ABSTRACT
Candida guilliermondii was isolated from sterile specimens with increasing frequency over a several-month period despite a paucity of clinical evidence suggesting true Candida infections. However, a health care-associated outbreak was strongly considered due to growth patterns in the microbiology laboratory that were more consistent with true infection than environmental contamination. Therefore, an extensive investigation was performed to identify its cause. With the exception of one case, patient clinical courses were not consistent with true invasive fungal infections. Furthermore, no epidemiologic link between patients was identified. Rather, extensive environmental sampling revealed C. guilliermondii in an anaerobic holding jar in the clinical microbiology laboratory, where anaerobic plates were prereduced and held before inoculating specimens. C. guilliermondii grows poorly under anaerobic conditions. Thus, we postulate that anaerobic plates became intermittently contaminated. Passaging from intermittently contaminated anaerobic plates to primary quadrants of aerobic media during specimen planting yielded a colonial growth pattern typical for true specimen infection, thus obscuring laboratory contamination. A molecular evaluation of the C. guilliermondii isolates confirmed a common source for pseudo-outbreak cases but not for the one true infection. In line with Reason's model of organizational accidents, active and latent errors coincided to contribute to the pseudo-outbreak. These included organism factors (lack of growth in anaerobic conditions obscuring plate contamination), human factors (lack of strict adherence to plating order, leading to only intermittent observation of aerobic plate positivity), and laboratory factors (novel equipment). All of these variables should be considered when evaluating possible laboratory-based pseudo-outbreaks.
KEYWORDS: Candida, pseudo-outbreak, anaerobic, contamination, epidemiology, guilliermondii, optical mapping, pulsed-field gel electrophoresis, swan neck
INTRODUCTION
The repeated isolation of rare organisms may indicate either a true cluster of health care-associated infections (HAIs) or a pseudo-outbreak. Pseudo-outbreaks result from specimen contamination during or after collection. Maki (1) and Weinstein and Stamm (2) suggest that pseudo-outbreaks should be considered when clusters of positive sterile cultures with an unusual organism and without compatible associated clinical symptoms and/or without an obvious endogenous source are identified. Pseudo-outbreaks can lead to adverse outcomes related to unnecessary antimicrobial use and inappropriate treatment (1).
Many organisms are ubiquitous in the environment and can directly contaminate microbiologic media. Practically, such contamination is usually readily identified based on the growth pattern. Because standard plating practices lead to successive organism dilution, in true infections, the largest number of colonies appears in the first streaked quadrant, followed by less growth successively in the second, third, and fourth quadrants. In contrast, contaminated specimens typically have no dilution effect, and growth is often seen on plate edges with no relationship to the specimen inoculation site or streak lines. If these typical patterns are not found, contamination can be masked, leading to confusion with true infection.
On 2 September 2012, the clinical microbiology laboratory notified the Division of Infection Control/Hospital Epidemiology (IC/HE) of five patients in whom Candida guilliermondii (teleomorph Meyerozyma guilliermondii) was isolated from sterile specimens during a 24-hour period. Although clinical infections caused by non-albicans Candida are increasing, C. guilliermondii accounts for only 1% of all Candida infections, raising the question of whether these isolates represented a true health care-associated outbreak or a pseudo-outbreak (3). Microbiology laboratory review of culture plate growth patterns was strongly suggestive of true infection. Thus, extensive epidemiologic, microbiologic, and molecular investigations into potential sources of the cluster were initiated. Here, we describe the investigations and characterize the identification of a novel source of laboratory contamination.
RESULTS
Epidemiologic investigation.
Twenty-five C. guilliermondii case patients (36 specimens) were identified; the first identified case occurred on 21 April 2012. No additional cases were identified until June. An epidemic curve for the outbreak is shown in Fig. 1A. Figure 1B shows the distribution of C. guilliermondii isolated at our institution from 1998 to 2012; no cases were detected during the 2-year period immediately preceding this outbreak. Prior to 2010, C. guilliermondii was identified, on average, one to two times per year. The majority of preoutbreak isolates were from blood cultures, followed by nail isolates. Isolates during the outbreak period were primarily from procedural specimens, with the exception of blood culture specimens from a single patient.
FIG 1.
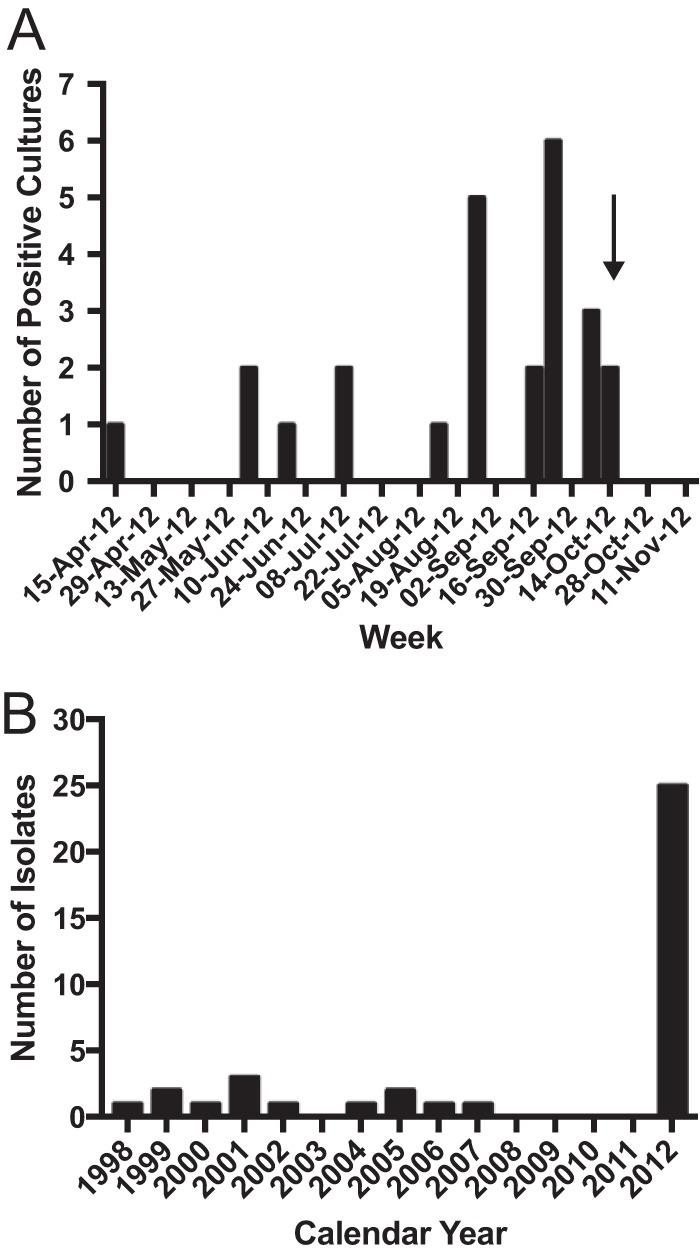
C. guilliermondii isolation over time at our institution. (A) Epidemic curve of C. guilliermondii cases by week during pseudo-outbreak period. Arrow demarcates end of outbreak period after which no additional cases were identified. (B) Historical isolation of C. guilliermondii by year.
A manual review of case patient records suggested one true C. guilliermondii infection in September in the middle of the peak of the pseudo-outbreak (the single blood culture isolate); the clinical courses of other affected patients were not consistent with a true Candida infection (Table 1). Specifically, the only patient who appeared to have a true infection was severely immunocompromised due to multiple myeloma and Crohn's disease. During the course of her hospitalization, she developed severe sepsis with five blood culture sets showing budding yeast identified as C. guilliermondii from Gram stain morphologies and culture growth. The first two sets were drawn through a central venous catheter and also grew Candida glabrata. The last three sets were obtained by venipuncture and grew C. guilliermondii in aerobic bottles only. The patient subsequently died. Autopsy cultures from the liver and lung were positive for both C. guilliermondii and C. neoformans.
TABLE 1.
Brief clinical courses of patients with cultures positive for C. guilliermondii during the pseudo-outbreak period
| Patient | Date (mo/day/yr) | Clinical history | Specimen type | Procedurea | Pathologyb | Procedure within 90 d prior to positive culture (mo/day/yr)a | Antifungal therapy |
|---|---|---|---|---|---|---|---|
| 1 | 4/21/2012 | Ascites | Peritoneal fluid | Paracentesis | N/A | Paracentesis (outside facility, 4/16/2016) | Micafungin |
| 2 | 6/8/2012 | Ascites | Peritoneal fluid | Paracentesis | N/A | Paracentesis (5/9/2012) | None |
| 3 | 6/8/2012, 8/22/2012 | Ascites | Peritoneal fluid | Paracentesis (performed by interventional radiology) | N/A | None | None after first positive culture; voriconazole after second |
| 4 | 6/22/2012 | Abscess | Abscess fluid | Aspiration of abdominal abscess (US guided) | N/A | None | None |
| 5 | 7/10/2012 | Knee pain, total joint replacement | Joint fluid | Right knee revision, total knee arthroplasty | Special stains for fungi negative | Fluoroscopy-guided right knee aspiration (4/24/2912) | None |
| 6 | 7/10/2012 | Shoulder pain | Bone | Bone biopsy (CT guided) | N/A | Right shoulder cortisone injection at outside facility, date unknown | None |
| 7 | 8/15/2012 | Pulmonary nodules | Lung tissue | Transbronchial biopsy (bronchoscopy) | Special stains for fungi negative | Electrophysiology study (8/7/2012) | None |
| 8 | 8/27/2012 | Pericardial effusion | Pericardial tissue | Pericardial window | GMS stain negative | Biopsy of hypopharyngeal mass and femur fracture repair (7/30/2012) | Micafungin |
| 9 | 8/27/2012 | Ascites | Peritoneal fluid | Paracentesis | N/A | Paracentesis (outside facility, 8/20/2012) | None |
| 10 | 8/28/2012 | Pericardial effusion | Pericardial fluid | Pericardial window | N/A | Pericardiocentesis (8/21/2012) | Voriconazole |
| 11 | 8/28/2012 | Wrist pain | Synovial tissue | Left wrist synovectomy | GMS stain negative | None | Fluconazole |
| 12 | 8/29/2012 | Hip pain and total joint replacement | Bone | Resection arthroplasty | Not performed | Drainage of right hip hematoma (8/22/2012) and right hip total arthroplasty (8/1/2012) | Fluconazole |
| 13 | 9/17/2012 | Pleural adhesions | Pleural tissue | Video-assisted thoracoscopy with exploration and washout | Special stains for fungi negative | Biopsy of mediastinal mass (6/21/2012), radical thymectomy (8/6/2012) | None |
| 14 | 9/18/2012 | Sepsis | Blood | None | N/A | Arterial line insertion (8/25/2012), peripherally inserted central catheter (7/16/2012) | Micafungin |
| 15 | 9/23/2012 | Ascites | Peritoneal fluid | Paracentesis (US guided) | N/A | Multiple paracenteses | None |
| 16 | 9/23/2012 | Sepsis, rash | Skin | Punch biopsy | N/A | None | None |
| 17 | 9/24/2012 | Hip pain and total joint replacement | Joint fluid aspirate | Left hip revision arthroplasty | N/A | Left hip aspiration (interventional radiology, 8/24/2012) | None |
| 18 | 9/26/2012 | Pleural effusion | Pleural fluid | Thoracentesis | N/A | Tunneled peripherally inserted central catheter (7/16/2012) | Voriconazole |
| 19 | 9/28/2012 | Ascites | Peritoneal fluid | Aspiration of abdominal fluid collection (US guided) | N/A | Pancreatic debridement (8/16/2012), exploratory laparotomy (9/12/2012) | Micafungin |
| 20 | 9/28/2012 | Mediastinal adenopathy | Bronchial tissue | Bronchoscopy | Special stains for fungi negative | Bone biopsy (CT-guided, 7/31/2012) | None |
| 21 | 10/9/2012 | Mediastinal adenopathy | Bronchial tissue | Bronchoscopy | Special stains for fungi negative | None | None |
| 22 | 10/11/2012 | Skin lesions | Skin | Punch biopsy | Special stains for fungi negative | None | None |
| 23 | 10/11/2012 | Mediastinal adenopathy | Bronchial tissue | Bronchoscopy | Special stains for fungi negative | None | None |
| 24 | 10/14/2012 | Ascites | Peritoneal fluid | Paracentesis | N/A | Multiple prior paracenteses (US guided), embolization procedure | None |
| 25 | 10/14/2012 | Pleural effusion | Pleural fluid | Thoracentesis | N/A | Pleurex catheter placement (US guided, 10/5/2012), video-assisted thoracic surgery (8/10/2012) | None |
CT, computed tomography; US, ultrasound.
N/A, not applicable; GMS, Gomori methenamine silver.
All other case patients were either clinically stable or demonstrated improvement despite limited or no directed antifungal therapy and thus were deemed unlikely to have a true clinical Candida infection. Eight patients received antifungal therapy. Four of seven patients with C. guilliermondii isolated from peritoneal fluid were not treated; an additional patient was treated after a second positive culture was reported, despite a benign course during the 2-month period between the positive cultures. Another patient had positive pericardial and pleural fluid cultures after a pericardial window, without significant associated morbidity. One patient with obvious clinical morbidity had an infected hip with 18 joint fluid and tissue cultures taken over the course of 4 days. However, although four of 18 cultures were positive for C. guilliermondii, almost all were positive for Proteus mirabilis, presumably the true etiologic agent of the joint infection.
In addition, no associations were found between a sterile site culture positive for C. guilliermondii and patient demographics, comorbidities, procedure types, or clinical staff members caring for the patient (Table 1). Nine (36%) case patients were male and the mean age was 59 years (range, 24 to 88). Case patients were admitted to a variety of services, including hospital medicine, oncology, cardiology, orthopedics, and general surgery. C. guilliermondii was isolated from twenty-four (96%) patients after unrelated procedures that were performed in diverse locations throughout the medical center (east campus, 28%; west campus, 56%; outpatients, 16%). The affected procedures included bedside procedures (8 procedures), operating room surgical procedures (5 procedures), interventional radiology procedures (5 procedures), bronchoscopy (4 procedures), dermatology biopsy (1 procedure), and interventional cardiology procedures (1 procedure).
The only common exposure identified was the receipt of lidocaine injections during procedures (24/25 patients [96%]). However, the vials of lidocaine were ruled out as the source on the basis of several factors. First, the lidocaine came from 11 different types of kits, including temporary central venous catheter insertion, peripherally inserted central venous catheter insertion, paracentesis, thoracentesis, arthrocentesis, and lumbar puncture kits. Second, multiple lots of lidocaine from different suppliers were used. Third, sampling from lidocaine vials used for sterile procedures and from single and multidose vials used throughout the institution was undertaken, and all cultures were sterile. Institutional findings were supported by communications with the Massachusetts Department of Public Health and the Centers for Disease Control and Prevention, who reported no other isolation of C. guilliermondii from lidocaine vials.
Finally, procedures during which sterile fluid was obtained were observed by IC/HE staff. No breaks in sterile technique were noted. Environmental cultures were obtained during and immediately following procedures, including from the automated pharmacy dispensing system that contained additional lidocaine vials for use, and all were negative for C. guilliermondii. Thus, the clinical epidemiologic investigation did not support a true outbreak.
Clinical microbiology laboratory investigation.
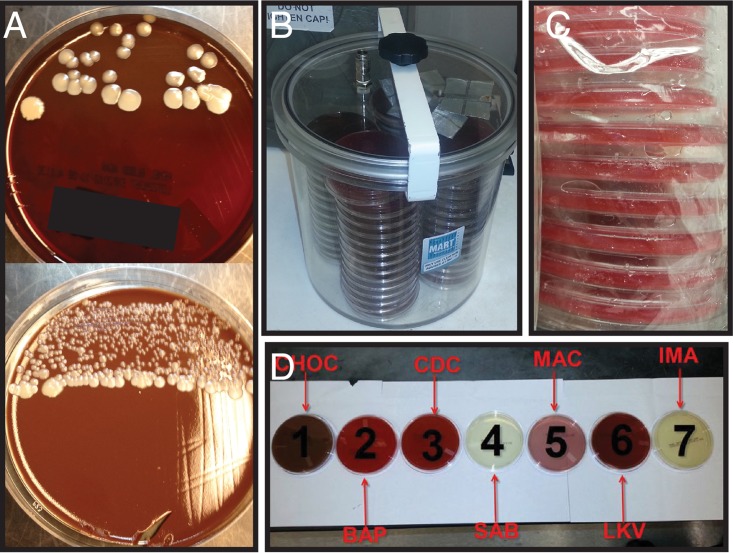
The growth patterns on culture plates were examined and initially found to be consistent with true infection rather than environmental contamination (Fig. 2A). Growth was always observed in the first inoculation quadrant, was occasionally also observed in the secondary quadrant, and was never observed outside streak lines. Culture growth was observed most frequently on blood and chocolate aerobic plates, was occasionally observed on Sabouraud dextrose and inhibitory mold agar when fungal cultures were planted, and was never observed on anaerobic culture plates. However, of note, Gram staining of primary specimens was consistently negative for yeast forms. Positive specimens were transported by either courier or pneumatic tubes from two different campuses and received in different types of transport tubes, including sterile cups, syringes, and swabs. They were processed in two biosafety cabinets on different shifts by a large variety of technologists.
FIG 2.
Laboratory investigation of C. guilliermondii outbreak. (A) Examples of C. guilliermondii growing in primary streak areas from two patient samples. (B) Anaerobic holding jar. (C) Example of excess moisture in unopened anaerobic media packages. (D) Proper plating order, beginning on the left, with the most-enriched medium and progressing to the most selective on the right (CHOC, chocolate agar; BAP, sheep blood agar; CDC, CDC anaerobic agar; SAB, Sabouraud dextrose agar; MAC, lactose MacConkey agar; LKV, laked-blood kanamycin vancomycin agar; IMA, inhibitory mold agar).
Multiple environmental cultures (n = 225) were obtained from the clinical microbiology laboratory for evaluating any area potentially in contact with specimens and/or technologists planting specimens. These included transfer pipettes, cotton swabs, plastic loops, flammable loop handles, incubators, biosafety cabinets, cold rooms, floors, countertops, the front specimen receipt desk, work cards, air vents, phones, gloves, media, door handles, plate racks, and surfaces where plates were stored. C. guilliermondii was isolated from four swabs obtained from the interior side, bottom, and rim of the anaerobic media holding jar (Fig. 2B). The identification of representative environmental and cluster isolates as C. guilliermondii was confirmed by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry.
The anaerobic holding jar used in our laboratory is part of the Advanced Anoxomat (Advanced Instruments, Inc., Norwood, MA). Plates were held in the anaerobic holding jar prior to their use in setting up anaerobic cultures. The Anoxomat system is a microprocessor-controlled instrument for rapidly establishing anaerobic environments inside airtight acrylic chambers. It achieves anaerobiosis by rapidly evacuating and injecting a gas mixture of 5% H2, 10% CO2, and 85% N2 in three separate cycles. A pouch is present in the chamber lid to hold palladium catalyst to aid the consumption of oxygen present in the chamber. Cultures of baked catalyst in line for future use were all negative. Cultures from gas line ports that connect the anaerobic gas cylinder to the Anoxomat and the Anoxomat to the anaerobic holding jar and additional Anoxomat anaerobic chambers used to incubate anaerobic plate cultures after specimen inoculation were all negative.
On the basis of these findings, the procedures for anaerobic plate processing were further investigated. The standard laboratory procedure is to streak out samples on a defined hierarchical order of plates: blood, chocolate, CDC anaerobe sheep blood, Sabouraud dextrose, lactose MacConkey, laked blood kanamycin vancomycin (LKV), and inhibitory mold agar (Fig. 2D). Interviews with the microbiology technologists revealed that the recommended hierarchy was not rigorously followed during the cluster period. In addition, the same anaerobic holding jar was used continuously during the outbreak period, without a regular defined cleaning schedule.
Moisture condensate on the inside walls of the holding jar was noted upon inspection and was attributed to the excessive moisture present in anaerobic agar plates (Fig. 2C). During the Anoxomat evacuation and injection process, water droplets were observed moving on the inside of the lids of these excessively moist anaerobic agar plates. Technologists noted excessive moisture in unopened anaerobic CDC anaerobe sheep blood and LKV agar media (Remel, Lenexa, KS) as early as April 2012, roughly coincident with the first isolation of C. guilliermondii during the pseudo-outbreak. Uninoculated anaerobic media, which had not been prereduced in the holding jar, were sampled extensively by aerobic incubation and by swabbing the surface of media onto plates, and C. guilliermondii was never isolated.
Molecular investigation.
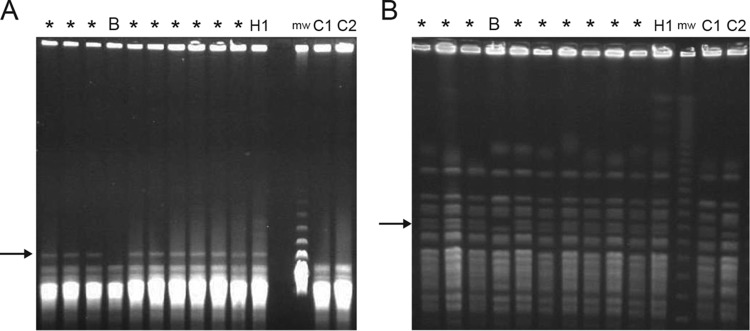
Pulsed-field gel electrophoresis was performed on representative environmental and patient samples. In BssHI, NotI, and SfiI restriction digests, isolates from procedural specimens and the anaerobic holding jar had identical banding patterns. The single blood culture isolate from the patient with a clinical course consistent with true infection had a different banding pattern, showing at least one band difference on BssHI-, NotI-, and SfiI-digested samples in comparison with the other pseudo-outbreak isolates (Fig. 3).
FIG 3.
Pulsed-field gel electrophoresis of pseudo-outbreak strains. (A) BssHI digest. (B) NotI digest. *, patient pseudo-outbreak isolates; B, the one blood culture outbreak isolate; H1, anaerobic holding jar isolate; mw, molecular weight marker; C1, C2, environmental isolates from the University of Iowa reference laboratory unrelated to outbreak isolates. Arrows highlight a single band difference between blood and patient/holding jar isolates. SfiI restriction digest is not shown.
Whole-genome mapping was then performed because of the increased resolution compared with traditional pulsed-field gel electrophoresis. Importantly, optical mapping patterns from holding jar isolates and procedural specimens were essentially identical. However, the blood culture isolate appeared to be distinct (Fig. 4). Specific and prominent differences between the blood culture isolate and a representative procedural isolate include an unaligned telomeric region in chromosome 1, a 7-kb deletion in chromosome 2, a 15-kb insertion in chromosome 4, and a 25-kb insertion in chromosome 8.
FIG 4.
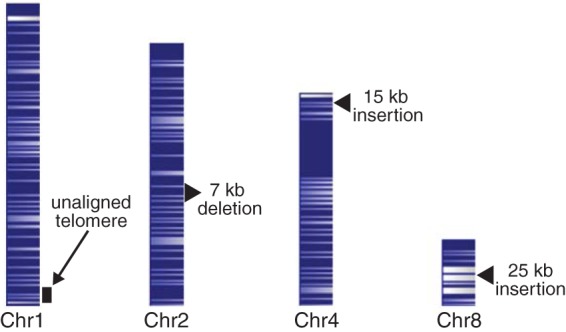
Whole-genome mapping. AflIII restriction maps of C. guilliermondii chromosomes from a representative pseudo-outbreak strain. Because of the high density of restriction sites and condensed nature of the chromosome maps, lines denoting restriction sites have merged into dark and light banding patterns corresponding to areas with greater or lesser restriction site density. Arrow and arrowheads indicate the differences found between the whole-genome map of the blood culture isolate and that of the whole-genome map pattern shared by the pseudo-outbreak and anaerobic holding jar strains.
Intervention and follow-up.
After the source of C. guilliermondii was identified, several steps were taken. First, the holding jar was thoroughly cleaned, and a daily disinfection protocol was implemented. Second, the importance of plating order was reinforced with technologists, and a pictorial representation of plating order was placed above each specimen-processing area as a reminder (Fig. 2D). Third, to prevent condensation from occurring inside the jar, suppliers of anaerobic media were changed to secure drier plates, and anaerobic plates were placed into an ambient air drying rack for several hours prior to prereduction. In the 18 months since these interventions, C. guilliermondii was not isolated from any additional patient specimens or environmental samples.
DISCUSSION
Pseudo-outbreaks caused by bacterial, fungal, and viral pathogens are well documented and have been traced to a variety of sources, including contaminated medical or laboratory equipment, tap water rinsing, use of nonsterile solutions in the microbiology laboratory, and specimen cross-contamination (4, 7–13). They have also been traced to highly contaminated environments, such as drinking water and ice machines (14, 15), which may introduce microorganisms into patients or clinical specimens during collection.
Candida pseudo-outbreaks are rare. Two pseudo-fungemias with C. guilliermondii were traced to the contaminated hands and nails of health care workers collecting blood cultures and to contaminated heparin used to flush butterfly needles prior to blood draws (16, 17). Three other pseudo-outbreaks were attributed to different Candida species: Candida versatilis associated with the supplementation of blood culture bottles with olive oil (18), Candida parapsilosis associated with the grinding of tissue in contaminated salt solution prior to plating (19), and C. parapsilosis associated with the manual aeration of an older blood culture system by a colonized technologist (20). There are no prior reports of pseudo-outbreaks definitively traced to anaerobic culture handling, although one report did suggest that a Clostridium sordellii pseudo-outbreak might be associated with the storage of prereduced plates in an anaerobic chamber (21).
Several simultaneous factors coincided to yield this pseudo-outbreak and to obscure the distinction between true infection and media contamination. In this sense, we invoke Reason's model of organizational accidents (22), with active errors (streaking plates out of order) and latent errors (handling of anaerobic plates and cleaning techniques) aligned for an adverse event to occur (23).
Specifically, we postulate that the forcible evacuation of the Anoxomat overcame the swan neck barrier of petri dishes, and forced aerosolized organisms into the interiors of the anaerobic plates. Normally, primary quadrants of different culture plate media are successively streaked with the same swab in a defined order during specimen planting (Fig. 2D). However, this order was not rigorously adhered to during the outbreak period. Therefore, organisms were inadvertently transferred sequentially during specimen planting from randomly contaminated anaerobic plates to the primary quadrants of other media. Because plates were only sometimes streaked out of order, the contamination of aerobic plates occurred only intermittently. C. guilliermondii does not grow anaerobically, obscuring the random localization of organisms on anaerobic media that otherwise would have facilitated the recognition of environmental contamination. The subsequent intermittent growth on aerobic medium in primary and also sometimes secondary quadrants simulated the pattern seen during true infections.
Additional contributing factors included the initial contamination of the surface of the holding jar coupled with the excessively moist anaerobic media used during the outbreak period, leading to condensation in the holding jar. This moisture presumably enhanced the growth of C. guilliermondii and the potential for aerosol dissemination during forcible evacuation and filling of the chamber by the Anoxomat. The potential for contamination was also enhanced by the lack of a scheduled disinfection protocol for the device. Of note, the manufacturer's operation manual specifies which types of disinfectant should be used, but does not provide guidance for the frequency of its application (24).
The unlikely occurrence during the height of the pseudo-outbreak of a rare true C. guilliermondii clinical infection with multiple positive blood and postmortem cultures also reinforced the initial concern that additional cases might represent a large outbreak of HAIs and led to a thorough epidemiologic investigation of all positive cultures. However, molecular diagnostics demonstrated that this one rare true infection was distinct from the clonal organisms isolated from the anaerobic holding jar and pseudo-outbreak specimens.
This pseudo-outbreak highlights the need for vigilance during the repeated isolation of an unusual organism. The late recognition of this unusual and unsuspected type of laboratory contamination led to unnecessary clinical workup, treatment, and follow-up and a large epidemiologic investigation, including input from public health agencies.
While several simultaneous events contributed to the pseudo-outbreak, other factors prompted its recognition. In particular, one astute microbiology technologist noted the first cluster of cases and reported to IC/HE the suspicion of laboratory contamination. Almost simultaneously, an infectious diseases consultant relayed the observation of serial cases lacking the clinical characteristics suggestive of deep Candida infection. In the microbiology laboratory, the lack of Gram stain positivity for yeast in all suspect cultures despite occasional moderate levels of culture growth also raised the concern about a pseudo-outbreak, rather than a true cluster of HAIs.
Our investigation also emphasizes several quality assurance considerations for clinical microbiology laboratories. First, the preemptive meticulous, regular, targeted, and documented disinfections of equipment and surfaces are prudent to minimize contamination risk related to any technology that alters airflow, such as the Anoxomat, because such technologies may inadvertently lead to unrecognized contamination through compromising swan neck barriers that normally keep media sterile. Second, stringent procedures for media acceptability must be followed (25). Third, a rigorous adherence to standard operating procedures, even for seemingly less-compelling practices, such as plating order, should be emphasized.
Altogether, these observations indicate how several deficiencies in practice and unrecognized technical vulnerabilities, perhaps minor by themselves, can align in a classic Reason's model of organizational accidents, also known as the “Swiss cheese” model, to lead to adverse events.
MATERIALS AND METHODS
Setting.
Beth Israel Deaconess Medical Center is a 672-bed academic tertiary care medical center in Boston, Massachusetts. The medical center includes east and west campuses situated approximately 0.25 miles apart. Patients are housed on both campuses, and procedures are performed at both sites.
Epidemiologic case definition and investigation.
An epidemiologic case was defined as any patient with a microbiologic culture from a sterile site (blood, body fluid, or tissue) that grew C. guilliermondii during the 6-month period before and 18 months after the outbreak identified on 2 September 2016. These dates were chosen to span a period significantly preceding and following the clinical cluster. The institution's microbiology database was queried for the 14-year period prior to the outbreak to identify baseline hospital incidence of C. guilliermondii-positive microbiologic cultures. An epidemic curve and a line list were then used to identify common exposures and guide further investigation during the time frame of the epidemiologic investigation. Additional investigations included: a manual review of all cases to determine the presence of true infection, an observation of common procedures by infection control staff, and an environmental culturing of key procedural areas and equipment in locations where C. guilliermondii-positive cultures had been collected. In the clinical microbiology laboratory, specimen processing was observed from the point of intake through the work-up of positive cultures. Standard methods of cleaning and quality assurance steps were reviewed in depth.
Microbiology procedures.
Standard culture practices for sterile source specimens (referred to here as “planting”) included plating on sheep blood, chocolate, and lactose MacConkey agar for aerobic cultures, CDC anaerobic agar and laked-blood kanamycin agar (LKV) plates for anaerobic cultures, and, for fungal cultures, when ordered, Sabouraud dextrose agar (SAB) and inhibitory mold agar (IMA). Per standard laboratory practice, planting involved lining up media plates in a HEPA-filtered biosafety cabinet (Fig. 2D), transferring a small volume of sample to each plate using a sterile disposable transfer pipette, and streaking the primary quadrant of each plate consecutively with a single cotton-tipped swab, moving from the least-selective to the most-selective medium. Secondary and tertiary quadrants were then streaked for each plate individually with a flame-sterilized metal loop.
C. guilliermondii was identified using the Vitek 2 yeast ID card (bioMérieux, Durham, NC). A subset of isolates was sent to the Mayo Clinic reference laboratories (Rochester, MN) for confirming the identification using MALDI-TOF (matrix-assisted laser desorption ionization–time of flight) mass spectrometry. Environmental cultures were plated on Sabouraud dextrose agar (SAB) (Remel, Lenexa, KS) and incubated at 30°C for 14 days.
Genome mapping.
Pulsed-field gel electrophoresis using standard techniques was performed at the University of Iowa reference laboratory (6). Whole-genome mapping was performed at OpGen (Gaithersburg, MD) as previously described (5). Specifically, yeast genomic DNA was adsorbed to a solid surface, digested with a six-base cutter, stained, and imaged. Restriction fragments were aligned with the eight large supercontigs of the C. guilliermondii ATCC 6260 reference genome sequence (26) using MapSolver (OpGen, Gaithersburg, MD). Each supercontig corresponded to a single chromosome numbered in decreasing order of size. Notably, whole-genome mapping detected a 400-kb repetitive region in supercontig 5 not previously resolved in the reference next-generation sequenced-based genome sequence. Therefore, to preserve the convention of numbering chromosomes according to their actual size, we reference the previously designated supercontig 5 sequence as chromosome 4, and supercontig 4 as chromosome 5.
Ethical considerations.
All studies were performed as part of normal quality assurance activities in our clinical microbiology and hospital infection control departments. Thus, the study was exempt from an institutional board review.
ACKNOWLEDGMENTS
We thank OpGen, Inc., for performing the whole-genome mapping experiments described in the paper at no cost.
REFERENCES
- 1.Maki DG. 1980. Through a glass darkly. Nosocomial pseudoepidemics and pseudobacteremias. Arch Intern Med 140:26–28. [PubMed] [Google Scholar]
- 2.Weinstein RA, Stamm WE. 1977. Pseudoepidemics in hospital. Lancet ii:862–864. doi: 10.1016/S0140-6736(77)90793-0. [DOI] [PubMed] [Google Scholar]
- 3.Pfaller MA, Diekema DJ, Mendez M, Kibbler C, Erzsebet P, Chang SC, Gibbs DL, Newell VA. 2006. Candida guilliermondii, an opportunistic fungal pathogen with decreased susceptibility to fluconazole: geographic and temporal trends from the ARTEMIS DISK antifungal surveillance program. J Clin Microbiol 44:3551–3556. doi: 10.1128/JCM.00865-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hallin M, Deplano A, Roisin S, Boyart V, De Ryck R, Nonhoff C, Byl B, Glupczynski Y, Denis O. 2012. Pseudo-outbreak of extremely drug-resistant pseudomonas aeruginosa urinary tract infections due to contamination of an automated urine analyzer. J Clin Microbiol 50:580–582. doi: 10.1128/JCM.06268-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clarridge JE III, Harrington AT, Roberts MC, Soge OO, Maquelin K. 2013. Impact of strain typing methods on assessment of relationship between paired nares and wound isolates of methicillin-resistant Staphylococcus aureus. J Clin Microbiol 51:224–231. doi: 10.1128/JCM.02423-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 33:2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cosgrove SE, Ristaino P, Caston-Gaa A, Fellerman DP, Nowakowski EF, Carroll KC, Perl TM, Maragakis LL. 2012. Caveat emptor: the role of suboptimal bronchoscope repair practices by a third-party vendor in a pseudo-outbreak of pseudomonas in bronchoalveolar lavage specimens. Infect Control Hosp Epidemiol 33:224–229. doi: 10.1086/664051. [DOI] [PubMed] [Google Scholar]
- 8.Balada-Llasat JM, Elkins C, Swyers L, Bannerman T, Pancholi P. 2010. Pseudo-outbreak of Cupriavidus pauculus infection at an outpatient clinic related to rinsing culturette swabs in tap water. J Clin Microbiol 48:2645–2647. doi: 10.1128/JCM.01874-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dundar D, Meric M, Vahaboglu H, Willke A. 2009. Pseudo-outbreak of Serratia marcescens in a tertiary care hospital. New Microbiol 32:273–276. [PubMed] [Google Scholar]
- 10.Blossom DB, Alelis KA, Chang DC, Flores AH, Gill J, Beall D, Peterson AM, Jensen B, Noble-Wang J, Williams M, Yakrus MA, Arduino MJ, Srinivasan A. 2008. Pseudo-outbreak of Mycobacterium abscessus infection caused by laboratory contamination. Infect Control Hosp Epidemiol 29:57–62. doi: 10.1086/524328. [DOI] [PubMed] [Google Scholar]
- 11.Ortiz A, Esteban J, Zamora N. 2007. Molecular identification by random amplified polymorphic DNA analysis of a pseudo-outbreak of Mycobacterium fortuitum due to cross-contamination of clinical samples. J Med Microbiol 56:871–872. doi: 10.1099/jmm.0.46959-0. [DOI] [PubMed] [Google Scholar]
- 12.Diederen BM, Verhulst C, van't Veen A, van Keulen PH, Kluytmans JA. 2006. Pseudo-outbreak of hepatitis B virus infection associated with contamination of a semiautomatic cap remover. Infect Control Hosp Epidemiol 27:1258–1260. doi: 10.1086/508844. [DOI] [PubMed] [Google Scholar]
- 13.Faden H, Ramani R, Lamson D, St George K. 2010. Pseudo-outbreak of adenovirus infection in a neonatal intensive care unit due to a false-positive antigen detection test. J Clin Microbiol 48:4251–4252. doi: 10.1128/JCM.01262-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lalande V, Barbut F, Varnerot A, Febvre M, Nesa D, Wadel S, Vincent V, Petit JC. 2001. Pseudo-outbreak of Mycobacterium gordonae associated with water from refrigerated fountains. J Hosp Infect 48:76–79. doi: 10.1053/jhin.2000.0929. [DOI] [PubMed] [Google Scholar]
- 15.Gebo KA, Srinivasan A, Perl TM, Ross T, Groth A, Merz WG. 2002. Pseudo-outbreak of Mycobacterium fortuitum on a human immunodeficiency virus ward: transient respiratory tract colonization from a contaminated ice machine. Clin Infect Dis 35:32–38. doi: 10.1086/340741. [DOI] [PubMed] [Google Scholar]
- 16.Medeiros EA, Lott TJ, Colombo AL, Godoy P, Coutinho AP, Braga MS, Nucci M, Brandt ME. 2007. Evidence for a pseudo-outbreak of Candida guilliermondii fungemia in a university hospital in Brazil. J Clin Microbiol 45:942–947. doi: 10.1128/JCM.01878-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yagupsky P, Dagan R, Chipman M, Goldschmied-Reouven A, Zmora E, Karplus M. 1991. Pseudooutbreak of Candida guilliermondii fungemia in a neonatal intensive care unit. Pediatr Infect Dis J 10:928–932. doi: 10.1097/00006454-199112000-00010. [DOI] [PubMed] [Google Scholar]
- 18.Brandt ME, Benjamin LE, Steinkraus GE. 2003. Pseudooutbreak of Candida versatilitis fungemia in a microbiology laboratory. Diagn Microbiol Infect Dis 46:73–75. doi: 10.1016/S0732-8893(02)00573-4. [DOI] [PubMed] [Google Scholar]
- 19.Deresinski SC, Clemons KV, Kemper CA, Roesch K, Walton B, Stevens DA. 1995. Genotypic analysis of pseudoepidemic due to contamination of Hanks' balanced salt solution with Candida parapsilosis. J Clin Microbiol 33:2224–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kappstein I, Krause G, Hauer T, Jonas D. 1998. Pseudo-outbreak of candidaemia with Candida parapsilosis. J Hosp Infect 40:164–165. doi: 10.1016/S0195-6701(98)90100-1. [DOI] [PubMed] [Google Scholar]
- 21.Aronoff DM, Thelen T, Walk ST, Petersen K, Jackson J, Grossman S, Rudrik J, Newton DW, Chenoweth CE. 2010. Pseudo-outbreak of Clostridium sordellii infection following probable cross-contamination in a hospital clinical microbiology laboratory. Infect Control Hosp Epidemiol 31:640–642. doi: 10.1086/652774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reason J. 2000. Human error: models and management. BMJ 320:768–770. doi: 10.1136/bmj.320.7237.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reason JT, Carthey J, de Leval MR. 2001. Diagnosing “vulnerable system syndrome”: an essential prerequisite to effective risk management. Qual Health Care 10 Suppl 2:ii21–ii25. doi: 10.1136/qhc.0100021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mart Microbiology BV. Operation Manual Anoxomat Mark II CTS V15, SW 3.5.4, p 60. Mart Microbiology BV, Drachten, Netherlands. [Google Scholar]
- 25.NCCLS. 2004. Quality control for commercially prepared microbiological culture media. Approved standard, 3rd ed, M22-A3 NCCLS, Wayne, PA. [Google Scholar]
- 26.Butler G, Rasmussen MD, Lin MF, Santos MA, Sakthikumar S, Munro CA, Rheinbay E, Grabherr M, Forche A, Reedy JL, Agrafioti I, Arnaud MB, Bates S, Brown AJ, Brunke S, Costanzo MC, Fitzpatrick DA, de Groot PW, Harris D, Hoyer LL, Hube B, Klis FM, Kodira C, Lennard N, Logue ME, Martin R, Neiman AM, Nikolaou E, Quail MA, Quinn J, Santos MC, Schmitzberger FF, Sherlock G, Shah P, Silverstein KA, Skrzypek MS, Soll D, Staggs R, Stansfield I, Stumpf MP, Sudbery PE, Srikantha T, Zeng Q, Berman J, Berriman M, Heitman J, Gow NA, Lorenz MC, Birren BW, Kellis M, Cuomo CA. 2009. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459:657–662. doi: 10.1038/nature08064. [DOI] [PMC free article] [PubMed] [Google Scholar]