polypeptide N-acetylgalactosaminyltransferase family, GalNAc-T1, -T2, and -T3. J. Biol. Chem. 272, 23503–23514 [DOI] [PubMed] [Google Scholar]
- 61. Schjoldager K. T., Vester-Christensen M. B., Bennett E. P., Levery S. B., Schwientek T., Yin W., Blixt O., and Clausen H. (2010) O-Glycosylation modulates proprotein convertase activation of angiopoietin-like protein 3: possible role of polypeptide GalNAc-transferase-2 in regulation of concentrations of plasma lipids. J. Biol. Chem. 285, 36293–36303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yang Z., Wang S., Halim A., Schulz M. A., Frodin M., Rahman S. H., Vester-Christensen M. B., Behrens C., Kristensen C., Vakhrushev S. Y., Bennett E. P., Wandall H. H., and Clausen H. (2015) Engineered CHO cells for production of diverse, homogeneous glycoproteins. Nat. Biotechnol. 33, 842–844 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.