LETTER
The identification, treatment, and control of carbapenem-resistant Enterobacteriaceae (CRE) infections are a major challenge for health care institutions and diagnostic laboratories worldwide. Those producing plasmid-mediated OXA-48, KPC, NDM, or VIM-like carbapenemases (CPE) are most concerning, being frequently involved in nosocomial outbreaks that are difficult and very costly to manage (1). A simple and rapid test able to provide sensitive and specific identification of CRE and also the carbapenemase present is critical in any strategy aimed at addressing this problem.
Numerous phenotypic (MIC determination, disc diffusion, selective culture media, acidimetric) and genotypic (PCR amplification, microarrays, DNA sequencing) methods have been used in the laboratory isolation, diagnosis, and confirmation of CRE (2). Recently, two novel immunochromatographic lateral flow assays (Coris BioConcept, Gembloux, Belgium) were developed for the specific detection of OXA-48 (OXA-48 K-SeT) and KPC-like (KPC K-SeT) carbapenemase producers. These assays have been evaluated in multiple laboratories, using diverse sets of organisms (Escherichia coli, Klebsiella, Enterobacter, Serratia, Providencia, Pseudomonas spp.) carrying multiple β-lactamase, OXA-48 (OXA-48, -162, -181, -204, -232, -242), and KPC (KPC-2/3/4) allelic variants (3, 4, 5, 6). Both have a reported sensitivity and specificity of 100% compared to the results of molecular detection of carbapenemase genes as the gold standard (3, 4, 5, 6). They have also been shown to be compatible with organisms recovered from most unsupplemented, selective, solid, and liquid culture media currently in use in diagnostic laboratories and with bacteria taken directly from positive blood culture bottles (BacT/Alert; bioMérieux, Macy L'Etoile, France) or culture-positive urinary samples (3, 4). A modification of this system, Resist-3 O.K.N. (Coris BioConcept) designed for the simultaneous detection of OXA-48, KPC, and NDM-like enzymes using a single disposable cartridge, has now been manufactured. Here we assessed the ability of the Resist-3 O.K.N. assay to detect OXA-48, KPC, and NDM coproduction in a collection of 112 nonreplicate well-defined CRE isolates received in our laboratories.
All isolates were carbapenem-resistant Enterobacteriaceae (ertapenem MIC, >1 μg/ml) recovered from routine clinical samples submitted to Barts Health NHS Trust or the Antimicrobial Research Laboratory at Queen Mary University of London between January 2011 and December 2016. Identification was performed using the Bruker matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry system (Bruker UK Ltd., Coventry, UK), with carbapenem MICs determined using MicroScan WalkAway Negative Combo 36 panels (Siemens Healthcare Diagnostics, Deerfield, IL) and confirmed with Etest gradient strips (bioMérieux). Multiplex PCRs were used to confirm the presence of class A (KPC, BKC, SME, VEB, PER, GES), B (IMP, VIM, NDM, SIM, SPM, DIM, GIM, KHM, FIM, AIM, DIM, TMB, FRI), and D (OXA-like) carbapenemase genes, and all allelic variants identified were confirmed by Sanger sequencing of the entire coding regions (7, 8). The collection consisted of carbapenemase-producing Klebsiella pneumoniae (n = 57), E. coli (n = 17), Enterobacter spp. (n = 14), and Providencia stuartii (n = 3) and single isolates of Citrobacter koseri, Morganella morganii, and Proteus mirabilis (Table 1). Carbapenemases produced by the isolates were identified as KPC-2/4, NDM-1/5/7, VIM-1/4, IMP-1, OXA-48, and OXA-232. Nine isolates produced 2 carbapenemases, either OXA-48 in combination with NDM-1 (n = 6) or KPC-2 in combination with VIM-1 (n = 3).
TABLE 1.
Detection of carbapenemase production using the Resist-3 O.K.N. assay
| Isolate | Carbapenemase(s) carried (no. of isolates) | Resist-3 O.K.N. result |
||
|---|---|---|---|---|
| OXA-48-like | KPC-like | NDM-like | ||
| Klebsiella pneumoniae | OXA-48 (23) | 23 | 0 | 0 |
| OXA-48 + NDM-1 (4) | 4 | 0 | 4 | |
| OXA-232 (6) | 6 | 0 | 0 | |
| KPC-2 (8) | 0 | 8 | 0 | |
| KPC-2 + VIM-1 (1) | 0 | 1 | 0 | |
| NDM-1 (14) | 0 | 0 | 14 | |
| VIM-4 (1) | 0 | 0 | 0 | |
| Escherichia coli | OXA-48 (1) | 1 | 0 | 0 |
| KPC-2 (3) | 0 | 3 | 0 | |
| KPC-2 + VIM-1 (1) | 0 | 1 | 0 | |
| NDM-1 (6) | 0 | 0 | 6 | |
| NDM-5 (3) | 0 | 0 | 3 | |
| NDM-7 (2) | 0 | 0 | 2 | |
| VIM-1 (1) | 0 | 0 | 0 | |
| Enterobacter cloacae | OXA-48 + NDM-1 (2) | 2 | 0 | 2 |
| NDM-1 (2) | 0 | 0 | 2 | |
| KPC-4 (2) | 0 | 2 | 0 | |
| IMP-1 (1) | 0 | 0 | 0 | |
| VIM-1 (4) | 0 | 0 | 0 | |
| Enterobacter aerogenes | KPC-2 (2) | 0 | 2 | 0 |
| NDM-1 (1) | 0 | 0 | 1 | |
| Citrobacter koseri | KPC-2 + VIM-1 (1) | 0 | 1 | 0 |
| Providencia stuartii | NDM-1 (1) | 0 | 0 | 1 |
| VIM-1 (2) | 0 | 0 | 0 | |
| Morganella morganii | VIM-1 (1) | 0 | 0 | 0 |
| Proteus mirabilis | VIM-1 (1) | 0 | 0 | 0 |
Detection of OXA-48, KPC, and NDM using the Resist-3 O.K.N. cassettes was carried out according to the manufacturer's protocol. Bacteria were grown for 18 h at 37°C on Mueller-Hinton II agar plates (Oxoid), and a single colony was emulsified in 5 drops of the lysis buffer. Cassettes were loaded with 3 drops of lysate and read within 5 min. Six carbapenem-susceptible Gram-negative type strains (K. pneumoniae NCTC 9633, E. coli NCTC 12241, Enterobacter cloacae NCTC 13380, Enterobacter aerogenes NCTC 9375, Pseudomonas aeruginosa ATCC 27852, Acinetobacter baumannii ATCC 19606) were used as negative controls. Eighteen additional Enterobacteriaceae isolates (K. pneumoniae [n = 8] or Klebsiella oxytoca [n = 1], E. coli [n = 4], E. cloacae [n = 2] or E. aerogenes [n = 2], Serratia marcescens [n = 1]) with phenotypic resistance to ertapenem but without a known carbapenemase were also used as negative controls.
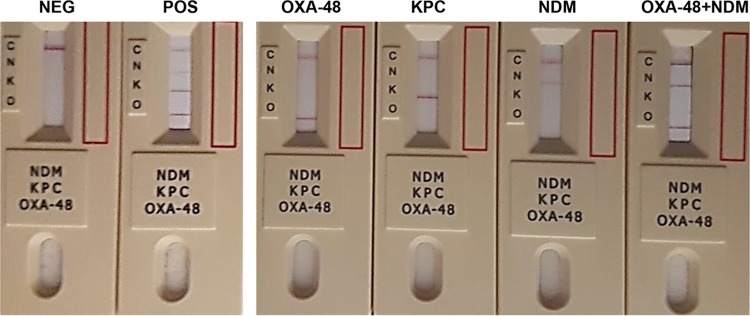
There was complete agreement between the carbapenemases detected by PCR and the results obtained with Resist-3 O.K.N. (Table 1). The lateral flow device was able to correctly identify and differentiate OXA, KPC, and NDM production among all CRE species and enzyme variants tested, including those carrying more than one carbapenemase. No cross-reactions were observed for strains with carbapenem resistance due to IMP-1 or VIM-1/4 (Table 1) with any of the susceptible type strains or with the 18 CRE that had no OXA, KPC, or NDM carbapenemase detectable by PCR (Fig. 1).
FIG 1.
Detection of OXA-48, KPC, and NDM-like carbapenemases in isolates producing single and dual enzymes using the Resist-3 O.K.N. assay. NEG, negative; POS, positive.
We found the Resist-3 O.K.N. assay to be 100% sensitive and specific in the detection and differentiation of OXA-48, KPC, and NDM-like carbapenemases among CRE recently referred to our laboratory. Although the assay does not currently extend to the detection of VIM or IMP-like metallo-β-lactamases, the ability to detect 3 of the 5 most prevalent and transmissible carbapenemases found worldwide is a significant advantage. The ease of use, speed, and low cost (less than $15) of the cassettes make it attractive as a diagnostic tool for use in the management of CRE outbreaks. Its role either as a primary-screening, confirmatory, or rapid point-of-care test should be assessed further, ideally prospectively in the setting of a polyclonal nosocomial CRE outbreak.
ACKNOWLEDGMENTS
Resist-3 O.K.N. assays were supplied free of charge for evaluation by BioConnections (Knypersley, UK). No other specific funding was used to undertake this study, as it was performed as part of our routine activities.
We declare no conflict of interest and have no association with Coris BioConcept.
REFERENCES
- 1.Otter JA, Burgess P, Davies F, Mookerjee S, Singleton J, Gilchrist M, Parsons D, Brannigan ET, Robotham J, Holmes AH. 13 October 2016. Counting the cost of an outbreak of carbapenemase-producing Enterobacteriaceae: an economic evaluation from a hospital perspective. Clin Microbiol Infect doi: 10.1016/j.cmi.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Lutgring JD, Limbago BM. 2016. The problem of carbapenemase-producing-carbapenem-resistant-Enterobacteriaceae detection. J Clin Microbiol 54:529–534. doi: 10.1128/JCM.02771-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wareham DW, Shah R, Betts JW, Phee LM, Momin MH. 2016. Evaluation of an immunochromatographic lateral flow assay (OXA-48 K-SeT) for rapid detection of OXA-48-like carbapenemases in Enterobacteriaceae. J Clin Microbiol 54:471–473. doi: 10.1128/JCM.02900-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glupczynski Y, Evrard S, Ote I, et al. 2016. Evaluation of two new commercial immunochromatographic assays for the rapid detection of OXA-48 and KPC carbapenemases from cultured bacteria. J Antimicrob Chemother 71:1217–1222. doi: 10.1093/jac/dkv472. [DOI] [PubMed] [Google Scholar]
- 5.Dortet L, Jousset A, Sainte-Rose V, Cuzon G, Naas T. 2016. Prospective evaluation of the OXA-48 K-SeT assay, an immunochromatographic test for the rapid detection of OXA-48-type carbapenemases. J Antimicrob Chemother 71:1834–1840. doi: 10.1093/jac/dkw058. [DOI] [PubMed] [Google Scholar]
- 6.Meunier D, Vickers A, Pike R, Hill RL, Woodford N, Hopkins KL. 2016. Evaluation of the K-SeT R.E.S.I.S.T. immunochromatographic assay for the rapid detection of KPC and OXA-48-like carbapenemases. J Antimicrob Chemother 71:2357–2359. doi: 10.1093/jac/dkw113. [DOI] [PubMed] [Google Scholar]
- 7.Oikonomou O, Liakopoulos A, Phee LM, Betts J, Mevius D, Wareham DW. 2016. Providencia stuartii isolates from Greece: co-carriage of cephalosporin (blaSHV-5, blaVEB-1), carbapenem (blaVIM-1), and aminoglycoside (rmtB) resistance determinants by a multidrug-resistant outbreak clone. Microb Drug Resist 22:379–386. doi: 10.1089/mdr.2015.0215. [DOI] [PubMed] [Google Scholar]
- 8.Dallene C, Da Costa A, Decre D, Favier C, Arlet G. 2010. Development of a set of multiplex PCR assays for the detection of genes encoding β-lactamases in Enterobacteriaceae. J Antimicrob Chemother 65:490–495. doi: 10.1093/jac/dkp498. [DOI] [PubMed] [Google Scholar]