Abstract
An on-going surveillance program on Crimean-Congo haemorrhagic fever virus (CCHFV) in Iran has been launched since 2000. An outbreak of CCHF occurred in northern Iran between June and July 2015. Three cases were involved in this outbreak. One patient died after admission to hospital, and the others were treated successfully. Phylogenetic analysis showed that three sequences obtained from Iranian patients grouped within clade IV (Asia-1), clade V (Europe) and clade VI (Greece). The partial sequence of the strain Noshahr59 (KT588642) showed the highest similarity with other strains isolated from Russia, Kosovo and Turkey (Clade V, Europe). The genome sequence of the strain Chalous65 (KT588640) showed 100% homology to the strain AP29 isolated from Greece (DQ211638). The genome sequence of the strain Noshahr43 (KT588641) showed 88% similarity to the Pakistani and previously reported Iranian strains (AF527810, AJ538198, AY366379 and AY366373). These data support previous studies, which showed a distinct similarity between Iranian S segments of CCHFV strains with other strains within clade IV (Asia-1) and clade V (Europe). In addition, clade VI was detected for the first time in Iran. Moreover, strain Chalous65 with similar genetic characteristics to strain AP29 from Greece was isolated from a fatal human case.
Electronic supplementary material
The online version of this article (doi:10.1007/s13337-016-0359-z) contains supplementary material, which is available to authorized users.
Keywords: CCHFV, RT-PCR, Bunyavirus, Genome, Sequence, Phylogenetic, Iran
Introduction
Crimean-Congo haemorrhagic fever virus (CCHFV) is a tick-borne disease. The virus is a member of the Bunyaviridae family (Nairovirus genus) that causes severe haemorrhagic disease in humans, with a case fatality rate as high as 30–40%. Abattoir workers, shepherds, veterinarians and other persons in close contact with livestock and ticks are at risk of infection [3]. Different hard ticks including the genera Hyalomma, Rhipicephalus, and Haemaphysalis, as well as a low number of soft ticks (genus Argas) were shown to be the main vectors and reservoirs of this virus in Iran [2, 11, 12]. In addition to zoonotic transmission, CCHFV can spread from person to person through blood and is one of the rare haemorrhagic fever viruses able to cause nosocomial outbreaks in hospitals among health care workers [4, 5]. CCHF symptoms can include fever, chills, severe headache, dizziness, back, and abdominal pains, vomiting, nausea, diarrhea and hemorrhagic manifestations [14].
CCHFVs are relatively divergent in its genome sequence and the viruses are grouped in seven distinct clades based on the S segment sequence analysis. Clade I in West-Africa, clade II in central Africa, clade III in South- and West-Africa, clade IV in the Middle-East and Asia, clade V in Europe and clade VI in Greece. The clade IV may be divided into two distinct clades, Asia-1 and Asia-2 [6, 14].
In 2015, three human cases were hospitalised in the northern part of Iran with suspected symptoms for CCHF. These cases were interesting as ‘suspect’ infection with CCHFV in such a short period of time is rare in this region of Iran. The principal objective of this study was the genetic characterisation including a phylogenetic analysis of CCHFV strains from human cases in northern Iran.
Materials and methods
Laboratory tests
This study was conducted on samples received based on a surveillance and control program of CCHF in Iran between June and July 2015. Three human cases were hospitalised after suffering from fever and haemorrhage. Human serum samples were taken according to the protocol of National Expert Committee (NEC) on viral haemorrhagic fevers in Iran. To investigate the serum samples an ELISA was initially used to detect CCHFV-specific IgM and IgG in human serum. For IgM detection, the ELISA plate was coated with the goat IgG fraction to human IgM. Sera sample were added to plates and incubated. Then, immuno ascites was added and the plate was incubated for 1 h at 37 °C. Peroxidase labelled anti-mouse immunoglobulin was added and the plate was incubated again. After washing, hydrogen peroxide and TMB were added and the plate was incubated at room temperature. Finally, plate was read by ELISA reader. IgG detection involved coating the ELISA plate with mouse hyper immune ascetic fluid in PBS and incubating overnight. Following washing step, the recombinant antigen and skimmed milk were added, and the plate was incubated for 3 h. Diluted serum was added and the plate was incubated. Peroxidase-labelled anti-animal immunoglobulin was added and finally, hydrogen peroxide (H2O2) and 3,3,5,5` tetramethyl benzidine (TMB) was added and the plate was incubated for 15 min. The enzymatic reaction was stopped and the plate was read by the ELISA reader [7]. Viral RNA was extracted by using a Viral RNA extraction kit (Qiagen GmbH, Hilden, Germany). The RNA was analysed by one-step RT-PCR, which amplifies a 536-bp fragment of the S segment of the CCHFV genome. PCR machine was planned on 30 min at 50 °C for reverse transcription reaction (cDNA synthesis); followed by 35 cycles of 95 °C for 30 s, 50 °C for 30 s, 72 °C for 45 s, and a final extension at 72 °C/5 min. The positive molecular result for CCHFV demonstrates an acute CCHFV infection due to detection of viral RNA during the viraemic phase. The amplified fragments from the RT-PCR were purified and Sanger sequenced. The partial sequences described in this study have been deposited in the GenBank database under Accession Nos. KT588640, KT588641 and KT588642 [8].
Bioinformatic analysis
In addition to the three CCHFV virus sequences obtained from these Iranian patients, previously available S-segment sequences representing all seven clades of CCHFV were retrieved from GenBank at www.ncbi.nih.gov and incorporated into the alignments for the phylogenetic analyses. The sequence alignment was undertaken using Clustal W and phylogenetic trees were generated by the maximum likelihood (ML) method with Kimura 2-parameter distance using Mega 6 software. Bootstrap confidence limits were based on 1000 replicates. This method evaluates the topologies of different trees and chooses the best tree based on a specified model. This model is based on the evolutionary process that can account for the conversion of one sequence into another [8].
Results and discussion
Case Chalous65 died, but two other cases (Noshahr59 and Noshahr43) were successfully rehabilitated. The IgM test for cases Noshahr59 and Noshahr43 were positives, and negative for the Chalous65. IgG was not detected in any of the serum samples tested, as all samples were taken at initial phase of infection. (Table 1).
Table 1.
Patient status and diagnosis details
| Strain | Date | Diagnostic results |
|---|---|---|
| Noshahr59 | 2015 | RT-PCR +/IgM+/IgG− |
| Chalous65 | 2015 | RT-PCR +/IgM−/IgG− |
| Noshahr43 | 2015 | RT-PCR+/IgM+/IgG− |
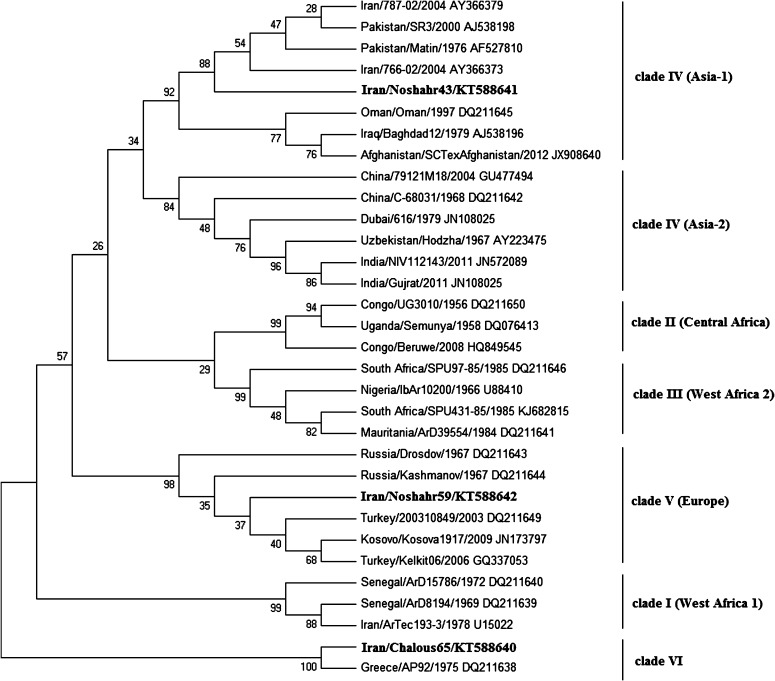
All three cases tested positive for the presence of CCHFV RNA. In the S segment, the phylogenetic tree based on ML demonstrated that three sequences obtained from the Iranian patients grouped within clade IV (Asia-1), clade V (Europe) and clade VI (Greece). Specifically, the Noshahr59 (KT588642) sequence showed the highest similarity to the sequences from strains isolated in Russia, Kosovo and Turkey (clade V, Europe). The Chalous65 (KT588640) sequence showed 100% homology to the sequence from strain AP29 isolated in Greece (DQ211638). Interestingly, the Noshahr43 (KT588641) sequence presented the highest identity (88%) to a Pakistani and previously reported Iranian strains (AF27810, AJ538198, AY366379, AY366373) (Fig. 1).
Fig. 1.
Phylogenetic relationships between sequences obtained from this study and respective representative CCHFV strains from GenBank. The GenBank accession number and the geographic origin are given for each isolate. The numerical values in the tree represent bootstrap results. The sequences obtained from this study are in bold
The number of CCHF patients among the Iranian population is of concern and is being more regularly monitored by the Iranian health authorities. By advancing the knowledge of CCHF and based on our previous studies of the molecular epidemiology and genetic diversity of CCHFV in Iran [9, 10], this investigation focused on recent samples and provided an insight into the genetic origins of CCHFV that circulates in Iran. Thus, phylogenetic analysis of the CCHFV strains in the northern of Iran was undertaken in order to trace the potential origin of CCHFV strains from symptomatic patients. An interesting observation from this molecular survey at the nucleotide level is evidence for the detection of three different CCHFV genome variants in northern Iran. It was previously observed that European CCHFV strain only circulate in northern Iran [6]. Although, Iran has been known to be a CCHF-endemic country since 1999, the northern part of Iran was not considered as an area with high prevalence of human CCHF cases. Therefore, the detection of three human CCHF cases with different genomic origins in a short period of time in Northern province of Iran is noteworthy and reflects the fact that different lineages of CCHFV exist in Iran (Asia-1, Asia-2, Europe and Greece) and circulate by infected hosts and vectors.
These data reported in this study are in conformity with previous studies, which showed similarity between Iranian S segments of CCHFV strains with other strains within clade IV (Asia-1) and clade V (Europe) [1]. In addition, clade VI was detected for the first time in Iran. Clade VI contains strains isolated from Rhipicephalus bursa ticks in Greece (including the prototype strain AP29) and from human in Turkey. Moreover, strain Chalous65 with similar genetic characteristics to strain AP29 from Greece caused a fatality. Our data supports previous studies that reported strains in clade VI that are currently associated with clinical symptoms in human cases. It was suggested that the large genetic difference for AP29 is related to the different genus of the tick vector. Thus, the adaptation of CCHFV to region-specific vectors and hosts leads to the emergence of local virus variants with different pathogenicity for humans [13].
The genetic diversity of CCHFV strains among Iranian CCHF patients in Northern Province of Iran is reported. The Iranian public health authorities should consider how to implement preventive measures in order to reduce the risk of CCHFV cases in Iran and clinicians should include CCHFV in the differential diagnosis of febrile syndromes accompanied by thrombocytopenia and haemorrhagic, especially in endemic regions.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgement
We thank all technical assistance for laboratory analysis.
References
- 1.Biglari P, Chinikar S, Belqeiszadeh H, Telmadarraiy Z, Mostafavi E, Ghaffari M, et al. Phylogeny of tick-derived Crimean-Congo hemorrhagic fever virus strains in Iran. Ticks Tick Borne Dis. 2016;7:1216–1221. doi: 10.1016/j.ttbdis.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 2.Champour M, Chinikar S, Mohammadi G, Razmi G, Shah-Hosseini N, Khakifirouz S, et al. Molecular epidemiology of Crimean–Congo hemorrhagic fever virus detected from ticks of one humped camels (Camelus dromedarius) population in northeastern Iran. J Parasit Dis. 2014;40:110–115. [DOI] [PMC free article] [PubMed]
- 3.Champour M, Mohammadi GR, Chinikar S, Razmi G, Shah-Hosseini N, Khakifirouz S, et al. Seroepidemiology of Crimean-Congo hemorrhagic fever virus in one-humped camels (Camelus dromedarius) population in northeast of Iran. J Vector Borne Dis. 2014;51:62–65. [PubMed] [Google Scholar]
- 4.Chinikar S, Ghiasi SM, Naddaf S, Piazak N, Moradi M, Razavi MR, et al. Serological evaluation of Crimean-Congo hemorrhagic fever in humans with high-risk professions living in enzootic regions of Isfahan Province of Iran and genetic analysis of circulating strains. Vector Borne Zoonotic Dis. 2012;12:733–738. doi: 10.1089/vbz.2011.0634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chinikar S, Shayesteh M, Khakifirouz S, Jalali T, Rasi Varaie FS, Rafigh M, et al. Nosocomial infection of Crimean-Congo haemorrhagic fever in eastern Iran: case report. Travel Med Infect Dis. 2013;11:252–255. doi: 10.1016/j.tmaid.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 6.Chinikar S, Shah-Hosseini N, Bouzari S, Jalali T, Shokrgozar MA, Mostafavi E. New circulating genomic variant of Crimean-Congo hemorrhagic fever virus in Iran. Arch Virol. 2013;158:1085–1088. doi: 10.1007/s00705-012-1588-0. [DOI] [PubMed] [Google Scholar]
- 7.Chinikar S, Shah-Hosseini N, Bouzari S, Shokrgozar MA, Mostafavi E, Jalali T, et al. Assessment of recombination in the S-segment genome of Crimean-Congo hemorrhagic fever virus in Iran. J Arthropod Borne Dis. 2016;10:12–23. [PMC free article] [PubMed] [Google Scholar]
- 8.Chinikar S, Bouzari S, Shokrgozar MA, Mostafavi E, Jalali T, Khakifirouz S, et al. Genetic diversity of Crimean Congo hemorrhagic fever virus strains from Iran. J Arthropod Borne Dis. 2016;10:127–140. [PMC free article] [PubMed] [Google Scholar]
- 9.Farhadpour F, Telmadarraiy Z, Chinikar S, Akbarzadeh K, Moemenbellah-Fard M, Faghihi F, et al. Molecular detection of Crimean-Congo haemorrhagic fever virus in ticks collected from infested livestock populations in a New Endemic Area, South of Iran. Trop Med Int Health. 2016;21:340–347. doi: 10.1111/tmi.12667. [DOI] [PubMed] [Google Scholar]
- 10.Kayedi MH, Chinikar S, Mostafavi E, Khakifirouz S, Jalali T, Hosseini-Chegeni A, et al. Crimean-Congo hemorrhagic fever virus clade IV (Asia 1) in ticks of Western Iran. J Med Entomol. 2015;52:1144–1149. doi: 10.1093/jme/tjv081. [DOI] [PubMed] [Google Scholar]
- 11.Mehravaran A, Moradi M, Telmadarraiy Z, Mostafavi E, Moradi AR, Khakifirouz S, et al. Molecular detection of Crimean-Congo haemorrhagic fever (CCHF) virus in ticks from southeastern Iran. Ticks Tick Borne Dis. 2013;4:35–38. doi: 10.1016/j.ttbdis.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 12.Mohammadian M, Chinikar S, Telmadarraiy Z, Vatandoost H, Oshaghi MA, Hanafi-Bojd AA, et al. Molecular assay on crimean congo hemorrhagic fever virus in ticks (ixodidae) collected from Kermanshah Province, Western Iran. J Arthropod Borne Dis. 2016;10:383–393. [PMC free article] [PubMed] [Google Scholar]
- 13.Papa A, Dalla V, Papadimitriou E, Kartalis G, Antoniadis A. Emergence of Crimean-Congo haemorrhagic fever in Greece. Clin Microbiol Infect. 2010;16:843–847. doi: 10.1111/j.1469-0691.2009.02996.x. [DOI] [PubMed] [Google Scholar]
- 14.Whitehouse CA. Crimean-Congo hemorrhagic fever. Antivir Res. 2004;64:145–160. doi: 10.1016/S0166-3542(04)00163-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.