ABSTRACT
The rapid aminoglycoside NP (Nordmann/Poirel) test was developed to rapidly identify multiple aminoglycoside (AG) resistance in Enterobacteriaceae. It is based on the detection of the glucose metabolism related to enterobacterial growth in the presence of a defined concentration of amikacin plus gentamicin. Formation of acid metabolites was evidenced by a color change (orange to yellow) of the red phenol pH indicator. The rapid aminoglycoside NP test was evaluated by using bacterial colonies of 18 AG-resistant isolates producing 16S rRNA methylases, 20 AG-resistant isolates expressing AG-modifying enzymes (acetyl-, adenyl-, and phosphotransferases), and 10 isolates susceptible to AG. Its sensitivity and specificity were 100% and 97%, respectively, compared to the broth dilution method, which was taken as the gold standard for determining aminoglycoside resistance. The test is inexpensive, rapid (<2 h), and implementable worldwide.
KEYWORDS: rapid diagnostic test, 16S rRNA methylases, antibiotic, susceptibility testing, gentamicin, amikacin, plazomicin, tobramycin, netilmicin, kanamycin
INTRODUCTION
Extended-spectrum β-lactamases that hydrolyze extended-spectrum cephalosporins and carbapenemases that also hydrolyze carbapenems are disseminating worldwide in Enterobacteriaceae, and therapeutic options are becoming limited (1). In those multidrug resistant isolates, aminoglycosides (AG) may still be considered valuable treatment options (2), but their efficacy has been reduced by the surge and the dissemination of resistance.
The most prevalent mechanisms of resistance to AG in Enterobacteriaceae are enzymes that modify the chemical structure of aminoglycosides and can be nucleotidyltransferases, phosphotransferases, or acetyltransferases (3). The genetic determinants coding for those aminoglycoside-modifying enzymes are often present in various combinations in a given strain and are typically found on plasmids. They do not confer cross-resistance to all AG molecules.
However, plasmid-mediated 16S rRNA methyltransferases conferring a high level of resistance to multiple AG is also reported (4), and they are identified at a high frequency, especially among producers of NDM-like carbapenemases (5). Enzymes encoded by these genes methylate the aminoglycoside tRNA recognition site (A-site) of the 16S rRNA, which is the intracellular target of the AG. The 16S rRNA methylases identified in Enterobacteriaceae are ArmA, RmtB to RmtF, and NpmA, with ArmA being the most frequently identified methylase (4). Those methylases exhibit a significant amino acid heterogeneity, sharing similar amino acid motifs in their active sites. They confer resistance to almost all AG (amikacin, gentamicin, tobramycin, and kanamycin, but not neomycin), whereas NpmA confers additional resistance traits to neomycin (given as a topical agent) and to apramycin (not used for humans) (6).
In an area of paucity of novel molecules, rapidly identifying multiple resistance to AG by a simple test may be useful for implementing antibiotic stewardship and containment of those multidrug-resistant bacteria.
Thus, we developed a rapid colorimetric test based on the detection of bacterial growth in the presence of a defined concentration of a mix of two AG molecules (amikacin and gentamicin). Bacterial growth detection was based on glucose metabolism (7). Acid formation related to glucose metabolism in Enterobacteriaceae may be observed through the color change of a pH indicator. This test is rapid (less than 4 h) and easy to handle.
RESULTS AND DISCUSSION
A total of 48 enterobacterial isolates were tested to evaluate the performance of the rapid aminoglycoside NP test, including 39 AG-resistant isolates and 10 AG-susceptible isolates (Table 1). Among the 38 AG-resistant isolates, 18 isolates produced 16S rRNA methylases and exhibited high MIC values for both amikacin and gentamicin, while 20 produced AG-modifying enzymes and exhibited various levels of resistance for amikacin and gentamicin (Table 1).
TABLE 1.
Rapid aminoglycoside NP test results for AG-resistant enterobacterial producers of 16S rRNA methylases and AG-modifying enzymes and for AG-susceptible enterobacterial isolates
| Isolate | Species | Mechanism of AG resistancea | Resistance findings forb: |
Rapid aminoglycoside NP resultc | |||
|---|---|---|---|---|---|---|---|
| Amikacin |
Gentamicin |
||||||
| MIC (μg/ml) | Phenotype | MIC | Phenotype | ||||
| AG susceptible | |||||||
| FR136 | Escherichia coli ATCC 25922 | 2 | S | 0.5 | S | − | |
| R2278 | Escherichia coli ATCC 35218 | 4 | S | 1 | S | − | |
| R1436 | Escherichia coli | 2 | S | 1 | S | − | |
| R1438 | Escherichia coli | 2 | S | 1 | S | − | |
| R347 | Klebsiella pneumoniae | 4 | S | 0.5 | S | − | |
| S155 | Klebsiella pneumoniae | 2 | S | 0.5 | S | − | |
| R1433 | Enterobacter cloacae | 4 | S | 2 | S | − | |
| R713 | Enterobacter cloacae | 4 | S | 0.5 | S | − | |
| HM | Enterobacter cloacae | 4 | S | 2 | S | − | |
| R2260 | Proteus mirabilis | 4 | S | 4 | S | − | |
| AG-resistant producers of methylases | |||||||
| R989 | Escherichia coli | armA | >256 | R | >256 | R | + |
| R1012 | Escherichia coli | armA | >256 | R | >256 | R | + |
| R500 | Klebsiella pneumoniae | armA | >256 | R | >256 | R | + |
| R108 | Klebsiella pneumoniae | armA | >256 | R | >256 | R | + |
| R2257 | Klebsiella pneumoniae | armA | >256 | R | >256 | R | + |
| R2258 | Enterobacter cloacae | armA | >256 | R | >256 | R | + |
| 62Carb | Escherichia coli | rmtB | >256 | R | >256 | R | + |
| E28 | Escherichia coli | rmtB | >256 | R | >256 | R | + |
| R107 | Escherichia coli | rmtB | >256 | R | >256 | R | + |
| R990 | Escherichia coli | rmtB | >256 | R | >256 | R | + |
| R262 | Escherichia coli | rmtC | >256 | R | >256 | R | + |
| R451 | Escherichia coli | rmtC | >256 | R | >256 | R | + |
| R265 | Providencia stuartii | rmtC | >256 | R | >256 | R | + |
| R38 | Providencia stuartii | rmtC | >256 | R | >256 | R | + |
| N34 | Klebsiella pneumoniae | rmtC | >256 | R | >256 | R | + |
| R502 | Klebsiella pneumoniae | rmtF | >256 | R | >256 | R | + |
| R2160 | Klebsiella pneumoniae | rmtG | >256 | R | >256 | R | + |
| R2161 | Escherichia coli TOP10 | npmA | 128 | R | 32 | R | + |
| AG-resistant producers of AG-modifying enzymes | |||||||
| pWP701 | Escherichia coli | AAC(3)-IV | 1 | S | 128 | R | − |
| pFCT4392 | Escherichia coli | AAC(3)-Ia | 1 | S | 32 | R | − |
| 390 | Escherichia coli | AAC(3)-IV | 16 | S | >128 | R | − |
| 22089 | Escherichia coli | AAC(3)-I, AAC(3)-V | 16 | S | 128 | R | − |
| 97 | Escherichia coli | AAC(3)-IV | 2 | S | 32 | R | − |
| 92/31 | Escherichia coli | AAC(3)-IV | 4 | S | 16 | R | − |
| 4000 | Escherichia coli | APH(3′)-I, AAC(3)-V | 4 | S | 64 | R | − |
| pFCT1163 | Escherichia coli | AAC(6′)-Ia | >128 | R | 4 | S | − |
| 1054 | Escherichia coli | AAC(6′)-I | 8 | S | 8 | I | − |
| 1488 | Escherichia coli | AAC(6′)-II | 4 | S | 16 | R | − |
| 1054 | Escherichia coli | AAC(6′)-I | 16 | S | 8 | I | − |
| RP4 | Escherichia coli | APH(3′)-Ib | 1 | S | 1 | S | − |
| 122971 | Escherichia coli | ANT(2″) | 4 | S | >128 | R | − |
| 1800 | Klebsiella pneumoniae | AAC(6′)-I + AAC(3)-I | 32 | I | 16 | R | − |
| 13000 | Klebsiella pneumoniae | APH(3′)-I, AAC(3)-V | 0.5 | S | >128 | R | − |
| 22233 | Klebsiella pneumoniae | AAC(3)-V | 16 | S | >128 | R | − |
| 1110 | Enterobacter cloacae | ANT(2″) | 1 | S | 32 | R | − |
| BM2667 | Providencia stuartii | APH(3′)-VI, AAC(2′) | 64 | R | 16 | R | − |
| 531 | Providencia rettgeri | AAC(2′) | 16 | S | 4 | S | − |
| 4290 | Serratia marcescens | AAC(3)-I | 4 | S | 64 | R | − |
AAC, N-acetyltransferases; ANT, O-adenyltransferases, APH, O-phosphotransferases.
MICs were determined by broth microdilution method. S, susceptible; R, resistant; I, intermediate.
Results of the rapid aminoglycoside NP test after 2 h of incubation at 35 ± 2°C under ambient air.
A good correlation was obtained between multiple AG resistance and positivity of the test and, conversely, non-multiple AG resistance and negativity of the test (Table 1). The sensitivity and the specificity of the test were 100% and 97%, respectively, compared with broth dilution, taken as the gold standard for determining susceptibility and resistance to AG.
The reading of the color change in the wells every hour during 4 h revealed that final results were obtained within 2 h after inoculation for positive and negative strains when the tray was incubated at 35 ± 2°C under an ambient atmosphere. The readings of the test should not be extended after 4 h of incubation. After 4 h, the dye reagent is not stable enough, and inconsistent color shifts were observed under our experimental incubation conditions. The required test incubation time was therefore set to 2 h. AG susceptibility results obtained with the rapid aminoglycoside NP test consequently were obtained at least 16 h sooner than with the disk diffusion method, Etest system, and broth dilution method.
Solutions containing other combinations of AG or other AG molecules (kanamycin, tobramycin, streptomycin, and apramycin) were compatible with the test in its current formulation. The test may be performed from cultured bacteria grown on media such as Luria-Bertani, Mueller-Hinton, Uriselect-4, eosin-methylene blue, blood agar, and chocolate agar. Interference may be observed with colonies grown on acidifying culture media, such as Drigalski, MacConkey, and bromocresol purple. The test, as proposed here, has been optimized, and other conditions, reagents, and culture media might lead to a decrease of efficiency or stability of the test.
The study has two main limitations. First, the positivity of the test does not always superimpose the presence of 16S rRNA methylases. Indeed, combined production of nucleotidyl-, phosphoryl-, and acetyltransferases in a given enterobacterial strain might confer resistance to both gentamicin and amikacin, although such combinations, with MICs of >30 μg/ml, are rare. Second, this test is not compatible with nonfermenters, such as Pseudomonas aeruginosa and Acinetobacter baumannii, which may also produce 16S rRNA methylases (8, 9).
The rapid aminoglycoside NP test can be implemented for determination of resistance to multiple AG, in particular in countries facing an epidemic spread of carbapenemase producers (1). The use of the rapid aminoglycoside NP test may also contribute to rapidly identifying carriers of multidrug-resistant (MDR) isolates producing plasmid-mediated 16S rRNA methylases, to rapidly implement infection control measures, and consequently to control their spread. In a world that is rapidly witnessing an evolution toward pandrug resistance, the rapid aminoglycoside test may guide prescription of novel broad-spectrum AG, such as plazomicin, by differentiating aminoglycoside-modifying enzymes and methylase producers. Plazomicin is indeed active against aminoglycoside-modifying enzymes, such as the nucleotidyl, acetyl, and phosphoryl enzymes (10), although plazomicin is inactive against those 16S rRNA methylase producers (11). This test may also support the development of such novel AG molecules, facilitating patient enrollment in pivotal clinical trials.
Conclusions.
The rapid aminoglycoside NP test is easy to perform, rapid (<2 h), sensitive, and specific. It detects resistance to multiple AG among Enterobacteriaceae from selective and nonselective media prior to obtaining any antibiotic susceptibility testing results.
MATERIALS AND METHODS
Isolate collection.
A total of 48 isolates from clinical samples of worldwide origin were used to evaluate the performance of the rapid aminoglycoside NP (Nordmann/Poirel) test, including 28 isolates resistant to AG and 10 isolates susceptible to AG (Table 1). The AG-resistant isolates were characterized at the molecular level for their resistance mechanisms, with 18 isolates harboring 16S rRNA methylases and 20 others producing different AG-modifying enzymes (acetyl, adenyl, and phosphoryl enzymes) (Table 1).
MIC determination.
MICs of amikacin and gentamicin were determined using the broth microdilution method in cation-adjusted Mueller-Hinton broth (MHB-CA), as recommended by the Clinical and Laboratory Standards Institute (CLSI) guidelines (12). Amikacin and gentamicin sulfate (Fluorochem, Derbyshire, United Kingdom) were tested over a range of dilutions from 0.25 to 256 μg/ml. A final inoculum of 5 × 105 CFU/ml of each strain was distributed in the 96-well tray (Sarstedt, Nümbrecht, Germany). All experiments were repeated in triplicate, and Escherichia coli ATCC 25922 was used as a quality control. According to the CLSI breakpoints (13), enterobacterial isolates with gentamicin MICs of ≤4 and amikacin MICs of ≤16 μg/ml were categorized as susceptible, while those with gentamicin MICs of ≥16 and amikacin MICs of ≥64 μg/ml were categorized as resistant.
Rapid aminoglycoside NP test. (i) Reagents and solutions.
(a) Stock solutions of AG. Amikacin and gentamicin were diluted into MHB-CA medium in glass tubes to obtain stock solutions at a concentration of 1.6 mg/ml each. The antibiotic powders and the stock solutions can be stored at 4°C and −20°C, respectively, before their use.
(b) Rapid aminoglycoside NP solution. To prepare 250 ml of the rapid aminoglycoside NP solution, the culture medium and the pH indicator were mixed in a glass bottle with the following ingredients: 6.25 g of MHB-CA powder, 0.0125 g of phenol red (Sigma-Aldrich), and 225 ml of distilled water. The pH of the solution was adjusted to 6.7 by adding drops of 1 M HCl. This solution was then autoclaved at 121°C for 15 min. After cooling the solution to room temperature, 25 ml of 10% d-(+)-glucose anhydre (Roth, Karlsruhe, Germany), sterilized by filtration, was added. The final concentrations in the solution were 2.5% MHB-CA powder, 0.005% phenol red indicator, and 1% d-(+)-glucose. This solution can be kept in 10-ml aliquots at 4°C for 1 week or at −20°C for 1 year. The thawed 10-ml aliquots shall be prewarmed for 10 min at 37°C before being used to prevent bacterial growth delay, resulting in a delay for obtaining the color change of the test.
(c) AG-containing rapid aminoglycoside NP solution. Amikacin and gentamicin stock solution was added to the rapid aminoglycoside NP solution, before use, to a final concentration of 40 μg/ml each.
(ii) Bacterial inoculum.
A standardized enterobacterial inoculum was prepared using freshly obtained (overnight) bacterial colonies grown on Luria-Bertani or Mueller-Hinton plates. The bacterial colonies were resuspended in sterile 0.85% NaCl to obtain a 3 to 3.5 McFarland optical density (ca. 109 CFU/ml) using a DEN-1B McFarland densitometer from Grant Instruments Ltd., England. A bacterial suspension was prepared for each isolate to be tested and for the susceptible and resistant isolates used as controls (Table 1). E. coli ATCC 25922 (FR136) was used as the negative control, and an ArmA-producing E. coli strain from our collection (R989) was used as the positive control. The bacterial suspensions should be used to a maximum of 60 min after their preparation.
(iii) Tray inoculation.
The tray inoculation test was performed in a 96-well polystyrene microtest plate (round base, with lid, sterile; reference 82.1582.001; Sarstedt, Nümbrecht, Germany). For each isolate, two wells were inoculated in parallel with the bacterial suspension with and without the AG mix, respectively. The rapid aminoglycoside NP test was performed with the following steps (Fig. 1): (i) 150 μl of AG-free rapid aminoglycoside NP solution was transferred in the four wells of the first row; (ii) 150 μl of rapid aminoglycoside NP solution supplemented with AG mix was transferred in the four wells of the second row; (iii) 50 μl of sterile 0.85% NaCl was added to the two wells of the first column; (iv) 50 μl of the AG-susceptible isolate suspension used as a negative control (E. coli ATCC 25922) was added to the two wells of the second column; (v) 50 μl of the AG-resistant isolate suspension used as positive control (16S rRNA methylase producer) was added to the two wells of the third column; (vi) 50 μl of the tested isolate suspension was added to the two wells of the last column.
FIG 1.
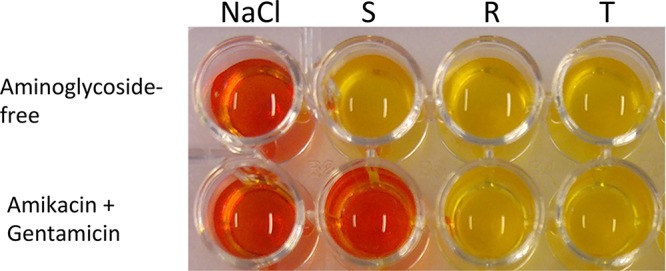
Representative results of the rapid aminoglycoside NP test. The rapid aminoglycoside NP test was performed with a reference susceptible isolate (S) in the second column and with a reference 16S rRNA methylase isolate (ArmA) (R) in the third column in a reaction medium without (upper) and with (lower) the AG mix (amikacin plus gentamicin). Noninoculated wells are shown in the first column (NaCl-containing wells). The tested isolate (T), in the fourth column, grew in the presence of the mix of aminoglycosides and therefore presented resistance to multiple aminoglycosides. The picture was taken after 2 h of incubation.
After mixing the bacterial suspension with the reactive medium by pipetting up and down, the final concentration of bacteria was ca. 108 CFU/ml in each well, and the final concentrations of amikacin and gentamicin were 30 μg/ml each.
(iv) Tray incubation.
The inoculated tray was incubated up to 4 h at 35 ± 2°C in ambient air, left unsealed and without agitation. Leaving the tray unsealed was done due to the oxygen requirement for glucose metabolism of Enterobacteriaceae.
(v) Tray reading.
Visual inspection of the tray was made after 10 min (to check for any spontaneous color change of the medium) and every hour up to 4 h. The test was considered positive (AG resistant) if the isolate grew in the presence of the AG mix, while it was considered negative (AG susceptibility) if the susceptible isolate did not grow in the presence of the AG mix. Cell growth was considered positive when the color of the assay reagent turned from orange to yellow. The color change indicates an acidification of the medium due to glucose metabolism that is used as a proxy for cell growth. Final reading was done when an assay of the second row turned yellow (positive) or at 4 h in case of no color change (negative). The test was considered interpretable when (i) NaCl wells remained orange, meaning the absence of medium contamination, (ii) susceptible and resistant wells in first row turned from orange to yellow, confirming the metabolism of glucose by the isolates, and (iii) the susceptible well in row 2 remained orange (no growth) and the resistant well in row 2 turned yellow (cell growth and glucose metabolism positive).
Other tested conditions.
The rapid aminoglycoside NP test was also performed by testing other concentrations of the same mix of AG molecules or another AG mix, such as tobramycin and kanamycin or amikacin and kanamycin. The test also was performed from overnight culture on different agar plates to determine the potential impact of the culture medium prior to the realization of the test. The following culture media were tested: nonselective culture medium, such as Columbia agar plus 5% sheep blood (bioMérieux), the nonselective chromogenic medium UriSelect 4 (Bio-Rad), chocolate agar plus PolyVitex (bioMérieux), eosin methylene blue agar (Sigma-Aldrich), Drigalski agar (Bio-Rad), MacConkey agar (VWR BDH Prolabo, Leuven, Belgium), and bromocresol purple (bioMérieux).
ACKNOWLEDGMENTS
This work was financed by the University of Fribourg, Switzerland.
We are grateful to T. Lambert for providing us with reference strains producing various AG-modifying enzymes.
A patent corresponding to the rapid aminoglycoside NP test has been submitted on behalf of the University of Fribourg (application number EP16194477.2).
REFERENCES
- 1.Nordmann P, Naas T, Poirel L. 2011. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 17:1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Falagas ME, Karageorgopoulos DE, Nordmann P. 2011. Therapeutic options for infections with Enterobacteriaceae producing carbapenem-hydrolyzing enzymes. Future Microbiol 6:653–666. doi: 10.2217/fmb.11.49. [DOI] [PubMed] [Google Scholar]
- 3.Ramirez MS, Tolmasky ME. 2010. Aminoglycoside modifying enzymes. Drug Resist Updat 13:151–171. doi: 10.1016/j.drup.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doi Y, Wachino J, Arakawa Y. 2016. Aminoglycoside resistance: the emergence of acquired 16S ribosomal RNA methyltransferases. Infect Dis Clin North Am 30:523–537. doi: 10.1016/j.idc.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bercot B, Poirel L, Nordmann P. 2011. Updated multiplex polymerase chain reaction for detection of 16S rRNA methylases: high prevalence among NDM-1 producers. Diagn Microbiol Infect Dis 71:442–445. doi: 10.1016/j.diagmicrobio.2011.08.016. [DOI] [PubMed] [Google Scholar]
- 6.Wachino J, Shibayama K, Kurokawa H, Kimura K, Yamane K, Suzuki S, Shibata N, Ike Y, Arakawa Y. 2007. Novel plasmid-mediated 16S rRNA m1A1408 methyltransferase, NpmA, found in a clinically isolated Escherichia coli strain resistant to structurally diverse aminoglycosides. Antimicrob Agents Chemother 51:4401–4409. doi: 10.1128/AAC.00926-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hugh R, Leifson E. 1953. The taxonomic significance of fermentative versus oxidative metabolism of carbohydrates by various gram negative bacteria. J Bacteriol 66:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rahman M, Prasad KN, Pathak A, Pati BK, Singh A, Ovejero CM, Ahmad S, Gonzalez-Zorn B. 2015. RmtC and RmtF 16S rRNA methyltransferase in NDM-1-producing Pseudomonas aeruginosa. Emerg Infect Dis 21:2059–2062. doi: 10.3201/eid2111.150271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou Y, Yu H, Guo Q, Xu X, Ye X, Wu S, Guo Y, Wang M. 2010. Distribution of 16S rRNA methylases among different species of Gram-negative bacilli with high-level resistance to aminoglycosides. Eur J Clin Microbiol Infect Dis 29:1349–1353. doi: 10.1007/s10096-010-1004-1. [DOI] [PubMed] [Google Scholar]
- 10.Haidar G, Alkroud A, Cheng S, Churilla TM, Churilla BM, Shields RK, Doi Y, Clancy CJ, Nguyen MH. 2016. Association between the presence of aminoglycoside-modifying enzymes and in vitro activity of gentamicin, tobramycin, amikacin, and plazomicin against Klebsiella pneumoniae carbapenemase- and extended-spectrum-beta-lactamase-producing Enterobacter species. Antimicrob Agents Chemother 60:5208–5214. doi: 10.1128/AAC.00869-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhanel GG, Lawson CD, Zelenitsky S, Findlay B, Schweizer F, Adam H, Walkty A, Rubinstein E, Gin AS, Hoban DJ, Lynch JP, Karlowsky JA. 2012. Comparison of the next-generation aminoglycoside plazomicin to gentamicin, tobramycin and amikacin. Expert Rev Anti Infect Ther 10:459–473. doi: 10.1586/eri.12.25. [DOI] [PubMed] [Google Scholar]
- 12.Clinical and Laboratory Standard Institute. 2012. Methods for dilution of antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard, 9th ed Document M07-A9. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 13.Clinical and Laboratory Standard Institute. 2014. Performance standards for antimicrobial susceptibility testing, 24th informational supplement. Document M100-S24. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]


