ABSTRACT
The FilmArray gastrointestinal (GI) panel (BioFire Diagnostics, Salt Lake City, UT) is a simple, sample-to-answer, on-demand, multiplex, nucleic acid amplification test for syndromic diagnosis of infectious gastroenteritis. The aim of this study was to measure the yield of follow-up testing with FilmArray GI panel within 4 weeks of an initial test. Consecutive adult and pediatric patients tested at an academic institution between August 2015 and June 2016 were included in this study. Of 145 follow-up tests in 106 unique patients with an initial negative result, 134 (92.4%) tests and 98 (92.5%) patients remained negative upon follow-up testing. Excluding targets that are not reported at this institution (Clostridium difficile, enteroaggregative Escherichia coli, enteropathogenic E. coli, and enterotoxigenic E. coli), 137 (94.5%) follow-up tests and 101 (95.3%) patients remained negative. Weekly conversion rates were not significantly different across the 4-week follow-up interval. No epidemiological or clinical factors were significantly associated with a negative to positive conversion. Of 80 follow-up tests in patients with an initial positive result, 43 (53.8%) remained positive for the same target, 34 (42.5%) were negative, and 3 were positive for a different target (3.8%). Follow-up testing with FilmArray GI panel within 4 weeks of a negative result rarely changed the initial result, and the follow-up test reverted to negative less than half the time after an initial positive result. In the absence of clinical or epidemiological evidence for a new infection, follow-up testing should be limited and FilmArray GI panel should not be used as a test of cure.
KEYWORDS: FilmArray gastrointestinal, GI PCR, repeat, syndromic
INTRODUCTION
The FilmArray gastrointestinal (GI) panel (BioFire Diagnostics, Salt Lake City, UT) is a novel sample-to-answer, on-demand, multiplex, real-time PCR assay for the syndromic diagnosis of infectious gastroenteritis (1). The FilmArray GI panel has been adopted by many academic and community hospitals in the United States. Studies indicate that the FilmArray GI panel has a sensitivity and specificity of >90 and >97%, respectively, for the various targets, although some require further validation (1, 2). What remains to be determined is the utility of follow-up testing with the FilmArray GI panel when the initial result is negative. Prior studies on Clostridium difficile have shown that follow-up testing within 7 days of a negative result rarely yields new information (3, 4), and it was hypothesized that, in a similar fashion, repetition of the FilmArray GI panel following an initial negative result would be of limited clinical utility. The main aim of this retrospective study was to investigate the value of follow-up testing with FilmArray GI panel within 4 weeks when the initial result is negative, although the yield of follow-up testing after an initial positive result was also investigated.
RESULTS
Between August 2015 to June 2016, 3,168 stools from 2,633 patients with suspected infectious gastroenteritis were tested with the FilmArray GI panel. Patient ages ranged from <1 to 100 years with 22.6% under 18 years of age. The majority (n = 2,294 [72.4%]) of the specimens were collected in the outpatient setting. FilmArray GI panel was successfully performed on 3,132 (98.9%) stool samples. There were 36 invalid results, 16 (44.4%) of which were due to a single malfunctioning instrument. All invalid results were resolved by retesting the same stool sample. In total, 1,042 (32.9%) stool samples were positive for at least one pathogen, with 241 (7.6%) positive for two or more pathogens. The distribution of pathogens detected per sample and per patient is summarized in Table 1.
TABLE 1.
Pathogens detected with the FilmArray GI panel during the study period
| Pathogena | No. of stools (% of total stools) (n = 3,168) | No. of patients (% of total patients) (n = 2,633) | No. of stools with mixed pathogens (% of positive stools) |
|---|---|---|---|
| Campylobacter spp. | 93 (2.9) | 87 (3.3) | 36 (38.7) |
| Clostridium difficile | 308 (9.7) | 276 (10.5) | 75 (24.4) |
| Plesiomonas shigelloides | 9 (0.3) | 9 (0.3) | 7 (77.8) |
| Salmonella spp. | 52 (1.6) | 51 (1.9) | 25 (48.1) |
| Vibrio cholerae | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Vibrio spp. | 5 (0.2) | 5 (0.2) | 2 (40.0) |
| Yersinia enterocolitica | 15 (0.5) | 15 (0.6) | 2 (13.3) |
| EAEC | 126 (4.0) | 121 (4.6) | 93 (73.8) |
| EPEC | 205 (6.5) | 195 (7.4) | 121 (59.0) |
| ETEC | 62 (2.0) | 60 (2.3) | 47 (75.8) |
| STEC | 38 (1.2) | 37 (1.4) | 20 (52.6) |
| E. coli O157 | 5 (0.2) | 5 (0.2) | 2 (40.0) |
| Shigella/EIEC | 29 (0.9) | 29 (1.1) | 15 (51.7) |
| Cryptosporidium spp. | 14 (0.4) | 13 (0.5) | 6 (42.9) |
| Cyclospora cayetanensis | 4 (0.0) | 4 (0.2) | 1 (25.0) |
| Entamoeba histolytica | 1 (0.0) | 1 (0.0) | 0 (0.0) |
| Giardia lamblia | 32 (1.0) | 30 (1.1) | 9 (28.1) |
| Adenovirus 40/41 | 39 (1.2) | 37 (1.4) | 9 (23.1) |
| Astrovirus | 35 (1.1) | 35 (1.3) | 7 (20.0) |
| Norovirus GI/GII | 214 (6.8) | 185 (7.0) | 56 (26.2) |
| Rotavirus A | 22 (0.7) | 21 (0.8) | 8 (36.4) |
| Sapovirus | 60 (1.9) | 55 (2.1) | 21 (35.0) |
| Total pathogens detected | 1,368 | 1,272 | 562 |
EAEC, enteroaggregative E. coli; EPEC, enteropathogenic E. coli; ETEC, enterotoxigenic E. coli; STEC, Shiga-like toxin-producing E. coli (stx1/stx2); EIEC, enteroinvasive E. coli.
In total, 306 (11.6%) patients had at least one follow-up test. There were 159 (6.0%) unique patients who had 225 follow-up tests performed within 4 weeks of the index test with an average of 1.4 follow-up tests per patient (range: 1 to 6 repeats). The average interval between tests was 13.1 days (range, 0 to 28 days; standard deviation [SD], 7.9 days). In three cases the test was repeated on a separate sample the same day as the index test, although the reason for repetition was unclear upon review of the clinical documentation. Of the 145 follow-up tests in the 106 unique patients with an initial negative result, 134 (92.4%) tests and 98 (92.5%) patients remained negative upon follow-up testing. Weekly conversion rates were 9.3% (4/43), 6.1% (3/49), 12.5% (3/24), and 3.4% (1/29) during weeks 1 to 4, respectively (P > 0.05 for all comparisons by Pearson chi-square test; Fig. 1A). The pathogens detected on follow-up testing included norovirus GI/II (n = 3), enterotoxigenic E. coli (ETEC; n = 2), Campylobacter spp. (n = 1), C. difficile (n = 1), Salmonella spp. (n = 1), enteroaggregative E. coli (EAEC; n = 1), enteropathogenic E. coli (EPEC; n = 1), Shiga-like toxin-producing E. coli (STEC; n = 1), adenovirus (n = 1), and astrovirus (n = 1). One sample was positive for EAEC, EPEC, and ETEC. Excluding C. difficile, EAEC, EPEC, and ETEC results, which are not reported at this institution, 137 (94.5%) follow-up tests and 101 (95.3%) patients remained negative within 4 weeks of an initial negative result.
FIG 1.
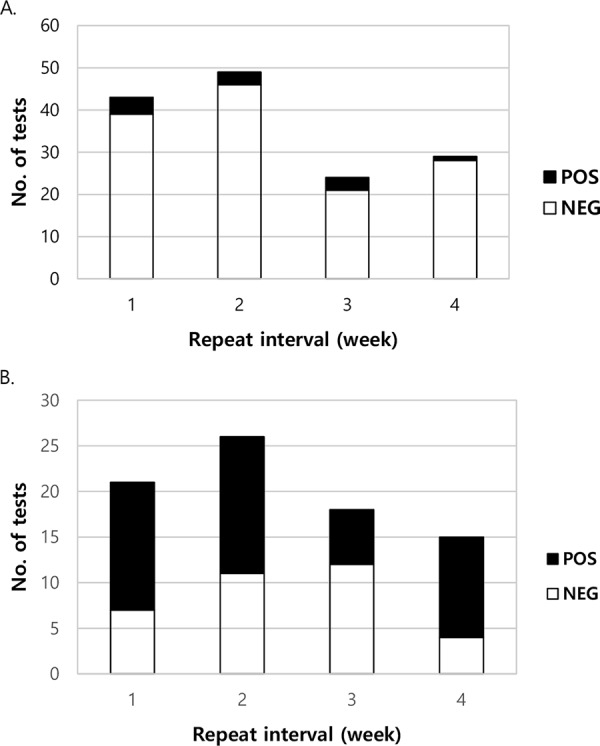
Conversion and reversion rates for follow-up tests with FilmArray GI panel within a 4-week interval. Results are shown for patients who underwent follow-up testing within 4 weeks after a negative (A) and a positive result (B). Weekly conversion and reversion rates, respectively, were not significantly different (P > 0.05 for all comparisons by Pearson chi-square test).
Chart review was performed in all 145 discrete episodes with an initial negative result, followed by an additional FilmArray GI panel sent within 4 weeks of the index episode to evaluate potential differences between nonconverters and converters. As noted above, in 134 cases both tests were negative. In 11 cases the second FilmArray GI panel converted to a positive result. These tests were performed in 106 unique patients; 3 patients fulfilled criteria as both a converter and a nonconverter at separate times and were included in the analyses for both groups. In comparing the two groups, there were no statistically significant differences, including age, sex, immunocompromised status, laboratory findings at the time of the index diarrheal illness, severity of illness at presentation, rate of hospitalization, or receipt of antibiotics, either given specifically for the diarrheal illness or for other reasons (Table 2). There was a trend toward more rapid improvement in the diarrhea in converters, although not quite reaching statistical significance (Table 2), and this result is likely skewed downward due to missing data as the time to resolution could not be determined in 6 of 11 (54.5%) converters (data not shown). There was also no difference in the time between testing, relapse following documented resolution of diarrheal symptoms as the reason for retesting, or whether there was ultimately an infectious or noninfectious etiology suspected or confirmed by retrospective chart review as the cause of the index diarrheal illness (Table 2). However, in the granular case details of the converters, of the 4 with a presumptive infectious diagnosis (both at the time of presentation and on retrospective chart review) for the index diarrheal illness, all were retested within 2 days, 3 were outpatients, 3 had no change in their diarrheal symptoms between tests, and 1 had no clinical data available for review (Table 3). Of the 7 with a presumptive noninfectious cause for the index diarrheal illness, follow-up tests were sent 8 to 28 days later, and 6 of 7 (85.7%) patients showed clinical improvement in between testing and a clear relapse of diarrheal symptoms (Table 3), suggesting the presence of a separate cause for the symptoms that prompted retesting. The one patient (patient 9 in Table 3) who had no improvement in diarrheal symptoms between tests was positive for C. difficile on the second FilmArray GI panel, but the Xpert C.diff Epi tcdB PCR that was sent simultaneously was negative. This suggests that the FilmArray GI panel result may have been a false positive since Xpert is the most sensitive nucleic acid test for C. difficile detection (5), although it is possible that this isolate contained only tcdA since FilmArray GI panel detects both toxin genes.
TABLE 2.
Comparison of clinical and epidemiological data between nonconverters and converters on follow-up testing with the FilmArray GI panel within 4 weeks of a negative result
| Parametera | No. (%) of patientsb |
P | |
|---|---|---|---|
| Nonconverters (n = 134) | Converters (n = 11) | ||
| Sex (n = 145) | |||
| Male | 67 (50) | 6 (54.5) | 0.77 |
| Female | 67 (50) | 5 (45.5) | |
| Mean age in yrs (n = 145) | 35.2 [23.8] | 41.6 [23.1] | 0.44 |
| Age category (n = 145) | |||
| Adult (age > 18 yrs) | 88 (65.7) | 8 (72.7) | 0.75 |
| Pediatric (age ≤ 18 yrs) | 46 (34.3) | 3 (27.3) | |
| Patient location (n = 145) | |||
| Inpatient | 94 (70.1) | 6 (54.5) | 0.13 |
| Outpatient | 28 (20.9) | 5 (45.5) | |
| Emergency department | 12 (9) | 0 | |
| Mean WBC (K/μl) (n = 136) | 6.5 [5.8] | 6.4 [6.5] | 0.92 |
| Mean lactate (mmol/liter) (n = 30) | 1.3 [0.7] | 1.5 [0.2] | 0.25 |
| Mean creatinine (mg/dl) (n = 137) | 1.1 [1.0] | 1.6 [1.5] | 0.03 |
| Mean stools/day on presentation (n = 86) | 5.5 [3.4] | 6.2 [2.4] | 0.36 |
| Hospitalized for diarrheal illness (n = 143) | |||
| Yes | 32 (24.1) | 1 (9.1) | 0.45 |
| No | 101 (75.9) | 10 (90.9) | |
| Any antibiotics given (n = 143) | |||
| Yes | 86 (64.7) | 4 (40) | 0.17 |
| No | 47 (35.3) | 6 (60) | |
| Antibiotics given for diarrheal illness (n = 90) | |||
| Yes | 27 (31.4) | 0 | 0.31 |
| No | 59 (68.6) | 4 (100) | |
| Mean days to resolution (n = 61) | 6.3 [5.5] | 2.2 [1.6] | 0.01 |
| Interval change (n = 136) | |||
| Better | 73 (57.9) | 6 (60) | 0.89 |
| Worse | 3 (2.4) | 0 | |
| Same | 50 (39.7) | 4 (40) | |
| Mean days between tests (n = 145) | 13.0 [8.0] | 10.5 [9.1] | 0.35 |
| Relapse as reason for second test (n = 139) | |||
| Yes | 52 (40.3) | 5 (50) | 0.74 |
| No | 77 (59.7) | 5 (50) | |
| Confirmed/suspected noninfectious etiology (n = 141) | |||
| Yes | 98 (75.4) | 7 (63.6) | 0.47 |
| No | 32 (24.6) | 4 (36.4) | |
| Immunocompromised (BMT or SOT) (n = 144) | |||
| Yes | 79 (59) | 5 (50) | 0.74 |
| No | 55 (41) | 5 (50) | |
BMT, bone marrow transplant; SOT, solid organ transplant; WBC, white blood cell count. “Relapse” was defined as the return of diarrheal symptoms following documented resolution as determined by chart review.
Except as noted otherwise in column 1. Standard deviations are indicated within brackets.
TABLE 3.
Clinical characteristics of converters on follow-up testing with the FilmArray GI panel within 4 weeks of a negative result
| ID | Sex | Age (yr) | Interval change | Day(s) between tests | Presumptive cause of diarrheaa | Result of second FilmArray analysis |
|---|---|---|---|---|---|---|
| 1 | M | 64 | Same | 1 | Norovirus | Norovirus |
| 2 | F | 0.9 | Better | 20 | Medication effect | Norovirus |
| 3 | F | 17 | Better | 28 | Ulcerative colitis | ETEC |
| 4 | F | 60 | Better | 18 | GVHD | Campylobacter sp. |
| 5 | M | 53 | Same | 1 | Traveler's diarrhea | EAEC, EPEC, ETEC |
| 6 | F | 17 | Better | 15 | Medication effect | Astrovirus |
| 7 | M | 64 | Better | 10 | Medication effect | Salmonella enterica |
| 8 | M | 55 | Same | 2 | Adenovirus colitis | Adenovirus F 40/41 |
| 9 | M | 19 | Same | 8 | Medication effect | Clostridium difficile |
| 10 | M | 57 | Better | 12 | Chemotherapy effect | Norovirus |
| 11 | F | 51 | Unknown | 1 | Shiga toxin E. coli colitis | Shiga toxin E. coli, non-O157 |
Based on a retrospective chart review. GVHD, graft-versus-host disease.
In 80 follow-up tests performed within 4 weeks of an initial positive result, 34 (42.5%) became negative, 43 (53.8%) remained positive for one or more of the same targets as the initial results, and 3 (3.8%) showed clearance of the index target but were positive for a different one. The latter three cases were excluded from further evaluation since they could not be classified as either a reverter or a nonreverter. The 34 tests that turned negative on follow-up testing represented 33 unique patients. Eleven (32.4%) of these initial tests were positive for multiple targets. Weekly reversion rates were 33.3% (7/21), 42.3% (11/26), 66.7% (12/18), and 26.7% (4/15) during weeks 1 to 4, respectively (P > 0.05 for all comparisons by Pearson chi-square test; Fig. 1B). The 49 reversion pathogens included C. difficile (n = 13), EPEC (n = 5), Campylobacter spp. (n = 4), Salmonella spp. (n = 4), norovirus (n = 4), EAEC (n = 3), ETEC (n = 3), Giardia lamblia (n = 3), STEC (n = 2), astrovirus (n = 2), Plesiomonas shigelloides (n = 1), Shigella/enteroinvasive E. coli (EIEC; n = 1), Vibrio spp. (n = 1), Yersinia enterocolitica (n = 1), adenovirus (n = 1), and rotavirus (n = 1). Of the initial targets with a confirmatory or parallel testing modality, including reflex cultures for Campylobacter spp., Salmonella spp., and Shigella/EIEC and Xpert C.diff Epi tcdB PCR for suspected C. difficile colitis, 12 of 24 (50%) FilmArray GI panel results were confirmed. Eleven targets positive by FilmArray GI panel were determined to be negative by alternative testing, 10 of which were culture and one was C. difficile PCR.
The 43 tests that remained positive for one or more of the same targets as the initial results were performed in 31 unique patients. In 15 (34.9%) cases the initial test was positive for multiple targets. The initial tests were positive for 58 total targets, including C. difficile (n = 18), norovirus (n = 16), EPEC (n = 5), sapovirus (n = 4), Campylobacter spp. (n = 4), adenovirus (n = 2), EAEC (n = 2), Salmonella spp. (n = 2), STEC (n = 1), Giardia lamblia (n = 1), ETEC (n = 1), Cryptosporidium spp. (n = 1), and Shigella/EIEC (n = 1). On follow-up testing in these 43 cases, 54 targets were identified. Eleven (25.6%) follow-up tests were positive for multiple targets. Of these 54 targets, 50 (92.6%) were the same as on the initial test. Eight (13.8%) targets that were positive on initial testing were cleared, all of which occurred in the setting of a previous test positive for multiple targets. In 5 (62.5%) of these cases, directed antibiotics were administered between tests. The targets that were cleared included C. difficile (n = 3), Salmonella spp. (n = 2), Shigella/EIEC (n = 1), Campylobacter spp. (n = 1), and EPEC (n = 1). In four instances a different target was identified that was not seen on the initial test, including C. difficile (n = 3) and Vibrio spp. (n = 1). In three (75%) of these cases antibiotics were given in the time between testing, though in only two (66.7%) were these antibiotics directed against the target identified on FilmArray GI panel (data not shown).
Chart review was also performed in all 77 cases of follow-up testing after an initial positive result to evaluate for clinical or epidemiological differences between reverters and nonreverters. There was no difference in sex, immunocompromised status, days between tests, or reason for retesting, but those whose tests remained positive were significantly less likely to have been given antibiotics between tests (Table 4). There was a trend toward a higher likelihood for the presence of a viral target on the initial test and a younger age in nonreverters, although these were not significant with correction for multiple comparisons (Table 4).
TABLE 4.
Comparison of clinical and epidemiological data between nonreverters and reverters after retesting with the FilmArray GI panel within 4 weeks of a positive result
| Parametera | No. (%) of patientsb |
P | |
|---|---|---|---|
| Nonreverters (n = 43) | Reverters (n = 34) | ||
| Sex | |||
| Male | 23 (53.5) | 16 (47.1) | 0.58 |
| Female | 20 (46.5) | 18 (52.9) | |
| Mean age (yrs) | 29.8 [31.3] | 39.8 [23.5] | 0.02 |
| Age | |||
| >18 yrs | 19 (44.2) | 25 (73.5) | 0.01 |
| ≤18 yrs | 24 (55.8) | 9 (26.5) | |
| Mean days between tests | 12.7 [8.1] | 13.6 [6.7] | 0.59 |
| Immunocompromised (BMT or SOT) | |||
| Yes | 22 (51.2) | 14 (41.2) | 0.38 |
| No | 21 (48.8) | 20 (58.8) | |
| Viral target positive | |||
| Yes | 22 (51.2) | 8 (23.5) | 0.01 |
| No | 21 (48.8) | 26 (76.5) | |
| Antibiotics given | |||
| Yes | 14 (32.6) | 25 (73.5) | <0.01 |
| No | 29 (67.4) | 9 (26.5) | |
| Reason for retesting | |||
| Test of cure | 15 (36.6) | 11 (34.4) | 0.08 |
| Recurrence | 10 (24.4) | 15 (46.9) | |
| Persistence | 16 (39.0) | 6 (18.7) | |
BMT, bone marrow transplant; SOT, solid organ transplant. “Recurrence” is defined as a relapse of diarrheal symptoms following documented resolution. “Persistence” is defined as a lack of resolution of diarrheal symptoms between tests.
Except as noted otherwise in column 1. Standard deviations are indicated within brackets.
DISCUSSION
This is the first study to investigate the value of follow-up testing with the FilmArray GI panel. It has been previously shown that repeating C. difficile PCR in patients with a prior negative result rarely yields new information (3, 4). In this study, this observation was extended to a panel of pathogens detected by the FilmArray GI panel. This study shows that follow-up testing with the FilmArray GI panel within 4 weeks of a negative result is unlikely to produce a different result, and even more unlikely if the results for targets such as C. difficile, EAEC, EPEC, and ETEC—which are not routinely reported at this institution—are excluded. Therefore, follow-up testing is rarely clinically useful and adds to health care costs. The rare conversions upon follow-up testing may have been due to suboptimal stool sampling or timing of stool collection for the initial test, as suggested by the four patients with conversions when retested within 2 days, and if these are suspected, or if there is a significantly high index of suspicion for an infectious gastroenteritis and concern for a false-negative result, then repetition of the test may be warranted. However, new infection or a false-positive result as the cause of conversion cannot be ruled out. The latter cause is suggested, in some instances, by the fact that during the study period, 41.7, 43.2, and 53.2% of stool samples positive by the FilmArray GI panel for Campylobacter spp., Salmonella spp., and Shigella/EIEC, respectively, did not yield a positive culture for antibiotic susceptibility testing (data not shown). Much of the discrepancy is likely related to the higher sensitivity of nucleic acid-based testing, but these yields are lower than those reported in a recent evaluation of the FilmArray (6), suggesting the possibility of false-positive FilmArray results. Furthermore, in April 2016, our laboratory experienced the highest monthly number of positives for Campylobacter spp. in a single month. Thirteen of fourteen Campylobacter-positive samples determined by the FilmArray GI panel in that month did not yield a viable culture and were presumed to represent false-positive results (data not shown).
Consistent with other nucleic acid amplification tests (7, 8), follow-up testing after a positive FilmArray GI panel result was of limited clinical utility. Nearly half of the patients with an initial positive result remained positive for the same target within 4 weeks of the index test. Based on these results, follow-up testing after a positive result, especially if used as a test of cure, is not recommended, similar to the recommendations for C. difficile infection (9–11). This institution has previously used an electronic alert during order entry to educate providers and reduce repeat C. difficile tests within 7 days of a prior test (12). A similar electronic alert can be implemented to educate providers about the limited utility of repeated FilmArray GI panel testing and prevent ordering of additional tests on the same patient. Implementation of exclusion criteria based on duration of hospitalization can also be considered since, in addition to C. difficile and norovirus, the FilmArray GI panel detects pathogens that are most likely to be acquired outside the hospital.
This study had several limitations. Since this was an observational study, the stool samples that converted to a positive result on follow-up testing were not systematically investigated at the time of conversion to determine whether the result could be confirmed with a reference method. Second, due to a low conversion rate on follow-up testing, the small sample size limits any conclusions that can be drawn from this data set. Although potential epidemiological and clinical factors associated with these conversions and reversions were investigated in the present study, larger studies that are organized prospectively are needed to further understand whether any clinical factors may be predictive.
In conclusion, without clinical and/or epidemiological suspicion of a new-onset infectious gastroenteritis, follow-up testing with the FilmArray GI panel within 4 weeks of a negative test result is rarely informative, and the FilmArray GI panel should not be repeated as a test of cure following a positive result.
MATERIALS AND METHODS
Ethics.
In accordance with the Stanford University Institutional Review Board (IRB), this project was determined to constitute a quality improvement project and was exempt from IRB approval.
Study subjects and stool specimens.
The Stanford Hospital clinical microbiology laboratory serves adult and pediatric teaching hospitals and their affiliated clinics. Stool samples were received in a Cary-Blair transport vial and tested on-demand with the FilmArray GI panel. The laboratory began to offer the FilmArray GI panel in August 2015. Orders were accepted without any restrictions on symptoms, stool output, or the number of times the test had been performed previously for a given patient. Stool samples positive for Campylobacter spp., Salmonella spp., and Shigella/EIEC were cultured using conventional media (BBL Campy CVA agar [Becton-Dickinson; Franklin Lakes, NJ] incubated under microaerophilic gas at 42°C and BBL Hektoen enteric agar [Becton-Dickinson] incubated in room air at 37°C) for the purposes of pathogen isolation and antibiotic susceptibility testing. All stool samples (n = 3,168) tested with the FilmArray GI panel between August 2015 and June 2016 were included in this retrospective study. The results for each target per test were obtained using an electronic report generated from the laboratory information system. Patients with a follow-up result for the FilmArray GI panel within 4 weeks of an index test were identified. The 4-week time frame was reset after each test, allowing an individual patient to be included multiple times if they underwent serial testing with each follow-up test occurring within 4 weeks of the previous test. Depending on the permutations of the results on serial testing, an individual patient could be included in multiple analyses. Chart review was performed by M.M.H. and C.A.G. to obtain demographic and clinical data. The cause of the index diarrheal illness was retrospectively evaluated by synthesizing the clinical data, including clinical suspicions of the treating physicians, medications given to the patient prior to the onset of diarrhea, pathology results from colonic biopsy samples, FilmArray GI panel results (index and follow-up), and additional test results such as C. difficile testing, evaluated using Xpert C.diff Epi tcdB PCR (Cepheid, Sunnyvale, CA) on fresh stool samples, to determine the most likely cause of the diarrheal symptoms. The duration of diarrheal symptoms was determined by stool output recorded in physician notes. Diarrhea was considered to have been resolved when <3 stools were recorded in 24 h. Cases where stool output was not clearly recorded were not included in the calculation of time to resolution. The interval change in diarrheal symptoms was also based on stool output recorded in physician notes. A >50% decrease in stool output compared to the index symptoms was considered improvement and a >50% increase was considered worsening. In cases where stool output was not clearly documented, interval improvement was inferred if a recorded relapse of diarrheal symptoms was the reason for follow-up FilmArray GI panel testing. All antibiotic usage in the interval from the index to the follow-up test was included in the analysis.
FilmArray GI panel.
Stools in Cary-Blair transport media were tested with the FilmArray GI panel per the package insert. Positive results were conveyed to the ordering provider. The following targets were not reported to the provider or in the electronic medical record: C. difficile, EAEC, EPEC, and ETEC. However, these targets were included in this analysis.
Statistical analysis.
Clinical data were summarized and evaluated for differences between patients with an initial negative test whose follow-up test remained negative (nonconverter) and those whose subsequent test became positive (converter). A similar evaluation was done for patients with an initial positive test, followed by a negative test (reverter) or a subsequent positive test for the same target (nonreverter). The Mann-Whitney U test was used to compare nonparametrically distributed, continuous variables, the Student t test was used to compare normally distributed, continuous variables, and the chi-square and Fisher exact tests were used to compare categorical variables. To protect against random error from testing multiple hypotheses, the Bonferroni correction was used and α was set at 0.01. Weekly conversion and reversion rates were tabulated, and confidence intervals for proportions were calculated from the binomial distribution. Statistical analyses were performed using SPSS v23 (IBM Analytics, Armonk, NY).
REFERENCES
- 1.Buss SN, Leber A, Chapin K, Fey PD, Bankowski MJ, Jones MK, Rogatcheva M, Kanack KJ, Bourzac KM. 2015. Multicenter evaluation of the BioFire FilmArray gastrointestinal panel for etiologic diagnosis of infectious gastroenteritis. J Clin Microbiol 53:915–925. doi: 10.1128/JCM.02674-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khare R, Espy MJ, Cebelinski E, Boxrud D, Sloan LM, Cunningham SA, Pritt BS, Patel R, Binnicker MJ. 2014. Comparative evaluation of two commercial multiplex panels for detection of gastrointestinal pathogens by use of clinical stool specimens. J Clin Microbiol 52:3667–3673. doi: 10.1128/JCM.01637-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aichinger E, Schleck CD, Harmsen WS, Nyre LM, Patel R. 2008. Non-utility of repeat laboratory testing for detection of Clostridium difficile by use of PCR or enzyme immunoassay. J Clin Microbiol 46:3795–3797. doi: 10.1128/JCM.00684-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luo RF, Banaei N. 2010. Is repeat PCR needed for diagnosis of Clostridium difficile infection? J Clin Microbiol 48:3738–3741. doi: 10.1128/JCM.00722-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crobach MJT, Planche T, Eckert C, Barbut F, Terveer EM, Dekkers OM, Wilcox MH, Kuijper EJ. 2016. European Society of Clinical Microbiology and Infectious Diseases: update of the diagnostic guidance document for Clostridium difficile infection. Clin Microbiol Infect 22:S63–S81. doi: 10.1016/j.cmi.2016.03.010. [DOI] [PubMed] [Google Scholar]
- 6.Murphy CN, Fowler RC, Iwen PC, Fey PD. 12 December 2016. Evaluation of the BioFire FilmArray gastrointestinal panel in a midwestern academic hospital. Eur J Clin Microbiol Infect Dis Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 7.Sunkesula VCK, Kundrapu S, Muganda C, Sethi AK, Donskey CJ. 2013. Does empirical Clostridium difficile infection (CDI) therapy result in false-negative CDI diagnostic test results? Clin Infect Dis 57:494–500. doi: 10.1093/cid/cit286. [DOI] [PubMed] [Google Scholar]
- 8.Nicol MP. 2013. Xpert MTB/RIF: monitoring response to tuberculosis treatment. Lancet Respir Med 1:427–428. doi: 10.1016/S2213-2600(13)70133-4. [DOI] [PubMed] [Google Scholar]
- 9.Surawicz CM, Brandt LJ, Binion DG, Ananthakrishnan AN, Curry SR, Gilligan PH, McFarland LV, Mellow M, Zuckerbraun BS. 2013. Guidelines for diagnosis, treatment, and prevention of Clostridium difficile infections. Am J Gastroenterol 108:478–498. doi: 10.1038/ajg.2013.4. [DOI] [PubMed] [Google Scholar]
- 10.Bagdasarian N, Rao K, Malani PN. 2015. Diagnosis and treatment of Clostridium difficile in adults: a systematic review. JAMA 313:398–408. doi: 10.1001/jama.2014.17103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen SH, Gerding DN, Johnson S, Kelly CP, Loo VG, McDonald LC, Pepin J, Wilcox MH. 2010. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the Society for Healthcare Epidemiology of America (SHEA) and the Infectious Diseases Society of America (IDSA). Infect Control Hosp Epidemiol 31:431–455. doi: 10.1086/651706. [DOI] [PubMed] [Google Scholar]
- 12.Luo RF, Spradley S, Banaei N. 2013. Alerting physicians during electronic order entry effectively reduces unnecessary repeat PCR testing for Clostridium difficile. J Clin Microbiol 51:3872–3874. doi: 10.1128/JCM.01724-13. [DOI] [PMC free article] [PubMed] [Google Scholar]


