Abstract
Dihydrofolate reductase (DHFR) plays a key role in folate metabolism and is a target molecule of methotrexate. An increase in the cellular expression level of DHFR is one of the mechanisms of tumor resistance to methotrexate. The present study investigated the possibility that adenosine-to-inosine RNA editing, which causes nucleotide conversion by adenosine deaminase acting on RNA (ADAR) enzymes, might modulate DHFR expression. In human breast adenocarcinoma-derived MCF-7 cells, 26 RNA editing sites were identified in the 3′-UTR of DHFR. Knockdown of ADAR1 decreased the RNA editing levels of DHFR and resulted in a decrease in the DHFR mRNA and protein levels, indicating that ADAR1 up-regulates DHFR expression. Using a computational analysis, miR-25-3p and miR-125a-3p were predicted to bind to the non-edited 3′-UTR of DHFR but not to the edited sequence. The decrease in DHFR expression by the knockdown of ADAR1 was restored by transfection of antisense oligonucleotides for these miRNAs, suggesting that RNA editing mediated up-regulation of DHFR requires the function of these miRNAs. Interestingly, we observed that the knockdown of ADAR1 decreased cell viability and increased the sensitivity of MCF-7 cells to methotrexate. ADAR1 expression levels and the RNA editing levels in the 3′-UTR of DHFR in breast cancer tissues were higher than those in adjacent normal tissues. Collectively, the present study demonstrated that ADAR1 positively regulates the expression of DHFR by editing the miR-25-3p and miR-125a-3p binding sites in the 3′-UTR of DHFR, enhancing cellular proliferation and resistance to methotrexate.
Keywords: anticancer drug, breast cancer, cancer biology, microRNA (miRNA), RNA editing, dihydrofolate reductase
Introduction
Dihydrofolate reductase (DHFR)3 is a key enzyme in folate metabolism. DHFR catalyzes the reduction of dihydrofolate to tetrahydrofolate using NADPH as a cofactor. Because tetrahydrofolate is essential for de novo purine and thymidylate synthesis required for DNA synthesis, cell growth, and proliferation, DHFR is a target of chemotherapeutic agents such as methotrexate and pemetrexed (1–3). The clinical efficacy of methotrexate is often limited by the acquisition of resistance in cancer cells. As mechanisms of methotrexate resistance, mutations in the DHFR gene leading to a decreased affinity of DHFR protein to methotrexate (4–6) and decreased uptake of methotrexate due to impaired transport (7–9) are known. In addition, overexpression of DHFR protein in methotrexate-resistant cells (10) has been considered. DHFR expression is regulated by multiple mechanisms (11), including gene amplification (12, 13), Sp1 and E2F1-mediated transcriptional up-regulation (14, 15), as well as microRNA (miRNA)-mediated post-transcriptional repression (16–18). In this study, we sought to investigate a possibility that RNA editing might also underlie the regulatory mechanism.
RNA editing is a post-transcriptional process that alters the nucleotide sequence of RNA transcripts. In animals, the most common type of RNA editing is deamination of adenosine (A) into inosine (I), A-to-I RNA editing (19, 20). Because much of the cellular machinery treats inosine as a guanine nucleotide, the conversion of nucleotides potentially changes the amino acid sequence, splicing, miRNA targeting, or miRNA maturation (21). A-to-I RNA editing is catalyzed by adenosine deaminases acting on RNA (ADAR) enzymes (22, 23). They convert adenosines in double-stranded RNA (dsRNA) structures into inosines by hydrolytic deamination. There are three members of the ADAR family in vertebrates: ADAR1, ADAR2, and ADAR3 (also called ADAR, ADARB1, and ADARB2, respectively) (24). ADAR1 and ADAR2 are ubiquitously expressed and have RNA editing activity. In contrast, there is no evidence to support the enzymatic activity of ADAR3 (25). The ADAR1 gene produces two protein isoforms, ADAR1 p110 (110-kDa protein) and ADAR1 p150 (150-kDa protein), from different transcription initiation sites and start codons. The former is constitutively expressed in the nucleus, whereas the latter is localized in both nuclear and cytoplasmic compartments and induced by interferon (26, 27).
Early studies revealed that RNA editing plays important roles in the central nervous system (28). For example, glutamate receptor subtype A2 and 5-hydroxytryptamine receptor subtype 2C were known to be subjected to RNA editing (29–31), and the disruption of RNA editing in these genes leads to amyotrophic lateral sclerosis and Prader-Willi syndrome, respectively (32, 33). In these cases, A-to-I editing occurs within exons, changing crucial amino acids for protein function. Accumulating evidence has revealed that aberrant ADAR expression and disrupted RNA editing levels are associated with diseases including cancer, metabolic diseases, viral infections, autoimmune disorders, and neurological disorders (34). Recent progress in next-generation sequencing with RNA-Seq has resulted in the identification of global RNA editing sites in non-coding as well as coding regions, supporting broader roles of RNA editing in the body (35, 36). Information regarding RNA editing sites in the transcriptome is compiled in databases such as RADAR, but the biological significance of RNA editing in humans has not been completely elucidated. We noticed that DHFR has been included in RADAR, as a target of RNA editing. Multiple RNA editing sites were identified at introns 3, 4, and 5 as well as 3′-UTR of DHFR mRNA in the lymphoblastoid cell line and brain (37, 38). However, it is uncertain whether the DHFR is edited or not in breast cancer, in which ADAR1 has been reported to function as an oncogene (39). In the present study, we investigated the possibility that RNA editing might modulate DHFR expression and subsequently affect the proliferation and sensitivity of breast cancer cells to methotrexate.
Results
Knockdown of ADAR1 Results in a Dramatic Decrease in DHFR Expression in MCF-7 Cells
The effects of knockdown of ADAR1 or ADAR2 on human DHFR expression in MCF-7 cells were investigated. The siADAR1 targets both ADAR1 p110 and ADAR1 p150. As shown in Fig. 1, A and B, there was a significant decrease in ADAR1 (both p110 and p150) and ADAR2 protein levels by transfection of siADAR1 and siADAR2, respectively. Regarding the DHFR transcripts, four different variants (v1–v4) are registered in the NCBI database (Fig. 1C), and it is conceivable that the functional protein is produced only from the v1 transcript. Taking potential effects of RNA editing within introns on splicing into consideration, the expression levels of the four DHFR transcripts in the siRNA-transfected cells were determined. The knockdown of ADAR1 resulted in a significant decrease in the levels of all types of DHFR transcripts (Fig. 1, D–G). In contrast, the knockdown of ADAR2 significantly decreased only the DHFR v2 mRNA level. Furthermore, a significant decreased in DHFR protein levels by siADAR1 was also observed (Fig. 1H). These results indicated that ADAR1 has a role in up-regulating DHFR expression.
FIGURE 1.
Effects of ADAR1 or ADAR2 knockdown on DHFR expression levels in MCF-7 cells. ADAR1 p110 and ADAR1 p150 protein (A) and ADAR2 protein (B) in MCF-7 cells 72 h after transfection with 5 nm siADAR1, siADAR2, or siControl were determined by Western blotting and were normalized to GAPDH shown in H. C, human DHFR transcript variants (v1–v4) are schematically represented. Open rectangles indicate exons. DHFR v1 (D), v2 (E), v3 (F), v4 (G) mRNA, and DHFR protein (H) levels in MCF-7 cells 72 h after transfection with 5 nm siADAR1, siADAR2, or siControl were determined and were normalized to GAPDH. The Ct values of v1, v2, v3, and v4 in siControl-transfected MCF-7 cells were 21.4, 25.2, 28.9, and 29.9, respectively. The values represent the levels relative to the siControl. Each column represents the mean ± S.D. of three independent experiments. The values are expressed relative to the siControl. Each point represents the mean ± S.D. (n = 3). *, p < 0.05; **, p < 0.01; and ***, p < 0.001.
ADAR1 Edits the 3′-UTR of DHFR
The RADAR, a database for RNA editing sites, includes human DHFR, the transcript of which are subjected to RNA editing. The editing sites were identified in introns 3, 4, and 5 and the 3′-UTR of DHFR in the lymphoblastoid cell line and brain. It is unknown whether the DHFR transcript is subjected to RNA editing in other tissues including breast. We sought to investigate whether the 3′-UTR, rather than introns, of DHFR in MCF-7 cells is subjected to RNA editing because ADAR1 positively regulates DHFR expression regardless of the transcript variants. We performed direct sequence analyses of the 3′-UTR of DHFR using genomic DNA and cDNA from MCF-7 cells as templates. Because inosine is recognized as guanosine by sequence analysis, A-to-I RNA edited sites are identified as overlapping peaks of adenosine and guanosine. We found 26 edited sites in the 3′-UTR of DHFR with editing levels ranging from 2.9 to 71.4% (Fig. 2, A and B). They were located within inverted Alu repeats, which can form a dsRNA structure, a typical target of ADAR enzymes. To investigate whether ADAR1 or ADAR2 enzyme are responsible for the RNA editing events, we analyzed the sequence of the DHFR cDNA from MCF-7 cells transfected with siADAR1 or siADAR2. Typical sequencing electropherograms of these RNA-edited sites are shown in Fig. 2C. The RNA editing levels of all editing sites were significantly reduced by ADAR1 knockdown, but not by ADAR2 knockdown (Fig. 2B). These results indicated that ADAR1 plays a critical role in editing of the 3′-UTR of DHFR.
FIGURE 2.
Effects of ADAR1 or ADAR2 knockdown on RNA editing levels in the 3′-UTR of DHFR. A, human DHFR v1 mRNA and RNA editing sites in the 3′-UTR of DHFR are schematically represented. Open arrows indicate the direction of the Alu elements. The nucleotide numbering refers to the 5′ end of the mRNA as 1. Vertical arrows show the RNA editing sites, numbered as 1–26 from 5′ to 3′. B, RNA editing levels in the 3′-UTR of DHFR in MCF-7 cells 72 h after transfection with 5 nm siADAR1, siADAR2, or siControl. The editing level is reported as a percentage, which was calculated by dividing the peak height of “G” by the sum of the peak heights of “G” and “A” in the electropherograms, resulting from direct sequencing of PCR amplicons that used cDNA from these cells as a template. C, representative electropherograms from direct sequencing that used genomic DNA and cDNA from MCF-7 cells as a template. Each column represents the mean ± S.D. of three independent experiments. *, p < 0.05; **, p < 0.01; and ***, p < 0.001.
RNA Editing Stabilizes DHFR mRNA
To determine whether the decrease in DHFR mRNA by ADAR1 knockdown is due to accelerated mRNA degradation, we examined the stability of the DHFR mRNA in the siRNA-transfected cells. In the presence of α-amanitin, the DHFR mRNA level was gradually decreased. The half-life of the DHFR mRNA in the siControl-transfected cells was 28.8 h, whereas that in siADAR1-transfected cells was 9.4 h (Fig. 3A). These results indicated to us that the non-edited DHFR mRNA is more unstable than edited mRNA. To investigate this possibility, we performed direct sequence analysis of cDNA from siControl-transfected cells 6, 12, and 24 h after treatment of α-amanitin. As shown in Fig. 3B as a typical example (RNA editing site numbers 15 and 16), the heights of G were increased and those of A were decreased by treatment of α-amanitin for 24 h. Such changes were observed in the other RNA editing sites (data not shown). To evaluate the degradation rates of non-edited and edited DHFR mRNAs, their levels were calculated by multiplying the DHFR mRNA levels at each time point (Fig. 3A) by the ratio of the peak height of A and G, respectively, at corresponding time points (Fig. 3C). As shown in Fig. 3D, it was demonstrated that the non-edited DHFR mRNA was dramatically decreased by treatment of α-amanitin for 24 h, whereas the edited DHFR was not changed. Collectively, we found that DHFR mRNA was stabilized by ADAR1-mediated RNA editing.
FIGURE 3.
Effects of RNA editing on DHFR mRNA stability. A, MCF-7 cells 36 h after transfection with 5 nm siADAR1 or siControl were treated with 10 μg/ml of α-amanitin. Total RNA was prepared after 0, 6, 12, and 24 h. The DHFR mRNA levels were determined using real-time RT-PCR. The DHFR mRNA levels at time 0 (the time of addition of α-amanitin) in each treatment were assigned 100%. Each column represents the mean ± S.D. of three independent experiments. *, p < 0.05 and **, p < 0.01 compared with siControl. B, representative electropherograms from the direct sequencing that used cDNA from α-amanitin-treated MCF-7 cells as a template. C and D, the expression levels of DHFR mRNA, which is non-edited or edited in each editing sites were calculated by multiplying DHFR mRNA expression by the ratio of the peak height of “A” or “G,” respectively, in the electropherograms resulting from the direct sequencing of PCR amplicons that used cDNA from these cells as a template. The amounts of total DHFR mRNA at time 0 (the time of addition of α-amanitin) in each treatment were assigned 100%. Each point represents the mean of three independent experiments.
RNA Editing of the DHFR Transcript Destroys miRNA Recognition Elements in the 3′-UTR
We surmised that the decrease in extent of RNA editing by ADAR1 knockdown would augment the binding of miRNA(s) to the 3′-UTR of DHFR to decrease its expression, because the 3′-UTR extensively includes binding sites for miRNAs, which down-regulate gene expression via translational repression or mRNA degradation (40). By using a database miRediTar, we searched for miRNAs that would bind to the non-edited sequence in the 3′-UTR of DHFR but not to edited sequence, considering their substantial expression in MCF-7 cells (41). As a result, miR-25-3p and miR-125a-3p were raised as candidate miRNAs (Fig. 4A). To examine whether RNA editing affects the recognition of miR-25-3p and miR-125a-3p, we performed luciferase assays using reporter plasmids carrying a non-edited or edited microRNA recognition element (MRE) for miR-25-3p or miR-125a-3p. They were named pGL3p/MRE25(A) (non-edited MRE for miR-25–3p), pGL3p/MRE25(G) (edited MRE for miR-25-3p), pGL3p/MRE125(A) (non-edited MRE for miR-125-3p), and pGL3p/MRE125(G) (edited MRE for miR-125-3p). The luciferase activities of the pGL3p/MRE25(A) and pGL3p/MRE125(A) plasmids were significantly reduced (68 and 71% of the control, respectively) by the overexpression of miR-25 and miR-125a-3p, respectively, whereas the decrease in luciferase activities of the pGL3p/MRE25(G) and pGL3p/MRE125(G) plasmids were marginal (88 and 91% of the control, respectively) (Fig. 4B). Because a slight reduction was observed in the activities of empty plasmid, the decrease in activity of latter plasmids would likely be independent of the inserted fragment. Thus, it was suggested that miR-25-3p and miR-125a-3p recognize the non-edited sequences in the 3′-UTR of DHFR.
FIGURE 4.
Role of miR-25–3p and miR-125a-3p in ADAR1-dependent regulation of DHFR in MCF-7 cells. A, human DHFR mRNA, and the predicted MREs of miR-25-3p and miR-125a-3p (termed MRE25 and MRE125, respectively) in non-edited DHFR mRNA are schematically represented. The numbering denotes the 5′ end of the mRNA as 1. Open arrows indicate the direction of the Alu elements. Bold letters represent seed sequences. RNA editing sites are shown by numbered (refer to Fig. 2A) arrows. B, luciferase assay using plasmids containing a fragment of the 3′-UTR of DHFR. MCF-7 cells were transfected with the reporter plasmids (190 ng) along with the phRL-TK plasmid (10 ng) and 30 nm miR-25-3p, miR-125a-3p mimic, or miControl. Firefly luciferase activity for each construct was normalized to Renilla luciferase activity. The values are expressed relative to the pGL3-p plasmid. DHFR mRNA (C and E) and protein (D and F) levels in MCF-7 cells 72 h after co-transfection with 10 nm siRNA (siADAR1 or siControl) and 10 nm miRNA mimic (miR-25-3p, miR-125a-3p mimic or miControl) (C and D) or co-transfection with 10 nm siRNA and 10 nm AsO (As-miR-25-3p, As-miR-125a-3p, or AsControl) (E and F) were determined by real-time PCR or Western blot analysis and were normalized to GAPDH. The values are expressed relative to the control. Each column represents the mean ± S.D. of three independent experiments. *, p < 0.05; **, p < 0.01; and ***, p < 0.001.
To further confirm that siADAR1-mediated down-regulation of DHFR is because of binding of these miRNAs to the non-edited sequence of DHFR, overexpression or an inhibition experiment of miRNAs was performed. When miR-25-3p or miR-125a-3p were co-transfected with siADAR1, the decrease in DHFR mRNA (Fig. 4C) and protein (Fig. 4D) levels by knockdown of ADAR1 were augmented. However, inhibition of these miRNA functions attenuated the decrease in DHFR expression by ADAR 1 knockdown (Fig. 4, E and F). These results indicated that the non-edited DHFR-specific down-regulation by miR-25-3p and miR-125a-3p would be responsible for reduction of DHFR expression by ADAR1 knockdown.
Suppression of DHFR Expression by ADAR1 Knockdown Decreases Cell Viability and Increases Sensitivity to Methotrexate of MCF-7 Cells
We noticed that the viability of MCF-7 cells was significantly decreased when ADAR1 was knocked down (Fig. 5A). Because DHFR has a key role in regulating the DNA synthesis by converting dihydrofolate into tetrahydrofolate, we postulated that the decrease in cell viability would be due to the decrease in DHFR protein level. To test this hypothesis, folinic acid, which is readily converted into tetrahydrofolate, was added to the siADAR1-transfected MCF-7 cells (42). As shown in Fig. 5B, the decreased cell viability by ADAR1 knockdown was partially restored by folinic acid treatment, suggesting that suppression of DHFR protein is one of the underlying causes of decreased cell viability by ADAR1 knockdown.
FIGURE 5.
Effects of ADAR1-dependent regulation of DHFR expression on viability and sensitivity of MCF-7 cells to methotrexate. A, the viability of MCF-7 cells 72 h after transfection with 5 nm siADAR1, siADAR2, and siControl. The values are expressed relative to the siControl. B, the viability of siADAR1-transfected MCF-7 cells incubated for 72 h in the presence of 200 μm folinic acid. The values are expressed relative to the siControl. C, MCF-7 cells 24 h after transfection with 0.5 nm siADAR1 or siControl were treated with 50 nm methotrexate. After 0, 24, 48, and 72 h, the cell number was evaluated by the WST-8 assay. The values are expressed relative to 0 h. Each column and data point represents the mean ± S.D. of three independent experiments. *, p < 0.05; **, p < 0.01; and ***, p < 0.001.
Next, we investigated whether ADAR1 knockdown increases the sensitivity of MCF-7 cells to methotrexate because it is known that tumors with reduced expression of DHFR are more sensitive to methotrexate treatment (43, 44). In this experiment, the transfected siADAR1 was decreased to 0.5 nm to make it easier to detect the efficacy of methotrexate. The increase in cell number of siControl-transfected cells was decreased 48 and 72 h after methotrexate treatment, which was further decreased by ADAR1 knockdown, suggesting that the suppression of DHFR by ADAR1 knockdown can increase the efficacy of methotrexate (Fig. 5C).
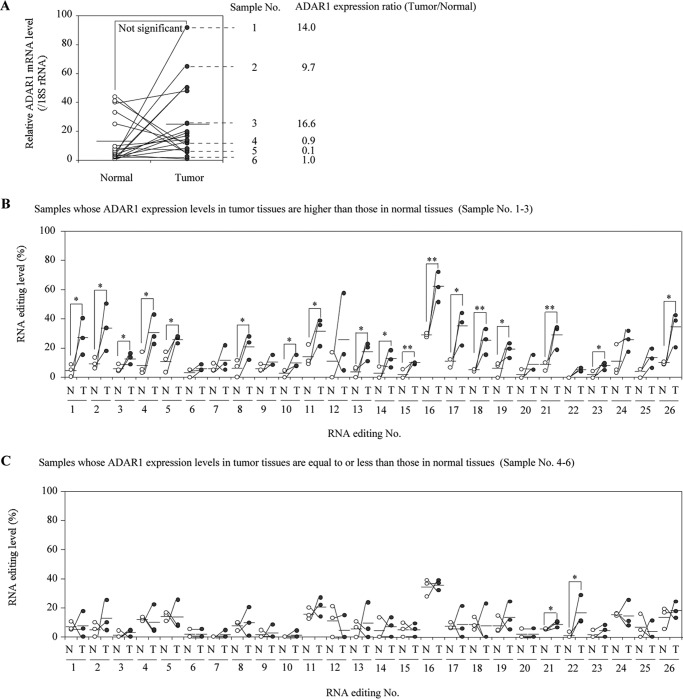
ADAR1 Expression and RNA Editing Levels in DHFR mRNA Are Higher in Breast Cancer Tissues Than in Normal Tissue
To examine the role of ADAR1 in RNA editing of DHFR in vivo, ADAR1 expression and RNA editing levels of the DHFR transcript in breast cancer tissues and adjacent normal tissues from 19 patients were determined. The ADAR1 mRNA levels in breast cancer tissues tended to be higher than those in adjacent normal tissues (Fig. 6A), consistent with a previous report (39). Next, by selecting 3 samples of which ADAR1 levels in cancer tissue were higher than in normal tissues (ADAR1 expression tumor/normal ratios of 9.7, 14.0, and 16.7) and 3 samples of which ADAR1 levels in cancer tissues were equal to or lower than in normal tissues (0.1, 0.9, and 1.0), the RNA editing levels in the 3′-UTR of DHFR were evaluated. In the former samples, the RNA editing levels of the DHFR transcript were higher in cancer tissues than in adjacent normal tissues (Fig. 6B), but in the latter samples (Fig. 6C), this phenomenon was not observed. These results suggest that the increased expression of ADAR1 in breast cancer contributes to hyper-editing of the DHFR transcript, which would result in an increase in DHFR expression.
FIGURE 6.
Expression levels of ADAR1 mRNA and RNA editing levels in the 3′-UTR of DHFR in human breast cancer tissues. A, expression levels of ADAR1 mRNA in breast cancer tissues and adjacent normal tissues obtained from 19 patients were determined by real-time RT-PCR. The expression levels were normalized to the 18S rRNA level. Data represent the mean of two independent experiments. The values are expressed relative to the sample with the lowest levels. Horizontal bars represent the mean. The RNA editing levels in the 3′-UTR of DHFR in breast cancer tissues of which ADAR1 expression is higher than (No. 1–3) (B) and equal to or lower than (No. 4–6) (C) in adjacent normal tissues. The editing level is represented as a percentage, which was calculated by dividing the peak height of “G” by the sum of peak heights of “G” and “A” in the sequencing electropherograms. Each column represents the mean of two independent experiments. *, p < 0.05 and **, p < 0.01.
Discussion
It has been proposed that RNA editing may alter miRNA recognition (45, 46), but the evidence is very limited (47–49). In this study, we examined the possibility that RNA editing may regulate DHFR expression by altering nucleotides in miRNA binding sites in the 3′-UTR.
We found that ADAR1 catalyzes RNA editing in the 3′-UTR of DHFR in MCF-7 cells and breast cancer tissues (Figs. 2, B and C, and 6, B and C). The identified RNA editing sites are in the inverted Alu repeats, which can form a perfectly matched dsRNA structure. This is consistent with a recent report showing that ADAR1, rather than ADAR2, is the main RNA editing enzyme for reversely oriented Alu repeats (50). As shown in Fig. 2B, knockdown of ADAR2 slightly increased RNA editing levels in the 3′-UTR of DHFR. It is known that homodimerization is important for the functions of both human ADAR1 and ADAR2, whereas heterodimerization between ADAR1 and ADAR2 has been shown to decrease editing activity (51). Therefore, a possible reason for the increase in RNA editing levels by ADAR2 knockdown may be due to an increase in ADAR1 homodimerization along with a decrease in ADAR2 heterodimerization with ADAR1.
We found that ADAR1 knockdown facilitated the degradation of DHFR mRNA in MCF-7 cells (Fig. 3A). Supporting this result, an increase in the instability of non-edited DHFR mRNA than edited mRNA was demonstrated (Fig. 3D), indicating that RNA editing is a critical factor for determining the DHFR mRNA degradation rate. As an underlying mechanism of the RNA editing-dependent regulation of DHFR expression, we focused on miRNA. By computer analysis, miR-25-3p and miR-125a-3p were predicted to bind to the non-edited but not edited DHFR (Fig. 4A). The non-edited sequence-specific recognition by these miRNAs was experimentally proven by the luciferase assay (Fig. 4B). Through co-transfection of siADAR1 with miRNA mimics or AsO, it was demonstrated that these miRNAs mediate the editing-associated modulation of DHFR expression (Fig. 4, C–F). The miR-25-3p and miR-125a-3p have been reported to regulate Smad7 and breast cancer early onset gene 1 in MCF-7 cells, respectively (52, 53), suggesting the functional expression of these miRNAs in this cell lines. In addition, it has been reported that these miRNAs are expressed also in breast cancer tissues (54, 55). Our study indicated that RNA editing in the 3′-UTR of DHFR would allow DHFR mRNA to escape from repression by these miRNAs, leading to an increase in DHFR expression in breast cancer cells. To examine whether the decrease in DHFR expression by ADAR1 knockdown was completely due to the function of miR-25-3p and miR-125a-3p, double transfection of AsOs for these miRNAs with siADAR1 was performed. As the result, double inhibition of these miRNAs could not completely restore the decreased DHFR expression level by knockdown of ADAR1 (data not shown), suggesting that other factors may be involved in this regulatory mechanism. miR-92a-3p and miR-92b-3p may be involved because they have the same seed sequence with miR-25-3p. In addition to miRNAs, a recent study revealed that RNA editing within reversely oriented Alu repeats enables the recruitment of an RNA-binding protein human antigen R to the 3′-UTR to increase the mRNA stability of cathepsin S (50). It would be of interest to investigate whether such factors are involved in the regulation of DHFR expression.
Another possible mechanism for the decrease in DHFR expression by ADAR1 knockdown is induction of negative regulators such as miRNA for DHFR expression, because A-to-I editing can alter the processing of miRNA, thereby affecting miRNA expression (56, 57). Further studies are warranted to explore the effects of RNA editing on expression levels of miRNAs including miR-24, miR-192, and miR-215, which have been reported to regulate DHFR expression (16–18).
By ADAR1 knockdown, the viability of MCF-7 cells was decreased (Fig. 5A), which is consistent with a previous report (39) and supports the oncogenic ability of ADAR1 in breast cancer cells. The decrease was partially restored by treatment with folinic acid, which can supply tetrahydrofolate (Fig. 5B). These results suggest that DHFR is likely one of the targets that are responsible for the oncogenic function of ADAR1. In addition, as shown in Fig. 5C, ADAR1 knockdown enhanced the sensitivity of MCF-7 cells to methotrexate, although the extent was not so dramatic (17.2 and 30.4% decrease in cell number by treatment with methotrexate in siControl- and siADAR1-transfected cells, respectively). A previous study has reported that MCF-7 is more resistant to methotrexate (IC50 value: 114 nm) compared with the other cell lines (IC50 value: 6–38 nm) because the MCF-7 cells have a mutated reduced folate career gene, leading to a decrease in methotrexate uptake (58). Thereby, we used another cell line A549, which has a relatively lower IC50 value of methotrexate (38 nm) than MCF-7, to examine the effects of ADAR1 knockdown on the efficiency of methotrexate. In accordance with the report by Yoon et al. (58), the reduction in cell viability of A549 cells by treatment with methotrexate (52.9% of control at 72 h) (Fig. 7) was larger than that of MCF-7 cells (82.8% of control at 72 h) (Fig. 5C). ADAR1 knockdown enhanced the sensitivity of A549 cells to methotrexate, but the effects (27.0% decrease: siControl + methotrexate versus siADAR1 + methotrexate) were small compared with those in MCF-7 cells (22.9% decrease), implying a low contribution of DHFR to cellular toxicity of methotrexate. It has been reported that dihydrofolate reductase like 1, which is encoded by a pseudogene DHFRP4, is expressed and has the same function as DHFR in folate metabolism (59). It is possible that this enzyme may complement the decreased expression of DHFR by ADAR1 knockdown, producing resistance to methotrexate.
FIGURE 7.

Effects of ADAR1-dependent regulation of DHFR expression on sensitivity of A549 cells to methotrexate. A549 cells 24 h after transfection with 0.5 nm siADAR1 or siControl were treated with 50 nm methotrexate. After 0, 24, 48, and 72 h, the cell number was evaluated by the WST-8 assay. The values are expressed relative to 0 h. Each point represents the mean ± S.D. of three independent experiments. ***, p < 0.001.
Accumulating evidence has revealed that ADAR-mediated A-to-I RNA editing is involved in cancer development and progression. It is of interest to note that it differs among cancer types whether ADARs function as oncogenes or tumor suppressive genes (34). In brain tumors, ADAR1 and ADAR2 are down-regulated, leading to a global reduction in A-to-I editing, which is functionally required for brain tumor development (60). In contrast, ADAR1 has been reported to be up-regulated in hepatocellular carcinoma (61) and esophageal squamous cell carcinoma (62), and cell-based assays have revealed ADAR1-induced tumorigenic phenotypes in these cancer cells. More recently, it was revealed that ADAR1 expression and the editing frequency of global transcripts were higher in breast tumors compared with normal tissues (39). In agreement with the report, higher expression levels of ADAR1 in breast cancer tissues than in adjacent normal tissues was observed in our study, although the difference was not significant due to the large interindividual variability in the expression (Fig. 6A). The ADAR1 gene is located on chromosome 1q, of which amplification has often been observed in breast cancer tissues (63). Fumagalli et al. (39) proposed that ADAR1 expression levels in breast cancer were determined in part by the extent of gene amplification as well as the expression of STAT1 (as a proxy for interferon response). In tumor tissues with high ADAR1 expression, RNA editing levels of the DHFR transcript were higher than those in normal tissues (Fig. 6B). It is conceivable that the enhanced RNA editing would result in high DHFR expression to facilitate the proliferation of cancer cells. Hence, ADAR1-mediated regulation of DHFR would be an important factor in cancer development and progression.
In conclusion, we found that DHFR is post-transcriptionally regulated through ADAR1-mediated RNA editing, affecting cell proliferation and sensitivity of breast cancer cells to methotrexate. This study could provide new insights into the regulatory mechanism of DHFR expression and the role of RNA editing in methotrexate response. ADAR1 may be a potential anti-tumor target for use with anti-folate compounds including methotrexate.
Experimental Procedures
Chemicals and Reagents
Methotrexate hydrate and folinic acid calcium salt hydrate were from Tokyo Kasei (Tokyo, Japan) and Sigma, respectively. The pGL3-promoter vector, phRL-TK vector, and Dual-Luciferase Reporter Assay System were purchased from Promega (Madison, WI). Lipofectamine RNAiMAX, Lipofectamine 2000, Silencer Select siRNA for human ADAR1 (s1007) (siADAR1), human ADAR2 (s1010) (siADAR2), negative control #1 (siControl), and miRNA mimics for miR-25–3p and miR-125a-3p and negative control #1 (miControl) were purchased from Life Technologies. Antisense locked nucleic acid/DNA oligonucleotides (AsO) for miR-25-3p (5′-TCA GTC CGA GAC AAG TGC AAT G-3′; locked nucleic acids are underlined), miR-125a-3p (5′-GGC TCC CAA GAA CCT CAC CTG T-3′) and the negative control (5′-AGA CUA GCG GUA UCU UAA ACC-3′) (AsControl) were commercially synthesized by Gene Design (Osaka, Japan). RNAiso, random hexamers, and SYBR Premix Ex Taq were from Takara (Shiga, Japan). ROX was purchased from Stratagene (La Jolla, CA). ReverTra Ace was purchased from Toyobo (Osaka, Japan). All primers were commercially synthesized at RIKAKEN (Nagoya, Japan). α-Amanitin was purchased from Calbiochem. The rabbit anti-human DHFR polyclonal antibody (ab49881) was from Abcam (Cambridge, MA), and mouse anti-human ADAR1 monoclonal antibody (sc-5579) and mouse anti-human ADAR2 monoclonal antibody (sc-73408) were from Santa Cruz Biotechnology (Santa Cruz, CA). The rabbit anti-human GAPDH polyclonal antibody (IMG-5143A) was purchased from IMGENIX (San Diego, CA). IRDye 680 goat anti-rabbit IgG and goat anti-mouse IgG were from LI-COR Biosciences (Lincoln, NE). Cell Counting Kit-8 (CCK-8) was from Dojin Chemical Laboratories (Kumamoto, Japan). Restriction enzymes were from New England Biolabs (Ioswich, MA). All other chemicals and solvents were of the highest grade commercially available.
Cell Cultures
MCF-7, a human breast adenocarcinoma cell line, was obtained from the American Type Culture Collection (Manassas, VA). A549, a human lung carcinoma cell line, was obtained from the Riken Gene Bank (Tsukuba, Japan). The MCF-7 cells were cultured in DMEM (Nissui Pharmaceutical, Tokyo, Japan) supplemented with 0.1 mm nonessential amino acids (Invitrogen) and 10% FBS (Invitrogen). All cells were maintained at 37 °C under an atmosphere of 5% CO2 and 95% air.
Transfection of siRNA, miRNA Mimic, or AsO into MCF-7 Cells and Preparation of Cell Homogenates and Total RNA
The MCF-7 cells were seeded into 6-well plates and 5 nm siRNA-Lipofectamine RNAiMAX complexes were added. For double transfection, 10 nm miRNA mimic or 10 nm AsO was co-transfected with 10 nm siRNA into the cells. After 72 h, the cells were harvested, suspended in a small amount of TGE buffer (10 mm Tris-HCl, 20% glycerol, and 1 mm EDTA, pH 7.4), and disrupted by freeze-thawing three times. Total RNA was prepared using RNAiso.
SDS-PAGE and Western Blot Analysis
To analyze DHFR (21 kDa), ADAR1 (110 and 150 kDa), and GAPDH (36 kDa) protein levels, cell homogenates from MCF-7 cells were separated by 15, 7.5, and 10% SDS-PAGE, respectively, and transferred to an Immobilon-P transfer membrane (Millipore, Billerica, MA). To analyze ADAR2 (90 kDa) protein levels, the cell homogenates were separated by 7.5% SDS-PAGE and transferred to a Protran nitrocellulose membrane (Whatman GmbH, Dassel, Germany). The membranes were probed with rabbit anti-human DHFR polyclonal, mouse anti-human ADAR1 monoclonal, mouse anti-human ADAR2 monoclonal, or rabbit anti-human GAPDH polyclonal antibodies and the corresponding fluorescent dye-conjugated secondary antibodies. The band densities were quantified with an Odyssey Infrared Imaging system (LI-COR Biosciences). The DHFR, ADAR1, and ADAR2 protein levels were normalized to GAPDH.
Real-time RT-PCR for DHFR Transcript Variants and ADAR1 mRNA
cDNA was synthesized from total RNA using ReverTra Ace. The sequences of the used primers are shown in Table 1. A 1-μl portion of the reverse transcription mixture was added to a PCR mixture containing 10 pmol of each primer, 12.5 μl of the SYBR Premix Ex Taq solution, and 75 nm Rox in a final volume of 25 μl. Real-time PCR was performed using Mx3000P (Stratagene, La Jolla, CA) with the MxPro QPCR software. The PCR conditions for DHFR v1 and v4 were as follows: after an initial denaturation at 95 °C for 30 s, amplification was performed by denaturation at 94 °C for 20 s, followed by annealing/extension at 62 °C for 20 s for 40 cycles. The PCR conditions for DHFR v2 and v3 were as follows: after an initial denaturation at 95 °C for 30 s, amplification was performed by denaturation at 94 °C for 20 s, followed by annealing at 53 °C for 20 s and extension at 72 °C for 20 s for 40 cycles. The PCR conditions for ADAR1 were as follows: after an initial denaturation at 95 °C for 30 s, amplification was performed by denaturation at 94 °C for 20 s, followed by annealing/extension at 64 °C for 20 s for 40 cycles. The levels of DHFR variant transcripts were normalized to those of GAPDH mRNA (64). The ADAR1 mRNA levels in human breast cancer tissues described below were normalized to those of 18S rRNA (65).
TABLE 1.
Primers for real-time RT-PCR
| Target gene | Accession No. | Primer | Sequence (5′ to 3′) |
|---|---|---|---|
| DHFR v1 | NM_000791 | Forward | TCC AGA GAA TGA CCA CAA CC |
| Reverse | ACG TGT CAC TTT CAA AGT CT | ||
| DHFR v2 | NM_001290354 | Forward | TGG CCA CCG CTC AGG TAA AC |
| Reverse | GTG ATT CAT GGC TTC CTT AT | ||
| DHFR v3 | NM_001290357 | Forward | TCC AGA GAA TGA CCA CAA CC |
| Reverse | GGG TGA TTC ATG GCT TCT TG | ||
| DHFR v4 | NR_110936 | Forward | TGG CCA CCG CTC AGG TAA AC |
| Reverse | AGA ACA CCT GGG TAT CTT AT | ||
| ADAR1 | NM_001025107 | Forward | GCT TGG GAA CAG GGA ATC G |
| Reverse | CTG TAG AGA AAC CTG ATG AAG CC | ||
| GAPDH | NM_002046 | Forward | CCA GGG CTG CTT TTA ACT C |
| Reverse | GCT CCC CCC TGC AAA TGA | ||
| 18S rRNA | NR_003286 | Forward | GGC CCT GTA ATT GGA ATG AGT C |
| Reverse | GAC ACT CAG CTA AGA GCA TCG |
Evaluation of RNA Editing Levels in the 3′-UTR of DHFR
To analyze RNA editing levels, direct sequencing of PCR products was performed. The 3′-UTR of DHFR was amplified by PCR using genomic DNA or cDNA as a template. The following primer sets for genomic DNA or cDNA were used to amplify the 3′-UTR of DHFR: the forward primer 5′-TTT ATC CAA CTT GAC AGT GG-3′ and the reverse primer 5′-CTT CAC CCT TGA TTA TTT GG-3′, or the forward primer 5′-AGC TGC TCT ATA GCA AGT CT-3′ and the reverse primer 5′-CTT CAC CCT TGA TTA TTT GG-3′. The PCR mixture consists of the genomic DNA or cDNA, 1× PCR buffer, 0.4 μm of each primer and 0.5 units of Gflex polymerase (TAKARA, Shiga, Japan) in a final volume of 25 μl. After an initial denaturation at 94 °C for 1 min, amplification was performed with denaturation at 98 °C for 10 s, annealing at 54 °C for 15 s, and extension at 72 °C for 70 (genomic DNA) or 40 s (cDNA) for 35 cycles, followed by a final extension at 72 °C for 5 min. The PCR products were subjected to electrophoresis using a 0.8% agarose gel. Control experiments without the reverse transcriptase were conducted to verify that the amplified products were from the reverse-transcribed cDNA rather than from contaminating genomic DNA. Specific products were purified and subjected to direct sequencing. The extent of editing was represented as a percentage calculated from the ratio of the peak height of “G” over the sum of the peak heights of “G” and “A” in the sequencing electropherograms.
Evaluation of the Stability of DHFR mRNA
MCF-7 cells were transfected with siRNA, as described above. After 36 h, the cells were treated with 10 μg/ml of α-amanitin, an inhibitor of transcription. Total RNA was prepared 0, 6, 12, and 24 h later. The DHFR mRNA level was determined by real-time RT-PCR, as described above.
Reporter Plasmids Construction
A fragment from +1901 to +2153 containing the MRE for miR-25-3p (MRE25) and that from +2443 to +2569 containing the MRE for miR-125a-3p (MRE125) of the non-edited 3′-UTR of DHFR were amplified by PCR using MCF-7 genomic DNA as a template and were inserted into the pGL3p vector at the XbaI site downstream of the luciferase gene. The constructed luciferase reporter plasmids were termed pGL3p/MRE25(A) and pGL3p/MRE125(A), respectively. Two corresponding fragments were also amplified by PCR using MCF-7 cDNA as a template and were inserted into the pGL3p vector. A clone containing MRE25 with edited nucleotides (guanosines) at site numbers 1, 2, 3, 5, 7, 8, 10, and 11 and that contained MRE125 with guanosines at numbers 16, 17, 18, 19, and 20 (refer to Fig. 2A) were selected and used as reporter plasmids (pGL3p/MRE25(G) and pGL3p/MRE125(G), respectively). By direct sequence analysis, we confirmed that RNA editing events did not occur in the transcripts of reporter genes in MCF-7 cells transfected with pGL3p/MRE25(A) or pGL3p/MRE25(G).
Luciferase Assay
MCF-7 cells were transiently transfected with various pGL3 luciferase reporter plasmids and the phRL-TK plasmid. Briefly, the day before transfection, the cells were seeded into 24-well plates. After 24 h, the cells were transfected with 190 ng of pGL3p plasmid, 10 ng of phRL-TK plasmid, and 30 nm miR-25-3p, miR-125a-3p mimic, or miControl using Lipofectamine 2000. After incubation for 48 h, the cells were resuspended in passive lysis buffer, and luciferase activity was measured on a luminometer using the Dual-Luciferase Reporter Assay System.
Assessment of Cell Viability
To evaluate cell viability, a WST-8 (2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium monosodium salt) assay, which is a modified 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay, was performed. MCF-7 cells were transfected with 5 nm siRNA as described above and incubated for 72 h. The viability of siADAR1-transfected MCF-7 cells was evaluated in the presence or absence of 200 μm folinic acid. The effects of ADAR1 knockdown on the sensitivity of MCF-7 or A549 cells to methotrexate were investigated as follows; MCF-7 or A549 cells were transfected with 0.5 nm siRNA. After 24 h, the cells were treated with 50 nm methotrexate for 24, 48, and 72 h. CCK-8 reagent was then added, and absorbance of the medium at 450 nm was measured. The percent cell viability was calculated by comparing the absorbance of the treated cells with the control cells.
Human Breast Cancer and Adjacent Normal Tissues
Breast cancer and adjacent normal tissues obtained as surgical samples from 19 Japanese patients (42–77 years old, did not undergo chemotherapy) with primary breast carcinoma (66) were used. This study was approved by the Ethics Committee of Kanazawa University (Kanazawa, Japan). Written informed consent was obtained from patients. The samples were obtained immediately after resection, divided into breast cancer and adjacent normal tissues, and immediately frozen with liquid nitrogen. The samples were stored at −80 °C until use. Total RNA was prepared using RNAiso.
Statistical Analyses
Statistical significance was determined by analysis of variance followed by Dunnett's multiple comparisons test or Tukey's method test. The comparison of two groups was performed using an unpaired, two-tailed Student's t test. Correlation analyses were performed by Spearman's rank method. A value of p < 0.05 was considered statistically significant.
Author Contributions
Ma. N., T. F., S. G., and Mi. N. designed the research. Ma. N. conducted most of the experiments and analyzed the results. Ma. N. and Mi. N. wrote the paper.
Acknowledgment
We thank Dr. Takano Taniya (Futaba Breast Clinic, Japan) for providing human breast cancer samples.
This work was supported in part by Grant-in-Aid for Scientific Research (B) from Japan Society for the Promotion of Science number 15H04663. The authors declare that they have no conflicts of interest with contents of this article.
- DHFR
- dihydrofolate reductase
- ADAR
- adenosine deaminase acting on RNA
- AsO
- antisense locked nucleic acid/DNA oligonucleotides
- A-to-I
- adenosine-to-inosine
- dsRNA
- double-stranded RNA
- miRNA
- microRNA
- MRE
- microRNA recognition element
- WST-8
- 2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium monosodium salt.
References
- 1. Schweitzer B. I., Dicker A. P., and Bertino J. R. (1990) Dihydrofolate reductase as a therapeutic target. FASEB J. 4, 2441–2452 [DOI] [PubMed] [Google Scholar]
- 2. Fowler B. (2001) The folate cycle and disease in humans. Kidney Int. Suppl. 78, 221–229 [DOI] [PubMed] [Google Scholar]
- 3. Nazki F. H., Sameer A. S., and Ganaie B. A. (2014) Folate: metabolism, genes, polymorphisms and the associated diseases. Gene 533, 11–20 [DOI] [PubMed] [Google Scholar]
- 4. Albrecht A. M., Biedler J. L., and Hutchison D. J. (1972) Two different species of dihydrofolate reductase in mammalian cells differentially resistant to amethopterin and methasquin. Cancer Res. 32, 1539–1546 [PubMed] [Google Scholar]
- 5. Jackson R. C., Hart L. I., and Harrap K. R. (1976) Intrinsic resistance to methotrexate of cultured mammalian cells in relation to the inhibition kinetics of their dihydrololate reductases. Cancer Res. 36, 1991–1997 [PubMed] [Google Scholar]
- 6. Goldie J. H., Krystal G., Hartley D., Gudauskas G., and Dedhar S. (1980) A methotrexate insensitive variant of folate reductase present in two lines of methotrexate-resistant L5178Y cells. Eur. J. Cancer 16, 1539–1546 [DOI] [PubMed] [Google Scholar]
- 7. Hill B. T., Bailey B. D., White J. C., and Goldman I. D. (1979) Characteristics of transport of 4-amino antifolates and folate compounds by two lines of L5178Y lymphoblasts, one with impaired transport of methotrexate. Cancer Res. 39, 2440–2446 [PubMed] [Google Scholar]
- 8. Moscow J. A., Gong M., He R., Sgagias M. K., Dixon K. H., Anzick S. L., Meltzer P. S., and Cowan K. H. (1995) Isolation of a gene encoding a human reduced folate carrier (RFC1) and analysis of its expression in transport-deficient, methotrexate-resistant human breast cancer cells. Cancer Res. 55, 3790–3794 [PubMed] [Google Scholar]
- 9. Kobayashi H., Takemura Y., and Ohnuma T. (1998) Variable expression of RFC1 in human leukemia cell lines resistant to antifolates. Cancer Lett. 124, 135–142 [DOI] [PubMed] [Google Scholar]
- 10. Assaraf Y. G. (2007) Molecular basis of antifolate resistance. Cancer Metastasis Rev. 26, 153–181 [DOI] [PubMed] [Google Scholar]
- 11. Abali E. E., Skacel N. E., Celikkaya H., and Hsieh Y. C. (2008) Regulation of human dihydrofolate reductase activity and expression. Vitam. Horm. 79, 267–292 [DOI] [PubMed] [Google Scholar]
- 12. Alt F. W., Kellems R. E., Bertino J. R., and Schimke R. T. (1978) Selective multiplication of dihydrofolate reductase genes in methotrexate-resistant variants of cultured murine cells. J. Biol. Chem. 253, 1357–1370 [PubMed] [Google Scholar]
- 13. Dolnick B. J., Berenson R. J., Bertino J. R., Kaufman R. J., Nunberg J. H., and Schimke R. T. (1979) Correlation of dihydrofolate reductase elevation with gene amplification in a homogeneously staining chromosomal region in L5178Y cells. J. Cell Biol. 83, 394–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dynan W. S., Sazer S., Tjian R., and Schimke R. T. (1986) Transcription factor Sp1 recognizes a DNA sequence in the mouse dihydrofolate reductase promoter. Nature 319, 246–248 [DOI] [PubMed] [Google Scholar]
- 15. Slansky J. E., Li Y., Kaelin W. G., and Farnham P. J. (1993) A protein synthesis-dependent increase in E2F1 mRNA correlates with growth regulation of the dihydrofolate reductase promoter. Mol. Cell. Biol. 13, 1610–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mishra P. J., Humeniuk R., Mishra P. J., Longo-Sorbello G. S., Banerjee D., and Bertino J. R. (2007) A miR-24 microRNA binding-site polymorphism in dihydrofolate reductase gene leads to methotrexate resistance. Proc. Natl. Acad. Sci. U.S.A. 104, 13513–13518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Song B., Wang Y., Kudo K., Gavin E. J., Xi Y., and Ju J. (2008) miR-192 Regulates dihydrofolate reductase and cellular proliferation through the p53-microRNA circuit. Clin. Cancer Res. 14, 8080–8086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Song B., Wang Y., Titmus M. A., Botchkina G., Formentini A., Kornmann M., and Ju J. (2010) Molecular mechanism of chemoresistance by miR-215 in osteosarcoma and colon cancer cells. Mol. Cancer 9, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wagner R. W., Smith J. E., Cooperman B. S., and Nishikura K. (1989) A double-stranded RNA unwinding activity introduces structural alterations by means of adenosine to inosine conversions in mammalian cells and Xenopus eggs. Proc. Natl. Acad. Sci. U.S.A. 86, 2647–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gallo A., and Galardi S. (2008) A-to-I RNA editing and cancer: from pathology to basic science. RNA Biol. 5, 135–139 [DOI] [PubMed] [Google Scholar]
- 21. Farajollahi S., and Maas S. (2010) Molecular diversity through RNA editing: a balancing act. Trends Genet. 26, 221–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim U., Wang Y., Sanford T., Zeng Y., and Nishikura K. (1994) Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proc. Natl. Acad. Sci. U.S.A. 91, 11457–11461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gerber A., O'Connell M. A., and Keller W. (1997) Two forms of human double-stranded RNA-specific editase 1 (hRED1) generated by the insertion of an Alu cassette. RNA 3, 453–463 [PMC free article] [PubMed] [Google Scholar]
- 24. Bass B. L., Nishikura K., Keller W., Seeburg P. H., Emeson R. B., O'Connell M. A., Samuel C. E., and Herbert A. (1997) A standardized nomenclature for adenosine deaminases that act on RNA. RNA 3, 947–949 [PMC free article] [PubMed] [Google Scholar]
- 25. Nishikura K. (2010) Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 79, 321–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Patterson J. B., and Samuel C. E. (1995) Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: evidence for two forms of the deaminase. Mol. Cell. Biol. 15, 5376–5388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Desterro J. M., Keegan L. P., Lafarga M., Berciano M. T., O'Connell M., and Carmo-Fonseca M. (2003) Dynamic association of RNA-editing enzymes with the nucleolus. J. Cell Sci. 116, 1805–1818 [DOI] [PubMed] [Google Scholar]
- 28. Tariq A., and Jantsch M. F. (2012) Transcript diversification in the nervous system: A to I RNA editing in CNS function and disease development. Front. Neurosci. 6, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sommer B., Köhler M., Sprengel R., and Seeburg P. H. (1991) RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell 67, 11–19 [DOI] [PubMed] [Google Scholar]
- 30. Lomeli H., Mosbacher J., Melcher T., Höger T., Geiger J. R., Kuner T., Monyer H., Higuchi M., Bach A., and Seeburg P. H. (1994) Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science 266, 1709–1713 [DOI] [PubMed] [Google Scholar]
- 31. Burns C. M., Chu H., Rueter S. M., Hutchinson L. K., Canton H., Sanders-Bush E., and Emeson R. B. (1997) Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature 387, 303–338 [DOI] [PubMed] [Google Scholar]
- 32. Kawahara Y., Ito K., Sun H., Aizawa H., Kanazawa I., and Kwak S. (2004) Glutamate receptors: RNA editing and death of motor neurons. Nature 427, 801. [DOI] [PubMed] [Google Scholar]
- 33. Morabito M. V., Abbas A. I., Hood J. L., Kesterson R. A., Jacobs M. M., Kump D. S., Hachey D. L., Roth B. L., and Emeson R. B. (2010) Mice with altered serotonin 2C receptor RNA editing display characteristics of Prader-Willi syndrome. Neurobiol. Dis. 39, 169–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Slotkin W., and Nishikura K. (2013) Adenosine-to-inosine RNA editing and human disease. Genome Med. 5, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wahlstedt H., Daniel C., Ensterö M., and Ohman M. (2009) Large-scale mRNA sequencing determines global regulation of RNA editing during brain development. Genome Res. 19, 978–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu H., Ma C. P., Chen Y. T., Schuyler S. C., Chang K. P., and Tan B. C. (2014) Functional Impact of RNA editing and ADARs on regulation of gene expression: perspectives from deep sequencing studies. Cell Biosci. 4, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ramaswami G., Lin W., Piskol R., Tan M. H., Davis C., and Li J. B. (2012) Accurate identification of human Alu and non-Alu RNA editing sites. Nat. Methods 9, 579–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ramaswami G., Zhang R., Piskol R., Keegan L. P., Deng P., O'Connell M. A., and Li J. B. (2013) Identifying RNA editing sites using RNA sequencing data alone. Nat. Methods 10, 128–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fumagalli D., Gacquer D., Rothé F., Lefort A., Libert F., Brown D., Kheddoumi N., Shlien A., Konopka T., Salgado R., Larsimont D., Polyak K., Willard-Gallo K., Desmedt C., Piccart M., Abramowicz M., Campbell P. J., Sotiriou C., and Detours V. (2015) Principles governing A-to-I RNA editing in the breast cancer transcriptome. Cell Rep. 13, 277–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bartel D. P. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 41. Fix L. N., Shah M., Efferth T., Farwell M. A., and Zhang B. (2010) MicroRNA expression profile of MCF-7 human breast cancer cells and the effect of green tea polyphenon-60. Cancer Genomics Proteomics 7, 261–277 [PubMed] [Google Scholar]
- 42. Lladó V., Terés S., Higuera M., Alvarez R., Noguera-Salva M. A., Halver J. E., Escribá P. V., and Busquets X. (2009) Pivotal role of dihydrofolate reductase knockdown in the anticancer activity of 2-hydroxyoleic acid. Proc. Natl. Acad. Sci. U.S.A. 106, 13754–13758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Salonga D., Danenberg K. D., Johnson M., Metzger R., Groshen S., Tsao-Wei D. D., Lenz H. J., Leichman C. G., Leichman L., Diasio R. B., and Danenberg P. V. (2000) Colorectal tumors responding to 5-fluorouracil have low gene expression levels of dihydropyrimidine dehydrogenase, thymidylate synthase, and thymidine phosphorylase. Clin. Cancer Res. 6, 1322–1327 [PubMed] [Google Scholar]
- 44. Banerjee D., Mayer-Kuckuk P., Capiaux G., Budak-Alpdogan T., Gorlick R., and Bertino J. R. (2002) Novel aspects of resistance to drugs targeted to dihydrofolate reductase and thymidylate synthase. Biochim. Biophys. Acta. 1587, 164–173 [DOI] [PubMed] [Google Scholar]
- 45. Liang H., and Landweber L. F. (2007) Hypothesis: RNA editing of microRNA target sites in humans? RNA 13, 463–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Peng Z., Cheng Y., Tan B. C., Kang L., Tian Z., Zhu Y., Zhang W., Liang Y., Hu X., Tan X., Guo J., Dong Z., Liang Y., Bao L., and Wang J. (2012) Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat. Biotechnol. 30, 253–260 [DOI] [PubMed] [Google Scholar]
- 47. Borchert G. M., Gilmore B. L., Spengler R. M., Xing Y., Lanier W., Bhattacharya D., and Davidson B. L. (2009) Adenosine deamination in human transcripts generates novel microRNA binding sites. Hum. Mol. Genet. 18, 4801–4807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang Q., Hui H., Guo Z., Zhang W., Hu Y., He T., Tai Y., Peng P., and Wang L. (2013) ADAR1 regulates ARHGAP26 gene expression through RNA editing by disrupting miR-30b-3p and miR-573 binding. RNA 19, 1525–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nakano M., Fukami T., Gotoh S., Takamiya M., Aoki Y., and Nakajima M. (2016) RNA editing modulates human hepatic aryl hydrocarbon receptor expression by creating microRNA recognition sequence. J. Biol. Chem. 291, 894–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stellos K., Gatsiou A., Stamatelopoulos K., Perisic Matic L., John D., Lunella F. F., Jaé N., Rossbach O., Amrhein C., Sigala F., Boon R. A., Fürtig B., Manavski Y., You X., Uchida S., et al. (2016) Adenosine-to-inosine RNA editing controls cathepsin S expression in atherosclerosis by enabling HuR-mediated post-transcriptional regulation. Nat. Med. 22, 1140–1150 [DOI] [PubMed] [Google Scholar]
- 51. Deffit S. N., and Hundley H. A. (2016) To edit or not to edit: regulation of ADAR editing specificity and efficiency. Wiley Interdiscip. Rev. RNA 7, 113–127 [DOI] [PubMed] [Google Scholar]
- 52. Smith A. L., Iwanaga R., Drasin D. J., Micalizzi D. S., Vartuli R. L., Tan A. C., and Ford H. L. (2012) The miR-106b-25 cluster targets Smad7, activates TGF-β signaling, and induces EMT and tumor initiating cell characteristics downstream of Six1 in human breast cancer. Oncogene 31, 5162–5171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xu X., Lv Y. G., Yan C. Y., Yi J., and Ling R. (2016) Enforced expression of hsa-miR-125a-3p in breast cancer cells potentiates docetaxel sensitivity via modulation of BRCA1 signaling. Biochem. Biophys. Res. Commun. 479, 893–900 [DOI] [PubMed] [Google Scholar]
- 54. Wu X., Zeng R., Wu S., Zhong J., Yang L., and Xu J. (2015) Comprehensive expression analysis of miRNA in breast cancer at the miRNA and isomiR levels. Gene 557, 195–200 [DOI] [PubMed] [Google Scholar]
- 55. He H., Xu F., Huang W., Luo S. Y., Lin Y. T., Zhang G. H., Du Q., and Duan R. H. (2015) miR-125a-5p expression is associated with the age of breast cancer patients. Genet. Mol. Res. 14, 17927–17933 [DOI] [PubMed] [Google Scholar]
- 56. Yang W., Chendrimada T. P., Wang Q., Higuchi M., Seeburg P. H., Shiekhattar R., and Nishikura K. (2006) Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 13, 13–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kawahara Y., Megraw M., Kreider E., Iizasa H., Valente L., Hatzigeorgiou A. G., and Nishikura K. (2008) Frequency and fate of microRNA editing in human brain. Nucleic Acids Res. 36, 5270–5280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yoon S. A., Choi J. R., Kim J. O., Shin J. Y., Zhang X., and Kang J. H. (2010) Influence of reduced folate carrier and dihydrofolate reductase genes on methotrexate-induced cytotoxicity. Cancer Res. Treat. 42, 163–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. McEntee G., Minguzzi S., O'Brien K., Ben Larbi N., Loscher C., O'Fágáin C., and Parle-McDermott A. (2011) The former annotated human pseudogene dihydrofolate reductase-like 1 (DHFRL1) is expressed and functional. Proc. Natl. Acad. Sci. U.S.A. 108, 15157–15162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Paz N., Levanon E. Y., Amariglio N., Heimberger A. B., Ram Z., Constantini S., Barbash Z. S., Adamsky K., Safran M., Hirschberg A., Krupsky M., Ben-Dov I., Cazacu S., Mikkelsen T., Brodie C., Eisenberg E., and Rechavi G. (2007) Altered adenosine-to-inosine RNA editing in human cancer. Genome Res. 17, 1586–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chan T. H., Lin C. H., Qi L., Fei J., Li Y., Yong K. J., Liu M., Song Y., Chow R. K., Ng V. H., Yuan Y. F., Tenen D. G., Guan X. Y., and Chen L. (2014) A disrupted RNA editing balance mediated by ADARs (adenosine deaminases that act on RNA) in human hepatocellular carcinoma. Gut 63, 832–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Qin Y. R., Qiao J. J., Chan T. H., Zhu Y. H., Li F. F., Liu H., Fei J., Li Y., Guan X. Y., and Chen L. (2014) Adenosine-to-inosine RNA editing mediated by ADARs in esophageal squamous cell carcinoma. Cancer Res. 74, 840–851 [DOI] [PubMed] [Google Scholar]
- 63. Chen L. C., Dollbaum C., and Smith H. S. (1989) Loss of heterozygosity on chromosome 1q in human breast cancer. Proc. Natl. Acad. Sci. U.S.A. 86, 7204–7207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tsuchiya Y., Nakajima M., Kyo S., Kanaya T., Inoue M., and Yokoi T. (2004) Human CYP1B1 is regulated by estradiol via estrogen receptor. Cancer Res. 64, 3119–3125 [DOI] [PubMed] [Google Scholar]
- 65. Komagata S., Nakajima M., Takagi S., Mohri T., Taniya T., and Yokoi T. (2009) Human CYP24 catalyzing the inactivation of calcitriol is post-transcriptionally regulated by miR-125b. Mol. Pharmacol. 76, 702–709 [DOI] [PubMed] [Google Scholar]
- 66. Tsuchiya Y., Nakajima M., Takagi S., Taniya T., and Yokoi T. (2006) MicroRNA regulates the expression of human cytochrome P450 1B1. Cancer Res. 66, 9090–9098 [DOI] [PubMed] [Google Scholar]