Abstract
Mycobacterium tuberculosis and related Corynebacterineae synthesize a family of lipomannans (LM) and lipoarabinomannans (LAM) that are abundant components of the multilaminate cell wall and essential virulence factors in pathogenic species. Here we describe a new membrane protein, highly conserved in all Corynebacterineae, that is required for synthesis of full-length LM and LAM. Deletion of the Corynebacterium glutamicum NCgl2760 gene resulted in a complete loss of mature LM/LAM and the appearance of a truncated LM (t-LM). Complementation of the mutant with the NCgl2760 gene fully restored LM/LAM synthesis. Structural studies, including monosaccharide analysis, methylation linkage analysis, and mass spectrometry of native LM species, indicated that the ΔNCgl2760 t-LM comprised a series of short LM species (8–27 residues long) containing an α1–6-linked mannose backbone with greatly reduced α1–2-mannose side chains and no arabinose caps. The structure of the ΔNCgl2760 t-LM was similar to that of the t-LM produced by a C. glutamicum mutant lacking the mptA gene, encoding a membrane α1–6-mannosyltransferase involved in extending the α1–6-mannan backbone of LM intermediates. Interestingly, NCgl2760 lacks any motifs or homology to other proteins of known function. Attempts to delete the NCgl2760 orthologue in Mycobacterium smegmatis were unsuccessful, consistent with previous studies indicating that the M. tuberculosis orthologue, Rv0227c, is an essential gene. Together, these data suggest that NCgl2760/Rv0227c plays a critical role in the elongation of the mannan backbone of mycobacterial and corynebacterial LM, further highlighting the complexity of lipoglycan pathways of Corynebacterineae.
Keywords: cell wall, glycolipid, lipid synthesis, membrane protein, Mycobacterium tuberculosis, Corynebacterium glutamicum, lipoarabinomannan, lipomannan
Introduction
The bacterial suborder Corynebacterineae includes important human and animal pathogens such as Mycobacterium tuberculosis (tuberculosis), Mycobacterium leprae (leprosy), and Corynebacterium diphtheriae (diphtheria). These bacteria synthesize a complex and highly distinctive cell wall, which confers intrinsic resistance to host antibacterial factors, antibiotics, and adverse environmental conditions (1). The inner layers of the mycobacterial/corynebacterial cell wall comprise peptidoglycan and arabinogalactan polysaccharide as well as covalently linked long chain mycolic acids that form the inner leaflet of an outer membrane (2, 3). A diverse range of non-covalently linked glycolipids and waxes form the outer leaflet and define the surface properties of these bacteria. Many of the cell wall components are essential for the growth of pathogenic mycobacteria species, hampering efforts to functionally characterize genes involved in cell wall assembly via standard gene deletion approaches (4–7). In contrast, the non-pathogenic Corynebacterium glutamicum can tolerate the loss of major cell wall components, making it a widely used model organism for studying pathways involved in core cell wall biosynthesis (8–18).
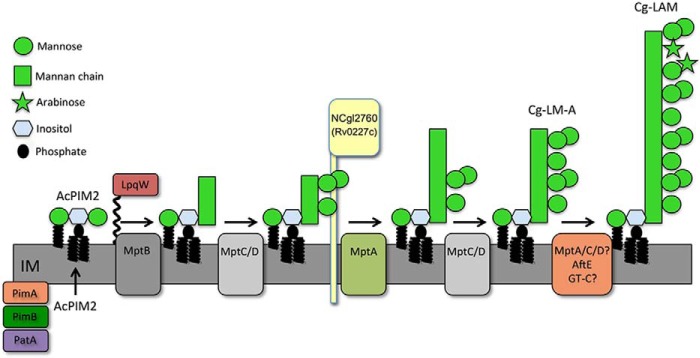
One class of non-covalently bound glycolipids synthesized by all mycobacteria/corynebacteria are the phosphatidyl-myo-inositol mannosides (PIMs).5 The PIMs are abundant cell wall components in their own right and are membrane anchors for the lipomannans (LM) and lipoarabinomannans (LAM). These lipoglycans are essential for both the viability and in vivo survival of pathogenic mycobacterial species, due to their capacity to modulate the host immune response during infection (7, 19–24). Many steps of the PIM → LM → LAM biosynthetic pathway are known (Fig. 1A; reviewed in Refs. 2 and 7). Phosphatidylinositol (PI) is mannosylated by GDP-mannose (GDP-Man)-dependent transferases PimA (26) and PimB (16) and acylated by PatA (27) in cytoplasmic reactions that form AcPIM2 and Ac2PIM2. These acylated PIM2 species can undergo additional mannosylation to form acylated PIM6 as an end product or to form hyperglycosylated LM and LAM in periplasmic reactions requiring polyprenol phosphomannose (PPM) donors. In mycobacteria, PimE uses PPM to elongate a population of AcPIM4 to form AcPIM6 (28), whereas studies in C. glutamicum show that MptB serves as the α1–6-mannosyltransferase that extends AcPIM2 to form the initial mannan backbone of LM (17) following its activation by lipoprotein LpqW (29, 30). However, the role of MptB in mycobacteria is unclear, possibly due to gene redundancy (17). PPM-dependent mannosyltransferases MptC and MptD add α1–2-mannose side chains (31) essential for virulence (32), whereas further extension of the α1–6 backbone is performed by MptA (12), forming LM. LM is converted to LAM by a series of α1–5-arabinosyltransferases, including EmbC (33, 34), AftB, AftC (15), and AftD (5), and then capped with additional sugars or organic acids in pathogenic species (35, 36).
FIGURE 1.
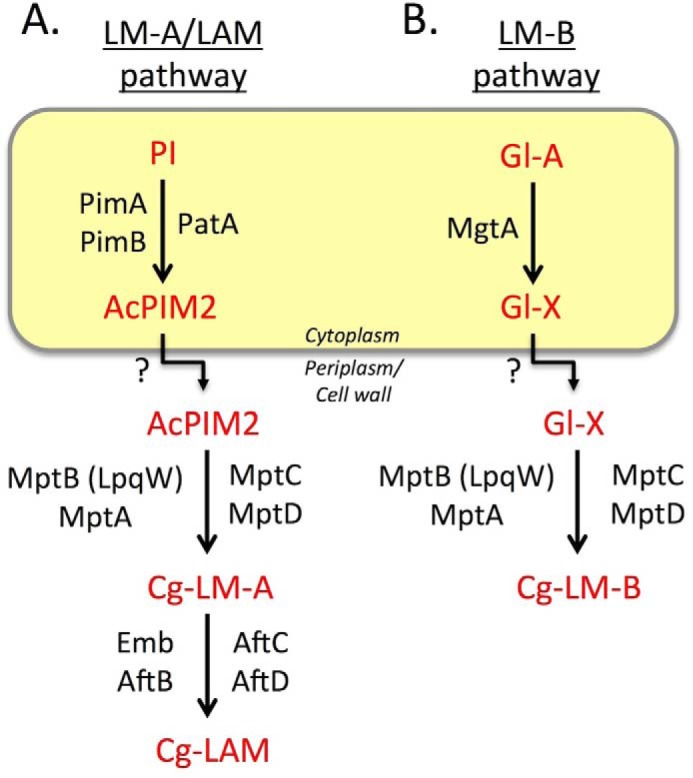
Summary of key steps of lipoglycan biosynthesis in C. glutamicum. A, the LM-A/LAM pathway. The early PIM intermediate AcPIM2 is assembled on the cytoplasmic leaflet of the cell membrane by GDP-Man-dependent mannosyltransferases PimA/PimB and acyltransferase PatA before being transported across the membrane for further elongation by polyprenol-phosphate-mannose-dependent mannosyltransferases (LpqW-activated MptB and MptA that form the main mannan chain; MptC and MptD that add mannose side chains) to form Cg-LM-A. Cg-LM-A is further modified by glycosyltransferases to form Cg-LAM. In mycobacteria, additional enzymes (e.g. EmbC, AftC, AftD, and CapA) are involved in the formation of a significantly larger and more complex arabinan structure. B, LM-B pathway. In C. glutamicum, a second family of lipomannans, termed Cg-LM-B, are assembled on a structurally distinct glycolipid anchor that contains the core structure Gl-A. Gl-A is extended with a single mannose residue by the cytoplasmic enzyme, MgtA, forming Gl-X, which is thought to be flipped and then elongated by MptA/B/C/D to form the end product, Cg-LM-B. This abundant species forms the bulk of LM in C. glutamicum. Whether this pathway exists in mycobacteria is unknown.
Studies in C. glutamicum have revealed a second lipoglycan pathway, termed LM-B, which is distinct from the conserved LM-A/LAM pathway (Fig. 1B). Although both pathways share many common enzymes, LM-A species are based on a glucopyranosyluronic acid diacylglycerol (Gl-A) anchor rather than PI (16, 18). On the cytoplasmic side, Gl-A is mannosylated by MgtA (previously named PimB), producing mannosyl-glucuronic acid diacylglycerol (Gl-X), which is extended by MptB (17, 29) and other enzymes common to both pathways to produce LM-B, the major LM pool of C. glutamicum (37, 38).
Here we show that deletion of NCgl2760, the orthologue of the essential M. tuberculosis Rv0227c gene (39, 40), results in a defect in the elongation of both LM-A and LM-B intermediates and the loss of mature LAM (12). Interestingly, this gene sits within a group of genes involved in synthesis of trehalose corynomycolates, including the recently described tmaT/NCgl2759 gene (Fig. 2A). We conclude that NCgl2760 has a direct role in regulating LM-A/LAM and LM-B synthesis, possibly by regulating the activity of LM mannosyltransferases and/or access of PIM intermediates to these enzymes, and that it may be a therapeutic target in pathogenic mycobacteria species.
FIGURE 2.
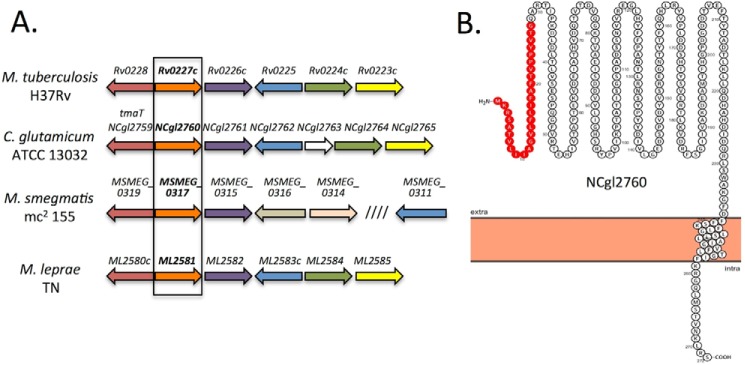
NCgl2760 is a predicted membrane protein encoded within a conserved genetic locus. A, the tmaT locus of Corynebacterineae. Orthologous genes in the four species are shown using the same color. The focus of the current study, NCgl2760, and its likely mycobacterial orthologues are indicated by a black box. B, Protter prediction for NCgl2760 (48). The protein is predicted to contain an N-terminal signal sequence (red) and a single transmembrane domain.
Results
Identification of a Corynebacterial Orthologue of M. tuberculosis Gene, Rv0227c
We have recently shown that the C. glutamicum gene NCgl2759 (orthologue of the essential M. tuberculosis gene, Rv0228) encodes an acetyltransferase that converts trehalose monocorynomycolate (TMCM) to an acetylated form (AcTMCM) and is essential for efficient transport of TMCM across the inner membrane for conversion to trehalose dicorynomycolate (TDCM) (41). To determine whether other genes in this locus are involved in trehalose corynomycolate biosynthesis, we investigated the function of NCgl2760, the next gene in the genome of C. glutamicum ATCC 13032. NCgl2760 encodes a putative protein of 272 residues and is the orthologue of M. tuberculosis Rv0227c, sharing 16.4% identity and 24.9% similarity (supplemental Fig. S1) and synteny to Rv0227c and related genes in other mycobacterial genomes (Fig. 2A). NCgl2760 is predicted to contain a single transmembrane domain and a large periplasmic domain (Fig. 2B).
Inactivation of the NCgl2760 Gene
To investigate NCgl2760 function, a two-step recombination strategy was used to create an NCgl2760 mutant for phenotypic characterization (Fig. 3). Deletion of the gene was achieved using the suicide vector pK18mobsacB (42), which carries a kanamycin resistance gene (aph) and Bacillus subtilis sacB gene, conferring sensitivity to sucrose. Potential deletion mutants were identified by PCR screening (data not shown) and then confirmed by Southern blotting analyses (Fig. 3). In contrast to the NCgl2759 null mutant (41), the ΔNCgl2760 strain grew at the same rate as the wild-type (parental) strain on solid media. Growth curves performed in liquid brain heart infusion (BHI) medium confirmed the absence of any detectable growth defect arising from deletion of the NCgl2760 gene (data not shown). This contrasts with the situation in M. tuberculosis, where the gene is regarded as essential (39, 40).
FIGURE 3.
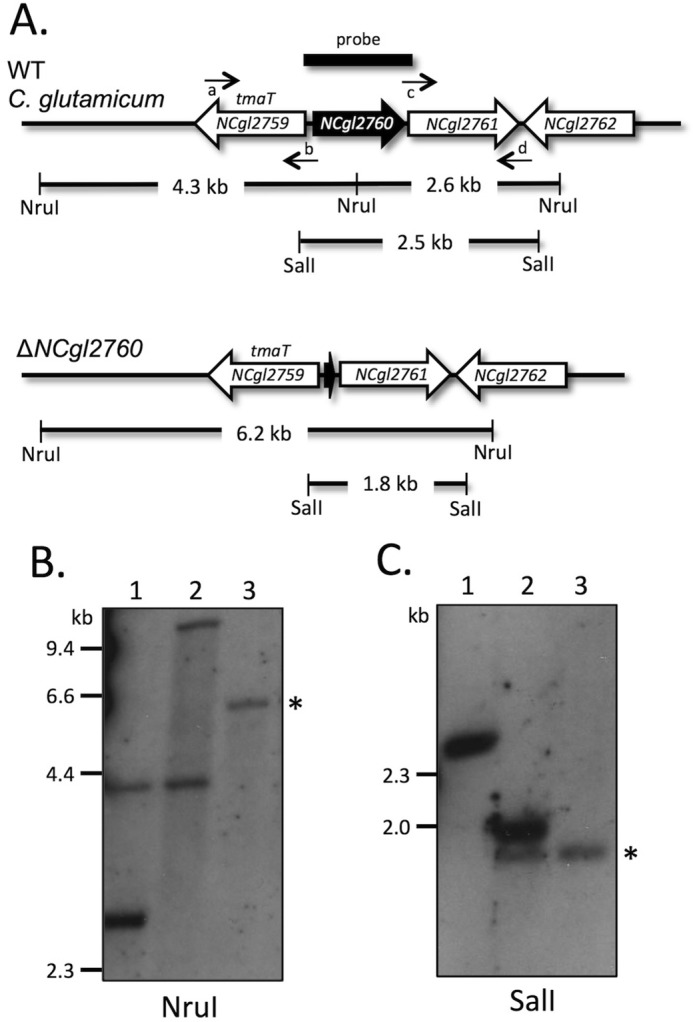
Disruption strategy and analysis of ΔNCgl2760. A, diagram showing the arrangement of genes in the NCgl2760 region of WT C. glutamicum (top) and ΔNCgl2760 mutant (bottom). NruI and SalI restriction sites, sizes of expected bands on Southern blotting, and position of the probe used are indicated. Small horizontal arrows indicate the binding sites for the four primers used to construct the ΔNCgl2760 mutant (a, NCgl2760-left_F; b, NCgl2760-left_R; c, NCgl2760-right_F; d, NCgl2760-right_R). B, Southern blotting analysis of NruI-digested DNA of C. glutamicum WT (lane 1), a single crossover strain (lane 2), and the ΔNCgl2761 mutant (lane 3). C, Southern blotting analysis of SalI-digested DNA of C. glutamicum WT (lane 1), a single crossover strain (lane 2), and the ΔNCgl2760 mutant (lane 3). Positions of DIG-labeled λ DNA standards digested with HindIII are indicated in kb. Bands showing that the NCgl2760 gene has been deleted (a 6.2-kb NruI fragment in B and a 1.8-kb SalI fragment in C) are indicated by asterisks.
Complementation of C. glutamicum ΔNCgl2760
Before phenotypic characterization of the mutant, complementation and control strains were created to determine whether any cell wall defect was due solely to disruption of NCgl2760. To complement the mutant, ΔNCgl2760 was transformed with pSM22:NCgl2760, a plasmid carrying a full-length NCgl2760 gene plus 180 bp of upstream sequence, which may include the native promoter. The empty pSM22 vector was also introduced into ΔNCgl2760 as a control.
Disruption of NCgl2760 Results in Accumulation of a Truncated LM Species
Cell wall components of ΔNCgl2760 and the complementation strains were analyzed and compared with the parental WT strain, C. glutamicum ATCC 13032. No changes were found in the levels or composition of free lipids, including the major PIMs and trehalose corynomycolates (supplemental Fig. S2). However, marked differences were observed in the LM/LAM profile of the ΔNCgl2760 mutant, following analysis of purified LM/LAM by PAGE (Fig. 4A). As expected, WT C. glutamicum produced two distinct populations of lipoglycans corresponding to LM (LM-A and LM-B co-migrate) and LAM (Fig. 4A, lane 1). In contrast, the ΔNCgl2760 strain (lane 3) and ΔNCgl2760 bearing empty pSM22 (lane 5) lacked mature LM or LAM but synthesized a faster-migrating “LM” species that we designated t-LM (truncated LM). Synthesis of mature LM and LAM was fully restored by complementation of ΔNCgl2760 with a plasmid-encoded copy of the gene (Fig. 4A, lane 4), confirming that the loss of LM/LAM and appearance of t-LM were due solely to deletion of the NCgl2760 gene.
FIGURE 4.
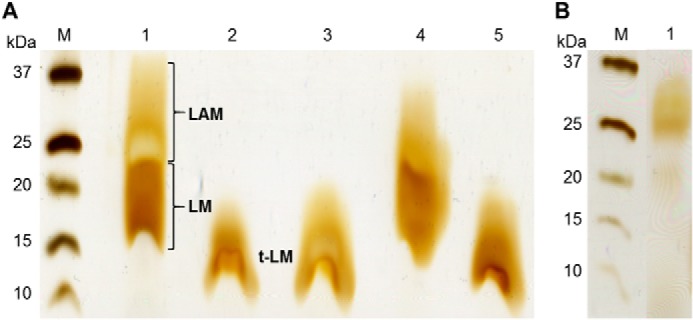
C. glutamicum ΔNCgl2760 synthesizes a novel truncated LM species. A, SDS-PAGE analysis of LM/LAM fractions from WT (lane 1), ΔmptA mutant (lane 2), ΔNCgl2760 mutant (lane 3), ΔNCgl2760 mutant complemented with a functional copy of NCgl2760 (lane 4), or plasmid alone (lane 5). The LM/LAM phenotypes of the ΔmptA and ΔNCgl2760 mutant lines are very similar. B, SDS-PAGE analysis of lipoglycans purified from a ΔmgtA mutant (lane 1).
ΔNCgl2760 and ΔmptA Mutants Synthesize a Similar Truncated LM
The LM/LAM phenotype of ΔNCgl2760 appeared to be very similar to that of a previously described C. glutamicum cell wall mutant lacking the mptA gene, which encodes an α(1→6)-mannosyltransferase responsible for extending the mannan backbone of LM-A and LM-B precursors (12). Disruption of mptA results in the synthesis of truncated LM-A- and LM-B-based lipoglycans that have a short mannan backbone and lack arabinose side chain elaborations (12). To allow direct comparison of these two lines, we deleted the mptA gene in C. glutamicum 13032 (see supplemental Fig. S3). As expected, LM/LAM analysis of the ΔmptA strain revealed a lack of full-length LM and LAM and the presence of a faster migrating lipoglycan species on SDS-PAGE (Fig. 4A, lane 2). Significantly, the faster migrating LM lipoglycan extracted from ΔmptA cells had the same migration on SDS-PAGE gels as the ΔNCgl2760 t-LM species (Fig. 4A, lanes 2 and 3), supporting the conclusion that the ΔNCgl2760 mutant has a defect in LM elongation.
To further define the nature of the LM defect in ΔNCgl2760, purified lipoglycans from WT and mutant lines were subjected to monosaccharide composition analysis. Whereas the WT LM/LAM fraction contained an average molar ratio of myo-inositol/mannose/arabinose of 1:100:7, the LM/LAM fractions of ΔNCgl2760 and ΔmptA had molar ratios of 1:30:0.13 and 1:23:0.17, respectively. Complementation of ΔNCgl2760 with the plasmid containing NCgl2760, but not empty pSM22, restored the molar ratio of myo-inositol/mannose/arabinose to 1:83:6. These analyses are consistent with loss of NCgl2760 being associated with a complete loss of LM arabinose side chains as well as an overall reduction in mannan backbone chain length.
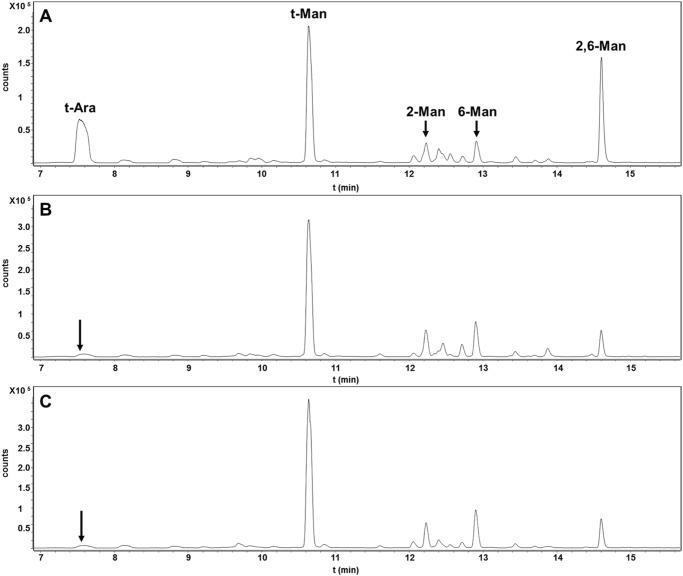
Methylation linkage analysis of the lipoglycan fractions from WT and mutant bacteria confirmed that the t-LM of the ΔNCgl2760 mutant lacked the terminal arabinose side chains with a concomitant increase in terminal mannose residues (Fig. 5). Interestingly, these analyses showed that the ΔNCgl2760 t-LM had fewer 2,6-linked mannose residues (with associated increase in 6-linked mannose), indicating fewer mannose side chains branching off the α1–6-linked mannan backbone (Fig. 5C). Analysis of the ΔmptA LM fraction (Fig. 5B) gave an almost identical branching pattern, consistent with both mutant lines producing short LM species containing an α1,6-linked mannan backbone with few mannose side chains and no arabinose capping.
FIGURE 5.
Linkage analyses of C. glutamicum WT, ΔmptA, and ΔNCgl2760 mutant LM/LAM. LM/LAM samples were methylated, hydrolyzed, and reduced, and the resultant PMAA sugars were analyzed by GC-MS. A–C, PMAA of LM/LAM preparations from C. glutamicum WT (A), ΔmptA (B), and ΔNCgl2760 (C) bacteria. The mutant LM/LAM lacked PMAA corresponding to terminal Araf and had altered abundance of PMAA corresponding to branched (2,6-disubstituted Man) and unbranched (6- and 2-monosubstituted Man).
LC-ESI-TOF-MS analysis of the LM fractions of WT and mutant bacteria provided further information on the nature of the defect in LM maturation. Direct analysis of WT LM by LC-ESI-TOF-MS allowed the detection of a polydisperse mixture of 117 different LM species (differing by 162 mass units) in the 2712–9190 Da size range (after deconvolution; Fig. 6A). The masses of these species were consistent with the presence of a mannan chain containing 10–50 mannose residues and a lipid anchor comprising a PI moiety with 2–3 fatty acyl chains. The plots in Fig. 6 show the masses (m/z, after deconvolution) of all detected LM species (from low to high molecular weight) and predicted mannan chain length (number of mannose residues). Supplemental Fig. S5 explains, step by step, the development of Fig. 6. In marked contrast, the ΔNCgl2760 t-LM fraction completely lacked LM species above 5500 Da and instead accumulated a range of LM species with masses of 2626–5500 Da (8–27 mannose residues; Fig. 6C). As in the WT fraction, a number of different series (91 species in total) were detected in this fraction that differed by m/z 162 and acyl composition of the lipid anchor. Similarly, 112 LM species were detected in the ΔmptA mutant (Fig. 6B), with a size distribution of 2206–8179 Da (8–43 mannose residues). However, the majority of these lipoglycan species had a size between 2200 and 4900 Da, very similar to ΔNCgl2760. Interestingly, up to 72% of LM species detected in the ΔNCgl2760 and ΔmptA mutants had identical molecular weights, indicating a very similar LM phenotype. An ΔmgtA mutant, in which the LM-B pathway is inactive (38) (Fig. 4B), also produced a series of LM species (78 species; Fig. 6D), with a size distribution of 3222–8859 Da (13–48 mannose residues), 32 of which were common to ΔNCgl2760. Interestingly, the C. glutamicum NCgl2759 mutant, lacking the acetyltransferase involved in TMCM transport, synthesized LM that was indistinguishable from WT LM (data not shown), indicating that the LM phenotype observed in ΔNCgl2760 bacteria is not an indirect consequence of defects in TMCM transport. Collectively, these analyses indicate that loss of NCgl2760 is associated with a defect in LM elongation, leading to an accumulation of early LM precursors containing an α1–6-linked mannan backbone (∼20 residues) with minimal mannose side chains.
FIGURE 6.
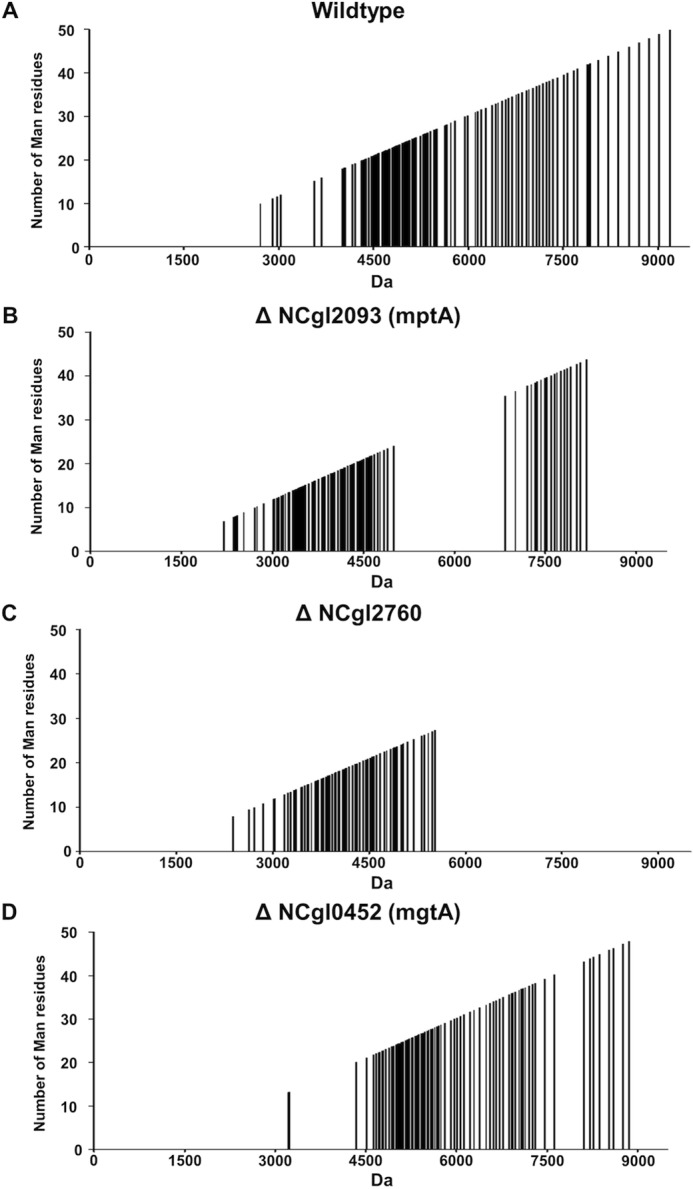
Q-TOF-MS profiling of LM samples from WT and ΔmptA and ΔNCgl2760 mutant lines. Purified LM/LAM samples from WT (A), ΔmptA (B), ΔNCgl2760 (C), and ΔmgtA (D) were analyzed by direct infusion Q-TOF-MS in negative ion mode. Individual LM species were identified based on their exact mass after deconvolution and mass difference (by m/z 162 or 324) from related LM species containing the same lipid moieties. The plots show the mass (m/z) of all detected species and predicted mannan chain length (number of mannose residues). C. glutamicum WT bacteria contained 117 clearly resolved LM species with masses between 2712 and 9190 Da (10–50 mannose residues). In contrast, LM species of the ΔmptA mutant (112 LM species) and ΔmgtA mutant (78 LM species) had masses between 2200 and 4900 Da (with a minor cluster at higher molecular mass) and between 3222 and 8859 Da (13–48 mannose residues), respectively. ΔNCgl2760 LM (91 lipoglycan species detected) had a size distribution between 2626 and 5520 Da (8–27 mannose residues), and no higher mass lipoglycan species were detected at all. ΔNCgl2760 shared 66 lipoglycan species with the ΔmptA mutant and 32 LM species with the ΔmgtA mutant, indicating that both LM pathways (LM-A and LM-B) are still active in the ΔNCgl2760 knock-out mutant.
Evidence That the Orthologue of NCgl2760 in M. smegmatis, MSMEG_0317, Is an Essential Gene
To investigate whether the putative orthologue in M. smegmatis (Fig. 2 and supplemental Fig. S1) also functioned in LM/LAM biosynthesis, we attempted to delete the MSMEG_0317 gene. A DNA fragment was synthesized containing 226 bp from the left and right flanking regions of MSMEG_0317 and cloned into an M. smegmatis suicide vector encoding streptomycin resistance and sucrose sensitivity via a sacB gene. This construct was electroporated into M. smegmatis mc2155, selecting for single crossover (SCO) clones in which the plasmid had integrated into the chromosome. Southern blotting confirmed integration at the MSMEG_0317 locus, and this SCO strain was cultured on Middlebrook 7H10 plates containing 10% (w/v) sucrose to select for potential double crossover (DCO) strains. PCR analysis of 50 clones revealed that all were WT revertents resulting from ejection of the plasmid from the chromosome, which restored the MSMEG_0317 gene (data not shown). The lack of any ΔMSMEG_0317 mutants raised the possibility that the gene is essential in M. smegmatis.
If MSMEG_0317 is essential, introduction of a second copy at another site in the chromosome of the SCO strain should allow a DCO strain to be derived. The MSMEG_0317 gene was cloned into the integrative expression vector, pMV361 (43), and then electroporated into MSMEG_0317 SCO. The resultant strain was cultured as described above to select potential DCO strains. This time, in 5 of 17 strains tested, the MSMEG_0317 gene had been replaced by the deleted copy (Fig. 7). These differences are statistically significant (χ2 p value = 0.000067 (p < 0.05)), providing strong evidence of essentiality. Our findings suggest that, in contrast to the situation in C. glutamicum, M. smegmatis may require the presence of full-length LM and/or LAM for viability.
FIGURE 7.
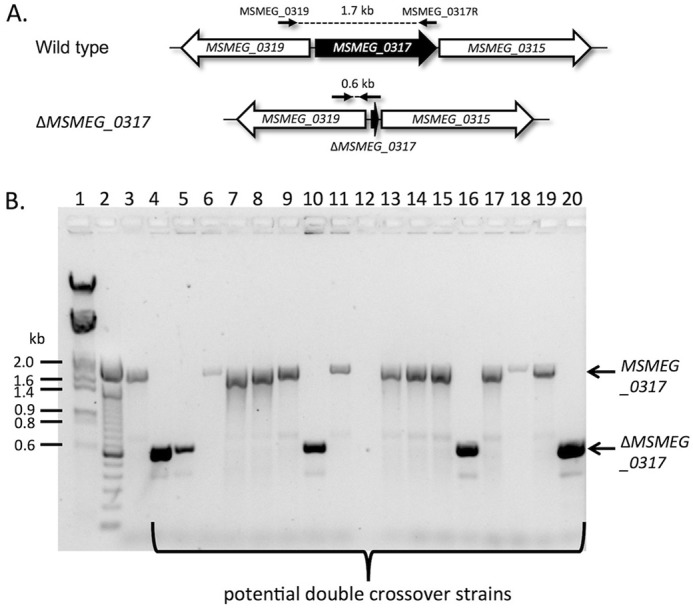
The M. smegmatis orthologue of NCgl2760, MSMEG_0317, is an essential gene. A, the MSMEG_0317 locus. Primer binding sites are shown by small horizontal arrows, and expected PCR product sizes for the MSMEG_0317 gene and ΔMSMEG_0317 allele are indicated in kb. B, potential DCO clones derived from MSMEG_0317 SCO containing an integrated pMV361:MSMEG_0317 plasmid were analyzed by PCR using primers MSMEG_0319 and MSMEG_0317R. Lane 1, λHindIII/EcoRI molecular weight DNA markers (sizes shown in kb); lane 2, 100-bp DNA ladder; lane 3, wild-type M. smegmatis genomic DNA as template; lanes 4–20, potential DCO strain genomic DNA as templates. Clones in lanes 4, 5, 10, 16, and 20 gave the expected size band (0.6 kb) for a DCO, whereas the remaining 12 clones were WT revertents (1.7 kb). The reaction shown in lane 12 failed to give a detectable product.
Discussion
We have recently shown that the C. glutamicum acetyltransferase TmaT (TMCM mycolyl acetyltransferase; NCgl2759) catalyzes the acetylation of the major cell wall glycolipid TMCM and that this modification is important for periplasmic transport of this glycolipid (41). A subsequent study showed that the mycobacterial orthologue has the same function (44). Based on these findings, we hypothesized that the adjacent gene, NCgl2760, which is syntenic in both corynebacteria and mycobacteria, would also have a role in corynomycolate biosynthesis. However, deletion of NCgl2760 had no effect on the steady state levels of TMCM or TDCM. Further analyses revealed that loss of NCgl2760 leads to a profound defect in the elongation of LM precursors and the synthesis of mature LM and LAM. This defect was completely reversed by genetic complementation with NCgl2760. Thus, NCgl2760 appears to have a role in the synthesis of lipoglycans that are structurally unrelated to TMCM, despite the location of its encoding gene.
The LM/LAM phenotype observed following disruption of NCgl2760 was very similar to that generated by deletion of the C. glutamicum gene encoding MptA, a polyprenol-phosphate-mannose-dependent α1–6-mannosyltransferase that extends the mannan backbone of LM-A and LM-B intermediates (12) following initial α1–6-Man extension by MptB (17) and α1–2-Man side branching by MptC/D (31). Direct comparison of the ΔmptA and ΔNCgl2760 mutants revealed truncated species with similar SDS-PAGE migration and similar LM/LAM profiles (Figs. 4 and 6). Lipoglycans from both mutants lacked arabinose, as expected, and had significantly less 2,6-linked Man, suggesting that Man side branches are normally added after full extension of the α1–6-linked Man backbone and are concentrated at the distal end of LM, consistent with previous findings (12).
Interestingly, neither MptA nor NCgl2760 are required for normal growth of C. glutamicum, in marked contrast to the essentiality of these genes in pathogenic and non-pathogenic species of mycobacteria (12). Notwithstanding the similar mutant phenotypes, the encoded MptA and NCgl2760 proteins share no other common characteristics. MptA is an integral membrane protein with 13 predicted transmembrane domains and a DXD motif of a GT-C glycosyltransferase, consistent with transfer of mannose from a PPM donor. NCgl2760 has a single transmembrane domain, placing the bulk of the protein in the periplasm (Fig. 2B), where a large loop of MptA, including the DXD motif, is also located (12). However, it has no motifs suggesting an enzymatic function, making NCgl2760 the second protein without an obvious glycosyltransferase function to be assigned to these pathways. The first was LpqW, a lipoprotein proposed to serve as an activator of the MptB mannosyltransferase (29).
Based on these very similar phenotypes, we propose that NCgl2760 plays a key role in regulating steps in the LM/LAM biosynthetic pathway. The fact that this mutant produces LM species with a more restricted molecular size distribution than the ΔmptA mutant suggests that NCgl2760 may act just before the elongation steps catalyzed by MptA (Fig. 8). NCgl2760 could either directly or indirectly influence MptA function and/or regulate access of the LM substrate or the PPM donor to MptA. Structural studies may provide important insights into the role of NCgl2760 and its mycobacterial orthologues.
FIGURE 8.
NCgl2760 is required for elongation of LM-A/LAM lipoglycans in C. glutamicum. Only the LM-A-based lipoglycan pathway (assembly on PI) is shown for clarity, although the LM-B pathway (assembly on GlcA-DAG) is also defective in the mutant.
In contrast to the situation in C. glutamicum, the mycobacterial orthologues are essential for normal growth (39, 40). It has previously been proposed that the M. tuberculosis orthologue, Rv0227c, may have a role in invasion and infection of host cells (45). Specifically, evidence was proposed for surface expression of the Rv0227c protein based on electron microscopy and Western blotting studies, consistent with the presence of a putative signal sequence and membrane anchor in the corynebacterial orthologue (Fig. 2B). High activity binding peptides were identified within Rv0227c, some of which inhibited entry to the cell lines tested and could be considered candidates for inclusion in an anti-tuberculosis vaccine. Although our findings do not rule out a direct role for Rv0227c in host cell invasion, our data suggest that Rv0227c has a role in the synthesis of LM and ultimately LAM, which are ligands for host cell receptors (e.g. Toll-like receptors and C-type lectins, including dendritic cell-specific intercellular adhesion molecule-3 grabbing nonintegrin (DC-SIGN) and the macrophage mannose receptor (MMR) (for a review, see Ref. 7)) with binding influencing pathogen uptake and subsequent interactions between pathogen and host.
C. glutamicum has proved a useful model system for identifying pathways of cell wall biosynthesis in the actinomycetes, because it is more permissive to loss of cell wall components than mycobacteria. Indeed, Rv0227c is reported to be essential for in vitro growth of M. tuberculosis H37Rv based on high-density mutagenesis and sequencing studies (39, 40). Furthermore, we show here that the M. smegmatis orthologue is essential under standard growth conditions (Fig. 7). C. glutamicum may be more tolerant to loss of enzymes involved in LM/LAM biosynthesis because it has two pathways of glycolipid anchor biosynthesis: the PI-based LM-A pathway and the GlcA-Gro-Ac2-based LM-B pathway (Fig. 1). However, our findings indicate that NCgl2760 influences both the LM-A and LM-B pathways. This is also the case for a number of previously characterized GT-C type mannosyltransferases involved in synthesis of the mannan backbone and mannose side branches (12, 17, 31), showing that the same reactions occur on different lipid anchors. It remains unclear why LM-B species fail to progress further down the pathway to form LAMs.
Based on our findings in C. glutamicum, it is likely that M. tuberculosis Rv0227c fulfills a similar function in LM/LAM biosynthesis and is a possible drug target (39, 40). Indeed, interfering with Rv0227c activity should affect M. tuberculosis viability while also blunting the immunosuppressive properties of bacteria in vivo. The predicted location of Rv0227c within the periplasmic space may also increase access of the functional domain of this protein to inhibitors.
Experimental Procedures
Bacterial Strains, Culture Conditions, Transformation, and Genetic Manipulation
Escherichia coli DH5α was grown in Luria-Bertani (LB) medium at 37 °C with aeration. C. glutamicum ATCC 13032 was grown in BHI medium (Oxoid) or LBHIS (LB, BHI, sorbitol) (46) at 30 °C with aeration. When necessary, ampicillin was added to a final concentration of 100 μg ml−1 and kanamycin at 50 μg ml−1. E. coli plasmid DNA was isolated from 10 ml of an overnight culture using the High Pure plasmid isolation kit (Roche Applied Science), and C. glutamicum genomic DNA was extracted from ∼0.5 g of cells using the Illustra DNA extraction kit (GE Healthcare), according to the manufacturer's instructions. When necessary, DNA was purified using an UltraClean 15 DNA purification kit (MoBio). PCRs were performed in a PTC-200 thermal cycler (MJ Research) using Taq polymerase (Roche Applied Science) or ProofStart DNA polymerase (Qiagen). Initial denaturation of template DNA was done at 95 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 1 min, a 1-min primer-specific annealing step, and a 1-min/kb extension step at 72 °C. The program included a 10-min final extension step at 72 °C. PCR products were purified by extraction from 1% agarose gels using an UltraClean 15 DNA purification kit (MoBio). Endonucleases, T4 ligase, polynucleotide kinase system, and alkaline phosphatase were obtained from New England Biolabs and used according to the manufacturer's instructions.
Bioinformatic Identification and Analysis of NCgl2760
The corynebacterial orthologue of mycobacterial Rv0227c was found using the BLASTp (47) algorithm. Protein topology predictions were performed using Protter version 1.0 (48). Amino acid sequence alignments were produced using Clustal Omega.
Construction of C. glutamicum ΔNCgl2760 and Complementation Strains
The NCgl2760 gene was deleted using a two-step allelic replacement strategy previously used in our laboratory to create other cell wall mutants (11, 16, 29, 41). A 1.0-kb fragment containing sequence from the left side of the NCgl2760 gene was amplified using ProofStart DNA polymerase (Qiagen) and the primers NCgl2760-left_F (5′-GTCACCCGGGTAAATAAAGGCAACC) and NCgl2760-left_R (5′-GATAGGATCCCAATAATGAGGACCG) and cloned into the XmaI/BamHI sites (underlined) of pUC19 (49), creating plasmid pUC-NCgl2760left. A 1.0-kb fragment containing sequence from the right side of the NCgl2760 gene was amplified using primers NCgl2760-right_F (5′-TAGCGGATCCAATTTTCAAGCGCGGCCAGCTGATGAGC) and NCgl2760-right_R (5′-GATATCTAGATTTTCACCCCGGCGG) and cloned into the BamHI/XbaI sites (underlined) of pUC19, creating plasmid pUC-NCgl2760right. The right flanking sequence was then liberated from pUC-NCgl2760right using BamHI/XbaI and subcloned into Xba/BamHI-digested pUC-NCgl2760left, fusing the left and right flanking sequences to create plasmid pUC-ΔNCgl2760. This entire insert was then liberated using XmaI/XbaI and subcloned into XmaI/XbaI-digested pK18mobsacB, a suicide plasmid for C. glutamicum (50) that contains kanamycin and sucrose selection markers. The resultant plasmid, pK18mobsacB:ΔNCgl2760, was sequenced and then electroporated into electrocompetent C. glutamicum cells, prepared as described previously (46), using an ECM 630 electroporator (BTX). Clones resulting from single homologous recombination events were selected on kanamycin. These were grown overnight without antibiotic selection and then serially diluted and plated onto LBHIS plates containing 10% sucrose to select for a second crossover event. Sucrose-resistant, kanamycin-sensitive colonies were screened by PCR using NCgl2760-left_F and NCgl2760-right_R primers (data not shown). Potential NCgl2760 mutants were confirmed using Southern blotting hybridization.
To complement the ΔNCgl2760 strain, the entire NCgl2760 gene plus 180 bp of upstream sequence was PCR-amplified using primers NCgl2760-comp_F (5′-TCGGATATCGCGAGTACTGCACCG) and NCgl2760-comp_R (5′-TCGGATATCGGGTCGGGGTCATAAGGATTTAACTCC), digested with EcoRV (underlined), and cloned into the unique PvuII site of pSM22 (42), which contains the corynebacterial origin of replication repA and kanamycin resistance gene aphA3. A sequenced complementation plasmid (pSM22:NCgl2760) and pSM22 control plasmid were electroporated into the C. glutamicum ΔNCgl2760 deletion strain, followed by selection on kanamycin-supplemented BHI plates.
Construction of NCgl2093 (mptA) and NCgl0452 (mgtA) Mutants
Both genes were deleted using synthetic gBlock gene fragments (Integrated DNA Technologies), whereby a 500-bp DNA sequence was custom-designed from ∼220 bp of sequence upstream and downstream of the genes plus 48 bp containing the 5′- and 3′-end of each gene. Both synthetic DNA fragments were designed with a BamHI site at one end and an XbaI site at the other end for cloning into pK18mobsacB. These plasmid constructs were then electroporated into C. glutamicum ATCC 13032. Kanamycin-resistant colonies were tested for the presence of integration of the plasmid via a single crossover event by PCR. Confirmed single crossover clones were then cultured on plates containing 10% sucrose to generate double crossover strains that were confirmed by PCR and Southern blotting hybridization (supplemental Figs. S3 and S4).
Southern Hybridization
For Southern blotting analysis, 2 μg of genomic DNA was digested with appropriate restriction enzymes under optimal conditions for 16 h. Purified samples and digoxygenin (DIG)-labeled, HindIII-digested λ DNA markers were separated on a 1% agarose gel followed by depurination, denaturation, neutralization, and capillary transfer onto a nylon membrane. The membrane was then hybridized at 65 °C with a gene-specific probe prepared by DIG labeling a 1.0-kb PCR product obtained using primers NCgl2760-comp_F and NCgl2760-comp_R.
Compositional Cell Wall Analysis
WT and mutant cells were harvested by centrifugation at logarithmic growth phase (A600 of 1–3) from 100 ml of culture in BHI. Free lipids, including PIMs and trehalose mycolates, were extracted and purified as described previously (41). The delipidated cell pellet was then subjected to ethanol reflux by suspending in 5 ml of 50% (v/v) ethanol and incubating at 100 °C for 2 h, with occasional vortex mixing. Samples were centrifuged at 800 × g for 5 min, the supernatant was collected, and the procedure was repeated two more times. Pellets were used for corynomycolic acid extraction. The supernatant was pooled, and ethanol was removed under a stream of N2 followed by freeze-drying. The dried pellets were resuspended in 400 μl of water containing 0.02% (w/v) CaCl2 and 10 units of proteinase K, followed by incubation at 37 °C for 2–3 h. The digest was diluted with 50% propan-1-ol (50 μl) and 1 m ammonium acetate (50 μl) and loaded onto a column of octyl-Sepharose (1 ml) equilibrated in 5% 1-propanol and 50 mm ammonium acetate. After washing the column, lipoglycans were eluted with 30, 40, 50, and 60% 1-propanol (1-ml volumes), and carbohydrate-containing fractions were identified by spotting 5-μl aliquots on high-performance thin layer chromatography (HPTLC) sheets and stained with orcinol-H2SO4 (51). Lipoglycan-containing fractions (in 30–40% propan-1-ol fractions) were pooled, dried under vacuum, and resuspended in 30% (v/v) propan-1-ol (100 μl/100 mg pellet weight) for further analysis.
HPTLC Analyses
Extracted lipids were analyzed on aluminum-backed HPTLC silica sheets (Merck) in chloroform, methanol, 13 m ammonia, 1 m ammonium acetate, water (180:140:9:9:2.3, v/v/v/v/v). Glycolipids were detected with orcinol in HCl (100 °C, 5 min) followed by charring at 150 °C to detect all lipids.
Polyacrylamide Gel Electrophoresis of LM/LAM
Purified lipoglycan fractions were mixed with PAGE sample buffer and incubated at 98 °C for 5 min. Samples were then separated on a Mini-Protean®TGXTM precast gel (Bio-Rad), and LM/LAMs were visualized using the Pierce® silver stain kit according to the manufacturer's instructions (Thermo Scientific). In detail, the gel was washed for 5 min in ultrapure water (two times); fixed for 15 min in 30% (v/v) ethanol, 10% (v/v) acetic acid solution (two times); washed for another 5 min in 10% (v/v) ethanol (two times); and washed for 5 min in ultrapure water (two times). Next, the gel was sensitized for 1 min (50 μl of sensitizer with 25 ml of water) and subsequently washed for 1 min in water (two times). Next, the gel was stained for 30 min (0.5 ml Enhancer with 25 ml of stain), washed two times for 20 s in ultrapure water, and finally developed (0.5 ml of Enhancer with 25 ml of Developer) for 2–3 min until bands appeared. The reaction was stopped by adding a 5% (v/v) acetic acid solution. The sugar content was also quantified by GC-MS after methanolysis and TMS derivatization (51).
Methanolysis of Extracted LM/LAM Samples for GC-MS Analysis
LM/LAM samples (24 μl of stock, 1 mm) were transferred to a heat sealed capillary tube (250-μl Drummond microdispenser, Drummond Scientific Co.), together with 10 μl of 0.1 mm scyllo-inositol as an internal standard. All samples were subsequently washed with methanol, dried in a vacuum concentrator, and resuspended in 50 μl of 0.5 m methanolic HCl (Supelco). The glass tubes were flame-sealed under vacuum and subjected to solvolysis (80 °C, 12–18 h). Following transfer to GC vial inserts, the methanolysate was neutralized by adding fresh pyridine (10 μl) and acetic anhydride (10 μl). After 30 min (room temperature), samples were dried down again, and the methylesters were derivatized by adding 35 μl of BSTFA + 1% TMCS (Thermo Scientific). GC-MS analysis was performed on an Agilent HP 6890 GC system and an Agilent HP 5973 MSD (Agilent Technologies) in electron ionization mode as described previously (25).
LC/MS-ESI-TOF Analysis of LM/LAM
Purified LM/LAM was suspended in 5 mm ammonium carbonate/bicarbonate buffer (pH 8.16), and 4 μl was injected by direct infusion into a Agilent 6550 iFunnel Q-TOF LC/MS system (Agilent Technologies). The run was performed in negative ionization mode, with a mass range of 100–3200 Da. The reference nebulizer was set to 5 p.s.i.g. with a detection window of 100 ppm, a minimum height of 1000 counts, an acquisition rate of 0.4 spectra/s, and an acquisition time of 1589.4 ms/spectrum. The gas temperature was set to 220 °C with a drying gas of 7 liters/min, dual AJS ESI 3500 V, Fragmentor 365 V, and Skimmer 65 V. The flow rate was set to 0.2 ml/min, and a solvent consisting of 0.1 m formic acid, acetonitrile (1:1, v/v) was used to wash the lines for 25 min. Data were analyzed using MassHunter, and the length of the mannan chain was calculated, assuming the presence of a PI lipid anchor containing C16:1/18:1/16:0 fatty acids. Analyses were performed in duplicate.
Linkage Analysis of LM/LAM
Purified LM/LAM fractions were subjected to methylation linkage analysis as described previously (16). Permethylated, peracetylated sugars were analyzed on an Agilent HP 6890 GC and 5973 MSD (Agilent Technologies) in electron ionization mode, using a DB5 capillary column (J&W Scientific CP9013, 30 m, 250-μm inner diameter, 0.25-μm film thickness), with a 10-m inert duraguard. The injector insert and GC-MS transfer line temperatures were 270 and 250 °C, respectively. The oven temperature gradient was set to 140 °C (2 min), 140–230 °C at 5.0 °C/min and in the end held for 10 min. The run was also performed in SIM (selected ion monitoring) mode, selecting the ions 161, 162, 189, 190, and 205, which are specific for all partially methylated alditol acetates (PMAAs).
Construction of a Conditional Gene Knock-out of MSMEG_0317
A 500-bp synthetic DNA fragment (Integrated DNA Technologies) was designed containing 226 bp of sequence upstream and downstream of the gene plus 48 bp containing the 5′- and 3′-end of the gene. For cloning purposes, the synthetic DNA was designed with a SacI site at the 5′-end and a PstI site at the 3′-end. The fragment was cloned into pMSS (a suicide vector for M. smegmatis carrying sacB and streptomycin resistance genes) digested with SacI/PstI to create plasmid pMSS-ΔMSMEG_0317. This plasmid was then electroporated into M. smegmatis mc2155 selecting for streptomycin resistance. Colonies were screened for sucrose sensitivity, indicating integration of the entire plasmid via an SCO event at the MSMEG_0317 locus, and a confirmed SCO clone was isolated.
To construct a conditional gene knock-out from the MSMEG_0317 SCO strain, MSMEG_0317 was PCR-amplified from M. smegmatis genomic DNA using primers MSMEG_0317F (5′-GCGAATTCTTGAACCGCGCTGTGGCG) and MSMEG_0317R (5′-GCAAGCTTCAGATCGGTCGGTCCGG) containing EcoRI and HindIII sites, respectively (underlined). The amplified fragment was then cloned into the integrative vector pMV361 vector (43), and a sequence-verified clone was electroporated into electrocompetent MSMEG_0317 SCO cells, selecting on kanamycin plates. Streptomycin- sensitive, sucrose-resistant clones were derived from this strain as described above and analyzed by PCR to determine whether any were DCO strains using the primers MSMEG_0319 (5′-GGTTCGCCAGCCACACCG) and MSMEG_0317R.
Author Contributions
M. J. M., P. K. C., and R. L. C. designed the study and wrote the paper. T. J. C., R. B., Y. Y.-B., and A. K. R. contributed the experiments shown in Figs. 3–5 and supplemental Fig. S2. S. K. performed the experiments shown in Figs. 4–6 and supplemental Fig. S5 and prepared the figures. R. B. performed the experiments shown in supplemental Figs. S3 and S4. P. K. C. prepared Figs. 1–3, 7, and 8 and supplemental Fig. S1. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
This work was supported by National Health and Medical Research Council of Australia (NHMRC) Project Grant 1064466, Australian Research Council (ARC) Centre of Excellence in Structural and Functional Microbial Genomics Grant COE562063, and ARC Discovery Grant 140101244. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Figs. S1–S5.
- PIM
- phosphatidyl-myo-inositol mannoside
- LM
- lipomannan(s)
- LAM
- lipoarabinomannan(s)
- t-LM
- truncated LM
- PI
- phosphatidylinositol
- Man
- mannose
- PPM
- polyprenol phosphomannose
- TMCM
- trehalose monocorynomycolate
- Gl-A
- glucopyranosyluronic acid diacylglycerol
- Gl-X
- mannosyl-glucuronic acid diacylglycerol
- TDCM
- trehalose dicorynomycolate
- AcTMCM
- trehalose acetyl-monocorynomycolate
- HPTLC
- high-performance thin layer chromatography
- SCO
- single crossover
- DCO
- double crossover
- ESI
- electrospray ionization
- BHI
- brain heart infusion
- DIG
- digoxygenin
- PMAA
- partially methylated alditol acetate.
References
- 1. Barry C. E. 3rd, and Mdluli K. (1996) Drug sensitivity and environmental adaptation of mycobacterial cell wall components. Trends Microbiol. 4, 275–281 [DOI] [PubMed] [Google Scholar]
- 2. Jankute M., Cox J. A., Harrison J., and Besra G. S. (2015) Assembly of the mycobacterial cell wall. Annu. Rev. Microbiol. 69, 405–423 [DOI] [PubMed] [Google Scholar]
- 3. Brennan P. J., and Nikaido H. (1995) The envelope of mycobacteria. Annu. Rev. Biochem. 64, 29–63 [DOI] [PubMed] [Google Scholar]
- 4. Haites R. E., Morita Y. S., McConville M. J., and Billman-Jacobe H. (2005) Function of phosphatidylinositol in mycobacteria. J. Biol. Chem. 280, 10981–10987 [DOI] [PubMed] [Google Scholar]
- 5. Skovierová H., Larrouy-Maumus G., Zhang J., Kaur D., Barilone N., Korduláková J., Gilleron M., Guadagnini S., Belanová M., Prevost M. C., Gicquel B., Puzo G., Chatterjee D., Brennan P. J., Nigou J., and Jackson M. (2009) AftD, a novel essential arabinofuranosyltransferase from mycobacteria. Glycobiology 19, 1235–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barry C. E. 3rd, Lee R. E., Mdluli K., Sampson A. E., Schroeder B. G., Slayden R. A., and Yuan Y. (1998) Mycolic acids: structure, biosynthesis and physiological functions. Prog. Lipid Res. 37, 143–179 [DOI] [PubMed] [Google Scholar]
- 7. Mishra A. K., Driessen N. N., Appelmelk B. J., and Besra G. S. (2011) Lipoarabinomannan and related glycoconjugates: structure, biogenesis and role in Mycobacterium tuberculosis physiology and host-pathogen interaction. FEMS Microbiol. Rev. 35, 1126–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gande R., Gibson K. J., Brown A. K., Krumbach K., Dover L. G., Sahm H., Shioyama S., Oikawa T., Besra G. S., and Eggeling L. (2004) Acyl-CoA carboxylases (accD2 and accD3), together with a unique polyketide synthase (Cg-pks), are key to mycolic acid biosynthesis in Corynebacterianeae such as Corynebacterium glutamicum and Mycobacterium tuberculosis. J. Biol. Chem. 279, 44847–44857 [DOI] [PubMed] [Google Scholar]
- 9. Alderwick L. J., Radmacher E., Seidel M., Gande R., Hitchen P. G., Morris H. R., Dell A., Sahm H., Eggeling L., and Besra G. S. (2005) Deletion of Cg-emb in corynebacterianeae leads to a novel truncated cell wall arabinogalactan, whereas inactivation of Cg-ubiA results in an arabinan-deficient mutant with a cell wall galactan core. J. Biol. Chem. 280, 32362–32371 [DOI] [PubMed] [Google Scholar]
- 10. Alderwick L. J., Seidel M., Sahm H., Besra G. S., and Eggeling L. (2006) Identification of a novel arabinofuranosyltransferase (AftA) involved in cell wall arabinan biosynthesis in Mycobacterium tuberculosis. J. Biol. Chem. 281, 15653–15661 [DOI] [PubMed] [Google Scholar]
- 11. Lea-Smith D. J., Pyke J. S., Tull D., McConville M. J., Coppel R. L., and Crellin P. K. (2007) The reductase that catalyzes mycolic motif synthesis is required for efficient attachment of mycolic acids to arabinogalactan. J. Biol. Chem. 282, 11000–11008 [DOI] [PubMed] [Google Scholar]
- 12. Mishra A. K., Alderwick L. J., Rittmann D., Tatituri R. V., Nigou J., Gilleron M., Eggeling L., and Besra G. S. (2007) Identification of an α(1→6) mannopyranosyltransferase (MptA), involved in Corynebacterium glutamicum lipomanann biosynthesis, and identification of its orthologue in Mycobacterium tuberculosis. Mol. Microbiol. 65, 1503–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Seidel M., Alderwick L. J., Birch H. L., Sahm H., Eggeling L., and Besra G. S. (2007) Identification of a novel arabinofuranosyltransferase AftB involved in a terminal step of cell wall arabinan biosynthesis in Corynebacterianeae, such as Corynebacterium glutamicum and Mycobacterium tuberculosis. J. Biol. Chem. 282, 14729–14740 [DOI] [PubMed] [Google Scholar]
- 14. Seidel M., Alderwick L. J., Sahm H., Besra G. S., and Eggeling L. (2007) Topology and mutational analysis of the single Emb arabinofuranosyltransferase of Corynebacterium glutamicum as a model of Emb proteins of Mycobacterium tuberculosis. Glycobiology 17, 210–219 [DOI] [PubMed] [Google Scholar]
- 15. Birch H. L., Alderwick L. J., Bhatt A., Rittmann D., Krumbach K., Singh A., Bai Y., Lowary T. L., Eggeling L., and Besra G. S. (2008) Biosynthesis of mycobacterial arabinogalactan: identification of a novel α(1→3) arabinofuranosyltransferase. Mol. Microbiol. 69, 1191–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lea-Smith D. J., Martin K. L., Pyke J. S., Tull D., McConville M. J., Coppel R. L., and Crellin P. K. (2008) Analysis of a new mannosyltransferase required for the synthesis of phosphatidylinositol mannosides and lipoarbinomannan reveals two lipomannan pools in corynebacterineae. J. Biol. Chem. 283, 6773–6782 [DOI] [PubMed] [Google Scholar]
- 17. Mishra A. K., Alderwick L. J., Rittmann D., Wang C., Bhatt A., Jacobs W. R. Jr, Takayama K., Eggeling L., and Besra G. S. (2008) Identification of a novel alpha(1→6) mannopyranosyltransferase MptB from Corynebacterium glutamicum by deletion of a conserved gene, NCgl1505, affords a lipomannan- and lipoarabinomannan-deficient mutant. Mol. Microbiol. 68, 1595–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mishra A. K., Klein C., Gurcha S. S., Alderwick L. J., Babu P., Hitchen P. G., Morris H. R., Dell A., Besra G. S., and Eggeling L. (2008) Structural characterization and functional properties of a novel lipomannan variant isolated from a Corynebacterium glutamicum pimB′ mutant. Antonie Van Leeuwenhoek 94, 277–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chatterjee D., and Khoo K. H. (1998) Mycobacterial lipoarabinomannan: an extraordinary lipoheteroglycan with profound physiological effects. Glycobiology 8, 113–120 [DOI] [PubMed] [Google Scholar]
- 20. Strohmeier G. R., and Fenton M. J. (1999) Roles of lipoarabinomannan in the pathogenesis of tuberculosis. Microbes Infect. 1, 709–717 [DOI] [PubMed] [Google Scholar]
- 21. Vercellone A., Nigou J., and Puzo G. (1998) Relationships between the structure and the roles of lipoarabinomannans and related glycoconjugates in tuberculosis pathogenesis. Front. Biosci. 3, e149–163 [DOI] [PubMed] [Google Scholar]
- 22. Nigou J., Gilleron M., Rojas M., García L. F., Thurnher M., and Puzo G. (2002) Mycobacterial lipoarabinomannans: modulators of dendritic cell function and the apoptotic response. Microbes Infect. 4, 945–953 [DOI] [PubMed] [Google Scholar]
- 23. Maeda N., Nigou J., Herrmann J. L., Jackson M., Amara A., Lagrange P. H., Puzo G., Gicquel B., and Neyrolles O. (2003) The cell surface receptor DC-SIGN discriminates between Mycobacterium species through selective recognition of the mannose caps on lipoarabinomannan. J. Biol. Chem. 278, 5513–5516 [DOI] [PubMed] [Google Scholar]
- 24. Schlesinger L. S., Hull S. R., and Kaufman T. M. (1994) Binding of the terminal mannosyl units of lipoarabinomannan from a virulent strain of Mycobacterium tuberculosis to human macrophages. J. Immunol. 152, 4070–4079 [PubMed] [Google Scholar]
- 25. McConville M. J., Thomas-Oates J. E., Ferguson M. A., and Homans S. W. (1990) Structure of the lipophosphoglycan from Leishmania major. J. Biol. Chem. 265, 19611–19623 [PubMed] [Google Scholar]
- 26. Korduláková J., Gilleron M., Mikusova K., Puzo G., Brennan P. J., Gicquel B., and Jackson M. (2002) Definition of the first mannosylation step in phosphatidylinositol mannoside synthesis. PimA is essential for growth of mycobacteria. J. Biol. Chem. 277, 31335–31344 [DOI] [PubMed] [Google Scholar]
- 27. Korduláková J., Gilleron M., Puzo G., Brennan P. J., Gicquel B., Mikusová K., and Jackson M. (2003) Identification of the required acyltransferase step in the biosynthesis of the phosphatidylinositol mannosides of mycobacterium species. J. Biol. Chem. 278, 36285–36295 [DOI] [PubMed] [Google Scholar]
- 28. Morita Y. S., Sena C. B., Waller R. F., Kurokawa K., Sernee M. F., Nakatani F., Haites R. E., Billman-Jacobe H., McConville M. J., Maeda Y., and Kinoshita T. (2006) PimE is a polyprenol-phosphate-mannose-dependent mannosyltransferase that transfers the fifth mannose of phosphatidylinositol mannoside in mycobacteria. J. Biol. Chem. 281, 25143–25155 [DOI] [PubMed] [Google Scholar]
- 29. Rainczuk A. K., Yamaryo-Botte Y., Brammananth R., Stinear T. P., Seemann T., Coppel R. L., McConville M. J., and Crellin P. K. (2012) The lipoprotein LpqW is essential for the mannosylation of periplasmic glycolipids in Corynebacteria. J. Biol. Chem. 287, 42726–42738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kovacevic S., Anderson D., Morita Y. S., Patterson J., Haites R., McMillan B. N., Coppel R., McConville M. J., and Billman-Jacobe H. (2006) Identification of a novel protein with a role in lipoarabinomannan biosynthesis in mycobacteria. J. Biol. Chem. 281, 9011–9017 [DOI] [PubMed] [Google Scholar]
- 31. Mishra A. K., Krumbach K., Rittmann D., Appelmelk B., Pathak V., Pathak A. K., Nigou J., Geurtsen J., Eggeling L., and Besra G. S. (2011) Lipoarabinomannan biosynthesis in Corynebacterineae: the interplay of two α(1→2)-mannopyranosyltransferases MptC and MptD in mannan branching. Mol. Microbiol. 80, 1241–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stoop E. J., Mishra A. K., Driessen N. N., van Stempvoort G., Bouchier P., Verboom T., van Leeuwen L. M., Sparrius M., Raadsen S. A., van Zon M., van der Wel N. N., Besra G. S., Geurtsen J., Bitter W., Appelmelk B. J., and van der Sar A. M. (2013) Mannan core branching of lipo(arabino)mannan is required for mycobacterial virulence in the context of innate immunity. Cell. Microbiol. 15, 2093–2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Alderwick L. J., Lloyd G. S., Ghadbane H., May J. W., Bhatt A., Eggeling L., Fütterer K., and Besra G. S. (2011) The C-terminal domain of the arabinosyltransferase Mycobacterium tuberculosis EmbC is a lectin-like carbohydrate binding module. PLoS Pathog. 7, e1001299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shi L., Berg S., Lee A., Spencer J. S., Zhang J., Vissa V., McNeil M. R., Khoo K. H., and Chatterjee D. (2006) The carboxy terminus of EmbC from Mycobacterium smegmatis mediates chain length extension of the arabinan in lipoarabinomannan. J. Biol. Chem. 281, 19512–19526 [DOI] [PubMed] [Google Scholar]
- 35. Dinadayala P., Kaur D., Berg S., Amin A. G., Vissa V. D., Chatterjee D., Brennan P. J., and Crick D. C. (2006) Genetic basis for the synthesis of the immunomodulatory mannose caps of lipoarabinomannan in Mycobacterium tuberculosis. J. Biol. Chem. 281, 20027–20035 [DOI] [PubMed] [Google Scholar]
- 36. Kaur D., Obregón-Henao A., Pham H., Chatterjee D., Brennan P. J., and Jackson M. (2008) Lipoarabinomannan of Mycobacterium: mannose capping by a multifunctional terminal mannosyltransferase. Proc. Natl. Acad. Sci. U.S.A. 105, 17973–17977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mishra A. K., Batt S., Krumbach K., Eggeling L., and Besra G. S. (2009) Characterization of the Corynebacterium glutamicum ΔpimB′ ΔmgtA double deletion mutant and the role of Mycobacterium tuberculosis orthologues Rv2188c and Rv0557 in glycolipid biosynthesis. J. Bacteriol. 191, 4465–4472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tatituri R. V., Illarionov P. A., Dover L. G., Nigou J., Gilleron M., Hitchen P., Krumbach K., Morris H. R., Spencer N., Dell A., Eggeling L., and Besra G. S. (2007) Inactivation of Corynebacterium glutamicum NCgl0452 and the role of MgtA in the biosynthesis of a novel mannosylated glycolipid involved in lipomannan biosynthesis. J. Biol. Chem. 282, 4561–4572 [DOI] [PubMed] [Google Scholar]
- 39. Griffin J. E., Gawronski J. D., Dejesus M. A., Ioerger T. R., Akerley B. J., and Sassetti C. M. (2011) High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 7, e1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sassetti C. M., Boyd D. H., and Rubin E. J. (2003) Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48, 77–84 [DOI] [PubMed] [Google Scholar]
- 41. Yamaryo-Botte Y., Rainczuk A. K., Lea-Smith D. J., Brammananth R., van der Peet P. L., Meikle P., Ralton J. E., Rupasinghe T. W., Williams S. J., Coppel R. L., Crellin P. K., and McConville M. J. (2015) Acetylation of trehalose mycolates is required for efficient MmpL-mediated membrane transport in Corynebacterineae. ACS Chem. Biol. 10, 734–746 [DOI] [PubMed] [Google Scholar]
- 42. McKean S., Davies J., and Moore R. (2005) Identification of macrophage induced genes of Corynebacterium pseudotuberculosis by differential fluorescence induction. Microbes Infect. 7, 1352–1363 [DOI] [PubMed] [Google Scholar]
- 43. Stover C. K., de la Cruz V. F., Fuerst T. R., Burlein J. E., Benson L. A., Bennett L. T., Bansal G. P., Young J. F., Lee M. H., and Hatfull G. F. (1991) New use of BCG for recombinant vaccines. Nature 351, 456–460 [DOI] [PubMed] [Google Scholar]
- 44. Belardinelli J. M., Yazidi A., Yang L., Fabre L., Li W., Jacques B., Angala S. K., Rouiller I., Zgurskaya H. I., Sygusch J., and Jackson M. (2016) Structure-function profile of MmpL3, the essential mycolic acid transporter from Mycobacterium tuberculosis. ACS Infect. Dis. 2, 702–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rodríguez D. M., Ocampo M., Curtidor H., Vanegas M., Patarroyo M. E., and Patarroyo M. A. (2012) Mycobacterium tuberculosis surface protein Rv0227c contains high activity binding peptides which inhibit cell invasion. Peptides 38, 208–216 [DOI] [PubMed] [Google Scholar]
- 46. van der Rest M. E., Lange C., and Molenaar D. (1999) A heat shock following electroporation induces highly efficient transformation of Corynebacterium glutamicum with xenogeneic plasmid DNA. Appl. Microbiol. Biotechnol. 52, 541–545 [DOI] [PubMed] [Google Scholar]
- 47. Altschul S. F., Gish W., Miller W., Myers E. W., and Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 48. Omasits U., Ahrens C. H., Müller S., and Wollscheid B. (2014) Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 30, 884–886 [DOI] [PubMed] [Google Scholar]
- 49. Yanisch-Perron C., Vieira J., and Messing J. (1985) Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33, 103–119 [DOI] [PubMed] [Google Scholar]
- 50. Schäfer A., Tauch A., Jäger W., Kalinowski J., Thierbach G., and Pühler A. (1994) Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145, 69–73 [DOI] [PubMed] [Google Scholar]
- 51. McConville M. J., Homans S. W., Thomas-Oates J. E., Dell A., and Bacic A. (1990) Structures of the glycoinositolphospholipids from Leishmania major: a family of novel galactofuranose-containing glycolipids. J. Biol. Chem. 265, 7385–7394 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.