Abstract
The Jeotgalicoccus sp. peroxygenase cytochrome P450 OleTJE (CYP152L1) is a hydrogen peroxide-driven oxidase that catalyzes oxidative decarboxylation of fatty acids, producing terminal alkenes with applications as fine chemicals and biofuels. Understanding mechanisms that favor decarboxylation over fatty acid hydroxylation in OleTJE could enable protein engineering to improve catalysis or to introduce decarboxylation activity into P450s with different substrate preferences. In this manuscript, we have focused on OleTJE active site residues Phe79, His85, and Arg245 to interrogate their roles in substrate binding and catalytic activity. His85 is a potential proton donor to reactive iron-oxo species during substrate decarboxylation. The H85Q mutant substitutes a glutamine found in several peroxygenases that favor fatty acid hydroxylation. H85Q OleTJE still favors alkene production, suggesting alternative protonation mechanisms. However, the mutant undergoes only minor substrate binding-induced heme iron spin state shift toward high spin by comparison with WT OleTJE, indicating the key role of His85 in this process. Phe79 interacts with His85, and Phe79 mutants showed diminished affinity for shorter chain (C10–C16) fatty acids and weak substrate-induced high spin conversion. F79A OleTJE is least affected in substrate oxidation, whereas the F79W/Y mutants exhibit lower stability and cysteine thiolate protonation on reduction. Finally, Arg245 is crucial for binding the substrate carboxylate, and R245E/L mutations severely compromise activity and heme content, although alkene products are formed from some substrates, including stearic acid (C18:0). The results identify crucial roles for the active site amino acid trio in determining OleTJE catalytic efficiency in alkene production and in regulating protein stability, heme iron coordination, and spin state.
Keywords: crystal structure, cytochrome P450, decarboxylase, enzyme mechanism, fatty acid oxidation, CYP152L1, EPR spectroscopy, alkene, fatty acid hydroxylation, product analysis
Introduction
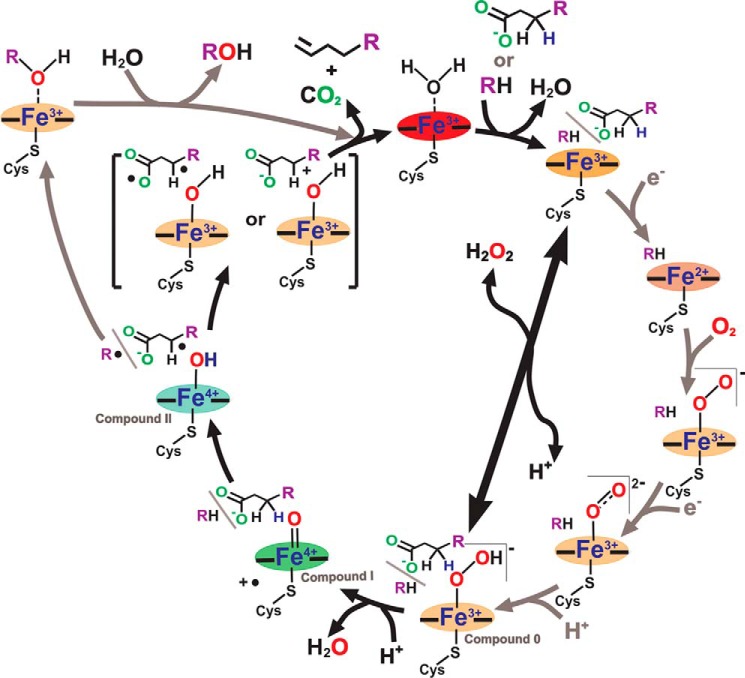
The cytochromes P450 (P450s or CYPs) are a superfamily of heme b-containing mono-oxygenase enzymes widespread in nature and spanning all three domains of life: archaea, bacteria, and eukaryota (1). Most P450s catalyze reductive scission of dioxygen bound to their heme iron, relying on the delivery of two electrons acquired from pyridine nucleotide cofactors (NADPH or NADH). In the canonical P450 catalytic cycle, two electrons are donated at discrete points in the cycle via a redox partner protein (2). The first electron is delivered to the substrate-bound, cysteine thiolate-coordinated ferric heme iron, reducing it to the ferrous form, allowing dioxygen to bind. The second electron reduces this ferric-superoxo complex to the ferric-peroxo state. As shown in Fig. 1, two successive protonations of iron-oxo species then occur. Protons are typically relayed from a conserved acid-alcohol amino acid pair (e.g. Asp251/Thr252 in the camphor hydroxylase P450cam, CYP101A1) to form first the ferric-hydroperoxo species (compound 0) and then the highly reactive ferryl-oxo, heme radical cation species compound I (with loss of a water molecule) (3, 4). The P450 compound I abstracts a hydrogen from the substrate to form the ferryl-hydroxo (compound II) form, prior to oxidizing the substrate to complete the catalytic process (5).
FIGURE 1.
The OleTJE catalytic cycle. OleTJE catalyzes the oxidative decarboxylation of fatty acids (major reaction) and can also hydroxylate fatty acids at the α- and β-positions (minor reaction). The canonical P450 catalytic cycle is shown in circular representation, initiating with the binding of fatty acid substrate (shown as RH, and also as a chemical structure) that converts the LS OleTJE P450 to a HS form. Shown in gray arrows from this point onwards are reduction (×2) and protonation (×1) steps that follow the canonical path and require one or more redox partners. The thick black arrow traversing the cycle illustrates the “peroxide shunt” pathway by which ferric, fatty acid-bound OleTJE can be converted directly to the transient ferric-hydroperoxo (compound 0) form using H2O2. At this point, the canonical and peroxide shunt pathways converge with a protonation step that results in dehydration of a transient intermediate to form the ferryl-oxo compound I. Compound I deprotonates the substrate, producing a substrate radical and the ferryl-hydroxo compound II. At this stage, the catalytic pathways for substrate hydroxylation and decarboxylation diverge. Hydroxylated fatty acids are formed by the “radical rebound” mechanism (5), as shown in the outer loop to the left of the main cycle (gray arrows). However, recent studies have indicated that that fatty acid decarboxylation to form terminal alkenes likely involves an unusually stable compound II and that reduction of compound II (or another oxidant) by the substrate radical could form a substrate diradical or carbocation species that decomposes with CO2 release to form the terminal alkene product (15, 39). The P450 heme (illustrated by the iron in the relevant oxidation state and equatorial/axial bonds) is shaded to approximate the color of the relevant heme intermediate species.
Although most P450s operate in this way, several are also known to catalyze substrate oxidation using the “peroxide shunt” mechanism, in which H2O2 or an organic hydroperoxide (e.g. meta-chloroperoxybenzoic acid) convert a substrate-bound resting state of the P450 directly to compound 0 (Fig. 1) (4, 6). As in the canonical cycle, a further protonation of compound 0 leads to formation of compound I and to oxidative catalysis. In many P450s, this reaction is inefficient, and oxidative damage to the protein and/or heme occurs alongside any productive turnover (7). However, certain P450s have evolved to use the peroxide shunt efficiently for substrate oxidation. The best characterized P450s are in the CYP152 family of peroxygenases. These enzymes may have evolved in ancient prokaryotes at a time when the environment was devoid of oxygen, but relatively rich in hydrogen peroxide and peroxygenated organic compounds. Although peroxygenase activity is likely to have evolved in archaea, it has been conserved in P450s from bacteria and eukaryotes, including humans (8).
Among the best characterized of the CYP152 family peroxygenases is the industrially relevant OleTJE (CYP152L1), first isolated from Jeotgalicoccus sp. ATCC 8456 in 2011 by Schirmer and co-workers (9). OleTJE acts primarily as a H2O2-dependent fatty acid decarboxylase to form terminal alkenes but also hydroxylates a proportion of these substrates to generate α- and β-hydroxylated fatty acids. Lipid substrates with chain lengths ∼C12-C20 were shown to be good substrates (9, 10). Later studies showed that the P450cam redox partner system (NADH-putidaredoxin reductase and putidaredoxin), along with a NADH recycling system) could also be used effectively with OleTJE to decarboxylate alkanoic acids as short as C4 (11). OleTJE also functions as a typical P450 mono-oxygenase when fused to the phthalate dioxygenase reductase-like domain of CYP116B2 (RhFRED) from Rhodococcus sp. NCIMB9784 (12) or by utilizing the flavodoxin/flavodoxin reductase system of Escherichia coli in vivo (12, 13). OleTJE is also active when used in conjunction with a light-driven, in situ H2O2-generating system (14).
Transient formation of P450 compound I in OleTJE was reported by Makris and co-workers (15) on the addition of H2O2 to OleTJE in complex with perdeuterated arachidic acid, and identified by the appearance of a characteristic 370-nm Soret maximum and an additional absorption band at 690 nm. These spectral data are similar to those reported by Rittle and Green (4) in studies with Sulfolobus acidocaldarius CYP119A1. The transient capture of compound I in the perdeuterated substrate-bound form of OleTJE likely results from a 2H-kinetic isotope effect on compound I decay. Compound I formation is considered to initiate fatty acid decarboxylation by abstracting a substrate hydrogen atom (15).
As an alkene-producing enzyme, OleTJE has attracted interest for industrial applications, including in the production of biofuels and petrochemicals (16). In vivo production of alkenes was reported in E. coli strains transformed with OleTJE (12). Yan et al. (17) also developed systems in which the tandem activities of a lipase and OleTJE were used to produce 1-alkenes from low cost triacylglycerol feedstocks.
Other prominent members of the CYP152 peroxygenase family include Bacillus subtilis P450 BSβ (CYP152A1, 41% sequence identity to OleTJE) and Sphingomonas paucimobilis P450 SPα (CYP152B1, 37% sequence identity to OleTJE) (10). Both enzymes are primarily fatty acid hydroxylases, with P450 SPα hydroxylating 100% at the Cα position, and P450 BSβ initially reported to hydroxylate at ∼43% in the Cα position and ∼57% in the Cβ position (18). However, studies by Rude et al. (9) showed that P450 BSβ produced 1-pentadecene at ∼15% of total product alongside α- and β-hydroxylated palmitic acid products. Crystal structure data show that the active sites of OleTJE and P450 BSβ are very similar, with the carboxylate group of fatty acid substrates being coordinated by a conserved arginine residue (Arg245 in OleTJE, Arg242 in P450 BSβ, and Arg241 in P450 SPα) (10, 18). An R242A mutant of P450 BSβ abolished fatty acid hydroxylase activity, whereas an R242K mutation substantially reduced substrate hydroxylation (19).
A key difference in the active sites of OleTJE and P450 BSβ is at position 85 (His85 in OleTJE and Gln85 in P450 BSβ) (10). In OleTJE, the His85 imidazole side chain points into the active site toward the heme iron. This orientation sandwiches the imidazole side chain between the heme and Phe79. It was postulated that the difference in the nature of the residue at position 85 may underlie the strong decarboxylase activity of OleTJE. His85 could act as a proton donor to the compound I intermediate, together with its reduction to compound II using an electron from the fatty acid substrate carboxylate group. Homolytic scission of the C-Cα bond of the fatty acid, concomitant with abstraction of a hydrogen from Cα to form compound II, could then produce the terminal alkene and CO2 products (10). Rude et al. investigated the role of the residue at position 85 in P450 BSβ by creating the Q85H mutant. During in vitro turnover experiments with hexadecanoic acid, the P450 BSβ Q85H mutation resulted in decreased α-hydroxy-hexadecanoic acid formation and in increased 1-pentadecene and β-hydroxy hexadecanoic acid formation. Despite this, the ratio of decarboxylation to hydroxylation only increased from 0.19 in WT P450 BSβ to 0.30 in the P450 BSβ Q85H mutant, compared with a ratio of 3.32 in OleTJE (9).
Through its interactions with His85 in OleTJE, Phe79 is also likely to be important for catalytic activity in OleTJE. A phenylalanine (Phe79) is retained at this position in P450 BSβ. However, the exclusive fatty acid α-hydroxylase P450 SPα has a leucine at the corresponding position (Leu79). In P450 BSβ, the F79L mutation resulted in increased α-hydroxylation, with levels of β-hydroxylation falling from 58 to 17% of the overall product (20).
In this study, we explore further the structure and mechanism of OleTJE by mutating the key active site residues His85, Phe79, and Arg245 and by analyzing their effects on the catalytic properties of this biotechnologically important alkene-producing enzyme. The H85Q mutant was characterized to establish whether His85 is crucial to OleTJE decarboxylase activity and also because its replacement by Glu85 in P450 BSβ results mainly in fatty acid hydroxylation (Fig. 2). The importance of Phe79 in its interactions with His85 and in regulating active site structure and catalytic activity were explored by characterization of F79A, F79W, and F79Y variants. Finally, the fatty acid carboxylate coordinating Arg245 was mutated to neutral (R245L) and charge reversal (R245E) variants to assess influence on activity and regioselectivity of substrate oxidation. Mutants were characterized by a combination of kinetic, spectroscopic, structural, and analytical methods to provide new insights into OleTJE structure/function.
FIGURE 2.
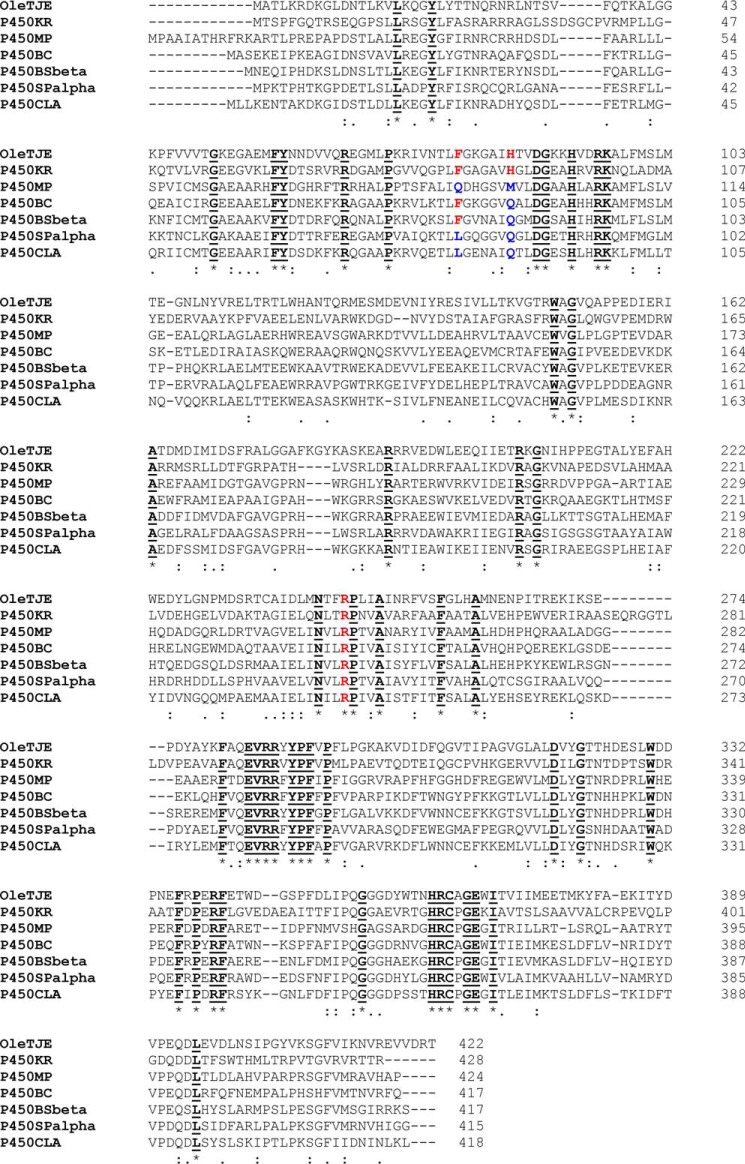
Alignment of OleTJE with other bacterial peroxygenase P450s. OleTJE (CYP152L1) is aligned with the peroxygenases P450 KR from K. rhizophila, CYP-MP (P450MP) from M. populi (30), a peroxygenase from Bacillus clausii (P450BC) (9), B. subtilis P450 BSβ (CYP152A1) (27), S. paucimobilis P450 SPα (CYP152B1) (28), and C. acetobutylicum P450 CLA (CYP152A2) (33). The OleTJE active site residues Phe79, His85 and Arg245 are in bold, red font. Non-conserved residues at these positions are shown in bold, blue font. Arg245 and the adjacent Pro246 are conserved in all the peroxygenases. Other completely conserved residues are indicated in bold, underlined black font.
Results
Expression and Purification of OleTJE and OleTJE Mutants
The expression, purification and structural characterization of the WT OleT P450 were reported by Belcher et al. (10). The structural data for the substrate-bound and substrate-free forms of OleTJE were used to design mutations to explore the fatty acid carboxylate binding site of the P450. The codon-optimized OleTJE (CYP152L1) gene was cloned into pET15b, and site-directed mutagenesis was done to create the H85Q, F79A, F79W, F79Y, R245E, and R245L mutants. All mutants were found to express reasonably well in the E. coli C41 (DE3) strain. Typical OleTJE mutant recovery was ∼15 mg/liter for H85Q and F79W, 8 mg/liter for F79Y and R245L, 2 mg/liter for F79A, and 1.5 mg/liter for the R245E OleTJE mutant, compared with ∼20 mg/liter for WT OleTJE. The heme precursor δ-aminolevulinic acid (500 μm) was added at the point of induction of OleTJE mutant gene expression with isopropyl 1-thio-β-d-galactopyranoside to aid heme incorporation. However, heme insertion levels were diminished in the R245E and R245L mutants (∼25–35% heme incorporation). The simultaneous addition of hemin (4 μg/ml) at the point of induction increased heme incorporation by ∼5–10% in these mutants.
All OleTJE mutants were purified in the same way, using nickel-iminodiacetic acid (Ni-IDA)2 chromatography. Samples for enzymatic/spectroscopic assays were frozen after ensuring a high level of purity by SDS-PAGE. Samples destined for crystallography were treated with TEV protease to cleave the N-terminal His tag and then passed through a Ni-IDA column to separate the untagged form from column-retained tagged OleTJE. Gel filtration chromatography was used to further purify untagged OleTJE proteins, and samples were then used directly in crystallographic trials. Fig. 3 shows an SDS-PAGE gel containing samples of purified WT and mutant OleTJE proteins. The F79W, F79Y, R245E, and R245L OleTJE mutants proved less stable than the WT and the H85Q and F79A mutants in that they were prone to aggregation and precipitation on refrigeration or during crystallization trials. However, the H85Q and F79A OleTJE P450s were successfully crystallized for structural analysis (see “Structural Properties of OleTJE Mutant Enzymes” below).
FIGURE 3.
Purification of WT and active site mutants of OleTJE. The gel image shows the purified forms (from left to right) of WT OleTJE and its active site mutants H85Q, F79A, F79W, F79Y, R245L and R245E. The outside lanes show protein markers with their associated molecular masses (kDa) indicated (NEB Broad Range Protein Ladder).
UV-visible Absorbance Properties of OleTJE Mutants
When purified from E. coli, the WT, ferric OleTJE exhibits a mixed low spin (LS)/high spin (HS) heme iron state that can be converted essentially completely to a LS form by the addition of H2O2. This likely indicates that fatty acids from E. coli are bound and retained by the enzyme during the purification process. The WT OleTJE was extensively dialyzed to remove bound fatty acids. However, this mixed spin character was absent in all OleTJE mutants generated, suggesting that the mutations may compromise affinity for mid-chain to long chain fatty acids through structural perturbation of the P450 active site.
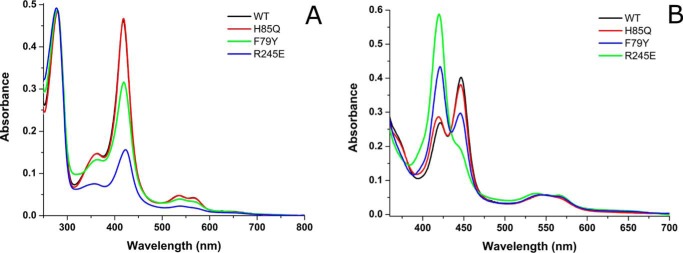
In WT OleTJE, the ferric heme Soret peak is at 418.5 nm, with the α and β bands at ∼567 and 535 nm, respectively. The H85Q mutant has a similar spectrum with a Soret peak at 418.5 nm, whereas the F79A/W/Y mutants have Soret maxima at 419/419/419.5 nm. In the R245E mutant, there is a substantial spectral change, with the Soret peak at ∼423 nm and a major Q-band feature at ∼538 nm. This phenomenon is not observed in the R245L OleTJE (Soret maximum at 417.5 nm, with α/β bands at ∼566/535 nm), indicating that the R245E mutation alters the heme environment in the distal pocket (Fig. 4A). A likely model is that Glu245 coordinates the R245E mutant heme iron, as was demonstrated previously in the A264E mutant of Bacillus megaterium P450 BM3 (CYP102A1) (21).
FIGURE 4.
UV-visible spectroscopy of OleTJE and key active site mutants. A shows typical UV-visible absorption spectra for the ferric, substrate-free forms of WT (black), H85Q (red), F79Y (green), and R245E (blue) OleTJE. The spectra are nearly identical at the heme Soret and protein (∼280 nm) peaks for the WT and H85Q forms (both at 5 μm) and reflect essentially complete heme incorporation into these proteins. However, the heme/protein (Reinheitszahl, Rz) ratio is much lower in the F79Y and R245E OleTJE mutants, indicating lower heme incorporation in these variants. B shows the FeII-CO complexes of the same proteins (corrected to 5 μm by heme content in all cases), showing that the WT and H85Q mutants form predominantly the cysteine thiolate-coordinated P450 form, with a proportion of the thiol-coordinated P420 state. There is a considerably greater proportion of the P420 species in the F79Y mutant, whereas the R245E mutant forms almost completely the P420 form. The P450 species have a Soret maximum at 448 ± 1 nm and the P420 species a Soret maximum at 422 ± 1 nm in all cases.
Spectral differences are observed between the dithionite-reduced WT and mutant forms of OleTJE. For WT OleTJE, reduction is associated with a decreased Soret peak intensity and a shift in absorption maximum to 415 nm, with merging of the α and β bands into a broad feature peaking at ∼540 nm (10). Similar spectral shifts occur on the reduction of OleTJE mutants H85Q and F79A (Soret shifts to 416.5 and 418 nm). However, for F79W and F79Y OleTJE, the Soret band is red-shifted to ∼422 nm with peak intensity less diminished than in the WT and the H85Q/F79A variants. The red shift suggests that cysteine thiolate protonation occurs in the F79Y/W variants (22). In reduced R245E OleTJE, the Soret peak shifts to 417.5 nm, with a Q-band peak at ∼547.5 nm, whereas in reduced R245L the Soret is at 420 nm with α/β bands at 559/536 nm.
Further comparative studies of the heme environments in WT and mutant OleTJE proteins were done using the FeII-CO complexes of these P450s. The WT OleTJE formed predominantly the P450 state with a Soret band at 449 nm and a smaller P420 feature at ∼423 nm. These bands originate from OleTJE FeII-CO complexes in which the proximal ligand is either a cysteine thiolate (P450) or a cysteine thiol (P420) (23, 24). The P450:P420 ratio differs considerably between WT OleTJE and the various mutants. Under the conditions used, the highest P450:P420 ratio occurs for the WT OleTJE with a P450 component of ∼75%. H85Q has a similar proportion of P450 (∼70%) but P450 content decreases in the F79Y mutant (∼45%, similar to the F79A/W mutants), and the R245E mutant forms almost completely the P420 form (∼10% P450) (Fig. 4B). These data show that active site mutations influence the OleTJE variant heme environments, including perturbation of the proximal ligand protonation state in the ferrous and/or FeII-CO forms. Although the heme iron of R245E OleTJE is likely coordinated by Glu245 carboxylate in the ferric state, CO can clearly displace this distal ligand in the ferrous state of the enzyme. The ferrous R245L/E mutant Soret spectral peaks are similarly positioned at 416.5/417 nm, suggesting displacement of a Glu245 distal ligand in the ferrous state for the R245E mutant. The R245L mutant also forms a much higher proportion of the P450 FeII-CO complex at ∼35% (not shown).
Interactions of WT and Mutant OleTJE Proteins with Fatty Acid Substrates
To examine the influence of lipid substrates on OleTJE ferric heme iron spin state equilibrium and to determine Kd values for substrate binding, fatty acids of different chain lengths were titrated with WT and mutant OleTJE P450s. For WT OleTJE, binding of arachidic acid (C20:0) gives an extensive shift toward HS ferric heme iron, with decreased Soret band intensity at ∼418 nm and increased HS band intensity at 389 nm. Development of HS character is further confirmed by the appearance of a thiolate to HS ferric heme iron charge transfer band at ∼650 nm. For WT OleTJE, the amount of HS heme formed decreases as the fatty acid chain length decreases. The Kd values remain low for saturated fatty acids tested in the chain length range from C20 to C14, but affinity decreases considerably for C12 and C10 substrates; e.g. Kd = 1.73 ± 0.08 μm for palmitic acid (C16:0) compared with 47.1 ± 2.1 μm for capric acid (C10:0) (Table 1).
TABLE 1.
Binding affinity of saturated fatty acids for WT and mutant forms of OleTJE
The dissociation constants (Kd values) for the binding of the saturated fatty acids decanoic acid (capric acid, C10:0), dodecanoic acid (lauric acid, C12:0), tetradecanoic acid (myristic acid, C14:0), hexadecanoic acid (palmitic acid, C16:0), octadecanoic acid (stearic acid, C18:0), and arachidic acid (eicosanoic acid, C20:0) were determined by UV-visible titration with WT OleTJE and with the H85Q, R245E, and F79A/W/Y mutants. NB indicates that no significant binding (indicated by lack of any obvious HS heme iron development) was detected for the indicated mutants. A limit of <2% HS heme iron accumulation is given in these cases. This was also the case for the R245L OleTJE mutant, where no significant optical binding could be detected for any fatty acid tested.
| OleTJE variant | Fatty acid Kd (HS) |
|||||
|---|---|---|---|---|---|---|
| C20:0 | C18:0 | C16:0 | C14:0 | C12:0 | C10:0 | |
| μm (%) | ||||||
| WT | 4.43 ± 0.19 (95) | 4.09 ± 0.16 (70) | 1.73 ± 0.08 (25) | 1.45 ± 0.05 (45) | 11.7 ± 0.43 (40) | 47.1 ± 2.1 (35) |
| H85Q | 5.27 ± 0.05 (25) | 5.92 ± 0.08 (<10) | 5.36 ± 0.18 (<10) | 10.5 ± 0.1 (<10) | 37.3 ± 2.5 (<10) | NB (<2%) |
| F79A | 3.4 ± 0.3 (<10) | 8.7 ± 0.5 (<10) | 22.6 ± 1.9 (<10) | 39.1 ± 2.5 (<10) | 55.0 ± 4.8 (<10) | 465 ± 11 (<10) |
| F79W | 4.41 ± 0.27 (20) | 4.52 ± 0.20 (15) | 12.7 ± 0.7 (<10) | 24.1 ± 0.9 (15) | 106 ± 9 (<10) | NB (<2%) |
| F79Y | 4.25 ± 0.21 (<10) | 8.91 ± 0.33 (<10) | 18.2 ± 0.7 (<10) | 30.7 ± 1.0 (20) | 196 ± 5 (25) | NB (<2%) |
| R245E | NB (<2%) | NB (<2%) | NB (<2%) | NB (<2%) | NB (<2%) | 1260 ± 160 (40) |
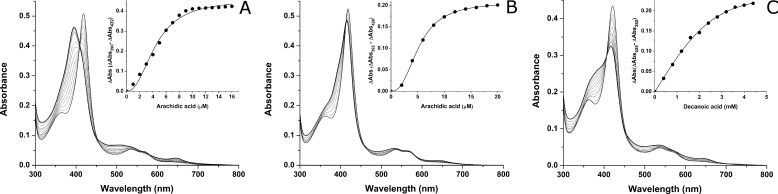
Binding of arachidic acid also induces HS heme formation in H85Q and F79A OleTJE mutants but with a much less extensive HS shift than seen in WT OleTJE. At apparent saturation with arachidic acid, the Soret band shifts to ∼415 nm for H85Q and to ∼414 nm for F79A OleTJE. Arachidic acid spectral binding data for WT and the H85Q OleTJE mutant are compared in (Fig. 5). Despite incomplete HS shifts in these mutants, substrate turnover data confirm that both the H85Q and F79A mutant P450s can oxidize fatty acids in the C10-C20 range (see “Products Formed by Novel OleTJE Mutants” below). Thus, productive substrate binding occurs for these fatty acids in both mutants. These mutations may influence the binding mode of the fatty acid and its carboxylate group, which in turn affects the efficiency with which the substrate can displace the distal water ligand to convert the P450s to a HS state. The reaction of H2O2 with substrate-bound OleTJE generates reactive heme species (ferryl-oxo and ferryl-hydroxo compounds I and II; Fig. 1) that oxidize the substrate, and the retention of catalytic activity in the H85Q/F79A mutants indicates that fatty acid binding modes remain close to this reactive iron-oxo species such that hydroxylation (at substrate α- and β-carbons) and decarboxylation remain feasible.
FIGURE 5.
Fatty acid binding titrations with WT and mutant OleTJE proteins. A shows a UV-visible spectral binding titration for WT OleTJE (5.5 μm) with arachidic acid (C20:0). There is extensive conversion of the LS heme iron (at 418.5 nm) toward HS (at 395 nm). HS heme iron development is also evident from the development of a cysteine thiolate-to-HS ferric heme iron charge transfer species at ∼650 nm. The inset shows the fitting of absorbance change versus [arachidic acid] using the Hill function, producing an apparent Kd of 4.43 ± 0.19 μm. B, shows a titration of the OleTJE H85Q mutant (5.6 μm) with arachidic acid, in this case producing a much smaller spectral conversion toward HS. The inset shows the fit of induced absorption change (418.5–415 nm) versus [arachidic acid], fitted as above to give a Kd of 5.27 ± 0.05 μm. C shows a titration of the OleTJE R245E mutant (4.7 μm) with capric acid (C10:0), with absorption change (∼422.5–384 nm) versus [capric acid] data fitted using a hyperbolic function to give a Kd of 1260 ± 160 μm.
It should be noted that for P450 BSβ and P450 SPα (where residue 85 is a glutamine), the HS shift upon substrate binding is very small (BSβ) or not observed at all (SPα) (25). The H85Q OleTJE mutant thus mimics the spectral properties of these other bacterial peroxygenases on binding fatty acid substrates. For both the H85Q and F79A OleTJE mutants, there is a pattern of increasing substrate Kd values as the fatty acid chain length decreases, consistent with the properties of WT OleTJE and likely reflecting the fewer hydrophobic interactions made by shorter chain substrates in the active site channel (Table 1).
The F79W and F79Y OleTJE proteins show modest spin state shifts with most fatty acids tested, suggesting again that the binding modes of these substrates are perturbed. However, in the F79Y OleTJE, there is a notable increase in HS content on saturation with lauric acid (C12:0, ∼25% high spin) and myristic acid (C14:0, ∼20% high spin) compared with longer chain fatty acids (<10% for arachidic acid, stearic acid (C18:0) and palmitic acid). This suggests that these shorter chain fatty acids can adopt binding modes in F79Y OleTJE that are compatible with more efficient displacement of the distal water molecule than can occur with longer chain substrates, probably in part because of their greater solubility. With the exception of the R245E OleTJE variant, the Kd values for the longest substrates (C20:0 and C18:0) are similar for the WT and mutant OleTJE enzymes (Table 1). As fatty acid chain length decreases from C16:0, there are wider variations in the substrate Kd values, with all mutants apart from R245E (see below) showing weaker binding of fatty acids from C16:0 to C10:0 by comparison with the WT OleTJE.
In the case of the R245L mutant, addition of the solvents ethanol, methanol, and DMSO resulted in the development of up to ∼10% HS heme iron. This phenomenon was not observed with the R245E mutant, likely because of the Glu245 carboxylate occupying the distal ligand position on the heme iron. This issue prevented any accurate assessment of the role of fatty acids in perturbing the R245L heme iron spin state equilibrium, because spectral changes induced by fatty acids dissolved in these solvents were of similar magnitude to those induced using solvents alone. However, addition of 1 mm lauric acid prepared in aqueous buffer A produced no significant change in the R245L UV-visible spectrum. Similarly, addition of a saturating amount of arachidic acid (in ethanol) did not induce any further R245L HS heme iron development over that caused by solvent alone. Thus, fatty acid substrate binding modes in this mutant may be particularly inefficient in displacement of the axial water ligand in ferric R245L OleTJE.
The R245E OleTJE mutant exhibits unusual behavior in that there is no apparent shift in Soret maximum from 423 nm or development of HS heme iron observed in optical titrations with any of the fatty acids from C20:0 to C12:0. However, titration with capric acid (C10:0), resulted in ∼40% HS heme iron accumulation at apparent saturation (Fig. 5C). These observations suggest that the longer chain fatty acids do not bind R245E OleTJE in a mode that enables HS development/displacement of a Glu245 axial ligand. However, the substantial HS heme accumulation in R245E OleTJE on binding capric acid suggests that this short chain substrate can displace the axial ligand, albeit with a very high Kd (1260 ± 160 μm). Coordination of heme iron by glutamate oxygen was also observed in the P450 BM3 A264E mutant, with arachidonic acid substrate binding causing nearly complete distal coordination by the Glu264 carboxylate (21, 26). However, the situation may be reversed for the OleTJE R245E mutant, with capric acid partially displacing a glutamate ligand at high concentrations.
Stopped Flow Kinetic Analysis of Fatty Acid Oxidation by WT and Mutant OleTJE P450s
Stopped flow absorption spectroscopy was used to probe the effects of mutations to His85 (H85Q) and Phe79 (F79A/W/Y) on the kinetics of OleTJE single turnover substrate oxidation reactions. WT, H85Q, and F79A/W variants were incubated with arachidic acid, whereas F79Y OleTJE was incubated with nearly saturating lauric acid to achieve the maximal heme iron spin state conversion for this mutant. The H2O2-dependent oxidation of substrates was observed by the rapid HS to LS conversion of the OleTJE heme iron in each case. Reaction progress was monitored at wavelengths close to the ferric, LS Soret maximum for WT OleTJE and the four mutants. Rate constants for the single turnover reactions (kobs) were determined using H2O2 concentrations up to 200 μm. Under the conditions used, plots of kobs versus [H2O2] were hyperbolic for WT OleTJE and for the H85Q and F79Y variants. The data were fitted using a hyperbolic function to derive the limiting rate constant (klim) and the apparent Kd for H2O2. The WT OleTJE had values of klim = 115 ± 8 s−1, Kd = 107 ± 16 μm, compared with 101 ± 16 s−1, 749 ± 141 μm for F79Y, and 286 ± 18 s−1, 327 ± 29 μm for H85Q. The dependence of kobs versus [H2O2] was linear for the F79A and F79W mutants, with second order rate constants of 0.188 ± 0.004 μm−1 s−1 and 0.107 ± 0.005 μm−1 s−1, respectively. The comparable second order rate constants (klim/Kd) for the other proteins are 1.075 ± 0.236 μm−1 s−1 (WT), 0.875 ± 0.133 μm−1 s−1 (H85Q), and 0.838 ± 0.507 μm−1 s−1 (F79Y).
Stopped flow kinetic data for the reactions of WT and mutant forms of fatty acid-bound OleTJE with H2O2 indicate that the WT, H85Q, and F79Y enzyme single turnovers are more efficient than are those of the other two Phe79 mutants. Although this is consistent with the superior catalytic properties of the WT and H85Q mutants in product formation (Table 3), the F79A OleTJE is a more productive enzyme for C10–C16 substrates than are F79Y/W, with more similar levels of product formation for the F79A/Y/W mutants with C18 and C20 substrates. However, in view of only minor development of HS heme iron in arachidic acid-bound F79Y OleTJE, lauric acid was used instead with the F79Y variant to enhance HS formation and to improve experimental data quality. In this case it appears that single turnover efficiency (i.e. the apparent second order rate constant for F79Y OleTJE-dependent substrate oxidation by H2O2) is at a level similar to that of the WT and H85Q OleTJE enzymes with arachidic acid. Despite this finding, percentage product formation for lauric acid with the F79Y mutant is much lower than observed for the WT/H85Q/F79A OleTJE enzymes with the same substrate and similar to that for the F79A OleTJE with arachidic acid. These data suggest that other factors (e.g. the rate constants for substrate association/product dissociation and/or efficiency of substrate oxidation with different chain substrates) have a strong influence on overall product formation.
TABLE 3.
Conversion of fatty acid substrates into alkene and hydroxylated fatty acid products by WT and mutant OleTJE enzymes
The WT, H85Q, R245E/L, and F79A/W/Y OleTJE enzymes were used to catalyze H2O2-dependent decarboxylation/hydroxylation of saturated fatty acid substrates (C10:0-C20:0) as described under “Experimental Procedures.” The percentage conversions of each fatty acid substrate into their respective (i) n-1 terminal alkene, (ii) 2-OH fatty acid, and (iii) 3-OH fatty acid products are shown for each OleTJE variant. The percentage of the various products formed in each reaction is given to the nearest 1% where these products form 10% or more of the total and to the nearest 0.1% where products form less than 10% of the overall yield. The total product formation for each OleTJE variant/substrate combination is presented in the same way. The data presented are average values from two sets of experiments differing by less than 10% in all cases.
| Substrate | Products | OleTJE variant and % products formed |
||||||
|---|---|---|---|---|---|---|---|---|
| WT | H85Q | F79A | F79W | F79Y | R254L | R245E | ||
| C10:0 | 1-Nonene | 51 | 65 | 37 | 1.5 | 0.3 | 0.1 | 0.2 |
| 2-OH capric acid | 5.2 | 8 | 1.4 | 2.1 | 0 | 9.3 | 0 | |
| 3-OH capric acid | 31 | 25 | 15 | 2.1 | 0 | 0 | 0 | |
| Total conversion | 87 | 98 | 53 | 5.7 | 0.3 | 9.4 | 0.2 | |
| C12:0 | 1-Undecene | 80 | 79 | 64 | 1.3 | 1.3 | 0 | 0 |
| 2-OH lauric acid | 5.2 | 7.8 | 7.4 | 3.6 | 3.4 | 2.7 | 2.6 | |
| 3-OH lauric acid | 9.0 | 9.2 | 8.5 | 1.6 | 1.4 | 1.1 | 1.1 | |
| Total conversion | 94 | 96 | 80 | 6.5 | 6.1 | 3.8 | 3.7 | |
| C14:0 | 1-Tridecene | 72 | 81 | 22 | 0 | 0 | 0 | 0 |
| 2-OH myristic acid | 3.3 | 3.1 | 1.4 | 1.0 | 1.2 | 0.1 | 1.1 | |
| 3-OH myristic acid | 25 | 16 | 19 | 2.0 | 3.1 | 0 | 0 | |
| Total conversion | 100 | 100 | 42 | 3.0 | 4.3 | 0.1 | 1.1 | |
| C16:0 | 1-Pentadecene | 27 | 22 | 9.3 | 1.2 | 0.8 | 0.5 | 0.6 |
| 2-OH palmitic acid | 3.6 | 3.2 | 2 | 2 | 1.6 | 1.2 | 1.2 | |
| 3-OH palmitic acid | 12 | 13 | 5.1 | 2.3 | 1.7 | 1.5 | 1.5 | |
| Total conversion | 43 | 38 | 16 | 5.5 | 4.1 | 3.2 | 3.3 | |
| C18:0 | 1-Heptadecene | 40 | 36 | 12 | 11 | 23 | 7.1 | 16 |
| 2-OH stearic acid | 7 | 1.4 | 2.3 | 0 | 2.7 | 0 | 0 | |
| 3-OH stearic acid | 0.4 | 0.1 | 5.9 | 0.9 | 0.7 | 0 | 0 | |
| Total conversion | 47 | 38 | 20 | 12 | 27 | 7.1 | 16 | |
| C20:0 | 1-Nonadecene | 14 | 11 | 5 | 3.4 | 2.5 | 0 | 0.1 |
| 2-OH arachidic acid | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 3-OH arachidic acid | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Total conversion | 14 | 11 | 5 | 3.4 | 2.5 | 0 | 0.1 | |
Characterization of OleTJE WT and Mutant P450 Heme Coordination by EPR
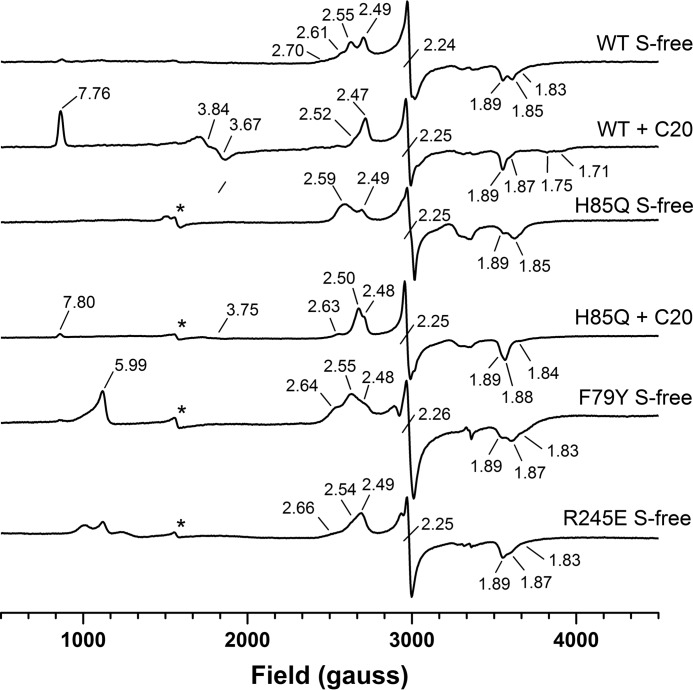
Continuous wave X-band EPR spectra were recorded for WT and all mutant forms of OleTJE in their substrate-free and arachidic acid-bound states to characterize the heme coordination state and the influence of mutations to the key residues in the heme distal pocket. As reported previously, the substrate-free form of WT OleTJE has a complex EPR spectrum with at least four LS (S = 1/2) ferric species with rhombic anisotropy; the most prominent of which have g values of gz = 2.49, gy = 2.24, and gx = 1.89 (2.49/2.24/1/89) and 2.55/2.24/1.85 (Fig. 6) (10). The g values for these species and those for all other mutants and their substrate-bound forms are presented in Table 2. These LS species are consistent with ferric heme iron coordinated by cysteine thiolate and a distal water ligand in a large water-filled active site, and with there being a variety of coordination geometries and hydrogen bonding interactions/networks with the distal water ligand. There is no major contribution from a HS (S = 5/2) species in the WT substrate-free form, but a substantial HS component appears on the addition of arachidic acid to WT OleTJE. This is split into two major species with g values of 7.76/3.84/1.71 and 7.76/3.67/1.75. As discussed previously, this heterogeneity likely results from, for example, changes in conformation of the heme and thiolate ligand geometry, rather than from any axial ligand switch in the HS, pentacoordinate state (10). The binding of arachidic acid also results in a dominant LS form with g values of 2.47/2.25/1.89, suggesting a much more ordered active site organization in the substrate-bound state.
FIGURE 6.
EPR spectroscopic properties of WT and mutant forms of OleTJE. X-band EPR spectra are shown for WT and H85Q OleTJE mutants in both ligand-free and arachidic acid substrate-bound (C20:0) forms and for the F79Y and R245E OleTJE mutants in their ligand-free forms. The WT and mutant forms of OleTJE have complex LS EPR spectra with several distinct species (see Table 2 for sets of g values). HS heme iron accumulation is seen following addition of arachidic acid to WT OleTJE (g = 7.76/3.84/1.71 and 7.76/3.67/1.75), but only a very small HS signal is seen for the arachidic acid-bound H85Q mutant (g = 7.80/3.75). The asterisks show signals for a proportion of a HS penta-coordinated, thiol-ligated, ferric heme species in the H85Q, F79Y, and R245E mutants.
TABLE 2.
EPR g values for substrate-free and fatty acid-bound forms of WT and mutant OleTJE proteins
X-band EPR data were collected at 10 K for the substrate-free and arachidic acid-bound forms of OleTJE. The g values for the LS species and for HS species (where evident) are presented. In the case of fatty acid-bound forms giving weak HS signals, only those component g values that can be accurately identified are shown. ND indicates not determined in those cases where no significant HS ferric heme iron signal could be identified or where signals were of insufficient intensity to be accurately assigned. For each of the OleTJE LS forms, two to four different rhombic species are identified in each case. The most prominent of these species is iidentified as the “Major species” in each case.
| OleTJE variant | Rhombic HS signals | Rhombic LS signals (gz/gy/gx) |
|
|---|---|---|---|
| Major species | Minor species | ||
| WT | ND | 2.49/2.24/1.89 | 2.70/2.24/1.83; 2.61/2.24/1.85; 2.55/2.24/1.85 |
| H85Q | ND | 2.59/2.25/1.85 | 2.49/2.25/1.89 |
| F79A | ND | 2.48/2.25/1.89 | 2.65/2.25/1.84; 2.58/2.25/1.86 |
| F79W | ND | 2.54/2.25/1.86 | 2.65/2.25/1.84; 2.47/2.25/1.89 |
| F79Y | ND | 2.55/2.26/1.87 | 2.64/2.26/1.83; 2.48/2.26/1.89 |
| R245L | ND | 2.54/2.25/1.87 | 2.65/2.25/1.85; 2.46/2.25/1.90 |
| R245E | ND | 2.49/2.25/1.89 | 2.66/2.25/1.83; 2.54/2.25/1.87 |
| WT+C20:0 | 7.76/3.84/1.71; 7.76/3.67/1.75 | 2.47/2.25/1.89 | 2.52/2.25/1.87 |
| H85Q+C20:0 | 7.80/3.75 | 2.50/2.25/1.88 | 2.63/2.25/1.84; 2.48/2.25/1.89 |
| F79A+C20:0 | ND | 2.49/2.26/1.89 | 2.60/2.26/1.84; 2.54/2.26/1.86 |
| F79W+C20:0 | ND | 2.54/2.25/1.86 | 2.67/2.25/1.83; 2.46/2.25/1.89 |
| F79Y+C20:0 | 7.79 | 2.56/2.26/1.86 | 2.65/2.26/1.83; 2.49/2.26/1.89 |
| R245L+C20:0 | 7.70, 3.74 | 2.53/2.25/1.87 | 2.65/2.25/1.85; 2.46/2.25/1.90 |
| R245E+C20:0 | ND | 2.53/2.25/1.87 | 2.67/2.25/1.85; 2.47/2.25/1.90 |
For the H85Q OleTJE mutant, the substrate-free LS spectrum is less complex than that for WT OleTJE. There are two major LS species at 2.59/2.25/1.85 and 2.49/2/25/1.89. The signal for the former is broad and may encompass two LS species. The addition of arachidic acid produces a much smaller HS signal than in WT OleTJE (with features at gz = 7.8 and gy = 3.75) and alters the LS spectrum, with two major species at 2.50/2.25/1.88 and 2.48/2.25/1.89. The LS heme iron signals for the H85Q mutant are quite distinct from those for WT OleTJE in both the substrate-free and arachidic acid-bound forms, indicating that the H85Q mutation alters the environment of the distal pocket around the heme iron, e.g. through altering the hydrogen bonding network to the distal water ligand in the LS substrate-free form (Fig. 6).
In F79A OleTJE the major LS species is at 2.48/2.25/1.89, similar to that of the main LS species in the WT enzyme. Minor species are at 2.65/2.25/1.84 and 2.58/2.25/1.86. The F79Y and F79W mutants have similar spectra to F79A OleTJE, although the central set of g values is predominant in these mutants (2.55/2.26/1.87 and 2.54/2.25/1.86, respectively). Binding of arachidic acid does not produce any significant HS signal in the F79A/W mutants, although a small HS signal is observed for substrate-bound F79Y OleTJE (gz = 7.79). However, substrate does cause some perturbations to the Phe79 mutants LS g values, particularly in the case of the F79A variant (Table 2). Notably, there is a large axial HS signal at g = 5.99 in both the F79W and F79Y OleTJE mutants that likely relates to HS penta-coordinated, thiol-ligated, ferric heme species in these mutants. This observation is consistent with some loss of heme in these variants, and UV-visible studies of these mutants also revealed a propensity for cysteine thiolate protonation of their heme iron to form the P420 state.
The R245E/L mutants result in less complex substrate-free EPR spectra than in WT OleTJE, with two major, similar sets of g values for both R245E (2.54/2.25/1.87 and 2.49/2.25/1.89) and R245L (2.54/2.25/1.87 and 2.46/2.25/1.90). The addition of arachidic acid has little effect on these LS g values but does produce a small rhombic HS signal (gz = 7.70, gy = 3.74) in the R245L mutant that mirrors the data from UV-visible titrations and results from effects of the solvent (Fig. 6 and Table 2). The R245E-capric acid complex proved unstable at the high protein concentration required for EPR analysis, aggregating and precipitating at ∼50 μm.
Products Formed by Novel OleTJE Mutants
Fatty acid substrate conversion reactions were set up using 1 μm WT, H85Q, F79A/W/Y, and R245L/E OleTJE enzymes and using 200 μm fatty acids (capric acid, lauric acid, myristic acid, palmitic acid, stearic acid, or arachidic acid), as described under “Experimental Procedures.”
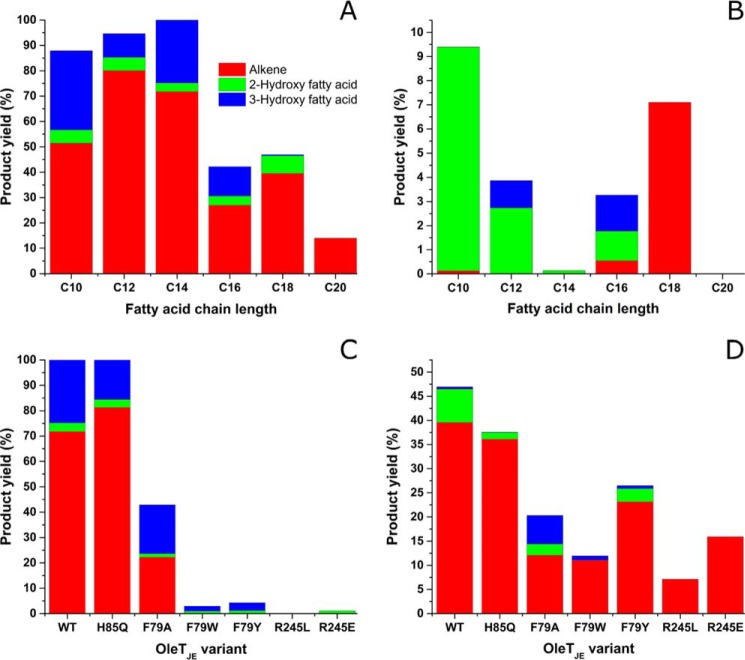
For WT OleTJE, activity was highest for shorter chain length substrates (C10:0 to C14:0), with essentially complete oxidation of myristic acid observed. These data are consistent with studies by Liu et al (12). Activity decreased with longer chain lengths, possibly because of the lower solubility of the substrates and/or the higher affinity of the terminal alkenes/hydroxylated products formed for the OleTJE active site (15). For all the fatty acids tested with WT OleTJE (C10:0 to C20:0), the major reaction was decarboxylation of the fatty acids to form terminal alkenes. For most substrates, 3-hydroxy fatty acid products were more prevalent than 2-hydroxy fatty acids. However, the reverse was the case for stearic acid (C18:0). When arachidic acid was used as a substrate, only 1-nonadecene was detected, with negligible amounts of 2-/3-hydroxy arachidic acid produced (Fig. 7A and Table 3).
FIGURE 7.
Products of fatty acid turnover from WT and mutant OleTJE enzymes. The bar charts show yields of different alkenes (red), 2-hydroxy fatty acids (green), and 3-hydroxy fatty acids (blue) resulting from catalytic turnover by WT and mutant forms of OleTJE, as described under “Experimental Procedures.” The data shown are averages from two sets of experiments, with values varying by <10% in all cases. A shows proportions of the different products formed in turnover of C10:0, C12:0, C14:0, C16:0, C18:0, and C20:0 substrates by WT OleTJE. B shows the products formed by the R245L OleTJE mutant with the same substrates, illustrating a switch from fatty acid hydroxylation toward fatty acid decarboxylation (terminal alkene formation) at longer substrate chain lengths. C shows products formed from the C14:0 substrate (myristic acid) by WT and all OleTJE mutants. D shows products formed from the C18:0 substrate (stearic acid) by WT and all OleTJE mutants.
OleTJE His85 was postulated as a proton donor to iron-oxo species in the WT OleTJE reaction (10). The P450 BSβ and SPα enzymes have a glutamine at this position, and catalyze predominantly (BSβ) or exclusively (SPα) fatty acid hydroxylation. The H85Q mutant was found to generate a product profile very similar to that for the wild type OleTJE for all substrates tested (Fig. 7B and Table 3). The n − 1 terminal alkenes are again the dominant products in all cases, with 3-hydroxy fatty acids formed to greater extents than 2-hydroxy fatty acids for substrates from C10:0-C16:0. As with WT OleTJE, the 2-hydroxy stearic acid product is formed to higher levels than 3-hydroxy stearic acid, and 1-nonadecene is the only significant product from arachidic acid with H85Q OleTJE.
The F79A OleTJE mutant exhibited decreased activity compared with the WT and H85Q enzymes for all fatty acids tested. F79A OleTJE mutant activity was highest with lauric acid, with 80% product conversion (compared with 94%/96% for WT/H85Q OleTJE), but overall product formation was much lower with myristic acid (42% for F79A compared with 100% with WT/H85Q). The F79A OleTJE mutant produced 3-hydroxy fatty acids in excess over 2-hydroxy fatty acids for all substrate chain lengths from C10-C18. For arachidic acid, the only product formed was 1-nonadecene, albeit with only 5% conversion (compared with 14%/11% for WT/H85Q OleTJE) (Table 3).
The F79W/Y mutants were generated to analyze effects of potentially more conservative mutations to Phe79. However it was found that, with one exception, both the F79W/Y OleTJE mutants had much lower activity than F79A OleTJE. The only outlier was the F79Y mutant with stearic acid substrate, where overall product formation was 27% compared with 20% for F79A OleTJE, because of a higher amount of 1-heptadecene produced by the F79Y mutant. Although amounts of alkene products from the F79A/W/Y mutants are broadly comparable for the C18:0 and C20:0 substrates, alkenes are formed in much lower amounts in the F79W/Y mutants compared with the F79A OleTJE for the C10:0-C16:0 substrates (Table 3).
Crystallographic data indicated that OleTJE Arg245 plays a key role in catalysis by coordinating the carboxylate group of the fatty acid substrate (10). The R245L and R245E OleTJE mutants were generated and purified to investigate effects of side chain charge removal and reversal. The R245L/E OleTJE mutants generally have much lower levels of activity than WT OleTJE and the two most productive mutants (H85Q and F79A). However, the R245L/E mutants convert 7.1%/16% stearic acid to 1-heptadecene, with no detectable hydroxylated products. Almost no decarboxylated product (1-nonadecene) was observed for these mutants with arachidic acid, and alkene production was in the range from 0 to 0.6% for the R245L/E mutants and C10:0-C16:0 substrates (Table 3). The unusual “switch” from mainly substrate hydroxylase activity at shorter chain lengths toward decarboxylase activity for stearic acid is shown in Fig. 7C, and Fig. 7D shows the product profiles from WT and all OleTJE mutant enzymes with stearic acid (C18:0) substrate. Alkenes are the major products formed by WT and all OleTJE mutants with this substrate.
Structural Properties of OleTJE Mutant Enzymes
Crystallization trials were undertaken for all OleTJE mutant enzymes. Crystals were obtained for both the H85Q and F79A mutant P450s in complex with arachidic acid substrate, as well as for the WT OleTJE in conditions containing formate. In the latter case, this produced a complex in which formate coordinates the WT OleTJE heme iron in the distal position. Structures for these mutant enzymes were determined to 1.44 Å (WT/formate), 1.80 Å (H85Q/arachidic acid) and 1.95 Å (F79A/arachidic acid) (Table 4). Attempts to crystallize other mutants were not successful, largely because of the propensity of other OleTJE mutants to aggregate at high concentrations.
TABLE 4.
Data reduction and final structural refinement statistics for OleTJE WT and mutant crystal structures
The data are presented for WT OleTJE bound to formate and for the arachidic acid-bound forms of the H85Q and F79A OleTJE mutants.
| OleTJE WT (formic acid) | OleTJE H85Q (C20:0) | OleTJE F79A (C20:0) | |
|---|---|---|---|
| Data collection | |||
| Space group | P43212 | P212121 | P212121 |
| Cell dimensions | |||
| a, b, and c (Å) | 60.170, 60.170, 241.70 | 48.928, 115.494, 163.247 | 51.88, 110.120, 164.850 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Rmerge (%) | 0.035 (0.625) | 0.051 (0.681) | 0.057 (0.842) |
| I/σI | 25.8 (2.9) | 11.4 (2.6) | 8.6 (1.8) |
| Completeness (%) | 99.9 (100.0) | 99.7 (99.6) | 99.6 (99.8) |
| Redundancy | 12.8 (6.5) | 6.1 (5.2) | 6.5 (6.6) |
| Refinement | |||
| Resolution (Å) | 48.21–1.44 (1.47–1.44) | 25.50–1.80 (1.83–1.80) | 55.06–1.95 (2.00–1.95) |
| No. reflections | 77,736 (5628) | 81,942 (5951) | 66,360 (4861) |
| Rwork/Rfree | 13.46/17.27 (21.4/28.0) | 18.27/21.99 (25.7/29.3) | 15.72/20.12 (27.1/33.1) |
| No. non-hydrogen atoms | 3798 | 7482 | 7704 |
| Mean B factor (A2) | 19.79 | 27.35 | 29.706 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.020 | 0.019 | 0.013 |
| Bond angles (°) | 2.020 | 1.875 | 1.270 |
| PDB numbers | 5M0N | 5M0O | 5M0P |
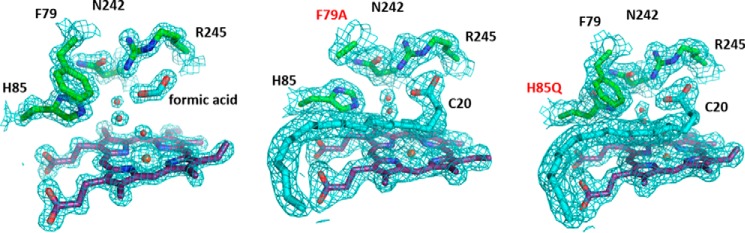
Few structural changes occur in the OleTJE structure because of the mutations. Arachidic acid substrate is bound similarly in both the H85Q and F79A mutants as was previously reported for the WT OleTJE (10). However, in the WT arachidic acid-bound structure, a sixth ligand water molecule could not be distinguished, likely because of the HS state of the WT OleTJE substrate-ligand complex. In contrast, for both OleTJE H85Q/F79A mutant structures, a sixth ligand water molecule is clearly visible. A similar situation is observed for the higher resolution structure of the WT OleTJE-formic acid complex, which remains LS in solution (Fig. 8). UV-visible substrate binding data for the H85Q/F79A mutants indicate much smaller HS shifts than observed for WT OleTJE, consistent with retention of the aqua-ligated LS state in the major fraction of the mutant P450s in the crystal structures.
FIGURE 8.
Active site structures for WT and mutant OleTJE proteins. The three panels show active site details from the crystal structures of WT OleTJE in complex with formic acid (left panel) and for the F79A (middle panel) and H85Q (right panel) OleTJE mutants bound to arachidic acid. Selected residues in the active site are shown as atom colored sticks (green carbons), whereas the heme group and associated ligands are shown with purple and cyan carbons, respectively. Positions of mutations are labeled in red. Electron density is shown as a blue mesh contoured at 2 σ for the WT OleTJE-formic acid complex, and at 1.5 σ for the two mutant structures. Three water molecules are clearly defined in the active site of these three structures (oxygens shown as red spheres), one of which acts as the sixth ligand to the heme iron.
The lack of any large scale structural changes in the H85Q/F79A OleTJE mutants suggests that that differences in product profiles for the WT and various OleTJE mutants are linked to subtle changes in the active site, possibly related to mobility of active site residues and/or substrates. Minor changes in the positioning and dynamic behavior of the α and β substrate hydrogens might have substantial effects on the product profile but are unlikely to be distinguished using medium resolution crystallography.
Discussion
The peroxygenase enzyme P450 BSβ (CYP152A1) was discovered in the late 1990s by Matsunaga et al. (27) and shown to catalyze the H2O2-dependent 1-(α) and 2-(β) hydroxylation of myristic acid, with 2-hydroxy myristic acid being the main product. Its counterpart P450 SPα (CYP152B1) was also identified by Matsunaga et al. and shown to catalyze the α-hydroxylation of lipids, producing 2-hydroxy myristic acid from the C14:0 substrate, as well as catalyzing 2-hydroxylation of longer chain saturated fatty acids and mono-unsaturated fatty acids (28, 29). However, only after studies by Rude et al. (9) was the capacity revealed of the related OleTJE enzyme (CYP152L1) to oxidatively decarboxylate fatty acids to form terminal alkenes. They also demonstrated that 1-pentadecene was formed not only by H2O2-dependent oxidative decarboxylation of palmitic acid in OleTJE but also by P450 BSβ and related P450s from Corynebacterium efficiens, Kocuria rhizophila, and Methylobacterium populi (9, 30). OleTJE is the most productive of the fatty acid peroxygenase P450s characterized to date with respect to its generation of alkene products over hydroxylated fatty acids. OleTJE was also shown to be functional in alkene production when driven by a redox partner system or using a photocatalytic system to generate H2O2 (11, 14). OleTJE is currently the most studied of the peroxygenase P450s, and the one with greatest potential for biotechnological applications in production of alkenes for use in biofuel production, for example (31).
The determination of the crystal structure of OleTJE revealed a high level of similarity with P450 BSβ in overall fold and active site structure (10). The OleTJE binding pocket is more extended than that of P450 BSβ, explaining its ability to accommodate fatty acids as long as arachidic acid (C20:0). The OleTJE structure highlights structural features common to other P450s (including the heme binding motif surrounding the conserved cysteine thiolate), as well as key residues involved in interacting with the substrate carboxylate. Amino acid alignment of OleTJE with other characterized P450 peroxygenases reveals that only 48 amino acids are in conserved positions in each of the seven P450 enzymes aligned, which vary in length from 415 to 428 amino acids (Fig. 2). The substrate carboxylate coordinating OleTJE Arg245 and its adjacent Pro246 are conserved in all these peroxygenases, replacing the acid-alcohol pair (e.g. Asp251/Thr252 in P450cam) found in most mono-oxygenase P450s and involved in protonation of ferric-peroxo and ferric-hydroperoxo catalytic cycle intermediates (3). These key mutations illustrate how these peroxygenases have diverged from mono-oxygenase P450s, exchanging a dipeptide involved in iron-oxo species protonation for one selected for binding the substrate carboxylate. The R245E/L mutants are severely catalytically compromised, consistent with a crucial role of Arg245 in orientating fatty acid carboxylate into a position compatible with efficient decarboxylation/hydroxylation of the substrate. However, these mutants do form small amounts of products (Table 3). The R245L/E mutants produced most alkene from the C18:0 substrate stearic acid but negligible amounts from the C20:0 substrate arachidic acid, potentially because of tighter product binding for the longer chain substrate. With one exception, the R245L/E mutants do not exhibit any significant substrate-induced HS ferric heme iron development. Only in the case of R245E with the C10:0 substrate capric acid is there any substantial HS accumulation (∼40%), albeit achieved at high substrate concentration (Kd = 1260 ± 160 μm) (Table 1). The HS conversion suggests that capric acid does displace Glu245 from a distal position on the heme iron, although only a small proportion of the substrate is converted to 1-nonene (0.2%), and no significant formation of hydroxylated forms of capric acid was observed (Fig. 5C and Table 3).
The H85Q mutant was generated to explore the role of the OleTJE His85 residue, which is replaced by a glutamine in the well characterized P450 BSβ, P450 SPα and Clostridium acetobutylicum P450 CLA (CYP152A2) enzymes, all of which catalyze predominantly or exclusively fatty acid hydroxylation (33) (Fig. 2). The OleTJE H85Q mutant gives a very similar product profile to WT OleTJE with each substrate tested. However, UV-visible binding studies demonstrate that substrate-induced H85Q HS heme accumulation is much lower than in the WT OleTJE in all cases (Tables 1 and 3). EPR studies confirms much lower accumulation of HS heme iron in arachidic acid-bound H85Q OleTJE compared with the WT enzyme (Fig. 6). However, heme spectral shifts are sufficient to determine Kd values for all substrates with H85Q OleTJE, showing that affinity is similar to WT OleTJE for longer chain substrates but significantly weaker for myristic acid and lauric acid substrates, with negligible heme spin state change observed on titration of H85Q OleTJE with capric acid (Table 1). The substantially diminished (or completely absent) extents of substrate-dependent HS shift in the H85Q mutant are consistent with the properties of the SPα, BSβ, and CLA peroxygenases (18–20, 33). The similarity in product profiles between WT and H85Q OleTJE suggests that His85 is not crucial for proton transfer to transient heme iron-oxo species to facilitate C–C bond scission and alkene production or that alternative pathway(s) exist in the H85Q mutant. A similar conclusion regarding the non-essentiality of a histidine in this position was also reached by Amaya et al. (30) in their studies of the M. populi OleTJE ortholog (CYP-MP), in which a methionine is located in the relevant position, although the CYP-MP enzyme is a less efficient decarboxylase than OleTJE.
OleTJE structural data showed that the His85 imidazole moiety is “sandwiched” between Phe79 and the heme edge (10). The F79A/Y/W mutants were produced and characterized to explore the effects of both removing the aromatic side chain and extending its size and chemical character. All these mutants showed reduced total activity compared with WT and H85Q OleTJE, with F79A OleTJE being the most active, forming between 36 and 43% of the product amount from WT OleTJE in the same unit time for the C14:0-C20:0 substrates but increasing to 61 and 85% of the amount for the shorter chain capric acid and lauric acid substrates. The alkenes are the major products with F79A OleTJE and all substrates, with the 3-hydroxylated fatty acid formed in excess over 2-hydroxyated fatty acids for the C10-C18 substrates. For the C20:0 arachidic acid, only 1-nonadecene was formed to a detectable level at 5% of the starting material (Table 3).
Despite the more “conservative” nature of the F79W and (particularly) the F79Y mutations, the activities of these variants were much lower than observed for F79A OleTJE with C10:0-C16:0 substrates. Within this group of substrates, hydroxylated fatty acids are formed in excess over alkenes in all cases except for the F79Y mutant with capric acid (where 1-nonene is the only product observed at 0.3% of the starting material), and total product formation does not exceed 6.5% for the F79Y/W mutants. However, with the C18:0 substrate stearic acid, the major product is 1-heptadecene (23%/11% products, respectively), and 1-nonadecene is the only product from arachidic acid (2.5%/3.4% products, respectively).
As is also the case for the H85Q mutant, each of the F79A/Y/W mutants give less substantial HS heme conversions than seen in WT OleTJE (Table 1). All Phe79 mutants also show a similar pattern of decreasing affinity as fatty acid chain length decreases, with capric acid not inducing any significant heme spin state change in the F79Y/W OleTJE mutants.
Many bacterial P450s exploit a substrate binding-dependent LS to HS heme shift to regulate catalytic activity through development of a more positive potential in the substrate-bound form of the heme iron. For example, arachidonic acid binding to P450 BM3 causes an ∼140-mV increase in ferric heme iron reduction potential, facilitating electron transfer from the linked cytochrome P450 reductase domain to drive catalysis (34). However, OleTJE shows negligible change in the redox potential of the heme FeIII/FeII couple on arachidic acid binding (−105 mV and −103 mV versus normal hydrogen electrode, respectively, for substrate-free and substrate-bound forms), consistent with its dependence on H2O2 for generating reactive iron-oxo species (10). The relatively positive potential in OleTJE is likely related in part to the structural arrangement around the proximal cysteinate ligand. Here, it is notable that a phenylalanine residue (typically seven amino acids before the cysteinate) heavily conserved in mono-oxygenase P450s is absent in all of the peroxygenases aligned in Fig. 2. Mutations to the relevant Phe393 residue in P450 BM3 cause substantial changes in heme iron potential (35, 36). Recent studies showed that OleTJE turnover can be driven by heterologous redox partner systems, and this is likely facilitated by the large driving force (>210 mV) for electron transfer from NAD(P)H through redox partner proteins and onto the substrate-bound OleTJE heme iron (11, 12).
In conclusion, we provide a detailed study of the properties of WT OleTJE and of important active site mutants of this alkene producing P450. We show that glutamine substitutes effectively for histidine in the OleTJE H85Q mutant, which has very similar product profiles to WT OleTJE. The major difference lies in the much weaker ability of fatty acid substrates to induce HS heme iron formation in the H85Q mutant (Fig. 5B), a property consistent with those of the P450 SPα and P450 BSβ peroxygenases, both of which have a glutamine in the same position (20, 38). The data appear to rule out a key role for His85 in proton donation to reactive iron-oxo species in OleTJE, and suggest that the system is robust and that alternative protonation pathway(s) may exist. Mutations to OleTJE Phe79 also diminish substrate-dependent heme HS conversion and weaken affinity for shorter chain (C10:0-C16:0) fatty acid substrates. This translates into much lower product formation for the F79Y/W mutants from these substrates, although F79A OleTJE is less severely affected, with between 36 and 85% overall product formation compared with that of WT OleTJE for C10:0-C20:0 substrates (Table 3). These data indicate that the more “conservative” F79Y/W mutations are actually more disruptive to active site structure and catalytic efficiency than is the F79A mutation, a conclusion consistent with the greater propensity of the F79Y/W mutants to aggregate/precipitate and their failure to crystallize. These properties are shared with the R245L/E variants, in which the substrate carboxylate tethering Arg245 is mutated. Each of the F79Y/W and R245E/L mutations show some heme depletion, with R245E OleTJE being the worst affected (Fig. 4B). The red-shifted (423 nm) Soret band of the oxidized R245E OleTJE protein suggests that the glutamate may coordinate the heme iron in the ferric state. The R245L/E mutants retain low activity, with stearic acid (C18:0) being the best substrate and forming the greatest amounts of alkene product for both mutants (Table 3). Crystal structure data for the H85Q and F79A mutants reveal only minor alterations to OleTJE active site structure, suggesting that quite subtle changes in the positions of active site amino acids, water molecule(s), and the substrate carboxylate region may be sufficient to trigger catalysis when H2O2 binds to initiate compound 0 formation. Makris's group have revealed how this could lead to substrate hydroxylation by the radical rebound mechanism, involving hydrogen atom abstraction from the substrate by compound I and subsequent “rebound” of the hydroxyl group to the substrate radical (5). However, through their identification and characterization of the transient ferryl-hydroxo compound II, a novel mechanism for substrate decarboxylation was proposed. In this model, compound II abstracts a further electron from the substrate radical to form a diradical or carbocation intermediate (Fig. 1), which decarboxylates to form CO2 and the n − 1 terminal alkene, with protonation of the ferric-hydroxo species restoring the water-coordinated resting form of the heme iron (15, 39). Further protein engineering studies on OleTJE are now clearly needed to improve its stability and catalytic performance as an alkene producing enzyme and to facilitate its biotechnological application.
Experimental Procedures
Expression and Purification of OleTJE and OleTJE Mutants
The gene encoding OleTJE from Jeotgalicoccus sp. ATCC 8456 was codon-optimized for expression in E. coli and cloned into a modified pET15b (GenScript, Cherwell, UK), incorporating a TEV cleavage site with an N-terminal polyhistidine tag. Rationally designed mutants (H85Q, F79A, F79W, F79Y, R245L, and R245E) were created by site-directed mutagenesis, using a QuikChange Lightning kit (Agilent Technologies LDA UK Ltd., Cheadle, UK). The oligonucleotide primers used were: H85Q forward, 5′-ttttaccatcaaccgtctg gattgcgcctttaccaaac-3′; H85Q reverse, 5′-gtttggtaaaggcgcaatccagacggttgatggtaaaa-3′; F79A forward, 5′-gtatggattgcgcctttaccagccagggtattcacaatacgttt-3′; F79A reverse, 5′-aaacgtattgtgaataccctggctggtaaaggcgcaatccatac-3′; F79W forward, 5′-attgcgcctttaccc cacagggtattcacaatacgtttcggc-3′; F79W reverse, 5′-gccgaaacgtattgtgaataccctgtggggtaaaggcgcatt-3′; F79Y forward, 5′ tgcgcctttaccatacagggtattcacaatacgtttcg-3′; F79Y reverse, 5′-cgaaacgtattgtgaataccctgtatggtaaaggcgca-3′; R245L forward, 5′-gatgaacacgttcctgccgctgattgcgatc-3′; R245L reverse, 5′-gatcgcaatcagcggcaggaacgtgttcatc-3′; R245E forward, 5′-ttgatctgatgaacacgttcgagccgctgattgcgatcaatcg-3′ and R245E reverse, 5′-cgattgatcgcaatcagcggctcgaacgtgttcatcagatcaa-3′. E. coli strain C41 (DE3) (Lucigen, Middleton, UK) was used as the expression host for WT OleTJE and all mutants. Cells transformed with the plasmids were grown at 37 °C with shaking at 200 rpm in total volumes of 500 ml of 2YT broth containing 50 μg/ml ampicillin. Expression of WT/mutant OleTJE genes was induced with 100 μm isopropyl 1-thio-β-d-galactopyranoside when an OD600 of 0.5 was reached. At point of induction, 500 μm δ-aminolevulinic acid was added to all mutants, and 4 μg/ml hemin was also added to the R245L and R245E expression cultures to aid heme incorporation. At this point, the incubation temperature was lowered to 25 °C, and the cells were grown for a further 16 h. The cells were harvested by centrifugation at 6000 rpm at 4 °C using a JLA-8.1000 rotor in an Avanti J-26 XP centrifuge. The cells were resuspended in 100 mm potassium phosphate (pH 8.0), combined, and centrifuged as before. The cell pellet was then frozen at −80 °C until required for purification.
Cells were thawed and resuspended in 200 g/liter extraction buffer (100 mm potassium phosphate, 1 m NaCl, 10% glycerol, pH 8.0) along with SigmaFAST protease inhibitor mixture tablets, EDTA-Free (Sigma-Aldrich) using 1 tablet/100 ml, and 100 μg/ml DNase I (bovine pancreas; Sigma-Aldrich). The cells were lysed using a cell disruptor (Constant Cell Disruption Systems, Daventry, UK) at a pressure of 15,000 p.s.i. The homogenate was then centrifuged at 20,000 rpm at 4 °C for 90 min using a JA-25.50 rotor (Beckman-Coulter Ltd., High Wycombe, UK). The supernatant was removed and incubated overnight at 4 °C with (10 ml/100 g of cell pellet) Ni-IDA chromatographic medium (Generon, Maidenhead, UK). The mixture was then transferred to a column, and the resin bed was washed with 20 column volumes of buffer A (100 mm KPi, 750 mm NaCl, 10% glycerol, pH 8.0) containing 50 mm imidazole. The resin was then washed with 2 column volumes of buffer A with 150 mm imidazole, and the protein was eluted in buffer A with 175 mm imidazole. For OleTJE samples not destined for protein crystallization, the protein was extensively dialyzed into buffer A to remove any imidazole and bound fatty acids retained from expression in E. coli and then concentrated by ultrafiltration in a Vivaspin (30,000 molecular weight cutoff; GE Healthcare) and transferred to buffer B (100 mm KPi, 750 mm NaCl, 20% glycerol, pH 8.0) using a PD 10 desalting column (GE Healthcare). The protein was then snap frozen in liquid nitrogen and stored at −80 °C until required. Protein destined for crystallography was not frozen at this point, and instead TEV protease (∼500 units) was added to the eluted protein to remove the His tag, and the sample was incubated at 4 °C overnight. The S219V variant of TEV protease was expressed using plasmid pRK793 (Addgene plasmid no. 8827) (37). The protein was then exchanged into buffer A using a PD10 desalting column and added to a bed of Ni-IDA resin. Cleaved protein was eluted with buffer A including 20 mm imidazole. This was then dialyzed extensively to remove imidazole and bound lipids. The protein was concentrated using a Vivaspin and then gel-filtered using a Superdex 200 16/600 (GE Healthcare) column pre-equilibrated with buffer C (100 mm KPi, 750 mm NaCl, pH 8.0).
UV-visible Spectroscopy of WT and Mutant OleTJE Hemoproteins
The UV-visible spectra of WT and mutant forms OleTJE were collected using a Cary 60 UV-visible spectrophotometer (Agilent). Spectra were recorded at ambient temperature for the proteins in buffer A, typically at concentrations of ∼4–6 μm. The ferrous and FeII-CO forms of the hemoproteins were produced under anaerobic conditions using degassed buffer A. Reduced enzymes were formed by the addition of a few grains of solid sodium dithionite to ferric enzymes. The FeII-CO complexes were then formed by slowly bubbling CO gas into reduced OleTJE proteins until no further spectral shift was observed.
Fatty Acid Binding Titrations with OleTJE Mutants
Binding of saturated fatty acids (C12:0, C14:0, C16:0, C18:0, and C20:0) to the OleTJE mutants was performed at 25 °C in buffer A. Substrates were dissolved in 100% ethanol, and additions of 0.5-μl aliquots of substrate stocks (of concentration from 1–100 mm, according to the particular substrate and its affinity for the P450s) were made until no further spectral shift was observed. Spectra (800–300 nm) were recorded using a Cary 60 UV-visible spectrophotometer. Difference spectra were obtained by subtracting the ligand-free spectrum of the P450 from the ligand-bound spectra at each substrate concentration. The absorbance difference maximum (ΔApeak) and minimum (ΔAtrough) were then identified from the individual data sets, and the maximal absorbance change (ΔAmax) was calculated by subtracting ΔAtrough from ΔApeak in each case, using the same wavelength pair throughout each individual titration. ΔAmax was then plotted against substrate concentration, and the data were fitted using either a hyperbolic (Michaelis-Menten or Morrison equations) (32) or a sigmoidal (Hill) function to determine the dissociation constants (Kd values) for fatty acid substrate binding.
Stopped Flow Kinetic Analysis of Substrate Oxidation by OleTJE WT and Mutant Enzymes
An Applied Photophysics SX18 MR stopped flow spectrophotometer with a photodiode array detector (Leatherhead, UK) was used to collect stopped flow absorption data. For the WT, H85Q, F79A, and F79W OleTJE mutants, a 2.5 molar excess of arachidic acid (C20:0) was mixed with these P450 samples (giving a final P450 concentration of 5 μm), and the mixture was passed through a 0.2-μm syringe filter (Sartorius UK Ltd., Epsom, UK) to remove excess lipids from the P450 protein solution. A nearly saturated aqueous solution of lauric acid (C12:0, 250 μm) was mixed with the F79Y mutant (again giving a final P450 concentration of 5 μm) and passed through a 0.2-μm syringe filter as above. The substrate-bound OleTJE proteins were then mixed with different concentrations of H2O2 (final concentration post-mixing, 6.25–200 μm) in the stopped flow instrument at 25 °C.
Conversion of the fatty acid-bound, HS ferric forms of the OleTJE enzymes to their LS forms was monitored at 421 nm for the OleTJE WT and OleTJE H85Q mutant enzymes and at 423 nm for the OleTJE F79A enzyme. The data were fitted using a single exponential function and using the Pro-Data SX software suite (Applied Photophysics). The observed reaction rate constants (kobs values) were plotted against the relevant H2O2 concentration, and the data were fitted using a hyperbolic (Michaelis-Menten) function (for the WT, F79Y, and H85Q OleTJE enzymes) or using a linear function in the case of the other OleTJE mutants.
EPR Analysis of OleTJE Mutants
Continuous wave X-band EPR spectra for the OleTJE WT and mutant P450s were obtained at 10 K using a Bruker ELEXSYS E500 EPR spectrometer with an ER 4122SHQ Super High Q cavity. Temperature was controlled with an ESR900 cryostat (Oxford Instruments, Abingdon, UK). EPR spectra were collected for WT and OleTJE mutants at concentrations of 100–200 μm for ligand-free, arachidic acid-bound, and imidazole-bound forms. Arachidic acid was added to dilute OleTJE protein until the UV-visible spectrum showed no further optical change toward high spin. The enzyme was then concentrated to an appropriate concentration (200 μm for WT, H85Q, and F79A OleTJE; 150 μm for R245L OleTJE; and 100 μm for the F79W, F79Y, and R245E mutants) (the latter in its substrate-free form), taking into consideration that some of the mutants are prone to precipitation at high concentrations) using a Vivaspin (30,000 molecular weight cutoff).
Crystallography of the OleTJE H85Q and OleTJE F79A Mutants
Crystallization trials for the OleTJE H85Q and OleTJE F79A mutant enzymes were carried out with 400 nl (200 nl of protein and 200 nl of precipitant) sitting drops in MRC 96-well plates (Molecular Dimensions, Newmarket, UK). Molecular Dimensions screens Clear Strategy Screen I, Clear Strategy Screen II, PACT premier, JCSG-plus, and Morpheus were used for OleTJE H85Q. Diffracting crystals were taken from well H10 of JCSG-plus (0.2 m ammonium acetate, 0.1 m Bis-Tris, 25% (w/v) PEG 3350, pH 5.5). For OleTJE F79A, C3 Shotgun Screen (SG1) and JCSG-plus screens were used. Crystals formed were used to make a seed stock, and further plates were set up using an optimized screen. Diffracting crystals were grown in 0.2 m MgCl2, 0.1 m HEPES, 20% PEG 6000 (pH 7.5). Plates were set up using a Mosquito nanoliter pipetting robot (TTP Labtech, Melbourn, UK). For OleTJE H85Q, ligand-free and arachidic acid-bound proteins were used for crystal trials at a concentration of 11.5 mg/ml. For the OleTJE F79A mutant, arachidic acid-bound enzyme was used at a protein concentration of 13.5 mg/ml. Arachidic acid was bound to the proteins in the same way as described for the EPR studies.
Fatty Acid Substrate Conversion Reactions with WT and Mutant OleTJE Enzymes
Fatty acid substrate conversion reactions were set up using 1 μm of wild-type, H85Q, F79A, F79W, F79Y, R245L, and R245E OleTJE enzymes. The proteins were incubated with 200 μm fatty acid (capric acid, lauric acid, myristic acid, palmitic acid, stearic acid, or arachidic acid), and the reactions were started by the addition of 400 μm H2O2 in a total volume of 500 μl of buffer A. The reactions were incubated at 32 °C with shaking for 30 min. Thereafter, reactions were stopped by acidifying with 20 μl of 37% HCl. For the C10:0, C12:0, and C16:0 reactions, the internal standards 1-undecene, myristic acid (C14:0), α-OH myristic acid, and β-OH myristic acid were added. For the C14:0, C18:0, and C20:0 reactions, internal standards 1-pentadecene, palmitic acid (C16:0), β-OH palmitic acid, and α-OH palmitic acid were added. After completion of the reactions, the mixtures were extracted with an equal volume of dichloromethane. The dichloromethane extract was then dried with anhydrous magnesium sulfate before the product sample was subjected to GC/MS analysis.
Characterization of Fatty Acid Oxidation Products Using GC/MS
Analysis of products formed by fatty acid oxidation using WT and mutant OleTJE enzymes was done using an Agilent 6977A series GC/MSD installed with an Agilent HP-5MS UI 30 m × 0.250 mm × 0.25 μm column (helium flow rate, 1.2 ml/min) (Agilent). A 1-μl sample was injected via sandwich injection with 1 μl of N,O-bis(trimethylsilyl)trifluoroacetamide containing 1% trimethylchlorosilane to derivatize fatty acids and hydroxylated fatty acids. The split ratio was 10:1, and the inlet was set to a temperature of 280 °C. The oven temperature was held at 40 °C for 1 min and then ramped at 10 °C/min to 290 °C, where it was held for 1 min. Electronic ionization was used, scanning ions in the range of 40–550 m/z at a temperature of 250 °C. For quantitation, external standard calibrations were generated, and the selected ion monitoring mode was used to identify appropriate m/z ion fragments for substrate and product analysis.
Author Contributions
A. W. M. and D. L. conceived and coordinated the study. S. M., D. L., and A. W. M. wrote the paper. S. M., J. D. B., and K. L. T. prepared expression constructs, purified enzymes, and performed substrate binding experiments. S. M., H. M. G., K. J. M., A. W. M., and S. E. J. R. collected and analyzed EPR data. S. M., K. J. M., and R. T. B. collected and analyzed GC-MS data from fatty acid oxidation experiments. S. M. crystallized P450 mutants, and C. W. L. and D.L. collected and analyzed crystallographic data. S. M. collected transient kinetic data. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgment
We thank Marina Golovanova (University of Manchester) for technical support.
This work was supported by the UK Biotechnology and Biological Sciences Research Council (BBSRC) and by BBSRC/Shell UK Ltd. for Research Grants BB/N006275/1 (to A. W. M. and D. L., supporting K. J. M. and H. M. G.) and BB/K017802/1 (to A. W. M. and D. L., supporting K. L. T.). This work was also supported by BBSRC and Agilent Technologies UK Ltd. for BBSRC Industrial CASE Ph.D. Studentship BB/J012645/1 (to A.W.M. and R.T.B., supporting S. M.). The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (codes 5M0N, 5M0O, and 5M0P) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- Ni-IDA
- nickel-iminodiacetic acid
- LS
- low spin
- HS
- high spin
- BBSRC
- Biotechnology and Biological Sciences Research Council.
References
- 1. Munro A. W., Girvan H. M., and McLean K. J. (2007) Variations on a(t) heme: novel mechanisms, redox partners and catalytic functions in the cytochrome P450 superfamily. Nat. Prod. Rep. 24, 585–609 [DOI] [PubMed] [Google Scholar]
- 2. Denisov I. G., Makris T. M., Sligar S. G., and Schlichting I. (2005) Structure and chemistry of cytochrome P450. Chem. Rev. 105, 2253–2277 [DOI] [PubMed] [Google Scholar]
- 3. Raag R., Martinis S. A., Sligar S. G., and Poulos T. L. (1991) Crystal structure of the cytochrome P-450CAM active site mutant Thr252Ala. Biochemistry 30, 11420–11429 [DOI] [PubMed] [Google Scholar]
- 4. Rittle J., and Green M. T. (2010) Cytochrome P450 compound I: capture, characterization, and C-H bond activation kinetics. Science 330, 933–937 [DOI] [PubMed] [Google Scholar]
- 5. Groves J. T. (2006) High-valent iron in chemical and biological oxidations. J. Inorg. Biochem. 100, 434–447 [DOI] [PubMed] [Google Scholar]
- 6. Joo H., Lin Z., and Arnold F. H. (1999) Laboratory evolution of peroxide-mediated cytochrome P450 evolution. Nature 399, 670–673 [DOI] [PubMed] [Google Scholar]
- 7. Li Q. S., Ogawa J., and Shimizu S. (2001) Critical role of the residue size at position 87 in H2O2-dependent substrate hydroxylation activity and H2O2-inactivation of cytochrome P450BM-3. Biochem. Biophys. Res. Commun. 280, 1258–1261 [DOI] [PubMed] [Google Scholar]
- 8. Hrycay E. G., and Bandiera S. M. (2012) The monooxygenase, peroxidase and peroxygenase properties of cytochrome P450. Arch. Biochem. Biophys. 522, 71–89 [DOI] [PubMed] [Google Scholar]
- 9. Rude M. A., Baron T. S., Brubaker S., Alibhai M., Del Cardayre S. B., and Schirmer A. (2011) Terminal olefin (1-alkene) biosynthesis by a novel P450 fatty acid decarboxylase from Jeotgalicoccus species. Appl. Environ. Microbiol. 77, 1718–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Belcher J., McLean K. J., Matthews S., Woodward L. S., Fisher K., Rigby S. E., Nelson D. R., Potts D., Baynham M. T., Parker D. A., Leys D., and Munro A. W. (2014) Structure and biochemical properties of the alkene producing cytochrome P450 OleTJE (CYP152L1) from the Jeotgalicoccus sp. 8456 bacterium. J. Biol. Chem. 289, 6535–6550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dennig A., Kuhn M., Tassoti S., Thiessenhusen A., Gilch S., Bülter T., Haas T., Hall M., and Faber K. (2015) Oxidative decarboxylation of short-chain fatty acids to 1-alkenes. Angew. Chem. Int. Ed. Engl. 54, 8819–8822 [DOI] [PubMed] [Google Scholar]
- 12. Liu Y., Wang C., Yan J., Zhang W., Guan W., Lu X., and Li S. (2014) Hydrogen peroxide-independent production of α-alkenes by OleTJE P450 fatty acid decarboxylase. Biotechnol. Biofuels 7, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McIver L., Leadbeater C., Campopiano D. J., Baxter R. L., Daff S. N., Chapman S. K., and Munro A. W. (1998) Characterisation of flavodoxin NADP+-oxidoreductase and flavodoxin: key components of electron transfer in Escherichia coli. Eur. J. Biochem. 257, 577–585 [DOI] [PubMed] [Google Scholar]
- 14. Zachos I., Gassmeyer S. K., Bauer D., Sieber V., Hollmann F., and Kourist R. (2015) Photobiocatalytic decarboxylation for olefin synthesis. Chem. Commun. (Camb.) 51, 1918–1921 [DOI] [PubMed] [Google Scholar]
- 15. Grant J. L., Hsieh C. H., and Makris T. M. (2015) Decarboxylation of fatty acids to terminal alkenes by cytochrome P450 compound I. J. Am. Chem. Soc. 137, 4940–4943 [DOI] [PubMed] [Google Scholar]
- 16. Zhang F., Rodriguez S., and Keasling J. D. (2011) Metabolic engineering of microbial pathways for advanced biofuels production. Curr. Opin. Biotechnol. 22, 775–783 [DOI] [PubMed] [Google Scholar]
- 17. Yan J., Liu Y., Wang C., Han B., and Li S. (2015) Assembly of lipase and P450 fatty acid decarboxylase to constitute a novel biosynthetic pathway for production of 1-alkenes from renewable triacylglycerols and oils. Biotechnol. Biofuels 8, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fujishiro T., Shoji O., Nagano S., Sugimoto H., Shiro Y., and Watanabe Y. (2011) Crystal structure of H2O2-dependent cytochrome P450SPα with its bound fatty acid substrate: insight into the regioselective hydroxylation of fatty acids at the α position. J. Biol. Chem. 286, 29941–29950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Matsunaga I., Ueda A., Sumimoto T., Ichihara K., Ayata M., and Ogura H. (2001) Site-directed mutagenesis of the putative distal helix of peroxygenase cytochrome P450. Arch. Biochem. Biophys. 394, 45–53 [DOI] [PubMed] [Google Scholar]
- 20. Lee D. S., Yamada A., Sugimoto H., Matsunaga I., Ogura H., Ichihara K., Adachi S., Park S. Y., and Shiro Y. (2003) Substrate recognition and molecular mechanism of fatty acid hydroxylation by cytochrome P450 from Bacillus subtilis: crystallographic, spectroscopic, and mutational studies. J. Biol. Chem. 278, 9761–9767 [DOI] [PubMed] [Google Scholar]
- 21. Girvan H. M., Marshall K. R., Lawson R. J., Leys D., Joyce M. G., Clarkson J., Smith W. E., Cheesman M. R., and Munro A. W. (2004) Flavocytochrome P450 BM3 mutant A264E undergoes substrate-dependent formation of a novel heme iron ligand set. J. Biol. Chem. 279, 23274–23286 [DOI] [PubMed] [Google Scholar]
- 22. McLean K. J., Warman A. J., Seward H. E., Marshall K. R., Girvan H. M., Cheesman M. R., Waterman M. R., and Munro A. W. (2006) Biophysical characterization of the sterol demethylase P450 from Mycobacterium tuberculosis, its cognate ferredoxin and their interactions. Biochemistry 45, 8427–8443 [DOI] [PubMed] [Google Scholar]
- 23. Perera R., Sono M., Sigman J. A., Pfister T. D., Lu Y., and Dawson J. H. (2003) Neutral thiol as a proximal ligand to ferrous heme iron: implications for heme proteins that lose cysteine thiolate ligation on reduction. Proc. Natl. Acad. Sci. U.S.A. 100, 3641–3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Driscoll M. D., McLean K. J., Levy C., Mast N., Pikuleva I. A., Lafite P., Rigby S. E., Leys D., and Munro A. W. (2010) Structural and biochemical characterization of Mycobacterium tuberculosis CYP142: evidence for multiple cholesterol 27-hydroxylase activities in a human pathogen. J. Biol. Chem. 285, 38270–38282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Matsunaga I., Yamada A., Lee D. S., Obayashi E., Fujiwara N., Kobayashi K., Ogura H., and Shiro Y. (2002) Enzymatic reaction of hydrogen peroxide dependent peroxygenase cytochrome P450s: kinetic deuterium isotope effect and analyses by resonance Raman spectroscopy. Biochemistry 41, 1886–1892 [DOI] [PubMed] [Google Scholar]
- 26. Joyce M. G., Girvan H. M., Munro A. W., and Leys D. (2004) A single mutation in cytochrome P450 BM3 induces the conformational rearrangement seen upon substrate binding in the wild-type enzyme. J. Biol. Chem. 279, 23287–23293 [DOI] [PubMed] [Google Scholar]
- 27. Matsunaga I., Ueda A., Fujiwara N., Sumimoto T., and Ichihara K. (1999) Characterization of the ybdT gene product of Bacillus subtilis: nove fatty acid β-hydroxylating cytochrome P450. Lipids 34, 841–846 [DOI] [PubMed] [Google Scholar]
- 28. Matsunaga I., Yokotani N., Gotoh O., Kusunose E., Yamada M., and Ichihara K. (1997) Molecular cloning and expression of fatty acid α-hydroxylase from Sphingomonas paucimobilis. J. Biol. Chem. 272, 23592–23596 [DOI] [PubMed] [Google Scholar]
- 29. Matsunaga I., Yamada M., Kusunose E., Miki T., and Ichihara K. (1998) Further characterization of hydrogen peroxide-dependent fatty acid α-hydroxylase from Sphingomonas paucimobilis. J. Biochem. 124, 105–110 [DOI] [PubMed] [Google Scholar]
- 30. Amaya J. A., Rutland C. D., and Makris T. M. (2016) Mixed regiospecificity compromises alkene synthesis by a cytochrome P450 peroxygenase from Methylobacterium populi. J. Inorg. Biochem. 158, 11–16 [DOI] [PubMed] [Google Scholar]
- 31. Herman N. A., and Zhang W. (2016) Enzymes for fatty acid-based hydrocarbon biosynthesis. Curr. Opin. Chem. Biol. 35, 22–28 [DOI] [PubMed] [Google Scholar]
- 32. Morrison J. F. (1969) Kinetics of the reversible inhibition of enzyme catalysed reactions by tight-binding inhibitors. Biochim. Biophys. Acta 185, 269–286 [DOI] [PubMed] [Google Scholar]
- 33. Girhard M., Schuster S., Dietrich M., Dürre P., and Urlacher V. B. (2007) Cytochrome P450 monooxygenase from Clostridium acetobutylicum: a new α-fatty acid hydroxylase. Biochem. Biophys. Res. Commun. 362, 114–119 [DOI] [PubMed] [Google Scholar]
- 34. Daff S. N., Chapman S. K., Turner K. L., Holt R. A., Govindaraj S., Poulos T. L., and Munro A. W. (1997) Redox control of the catalytic cycle of flavocytochrome P450 BM3. Biochemistry 36, 13816–13823 [DOI] [PubMed] [Google Scholar]
- 35. Ost T. W., Miles C. S., Munro A. W., Murdoch J., Reid G. A., and Chapman S. K. (2001) Phenylalanine 393 exerts thermodyamic control over the heme of flavocytochrome P450 BM3. Biochemistry 40, 13421–13429 [DOI] [PubMed] [Google Scholar]
- 36. Ost T. W., Clark J., Mowat C. G., Miles C. S., Walkinshaw M. D., Reid G. A., Chapman S. K., and Daff S. (2003) Oxygen activation and electron transfer in flavocytochrome P450 BM3. J. Am. Chem. Soc. 125, 15010–15020 [DOI] [PubMed] [Google Scholar]
- 37. Kapust R. B., Tozser J., Fox J. D., Anderson D. E., Cherry S., Copeland T. D., and Waugh D. S. (2001) Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 14, 993–1000. [DOI] [PubMed] [Google Scholar]
- 38. Imai Y., Matsunaga I., Kusunose E., and Ichihara K. (2000) Unique heme environment at the putative distal region of hydrogen peroxide-dependent fatty acid α-hydroxylase from Sphingomonas paucimobilis (peroxygenase P450SPα). J. Biochem. 128, 189–194 [DOI] [PubMed] [Google Scholar]
- 39. Grant J. L., Mitchell M. E., and Makris T. M. (2016) Catalytic strategy for carbon-carbon bond scission by the cytochrome P450 OleT. Proc. Natl. Acad. Sci. U.S.A. 113, 10049–10054 [DOI] [PMC free article] [PubMed] [Google Scholar]