Abstract
Mutation of the IDH1 gene is associated with differences in malignant astrocytoma growth characteristics that impact phenotypic severity, including cognitive impairment. We previously demonstrated greater cognitive impairment in patients with IDH1 wild type tumor compared to those with IDH1 mutant, and therefore we hypothesized that brain network organization would be lower in patients with wild type tumors. Volumetric, T1-weighted MRI scans were obtained retrospectively from 35 patients with IDH1 mutant and 32 patients with wild type malignant astrocytoma (mean age = 45 ± 14 years) and used to extract individual level, gray matter connectomes. Graph theoretical analysis was then applied to measure efficiency and other connectome properties for each patient. Cognitive performance was categorized as impaired or not and random forest classification was used to explore factors associated with cognitive impairment. Patients with wild type tumor demonstrated significantly lower network efficiency in several medial frontal, posterior parietal and subcortical regions (p < 0.05, corrected for multiple comparisons). Patients with wild type tumor also demonstrated significantly higher incidence of cognitive impairment (p = 0.03). Random forest analysis indicated that network efficiency was inversely, though nonlinearly associated with cognitive impairment in both groups (p < 0.0001). Cognitive reserve appeared to mediate this relationship in patients with mutant tumor suggesting greater neuroplasticity and/or benefit from neuroprotective factors. Tumor volume was the greatest contributor to cognitive impairment in patients with wild type tumor, supporting our hypothesis that greater lesion momentum between grades may cause more disconnection of core neurocircuitry and consequently lower efficiency of information processing.
Keywords: IDH1, Astrocytoma, Connectome, Cognition, MRI
Introduction
Malignant astrocytoma are an aggressive form of cerebral neoplasm, though proliferation and invasion characteristics can vary widely, even among tumors within the same histopathological grade [1]. Recently, tumor molecular markers have been identified as better predictors of growth kinetics than classical histology. Specifically, patients with mutation of the isocitrate dehydrogenase 1 (IDH1) gene [2] exhibit a marked survival benefit over patients with the wild-type tumor, independent of age and histologic grading [3, 4]. Such survival differences are believed to reflect growth characteristics unique to IDH1 subtypes. Mutant gliomas have a more diffuse pattern of growth and slower rate of cell proliferation, both of which are associated with better prognosis [1].
Differences in tumor proliferation and invasion characteristics, or “lesion momentum” [5] may also impact cognitive impairment, which is a frequent presenting problem in these patients [6]. We recently demonstrated that patients with wild type malignant gliomas show greater cognitive dysfunction compared to their mutant counterparts [7]. Additionally, greater lesion size was associated with worse cognition almost exclusively in patients with wild type tumors. Taken together, these findings suggest that the more severe cognitive impairment in patients with wild type malignant gliomas may relate to reduced neuroplasticity attributable to the greater lesion momentum characteristic of this molecular variant of malignant astrocytoma.
Measurements of brain connectivity can provide important insights regarding the neural mechanisms underlying cognitive function and dysfunction. Although tumors represent focal lesions within the brain, they tend to be associated with global cognitive deficits [8], suggesting both local and distant disruption of neurocircuitry. In fact, studies have demonstrated that cognitive performance is associated with reduced connectivity of large-scale, functional brain networks in both low and high grade gliomas [9–11].
Brain networks can be evaluated, among other methods, using coordinated variations in gray matter volumes. These structural covariance networks are believed to reflect underlying axonal connections as well as common genetic, neurodevelopmental and neuroplastic processes involved in the formation of functional neural communities [12, 13]. Accordingly, our group and others have shown that structural covariance networks are consistent with intrinsic functional and diffusion-derived white matter networks [14, 15] and are highly reproducible and reliable [16]. Furthermore, alterations in structural covariance networks are consistently observed in various neurologic syndromes [17, 18], including non-CNS cancer-related neurotoxicity [19].
Brain networks, or connectomes, are organized such that specialized regions are highly connected to their neighbors but sparsely connected to distant regions. This “small-world” architecture allows for efficient information exchange and parallel processing with minimal wiring cost [20, 21]. These complex network properties can be elegantly elucidated by applying graph theoretical analysis [22]. Previous studies employing this method have shown altered functional brain network organization in low and high grade glioma that is associated with cognitive status [23–26] and other studies have shown altered gray matter structure in glioma using voxel-based morphometry [27, 28]. However, no studies to date have examined potential differences in brain network connectivity related to tumor genotype. We therefore compared structural covariance networks between wild type and mutant malignant astrocytoma. We hypothesized that network organization, as indicated by efficiency, would be lower in patients with wild type tumors, reflecting greater lesion momentum, and that disrupted network connectivity would be associated with cognitive impairment.
Materials and methods
Participants
Patients with a centrally reviewed diagnosis of supratentorial, WHO grade III anaplastic astrocytoma or WHO grade IV glioblastoma, whose first therapeutic intervention was an open surgical resection at our institution from November 1993 to April 2009, with formalin-fixed, paraffin-embedded tissue, were cross-referenced with our neuropsychology database. A total of 69 patients were identified who met inclusion criteria [29], had detailed presurgical neuropsychological evaluation and anatomic MRI at the time of neuropsychological evaluation. Of these, 37 were mutant and 32 were wild type. There was no difference between the groups in education level, gender, tumor hemisphere or location but patients with mutant tumor demonstrated larger tumor volumes, less incidence of glioblastoma, younger age and higher Karnofsky performance status (Table 1). Tissue block acquisition, immunohistochemistry and DNA sequencing methods are detailed elsewhere [30]. The University of Texas MD Anderson Cancer Center Institutional Review Board approved this retrospective study.
Table 1.
Demographic and medical data
| IDH1 mutant | IDH1 wild type | Statistic | p | |
|---|---|---|---|---|
| Age | 38.8 (11.2) | 51.3 (14.3) | 16.1 | 0.001 |
| Education (years) | 14.9 (1.8) | 15.1 (2.5) | 0.049 | 0.83 |
| Male | 51% | 56% | 0.156 | 0.70 |
| Tumor | ||||
| Glioblastoma | 17% | 59% | 12.7 | 0.001 |
| T1 volume (cubic centimeters) | 65 (38) | 39 (27) | 10.3 | 0.001 |
| FLAIR volume (cubic centimeters) | 81 (46) | 70 (50) | 0.793 | 0.38 |
| Left hemisphere | 74% | 66% | 0.599 | 0.44 |
| Primary region (F, P, T)a | 40, 9, 51% | 53, 13, 34% | 1.99 | 0.37 |
| Mulitifocal | 53% | 47% | 0.494 | 0.48 |
| Karnofsky performance scale 90–100 | 97% | 72% | 2.86 | 0.004 |
| Karnofsky performance scale 70–80 | 3% | 28% | 2.87 | 0.004 |
| Karnofsky performance scale <60 | 0% | 0% | ||
Data are shown as mean (standard deviation) unless otherwise noted
F frontal, P parietal, T temporal
Frontal p = 0.25, parietal p = 0.50, temporal p = 0.11
Neuroimaging
We obtained the high resolution, T1-weighted MRI scans that were acquired for each patient as part of their presurgical standard of care. The median interval between neuropsychological evaluation and MRI was 1 day but there were a few outliers in the wild type group resulting in a significant group difference in this interval (W = 396, p = 0.02). Therefore, this interval (referred to hereafter as “testing interval”) was included as a covariate in relevant analyses.
MRIs were obtained using a GE Signa HDxt 1.5 or 3 T whole body scanner (General Medical Systems, Milwaukee, WI) with the following parameters: 1.2–1.5 mm slice thickness, 256 × 192 acquisition matrix, echo time = 2.1 or 4.2 ms, repetition time = minimum, field of view = 250 or 260 mm, flip angle = 20, 124 slices. There were slightly more patients in the wild type group who had 1.5 T scans (X2 = 2.74, p = 0.10) so this was used as a covariate in all neuroimaging analyses. Gadolinium-enhanced T1-weighted and T2 fluid-attenuated inversion recovery (FLAIR) images were acquired concurrently and used to determine lesion characteristics.
Lesion size and location
Tumor segmentation was semi-automated using the Vitrea 2 three-dimensional volumetric software (Vital Images, Minnetonka, MN) and was overseen by experienced, board certified neurosurgeons (GR, DC). Personnel scoring the tumor volumes were blinded to molecular stratification. Tumor volume was defined as the greater of the hypointense region on T1-weighted MRI, or the hyperintense area on the gadolinium-enhanced T1-weighted MRI. FLAIR volume was defined as the area of hyperintensity identified on the T2 FLAIR MRI sequence. Tumor location was categorized according to the primary location of the tumor. Multifocal tumors were assigned location based on the largest component of the tumor mass.
Cognitive assessment
Cognitive testing was conducted as part of a comprehensive, presurgical evaluation involving a flexible battery approach performed for clinical purposes (Table 2). Test scores were standardized using published normative data, all of which were stratified by patient age, in addition to gender, handedness, and level of education when appropriate, and converted into z-scores (mean = 0, standard deviation = 1).
Table 2.
Cognitive tests
| Test name | Cognitive domain | Normative data |
|---|---|---|
| Hopkins verbal learning test revised total recall | Memory | Benedict et al. [66] |
| Trail making test parts A&B | Executive function, attention, processing speed | Tombaugh [67] |
| Multilingual aphasia examination controlled oral word association | Language, executive function | Ruff et al. [68] |
| Multilingual aphasia examination token test | Language | Benton et al. [69] |
| Boston naming test | Language | Heaton [70] |
| WAIS-R/III | Wechsler [71, 72] | |
| Digit span | Attention | |
| Block design | Visual-spatial | |
| Symbol search | Processing speed | |
| Coding | Processing speed | |
| Similarities | Executive function | |
| Grooved pegboard | Motor | Heaton [70] |
| Grip strength | Motor | Heaton [70] |
WAIS-R/III Wechsler adult intelligence scale–revised or third edition
Structural covariance networks
MRIs were visually inspected and the scan of one participant in the mutant group was excluded for poor image quality. Gray matter volumes were segmented from T1-weighted MRI using voxel-based morphometry (VBM) via VBM8 Toolbox in Statistical Parametric Mapping 8 (SPM8) [31, 32]. Lesions can reduce the accuracy of spatial normalization techniques. We employed diffeomorphic anatomical registration through exponentiated lie algebra (DARTEL), which uses a large deformation framework to preserve topology and employs customized, sample-specific templates [33] resulting in superior image registration, even in lesioned brains, compared to other automated methods [34]. Successful normalization was confirmed via visual inspection using the check registration function in SPM8 as well as with whole volume slice montages. Normalized image quality was further evaluated with the check sample homogeneity function in VBM8 Toolbox.
Gray matter covariance networks were constructed for each patient using an innovative similarity-based extraction method [35].1 Network nodes were defined as 3 × 3 × 3 voxel cubes spanning the entire gray matter volume (i.e. 27 gray matter values per cube). A correlation matrix was calculated across all pairs of nodes and binarized based on a threshold estimated from a random network and false discovery rate (FDR) [35, 36]. We ensured that no binarized matrices were disconnected (i.e. had isolated nodes). Matrices were then submitted to graph theoretical analysis using Brain Connectivity Toolbox [37] and in-house code (https://github.com/srkesler/bNets.git) implemented in Matlab v2014b (Mathworks, Inc, Natick, MA). Connectome metrics were calculated as described previously [35, 38, 39]. Specifically, efficiency is defined as the inverse of the average shortest path between nodes and is high when nodes are able to interact directly. Degree refers to the number of connections a region has indicating how much that region is interacting with other regions in the network. To facilitate evaluation of regionally specific network effects, gray matter volumes were collapsed across 90 Automated Anatomical Labeling atlas [40].
Statistical analyses
Cognitive impairment
Cognitive performance was categorized as impaired if two or more tests had a z-score at or below −1.5 and/or one or more test had a z-score at or below −2.0 [39, 41]. Impairment could not be categorized for one participant in the mutant group due to missing data. Group difference in cognitive impairment was evaluated using logistic regression. T1 tumor volume was included as a covariate in the model since it differed between groups. We also included education level considering its significant independent contribution, as a proxy of cognitive reserve [42], to cognitive outcome following cancer [43] and other neurologic conditions. Age and histology were not included in this or any other models given that these variables reflect the known etiologic and pathologic differences associated with IDH1 subtypes [7, 44].
Brain network efficiencies
Global and local network efficiencies were evaluated separately using the general linear model. These models also included education as a covariate considering previous research showing that cognitive reserve mediates pathologic effects on brain network properties [39, 45, 46]. Total brain volume, network size and scanner field strength were also included. Degree is known to affect network properties in addition to network size [47] but including this variable resulted in a non-convergent model. This appeared to reflect the high correlation between degree and efficiency in both groups (r > 0.91, p < 0.0001). Therefore, degree was examined separately as an independent variable with the same covariates as above and also by fitting the power-law to the cumulative degree distribution [39, 48]. Since the effect of tumor volume on brain networks is unknown, models were compared with and without this variable. Nodal efficiencies were first corrected for covariates using linear regression and then group differences were measured multivariately with nonparametric permutation testing (2000 iterations) [38], FDR corrected.
Predictors of cognitive impairment
Given the large number of potential predictors, small sample size and presence of categorical variables as well as our prior observations of nonlinear interactions between brain metrics and cognitive outcome [38], we explored random forest models [49] of cognitive impairment. Global efficiency, tumor volume, scanner field strength, total brain volume, network size, testing interval, education level, Karnofsky Performance Scale score, tumor region (1 = frontal, 2 = temporal, 3 = parietal), multifocal tumor (0 = no, 1 = yes) and tumor hemisphere (1 = left, 2 = right) were included as potential features. Given the relationship of cognitive reserve (i.e. education level) with neural plasticity and repair [50, 51], we also examined a global efficiency*education interaction term. Variable importance was evaluated using mean decrease in Gini index [52]. Because classes (impaired vs. not impaired) were imbalanced in both groups, random minority over-sampling was employed [53, 54]. Predictions were made using the out-of-bag error estimate [55] complemented by leave-one-out cross-validation. Significance was evaluated using a two-sided exact binomial test of accuracy in addition to the area under the curve (AUC) of the receiver operating characteristic.
All statistical analyses were conducted using the R statistical package (R Foundation).
Results
Cognitive performance
Consistent with our previous study, the wild type group demonstrated significantly more frequent cognitive impairment (X2 = 3.3, p = 0.03) compared to mutant (Table 3).
Table 3.
Cognitive and brain network data
| IDH1 mutant | IDH1 wild type | Statistic | p | |
|---|---|---|---|---|
| Cognitive impairment | 65% | 84% | 3.34 | 0.03 |
| Testing interval (days) | 0.61 (0.87) | 1.9 (2.5) | 396 | 0.02 |
| MR field strength (1.5 T) | 19% | 38% | 2.74 | 0.10 |
| Global efficiency | 0.756 (0.009) | 0.750 (0.011) | 3.96 | 0.05 |
| Local efficiency | 0.860 (0.007) | 0.855 (0.008) | 3.54 | 0.06 |
| Network degree | 4415 (191) | 4287 (213) | 4.27 | 0.04 |
| Network size | 8595 (95) | 8557 (93) | 0.172 | 0.68 |
| Total brain volume (cubic centimeters) | 1450 (127) | 1417 (142) | 2.12 | 0.15 |
Data are shown as mean (standard deviation) unless otherwise noted
Brain network efficiencies
As a validation step, all participants demonstrated an expected small-world brain network organization as indicated by a small-worldness index greater than 1 [56]. Compared to mutant, the wild type group demonstrated lower global (F = 3.98, p = 0.05) and local (F = 3.54, p = 0.06) efficiencies (Table 3).
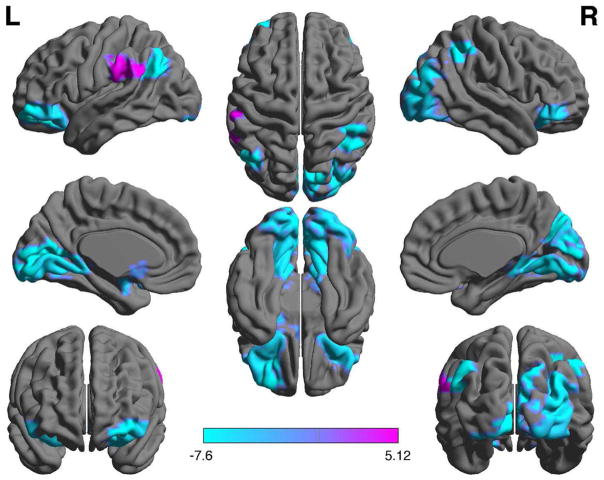
Regional analysis indicated significantly (p < 0.05, FDR corrected) lower efficiency in left amygdala, left angular gyrus, left calcarine sulcus, left caudate, right cuneus, bilateral inferior orbital gyri, left middle frontal orbital gyrus, bilateral lingual gyri, right inferior occipital gyrus, right middle occipital gyrus, and right inferior parietal lobule in the wild type group compared to mutant. The wild type group also showed significantly higher nodal efficiency in left supramarginal gyrus (Fig. 1).
Fig. 1.
Nodal (regional) efficiency. Patients with wild type tumor showed significantly (p < 0.05, FDR corrected) lower efficiency in multiple brain regions compared to mutant (blue) as well as one area of higher efficiency (purple). Color bar indicates log(p value). Regions are shown overlaid on a Montreal Neurological Institute 152 standard template from BrainNet Viewer [65]
Brain network degree
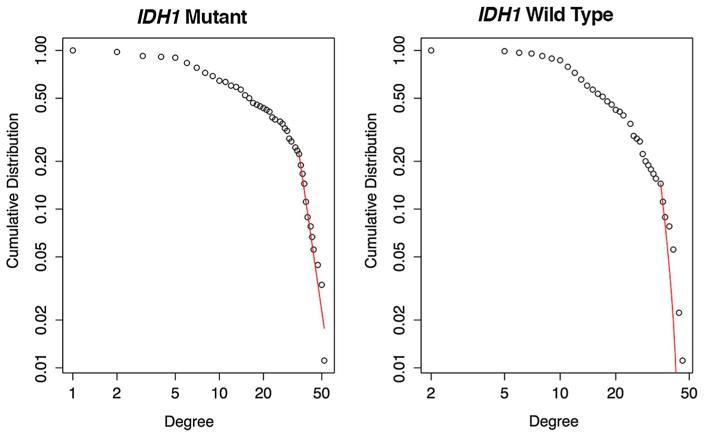
The wild type group showed significantly lower average degree (F = 4.27, p = 0.04) compared to mutant. While the cumulative degree distribution of the mutant group demonstrated goodness of fit with a power-law (X2 = 0.07, p = 0.95), wild type did not (X2 = 0.13, p = 0.0, Fig. 2). Power-law fit was lower in wild type compared to mutant (X2 = 2.19, p = 0.03).
Fig. 2.
Cumulative degree distribution power-law fit. Patients with mutant tumor demonstrated a good fit to the power-law (red line). However, patients with wild type tumor demonstrated a significantly poorer fit (p < 0.03) indicating fewer hubs and a disrupted information processing core
The inclusion of tumor volume in the models lowered significance somewhat (global efficiency p = 0.06; local efficiency p = 0.08; degree p = 0.06) but appeared only to reduce statistical power given that it was not a significant covariate (p > 0.94) so results are reported with tumor volume excluded.
Predictors of cognitive impairment
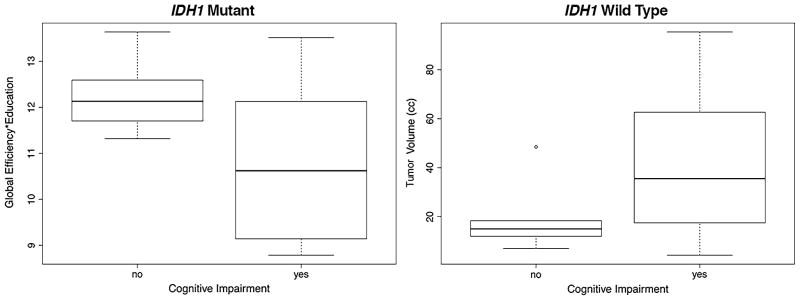
In the mutant group, random forest classification indicated a significant model of cognitive impairment that included, in order of importance, the global efficiency*education interaction term, education, global efficiency, total brain volume, and network size (out-of-bag accuracy = 84%, p < 0.0001, AUC = 0.96; leave-one-out cross validation accuracy = 84%, p < 0.0001, AUC = 0.96, Fig. 3).
Fig. 3.
Predictors of cognitive impairment. Random forest classification indicated that global efficiency (arbitrary units) and education (years) were the most important predictors of cognitive impairment in patients with mutant tumor while tumor volume (cc cubic centimeters) was the greatest contributor to cognitive impairment in patients with wild type tumor
In the wild type group, the final model included, in order of importance, network size, tumor volume, total brain volume and global efficiency (out-of-bag accuracy = 96%, p < 0.0001, AUC = 1.0, leave-one-out cross validation accuracy = 94%, AUC = 1.0, Fig. 3).
Discussion
We demonstrated that patients with IDH1 mutant tumor have significantly higher brain network global efficiency and degree compared to patients with wild type tumor, despite having larger tumor volumes. Lower efficiency indicates that, compared to mutant, brain network organization is less integrated in patients with wild type tumor and specifically, has inferior capacity for parallel information processing [57]. Regional analysis indicated diffusely lower connectivity of the wild type brain network including several medial frontal, posterior parietal and subcortical regions, which leaves fewer parallel paths, restricting information flow to longer-range paths. This dysconnectivity was confirmed by significantly lower network degree in patients with wild type tumor given that degree indicates a region’s total number of connections [37]. The slower growth of the mutant tumor likely allows the brain time to adapt to the tumor’s presence and reorganize critical neurocircuitry, similar to what is observed in low grade gliomas [58].
The resilience and plasticity of the brain network depends largely on a minimal set of regions. In this regard, the brain network tends to demonstrate a power-law distribution [48] wherein the majority of information processing is handled by a small number of core regions or hubs. The brain network is highly vulnerable to disruption of these regions [59] and accordingly, many neurologic and neurodegenerative conditions are associated with disconnection of these influential hubs [60]. For example, Ius et al. [61] proposed that surgical resection of certain core regions that they termed the “minimal common brain” has a high probability of permanent neurologic deficit. The cumulative degree distribution in patients with wild type tumor showed a significantly poorer power-law fit compared to patients with mutant tumor. Specifically, patients with wild type tumor showed a steeper exponential decay in the probability of high degree regions, which indicates fewer hubs.
Many of the hubs critical for brain network robustness are located within the default mode network [59, 62], which is important for a variety of cognitive functions. Given its relative metabolic demands and vulnerability to toxicity, this network is highly sensitive to various disease states [63]. Connectivity of these areas was significantly lower in patients with wild type tumor compared to mutant. These findings are consistent with a previous study demonstrating the sensitivity of default mode network connectivity to glioma WHO grade [9]. Our results indicate that higher cognitive impairment in patients with wild type tumor may reflect greater disruption of the brain’s structural core by wild type tumor characteristics. These may include metabolic disruptions, vascular effects and/or white matter infiltration, among others.
Unlike most previous studies of structural covariance networks, the similarity-based extraction method we applied [35] resulted in individual level networks that allowed us to examine correlates with cognitive outcome. We observed that global efficiency was significantly inversely, though nonlinearly, associated with cognitive impairment in patients with mutant tumor and this relationship was mediated by cognitive reserve, which is inherently tied to neural plasticity and repair [51]. A greater magnitude of neuropathology tends to be required for clinical effects to become manifest in individuals with higher cognitive reserve [42] and cognitive reserve is believed to increase neural reorganization following neurologic injury and disease [50].
In patients with wild type tumor, global efficiency was also inversely associated with cognitive impairment but tumor volume was a more important contributor. Additionally, education was not a significant factor. This finding suggests that the typical mediating effect of cognitive reserve on neuropathology and cognitive function may not be present in patients with wild type tumor, possibly because the neuroplastic mechanisms associated with cognitive reserve cannot keep pace with the greater lesion momentum of this molecular variant. Network size was actually the most important variable in the wild type cognitive impairment model. Given that high Gini index can sometimes reflect interaction effects [52], we explored, post hoc, the joint variable importance [64] of all pairs of features in the wild type cognitive impairment final model. This revealed that larger tumor volume tended to be associated with smaller network size in patients with wild type tumor (r = −0.30, p = 0.09). Thus, tumor volume is likely the greatest contributor to cognitive impairment in this group.
In addition to small sample size and the retrospective design, there are certain other limitations to our findings. The most significant is the combination of data from two different MRI field strengths. However, we conducted thorough image quality assurance and controlled for field strength in our analyses. Another caveat pertains to the comparison of networks of different size and degree as thresholding for networks remains an ongoing issue of debate [47]. We ensured that no nodes were disconnected after thresholding, covaried for size, and evaluated degree separately. Additionally, a non-cancer comparison group is necessary to more comprehensively evaluate the effects of IDH1 molecular variants on cognition and brain network organization. Lastly, we only controlled for the structural imaging variables that differed between the groups so that we were able to demonstrate that the structural network effects were independent of tumor or lesion size. We readily acknowledge the difficulties in delineating the tumor border from other pathological changes within the brain (e.g., edema) especially in the setting of non-enhancing lesions.
In conclusion, our findings suggest that IDH1 tumor status plays a significant role in the organization of the brain’s network topology and the associated cognitive function of patients with malignant astrocytoma. These findings provide additional support for diffuse effects of focal brain lesions and suggest that lesions impacting the brain’s structural core may reduce its ability to adapt to the presence of the tumor. Further research in this area may reveal additional insights regarding cognitive function in individual patients with brain tumor including novel information regarding neuroplastic response to tumor infiltration.
Acknowledgments
The authors would like to thank Vikram Rao, M.S., for assistance with image processing.
Funding
This work was supported by grants from the National Institutes of Health (1R01CA172145 to S.R.K, 1R03CA191559 to S.R.K, 1R01NR014195 to J.S.W./S.R.K. and K08NS070928 to G.R.).
Footnotes
Extract Individual GM Networks Toolbox v20150902 https://github.com/bettytijms/Single_Subject_Grey_Matter_Networks.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Baldock AL, Yagle K, Born DE, Ahn S, Trister AD, Neal M, Johnston SK, Bridge CA, Basanta D, Scott J. Invasion and proliferation kinetics in enhancing gliomas predict IDH1 mutation status. Neurooncol. 2014;16:779–786. doi: 10.1093/neuonc/nou027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hodges TR, Choi BD, Bigner DD, Yan H, Sampson JH. Isocitrate dehydrogenase 1 (IDH1): what it means to the neurosurgeon. J Neurosurg. 2013;118:1176. doi: 10.3171/2013.3.JNS122282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hartmann C, Hentschel B, Wick W, Capper D, Felsberg J, Simon M, Westphal M, Schackert G, Meyermann R, Pietsch T. Patients with IDH1 wild type anaplastic astrocytomas exhibit worse prognosis than IDH1-mutated glioblastomas, and IDH1 mutation status accounts for the unfavorable prognostic effect of higher age: implications for classification of gliomas. Acta Neuropathol. 2010;120:707–718. doi: 10.1007/s00401-010-0781-z. [DOI] [PubMed] [Google Scholar]
- 4.Cheng H-B, Yue W, Xie C, Zhang R-Y, Hu S-S, Wang Z. IDH1 mutation is associated with improved overall survival in patients with glioblastoma: a meta-analysis. Tumor Biol. 2013;34:3555–3559. doi: 10.1007/s13277-013-0934-5. [DOI] [PubMed] [Google Scholar]
- 5.Finger S. Lesion momentum and behavior. In: Finger S, editor. Recovery from brain damage. Springer; New York: 2013. pp. 135–164. [Google Scholar]
- 6.Tucha O, Smely C, Preier M, Lange KW. Cognitive deficits before treatment among patients with brain tumors. Neurosurgery. 2000;47:324–334. doi: 10.1097/00006123-200008000-00011. [DOI] [PubMed] [Google Scholar]
- 7.Wefel J, Noll K, Rao G, Cahill D. The neurocognitive phenotype of IDH1 genetic mutation status in malignant glioma prior to surgical resection. Neurooncol. 2016 doi: 10.1093/neuonc/now165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson SW, Damasio H, Tranel D. Neuropsychological impairments associated with lesions caused by tumor or stroke. Arch Neurol. 1990;47:397–405. doi: 10.1001/archneur.1990.00530040039017. [DOI] [PubMed] [Google Scholar]
- 9.Harris RJ, Bookheimer SY, Cloughesy TF, Kim HJ, Pope WB, Lai A, Nghiemphu PL, Liau LM, Ellingson BM. Altered functional connectivity of the default mode network in diffuse gliomas measured with pseudo-resting state fMRI. J Neurooncol. 2014;116:373–379. doi: 10.1007/s11060-013-1304-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Dellen E, de Witt Hamer PC, Douw L, Klein M, Heimans JJ, Stam CJ, Reijneveld JC, Hillebrand A. Connectivity in MEG resting-state networks increases after resective surgery for low-grade glioma and correlates with improved cognitive performance. NeuroImage Clin. 2012;2:1–7. doi: 10.1016/j.nicl.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maesawa S, Bagarinao E, Fujii M, Futamura M, Motomura K, Watanabe H, Mori D, Sobue G, Wakabayashi T. Evaluation of resting state networks in patients with gliomas: connectivity changes in the unaffected side and its relation to cognitive function. PLoS One. 2015;10:e0118072. doi: 10.1371/journal.pone.0118072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alexander-Bloch A, Giedd JN, Bullmore E. Imaging structural co-variance between human brain regions. Nat Rev Neurosci. 2013;14:322–336. doi: 10.1038/nrn3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mechelli A, Friston KJ, Frackowiak RS, Price CJ. Structural covariance in the human cortex. J Neurosci. 2005;25:8303–8310. doi: 10.1523/JNEUROSCI.0357-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hosseini SM, Kesler SR. Comparing connectivity pattern and small-world organization between structural correlation and resting-state networks in healthy adults. NeuroImage. 2013;78:402– 414. doi: 10.1016/j.neuroimage.2013.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gong G, He Y, Chen ZJ, Evans AC. Convergence and divergence of thickness correlations with diffusion connections across the human cerebral cortex. NeuroImage. 2012;59:1239–1248. doi: 10.1016/j.neuroimage.2011.08.017. [DOI] [PubMed] [Google Scholar]
- 16.Wang H, Jin X, Zhang Y, Wang J. Single-subject morphological brain networks: connectivity mapping, topological characterization and test–retest reliability. Brain Behav. 2016;6:e00448. doi: 10.1002/brb3.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tijms BM, Kate MT, Wink AM, Visser PJ, Ecay M, Clerigue M, Estanga A, Garcia Sebastian M, Izagirre A, Villanua J, Martinez Lage P, van der Flier WM, Scheltens P, Sanz Arigita E, Barkhof F. Gray matter network disruptions and amyloid beta in cognitively normal adults. Neurobiol Aging. 2015 doi: 10.1016/j.neurobiolaging.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 18.Liao W, Zhang Z, Mantini D, Xu Q, Wang Z, Chen G, Jiao Q, Zang YF, Lu G. Relationship between large-scale functional and structural covariance networks in idiopathic generalized epilepsy. Brain Connect. 2013;3:240–254. doi: 10.1089/brain.2012.0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hosseini SM, Koovakkattu D, Kesler SR. Altered small-world properties of gray matter networks in breast cancer. BMC Neurol. 2012;12:28. doi: 10.1186/1471-2377-12-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bassett DS, Bullmore E. Small-world brain networks. Neuroscientist. 2006;12:512–523. doi: 10.1177/1073858406293182. [DOI] [PubMed] [Google Scholar]
- 21.Bullmore E, Sporns O. The economy of brain network organization. Nat Rev Neurosci. 2012;13:336–349. doi: 10.1038/nrn3214. [DOI] [PubMed] [Google Scholar]
- 22.Bullmore E, Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neurosci. 2009;10:186–198. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- 23.Bosma I, Reijneveld JC, Klein M, Douw L, van Dijk BW, Heimans JJ, Stam CJ. Disturbed functional brain networks and neurocognitive function in low-grade glioma patients: a graph theoretical analysis of resting-state MEG. Nonlinear Biomed Phys. 2009;3:9. doi: 10.1186/1753-4631-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang Q, Zhang R, Hu X, Ding S, Qian J, Lei T, Cao X, Tao L, Qian Z, Liu H. Disturbed small-world networks and neurocognitive function in frontal lobe low-grade glioma patients. PLoS One. 2014;9:e94095. doi: 10.1371/journal.pone.0094095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu H, Ding S, Hu X, Yang K, Xiao C, Zou Y, Chen Y, Tao L, Liu H, Qian Z. Reduced efficiency of functional brain network underlying intellectual decline in patients with low-grade glioma. Neurosci Lett. 2013;543:27–31. doi: 10.1016/j.neulet.2013.02.062. [DOI] [PubMed] [Google Scholar]
- 26.van Dellen E, Douw L, Hillebrand A, Ris-Hilgersom IH, Schoonheim MM, Baayen JC, De Witt Hamer PC, Velis DN, Klein M, Heimans JJ, Stam CJ, Reijneveld JC. MEG network differences between low- and high-grade glioma related to epilepsy and cognition. PLoS One. 2012;7:e50122. doi: 10.1371/journal.pone.0050122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Misra A, Yuan R, Gohel S, Biswal B. Using MRI from 1000 subjects to identify abnormal grey matter in individual tumor subjects. 2014 40th Annual Northeast Bioengineering Conference (NEBEC); 2014. pp. 1–2. [Google Scholar]
- 28.Prust MJ, Jafari-Khouzani K, Kalpathy-Cramer J, Polaskova P, Batchelor TT, Gerstner ER, Dietrich J. Standard chemoradiation for glioblastoma results in progressive brain volume loss. Neurology. 2015 doi: 10.1212/WNL.0000000000001861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beiko J, Suki D, Hess KR, Fox BD, Cheung V, Cabral M, Shonka N, Gilbert MR, Sawaya R, Prabhu SS, Weinberg J, Lang FF, Aldape KD, Sulman EP, Rao G, McCutcheon IE, Cahill DP. IDH1 mutant malignant astrocytomas are more amenable to surgical resection and have a survival benefit associated with maximal surgical resection. Neuro Oncol. 2014;16:81–91. doi: 10.1093/neuonc/not159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ashburner J. Computational anatomy with the SPM software. Magn Reson Imaging. 2009;27:1163–1174. doi: 10.1016/j.mri.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 32.Kurth F, Gaser C, Luders E. A 12-step user guide for analyzing voxel-wise gray matter asymmetries in statistical parametric mapping (SPM) Nat Protoc. 2015;10:293–304. doi: 10.1038/nprot.2015.014. [DOI] [PubMed] [Google Scholar]
- 33.Ashburner J. A fast diffeomorphic image registration algorithm. NeuroImage. 2007;38:95–113. doi: 10.1016/j.neuroimage.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 34.Ripolles P, Marco-Pallares J, de Diego-Balaguer R, Miro J, Falip M, Juncadella M, Rubio F, Rodriguez-Fornells A. Analysis of automated methods for spatial normalization of lesioned brains. NeuroImage. 2012;60:1296–1306. doi: 10.1016/j.neuroimage.2012.01.094. [DOI] [PubMed] [Google Scholar]
- 35.Tijms BM, Series P, Willshaw DJ, Lawrie SM. Similarity-based extraction of individual networks from gray matter MRI scans. Cereb Cortex. 2012;22:1530–1541. doi: 10.1093/cercor/bhr221. [DOI] [PubMed] [Google Scholar]
- 36.Noble WS. How does multiple testing correction work? Nat Biotechnol. 2009;27:1135–1137. doi: 10.1038/nbt1209-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rubinov M, Sporns O. Complex network measures of brain connectivity: uses and interpretations. NeuroImage. 2010;52:1059–1069. doi: 10.1016/j.neuroimage.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 38.Kesler SR, Gugel M, Huston-Warren E, Watson C. Atypical structural connectome organization and cognitive impairment in young survivors of acute lymphoblastic leukemia. Brain Connect. 2016;6:273–282. doi: 10.1089/brain.2015.0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kesler SR, Watson CL, Blayney DW. Brain network alterations and vulnerability to simulated neurodegeneration in breast cancer. Neurobiol Aging. 2015;36:2429–2442. doi: 10.1016/j.neurobiolaging.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. NeuroImage. 2002;15:273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- 41.Wefel JS, Vardy J, Ahles T, Schagen SB. International Cognition and Cancer Task Force recommendations to harmonise studies of cognitive function in patients with cancer. Lancet Oncol. 2011;12:703–708. doi: 10.1016/S1470-2045(10)70294-1. [DOI] [PubMed] [Google Scholar]
- 42.Stern Y. Cognitive reserve. Neuropsychologia. 2009;47:2015– 2028. doi: 10.1016/j.neuropsychologia.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahles TA, Saykin AJ, McDonald BC, Li Y, Furstenberg CT, Hanscom BS, Mulrooney TJ, Schwartz GN, Kaufman PA. Longitudinal assessment of cognitive changes associated with adjuvant treatment for breast cancer: impact of age and cognitive reserve. J Clin Oncol. 2010;28:4434–4440. doi: 10.1200/jco.2009.27.0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hirose Y, Sasaki H, Abe M, Hattori N, Adachi K, Nishiyama Y, Nagahisa S, Hayashi T, Hasegawa M, Yoshida K. Subgrouping of gliomas on the basis of genetic profiles. Brain Tumor Pathol. 2013;30:203–208. doi: 10.1007/s10014-013-0148-y. [DOI] [PubMed] [Google Scholar]
- 45.Santarnecchi E, Rossi S, Rossi A. The smarter, the stronger: intelligence level correlates with brain resilience to systematic insults. Cortex. 2015;64:293–309. doi: 10.1016/j.cortex.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 46.Fischer FU, Wolf D, Scheurich A, Fellgiebel A. Association of structural global brain network properties with intelligence in normal aging. PLoS One. 2014;9:e86258. doi: 10.1371/journal.pone.0086258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Wijk BCM, Stam CJ, Daffertshofer A. Comparing brain networks of different size and connectivity density using graph theory. PLoS One. 2010;5:e13701. doi: 10.1371/journal.pone.0013701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Achard S, Salvador R, Whitcher B, Suckling J, Bullmore E. A resilient, low-frequency, small-world human brain functional network with highly connected association cortical hubs. J Neurosci. 2006;26:63–72. doi: 10.1523/JNEUROSCI.3874-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Breiman L. Random forests. Machine Learn. 2001;45:5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 50.Steffener J, Stern Y. Exploring the neural basis of cognitive reserve in aging. Biochim Biophys Acta. 2012;1822:467–473. doi: 10.1016/j.bbadis.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whalley LJ, Deary IJ, Appleton CL, Starr JM. Cognitive reserve and the neurobiology of cognitive aging. Ageing Res Rev. 2004;3:369–382. doi: 10.1016/j.arr.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 52.Wright MN, Ziegler A, Konig IR. Do little interactions get lost in dark random forests? BMC Bioinform. 2016;17:145. doi: 10.1186/s12859-016-0995-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Batista GEAPA, Prati RC, Monard MC. A study of the behavior of several methods for balancing machine learning training data. Sigkdd Explor. 2004;6:20–29. doi: 10.1145/1007730.1007735. [DOI] [Google Scholar]
- 54.del Río S, López V, Benítez JM, Herrera F. On the use of MapReduce for imbalanced big data using Random Forest. Inf Sci. 2014;285:112–137. doi: 10.1016/j.ins.2014.03.043. [DOI] [Google Scholar]
- 55.Liaw A, Wiener M. Classification and regression by random. Forest R News. 2002;2:18–22. (citeulike-article-id: 1121494) [Google Scholar]
- 56.Humphries MD, Gurney K. Network ‘small-world-ness’: a quantitative method for determining canonical network equivalence. PLoS One. 2008;3:e0002051. doi: 10.1371/journal.pone.0002051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Achard S, Bullmore E. Efficiency and cost of economical brain functional networks. PLoS Comput Biol. 2007;3:e17. doi: 10.1371/journal.pcbi.0030017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Duffau H. Diffuse low-grade gliomas and neuroplasticity. Diagn Interv Imaging. 2014;95:945–955. doi: 10.1016/j.diii.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 59.Alstott J, Breakspear M, Hagmann P, Cammoun L, Sporns O. Modeling the impact of lesions in the human brain. PLoS Comput Biol. 2009;5:e1000408. doi: 10.1371/journal.pcbi.1000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Crossley NA, Mechelli A, Scott J, Carletti F, Fox PT, McGuire P, Bullmore ET. The hubs of the human connectome are generally implicated in the anatomy of brain disorders. Brain. 2014;137:2382–2395. doi: 10.1093/brain/awu132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ius T, Angelini E, Thiebaut de Schotten M, Mandonnet E, Duffau H. Evidence for potentials and limitations of brain plasticity using an atlas of functional resectability of WHO grade II gliomas: towards a “minimal common brain”. NeuroImage. 2011;56:992–1000. doi: 10.1016/j.neuroimage.2011.03.022. [DOI] [PubMed] [Google Scholar]
- 62.Hagmann P, Cammoun L, Gigandet X, Meuli R, Honey CJ, Wedeen VJ, Sporns O. Mapping the structural core of human cerebral cortex. PLoS Biol. 2008;6:e159. doi: 10.1371/journal.pbio.0060159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kesler SR. Default mode network as a potential biomarker of chemotherapy-related brain injury. Neurobiol Aging. 2014;35(Suppl 2):S11–S19. doi: 10.1016/j.neurobiolaging.2014.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ishwaran H. Variable importance in binary regression trees and forests. Electron J Stat. 2007:519–537. doi: 10.1214/07-EJS039. [DOI] [Google Scholar]
- 65.Xia M, Wang J, He Y. BrainNet viewer: a network visualization tool for human brain connectomics. PLoS One. 2013;8:e68910. doi: 10.1371/journal.pone.0068910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Benedict RHB, Schretlen D, Groninger L, Brandt J. Hopkins verbal learning test—revised: normative data and analysis of inter-form and test-retest reliability. Clin Neuropsy. 1998;12(1):43– 55. doi: 10.1076/clin.12.1.43.1726. [DOI] [Google Scholar]
- 67.Tombaugh T. Trail Making Test A and B: normative data stratified by age and education. Arch Clin Neuropsych. 2004;19(2):203–214. doi: 10.1016/s0887-6177(03)00039-8. [DOI] [PubMed] [Google Scholar]
- 68.Ruff RM, Light RH, Parker SB, Levin HS. Benton controlled oral word association test: reliability and updated norms. Arch Clin Neuropsych. 1996;11(4):329–338. [PubMed] [Google Scholar]
- 69.Benton A, Hamsher Kd, Sivan AB. Multilingual aphasia examination. 3. Psychological Assessment Resources, Inc; Lutz, FL: 2000. [Google Scholar]
- 70.Heaton RK. Revised comprehensive norms for an expanded halstead-reitan battery: demographically adjusted neuropsychological norms for African American and Caucasian adults. Professional Manual: Psychological Assessment Resources 2004 [Google Scholar]
- 71.Wechsler D. Wechsler adult intelligence scale—revised. Harcourt Assessments; San Antonio, TX: 1981. [Google Scholar]
- 72.Wechsler D. Administration and Scoring Manual. 3. The Psychological Corporation; San Antonio: 1997. Wechsler adult intelligence scale. [Google Scholar]