Abstract
Deconstruction of cellulose, the most abundant plant cell wall polysaccharide, requires the cooperative activity of a large repertoire of microbial enzymes. Modular cellulases contain non-catalytic type A carbohydrate-binding modules (CBMs) that specifically bind to the crystalline regions of cellulose, thus promoting enzyme efficacy through proximity and targeting effects. Although type A CBMs play a critical role in cellulose recycling, their mechanism of action remains poorly understood. Here we produced a library of recombinant CBMs representative of the known diversity of type A modules. The binding properties of 40 CBMs, in fusion with an N-terminal GFP domain, revealed that type A CBMs possess the ability to recognize different crystalline forms of cellulose and chitin over a wide range of temperatures, pH levels, and ionic strengths. A Spirochaeta thermophila CBM64, in particular, displayed plasticity in its capacity to bind both crystalline and soluble carbohydrates under a wide range of extreme conditions. The structure of S. thermophila StCBM64C revealed an untwisted, flat, carbohydrate-binding interface comprising the side chains of four tryptophan residues in a co-planar linear arrangement. Significantly, two highly conserved asparagine side chains, each one located between two tryptophan residues, are critical to insoluble and soluble glucan recognition but not to bind xyloglucan. Thus, CBM64 compact structure and its extended and versatile ligand interacting platform illustrate how type A CBMs target their appended plant cell wall-degrading enzymes to a diversity of recalcitrant carbohydrates under a wide range of environmental conditions.
Keywords: carbohydrate metabolism, carbohydrate-binding protein, cellulase, cellulose, plant cell wall
Introduction
Plant cell walls are composed of structurally complex polysaccharides and comprise the most abundant source of organic carbon in the biosphere. Within plant cell walls, cellulose represents a primary target for enzymatic hydrolysis. Cellulose is synthesized as micrometer-long microfibrils consisting of parallel hydrogen-bonded β-1,4-glucan chains, containing both crystalline and less ordered regions. A wide range of microbial species and systems contribute to the degradation of plant cell walls through the production of an extensive repertoire of carbohydrate active enzymes (CAZymes).4 CAZymes acting on recalcitrant carbohydrates are frequently modular proteins containing a catalytic module connected via flexible linker sequences to a variable number of non-catalytic carbohydrate-binding modules (CBMs). CBMs potentiate the efficacy of the associated catalytic modules as they contribute to substrate targeting while promoting a close proximity between enzymes and structural polysaccharides (1–5). Thus, CAZymes are highly relevant biological players in the recycling of photosynthetically fixed carbon and have recently acquired industrial and environmental significance, in particular for the production of second generation lignocellulose-based biofuels (6, 7).
Based on primary sequence similarities, CAZymes and CBMs have been classified in families in the constantly updated CAZy database (8, 9). CBMs have been extensively characterized and are currently grouped into 80 sequence-based families in the CAZy database. Structure/function studies have revealed that the dominant fold assumed by CBMs is the β-sandwich. Based on the topology of their carbohydrate binding interfaces, CBMs have been classified into three types. Type A CBMs display a planar carbohydrate-binding interface that is adapted to bind the surface of crystalline polysaccharides. In contrast, type B proteins interact with internal regions of single glycan chains (endo-type) via protein surface clefts. Finally, type C modules recognize the termini of glycan chains (exo-type) through the fitting of a limited number of sugars in the binding pocket of the protein (5, 10). The planar binding site of type A CBMs generally contains three hydrophobic residues that, based on site-directed mutagenesis studies, are believed to interact with the crystalline surface of cellulose microfibrils (11–13). This class of CBMs includes members of CBM families 1, 2a, 3, 5, 10, 49, 63, 64, and 79, which bind to insoluble, highly crystalline, cellulose, and/or chitin. Thus, type A CBMs lack significant affinity for cellulo-oligosaccharides and chemically modified soluble forms of cellulose, whereas their recognition of crystalline ligands appears to be predominantly mediated by entropic forces (3, 14). The capacity of CBMs to bind a diversity of carbohydrates with a high degree of specificity and selectivity has promoted the exploration of a large number of biotechnological applications (15, 16). For example, fusion of CBMs with other proteins offers the possibility of targeted immobilization of antibodies (17–19), proteins (20), bacteriophages (21), and bacteria (22) onto cellulose matrices with the goal of developing sensor, microarray, and protein purification applications.
Type A CBMs are especially attractive tools for different biotechnological applications, because they selectively bind to crystalline carbohydrates present in versatile and abundant materials like paper, cotton, and nanocellulose. However, the use of these small molecules to target different peptides, proteins, enzymes, or antibodies into cellulosic supports is limited by our lack of knowledge of their biochemical properties. Here we generated and characterized a library of type A recombinant CBMs belonging to families 1, 2a, 3, 5, 9, 10, 49, 63, and 64. Of the 40 type A CBMs characterized, a CBM64 member was shown to display a high capacity to bind cellulose under different and extreme physical-chemical conditions. The structure of StCBM64C from Spirochaeta thermophila and a detailed biochemical characterization of this family revealed why CBM64 displays such versatile biochemical properties.
Results and Discussion
Production of a Library of 96 Recombinant Type A GFP-CBMs
Type A CBMs are unique because they bind to the crystalline surfaces of cellulose and chitin through a planar carbohydrate-binding platform rich in aromatic amino acid residues that reflects the topology of the ligand surface. To characterize the capacity of type A CBMs to recognize crystalline carbohydrates under different physicochemical conditions, 96 bacterial and fungal CBMs from families 1, 2a, 3, 5, 9, 10, 49, 63, and 64 were fused to the N terminus of the GFP and expressed in Escherichia coli. Although CBM9 are type C CBMs (5), one member of this family was included in the screen based on its capacity to bind crystalline cellulose. The proteins were purified through IMAC, and estimated molecular mass was confirmed through SDS-PAGE (supplemental Fig. S1). The data, presented in supplemental Table S1, revealed that 67 GFP-CBM fusions were expressed at high levels (>100 mg/liter of culture medium), with 8 of these 67 recombinant proteins being expressed at very high levels (>300 mg/liter of culture medium). In contrast, 12 proteins presented intermediate levels of expression (ranging from 50–100 mg/liter of culture medium), whereas 17 were produced at levels below 50 mg/liter of culture medium mainly as a result of accumulation in the form of inclusion bodies (supplemental Table S1). There was no correlation between the levels of expression and the CBM origin. Thus, CBMs expressed with lower (<50 mg/liter) and higher (>300 mg/liter) efficacies originated both from bacterial and fungal hosts.
Binding Efficacy of GFP-CBMs to Carbohydrates
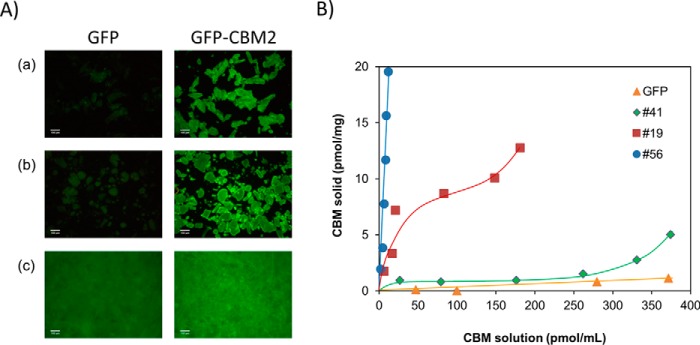
Binding efficacy of 40 GFP-CBM fusions that were expressed at higher levels and represented the widest biodiversity in terms of families was evaluated. Although the presence of an N-terminal GFP might affect the binding properties of the fused CBM, we have hypothesized that inclusion of linker sequences and the His6 tag separating the two fusion partners would minimize any steric interference of the GFP in the function of the C-terminal CBM. To probe this hypothesis, initial qualitative experiments compared the capacity of selected GFP-CBM fusions and of GFP to bind different insoluble polysaccharides by fluorescence microscopy. The qualitative data, exemplified for CBM2 from Pyrococcus furiosus (CBM #29) in Fig. 1A, suggest that the CBMs are fully functional and bind effectively to microcrystalline cellulose, chitosan, and paper. Thus, the binding of the 40 GFP-CBM fusions to Sigmacell cellulose type 20, microcrystalline cellulose powder, low molecular weight chitosan, potato starch, and chromatographic paper (Whatman no.1) was subsequently studied by determining the binding isotherms with 100 mm phosphate buffer, pH 7, at 25 °C. The equilibrium concentrations of GFP-CBM in the solid (pmol/mg) and liquid (pmol/ml) phases were determined on the basis of mass balance calculations performed with fluorescence data of solutions at loading and after 20-min incubations. As an example, Fig. 1B displays the isotherms representing the interaction of CBM64 from S. thermophila DSM 6192 (CBM #56), CBM2 from Ampullaria crossean (CBM #19), and CBM10 from Teredinibacter turnerae T7901 (CBM #41) to Sigmacell 20 cellulose, which are representative of high, medium, and low binding CBMs, respectively. The capacity of control GFP to interact with the polysaccharide was also evaluated (Fig. 1B). For the range of GFP-CBM loads tested (up to 20 pmol/mg of Sigmacell 20), maximum concentrations in the solid phase at equilibrium of 20, 13, and 5 pmol/mg were obtained from CBM #56, CBM #19, and CBM #41, respectively. For CBM #41, the binding to Sigmacell 20 is equivalent to the binding observed for the control GFP up to a concentration in the liquid phase of 300 pmol/ml. From here on, an increase in binding is detected. The amount of CBM #56, CBM #19, and CBM #41 in the solid phase at equilibrium corresponds to 95–99%, 63–89%, and 12–47% of the total amount of CBM in the solid/liquid system, respectively.
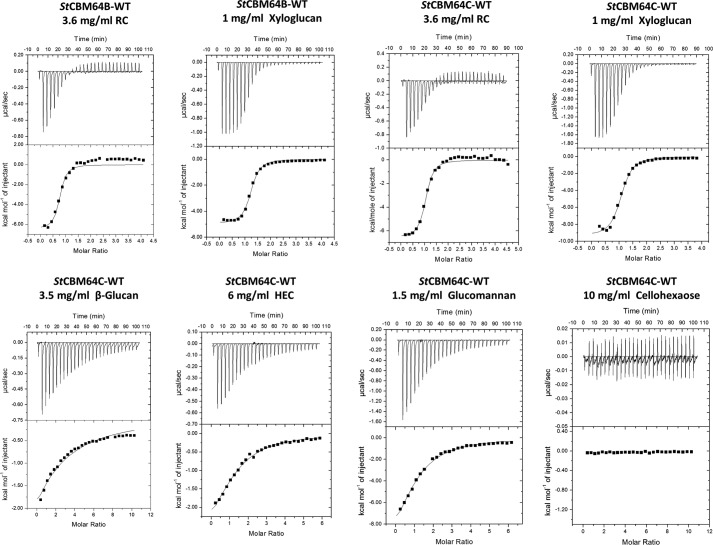
FIGURE 1.
Capacity of recombinant GFP-CBM fusions to bind insoluble cellulose. A, fluorescence microcopy images of microcrystalline cellulose (panel a), chitosan (panel b), and chromatographic paper (panel c) incubated with GFP (left column) and GFP-CBM2 from P. furiosus (CBM #29) (right column). Images were taken with a fluorescence microscope (10× magnification). B, binding isotherms are shown for a CBM64 from S. thermophila DSM 6192 (CBM #56), a CBM2 from A. crossean (CBM #19), and a CBM10 from T. turnerae T7901 (CBM #41) that are representative of high, medium, and low binding CBMs. The isotherm for GFP is also shown as a negative control. Isotherms were determined in 100 mm phosphate buffer, pH 7, 25 °C by contacting 10–100 pmol of GFP-CBM with 5 mg of Sigmacell 20 for 20 min. Experiments were performed in triplicate.
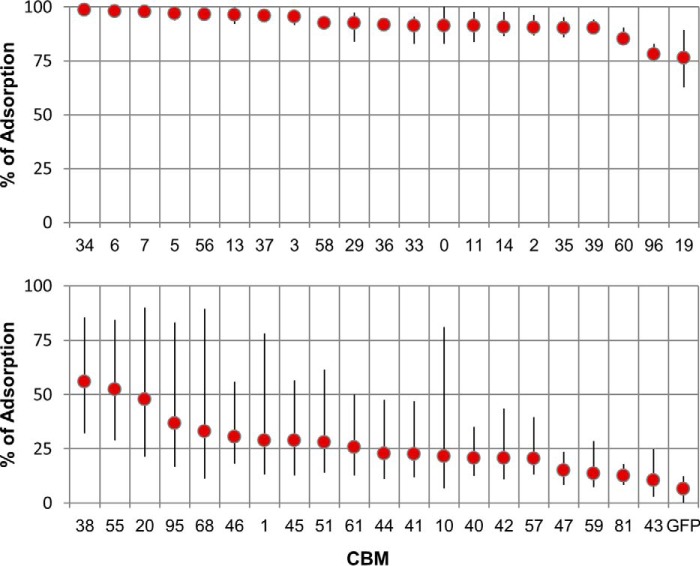
For each isotherm, the average percentage of the different amounts of GFP-CBM (10–100 pmol) initially loaded that bound to each substrate at equilibrium was calculated. These results are plotted in Fig. 2 for the case of Sigmacell cellulose powder. The extremes of the thinner bars superimposed on each data point correspond to the minimum and maximum percentages of adsorption measured across the range of loaded amounts of GFP-CBM. An analysis of the data shows that of the 40 CBMs tested, 20 exhibit high binding to Sigmacell 20 (more than 75% adsorption at loadings up to 100 pmol/5 mg). On the other side of the spectrum, 11 CBMs bind poorly to Sigmacell 20 (less than 25% average adsorption). The majority of these are CBMs of fungal origin from families 1 (CBMs #43–#47), 9 (CBM #51), and 10 (CBMs #40–#42). The largest variation in adsorption percentage across the range of loaded amounts is found for CBMs that display average adsorptions in the 25–50% range.
FIGURE 2.
Binding of GFP-CBMs to Sigmacell 20 cellulose powder. The average percentages of the different amounts of GFP-CBM (10–100 pmol) contacted with 5 mg of Sigmacell 20 that bound to the substrate after 20 min of incubation are shown. The extremes of the thinner bars superimposed on each data point correspond to the minimum and maximum percentages of adsorption measured across the range of loaded amounts of GFP-CBM. Triplicate experiments were performed in 100 mm phosphate buffer, pH 7, 25 °C.
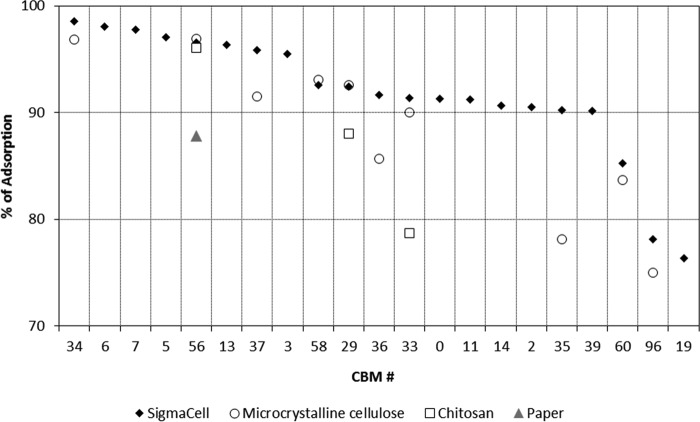
The binding specificity also varied considerably across the 40 CBMs studied when using the other four substrates (see supplemental Figs. S2–S5). For microcrystalline cellulose (supplemental Fig. S2), 10 CBMs displayed high binding (>75%) and 20 low binding (<25%). Again, the low binding CBMs are those from fungal origin of families 1 (CBMs #43–#47), 9 (CBM #51), and 10 (CBMs #40–#42). The affinity of CBMs to chromatographic paper (supplemental Fig. S3) and chitosan (supplemental Fig. S4) was in general lower when compared with Sigmacell cellulose and microcrystalline cellulose. Adsorption percentages higher than 75% were observed only in one (CBM #56) and three (CBM #56, CBM #29, and CBM #33) cases for paper and chitosan, respectively. The low performance of fungal CBMs was again the norm. The binding of CBMs to starch was in general very poor (<25%; supplemental Fig. S5). A comparison of the results obtained for the CBMs that displayed the highest binding (>75%) to Sigmacell, microcrystalline cellulose, paper, and chitosan is shown in Fig. 3. Here, CBM #56 (a CBM64 from S. thermophila) shows up as a high performer in all four substrates. A set of two CBMs also perform very well in three substrates other than paper: CBM #29 (a CBM2 from P. furiosus) and CBM #33 (a CBM2 from Thermobifida fusca YX). CBM #34 (a CBM2 from T. fusca YX), CBM #37 (a CBM2 from T. fusca YX), and CBM #58 (a CBM3 from Clostridium thermocellum) presented also very good binding both to SigmaCell and microcrystalline cellulose. The binding of the control GFP to the different substrates was always lower than 13% (supplemental Figs. S2–S5). The capacities of the CBMs to interact with their target ligands under different conditions were evaluated next for a diversity of temperatures, pH levels and ionic strengths and are as follows.
FIGURE 3.
High performing CBMs. The data show the average percentages of GFP-CBMs (10–100 pmol) contacted with 5 mg of Sigmacell 20 cellulose powder, microcrystalline cellulose, chromatographic paper, and chitosan that bound by more than 75% to the substrate after 20 min of incubation. Experiments were performed in 100 mm phosphate buffer, pH 7, 25 °C.
Effect of Temperature
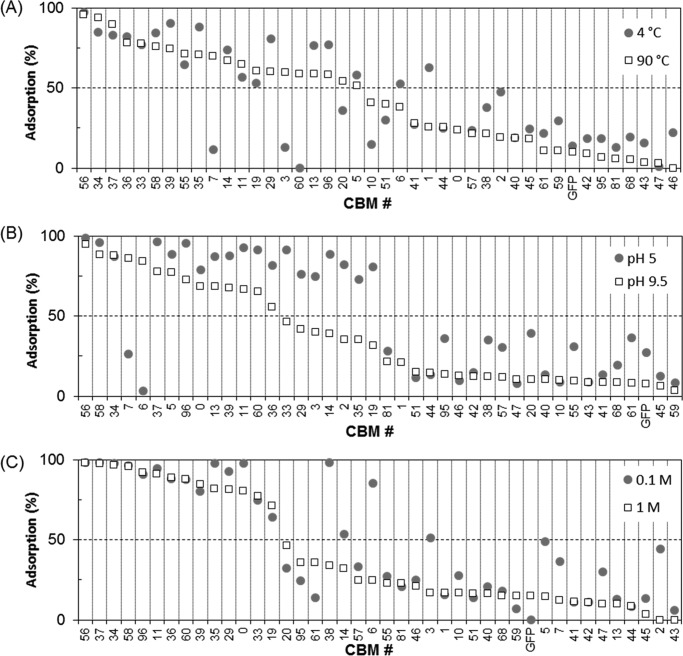
The effect of temperature on the binding of the 40 GFP-CBM fusions to Sigmacell 20 was studied in the 4–90 °C range. The experiments were performed by contacting 50 pmol of GFP-CBM with 5 mg of Sigmacell cell in 100 mm phosphate pH 7 buffer. The percentage of GFP-CBM adsorbed at equilibrium is shown in Fig. 4A for the two temperature extremes evaluated, 4 and 90 °C, and in supplemental Fig. S6 for the other temperatures (25, 35, 50, and 70 °C). An observation of Fig. 4A shows that seven CBMs perform very well at the temperature extremes (>75% binding) and across the temperature range studied (supplemental Fig. S6). Among these, we can again identify CBMs #33, #34, #37, #56, and #58, which had previously been shown to bind very well to Sigmacell, microcrystalline cellulose, paper, and chitosan at 25 °C (Figs. 3 and supplemental Figs. S2–S5). These results are consistent with the fact that these CBMs are derived from the thermophilic bacteria T. fusca YX (CBMs #33, #34, and #37) and C. thermocellum (CBMs #56 and #58). The largest differences in adsorption (>25%) across the temperature range studied were obtained for CBMs #3, #7, #10, and #60, which bound more effectively at 9 °C than at 4 °C, and for CBMs #1 and #2, which bound more effectively at 4 °C than at 90 °C (Fig. 4A).
FIGURE 4.
Effect of temperature, pH level, and ionic strength on the binding capacity of GFP-CBM fusions to Sigmacell 20 cellulose powder. A, adsorption percentage was determined in 100 mm phosphate buffer, pH 7, after contacting 50 pmol of GFP-CBM with 5 mg of Sigmacell 20 for 20 min at 4 °C and 90 °C. B, adsorption percentage was determined in 100 mm phosphate buffer, 25 °C after contacting 100 pmol of GFP-CBM with 5 mg of Sigmacell 20 for 20 min at pH 5 and pH 9.5. C, adsorption percentage was determined at 25 °C after contacting 50 pmol of GFP-CBM with 5 mg of Sigmacell 20 for 20 min in 100 mm and 1 m phosphate buffer, pH 7. Experiments were performed in triplicate.
Effect of pH
The effect of pH on the binding of the 40 GFP-CBM fusions to Sigmacell 20 was investigated. The experiments were performed by contacting 100 pmol of GFP-CBM with 5 mg of Sigmacell cell in 100 mm phosphate at 25 °C across the 5.0–9.5 pH range. The percentage of GFP-CBM adsorbed at equilibrium is shown in Fig. 4B for the two pH extremes evaluated and in supplemental Fig. S7 for the other pH values. Fig. 4B shows that four CBMs—#34, #37, #56, and #58—perform very well at the pH extremes (>75% binding) and across the pH range studied (supplemental Fig. S7). In general, the analyzed CBMs performed better at acidic pH levels, with 18 displaying more than 75% binding to Sigmacell (see Fig. 7). Notable exceptions are CBM #6, a CBM3 from Caldicellulosiruptor bescii, and CBM #7, a CBM3 from Clostridium cellulolyticum, which bind poorly to Sigmacell at pH 5 but perform very well at pH 9.5.
FIGURE 7.
Interaction of StCBM64B and StCBM64C with RC and a variety of soluble ligands as probed by ITC. Titrations were conducted in 50 mm NaHepes buffer (pH 7.5) containing 200 mm NaCl at 25 °C. The top portion of each panel shows the raw power data, whereas the bottom panel shows the integrated and heat of dilution corrected data.
Effect of Ionic Strength
The effect of ionic strength on the binding of the 40 GFP-CBM fusions to Sigmacell 20 was studied in the 100 mm to 1.5 m range. The experiments were performed by contacting 50 pmol of GFP-CBM with 5 mg of Sigmacell cell in phosphate buffer at the test ionic strength, pH 7 at 25 °C. The percentage of GFP-CBM adsorbed at equilibrium is shown in Fig. 4C for 100 mm and 1 m and in supplemental Fig. S8 for the other ionic strengths. In general, the performance of the analyzed CBMs varied little with the ionic strength (Fig. 4C). The largest differences in adsorption (>25%) were obtained for CBMs #2, #3, #5, #6, and #38, which bound more effectively at 0.1 m than at 1 m. Among the best performers across the ionic strength range tested, we can again identify CBMs #33, #34, #37, #56, and #58.
Crystal Structure of StCBM64C
The data presented above revealed that CBM #56 has a high capacity to bind a diversity of insoluble polysaccharides under a variety of physical-chemical conditions. CBM #56 is the C-terminal family 64 CBM from a putative GH10 xylanase (ADN02703) of S. thermophila DSM 6192 and was termed StCBM64B. CBM64s were initially identified at the C terminus of seven S. thermophila CAZymes, which have single GH5, GH9, GH10, or GH12 catalytic domains (38). Because attempts to crystalize StCBM64B failed, the structure of StCBM64C was solved to a resolution of 1.5 Å, as described under “Materials and Methods.” StCBM64C has 82% identity and 86% similarity to StCBM64B and is the C-terminal domain of the GH12 cellulase (AEJ60658/ADN02189) of S. thermophila DSM 6578. Data collection and refinement statistics for this final structure (deposited under PDB access code 5LU3) are presented in Table 1, as well as the data collection statistics of an isomorphous crystal of StCBM64C used for de novo S-SAD phasing. During the course of this project, the structure of a second member of CBM64, the C-terminal domain of S. thermophila DSM 6192 GH5 (ADN02996), here termed StCBM64A, became known (PDB code 5E9O) (39).
TABLE 1.
Crystallographic and refinement data of StCBM64C from Spirochaeta thermophile
Statistics for the highest resolution shell are shown in parentheses. 2NV, Jeffamine ED-2001; SSN, 4-oxobutanoic acid; FMT, formic acid.
| StCBM64C | StCBM64C sulfur-SAD | |
|---|---|---|
| Crystallographic data | ||
| Source | PXII (X06DA), SLS | PXII (X06DA), SLS |
| Detector | PILATUS 2M-F | PILATUS 2M-F |
| Wavelength (Å) | 1.0 | 1.8 |
| a (Å) = b (Å) | 56.70 | 56.72 |
| c (Å) | 66.74 | 66.74 |
| Space group | P43212 | P43212 |
| Molecules per ASU | 1 | 1 |
| Matthews coefficient (Å3·Dalton−1) | 2.71 | 2.71 |
| Mosaicity (°) | 0.18 | 0.11 |
| Resolution range (Å) | 43.21–1.53 (1.53–1.50) | 43.19–2.38 (2.38–2.30) |
| <I/σI> | 79.4 (16.0) | 129.0 (92.3) |
| Wilson B-factor | 13.3 | 14.3 |
| Rmerge (%)a | 2.8 (9.2) | 3.4 (4.4) |
| Rpim (%)b | 0.6 (4.0) | 0.5 (0.7) |
| Half-data set correlation CC½ | 1.00 (0.94) | 1.00 (1.00) |
| Multiplicity | 21.8 (6.8) | 45.6 (41.7) |
| No. of observed reflections | 394529 (5954) | 237504 (20201) |
| No. of unique reflections | 18085 (870) | 5212 (484) |
| Completeness (%) | 99.9 (99.0) | 99.9 (98.7) |
| Anomalous completeness (%) | 99.4 (89.4) | 99.8 (98.3) |
| Anomalous multiplicity | 11.6 (3.4) | 25.4 (22.3) |
| Figure of merit for two sulfur sites (before/after solvent flattening) | 0.40/0.91 | |
| Refinement statistics | ||
| Rworkc | 0.1237 | |
| Rfreed | 0.1511 | |
| Root mean square deviation bonds (Å) | 0.013 | |
| Root mean square deviation angles (°) | 1.349 | |
| Average B (Å2) | 17.20 | |
| Main chain | 13.3 | |
| Side chain | 16.1 | |
| Water molecules | 32.5 | |
| Ca2+ A/B | 36.1 (occe 0.35)/21.0 (occe 0.43) | |
| Ni2+ | 20.7 (occe 0.24) | |
| 2NV | 30.6 (occe 1.0) | |
| SSN | 41.3 (occe 1.0) | |
| FMT | 19.4 (occe 1.0) | |
| Amino acid residues per ASU | 88 | |
| Ligands per ASU | 2NV, Ca2+, Ni2+, SSN, FMT | |
| No. of water molecules | 87 | |
| Total no. of atoms | 888 | |
| Ramachandran favored (%) | 98.9 | |
| Ramachandran outliers (%) | 0.0 | |
a Rmerge = [Σ|I − <I>|]/Σ<I>, where I is the observed intensity, and <I> is the statistically weighted average intensity of multiple observations.
b Rpim = [Σ √(1/(n − 1)) Σ|I − <I>|]/Σ <I>, a redundancy-independent version of Rmerge.
c Rwork = Σ||Fcalc| − |Fobs||/Σ|Fobs| × 100, where Fcalc and Fobs are the calculated and observed structure factor amplitudes, respectively.
d Rfree is calculated for a randomly chosen 5% of the reflections.
e occ, occupancy.
StCBM64C adopts the classic β-sandwich jelly roll fold containing nine β-strands typical of the majority of CBM families (Fig. 5A). The order of the β-strands in β-sheet 1 is β1, β3, β8, β5, and β6, and the order of the β-strands in β-sheet 2 is β2, β9, β4, and β7. In the two β-sheets, the β-strands are anti-parallel with the exception of the small β-strand β1 in β-sheet 1, which runs parallel to β3. The β-strands are connected by short loops, which may reflect the enhanced stability presented by StCBM64C, although there is a small helix extending from residues Gln-57 to Val-60. The core of the β-sandwich is highly hydrophobic, with very few polar residues, and includes two phenylalanine, two tryptophan, two isoleucine, and two leucine residues. Both β-sheets present a flat, untwisted, surface typical of type A CBMs. Three-dimensional structural comparison using the SSM site revealed that the closest structural homologue of StCBM64C is StCBM64A from S. thermophila (PDB code 5E9O), with a Z score of 9.7, root mean square deviation of 0.55 Å over 84 aligned residues. An overlay of the two structures (Fig. 5B) emphasizes the high structural similarity between the two CBM64 members that have 78% primary sequence identity. The unique co-planar arrangement of the four exposed tryptophan side chains of StCBM64A is highly conserved inStCBM64C only changing the conformation of Trp-78, which can be explained by differences inherent to the crystal-packing environment. As observed for StCBM64A, the surface electrostatic potential of StCBM64C reveals an overall slightly acidic nature with the surface of the four exposed tryptophan side chains close to neutral. StCBM64C has a lower degree of homology with other functionally relevant CBMs with a β-sandwich fold, in particular with the CBM2s of Cellulomonas fimi (PDB codes 1HEJ and 1E5B), with a Z score of 3.2–4.1 and root mean square deviation of ∼3.3 Å over 68 aligned residues.
FIGURE 5.
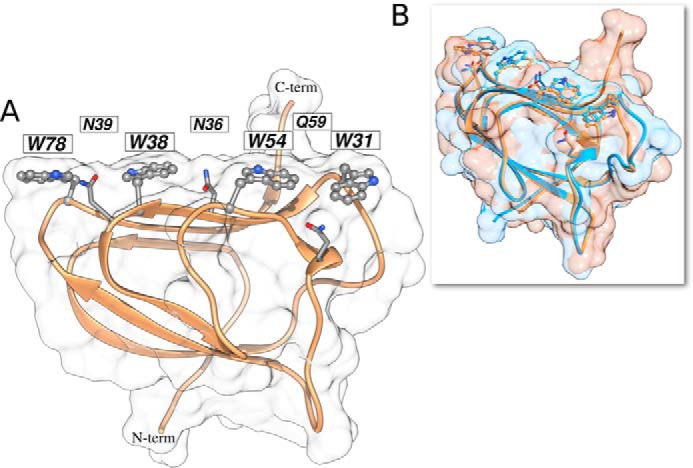
Structure of StCBM64C. A, StCBM64C shows a flat carbohydrate interacting platform, supported by four tryptophan residues (ball and stick) organized in a co-planar linear arrangement, interspersed with the three polar residues (in stick representation). B, overlay of StCBM64A (light blue) and StCBM64C (tan) and respective molecular surfaces reveals conservation in polysaccharide recognition. Molecular graphics performed with the UCSF Chimera package.
In general, type A CBMs have a β-sandwich fold similar to StCBM64C, and the carbohydrate binding interface has a flat surface that reflects the topology of the crystalline ligand (11–13). Usually, three aromatic residues, predominantly tryptophan amino acids, on the surface of type A CBMs participate in polysaccharide recognition (5). Here, however, the crystal structure of StCBM64C suggests that four residues, Trp-31, Trp-38, Trp-54, and Trp-78, which are linearly aligned at the surface of β-sheet 2, may constitute the ligand binding site (Fig. 5A). The fact that four instead of three tryptophan side chains, which form tighter sugar interactions than the tyrosine side chains identified in several type A CBMs, were recognized in the putative ligand interacting surface may explain the high binding efficacy revealed by CBM64. Arg-53, Glu-82, and Glu-86 are also located at the surface of the protein in the vicinity of the aromatic residues and thus may also play a role in ligand recognition. Strikingly, the later aromatic side chains alternate with the side chains of polar uncharged residues, namely Asn-39 (between Trp-78 and Trp-38), Asn-36 (between Trp-38 and Trp-54), and Gln-59 (between Trp-54 and Trp-31), as also observed in StCBM64A structure (39). These side chains could act both as hydrogen bond donors and acceptors, thus extending the potential number of contacts with the carbohydrate and the hydrophilicity of the interaction. The distances between Trp-31, Trp-54, Trp-38, and Trp-78 suggest that each one of the aromatic side chains will stack against one sugar ring, whereas the side chains of Asn-39, Asn-36, and Gln-59 would participate in the recognition of three intermediate glucose OH groups (Fig. 5). These observations suggest that CBM64 possesses a large carbohydrate binding platform, comprising an unprecedented large number of residues, organized in a unique structural arrangement.
Recently Georgelis et al. (37) reported the structure of EXLX1 CBM63 in complex with cellohexaose. Although the CBM, because it is characteristic of type A CBMs, does not interact with cellohexaose, the oligosaccharide was sandwiched between two CBM63s arranged in opposite polarity (37). The structure of cellohexaose bound to EXLX1 displays a conformation that is intermediate between the flat chains of cellulose and the twisted chains observed in oligosaccharides captured in complex with type B CBMs. When the structures of EXLX1 and StCBM64C are superposed, it is clear that although the two CBMs do not share a similar three-dimensional topology, the hydrophobic side chains of the aromatic residues of the two carbohydrate binding platforms line up in similar positions (Fig. 6). These observations enabled using CBM63 complexed structure to model a cellohexaose molecule into the putative StCBM64C carbohydrate-binding surface (Fig. 6A). The model (Fig. 6A) suggests that the pyranose rings of G2, G4, and G6 establish hydrophobic stacking interactions with the side chains of Trp-54, Trp-38, and Trp-78, respectively (numbering of the glucose residues starts at the reducing end of cellohexaose). In addition, Asn-39 and Asn-36 are within hydrogen bonding distance to C6 hydroxyl groups of G3 and G5. Gln-59 is positioned further away from C6 of G1, although it is likely that in a twisted cellohexaose conformation, it could also make polar interactions with the carbohydrate backbone. These observations suggest that, in contrast to other type A CBMs, StCBM64C can establish a hydrogen bond network with the crystalline ligand, which should be dominated by the side chains of Asn-39 and Asn-36 (see below). This analysis also suggests that StCBM64C contains an extended carbohydrate-binding interface that may comprise up to seven subsites. Thus, each one of the four aromatic side chains and the three polar residues that interdigitate the four tryptophans should bind to individual sugars of the polysaccharide chain, allowing for both hydrophobic and hydrophilic protein-carbohydrate interactions (Fig. 6A).
FIGURE 6.
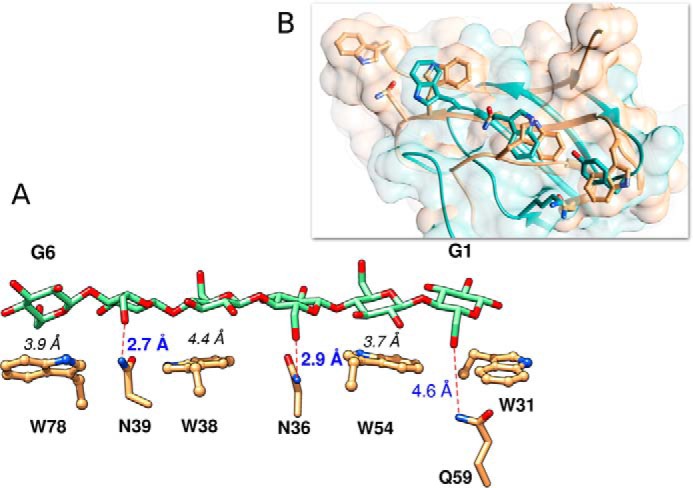
StCBM64C putative modeling with cellohexaose. A, StCBM64C active site residue side chain interactions with a model of a cellohexaose from CBM63 EXLX1 (PDB code 4FER). The six pyranose rings are numbered from the reducing end, as G1 to G6. The distance from the mass weighted ring centers of G2, G4, and G6 to, respectively, W54, W38, and W78, is given in black italic font. Putative H-bonds and their distances from N39/G5 and N36/G3, are marked as red dashed lines and bold blue font. The distance from Q59/G1 is also given. Relative to the PDB deposited structures, there is a 180° side chain flip for residues N36 and Q59. B, overlay of the active site from StCBM64C (tan) with CBM63 EXLX1 (turquoise) and respective molecular surfaces reveals conservation of the flat carbohydrate interacting platform. Molecular graphics performed with the UCSF Chimera package (41).
Structural Basis for Ligand Recognition by CBM64
Quantitative isothermal titration calorimetry (ITC) data, presented in Table 2 and exemplified in Fig. 7 showed that both StCBM64B and StCBM64C bound to regenerated (non-crystalline) insoluble cellulose (RC) very tightly, with a KA of ∼1.5–2.0 × 106 m−1. In addition, the two CBMs were unable to recognize cellohexaose, a property widely reported for type A CBMs and that result from the inability of the flat carbohydrate-binding platform to accommodate soluble, highly twisted oligosaccharides. The interaction of StCBM64B and StCBM64C with RC is enthalpically driven, although entropy makes a significant favorable contribution to ligand binding. Although type A CBMs bind regenerated cellulose through a predominantly endothermic process, which is driven by entropic forces, type B CBMs interact with soluble carbohydrates enthalpically. Thus, the thermodynamics of the binding of StCBM64B and StCBM64C to regenerated cellulose suggest that CBM64 displays an original mechanism of carbohydrate recognition that share properties with both type A and type B CBMs. A recent study revealed that some type A CBMs may interact with soluble polysaccharides, in particular with xyloglucan, a soluble β-1,4-glucan decorated with α-1,6-xylose side chains (40). Thus, the capacity of StCBM64B and StCBM64C to recognize a variety of soluble plant cell wall polysaccharides was initially evaluated by AGE. The data, presented in supplemental Fig. S9, revealed that both CBM64s bind different β-1,4- and β-1,3–1,4-glucan polysaccharides, in particular glucomannan, lichenan, hydroxyethyl-cellulose, carboxymethylcellulose, and the decorated glucan xyloglucan. The CBMs do not interact with β-1,3-glucan, xylan, galactomannan, galactan, arabinan, pullulan, and pectic polysaccharides. Thus, the binding of StCBM64B and StCBM64C to their soluble ligands was also investigated by ITC. The data, presented in Table 2 and exemplified in Fig. 7, confirmed that StCBM64B and StCBM64C bind to xyloglucan, hydroxyethylcellulose (HEC), glucomannan, and β-glucan. The two CBM64 members bound xyloglucan slightly less tightly than RC (Table 2). In addition, StCBM64B and StCBM64C bound HEC and glucomannan with a 20-fold lower affinity than xyloglucan and β-glucan more weakly (KA of ∼5 × 103 m−1). The capacity of typical type A CBMs, such as CBM2, CBM3 (40), and CBM64 (this study), to bind primarily to insoluble cellulose and, with a lower degree of affinity, also to soluble glucan polysaccharides suggests that the targeting role of type A CBMs is broader than previously anticipated. Binding to soluble cellulose may allow appended enzymes to initiate the attack on the plant cell wall by targeting first the exposed glucans and hemicelluloses, and once this activity uncovers the insoluble cellulose, the primary ligand for type A CBMs, the enzymes will target the crystalline regions of the polysaccharide.
TABLE 2.
Affinity and thermodynamic parameters of the binding of StCBM64B, StCBM64C, and its mutant derivatives to soluble and insoluble polysaccharides ligands
RC, regenerated cellulose; HEC, hydroxyethyl cellulose.
| Protein | Ligand | Ka | ΔG | ΔH | TΔS | n | |||
|---|---|---|---|---|---|---|---|---|---|
| m−1 × 104 | kcal mol−1 | kcal mol−1 | kcal mol−1 | ||||||
| StCBM64B-WT | RC | 144.00 ± 65.9 | −8.40 | −5.32 ± 0.26 | 3.07 | 1.10 ± 0.05 | |||
| Xyloglucan | 116.00 ± 12.2 | −8.29 | −4.95 ± 0.06 | 3.34 | 1.19 ± 0.01 | ||||
| Glucomannan | 1.03 ± 0.03 | −5.48 | −6.04 ± 0.08 | −0.56 | 1.02 ± 0.00 | ||||
| HEC | 1.10 ± 0.13 | −7.24 | −6.24 ± 0.14 | 1.00 | 1.00 ± 0.00 | ||||
| β−Glucan | 0.61 ± 0.23 | −5.17 | −1.87 ± 0.07 | 3.88 | 0.93 ± 0.02 | ||||
| Cellohexaose | No binding | No binding | No binding | No binding | No binding | ||||
| StCBM64C-WT | RC | 187.00 ± 48.70 | −8.66 | −6.51 ± 0.16 | 2.15 | 1.00 ± 0.02 | |||
| Xyloglucan | 69.60 ± 8.28 | −7.97 | −9.37 ± 0.17 | −1.43 | 1.05 ± 0.01 | ||||
| Glucomannan | 3.75 ± 0.32 | −6.25 | −13.55 ± 0.11 | −7.30 | 1.04 ± 0.07 | ||||
| HEC | 3.19 ± 0.29 | −8.09 | −3.55 ± 0.27 | 2.56 | 1.24 ± 0.07 | ||||
| β-Glucan | 0.53 ± 0.03 | −5.06 | −13.38 ± 0.48 | −8.32 | 0.99 ± 0.00 | ||||
| Cellohexaose | No binding | No binding | No binding | No binding | No binding | ||||
| StCBM64C-W31A | RC | 102.00 ± 11.30 | −7.44 | −11.19 ± 0.19 | −3.76 | 0.98 ± 0.01 | |||
| Xyloglucan | Binding too weak to accurately quantify | Binding too weak to accurately quantify | Binding too weak to accurately quantify | Binding too weak to accurately quantify | Binding too weak to accurately quantify | ||||
| StCBM64C-W38A | RC | 39.00 ± 5.29 | −7.63 | −7.15 ± 0.17 | 0.48 | 1.15 ± 0.02 | |||
| Xyloglucan | Binding too weak to accurately quantify | Binding too weak to accurately quantify | Binding too weak to accurately quantify | Binding too weak to accurately quantify | Binding too weak to accurately quantify | ||||
| StCBM64C-W54A | RC | 43.30 ± 9.04 | −7.65 | −11.14 ± 0.37 | −3.49 | 1.12 ± 0.04 | |||
| Xyloglucan | Binding too weak to accurately quantify | ||||||||
| StCBM64C-W78A | RC | 144.00 ± 34.30 | −8.41 | −3.49 ± 0.09 | 4.92 | 2.0 ± 0.00 | |||
| Xyloglucan | 1.09 ± 0.48 | −5.50 | −9.29 ± 0.22 | −3.79 | 1.0 ± 0.00 | ||||
| StCBM64C-W31A/W38A | RC | Binding too weak to accurately quantify | Binding too weak to accurately quantify | Binding too weak to accurately quantify | Binding too weak to accurately quantify | Binding too weak to accurately quantify | |||
| Xyloglucan | No binding | No binding | No binding | No binding | No binding | ||||
| StCBM64C-W31A/W54A | RC | Not determined | Not determined | Not determined | Not determined | Not determined | |||
| Xyloglucan | No binding | No binding | No binding | No binding | No binding | ||||
| StCBM64C-W38A/W54A | RC | Not determined | Not determined | Not determined | Not determined | Not determined | |||
| Xyloglucan | No binding | No binding | No binding | No binding | No binding | ||||
| StCBM64C-W31A/W38A/W54A | RC | Not determined | Not determined | Not determined | Not determined | Not determined | |||
| Xyloglucan | No binding | No binding | No binding | No binding | No binding | ||||
| RC | Binding too weak to accurately quantify | Binding too weak to accurately quantify | Binding too weak to accurately quantify | Binding too weak to accurately quantify | Binding too weak to accurately quantify | ||||
| Xyloglucan | 28.40 ± 8.48 | −7.42 | −2.77 ± 0.18 | 4.65 | 1.08 ± 0.05 | ||||
| RC | No binding | No binding | No binding | No binding | No binding | ||||
| Xyloglucan | 217.00 ± 41.90 | −8.64 | −7.15 ± 0.13 | 1.49 | 1.29 ± 0.01 | ||||
| StCBM64C-Q59A | RC | 177.00 ± 22.30 | −8.54 | −3.38 ± 0.05 | 5.16 | 0.99 ± 0.01 | |||
| Xyloglucan | 56.20 ± 6.52 | −7.84 | −3.96 ± 0.06 | 3.88 | 1.15 ± 0.01 | ||||
| RC | No binding | No binding | No binding | No binding | No binding | ||||
| Xyloglucan | 24.70 ± 2.40 | −7.36 | −8.79 ± 0.15 | −1.43 | 1.15 ± 0.01 | ||||
| RC | Binding too weak to accurately quantify | Binding too weak to accurately quantify | Binding too weak to accurately quantify | Binding too weak to accurately quantify | Binding too weak to accurately quantify | ||||
| Xyloglucan | 1.74 ± 0.164 | −5.78 | −10.34 ± 0.90 | −4.56 | 1.07 ± 0.06 | ||||
| RC | No binding | No binding | No binding | No binding | No binding | ||||
| Xyloglucan | 142.00 ± 11.60 | −5.17 | −6.66 ± 0.07 | 1.49 | 0.99 ± 0.00 | ||||
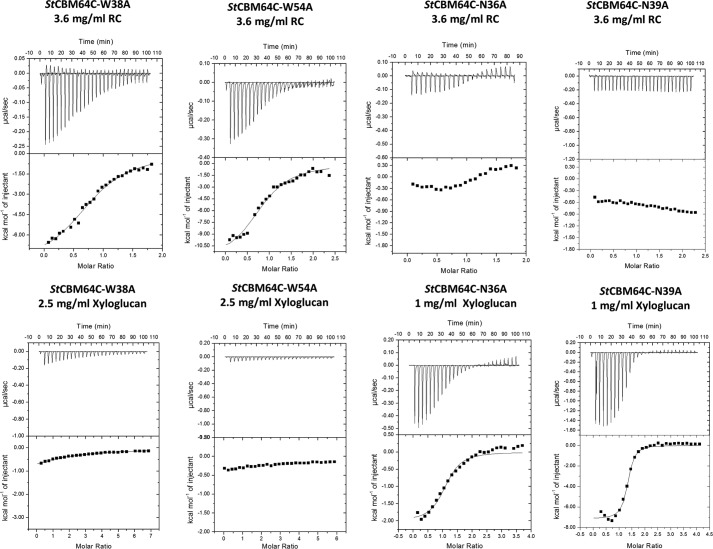
The location of CBM64 carbohydrate interacting platform was probed by analyzing the binding efficacy of different mutant derivatives of StCBM64B and StCBM64C. Initial pulldown qualitative assays performed with StCBM64C using Avicel as the insoluble ligand (supplemental Fig. S10) suggested that alanine substitution of any of the four tryptophan residues identified at the surface of β-sheet 2 had no effect on binding (supplemental Fig. S10). Thus, the interaction of StCBM64B and StCBM64C with RC was probed by ITC. StCBM64C W31A, W38A, and W54A derivatives displayed a moderate reduction in affinity to the insoluble ligand (Table 2 and Fig. 8). Complete abolition of RC recognition was only observed when double or triple tryptophan mutant derivatives were produced (Table 2). In contrast, single tryptophan alanine substitutions were sufficient to abolish xyloglucan recognition, which may be accounted for by the lower affinity observed against this polysaccharide. In addition, it is possible that the four aromatic side chains could straddle both the glucose and xylose units of xyloglucan, contributing to their exclusive binding importance. Strikingly, when Asn-36 and Asn-39 were singly substituted by alanine, the two CBM64 mutant derivatives lost their capacity to bind RC (Table 2), suggesting that the side chains of these residues hydrogen bond the C6 hydroxyls of glucose molecules as suggested by the model described above (Fig. 6A). In contrast, the side chain of Gln-59 does not seem to be relevant for hydrogen bonding the polysaccharide chains (Table 2). The critical contribution of the polar interactions for the recognition of insoluble ligands by CBM64 may result from their role in directing the CBM into a correct positioning in relation to the polysaccharide chain. Strikingly, N36A and N39A mutant derivatives display no diminished capacity to bind xyloglucan. Because xyloglucan is a decorated polysaccharide, with glucose residues substituted at C6 with xylose, there is no longer an hydrogen bond partner at this position of the polysaccharide. It is possible, however, that other residues of StCBM64C may bind the decorations of xyloglucan, although mutation of Arg-53 or Glu-84 had no effect in glycan recognition (supplemental Fig. S9). Consistently, Asn-36 and Asn-39, but particularly Asn-36, are critical for the recognition of the soluble glucans that act as ligands for CBM64 (supplemental Fig. S9). Thus, taken together, these data suggest that Asn-36 and Asn-39 play a critical role for the binding of non-decorated cellulose, both in the soluble and insoluble forms, whereas hydrophobic interactions dominate xyloglucan recognition. The four tryptophan residues and the two asparagine that comprise the carbohydrate binding platform of StCBM64C are completely conserved within CBM64, suggesting a common mechanism of ligand recognition in this CBM family.
FIGURE 8.
Representative ITC data of the interaction of mutant derivatives of StCBM64C with RC and xyloglucan. Titrations were conducted as described in the legend to Fig. 7. The top portion of each panel shows the raw power data, whereas the bottom panel shows the integrated and heat of dilution corrected data.
Conclusions
This report reveals that a wide range of type A CBMs have the capacity to recognize crystalline polysaccharides under a wide range of physiological conditions. The robust properties revealed by type A CBMs may have evolved as an adaptation to function under ecological niches imposing significant selective pressures, such as the mammal's gastrointestinal tract or the soil. From the CBMs of the various families analyzed, CBM64 displays a wider capacity to function under extreme conditions. The crystal structure of StCBM64C from S. thermophila combined with the site-directed mutagenesis experiments show that the flat surface identified in the protein structure constitutes the ligand-binding site and that both the side chains of Trp and Asn residues play a central role in carbohydrate recognition. The side chains of the four aromatic residues are co-planar and form an extensive hydrophobic platform that recognizes the predominantly apolar regions of the crystalline ligand. In addition, binding is assisted by polar interactions, particularly by Asn-36 and Asn-39. CBM64 has probably the most extensive binding platform described so far for a type A CBM, which is able to interact not only with crystalline carbohydrates, but also with diverse soluble polysaccharides, such as xyloglucan, glucomannan, β-1,4-glucan, and β-1,3–1,4-glucan. The structure and function of CBM64 illustrate how type A CBMs have evolved to increased ligand plasticity and biochemical stability, allowing potentiating plant cell wall degradation by complex cellulolytic systems.
Materials and Methods
Isolation of Microbial Genes through PCR and Gene Synthesis
A total of 96 genes encoding type A CBMs from different families were isolated either through PCR or gene synthesis. A high throughput pipeline was used for the efficient PCR assembly and cloning of the 96 constructs. The genes encoding 40 bacterial CBMs for which there was access to gDNA were isolated through PCR using NZYProof DNA polymerase (NZYTech Ltd., Lisboa, Portugal). Gene synthesis was used to produce 56 genes for which there was no access to a gDNA template. Artificial genes were designed with a codon usage table optimized for expression in E. coli and synthesized using standard procedures (23). Purified nucleic acids (∼50 ng) were subsequently cloned into pHTP9 E. coli expression vector using the NZYEasy cloning kit (NZYTech Ltd.), according to established protocols for the ligation independent cloning technology. In pHTP9, genes were expressed under the control of a T7 promoter. Encoded recombinant CBMs were fused to an N-terminal GFP and include an internal His6 tag for purification through IMAC. The 96 resulting recombinant plasmids were termed pHTP9_1 to pHTP9_96 and were sequenced to ensure that no mutations accumulated during gene isolation/synthesis and cloning. Recombinant fusion GFP-CBMs were termed CBM #1–CBM #96. The primary sequences of the 96 CBMs, their respective origins, and their families are presented in supplemental Table S1. For protein crystallography, a CBM #56 derivative lacking the GFP partner was obtained by isolating the respective gene from pHTP9_56 and cloning into pET21a, generating pVp1 (Novagen). The recombinant CBM #56 derivative encoded by the pET21a vector was termed StCBM64B. An homologue of StCBM64B, termed StCBM64C, was also recombinantly produced by synthesizing the artificial nucleic acid as described above and cloning the respective gene into pET21a, generating pVp2. StCBM64B and StCBM64C, encoded by pET21 derivatives pVp1 and Vp2, contain C-terminal His6 tags to allow purification through IMAC. The recombinant sequences of StCBM64B and StCBM64C are both displayed in supplemental Table S1.
Site-directed Mutagenesis
StCBM64B and StCBM64C mutant derivatives were generated by site-directed mutagenesis using plasmids pVp1 and Vp2 as templates. The primers used to generate these mutants are shown in supplemental Table S2. The generated nucleic acids were sequenced to ensure that only the appropriate mutations had been incorporated in the nucleic acids.
Expression of Recombinant CBMs in E. coli
The 96 pHTP9 plasmid derivatives were used to transform E. coli BL21 (DE3) cells. Recombinant E. coli cells were grown in NZY autoinduction LB medium (NZYTech Ltd.) supplemented with kanamycin (50 μg/ml) at 37 °C to early exponential phase (A600 nm = 1.5–2.0), and recombinant protein production occurred following a further incubation at 25 °C for 16 h. Recombinant E. coli cells were harvested by centrifugation at 1500 g at 4 °C for 15 min and lysed in NZY Bacterial Cell Lysis Buffer (NZYTech Ltd.). The His6-tagged recombinant GFP-CBMs were purified from cell-free extracts through IMAC in a high throughput pipeline as described previously (24). Recombinant proteins were eluted in 50 mm NaHepes, pH 7.5, 500 mm NaCl, and 300 mm imidazole. Homogeneity of purified proteins and molecular mass of recombinant CBMs were assessed by SDS-PAGE in 14% (w/v) acrylamide gels (supplemental Fig. S1). Protein concentration of GFP-CBM stock solutions varied between 0.5 and 3 mg/ml, as determined spectrophotometrically. For crystallographic studies, StCBM64B and StCBM64C were expressed as described above (except for the LB medium containing 100 mg/ml ampicillin that was used), purified through IMAC, and further purified by size exclusion chromatography. Following IMAC, fractions containing the purified proteins were buffer-exchanged, using PD-10 Sephadex G-25 M gel filtration columns (GE Healthcare), into 50 mm NaHepes buffer, pH 7.5, containing 200 mm NaCl and 5 mm CaCl2 and were then subjected to gel filtration using a HiLoad 16/60 Superdex75 column (GE Healthcare) at a flow rate of 1 ml/min. Purified StCBM64B and StCBM64C were concentrated using an Amicon 10-kDa molecular mass centrifugal concentrator and washed three times with 1 mm CaCl2. StCBM64B and StCBM64C purity was assessed by SDS/PAGE, and protein concentration was determined spectrophotometrically.
Oligosaccharides and Polysaccharides
All soluble sugars (barley mixed-linkage glucan (low viscosity), Konjac glucomannan, Tamarind xyloglucan, and carboxymethyl cellulose) and oligosaccharides were purchased from Megazyme International (Bray, Ireland). HEC, Sigmacell Cellulose Type 20, microcrystalline cellulose powder, low molecular weight chitosan, potato starch, and Whatman no.1 chromatography paper were obtained from Sigma, and Avicel was purchased from Merck. RC was prepared as described (25).
Biochemical Assays Using GFP-CBM Proteins
The interactions of GFP-CBM fusions with Sigmacell cellulose type 20, microcrystalline cellulose powder, low molecular weight chitosan, potato starch, and Whatman no. 1 chromatography paper were determined experimentally following a protocol directed to detect the fluorescence of GFP. First, solutions of GFP-CBM with concentrations between 0.06 and 0.6 μm were prepared by diluting the corresponding stock solutions with the binding buffers under study. Most experiments were performed using 100 mm phosphate buffer at pH 7. Citrate and carbonate buffers at 100 mm were used to cover the pH ranges 5–6 and 9.5–10, respectively. The effect of ionic strength was studied by varying the concentration of phosphate buffer in the 100–1500 mm range. In preparation for the binding experiments, 5 mg of the substrate under study were loaded onto the wells of MultiScreen-HV filter plates with 0.45-μm PVDF membranes (Merck-Millipore) and washed/equilibrated for 15 min with 3 × 200 μl of the relevant buffer. A MultiScreenHTS vacuum manifold (Merck-Millipore) was used to filter and discard spent buffer between washing steps. Then 200 μl of solutions containing 10, 20, 40, 60, 80, or 100 pmol of the GFP-CBM under study were loaded into each well and incubated for 20 min at the desired temperature (4–90 °C) in a well plate agitator/incubator. After incubation, the supernatants were directly transferred to the 96 wells of a white microplate by filtration with the vacuum manifold. The fluorescence of the recovered filtrates was measured using a Cary Eclipse fluorescence spectrophotometer (Varian) with excitation and emission set at 490 and 510 nm, respectively. The fluorescence of the solutions prior to loading was also measured in a microplate. Appropriate dilutions were used to ensure that measurements were performed in a region of linearity between fluorescence and concentration. Mass balance calculations were then performed to determine the equilibrium concentrations of GFP-CBM in the solid (pmol/mg) and liquid (pmol/ml) phases. Each binding condition was tested in triplicate. The binding of the GFP-CBM fusions was also analyzed qualitatively by examining the solid matrices recovered after the incubation procedure described above by fluorescence microscopy (Leica DMLB; Leica Microsystems). Control experiments were performed using GFP.
Affinity Gel Electrophoresis
Affinity gel electrophoresis was used to evaluate the capacity of StCBM64B and StCBM64C and their respective mutant derivatives to interact with soluble polysaccharides. The method used was essentially as described (26), using the polysaccharide ligands at a concentration of 0.25% (w/v). The nonbinding negative control protein was BSA.
Binding to Insoluble Polysaccharide
Qualitative assessment of the affinity of StCBM64B and StCBM64C and their mutant derivatives to insoluble cellulose (Avicel) was carried out as follows: 150 μl of ∼800 μg ml−1 solution of the recombinant CBMs in 50 mm NaHepes, pH 7.5, containing 100 mm NaCl, 5 mm CaCl2, and 0.05% (v/v) Tween 20 (Buffer A) was mixed with 12 mg of insoluble polysaccharide (Avicel). The reaction mixture was incubated for 1 h at room temperature with gentle shaking, after which time the insoluble ligand was collected by centrifugation at 13,000 × g for 10 min. The supernatant, comprising the unbound fraction, was removed, and the pellet was washed five times with 150 μl of buffer A. Bound and unbound fractions were analyzed by SDS-PAGE using a 16% acrylamide gel. Controls containing protein but no Avicel were included in parallel to ensure that no precipitation occurred during the assay period.
Isothermal Titration Calorimetry
ITC was performed essentially as described previously (27), using a Microcal VP-ITC calorimeter (Northampton, MA) at 25 °C. Before the experiment, purified proteins were buffer-exchanged against 50 mm phosphate buffer, pH 7.0, containing 0.1 mm CaCl2. The reaction cell contained protein at 30 μm, whereas the syringe contained either the oligosaccharides at 0.5–10 mm or the soluble polysaccharides at 1–6 mg/ml. The ligands were dissolved in the dialysis buffer (separately) to minimize heats of dilution. Titrations were performed by a first injection of 2 μl followed by 28 subsequent injections of 10-μl aliquots of either polysaccharide or oligosaccharide at 220-s intervals into ITC sample cell (volume, 1.4467 ml) containing different enzyme samples. The stirring speed and reference power were set at 307 rpm and 15 μcal/s, respectively. The heat background was measured under the same conditions by dropping the buffer only without ligand into the protein at the same concentration as in the cell. The molar concentration of CBM binding sites present in polysaccharide ligands was determined as described previously (28). Data analysis was performed by non-linear regression using a single binding model (Microcal Origin 7.0 software), and thermodynamic parameters, such as the association constant (Ka), number of binding sites in the protein (n), and the binding enthalpy change (ΔH) were determined. Gibbs free energy change (ΔG) and the entropy change (ΔS) were calculated according to the following equations: −RTlnKa = ΔG = ΔH − TΔS, where R is the gas constant, and T represents the absolute temperature (Luís et al., (42)). For experiments with RC, the ligand in the cell was at 3.6 mg/ml, and the protein (250–500 μm) but not the polysaccharide was injected.
Crystallization, Data Collection, Structure Determination, and Refinement
Crystallization experiments were performed using the hanging drop vapor diffusion method, and drops were prepared at 293.15 K by adding an equal volume (1 μl) of protein (35 mg/ml) and reservoir solution [1% (w/v) Jeffamine ED-2001, 0.1 m HEPES, pH 7.5, and 1 m succinic acid, pH 7.0). StCBM64C crystals were harvested from the crystallization drop and transferred into a cryo-stabilization solution mimicking the mother liquor supplemented with 30% (w/v) glycerol. Structure determination was done by the SAD method, using the anomalous diffraction of the sulfur atoms. The SAD experiment was conducted on Beamline PXIII (X06DA) of the Swiss Light Source (Paul-Scherrer Institut, Villigen, Switzerland). The crystals diffracted up to 1.5 Å resolution and belonged to P43212 space group. Complete native and sulfur-SAD data sets were collected and processed with XDS (29) and scaled with AIMLESS (30). The data collection and processing statistics are presented in Table 1.
De novo phasing was achieved by sulfur-SAD using synchrotron radiation with a wavelength of 1.8 Å on a single crystal using an inverse beam strategy with wedges of 10 degrees. At this wavelength, the crystal diffracted up to 2.3 Å resolution. Structure determination of StCBM64C was performed using AutoSol (31) from PHENIX (32). The SAD phasing, auto-building, and refinement using AutoSol found 2 sulfur sites and built a partial model with 83 residues and a final Rwork and Rfree of 0.209 and 0.225, respectively. This partial model was used as phase estimates for a higher (1.5 Å) resolution data set collected from a second isomorphous crystal at the Swiss Light Source, using radiation with 1.0 Å wavelength. The data collection and processing statistics for this second data set are also presented in Table 1. Restrained refinement was performed with PHENIX (32) using this higher resolution data set. Inspection of the electron density maps was carried out using COOT (33). In the final stages of refinement, the Rwork and Rfree converged to 11.9 and 14.5%. Geometrical validation and model improvement was carried out using PDB_REDO (34) and several validation programs such as PROCHECK (35) and MOLPROBITY (36). Analysis of the model showed that 98.9% of the protein residues are in the most favored or additionally allowed regions of the Ramachandran plot. Refinement statistics are summarized in Table 1. Coordinates and observed structure factor amplitudes have been deposited in the Protein Data Bank under the accession code 5LU3.
Modeling of the Cellohexaose from BsCBM63 onto the StCBM64C
Structure of a type A CBM from EXLX1, a bacterial expansin from Bacillus subtilis, with bound cellohexaose (BsCBM63, PDB code 4FER (37)) was used to analyze the potential putative binding site of StCBM64C. BsCBM63 was shown to bind carbohydrates through hydrophobic interactions of three co-planar linearly arranged aromatic residues: Trp-125, Trp-126, and Tyr-157. Because there is little sequence or structural homology between the two structures, the three co-linear aromatics from BsCBM63 were manually superposed on the four co-linear aromatics in StCBM64C: Trp-31, Trp-38, Trp-54, and Trp-78 based on their mass-weighted ring centers using CHIMERA (41). The aromatic rings of Trp-125, Trp-126, and Tyr-157 in BsCBM63 are nearly parallel to the planes of the pyranose rings of cellohexaose (CE6), which adopts the undistorted 4C1 chair conformation in an almost linear glycan chain. To optimize the interaction of the CE6 to StCBM64C, the CE6 position was tweaked to keep the distance between the aromatic and the pyranose rings between 3.0 and 4.5 Å (to account for CH–π interactions) and maximize the polar interactions between the CE6 and the Asn-36, Asn-39, and Gln-59. These residues are found on one side of the putative binding site between the co-linear tryptophans in StCBM64C but do not have an equivalent in the BsCBM63 structure.
Author Contributions
L. M. A. F., A. L. C., C. M. G. A. F., and D. M. P. conceived the experiments. P. M. M. P., V. M. R. P., J. L. A. B., and M. C. developed the majority of the experimental analysis. V. C., P. B., V. D. A., I. V., and M. J. R. assisted with data collection. C. M. G. A. F. and D. M. P. supervised the project, analyzed the data, and wrote the paper with contributions from the remaining authors.
Supplementary Material
Acknowledgments
We acknowledge access to the PXIII Beamline at the Swiss Light Source of the Paul Scherrer Institut (Villigen, Switzerland).
This work was supported by Qren, Grant 30270, and by Fundação para a Ciência e Tecnologia through projects PTDC/BIA-MIC/5947/2014, IF/01621/2013, RECI/BBB-BEP/0124/2012, PEst-C/EQB/LA0006/2013, and UID/Multi/04378/2013. This work was also supported by Fellowships IF/01621/2013 (to V. M. R. P.), SFRH/BPD/64917/2009 (to M. C.), SFRH/BD/86821/2012 (to P. B.), and SFRH/BDE/101026/2014 (to V. C.) and by Institute for Bioengineering and Biosciences through FCT-Portuguese Foundation for Science and Technology Grant UID/BIO/04565/2013 and by Programa Operacional Regional de Lisboa 2020 Project 007317. C. M. G. A. F. is financial beneficiary of the company that sells the cloning kits used in this study.

This article contains supplemental Tables S1 and S2 and Figs. S1–S10.
The atomic coordinates and structure factors (code 5LU3) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- CAZyme
- Carbohydrate-Active Enzyme
- PDB
- Protein Data Bank
- CBM
- carbohydrate-binding module
- IMAC
- immobilized metal affinity chromatography
- ITC
- isothermal titration calorimetry
- RC
- regenerated cellulose
- HEC
- hydroxyethylcellulose
- SAD
- single-wavelength anomalous dispersion.
References
- 1. Tomme P., Van Tilbeurgh H., Pettersson G., Van Damme J., Vandekerckhove J., Knowles J., Teeri T., and Claeyssens M. (1988) Studies of the cellulolytic system of Trichoderma reesei QM 9414: analysis of domain function in two cellobiohydrolases by limited proteolysis. Eur. J. Biochem. 170, 575–581 [DOI] [PubMed] [Google Scholar]
- 2. Gilbert H. J., Hall J., Hazlewood G. P., and Ferreira L. M. (1990) The N-terminal region of an endoglucanase from Pseudomonas fluorescens subspecies cellulosa constitutes a cellulose-binding domain that is distinct from the catalytic centre. Mol. Microbiol. 4, 759–767 [DOI] [PubMed] [Google Scholar]
- 3. Bolam D. N., Ciruela A., McQueen-Mason S., Simpson P., Williamson M. P., Rixon J. E., Boraston A., Hazlewood G. P., and Gilbert H. J. (1998) Pseudomonas cellulose-binding domains mediate their effects by increasing enzyme substrate proximity. Biochem. J. 331, 775–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boraston A. B., Kwan E., Chiu P., Warren R. A., and Kilburn D. G. (2003) Recognition and hydrolysis of noncrystalline cellulose. J. Biol. Chem. 278, 6120–6127 [DOI] [PubMed] [Google Scholar]
- 5. Gilbert H. J., Knox J. P., and Boraston A. B. (2013) Advances in understanding the molecular basis of plant cell wall polysaccharide recognition by carbohydrate-binding modules. Curr. Opin. Struct. Biol. 23, 669–677 [DOI] [PubMed] [Google Scholar]
- 6. Lamed R., and Zeikus J. G. (1980) Ethanol production by thermophilic bacteria: relationship between fermentation product yields of and catabolic enzyme activities in Clostridium thermocellum and Thermoanaerobium brockii. J. Bacteriol. 144, 569–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Demain A. L., Newcomb M., and Wu J. H. (2005) Cellulase, clostridia, and ethanol. Microbiol. Mol. Biol. Rev. 69, 124–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., and Henrissat B. (2009) The carbohydrate-active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lombard V., Golaconda Ramulu H., Drula E., Coutinho P. M., and Henrissat B. (2014) The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boraston A. B., Bolam D. N., Gilbert H. J., and Davies G. J. (2004) Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Raghothama S., Simpson P. J., Szabó L., Nagy T., Gilbert H. J., and Williamson M. P. (2000) Solution structure of the CBM10 cellulose binding module from Pseudomonas xylanase A. Biochemistry 39, 978–984 [DOI] [PubMed] [Google Scholar]
- 12. Tormo J., Lamed R., Chirino A. J., Morag E., Bayer E. A., Shoham Y., and Steitz T. A. (1996) Crystal structure of a bacterial family-III cellulose-binding domain: a general mechanism for attachment to cellulose. EMBO J. 15, 5739–5751 [PMC free article] [PubMed] [Google Scholar]
- 13. Xu G. Y., Ong E., Gilkes N. R., Kilburn D. G., Muhandiram D. R., Harris-Brandts M., Carver J. P., Kay L. E., and Harvey T. S. (1995) Solution structure of a cellulose-binding domain from Cellulomonas fimi by nuclear magnetic resonance spectroscopy. Biochemistry 34, 6993–7009 [PubMed] [Google Scholar]
- 14. Creagh A. L., Ong E., Jervis E., Kilburn D. G., and Haynes C. A. (1996) Binding of the cellulose-binding domain of exoglucanase Cex from Cellulomonas fimi to insoluble microcrystalline cellulose is entropically driven. Proc. Natl. Acad. Sci. U.S.A. 93, 12229–12234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shoseyov O., Shani Z., and Levy I. (2006) Carbohydrate binding modules: biochemical properties and novel applications. Microbiol. Mol. Biol. Rev. 70, 283–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Oliveira C., Carvalho V., Domingues L., and Gama F. M. (2015) Recombinant CBM-fusion technology: applications overview. Biotechnol. Adv. 33, 358–369 [DOI] [PubMed] [Google Scholar]
- 17. Cao Y., Zhang Q., Wang C., Zhu Y., and Bai G. (2007) Preparation of novel immunomagnetic cellulose microspheres via cellulose binding domain-protein A linkage and its use for the isolation of interferon alpha-2b. J. Chromatogr. A 1149, 228–235 [DOI] [PubMed] [Google Scholar]
- 18. Hussack G., Luo Y., Veldhuis L., Hall J. C., Tanha J., and Mackenzie R. (2009) Multivalent anchoring and oriented display of single-domain antibodies on cellulose. Sensors 9, 5351–5367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rosa A. M., Louro A. F., Martins S. A., Inácio J., Azevedo A. M., and Prazeres D. M. (2014) Capture and detection of DNA hybrids on paper via the anchoring of antibodies with fusions of carbohydrate binding modules and ZZ-domains. Anal. Chem. 86, 4340–4347 [DOI] [PubMed] [Google Scholar]
- 20. Ofir K., Berdichevsky Y., Benhar I., Azriel-Rosenfeld R., Lamed R., Barak Y., Bayer E. A., and Morag E. (2005) Versatile protein microarray based on carbohydrate-binding modules. Proteomics 5, 1806–1814 [DOI] [PubMed] [Google Scholar]
- 21. Tolba M., Minikh O., Brovko L. Y., Evoy S., and Griffiths M. W. (2010) Oriented immobilization of bacteriophages for biosensor applications. Appl. Environ. Microbiol. 76, 528–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang A. A., Mulchandani A., and Chen W. (2002) Specific adhesion to cellulose and hydrolysis of organophosphate nerve agents by a genetically engineered Escherichia coli strain with a surface-expressed cellulose-binding domain and organophosphorus hydrolase. Appl. Environ. Microbiol. 68, 1684–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sequeira A. F., Guerreiro C. I., Vincentelli R., Fontes C. M. (2016) T7 endonuclease I mediates error correction in artificial gene synthesis. Mol. Biotechnol. 58, 573–584 [DOI] [PubMed] [Google Scholar]
- 24. Saez N. J., and Vincentelli R. (2014) High-throughput expression screening and purification of recombinant proteins in E. coli. Methods Mol. Biol. 1091, 33–53 [DOI] [PubMed] [Google Scholar]
- 25. Boraston A. B. (2005) The interaction of carbohydrate-binding modules with insoluble non-crystalline cellulose is enthalpically driven. Biochem. J. 385, 479–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Henshaw J. L., Bolam D. N., Pires V. M., Czjzek M., Henrissat B., Ferreira L. M., Fontes C. M., and Gilbert H. J. (2004) The family 6 carbohydrate binding module CmCBM6–2 contains two ligand-binding sites with distinct specificities. J. Biol. Chem. 279, 21552–21559 [DOI] [PubMed] [Google Scholar]
- 27. Carvalho A. L., Goyal A., Prates J. A., Bolam D. N., Gilbert H. J., Pires V. M., Ferreira L. M., Planas A., Romão M. J., and Fontes C. M. (2004) The family 11 carbohydrate-binding module of Clostridium thermocellum Lic26A-Cel5E accommodates β-1,4- and β-1,3–1,4-mixed linked glucans at a single binding site. J. Biol. Chem. 279, 34785–34793 [DOI] [PubMed] [Google Scholar]
- 28. Bolam D. N., Xie H., Pell G., Hogg D., Galbraith G., Henrissat B., and Gilbert H. J. (2004) X4 modules represent a new family of carbohydrate-binding modules that display novel properties. J. Biol. Chem. 279, 22953–22963 [DOI] [PubMed] [Google Scholar]
- 29. Kabsch W. (2010) XDS. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 31. Terwilliger T. C., Grosse-Kunstleve R. W., Afonine P. V., Moriarty N. W., Zwart P. H., Hung L. W., Read R. J., and Adams P. D. (2008) Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 64, 61–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 34. Joosten R. P., Long F., Murshudov G. N., and Perrakis A. (2014) The PDB_REDO server for macromolecular structure model optimization. IUCrJ. 1, 213–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Laskowski R. A., MacArthur M. W., Moss D. S., and Thornton J. M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 36. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Georgelis N., Yennawar N. H., and Cosgrove D. J. (2012) Structural basis for entropy-driven cellulose binding by a type-A cellulose-binding module (CBM) and bacterial expansin. Proc. Natl. Acad. Sci. U.S.A. 109, 14830–14835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Angelov A., Loderer C., Pompei S., and Liebl W. (2011) Novel family of carbohydrate-binding modules revealed by the genome sequence of Spirochaeta thermophila DSM 6192. Appl. Environ. Microbiol. 77, 5483–5489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schiefner A., Angelov A., Liebl W., and Skerra A. (2016) Structural basis for cellulose binding by the type A carbohydrate-binding module 64 of Spirochaeta thermophila. Proteins 84, 855–858 [DOI] [PubMed] [Google Scholar]
- 40. Hernandez-Gomez M. C., Rydahl M. G., Rogowski A., Morland C., Cartmell A., Crouch L., Labourel A., Fontes C. M., Willats W. G., Gilbert H. J., and Knox J. P. (2015) Recognition of xyloglucan by the crystalline cellulose-binding site of a family 3a carbohydrate-binding module. FEBS Lett. 589, 2297–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 42. Luís A. S., Venditto I., Temple M. J., Rogowski A., Baslé A., Xue J., Knox J. P., Prates J. A., Ferreira L. M., Fontes C. M., Najmudin S., and Gilbert H. J. (2013) Understanding how noncatalytic carbohydrate binding modules can display specificity for xyloglucan. J. Biol. Chem. 288, 4799–4809 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.