Abstract
Chlorosis is one of the most common symptoms of plant diseases, including those caused by viruses and viroids. Recently, a study has shown that Peach latent mosaic viroid (PLMVd) exploits host RNA silencing machinery to modulate the virus disease symptoms through the silencing of chloroplast-targeted heat shock protein 90 (Hsp90C). To understand the molecular mechanisms of chlorosis in this viroid disease, we established an experimental system suitable for studying the mechanism underlying the chlorosis induced by the RNA silencing of Hsp90C in transgenic tobacco. Hairpin RNA of the Hsp90C-specific region was expressed under the control of a dexamethasone-inducible promoter, resulted in the silencing of Hsp90C gene in 2 days and the chlorosis along with growth suppression phenotypes. Time course study suggests that a sign of chlorosis can be monitored as early as 2 days, suggesting that this experimental model is suitable for studying the molecular events taken place before and after the onset of chlorosis. During the early phase of chlorosis development, the chloroplast- and photosynthesis-related genes were downregulated. It should be noted that some pathogenesis related genes were upregulated during the early phase of chlorosis in spite of the absence of any pathogen-derived molecules in this system.
Electronic supplementary material
The online version of this article (doi:10.1007/s13337-017-0361-0) contains supplementary material, which is available to authorized users.
Keywords: Chlorosis, Chloroplast Hsp90, Inducible silencing, Viroid
Introduction
Virus infection in plants causes a variety of symptoms resulting from the morphological and physiological disturbances of the host cells. Most common symptom, the leaf chlorosis incurs poor plant performance that leads to a significant loss in crop yield. The chlorosis symptoms are associated with reduced chlorophyll content of the leaves and structural changes in chloroplasts [13]. Thus, the effect of virus infection on chloroplast structure and function leads to the reduced photosynthetic activity. The loss of chlorophyll pigments and reduced photosynthetic activity results in decreased plant production. Till date, a lot of efforts have been made to understand the molecular basis of chlorosis symptoms development in virus-infected plants.
Studies have shown that virus infection down-regulates chloroplast- and photosynthesis-related proteins and genes (CPRPs and CPRGs, respectively) in virus-infected plants [11, 15, 22]. Rice stripe virus (RSV) downregulates CPRGs during infection and disturbs the photosynthesis [23]. The perturbation of photosynthesis by RSV might be caused by the regulation of a special set of miRNAs that target key genes in chloroplast zeaxanthin cycle, which can lead to impaired chloroplast structure and function [35]. However, it remains unknown which virus factors regulate the miRNA expression. By contrast, some viral factors were shown to reduce the expression level of CPRPs in the host by direct association with target proteins [10, 12, 29]. It remains unknown how the interaction of viral proteins with particular target proteins could lead the CPRGs transcriptome to global down-regulation. A recent study has shown that βC1 protein encoded by Geminivirus beta-satellite DNA has a role in vein chlorosis symptoms and inhibition of photosynthesis, which accompany the impaired chloroplasts structure and function and the reduced levels of CPRGs expression, in Nicotiana benthamiana plants [4]. It remains unclear how βC1 protein fulfills the pathogenic function.
Interestingly, studies have shown that the plant viruses exploit host RNA silencing machinery to modulate the virus disease symptoms through the silencing of CPRGs. In these cases, virus-derived small interfering RNAs (v-siRNAs) play pivotal roles in the viral pathogenesis. Cucumber mosaic virus-Y satellite (CMV-Y-sat) RNA was shown to induce bright yellow chlorosis through the mechanism in which a 22-nt v-siRNA derived from CMV-Y-sat RNA targets the magnesium protoporphyrin chelatase subunit I (CHLI) gene transcripts and down-regulates its expression by RNA silencing mechanism [26, 27]. The downregulation of CHLI protein, which is involved in chlorophyll biosynthesis, leads to the loss of chlorophyll production and severer chlorosis in a host-specific manner, which is dependent on the CHLI gene sequence [26, 27]. Besides the chlorosis, leaf twisting symptom has been attributed to the v-siRNA–mediated silencing of eIF4A in RSV-infected N. benthamiana [25].
A similar mechanism was shown in the pathogenesis of Peach latent mosaic viroid (PLMVd), which infects its natural host peach and causes albino phenotypes (albinism or peach calico, PC). An earlier study suggested that PLMVd replicates in the chloroplast, blocks the chloroplast development, and leads to the depletion of chloroplast-encoded proteins [21]. The PLMVd variants associated with PC contain an insertion of 12–14 nt that folds into a hairpin with a U-rich tetraloop, the sequence of which is critical for inciting the albino phenotype. The production of viroid-derived small RNAs (vd-sRNAs) upon infection by viroids is widely known [14]. Investigators analyzed the siRNA in PLMVd-infected plants and found that some vd-sRNAs encompassing the hairpin insertion would target mRNA of chloroplast-targeted heat shock protein 90 (Hsp90C). The vd-sRNAs were shown to induce the cleavage of Hsp90C mRNA, which is most likely to condition the PC symptoms [16].
These reports have suggested the involvement of different mechanisms for the induction of CPRGs down-regulation and subsequent leaf chlorosis. Besides those involving v-siRNA and vd-sRNA, however, the mechanisms underlying the CPRGs down-regulation remain to be studied. Furthermore, the molecular processes, which are triggered by the CPRGs down-regulation and lead plant leaves to chlorosis, are poorly understood. In previous studies, we have generated and analyzed the transgenic plants expressing Cauliflower mosaic virus transactivator/viroplasmin (Tav) protein under the control of a chemically inducible promoter (hereafter referred to as iTav-tobacco) [31, 32]. The iTav-tobacco shows a synchronous development of chlorosis after the treatment with a chemical inducer, and thus, provides a useful experimental system to study molecular processes in the chlorosis development.
The present study was undertaken to unravel the molecular mechanisms of chlorosis in viral and viroid diseases. Here we report the establishment of an experimental system suitable for the investigation of the mechanism underlying the chlorosis induced by the RNA silencing of Hsp90C. An induced expression of Hsp90C hairpin RNA (hpRNA) in transgenic tobacco resulted in the development of chlorosis after the treatment of a chemical inducer, supporting the finding in the previous study [16]. Leaf chlorosis in this new system was comparatively analyzed with that in iTav-tobacco.
Materials and methods
Amplification and sequencing of Nicotiana tabacum Hsp90C
Total RNA was extracted from about 100 mg of Nicotiana tabacum cv. SR1 leaf tissues using Sepasol-RNA I Super G (Nacalai Tesque, Inc., Kyoto, Japan) as described previously [32]. After treatment with the RNase-free recombinant DNase I (Takara Bio, Kusatsu, Japan) in the presence of recombinant ribonuclease inhibitor (Takara Bio) and subsequent purification using Sepasol-RNA I Super G, cDNA was synthesized from 2 µg total RNA using PrimeScript reverse transcriptase (Takara Bio) using random hexamer primers. The entire coding sequence of N. tabacum Hsp90C genes were obtained using the cDNA as a template and PCR primers listed in supplementary Table S1 as follows: firstly, a partial fragment was amplified using primers designed based on a partial sequence (Accession no. DV159063), then, upstream and downstream regions were amplified by 3′- and 5′-rapid amplification of cDNA ends (RACE) by using GeneRacer kit (Thermo Fisher Scientific, Yokohama, Japan) according to the manufacturer’s instructions, and finally, entire coding regions of two variants were amplified as single fragments using Hsp90C-150F and Hsp90C-3R-851R primers. The amplified fragments were cloned into Zero Blunt TOPO PCR cloning vector (Thermo Fisher Scientific, Yokohama, Japan) and sequenced.
Construction of Hsp90C-hpRNA Dexamethasone inducible vector
The 270 bp cDNA fragment, which is specific to Hsp90C but not found in genes for Hsp90 proteins localized in non-chloroplast subcellular compartments (Fig. 1a), was PCR amplified by using a cloned entire coding sequence as a template using the primers listed in supplementary Table S2. To form the hairpin loop structure, the coding and complementary sequences (270 bp each) were placed upstream and downstream of PCR amplified GUS spacer sequence (1025 bp), respectively. These fragments were introduced into XhoI and SpeI sites of pTA7200 vector [2] using Gibson assembly (New England Biolabs, Tokyo, Japan) according to the manufacturer’s instructions, resulting in pGVG-Hsp90C-hpRNA (Fig. 1b). After sequence confirmation using primers listed in supplementary Table S3, the recombinant plasmids were transformed into Agrobacterium tumefaciens GV3101.
Fig. 1.
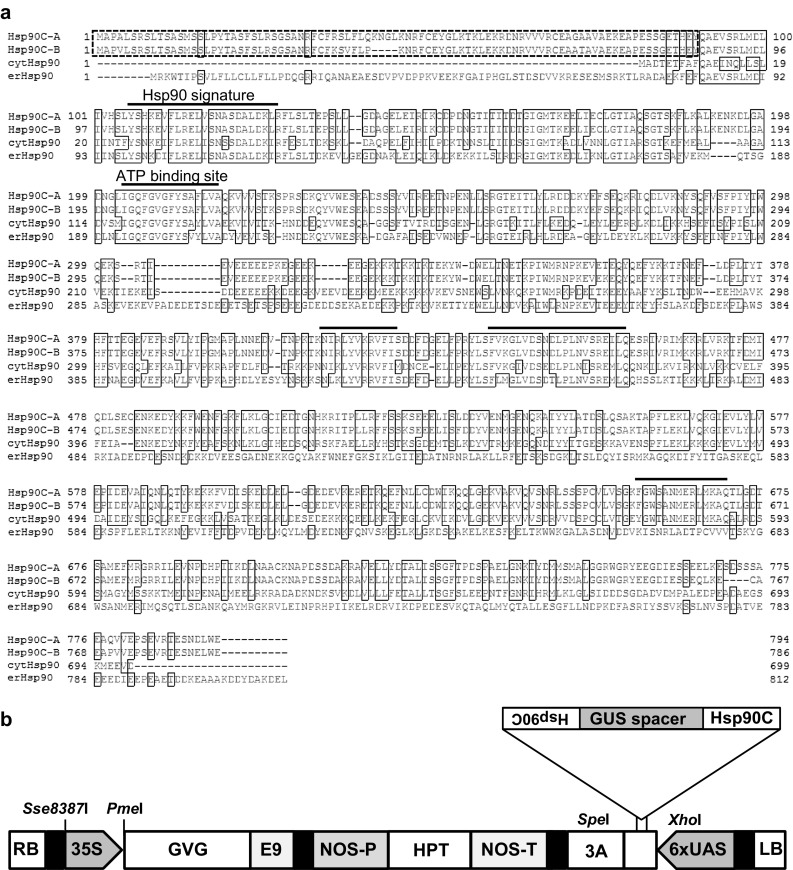
Amino acid sequences of Hsp90C protein products, a partial gene fragment of which were used for the construction of inducible silencing construct and schematic representation of the construct. a Alignment of the deduced amino acid sequences of Nicotiana tabacum chloroplast-targeted Hsp90 variants (Hsp90C-A and Hsp90C-B; Accession Nos. LC197928 and LC197929, respectively), cytosolic Hsp90 (cytHsp90; Accession No. BAE97400), and endoplasmic reticulum-targeted Hsp90 (erHsp90; Accession No. BAL42333). The N-terminal part of 90-amino acid residues that corresponds to the first 270 bp of the coding region, which was used for hpRNA construction, is shown with dashed line box. The Hsp90 family signature sequence, ATP binding site, and other four conserved motifs are denoted by thick lines. b Schematic representation of pGVG-Hsp90C-hpRNA vector used in this study. RB and LB, right and left borders; 6 × UAS six copies of the GAL4 upstream activating sequence and the −46 to +1 region of the 35S promoter; Hsp90C, 270 bp partial coding sequence of N. tabacum Hsp90CA; GUS spacer, 1025 bp partial coding sequence of GUS gene as a spacer; 3A, polyadenylation sequence of pea rbcS-3A; NOS-T, nopaline synthase terminator; HPT, hygromycin phosphotransferase gene; NOS-P, nopaline synthase gene promoter; E9, polyadenylation sequence of the pea rbcS-E9; GVG, a fusion gene of GAL4-binding domain, VP16 activation domain, and glucocorticoid receptor; 35S, Cauliflower mosaic virus 35S promoter
Plant transformation
The N. tabacum cv. SR1 plants were aseptically grown on an MS medium containing 0.8% agar but without any vitamins or plant hormones. Agrobacterium-mediated transformation of tobacco was carried out by leaf disc method using A. tumefaciens harboring pGVG-Hsp90C-hpRNA following the procedure described by Horsch et al. [8]. The transformants (T0) were selected on an MS medium supplemented with 50 µg/mL hygromycin B (Hyg). The presence of the transgene was confirmed by a PCR using Extract-N-Amp Plant PCR Kit (Sigma-Aldrich, Japan) according to the manufacturer’s instruction and primers GVGproF1 and Hsp90C-hp-286R (supplementary Table S2). PCR positive T0 plants were selected for the production of T1 and T2 seeds.
Screening of transgenic lines on a selective medium
The seeds from each transgenic line were surface sterilized with 1% (v/v) sodium hypochlorite solution and sown onto an MS medium plate containing 50 µg/mL Hyg and 20 µg/mL Meropenem as a control and another plate containing additional 1 µM Dexamethasone (Dex; Nacalai tesque, Kyoto, Japan). The plants were grown at 25 °C under the 16/8 h light/dark cycle condition. Fourteen days after sowing, the seedlings in the plates were observed for chlorosis and growth suppression phenotypes.
Growth of tobacco plants and the induction of Hsp90C silencing
T2 seeds of selected transgenic lines were sown in a Petri dish containing wet filter paper and grown under 16/8 h light/dark cycle condition at 25 °C. One week old seedlings were transferred to Jiffy 7 (Jiffy Products International, Kristiansand, Norway). The seedlings were grown for additional 2 or 3 weeks with watering every other day, but with 1000 times diluted Hyponex 6-10-5 (Hyponex Japan, Osaka, Japan) solution once a week. For induction of transgenes, transgenic tobacco plants were sprayed with freshly diluted 50 µM Dex solution containing 0.01% (v/v) Tween-20 using a spray bottle. Control plants were mock-treated by spraying with 2% ethanol solution containing 0.01% (v/v) Tween-20. The transgenic plants were observed every day for the phenotypic changes for a week and harvested at indicated time points (see below).
Estimation of total chlorophyll content
Leaf samples for total chlorophyll content estimation were collected at indicated time points. About 100 mg leaf tissues were ground to a fine powder in a centrifuge tube in the presence of liquid nitrogen by using bead- beater (Microsmash; Tomy, Tokyo, Japan) with two 5 mm stainless steel beads. The pigments were extracted with 1 mL 99.5% methanol by vortexing and subsequent centrifugation at 12,000 rpm for 10 min. The supernatant was collected in a fresh tube and absorbance was measured at 652, 665.2, and 750 nm using a spectrophotometer (UV-1800; Shimadzu, Kyoto, Japan). The total chlorophyll (a + b) content was calculated according to Porra et al. [19].
Quantitative Real Time-PCR
Total RNA was extracted, and about 10 µg of total RNA was treated with the RNase-free recombinant DNase I as described before to remove the contaminating genomic DNA. The cDNA was synthesized from the 2 µg total RNA by M-MuLV reverse transcriptase (New England Biolabs, Tokyo, Japan) using random hexamer primers. For qRT-PCR analysis of plant pathogenesis- and photosynthesis-related genes, primers listed in Supplementary Table S4 were designed by using IDT online qPCR primer designing tool (https://sg.idtdna.com/Primerquest/Home/Index). The Real-time qPCR was performed on an Applied Biosystems StepOnePlus Real-Time PCR (Applied Biosystems, USA) with KAPA SYBR FAST qPCR master mix (Nippon genetics, Tokyo, Japan). The qPCR conditions were as follows: initial holding at 95 °C for 20 s, 40 cycles of 95 °C for 3 s, 60 °C for 30 s, followed by 95 °C for 15 s, 60 °C for 1 min, 95 °C for 15 s. At the end, a melting curve was generated to analyze the specificity of the reactions. Each sample was tested in duplicates. The expression level of the EF1α gene was used as the endogenous control. The relative expression level of target genes was calculated by comparative CT (ΔΔCT) method.
Histological analysis
Leaf tissue samples were fixed with 4% glutaraldehyde in 60 mM HEPES (pH 7.0) containing 0.125 M sucrose. A post-fixation incubation was then performed with Dalton’s buffered 1% osmium tetroxide for 2 h at 4 °C. After dehydration, the tissues were embedded in Spurr resin (Nisshin EM, Tokyo, Japan). Semi-thin sections (3 µm) were prepared from at least five resin blocks and stained with 0.2% toluidine blue. Stained sections were examined using a BX-43 microscope (Olympus, Tokyo, Japan).
Results
Cloning and sequencing of N. tabacum chloroplast Hsp90
We determined nucleotide sequences for the coding regions of two Hsp90C variants through the RACE and amplification of entire coding regions. These two variants namely Hsp90C-A and Hsp90C-B coding regions consist of 2382 and 2358 nucleotides, respectively (Accession nos. LC197928 and LC197929, respectively). The ORFs of two Hsp90C variants are predicted to encode 794- and 786-amino acid proteins, respectively. Both proteins have N-terminal extensions that share homology with one another and with Hsp90Cs from other plants but are absent in cytoplasmic Hsp90. The extensions contained a feature of chloroplast transit peptide as predicted by chloroP 1.1 (http://www.cbs.dtu.dk/services/ChloroP/). Comparison with other Hsp90 family proteins as well as predicted domain structures including the transit peptide cleavage sites is described in supplementary Fig. S1.
Induced silencing of Hsp90C produces chlorosis and growth suppression phenotypes in transgenic plants
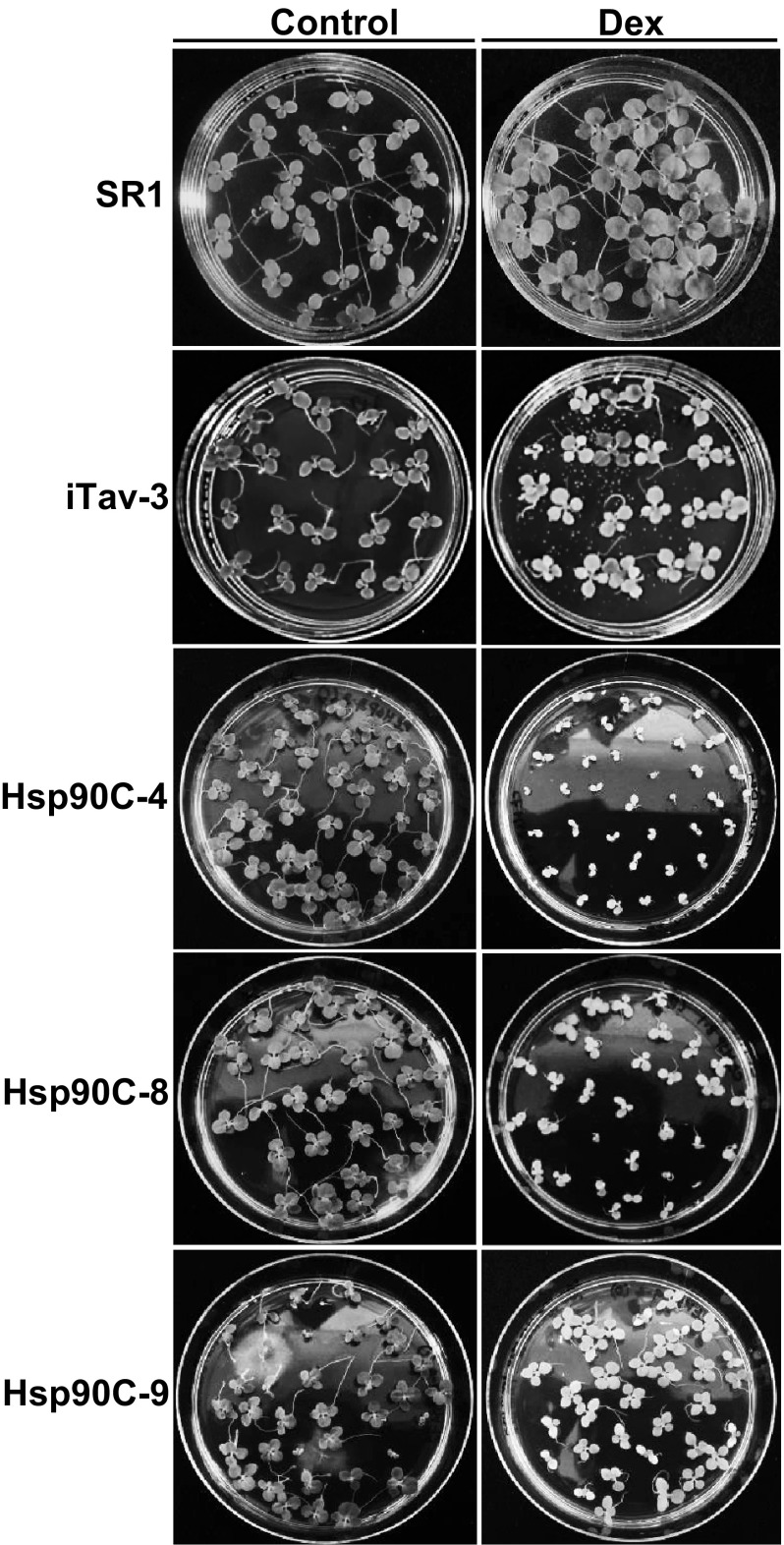
Agrobacterium-mediated transformation of tobacco brought 15 Hyg-resistant transgenic shoots and all the transgenic lines (T0) were confirmed to have the transgene by PCR (Supplementary Fig. S2). The T1 seeds from each transgenic line were screened on selection medium containing 1 µM Dex and observed for visible phenotypic changes. The transgenic lines that showed chlorosis and growth suppression phenotypes were selected to harvest T2 seeds. The chlorosis and growth suppression phenotypes were observed in the T2 generation of three transgenic lines grown on a Dex-containing medium (Fig. 2), suggesting the stable inheritance of inducible chlorosis in these lines. Chlorosis phenotypes of Hsp90C-silenced lines grown on a Dex-containing medium were more pronounced in cotyledons than in true leaves with varying degree of growth suppression among the transgenic lines. They all normally grew on a control medium that did not contain Dex. The non-transformed SR1 seedlings were grown normally on the both mediums and did not show any chlorosis.
Fig. 2.
Chlorosis and growth suppression phenotypes of T2 generation of transgenic lines grown on a medium containing Dex. The seeds of non-transformed control (SR1) were sown on an MS medium (Control) or a medium containing 1 µM Dex (Dex). Those of transgenic lines were sown on a selection medium containing 50 µg/mL Hyg alone (Control) or a medium containing both 50 µg/mL Hyg and 1 µM Dex (Dex). Previously reported iTav-3 served as a positive control. Three lines with inducible Hsp90C silencing constructs, Hsp90C-hpRNA-4, -8, and -9 (Hsp90C-4, -8, and -9, respectively), which showed different magnitudes of chlorosis and growth suppression are shown. The photographs were taken 2 weeks after sowing
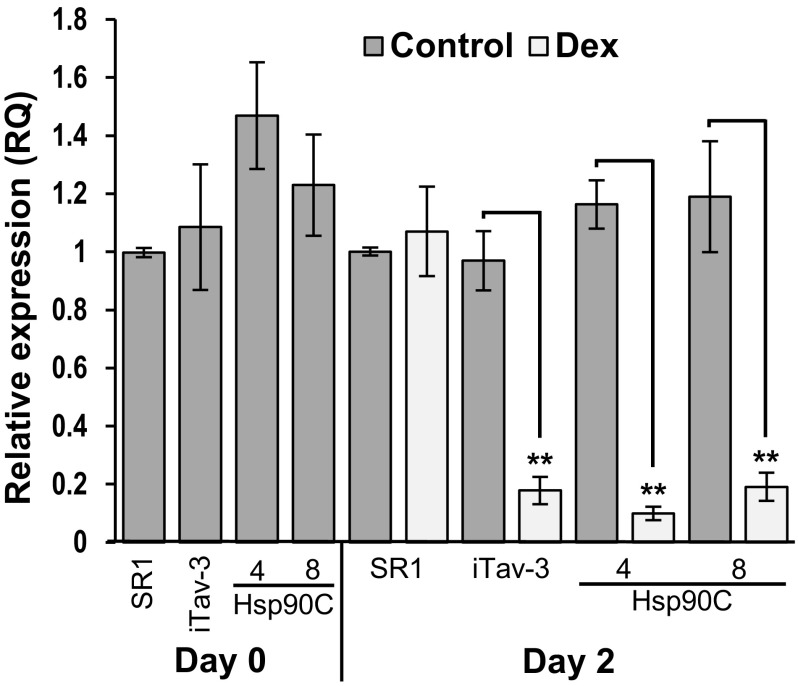
Two transgenic lines, Hsp90C-hpRNA-4 and Hsp90C-hpRNA-8 were selected for further characterization. Their 3-week-old T2 seedlings were examined for the expression of Hsp90C mRNA 2 days after spray-treated with 50 µM Dex. In mock-treated control plants, we did not find any significant difference in Hsp90C expression levels within transgenic lines and untransformed control SR1 plants (Fig. 3). It was also the case in untreated iTav-tobacco line 3 (hereafter iTav-3) and Dex-treated SR1 plants (Fig. 3). By contrast, a significant decrease in Hsp90C mRNA levels was observed not only in Hsp90C-silenced lines but also in iTav-3 (Fig. 3). The result indicates that Dex-induced expression of Hsp90C hpRNA successfully induced the silencing of Hsp90C gene expression. It also suggests that Hsp90C is downregulated in the Tav-induced chlorosis, which is consistent with our previous observation that some CPRGs were downregulated during Tav-induced chlorosis [32].
Fig. 3.
Quantitative RT-PCR analysis of Hsp90C gene expression. Non-transformed control (SR1), and three transgenic lines, iTav-3, Hsp90C-4, and -8 (see the legend for Fig. 2 for abbreviation) were analyzed for Hsp90C gene expression before (Day 0), 2 dpt with Dex (Day 2) by qRT-PCR in triplicate. The mean values of relative expression to untreated SR1 plants and their standard deviations are shown. The double asterisks show statistically significant difference between mock- and Dex-treated plants in Student t test (p < 0.01)
The mock- and Dex-treated plants were further grown and observed for their phenotypic changes up to 7 days post-treatment (dpt). Non-transformed SR1 plants did not show any phenotypic changes after Dex treatment (Fig. 4a). An additional control plant, Hsp90C-hpRNA-6, which harbors the same transgene as Hsp90C-hpRNA-4 and -8, did not show any phenotypic changes after Dex treatment (Fig. 4b). Not only iTav-3 [31, 32] but also Hsp90C-hpRNA-4 and -8 transgenic lines showed the chlorosis and growth suppression phenotypes with variation in symptom severity (Fig. 4c–e). Although iTav-3 showed a mosaic-like phenotype (Fig. 4f), Hsp90C-hpRNA-4 and -8 lines showed the uniform leaf chlorosis with leaf twisting and shrinking (Fig. 4d–h). These results indicate that the induced silencing of Hsp90C results in the chlorosis, confirming the previously reported notion that RNA silencing mediated downregulation of Hsp90C has a role in the induction of chlorotic symptom [16].
Fig. 4.
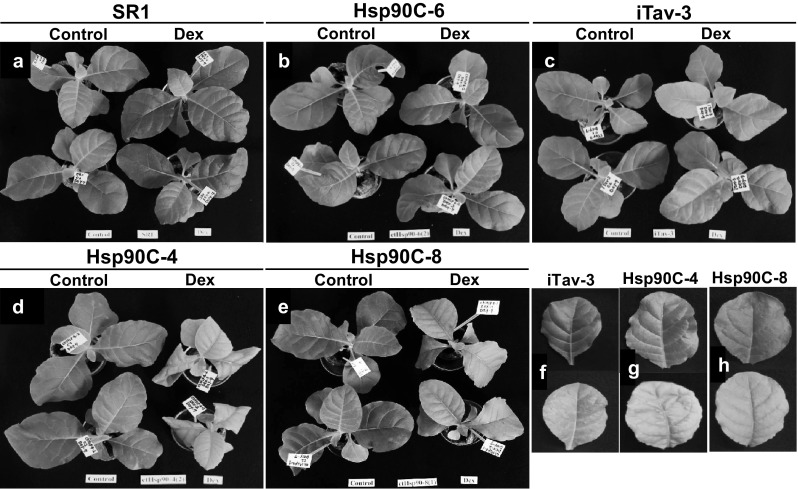
Chlorosis and growth suppression phenotypes observed in Dex-treated transgenic plants. Three-week-old plants were sprayed with 0.01% Tween-20 containing 2% ethanol (Control) or 0.01% Tween-20 containing 50 µM Dex (2% solution of 2.5 mM stock in ethanol) (Dex) every 24 h. a non-transformed control SR1. b Hsp90C-6, a Hsp90C-hpRNA transformant that did not show any response in the Dex-containing plate assay (Fig. 2), c iTav-3 as a positive control, d, e Hsp90C-4 and -8, selected Hsp90C-hpRNA transformed lines that showed clear chlorosis and growth suppression in the Dex-containing plate assay (Fig. 2). f–h Close-up photographs of leaves with symptom-like phenotypes from Dex-treated plants (lower panels) and control leaves from mock-treated plants (upper panels). f A typical chlorosis with mosaic patterns in Dex-treated iTav-3. g, h, chlorosis, leaf twisting and shrinking in Dex-treated Hsp90C-4 and Hsp90C-8, respectively. The photographs were taken at 7 dpt
Progression of chlorosis in iTav-tobacco and Hsp90C-silenced transgenic tobacco lines
Although clearly visible chlorosis was observed in iTav-3 and Hsp90C-silenced transgenic lines at 7 dpt, the progress of chlorosis in these transgenic lines might differ within the lines. To compare the progression of chlorosis in these lines, we measured total chlorophyll content in time course experiments: Total chlorophyll was measured before (day 0), and after 2, 5, and 7 days of Dex-treatment. The visible chlorosis symptoms were observed in some transgenic lines at 5 dpt or later but could not distinguish between control and Dex-treated plants at earlier time points. Although non-transformed control SR1 plants and the control transformant Hsp90C-hpRNA-6 plants showed no changes in chlorophyll content throughout the experiment, iTav-3 and Hsp90C-silenced transgenic lines showed a statistically significant decrease in total chlorophyll content as early as 2 dpt (Fig. 5a). Chlorophyll decrease in iTav-3 was pronounced during early (from day 0 to day 2) and late (from day 5 to day 7) phases of chlorosis development, whereas those in Hsp90C-silenced transgenic lines were more prominent during the middle phase (from day 2 to day 5) (Fig. 5b). The difference in kinetics of chlorophyll decrease may reflect the different mechanisms underlying the Tav-induced chlorosis and that induced by the silencing of Hsp90C.
Fig. 5.
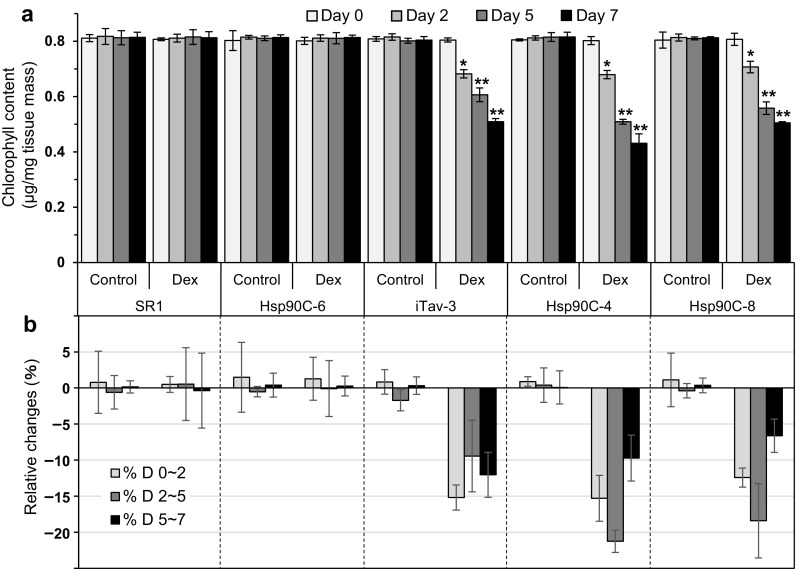
Decrease in total chlorophyll content during the progression of induced chlorosis. Three-week-old plants were mock- (Control) or Dex-treated (Dex) as in Fig. 4. Triplicate samples were collected from independent plants before (Day 0), and 2, 5, and 7 days of treatments (Day 2, Day 5 and Day 7, respectively) and chlorophyll content was measured in each of them. a Means of chlorophyll content in triplicate samples and standard deviations of the mean values are shown. Single and double asterisks indicate statistically significant difference between mock- and Dex-treated samples with p < 0.05 and p < 0.01, respectively in Student’s t-test. b Changes in chlorophyll content during early (from Day 0 to 2; % D 0–2), middle (from Day 2 to 5; % D 2–5), and late phases (from Day 5 to 7; % D 5–7) are shown in percentages to the mean values of Day 0 chlorophyll content with their standard deviations
Alterations in leaf tissue morphology by the silencing of Hsp90C
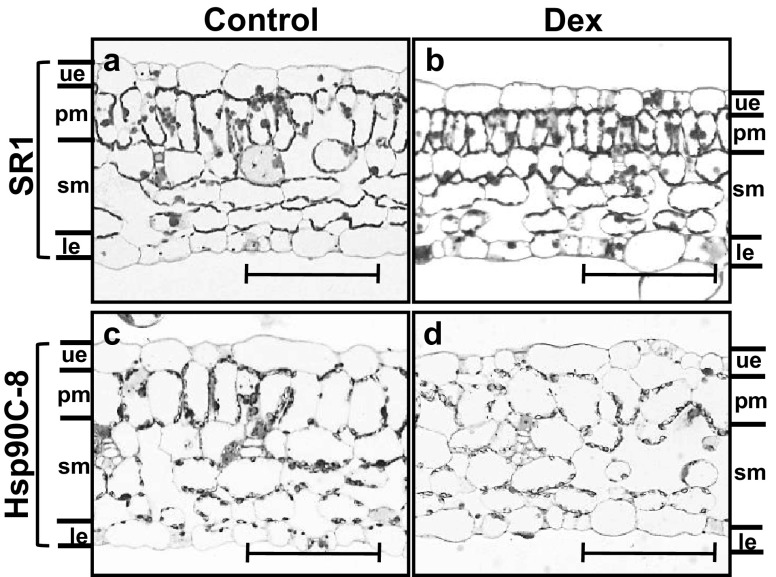
Because Hsp90C-silenced plants showed clear chlorosis, the morphology of leaf tissues was examined under a light microscope at 7 dpt in Dex- and mock-treated Hsp90C inducible silencing plants. Non-transformed control SR1 plants did not show any significant changes after Dex treatment (Fig. 6a, b). They exhibited typical epidermis (upper and lower), a palisade mesophyll cell layer, and a spongy mesophyll cell layers with some intercellular or apoplastic spaces (Fig. 6a, b). By contrast, Hsp90C inducible silencing lines showed altered leaf tissue morphology even without Dex-treatment: palisade mesophyll cells and spongy mesophyll cells were somewhat swollen as compared with SR1 leaf tissue (Fig. 6c). These morphological changes could be due to a leaky expression of the transgene and lower levels of RNA silencing of Hsp90C genes, although we failed to detect reduced Hsp90C transcript levels in mock-treated Hsp90C-hpRNA lines (Fig. 3). When treated with Dex, the Hsp90C-hpRNA lines showed less-ordered layers of further swollen mesophyll cells with reduced numbers of chloroplasts (Fig. 6d), which is consistent with aforementioned results of chlorosis development and reduced chlorophyll content (Figs. 4, 5). The morphological changes in Hsp90C-silenced lines could be attributed to an impaired chloroplast function resulted from the RNA silencing of Hsp90C genes, because developmental defects have been reported in Arabidopsis thaliana Hsp90C mutants and Hsp90C co-suppressed lines [17, 18, 30].
Fig. 6.
Histological analysis of chlorotic leaf tissue from transgenic plants with induced Hsp90C silencing. Leaf tissues from mock- and Dex-treated non-transformed control SR1, and Hsp90C-8 transgenic line were collected at 7 dpt. Transverse sections were observed under light microscope. ue upper epidermis, pm palisade mesophyll, sm spongy mesophyll, le, lower epidermis. Bar = 20 µm
Downregulation of CPRGs in transgenic tobacco plants during the early phase of chlorosis development
As mentioned before, downregulation of CPRGs is a common phenomenon in plants showing chlorosis. In this study, we found in Dex-treated iTav-3 plants the downregulation of Hsp90C (Fig. 3), which is unlikely a direct target of Tav, a nucleo-cytoplasmic shuttling protein [7]. On the other hand, previous studies have confirmed the role of Hsp90C in chloroplast biogenesis [10, 26, 27, 29]. Therefore, it is formally possible that the downregulation of Hsp90C alone could lead plants to chlorosis. To investigate whether or not the chlorosis mediated by the induced Hsp90C silencing accompanies the downregulation of other CPRGs, we examined the expression levels of ChlI, ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit (RSSU) and light harvesting chlorophyll a/b binding protein (LHCa/b) by qRT-PCR using EF1α mRNA as the internal reference. All three CPRGs were found to be significantly downregulated in Hsp90C-hpRNA-4 and -8 transgenic lines at 2 dpt, as well as in iTav-3 (Fig. 7a–c). Within Hsp90C-silenced lines, Hsp90C-hpRNA-4, which showed the severer chlorosis and morphological disorder, and the greater reduction in chlorophyll content (Figs. 4, 5), showed greater downregulation of CPRGs expression. The results suggest that the silencing of Hsp90C induces not only the chlorosis but also the downregulation of other CPRGs expression.
Fig. 7.
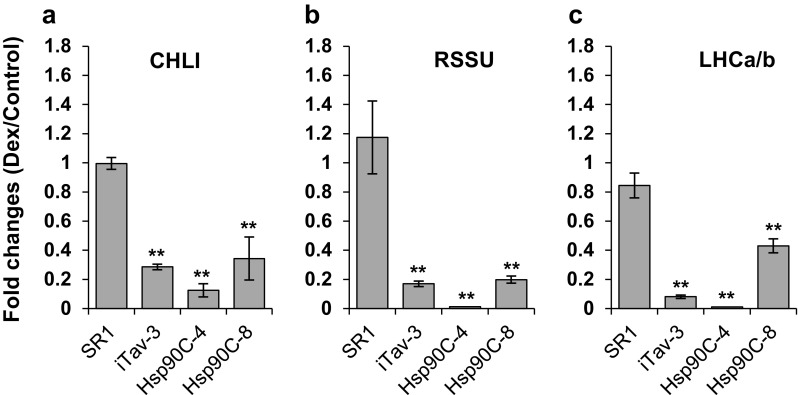
Quantitative RT-PCR analysis of chloroplast and photosynthesis-related gene expression. Non-transformed control (SR1), and three transgenic lines, iTav-3, Hsp90C-4, and -8 were mock- or Dex-treated for 2 days and analyzed for the expression of CHLI (a), RSSU (b), and LHCa/b (c) by qRT-PCR in triplicate. The mean values of relative expression in Dex-treated plants to that in mock-treated plants and the standard deviations are shown. The double asterisks show statistically significant difference between mock- and Dex-treated plants in Student t-test (p < 0.01)
Induction of plant defense genes in transgenic plants during the early phase of chlorosis development
We previously demonstrated that PR1a gene expression was upregulated in iTav-tobacco lines shortly after Dex-treatment [31, 32] as had been shown in transgenic tobacco plants constitutively expression Tav [28]. Unlike the chlorosis induced by Tav, which has been characterized as a viral pathogenesis factor [9], it seems unlikely that the silencing of a plant chloroplast protein gene, such as that of Hsp90C, activate the plant defense responses. To confirm this idea, we analyzed the expression of PR genes, including salicylic acid (SA)-responsive PR1a, ethylene-responsive PR4 [20] and jasmonic acid (JA)-responsive SAMDC [5] in SR1, iTav-3, and Hsp90C-hpRNA-4 and -8 transgenic lines. PR1a expression was upregulated 16-fold at 2 dpi with Dex in iTav-3 whereas SR1 did not show any PR1a gene activation after Dex treatment (Fig. 8a) as reported previously [31, 32]. Unexpectedly, PR1a expression was upregulated 85- and 57-folds in Hsp90C-hpRNA-4 and -8 transgenic lines at 2 dpt (Fig. 8a). PR4 expression was upregulated about 2.5-fold in iTav-3 and Hsp90C-hpRNA-4, but not in SR1 and Hsp90C-hpRNA-8 (Fig. 8b). Upregulation of SAMDC was observed only in Hsp90C-hpRNA-4 (Fig. 8c). The results indicate that not only the expression of viral pathogenesis factor such as Tav but also the silencing of Hsp90C induces the expression of PR genes, especially of PR1a, suggesting that a pathway is shared by the Tav-induced chlorosis and that induced by Hsp90C silencing.
Fig. 8.
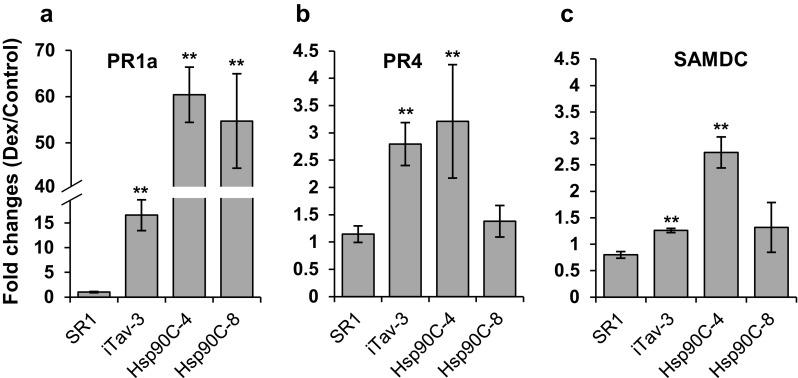
Quantitative RT-PCR analysis of plant defense gene expression. Non-transformed control (SR1), and three transgenic lines, iTav-3, Hsp90C-4, and -8 were mock- or Dex-treated for 2 days and analyzed for the expression of PR1a (a), PR4 (b), and SAMDC (c) by qRT-PCR in triplicate. The mean values of relative expression in Dex-treated plants to that in mock-treated plants and the standard deviations are shown. The double asterisks show statistically significant difference between mock- and Dex-treated plants in Student t-test (p < 0.01)
Discussion
The chlorotic symptom formation induced by DNA and RNA viruses, viroids, and satellite RNAs is a phenomenon which involves various interactions between pathogens’ and hosts’ factors [1, 11, 16, 26, 27]. In the present study, the role of the silencing of Hsp90C in chlorosis induction is established using an inducible silencing system. The silencing of Hsp90C gene was shown to have a role in the development of PC symptom in PLMVd-infected peach [16]. Although the downregulation of Hsp90C was clearly demonstrated in the report, they also demonstrated that various vd-sRNAs were produced in the infected plants and could target other host genes. Therefore, their conclusion, although quite reasonable, relies on the findings in A. thaliana mutants [30]. We generated a conditional silencing system for Hsp90C in tobacco and demonstrated that the silencing of Hsp90C alone leads plants to chlorosis, supporting the notion that Hsp90C silencing plays a major role in PLMVd pathogenesis. As discussed for iTav-tobacco in our previous report [32], the present system would also provide us with a good model system to analyze the dynamic molecular events leading to leaf chlorosis.
In this study, we selected two transgenic tobacco lines, Hsp90C-hpRNA-4 and -8 for characterization. The Hsp90C-hpRNA-4 showed severer symptom-like phenotypes—chlorosis, growth suppression, leaf twisting and shrinking—and a greater reduction in chlorophyll content than Hsp90C-hpRNA-8 (Fig. 4). The reason for these differences in symptom severity is unknown, but it might be due to the magnitude of silencing of Hsp90C genes. Indeed, the qRT-PCR analysis revealed that, after Dex treatment, the former line was more affected in the expression of Hsp90C mRNA than the latter. One may claim that the leaf twisting and shrinking phenotypes observed in the Dex-treated transgenic lines might be due to off-target effects of the induced expression of Hsp90C-hpRNA. This notion is unlikely because, firstly, we selected carefully the target region for hpRNA construction to include only the Hsp90C-specific region, which is not found in Hsp90 proteins located in different subcellular compartments, and secondly, developmental defects were reported in null mutants and co-suppression lines of A. thaliana [17, 30].
It should be noted that the leaf tissues of Hsp90C-hpRNA transgenic lines showed a somewhat affected morphology when examined under a light microscope (Fig. 6). Because the Dex-inducible expression system is known to show a leaky expression of the transgene [31], the mild morphological disorder should be attributed to the leaky expression of Hsp90C-hpRNA. The untreated transgenic lines grew normally and bore seeds as much as the untransformed parental plants, and the chlorophyll content in mock-treated transgenic lines was comparable to untransformed SR1 plants, and markedly decreased after Dex treatment (Fig. 5). Therefore, Dex-untreated and mock-treated transgenic lines can be regarded to be healthy. Nonetheless, it should be kept in mind that a lower level of Hsp90C silencing in the present system could affect plant physiology when the present system is used for precise mechanistic studies.
The expression analysis revealed that other CPRGs—CHLI, RSSU, and LHCa/b—were downregulated in Hsp90C-silenced plants as well as in Tav-expressing plants (Fig. 7). These results are consistent with the notion that the CPRGs downregulation is a common phenomenon in chlorosis mentioned before. Because Hsp90C has been proposed to have a role in the retrograde signaling from the chloroplast to the nucleus, the CPRGs downregulation could result from the signaling to the nucleus from malfunctioning chloroplast, which has been affected by the lack of Hsp90C. Also, the downregulation of Hsp90C was also observed in Tav-expressing plants (Fig. 3). The result raises a possibility that Hsp90C downregulation is a key event in the induction of chlorosis. This possibility should be tested by elucidating the mode of Hsp90C downregulation by Tav.
The expression analysis also revealed the high-level induction of PR1a gene in the Hsp90C-silenced transgenic tobacco plants. Because the PR1a is a solid marker of SA pathway activation, higher levels of SA production is anticipated in the Hsp90C-silenced plants. However, SA is known to be produced in the chloroplast [3] and it is not very likely that malfunctioning chloroplasts can produce SA efficiently. Moreover, we have previously suggested that PR1a induction during Tav-mediated chlorosis is independent of SA. Because Hsp90C is known to be involved in chloroplast biogenesis and retrograde signaling in A. thaliana and Chlamydomonas reinhardtti [6, 30, 33, 34], an unknown SA-independent signaling from the affected chloroplast may have a role in the regulation of the PR1a nuclear gene. On the other hand, higher production of ROS is anticipated in malfunctioning chloroplasts of Hsp90C-silenced plants [36]. The generation of ROS in the chloroplasts depends on multiple aspects of chloroplast physiology including photosynthesis, gene expression, chlorophyll (tetrapyrrole) biosynthesis, and hormonal control [3]. Elevated ROS inside the chloroplast results in transcriptional reprogramming through identified and unknown components of the retrograde signaling but also through hormone signaling, e.g., increased production of SA [24, 31]. Therefore, it is of great importance to quantify SA levels in the future study to analyze the molecular mechanisms underlying chlorosis mediated by the silencing of Hsp90C.
In this and our previous studies, the chlorosis along with growth suppression phenotypes was observed in two different inducible chlorosis systems: iTav and Hsp90C-hpRNA. In addition to this, the time course study for chlorosis progression revealed that the processes leading to chlorosis could be monitored as early as 2 dpt, which could provide a better experimental model to study the molecular events taken place before and after the onset of chlorosis.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Kazue Obara for technical assistance. This study was supported in part by The United Graduate School of Agricultural Sciences, Ehime University, and JSPS KAKENHI grants 26292026 and 15K14664 to Kobayashi. Bhor has been supported by Rotary Yoneyama Memorial Foundation.
References
- 1.Abbink TEM, Peart JR, Mos TNM, Baulcombe DC, Bol JF, Linthorst HJM. Silencing of a gene encoding a protein component of the oxygen-evolving complex of photosystem II enhances virus replication in plants. Virology. 2002;295:307–319. doi: 10.1006/viro.2002.1332. [DOI] [PubMed] [Google Scholar]
- 2.Aoyama T, Chua NH. A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 1997;11:605–612. doi: 10.1046/j.1365-313X.1997.11030605.x. [DOI] [PubMed] [Google Scholar]
- 3.Asada K. Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 2006;141:391–396. doi: 10.1104/pp.106.082040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhattacharyya D, Gnanasekaran P, Kumar RK, Kushwaha NK, Sharma VK, Yusuf MA, Chakraborty S. A geminivirus betasatellite damages the structural and functional integrity of chloroplasts leading to symptom formation and inhibition of photosynthesis. J Exp Bot. 2015;66:5881–5895. doi: 10.1093/jxb/erv299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biondi S, Scaramagli S, Capitani F, Altamura MM, Torrigiani P. Methyl jasmonate upregulates biosynthetic gene expression, oxidation and conjugation of polyamines, and inhibits shoot formation in tobacco thin layers. J Exp Bot. 2001;52:231–242. doi: 10.1093/jexbot/52.355.231. [DOI] [PubMed] [Google Scholar]
- 6.Cao D, Froehlich JE, Zhang H, Cheng CL. The chlorate-resistant and photomorphogenesis-defective mutant cr88 encodes a chloroplast-targeted HSP90. Plant J. 2003;33:107–118. doi: 10.1046/j.1365-313X.2003.016011.x. [DOI] [PubMed] [Google Scholar]
- 7.Haas M, Geldreich A, Bureau M, Dupuis L, Leh V, Vetter G, Kobayashi K, Hohn T, Ryabova L, Yot P, Keller M. The open reading frame VI product of Cauliflower mosaic virus is a nucleocytoplasmic protein: its N terminus mediates its nuclear export and formation of electron-dense viroplasms. Plant Cell. 2005;17:927–943. doi: 10.1105/tpc.104.029017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horsch RB, Fry JE, Hoffmann NL, Wallroth M, Eichholtz D, Rogers SG. A simple method for transferring genes into plants. Science. 1985;227:1229–1231. doi: 10.1126/science.227.4691.1229. [DOI] [PubMed] [Google Scholar]
- 9.Kobayashi K, Hohn T. The avirulence domain of Cauliflower mosaic virus transactivator/viroplasmin is a determinant of viral virulence in susceptible hosts. Mol Plant Microbe Interact. 2004;17:475–483. doi: 10.1094/MPMI.2004.17.5.475. [DOI] [PubMed] [Google Scholar]
- 10.Kong L, Wu J, Lu L, Xu Y, Zhou X. Interaction between Rice stripe virus disease-specific protein and host PsbP enhances virus symptoms. Mol Plant. 2014;7:691–708. doi: 10.1093/mp/sst158. [DOI] [PubMed] [Google Scholar]
- 11.Lehto K, Tikkanen M, Hiriart J-B, Paakkarinen V, Aro E-M. Depletion of the photosystem II core complex in mature tobacco leaves infected by the flavum strain of Tobacco mosaic virus. Mol Plant Microbe Interact. 2003;16:1135–1144. doi: 10.1094/MPMI.2003.16.12.1135. [DOI] [PubMed] [Google Scholar]
- 12.Lin L, Luo Z, Yan F, Lu Y, Zheng H, Chen J. Interaction between potyvirus P3 and ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO) of host plants. Virus Genes. 2011;43:90–92. doi: 10.1007/s11262-011-0596-6. [DOI] [PubMed] [Google Scholar]
- 13.Manfre A, Glenn M, Nuñez A, Moreau RA, Dardick C. Light quantity and photosystem function mediate host susceptibility to Turnip mosaic virus via a salicylic acid-independent mechanism. Mol Plant Microbe Interact. 2011;24:315–327. doi: 10.1094/MPMI-08-10-0191. [DOI] [PubMed] [Google Scholar]
- 14.Martínez de Alba AE Flores R, Hernández C. Two chloroplastic viroids induce the accumulation of small RNAs associated with posttranscriptional gene silencing. J Virol. 2002;76:13094–13096. doi: 10.1128/JVI.76.24.13094-13096.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mochizuki T, Ogata Y, Hirata Y, Ohki ST. Quantitative transcriptional changes associated with chlorosis severity in mosaic leaves of tobacco plants infected with Cucumber mosaic virus. Mol Plant Pathol. 2014;15:242–254. doi: 10.1111/mpp.12081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Navarro B, Gisel A, Rodio ME, Delgado S, Flores R, Di Serio F. Small RNAs containing the pathogenic determinant of a chloroplast- replicating viroid guide the degradation of a host mRNA as predicted by RNA silencing. Plant J. 2012;70:991–1003. doi: 10.1111/j.1365-313X.2012.04940.x. [DOI] [PubMed] [Google Scholar]
- 17.Oh SE, Yeung C, Babaei-Rad R, Zhao R. Cosuppression of the chloroplast localized molecular chaperone HSP90.5 impairs plant development and chloroplast biogenesis in Arabidopsis. BMC Res Notes. 2014;7:643. doi: 10.1186/1756-0500-7-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pogorelko GV, Kambakam S, Nolan T, Foudree A, Zabotina OA, Rodermel SR. Impaired chloroplast biogenesis in Immutans, an Arabidopsis variegation mutant, modifies developmental programming, cell wall composition and resistance to Pseudomonas syringae. PLoS ONE. 2016;11:1–19. doi: 10.1371/journal.pone.0150983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Porra RJ, Thompson WA, Kriedemann PE. Determination of accurate extinction coefficients and simultaneous-equations for assaying chlorophyll-a and chlorophyll-b extracted with 4 different solvents: verification of the concentration of chlorophyll standards by atomic-absorption spectroscopy. Biochim Biophys Acta. 1989;975:384–394. doi: 10.1016/S0005-2728(89)80347-0. [DOI] [Google Scholar]
- 20.Qin J, Zuo K, Zhao J, Ling H, Cao Y, Qiu C, Li F, Sun X, Tang K. Overexpression of GbERF confers alteration of ethylene-responsive gene expression and enhanced resistance to Pseudomonas syringae in transgenic tobacco. J Biosci. 2006;31:255–263. doi: 10.1007/BF02703918. [DOI] [PubMed] [Google Scholar]
- 21.Rodio M-E, Delgado S, De Stradis A, Gómez M-D, Flores R, Di Serio F. A viroid RNA with a specific structural motif inhibits chloroplast development. Plant Cell. 2007;19:3610–3626. doi: 10.1105/tpc.106.049775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodriguez A, Angel CA, Lutz L, Leisner SM, Nelson RS, Schoelz JE. Association of the P6 protein of Cauliflower mosaic virus with plasmodesmata and plasmodesmal proteins. Plant Physiol. 2014;166:1345–1358. doi: 10.1104/pp.114.249250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Satoh K, Kondoh H, Sasaya T, Shimizu T, Choi IR, Omura T, Kikuchi S. Selective modification of rice (Oryza sativa) gene expression by Rice stripe virus infection. J Gen Virol. 2010;91:294–305. doi: 10.1099/vir.0.015990-0. [DOI] [PubMed] [Google Scholar]
- 24.Shapiguzov A, Vainonen JP, Wrzaczek M, Kangasjärvi J. ROS-talk: how the apoplast, the chloroplast, and the nucleus get the message through. Front Plant Sci. 2012;3:292. doi: 10.3389/fpls.2012.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi B, Lin L, Wang S, Guo Q, Zhou H, Rong L, Li J, Peng J, Lu Y, Zheng H, Yang Y, Chen Z, Zhao J, Jiang T, Song B, Chen J, Yan F. Identification and regulation of host genes related to Rice stripe virus symptom production. New Phytol. 2016;209:1106–1119. doi: 10.1111/nph.13699. [DOI] [PubMed] [Google Scholar]
- 26.Shimura H, Pantaleo V, Ishihara T, Myojo N, Inaba J-I, Sueda K, Burgyan J, Masuta C. A viral satellite RNA induces yellow symptoms on tobacco by targeting a gene involved in chlorophyll biosynthesis using the RNA silencing machinery. PLoS Pathog. 2011;7:1–12. doi: 10.1371/journal.ppat.1002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith NA, Eamens AL, Wang MB. Viral small interfering RNAs target host genes to mediate disease symptoms in plants. PLoS Pathog. 2011;7:1–9. doi: 10.1371/journal.ppat.1002022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takahashi H, Shimamoto K, Ehara Y. Cauliflower mosaic virus gene VI causes growth suppression, development of necrotic spots and expression of defence-related genes in transgenic tobacco plants. Mol Gen Genet. 1989;216:188–194. doi: 10.1007/BF00334355. [DOI] [Google Scholar]
- 29.Tu Y, Jin Y, Ma D, Li H, Zhang Z, Dong J, Wang T. Interaction between PVY HC-Pro and the NtCF1β-subunit reduces the amount of chloroplast ATP synthase in virus-infected tobacco. Sci Rep. 2015;5:15605. doi: 10.1038/srep15605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vinti G, Fourrier N, Bowyer JR, López-Juez E. Arabidopsis cue mutants with defective plastids are impaired primarily in the photocontrol of expression of photosynthesis-associated nuclear genes. Plant Mol Biol. 2005;57:343–357. doi: 10.1007/s11103-004-7867-8. [DOI] [PubMed] [Google Scholar]
- 31.Waliullah S, Kosaka N, Yaeno T, Ali ME, Sekine KT, Atsumi G, Yamaoka N, Nishiguchi M, Takahashi H, Kobayashi K. Cauliflower mosaic virus Tav protein induces leaf chlorosis in transgenic tobacco through a host response to virulence function of Tav. J Gen Plant Pathol. 2015;81:261–270. doi: 10.1007/s10327-015-0600-4. [DOI] [Google Scholar]
- 32.Waliullah S, Mochizuki T, Sekine KT, Atsumi G, Ali ME, Yaeno T, Yamaoka N, Nishiguchi M, Kobayashi K. Artificial induction of a plant virus protein in transgenic tobacco provides a synchronous system for analyzing the process of leaf chlorosis. Physiol Mol Plant Pathol. 2014;88:43–51. doi: 10.1016/j.pmpp.2014.08.005. [DOI] [Google Scholar]
- 33.Willmund F, Dorn KV, Schulz-Raffelt M, Schroda M. The chloroplast DnaJ homolog CDJ1 of Chlamydomonas reinhardtii is part of a multichaperone complex containing HSP70B, CGE1, and HSP90C. Plant Physiol. 2008;148:2070–2082. doi: 10.1104/pp.108.127944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Willmund F, Schroda M. HEAT SHOCK PROTEIN 90C is a bona fide Hsp90 that interacts with plastidic HSP70B in Chlamydomonas reinhardtii. Plant Physiol. 2005;138:2310–2322. doi: 10.1104/pp.105.063578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang J, Zhang F, Li J, Chen JP, Zhang HM. Integrative analysis of the microRNAome and transcriptome illuminates the response of susceptible rice plants to Rice stripe virus. PLoS ONE. 2016;11:1–21. doi: 10.1371/journal.pone.0146946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao J, Zhang X, Hong Y, Liu Y. Chloroplast in plant-virus interaction. Front Microbiol. 2016;7:1–20. doi: 10.3389/fmicb.2016.01565. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.