Abstract
Fungal cell walls contain β-glucan polysaccharides that stimulate immune responses when recognized by host immune cells. The fungal pathogen Histoplasma capsulatum minimizes detection of β-glucan by host cells through at least two mechanisms: concealment of β-glucans beneath α-glucans and enzymatic removal of any exposed β-glucan polysaccharides by the secreted glucanase Eng1. Histoplasma yeasts also secrete the putative glucanase Exg8, which may serve a similar role as Eng1 in removing exposed β-glucans from the yeast cell surface. Here, we characterize the enzymatic specificity of the Eng1 and Exg8 proteins and show that Exg8 is an exo-β1,3-glucanase and Eng1 is an endo-β1,3-glucanase. Together, Eng1 and Exg8 account for nearly all of the total secreted glucanase activity of Histoplasma yeasts. Both Eng1 and Exg8 proteins are secreted through a conventional secretion signal and are modified post-translationally by O-linked glycosylation. Both glucanases have near maximal activity at temperature and pH conditions experienced during infection of host cells, supporting roles in Histoplasma pathogenesis. Exg8 has a higher specific activity than Eng1 for β1,3-glucans; yet despite this, Exg8 does not reduce detection of yeasts by the host β-glucan receptor Dectin-1. Exg8 is largely dispensable for virulence in vivo, in contrast to Eng1. These results show that Histoplasma yeasts secrete two β1,3-glucanases and that Eng1 endoglucanase activity is the predominant factor responsible for removal of exposed cell wall β-glucans to minimize host detection of Histoplasma yeasts.
Keywords: cell wall, fungi, macrophage, microbial pathogenesis, polysaccharide, Histoplasma, β-glucan, glucanase
Introduction
The dimorphic fungal pathogen Histoplasma capsulatum causes respiratory and systemic disease in mammals. Central to the pathobiology of Histoplasma is the ability of pathogenic yeasts to infect and parasitize host phagocytes (1). The cell wall is the first fungal structure encountered by host phagocytes, and thus features of the cell wall contribute to the outcome of the interaction between pathogen and host. Histoplasma's cell wall-localized Hsp60 protein drives internalization of the yeasts via CD11 family complement receptors (e.g. CR3 (2–4)) while limiting the host response (5–7). Even though this interaction is beneficial to yeasts in facilitating host cell infection, the close association of yeasts with the phagocyte plasma membrane can also activate host receptors that trigger immune responses. Given the polysaccharide nature of the fungal cell wall, many of these pathogen pattern-recognition receptors act as lectins. For example, Dectin-1 is the primary receptor for fungal β-glucans, and Dectin-1 recognition of this polysaccharide stimulates pro-inflammatory responses (8–11) leading to control and elimination of fungal cells. To reduce its detection by Dectin-1, Histoplasma yeasts produce an α-linked glucan polysaccharide that overlays the β-glucans, effectively masking the β-glucan from Dectin-1 (12–14). In addition, Histoplasma yeasts secrete a number of glycanases that can potentially remove exposed immunostimulatory polysaccharides from the yeast cell surface (15, 16).
Two of the glycanases abundantly secreted by Histoplasma are Eng1 and Exg8 (16). Both proteins are highly secreted by yeast cells, but not by avirulent mycelial phase cells, suggesting functional roles in pathogenesis. Histoplasma Eng1 belongs to the glycosyl hydrolase family 81 (GH81), which includes the Eng1 homologs of Candida albicans and Saccharomyces cerevisiae. In C. albicans and S. cerevisiae, Eng1 enzymes participate in cell separation during budding (17, 18). However, in Histoplasma, Eng1 does not contribute to cell separation and instead removes surface-exposed β-glucan molecules to reduce Dectin-1 detection of yeast cells (15). Histoplasma Exg8 is member of the GH55 family. The S. cerevisiae and C. albicans genomes do not contain any genes encoding GH55 proteins, but Aspergillus fumigatus encodes six GH55 proteins (19), including an Exg8 homolog. However, the cellular role for A. fumigatus Exg8 has not been defined. In this study, we provide biochemical characterization of the Histoplasma Exg8 and Eng1 proteins, defining their glucan substrate specificity and their endo- or exo-directed hydrolytic activity, and we investigate the role for Exg8 in Dectin-1 recognition and Histoplasma virulence.
Results
Eng1 and Exg8 Protein Features
Eng1 and Exg8 were identified initially as members of the secreted proteome of Histoplasma yeasts, the genes of which were transcriptionally up-regulated in pathogenic phase yeasts compared with avirulent mycelia (16). The ENG1 gene encodes a GH81 family protein with β-1,3-glucanase activity (supplemental Fig. 1A) (15). The Histoplasma Eng1 protein has high amino acid similarity with other fungal endoglucanases (60% similar to A. fumigatus Engl1), with a series of conserved aspartic acid and glutamic acid residues known to be important for activity (supplemental Fig. 1A) (20). The EXG8 gene encodes a protein that is most similar to the A. fumigatus Exg8 protein (69% amino acid similarity) and is predicted to be in the GH55 family. Similar to other proteins in this family, the Histoplasma Exg8 amino acid sequence contains two pectate lyase domains (supplemental Fig. 1B), as well as a conserved aspartic acid and two conserved glutamic acid residues important for glucanase activity in this family (supplemental Fig. 1B) (21, 22). Both Eng1 and Exg8 have predicted N-terminal secretion signals and N-linked and O-linked mucin-like glycan attachment sites (supplemental Fig. 1, A and B) consistent with extracellular localization.
To determine whether these two proteins are glycosylated, culture filtrates from Histoplasma expressing Eng1-FLAG and Exg8-FLAG fusion proteins were treated with PNGaseF (for removal of N-linked glycans) and analyzed by anti-FLAG immunoblot for a reduction in molecular weight due to glycan removal (Fig. 1). The electrophoretic mobility of Eng1 and Exg8 did not change after PNGaseF treatment, suggesting the Eng1 and Exg8 proteins lack N-linked glycosylation. To test for O-linked glycosylation, Eng1 and Exg8 were expressed in a Histoplasma strain lacking an O-linked protein mannosyltransferase (Pmt2), which is required for the O-linked addition of the initial mannose onto proteins.4 When expressed in this strain, both Eng1 and Exg8 have reduced electrophoretic mobility consistent with a reduction in molecular weight due to loss of O-linked glycans. The 99-kDa Eng1 protein migration is reduced to 89 kDa when lacking O-glycans. Similarly, the apparent molecular mass of Exg8 is reduced from 77 to 73 kDa when Pmt2-dependent addition of O-glycans is lost. There was no further reduction in molecular weight when these O-linked glycan-deficient proteins were treated with PNGaseF. Together, these data indicate both Eng1 and Exg8 are O-glycosylated but not N-glycosylated (Fig. 1).
FIGURE 1.
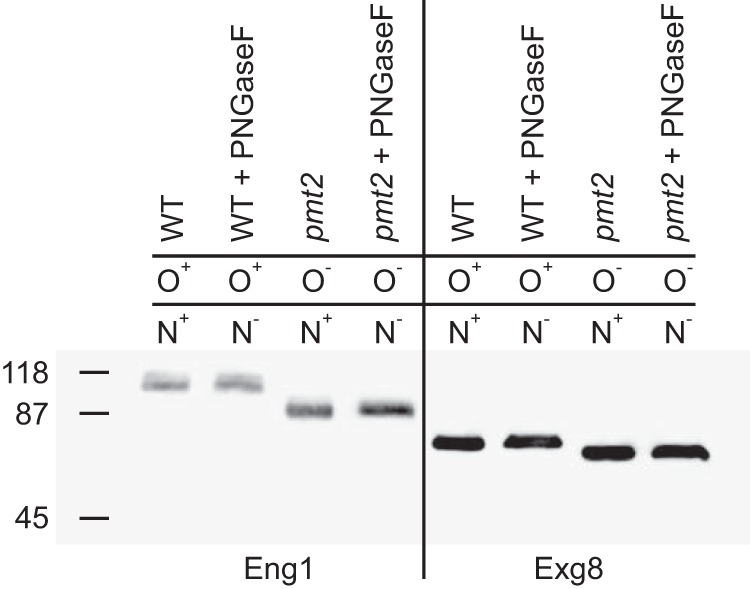
Eng1 and Exg8 have O-linked but not N-linked glycans. Glycan attachment to Eng1 and Exg8 proteins was determined by increased electrophoretic mobility of proteins when O-linked or N-linked glycans were absent. Culture filtrates from Histoplasma expressing Eng1-FLAG- or Exg8-FLAG-tagged proteins were prepared, and the Eng1 and Exg8 proteins were detected by immunoblotting for the FLAG epitope following electrophoretic separation. To prevent O-linked glycan attachment, fusion proteins were expressed in a mutant lacking O-linked mannosylation (pmt2) instead of wild type (WT). N-Linked glycans were removed by treatment of culture filtrate proteins with PNGaseF. The prevention of O-linked glycan attachment or removal of N-linked glycans are indicated by “−” in contrast to normal presence of the glycans (“+”).The molecular mass (kDa) of protein standards is indicated on the left of the immunoblot.
Eng1 and Exg8 are β1,3-Glucanases
We previously showed Histoplasma Eng1 can hydrolyze β1,3-glucan (15). To determine the specificity of Eng1 for β1,3-linked glucan and to determine the activity of Exg8, we quantified the hydrolytic activities of Eng1 and Exg8 on polysaccharides with different glycosidic linkages (Fig. 2). The polysaccharides tested include β1,3-glucan (laminarin), β1,6-glucan (pustulan), β1,3;1,6-glucan (scleroglucan), β1,4-glucose (cellulose), α1,4-glucan (amylose), α1,6-glucan (dextran), α1,4;1,6-glucan (pullulan), α1,2-mannose (mannan), and β1,4-N-acetylglucosamine (chitin). Substrates were incubated with purified Histoplasma Eng1 and Exg8 proteins, and the released reducing sugars were measured by reaction with dinitrosalicylic acid (23). Glycanase activity was below the limit of detection after 120 min of incubation for all substrates except laminarin (Fig. 2). Extending the reaction time to 24 h still did not reveal any significant glycanase activity for all substrates except laminarin (data not shown), indicating Eng1 and Exg8 enzymatic activities are specific to β1,3-linked glucans. The specific activity of Exg8 was almost 2-fold higher than Eng1 (Table 1). Exg8 was also more efficient than Eng1, as the Kcat for Exg8 was 2.7-fold higher than Eng1, and the Kcat/Km was 4.4-fold higher because of the greater substrate affinity of Exg8 for β1,3-glucan.
FIGURE 2.
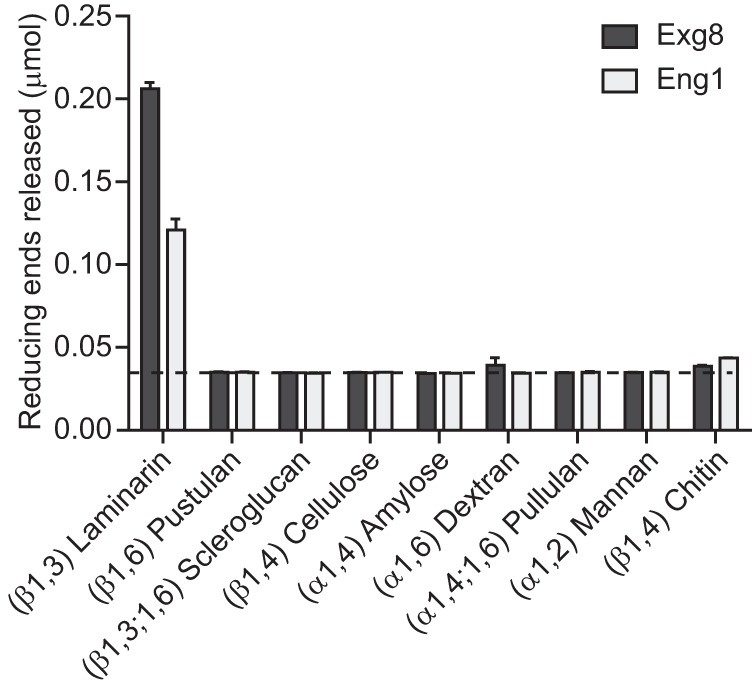
Eng1 and Exg8 are glucanases specific for β1,3-glucosidic linkages. Glucan hydrolysis activity of purified Exg8 (dark bars) and Eng1 (light bars) is shown. Data represent the glucose or N-acetylglucosamine (for chitin substrate) equivalents (micromoles) of reducing sugars released after incubation of Eng1 or Exg8 with glycan substrates. Glycosidic linkages of the individual substrates are indicated in parentheses. Dashed line represents the limit of detection, and error bars represent the standard deviation among replicate reactions (n = 3).
TABLE 1.
Enzyme kinetics for Histoplasma Eng1 and Exg8
Steady-state kinetic parameters were determined for purified Eng1 and Exg8 hydrolyses of laminarin substrate. For each, two independent protein purifications were assayed by incubation with laminarin substrate and quantification of the reducing sugars released. Vmax, Km, and Kcat values were determined by curve fitting of the data obtained from two different enzyme concentrations while varying the laminarin substrate concentration.
| Vmax | Km | Kcat | Kcat/Km | |
|---|---|---|---|---|
| units/mg | mg/ml | 1/s | ml/(mg s) | |
| Eng1 | 17.9 ± 1.1 | 1.62 ± 0.38 | 28.2 ± 1.7 | 17.3 ± 4.4 |
| Exg8 | 48.1 ± 2.5 | 1.00 ± 0.24 | 75.8 ± 3.9 | 75.8 ± 16.4 |
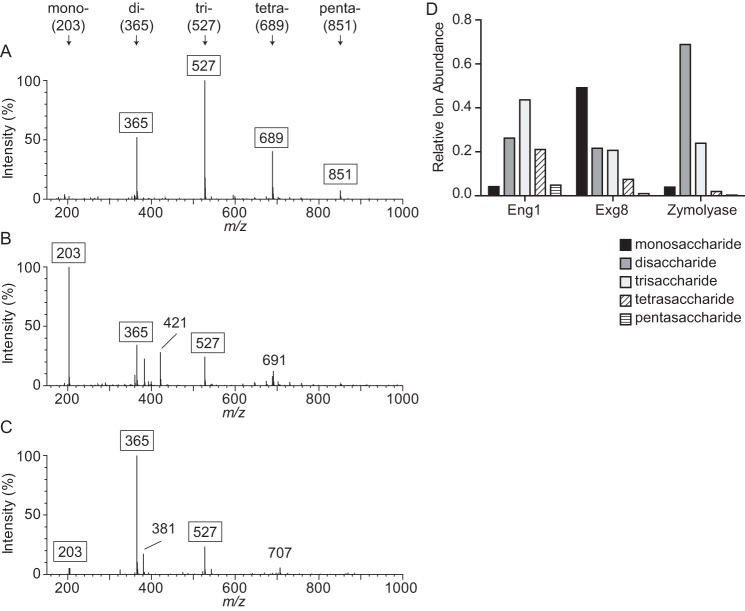
Of the characterized GH81 family proteins, all are endo-acting glucanases; however, enzymes in the GH55 family can have either endo- or exoglucanase activity (24). To validate Eng1 as an endoglucanase and to determine the action of Histoplasma Exg8, laminarin was digested for 30 min with purified enzymes, and the mono- and oligosaccharide products were analyzed by mass spectrometry (Fig. 3, A and B). The major products identified were monosaccharides (203 m/z), disaccharides (365 m/z), trisaccharides (527 m/z), tetrasaccharides (689 m/z), and pentasaccharides (851 m/z). Eng1 produced predominantly disaccharides, as well as some tri-, tetra-, and pentasaccharides (Fig. 3A). Very little monosaccharide was produced, consistent with the enzymatic activity acting on internal glycosidic linkages. Extending the digestion time overnight reduced the tetrasaccharide product, leaving predominantly di- and trisaccharides with a very minor amount of mono-, tetra-, and pentasaccharide products (data not shown). This pattern was quite similar to digestion of laminarin with the known endoglucanase zymolyase (Fig. 3C). Conversely, the predominant product of Exg8 hydrolysis of laminarin was monosaccharides, with minor portions of di- and trisaccharide (Fig. 3B). This pattern is consistent with Exg8 being able to hydrolyze terminal glucose from a glucan chain, indicating exoglucanase activity. Extending digestion of Exg8 overnight results in a similar profile of predominantly monosaccharides with di- and trisaccharides still present, suggesting inefficient hydrolysis of glucose oligomers smaller than 4 units in length (data not shown). To quantify the Eng1 and Exg8 digestion products, the ion intensity of the [M + Na]+ ion for each saccharide length was calculated relative to the sum of all [M + Na]+ ion intensities (monosaccharide to octosaccharide; Fig. 3D). For Exg8, 49% of the saccharide ions were monosaccharides, compared with only 4% for Eng1 and zymolyase. No quantifiable saccharides longer than 5 units were detected for any enzyme digest after 30 min, expect for zymolyase in which hexasaccharides were calculated at less than 1% of the total saccharide content. These data establish that Eng1 is an endoglucanase and Exg8 is an exoglucanase acting on β1,3-glucan.
FIGURE 3.
Eng1 is an endoglucanase and Exg8 is an exoglucanase. Hydrolysis products of Eng1 and Exg8 activity on laminarin (β1,3-glucan) are shown. Laminarin was digested for 30 min with 30 ng of purified Eng1 (A), Exg8 (B), or the endo-β-glucanase zymolyase (C), and the products were detected by mass spectrometry. The relative ion intensities for ions between 150 and 1000 m/z are shown with peaks for monosaccharides (mono-, 203 m/z), disaccharides (di-, 365 m/z), trisaccharides (tri-, 527 m/z), tetrasaccharides (tetra-, 689 m/z), and pentasaccharides (penta-, 851 m/z) indicated with arrows. The m/z values for each saccharide peak represent sodiated glucose minus one water per each additional glucose unit. D, relative ion intensities for singly charged monosaccharide through pentasaccharide peaks. Data shown are representative of three independent experiments.
Eng1 and Exg8 Are the Major Secreted Histoplasma Glucanases
Examination of the Histoplasma genome identified additional glycosyl hydrolase family proteins in addition to Eng1 and Exg8 (data not shown) raising the possibility of additional secreted glucanases. To determine the relative contribution of Eng1 and Exg8 to the total extracellular glucanase activity of Histoplasma yeasts, we depleted Eng1 and Exg8 activity from Histoplasma yeasts either singly or combined using a gfp sentinel RNAi system (25); Eng1 and/or Exg8 knockdown was monitored by simultaneous knockdown of GFP fluorescence. Transformed yeasts showed at least 85% reduction in GFP fluorescence indicating efficient depletion of the co-targeted ENG1 and/or EXG8 transcripts (data not shown). With depletion lines established, the contribution of each enzyme to the extracellular glucanase activity on β1,3-glucan was determined. Loss of Eng1 function yielded culture filtrates that had 30% less β-glucanase activity compared with wild-type culture filtrates (derived from gfp-RNAi) (Fig. 4A), consistent with previous results (15). Loss of Exg8 function caused a greater reduction in extracellular glucanase activity (66% reduction; Fig. 4A). Simultaneous depletion of both Eng1 and Exg8 depletes 93% of extracellular β-glucanase activity, showing Eng1 and Exg8 constitute the major extracellular β-glucanases. Examination of the β-glucanase activity associated with the surface of Histoplasma yeasts showed slight activity above the limit of detection with wild-type yeasts, and this small amount of activity decreased a statistically significant amount when Exg8 but not Eng1 was depleted from Histoplasma (Fig. 4A). Thus, the major extracellular β-glucanase activity is in the soluble fraction with little associated with the surface of yeast cells. Consistent with this conclusion, abundant Exg8 protein was detected in the culture filtrate; however, only a minute amount of Exg8 protein was in the cell wall fraction (Fig. 4B). Together with the previous demonstration that Eng1 is not associated with the surface of yeast cells (15), these data indicate that Eng1 and Exg8 are responsible for the vast majority of extracellular β-glucanase activity and that this activity is secreted but not associated with the yeast cells themselves.
FIGURE 4.
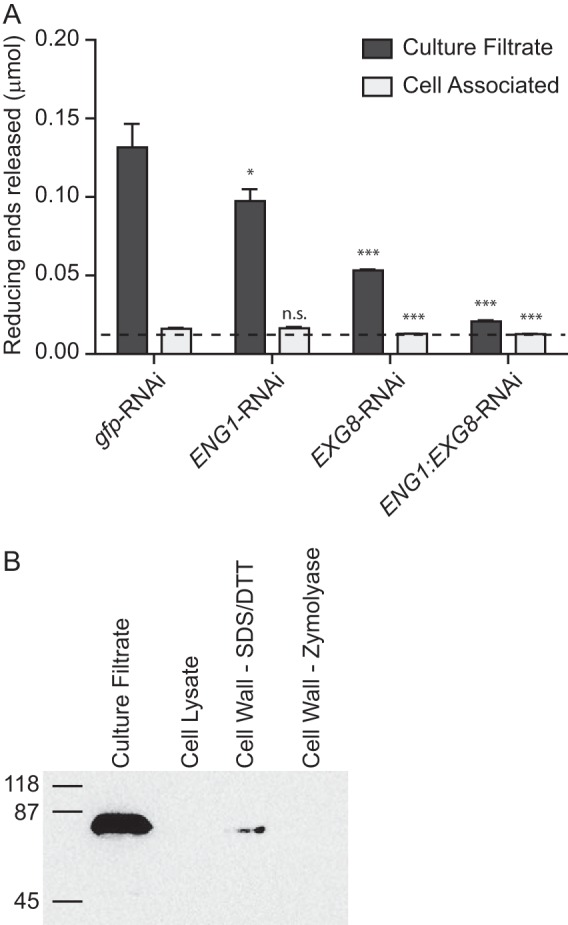
Eng1 and Exg8 are responsible for the majority of extracellular glucanase activity produced by yeasts. A, extracellular glucanase activity in the soluble fraction or associated with yeast cells. Culture supernatants were separated from yeast cells, and both fractions were tested for hydrolysis of laminarin to determine the activity in culture filtrates (dark bars) or activity associated with the yeast cells (light bars) of wild-type (gfp-RNAi) or glucanase-deficient strains (ENG1-RNAi, EXG8-RNAi, and ENG1/EXG8-RNAi strains). Data represent the glucose equivalents (micromoles) of reducing sugars released after 90 min of incubation with laminarin. Dashed line represents the limit of detection of the assay, and bars represent the mean reducing sugars released ± S.D. among replicate assays (n = 3). B, localization of Exg8 in extracellular fractions. Soluble and cell-associated fractions were tested for Exg8-FLAG protein by immunoblot of the FLAG epitope. Proteins in the culture filtrate, the soluble yeast cell lysate, and proteins extracted by SDS + DTT or by zymolyase + chitinase digestion of the insoluble cell wall fraction were separated by electrophoresis and probed for the FLAG epitope. Protein samples were derived from equivalent numbers of yeast cells. Numbers represent the molecular mass (kDa) of protein standards. n.s., not significant. *, p < 0.05; ***, p < 0.001.
Despite the reduction in secreted glucanase activity, depletion of Exg8 or depletion of Exg8 and Eng1 together does not increase sensitivity to cell wall-disrupting agents (Table 2); EXG8-RNAi and ENG1:EXG8-RNAi Histoplasma yeasts had no significant change in the 50% inhibitory concentrations (IC50) for cell wall-binding dyes (Congo Red, Uvitex, and Calcofluor White), detergent (SDS), and cell wall glycan-targeting antifungal drugs (Nikkomycin Z and Caspofungin, which impair chitin and β-glucan synthesis, respectively). These results suggest Exg8, like Eng1 (15), is not a glucanase required for major structural modifications or cell intrinsic function of the cell wall.
TABLE 2.
Sensitivity of glucanase-deficient Histoplasma yeasts to cell wall-destabilizing compounds
The mean concentration for 50% inhibition of yeast growth (IC50) ± S.D. among replicate assays (n = 3) are listed. The following abbreviations are used: CFW, Calcofluor White; CR, Congo red; Uvtx; Uvitex 3BSA; Cfg, caspofungin; Flc, fluconazole; NikZ, nikkomycin Z.
| CFW | CR | Uvtx | SDSa | Cfn | Flc | NikZ | |
|---|---|---|---|---|---|---|---|
| μg/ml | μg/ml | m % | m % | μg/ml | μg/ml | μg/ml | |
| gfp-RNAi | 6.1 ± 2.4 | 1.3 ± 0.1 | 11.0 ± 1.9 | 3.1 ± 1.5 | 10.9 ± 4.2 | 0.68 ± 0.07 | 4.3 ± 1.2 |
| EXG8-RNAi | 5.2 ± 2.0 | 1.1 ± 0.2 | 11.3 ± 0.3 | 4.8 ± 0.2 | 11.4 ± 4.5 | 0.52 ± 0.01 | 5.1 ± 0.4 |
| ENG1/EXG8-RNAi | 5.7 ± 1.8 | 1.1 ± 0.2 | 10.6 ± 1.5 | 4.4 ± 0.6 | 11.7 ± 4.7 | 0.59 ± 0.24 | 4.8 ± 1.6 |
a Concentrations were expressed as milli-percent by volume (m %).
Eng1 and Exg8 Are Functional in Temperature and pH Conditions of the Phagosome
The pathogenic phase-specific expression of Eng1 and Exg8 suggests these glycans have a function related to pathogenesis. Histoplasma yeasts normally reside within the phagosomal compartment of host macrophages at mammalian temperatures. While in this compartment, the Histoplasma yeast prevents complete acidification and maintains a luminal pH around 5.5 to 6.0 (26–28). To determine whether Eng1 and Exg8 are enzymatically active under conditions that Histoplasma yeasts experience during infection of macrophages, we quantified the hydrolysis of laminarin at various pH and temperatures (Fig. 5, A and B). With respect to pH, Eng1 had maximal activity from pH 5 to pH 8, decreasing only at high alkalinity (pH > 10) and acidity (pH < 3) (Fig. 5A). Exg8 is active from pH 4 to pH 8 with optimal activity at pH 4 to pH 7 (Fig. 5A). Increasing temperature continues to increase activity up to 45 °C for both enzymes (Fig. 5B). At temperatures above 45 °C, Exg8 activity decreases significantly; however, Eng1 activity remains high up to 70 °C. The high glucanase activity at 37 °C and under moderately acidic conditions (pH 5 to 7) indicates Eng1 and Exg8 would be active in the environment within the phagosome.
FIGURE 5.
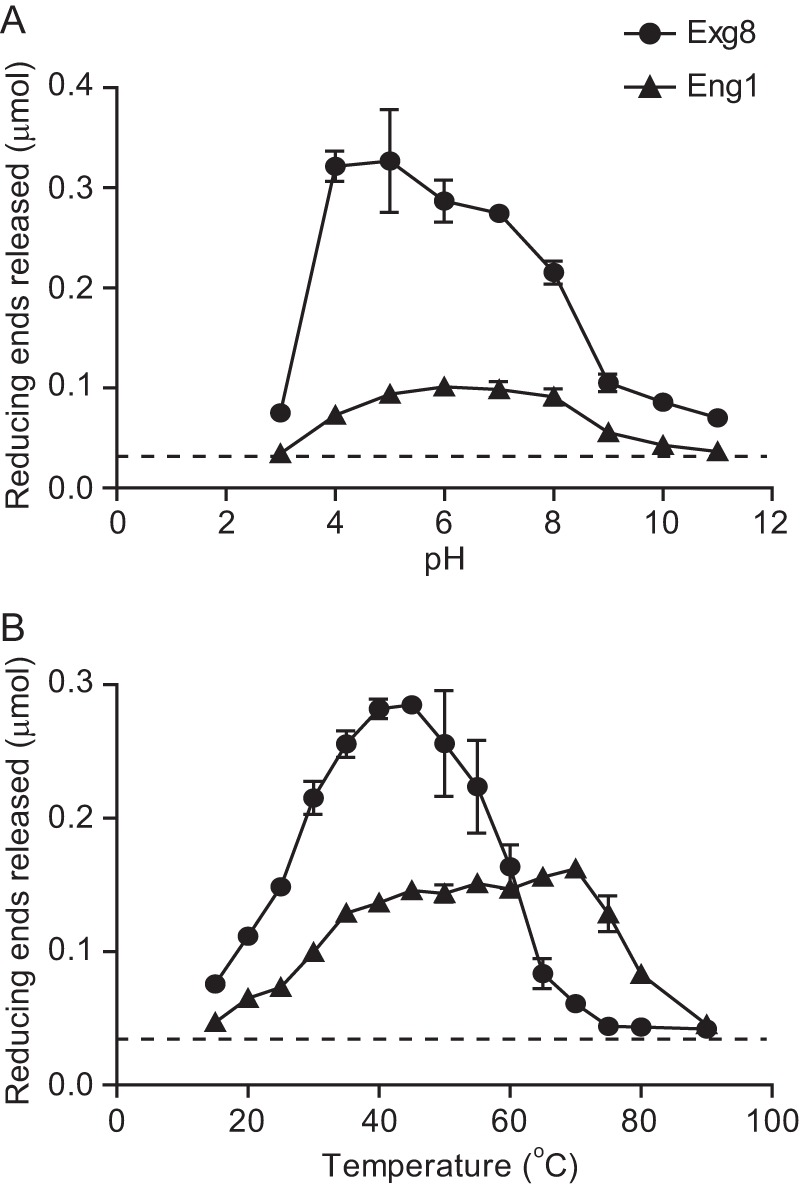
Eng1 and Exg8 are active under phagosome-like conditions. Glucanase activity is shown for Eng1 (circles) and Exg8 (triangles) at different ranges of pH (A) and temperature (B). Data represent reducing sugar equivalents of glucose released from laminarin after digestion with purified Eng1 or Exg8 for 2 h. Dashed line represents the limit of detection. Data points are the mean amount of reducing sugars released, and error bars represent the standard deviation among replicates (n = 3). Data are representative of two independently purified protein samples of Eng1 and Exg8.
Exg8 Does Not Reduce Detection of Histoplasma Yeasts
To determine the functional and possible additive roles of Eng1 and Exg1 in virulence, we tested yeasts depleted of Eng1 and/or Exg1 in aspects of Histoplasma pathogenesis. Overall association of yeasts with primary macrophages was unchanged in the absence of either glucanase (Fig. 6A). The response of macrophages to yeasts, as measured by production of the pro-inflammatory cytokines TNFα and IL-6, was unaffected by loss of Exg8; however, loss of Eng1 singly or in combination with Exg8 significantly increased both TNFα and IL-6 (Fig. 7, A and B). This is consistent with Eng1 but not Exg8 affecting recognition of yeasts by signaling receptors but not binding of yeasts to phagocytic receptors on macrophages (i.e. CR3 (2–4)). We previously showed that the Eng1 glucanase activity decreases surface β1,3-glucan exposure on yeasts, thereby enabling yeasts to avoid Dectin-1-dependent recognition by macrophages (15). As both Eng1 and Exg8 are active on β1,3-glucan, we measured the Dectin-1 recognition of yeasts lacking single glucanases or both. To avoid the contribution of other macrophage receptors, yeasts were added to fibroblasts in which only Dectin-1 was expressed. Despite Exg8 having more activity than Eng1, loss of Exg8 had no significant effect on β1,3-glucan recognition, whereas the loss of Eng1 resulted in a nearly 6-fold increase (Fig. 6B). No additional change was seen when both Eng1 and Exg8 were depleted. In the G186A background, in which α-glucan polysaccharides largely mask β-glucans (12, 14), loss of Exg8 similarly had negligible effects on Dectin-1 recognition of yeasts (supplemental Fig. 2). In this same background, depletion of Eng1 results in a 4-fold increase. Again, no additional β1,3-glucans are recognized when both Eng1 and Exg8 are depleted. Pre-incubation of Dectin-1-expressing fibroblasts with laminarin reduced the recognition of yeasts at least 10-fold, confirming the specificity as recognition of yeast β-glucan (Fig. 6B). Together, these data show that Eng1 but not Exg8 reduces β-glucan exposure on yeasts, which minimizes their detection by Dectin-1.
FIGURE 6.
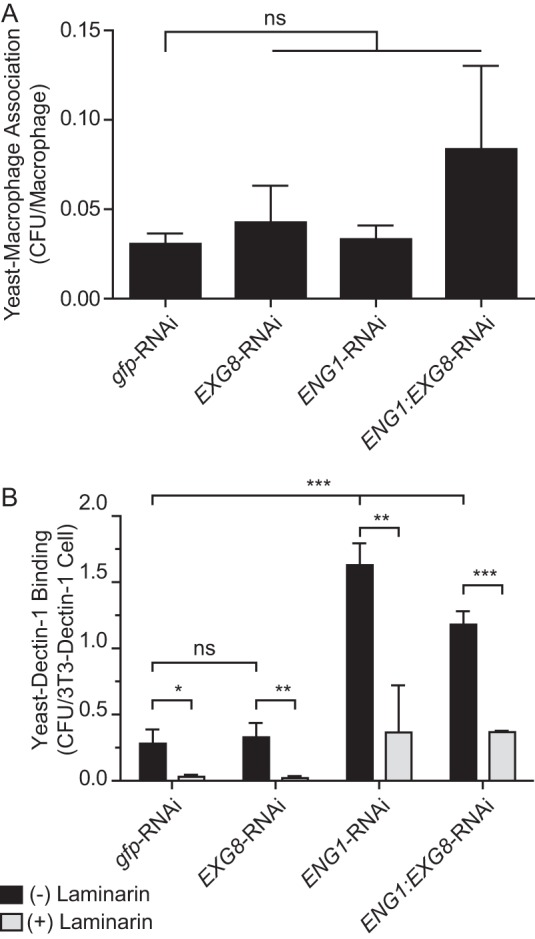
Eng1 and Exg8 do not affect yeast-macrophage association but Eng1 reduces yeast β-glucan recognition. A, association of yeast with macrophages. Yeasts lacking Exg8 (EXG8-RNAi), Eng1 (ENG1-RNAi), or both Exg8 and Eng1 (ENG1:EXG8-RNAi) were added to peritoneal macrophages (m.o.i. 0.5:1), and association after 2 h was determined by plating yeasts released from macrophages onto solid medium for cfu enumeration. B, relative recognition of glucanase-deficient Histoplasma yeasts by Dectin-1. Yeasts were added to Dectin-1-expressing 3T3 fibroblasts in the absence (black bars) or presence (gray bars) of the β-glucan competitor laminarin (1 mg/ml). Adherent yeasts were quantified by cfu determination. Data represent the mean cell-associated yeast per 2 × 105 macrophages (A) or 2 × 105 Dectin-1-expressing fibroblast (B) cells with error bars indicating standard deviation among replicate assays (n = 3). Asterisks indicate statistically significant differences in assay results between wild-type (gfp-RNAi) and glucanase-deficient yeasts as determined by Student's t test (*, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, not significant).
FIGURE 7.
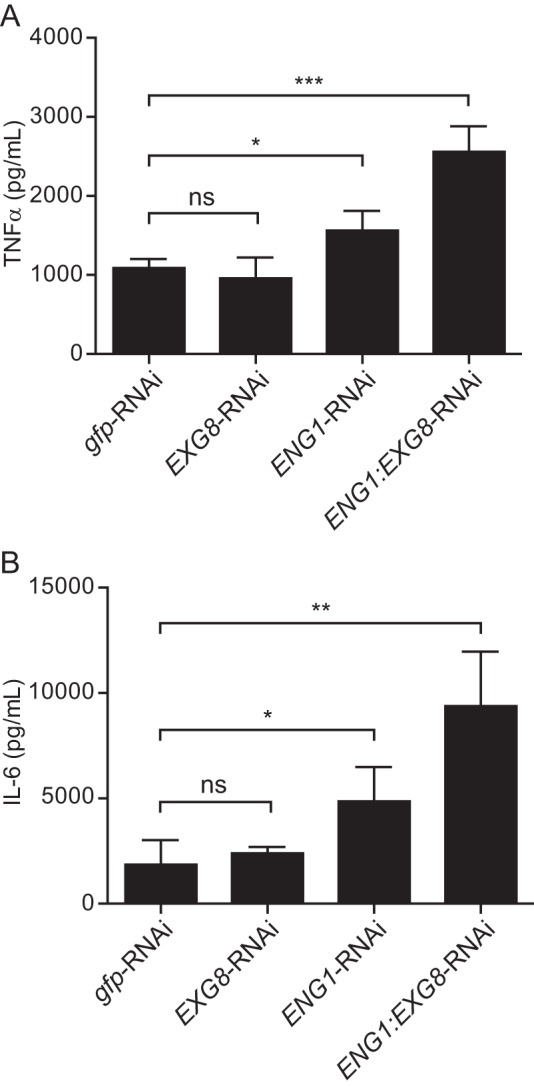
Eng1 but not Exg8 affects the macrophage proinflammatory cytokine response to Histoplasma. Macrophage production of TNFα (A) and IL-6 (B) 8 h after infection with wild-type (gfp-RNAi), Exg8-, Eng1-, or Eng1-Exg8 glucanase-deficient yeasts (m.o.i. 0.5:1). Cytokines were measured by ELISA and levels determined by comparison with cytokine standards. Asterisks indicate statistically significant differences between wild-type and glucanase-deficient yeasts as determined by Student's t test (ns, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001).
Eng1 Is the Primary Secreted Glucanase That Enhances Histoplasma Pathogenesis
Yeast survival against macrophages in culture was not affected by loss of either glucanase (Fig. 8A). In a murine model of respiratory histoplasmosis, loss of Exg8 function showed only a slight decrease (2-fold) in lung infection (Fig. 8B). Removal of Eng1, however, resulted in an 8-fold reduction compared with wild type (Fig. 8B). Similar to the in vitro effects on Dectin-1 binding, depletion of Exg8 did not further decrease the virulence of yeasts from that caused by single loss of Eng1 function. Thus, the Eng1 endoglucanase functionally contributes more than Exg8 to reducing the β1,3-glucan exposure of yeasts and facilitating infection.
FIGURE 8.
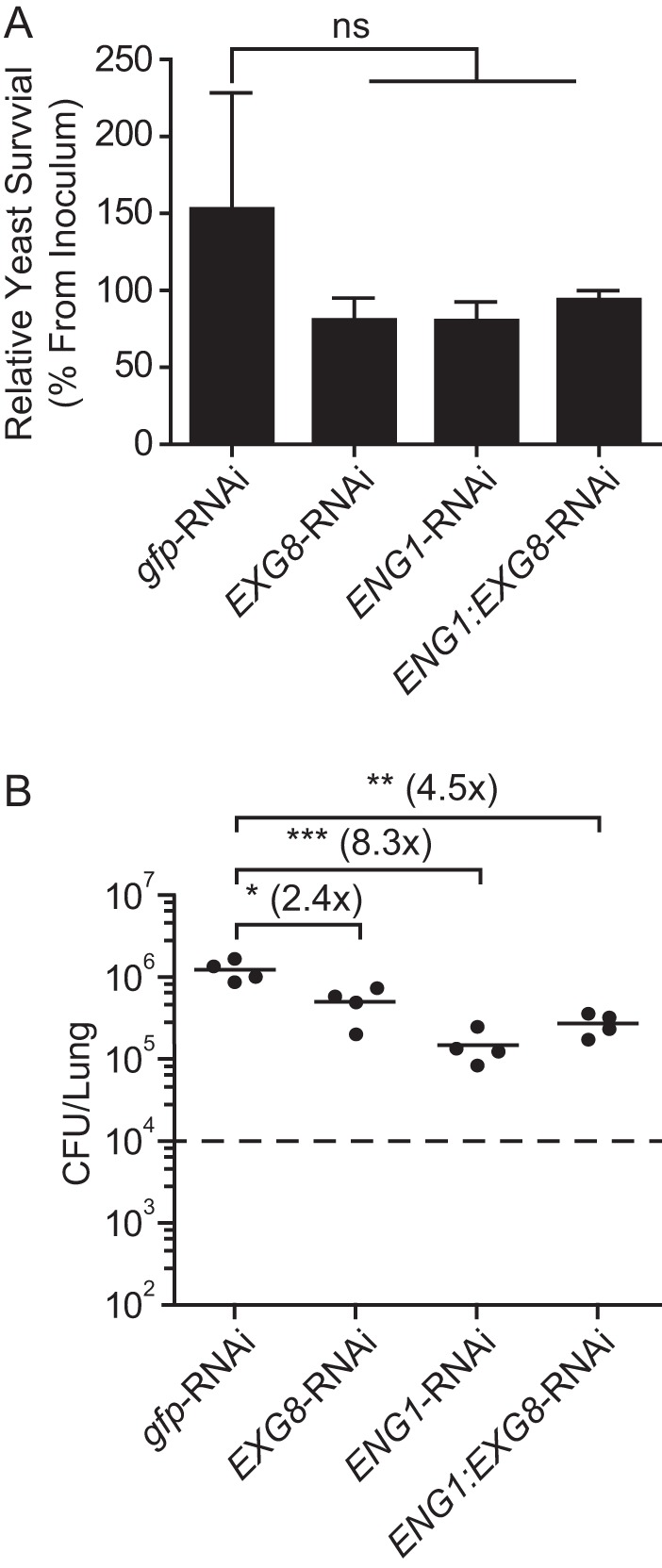
Eng1 but not Exg8 is the major glucanase facilitating Histoplasma virulence. A, survival of yeast following infection of peritoneal macrophages. Total viable yeasts (cfu) were quantified 8 h after infection with wild-type (gfp-RNAi), Exg8-, Eng1-, or Eng1-Exg8 glucanase-deficient yeasts and compared with the cfu of yeasts in the absence of macrophages. B, Histoplasma burden in the lungs 8 days following respiratory infection with glucanase-producing (gfp-RNAi) and single glucanase-deficient yeasts (EXG8-RNAi and ENG1-RNAi) or yeasts lacking both glucanases (ENG1:EXG8-RNAi). Data points represent the cfu counts from individual mouse lungs (n = 4), and horizontal bars indicate the mean fungal burden. The dashed horizontal line indicates the inoculum given. Fold change of the mean cfu value from wild type is indicated. Asterisks indicate statistically significant differences between wild-type and glucanase-deficient yeasts as determined by Student's t test (ns, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001).
Discussion
The fungal cell wall features centrally in the pathogenesis of H. capsulatum. Unlike many of the opportunistic fungal pathogens, Histoplasma yeast cells have reduced detection of cell wall pathogen-associated molecular patterns (14, 15). Although the cell wall is essential for fungal viability, Histoplasma yeasts effectively minimize cell wall β-glucans that are exposed to host phagocytes, either through masking or by removal from the exposed surface. This study characterizes two secreted glucanases (Eng1 and Exg8) important for this latter mechanism. Eng1 is important for pruning exposed β-glucans (15), and consistent with this role, we demonstrate that Eng1 is an endo-acting glucanase specific for β1,3-glucans (EC 3.2.1.39). Exg8 is an exo-acting glucanase, also specific for β1,3-glucans (EC 3.2.1.58). Together, the action of these enzymes can effectively hydrolyze β1,3-glucans into monomeric glucose, and removal of both enzymes from Histoplasma effectively eliminates extracellular glucanase activity. The enzymes are highly active in conditions that Histoplasma yeasts experience during infection, 37 °C and pH between 5 and 6, which characterizes the lumen of the host cell phagosome. Thus, Histoplasma Eng1 and Exg8 are factors produced during infection, with the ability to alter the surface of the yeast cell, specifically in reducing immunostimulatory β-glucans of the cell wall.
Although both Eng1 and Exg8 are produced by pathogenic yeasts, Eng1 is the major β-glucanase that removes exposed β-glucans from the yeast cell wall. Functional tests show that loss of Eng1 activity significantly increases Dectin-1 recognition of yeast cell β-glucans, and this translates into reduced virulence in a murine model of respiratory histoplasmosis (15). By comparison, loss of Exg8 activity has only minor effects on Dectin-1 recognition of the yeast cells (Fig. 6B) and minimal impact on Histoplasma virulence (Fig. 8B). Although Eng1 reduces recognition of yeasts by Dectin-1, overall binding of yeasts to macrophages is unaffected by either Eng1 or Exg8. This is not surprising, as association and phagocytosis of Histoplasma yeasts are primarily mediated through binding of yeasts to the complement receptor (2–4). Thus, glucanase removal of β-glucans minimizes immune cell detection of infecting yeasts but preserves invasion mechanisms to bind and enter host macrophages.
In light of the altered Dectin-1 recognition due to Eng1 but not Exg8, and with the demonstrated endo- versus exo-activity of Eng1 and Exg8, respectively, we propose a model in which exposed β-glucans on the Histoplasma cell wall are not simply the exposed ends of glucan chains but instead are polysaccharide “loops” exposed on the surface of the cell wall. In this structure, only an endo-acting glucanase could hydrolyze the β-glucan molecule, whereas an exoglucanase would be ineffective as the exposed β-glucan lacks any terminal ends. Intriguingly, loss of both enzymes is no greater than loss of Eng1 alone with respect to both Dectin-1 recognition and Histoplasma virulence. This indicates hydrolysis of cell wall β-glucans by Eng1 is sufficient to render them undetectable by Dectin-1 without requiring the remaining mono- or disaccharide exposed termini to be hydrolyzed by Exg8. This model fits with the described ligand specificity of Dectin-1 being oligosaccharides of at least seven glucose units (29).
In the environment, secreted glucanases are positioned potentially to hydrolyze polysaccharides of other fungal cell walls, perhaps as a source of glucose that can serve metabolic needs. Although we cannot exclude such a role for Histoplasma Eng1 and/or Exg8, our data indicate that Eng1 and Exg8 have likely evolved to facilitate Histoplasma pathogenesis. First, Eng1 and Exg8 are expressed at high levels by pathogenic yeasts but are only minimally expressed by the environmental mycelial form (16) suggesting Eng1 and Exg8 function specifically in yeast cell biology (i.e. pathogenesis as opposed to the saprobic mycelia). Second, during infection, Histoplasma yeasts are found almost exclusively within the phagosome of host phagocytes in which Histoplasma is the only fungal cell present, making it unlikely that Eng1 and Exg8 act on other fungi. Third, Eng1 and Exg8 have evolved to be compatible with conditions within the host phagosome, whereas glucanase activity is reduced in acidic conditions, common to Histoplasma-containing soils. Finally, Eng1 in particular has been shown to be important for pathogenesis by reducing surface exposure of yeast cell wall β-glucans (15). This does not preclude uptake of released mono- and oligosaccharides by yeasts, but together the biochemical and cellular characteristics support these β-glucanases functioning in Histoplasma pathogenesis, primarily by removing immunostimulatory β-glucans from the yeast cell surface to reduce host detection.
Experimental Procedures
Histoplasma Strains and Culture
H. capsulatum strains were derived from the wild-type clinical isolates G186A (ATCC 26029) and G217B (ATCC 26032) and are listed in Table 3. Histoplasma yeasts were grown in Histoplasma-macrophage medium (HMM)5 (30) and supplemented with 100 μg/ml uracil when necessary. Hemocytometer counts were used for precise enumeration of yeasts for infection studies.
TABLE 3.
Histoplasma strains
| Strain | Genotypea | Other designation |
|---|---|---|
| WU8 | (G186A) ura5-32Δ | WT |
| OSU22 | (G186A) ura5-32Δ zzz::pCR482 [hph, gfp] | RNAi sentinel |
| OSU244 | (G186A) ura5-32Δ zzz::pCR482 [hph, gfp] zzz::pED02 [URA5, gfp-RNAi] | gfp-RNAi |
| OSU245 | (G186A) ura5-32Δ zzz::pCR482 [hph, gfp] zzz::pED06 [URA5, gfp:ENG1-RNAi] | ENG1-RNAi |
| OSU205 | (G186A) ura5-32Δ zzz::pCR482 [hph, gfp] zzz::pAG26 [URA5, gfp:EXG8-RNAi] | EXG8-RNAi |
| OSU214 | (G186A) ura5-32Δ zzz::pCR482 [hph, gfp] zzz::pAG37 [URA5, gfp:ENG1:EXG8-RNAi] | ENG1:EXG8-RNAi |
| WU15 | (G217B) ura5-42Δ | WT |
| OSU194 | (G217B) ura5-42Δ zzz::pAG21 [G418R, gfp] | RNAi sentinel |
| OSU247 | (G217B) ura5-42Δ zzz::pAG21 [G418R, gfp] zzz::pED02 [URA5, gfp-RNAi] | gfp-RNAi |
| OSU248 | (G217B) ura5-42Δ zzz::pAG21 [G418R, gfp] zzz::pED06 [URA5, gfp:ENG1-RNAi] | ENG1-RNAi |
| OSU273 | (G217B) ura5-42Δ zzz::pAG21 [G418R, gfp] zzz:pAG26 [URA5, gfp:EXG8-RNAi] | EXG8-RNAi |
| OSU275 | (G217B) ura5-42Δ zzz::pAG21 [G418R, gfp] zzz:pAG37 [URA5, gfp:ENG1:EXG8-RNAi] | ENG1:EXG8-RNAi |
| OSU265 | (G217B) ura5-42Δ zzz:pQS14 [URA5, PH2B-ENG1:FLAG] | ENG1:FLAG |
| OSU266 | (G217B) ura5-42Δ zzz:pAG39 [URA5, PH2B-ENG1:6xHIS] | ENG1:6xHIS |
| OSU313 | (G217B) ura5-42Δ zzz::pKD02 [URA5, PH2B-EXG8:FLAG] | EXG8:FLAG |
| OSU314 | (G186A) ura5-32Δ zzz::pKD01 [URA5, PH2B-EXG8:6xHIS] | EXG8: HIS6 |
| OSU328 | (G217B) ura5-42Δ pmt2::T-DNA [hph] zzz:pQS14 [URA5, PH2B-ENG1:FLAG] | pmt2;ENG1:FLAG |
| OSU329 | (G217B) ura5-42Δ pmt2::T-DNA [hph] zzz:pKD02 [URA5, PH2B-EXG8:FLAG] | pmt2;EXG8:FLAG |
a Strains were constructed in the G186A (ATCC no. 26029) or G217B (ATCC no. 26032) Histoplasma strain backgrounds.
β-Glucanase Protein Sequence Analysis
Fungal homologs of the Histoplasma G186A Eng1 and Exg8 protein sequences were identified by BLAST search of C. albicans, S. cerevisiae, and Aspergillus genome databases and the derived protein sequences aligned using ClustalW with the BLOSUM62 matrix. Protein features were predicted using SignalP (version 4.0) (31) and the GPI-SOM prediction server (32) for glycosylphosphatidylinositol (GPI) anchor signals. N-Linked glycosylation sites were predicted using the consensus motif (NX(S/T)). For prediction of O-linked mucin sites, protein sequences were scanned for regions with at least 40% serine or threonine density within a 40-amino acid window (33).
Eng1 and Exg8 Localization
The ENG1 and EXG8 genes were amplified by high fidelity PCR and fused to the FLAG epitope (in plasmid pCR628 (34)) or a hexahistidine tag at the C terminus. Expression plasmids were transformed into Histoplasma uracil-auxotroph strains WU15 or WU8 (see Table 3) via Agrobacterium tumefaciens-mediated transformation (35). Expression, electrophoretic mobility, and subcellular localization of proteins was monitored by epitope tag immunoblotting (FLAG: M2 clone (Sigma), HIS6: 6G2A9 clone (GenScript)) of culture filtrates following electrophoretic separation of proteins through 10% polyacrylamide with SDS (SDS-PAGE). Potential N-linked glycans were removed by PNGaseF (New England Biolabs) prior to SDS-PAGE. O-Linked mannosylation was tested by expression of proteins in a Histoplasma strain that lacks the Pmt2 protein mannosyltransferase, which is required for O-linked glycosylation.5
For subcellular fractionation, 5 × 108 yeast cells expressing FLAG-tagged Eng1 or Exg8 were separated from culture filtrates by centrifugation and filtration. Cellular lysates were prepared from the yeast by mechanical disruption with 0.5-mm diameter glass beads. The cytosolic fraction was separated from cellular debris by centrifugation (10 min at 14,000 × g). Insoluble cellular material was treated with 1% SDS and 0.1 m dithiothreitol (DTT) to extract cell wall-associated proteins or incubated with 3 milliunits/μl zymolyase (GBiosciences) to release glycan-embedded cell wall proteins. Subcellular fractions representing secreted or cellular material from 2 × 107 yeasts were probed by immunoblotting.
Eng1 and Exg8 Protein Purification
Histoplasma yeasts expressing Eng1 or Exg8 with the C-terminal hexahistidine tag were grown in liquid HMM, and tagged proteins were purified by affinity chromatography (HisPur Co2+ Resin, Thermo Fisher Scientific). Protein homogeneity of the eluted fraction was determined by SDS-PAGE followed by silver staining of the protein gel. Purified proteins were stored at −20 °C in 50% glycerol. No significant loss of activity of the protein preparations was observed over at least 6 months of storage.
Determination of Glucanase Activity
Proteins were incubated with polysaccharide substrates in a 30-μl reaction volume (20 μl of substrate and 10 μl of protein solution). Substrates were prepared at 5 mg/ml in PBS, which included laminarin (Sigma), pustulan (Invivogen), carboxymethylcellulose (Sigma), amylose (Sigma), mannan (Sigma), swollen chitin (Sigma), scleroglucan (Invivogen), dextran (Spectrum Chemical), and pullulan (Tokyo Chemical Industry). Polysaccharide hydrolysis was measured by the addition of 3 volumes of dinitrosalicylic acid solution (0.687% (w/v) 3,5-dinitrosalicylic acid, 1.28% (v/v) phenol, 19.92% (w/v) potassium/sodium/tartrate, 1.226% (w/v) sodium hydroxide) and incubation at 95 °C for 5 min (23, 36). Saccharide-dependent reduction of 3,5-dinitrosalicylic acid to 3-amino-5-nitrosalicylic acid was quantified by absorbance at 540 nm and compared with a standard curve created from glucose or N-acetylglucosamine. Total extracellular and cell-associated glucanase activity was determined using culture filtrate equivalent to media from 2 × 107 cells or whole yeasts (2 × 107 cells), respectively.
Optimal temperatures for glucanase activities were determined by enzyme reactions carried out between 15 and 90 °C in 5 °C intervals. 30 ng of Eng1 or Exg8 protein was added to 5 mg/ml laminarin for 120 min, and reducing sugars were quantified. Similarly, the pH optimum was carried out in reactions incubated for 120 min at 37 °C using 25 mm buffer to control pH: sodium citrate (pH 3.0–5.0), MES (pH 6.0), HEPES (pH 7.0–9.0), and glycine (pH 10.0–11.0).
Kinetic Analysis of Eng1 and Exg8 Activity
Steady-state kinetic values were estimated using increasing concentrations (0, 0.5, 1, 2.5, 5, 10, 15, and 20 mg/ml) of laminarin. Reactions were carried out in a 30-μl reaction volume in 25 mm MES buffer, pH 6.0. Reducing sugars were quantified after 0, 1, 2, 5, and 15 min, and the slope of the linear region of the regression curve was calculated for each substrate concentration. Kinetic values were calculated using nonlinear least squares fitting to the expression ν = Vmax [S]/(Km + [S]) using Prism 5.03 software (GraphPad).
Mass Spectrometry of Digestion Products
Digests of the laminarin samples were subjected to nano-electrospray mass spectrometry analysis using a Waters Synapt G2 or Synapt G2-S instrument (Waters). 30 ng of protein was incubated with 5 mg/ml laminarin in water for 30 min at 37 °C and then diluted 20-fold with 50:50 water/LC-MS grade acetonitrile (Fisher). Samples were sprayed into the mass spectrometer using a pulled fused silica capillary fitted around a platinum electrode posed at the capillary voltage from the MassLynx 4.1 software (Waters). Acquisitions were from 50 to 5000 m/z. Typical voltages and amplitudes are as follows: capillary, 1.1 kV; source temperature, 30 °C; cone, 20 V; source offset, 5 V; trap collision energy, 4 V; transfer collision energy, 2 V. Ion mobility parameters as follows: helium gas flow, 180 ml/min; ion mobility gas flow, 45 ml/min; wave height, 7 V; wave velocity, 450 m/s.
After collection of the mass spectra, relative quantitation of the [M + Na]+ versions of glucose oligomers (mono- to octomers) was performed by using ion mobility drift times of the first four isotopic peaks of the ion of interest using the DriftScope 2.0 software (Waters). The ratios of the individual saccharides to the sum intensity of all saccharides (mono- through octosaccharides) were then determined for the specific enzymatic digestions.
Depletion of Glucanase Function(s)
Eng1 and/or Exg8 function was depleted from Histoplasma yeasts by RNA interference (RNAi) (25) using coding regions as follows: ENG1 = nucleotides (nts) 445-2091; EXG8 = nts 1744–2561; ENG1:EXG8 double knockdown fused ENG1 (nts 445–1521) and EXG8 (nts 1744–2561). RNAi plasmids were transformed into the RNAi sentinel strains OSU22 and OSU194 using A. tumefaciens-mediated transformation (35, 37), and Ura+ transformants were selected. GFP fluorescence of individual transformants was quantified using a modified gel documentation system (25) and ImageJ software (version 1.44p).
Cell Wall Sensitivity Assays
Yeast cells were grown in 96-well microtiter plates (38) with graded concentrations of antifungal compounds (fluconazole (Glenmark Generics); caspofungin (Merck); and nikkomycin Z (gift from John Galgiani)) and cell wall-destabilizing compounds (Calcofluor White (Sigma); Congo red (MP Biomedicals); SDS; and Uvitex) (39). Growth after 4 days was measured by optical density at 595 nm and compared with the turbidity of wells lacking inhibitors. 50% inhibitory concentration (IC50) values were computed by nonlinear regression of the dose-response data.
Yeast-Macrophage Interaction Assays
Peritoneal macrophages were harvested from C57BL/6 mice (Charles River) by peritoneal lavage and infected with yeast at 0.5:1 m.o.i. (yeast/macrophage). For association, nonadherent yeast were removed after 2 h. Yeasts were quantified by lysis of mammalian cells in water and plating of the lysate on solid HMM for colony-forming units (cfu). For survival, yeast cfu were determined after 8 h.
Cytokine Analysis
Peritoneal macrophages were infected at 0.5:1 m.o.i. with yeast, and supernatant was collected after 8 h. Cytokines (TNFα and IL-6) were quantified by ELISA (R&D Systems) and compared with standard curves.
Murine Model of Respiratory and Disseminated Histoplasmosis
C57BL/6 mice (Charles River) were infected with Histoplasma yeast by intranasal delivery of ∼1 × 104 yeast cells (15). At 8 days post-infection, lungs and spleens were collected and homogenized. Serial dilutions of the homogenates were plated on solid HMM to determine the fungal burden (cfu) in each organ.
Dectin-1 Binding Assay
Dectin-1-expressing 3T3 fibroblasts (3T3-D1) (8, 12) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) at 37 °C in 5% CO2, 95% air. 3T3-D1 cells were adhered to wells of a 24-well plate (3 × 104 cells/well). Yeast cells were added to the 3T3-D1 cells for 2 h at 37 °C at an m.o.i. of 50:1 (yeast/3T3 cells) in the absence or presence of 1 mg/ml laminarin. Unbound yeasts were removed by washing with PBS. Fibroblast-associated yeasts were released by lysing the 3T3-D1 cells with water and the yeasts quantified by plating of the lysate to enumerate cfu.
Statistical Analyses
All assays were performed with independent biological replicates (n = 3 unless otherwise noted) and statistically significant differences from wild type or other parental backgrounds (p < 0.05) as determined by one-tailed Student's t test.
Ethics Statement
All animal experiments were performed in compliance with the National Research Council's Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee at Ohio State University (protocol 2007A0241). Animal infections were performed under anesthesia, and all efforts were made to minimize suffering.
Author Contributions
A. L. G. and C. A. R. conceived of the study and were responsible for overall design of the experiments. A. L. G. performed and analyzed data for Figs. 1, 2, 4, and 6–8, supplemental Figs. S1 and S2, and Table 1. K. L. D. produced data for Figs. 2 and 5 and Table 2. A. D. V. and V. H. W. designed, produced, and analyzed experiments for the data in Fig. 3. A. L. G. and C. A. R. wrote the manuscript. All authors reviewed the results and approved the final version.
Supplementary Material
Acknowledgments
We thank Shahr Hashmi for assistance with Histoplasma growth experiments; Qian Shen for assistance with the Dectin-1 binding experiment; and Kristie Goughenour for isolating Histoplasma expression isolates. The antifungal drug nikkomycin was kindly provided by John Galgiani (University of Arizona).
This work was supported in part by National Institutes of Health Award R21-AI117122 from NIAID (to C. A. R.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Figs. 1 and 2.
A. L. Garfoot, unpublished data.
- HMM
- Histoplasma-macrophage medium
- GPI
- glycosylphosphatidylinositol
- m.o.i.
- multiplicity of infection
- nt
- nucleotide
- PNGaseF
- peptide:N-glycosidase F.
References
- 1. Garfoot A. L., and Rappleye C. A. (2016) Histoplasma capsulatum surmounts obstacles to intracellular pathogenesis. FEBS J. 283, 619–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bullock W. E., and Wright S. D. (1987) Role of the adherence-promoting receptors, CR3, LFA-1, and p150,95, in binding of Histoplasma capsulatum by human macrophages. J. Exp. Med. 165, 195–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lin J.-S., Huang J.-H., Hung L.-Y., Wu S.-Y., and Wu-Hsieh B. A. (2010) Distinct roles of complement receptor 3, Dectin-1, and sialic acids in murine macrophage interaction with Histoplasma yeast. J. Leukocyte Biol. 88, 95–106 [DOI] [PubMed] [Google Scholar]
- 4. Newman S. L., Bucher C., Rhodes J., and Bullock W. E. (1990) Phagocytosis of Histoplasma capsulatum yeasts and microconidia by human cultured macrophages and alveolar macrophages. Cellular cytoskeleton requirement for attachment and ingestion. J. Clin. Invest. 85, 223–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Guimarães A. J., Frases S., Gomez F. J., Zancopé-Oliveira R. M., and Nosanchuk J. D. (2009) Monoclonal antibodies to heat shock protein 60 alter the pathogenesis of Histoplasma capsulatum. Infect. Immun. 77, 1357–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Habich C., Kempe K., Gomez F. J., Lillicrap M., Gaston H., van der Zee R., Kolb H., and Burkart V. (2006) Heat shock protein 60: identification of specific epitopes for binding to primary macrophages. FEBS Lett. 580, 115–120 [DOI] [PubMed] [Google Scholar]
- 7. Long K. H., Gomez F. J., Morris R. E., and Newman S. L. (2003) Identification of heat shock protein 60 as the ligand on Histoplasma capsulatum that mediates binding to CD18 receptors on human macrophages. J. Immunol. 170, 487–494 [DOI] [PubMed] [Google Scholar]
- 8. Brown G. D., and Gordon S. (2001) Immune recognition: A new receptor for β-glucans. Nature 413, 36–37 [DOI] [PubMed] [Google Scholar]
- 9. Gantner B. N., Simmons R. M., Canavera S. J., Akira S., and Underhill D. M. (2003) Collaborative induction of inflammatory responses by dectin-1 and Toll-like receptor 2. J. Exp. Med. 197, 1107–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Underhill D. M., Rossnagle E., Lowell C. A., and Simmons R. M. (2005) Dectin-1 activates Syk tyrosine kinase in a dynamic subset of macrophages for reactive oxygen production. Blood 106, 2543–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wheeler R. T., and Fink G. R. (2006) A drug-sensitive genetic network masks fungi from the immune system. PLoS Pathog. 2, e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Edwards J. A., Alore E. A., and Rappleye C. A. (2011) The yeast-phase virulence requirement for α-glucan synthase differs among Histoplasma capsulatum chemotypes. Eukaryot. Cell 10, 87–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rappleye C. A., Engle J. T., and Goldman W. E. (2004) RNA interference in Histoplasma capsulatum demonstrates a role for α-(1,3)-glucan in virulence. Mol. Microbiol. 53, 153–165 [DOI] [PubMed] [Google Scholar]
- 14. Rappleye C. A., Eissenberg L. G., and Goldman W. E. (2007) Histoplasma capsulatum α-(1,3)-glucan blocks innate immune recognition by the β-glucan receptor. Proc. Natl. Acad. Sci. 104, 1366–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Garfoot A. L., Shen Q., Wüthrich M., Klein B. S., and Rappleye C. A. (2016) The Eng1 β-glucanase enhances Histoplasma virulence by reducing β-glucan exposure. mBio. 7, e01388–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Holbrook E. D., Edwards J. A., Youseff B. H., and Rappleye C. A. (2011) Definition of the extracellular proteome of pathogenic-phase Histoplasma capsulatum. J. Proteome Res. 10, 1929–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Baladrón V., Ufano S., Dueñas E., Martín-Cuadrado A. B., del Rey F., and Vázquez de Aldana C. R. (2002) Eng1p, an endo-1,3-β-glucanase localized at the daughter side of the septum, is involved in cell separation in Saccharomyces cerevisiae. Eukaryot. Cell. 1, 774–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Esteban P. F., Ríos I., García R., Dueñas E., Plá J., Sánchez M., de Aldana C. R., and Del Rey F. (2005) Characterization of the CaENG1 gene encoding an endo-1,3-β-glucanase involved in cell separation in Candida albicans. Curr. Microbiol. 51, 385–392 [DOI] [PubMed] [Google Scholar]
- 19. Mouyna I., Hartl L., and Latgé J.-P. (2013) β-1,3-Glucan modifying enzymes in Aspergillus fumigatus. Front. Microbiol. 4, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martín-Cuadrado A.-B., Fontaine T., Esteban P.-F., del Dedo J. E., de Medina-Redondo M., del Rey F., Latgé J. P., and de Aldana C. R. (2008) Characterization of the endo-β-1,3-glucanase activity of S. cerevisiae Eng2 and other members of the GH81 family. Fungal Genet. Biol. 45, 542–553 [DOI] [PubMed] [Google Scholar]
- 21. Ishida T., Fushinobu S., Kawai R., Kitaoka M., Igarashi K., and Samejima M. (2009) Crystal structure of glycoside hydrolase family 55 β-1,3-glucanase from the Basidiomycete Phanerochaete chrysosporium. J. Biol. Chem. 284, 10100–10109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bianchetti C. M., Takasuka T. E., Deutsch S., Udell H. S., Yik E. J., Bergeman L. F., and Fox B. G. (2015) Active site and laminarin binding in glycoside hydrolase family 55. J. Biol. Chem. 290, 11819–11832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Miller G. L. (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31, 426–428 [Google Scholar]
- 24. Lombard V., Golaconda Ramulu H., Drula E., Coutinho P. M., and Henrissat B. (2014) The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Youseff B. H., and Rappleye C. A. (2012) in Host-Fungus Interactions (Brand A. C., and MacCallum D. M., eds) pp. 151–164, Humana Press Inc., Totowa, NJ [Google Scholar]
- 26. Eissenberg L. G., Goldman W. E., and Schlesinger P. H. (1993) Histoplasma capsulatum modulates the acidification of phagolysosomes. J. Exp. Med. 177, 1605–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Strasser J. E., Newman S. L., Ciraolo G. M., Morris R. E., Howell M. L., and Dean G. E. (1999) Regulation of the macrophage vacuolar ATPase and phagosome-lysosome fusion by Histoplasma capsulatum. J. Immunol. 162, 6148–6154 [PubMed] [Google Scholar]
- 28. Taylor M. L., Espinosa-Schoelly M. E., Iturbe R., Rico B., Casasola J., and Goodsaid F. (1989) Evaluation of phagolysosome fusion in acridine orange stained macrophages infected with Histoplasma capsulatum. Clin. Exp. Immunol. 75, 466–470 [PMC free article] [PubMed] [Google Scholar]
- 29. Adams E. L., Rice P. J., Graves B., Ensley H. E., Yu H., Brown G. D., Gordon S., Monteiro M. A., Papp-Szabo E., Lowman D. W., Power T. D., Wempe M. F., and Williams D. L. (2008) Differential high-affinity interaction of dectin-1 with natural or synthetic glucans is dependent upon primary structure and is influenced by polymer chain length and side-chain branching. J. Pharmacol. Exp. Ther. 325, 115–123 [DOI] [PubMed] [Google Scholar]
- 30. Worsham P. L., and Goldman W. E. (1988) Quantitative plating of Histoplasma capsulatum without addition of conditioned medium or siderophores. J. Med. Vet. Mycol. 26, 137–143 [PubMed] [Google Scholar]
- 31. Petersen T. N., Brunak S., von Heijne G., and Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods. 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 32. Fankhauser N., and Mäser P. (2005) Identification of GPI anchor attachment signals by a Kohonen self-organizing map. Bioinformatics 21, 1846–1852 [DOI] [PubMed] [Google Scholar]
- 33. Fernández-Álvarez A., Marín-Menguiano M., Lanver D., Jiménez-Martín A., Elías-Villalobos A., Pérez-Pulido A. J., Kahmann R., and Ibeas J. I. (2012) Identification of O-mannosylated virulence factors in Ustilago maydis. PLoS Pathog. 8, e1002563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Edwards J. A., Chen C., Kemski M. M., Hu J., Mitchell T. K., and Rappleye C. A. (2013) Histoplasma yeast and mycelial transcriptomes reveal pathogenic-phase and lineage-specific gene expression profiles. BMC Genomics 14, 695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zemska O., and Rappleye C. A. (2012) in Host-Fungus Interactions (Brand A. C., and MacCallum D. M., eds) pp. 51–66, Humana Press Inc., Totowa, NJ [Google Scholar]
- 36. Ramada M. H., Lopes F. A., Ulhoa C. J., and Silva R. do N. (2010) Optimized microplate β-1,3-glucanase assay system for Trichoderma spp. screening. J. Microbiol. Methods 81, 6–10 [DOI] [PubMed] [Google Scholar]
- 37. Kemski M. M., Stevens B., and Rappleye C. A. (2013) Spectrum of T-DNA integrations for insertional mutagenesis of Histoplasma capsulatum. Fungal Biol. 117, 41–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Goughenour K. D., Balada-Llasat J.-M., and Rappleye C. A. (2015) Quantitative microplate-based growth assay for determination of antifungal susceptibility of Histoplasma capsulatum yeasts. J. Clin. Microbiol. 53, 3286–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Edwards J. A., Kemski M. M., and Rappleye C. A. (2013) Identification of an aminothiazole with antifungal activity against intracellular Histoplasma capsulatum. Antimicrob. Agents Chemother. 57, 4349–4359 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.