Abstract
A plethora of mutations in chromatin regulators in diverse human cancers is emerging, attesting to the pivotal role of chromatin dynamics in tumorigenesis. A recurrent theme is inactivation of the chromodomain helicase DNA-binding (CHD) family of proteins—ATP-dependent chromatin remodelers that govern the cellular machinery’s access to DNA, thereby controlling fundamental processes, including transcription, proliferation, and DNA damage repair. This review highlights what is currently known about how genetic and epigenetic perturbation of CHD proteins and the pathways that they regulate set the stage for cancer, providing new insight for designing more effective anti-cancer therapies.
Chromodomain helicase DNA-binding (CHD) proteins control the access of cellular machinery to DNA. CHD subfamily II members are tumor suppressors; subfamilies I and III may suppress or promote tumorigenesis, depending on context.
The advent of high-throughput sequencing technologies has made it increasingly straightforward to interrogate the genomes of human tumors in an effort to identify cancer-driving mutations, diagnose tumor subtypes, and implement regimens for personalized therapies. The picture that is emerging from these studies is that lesions in genes encoding chromatin regulators are among the most prevalent mutations, underscoring the importance of chromatin structure and function in tumorigenesis. Indeed, efforts of The Cancer Genome Atlas Consortium have established that chromatin regulators are some of the most frequent mutations in 12 major types of human cancer, including glioma and leukemia, as well as tumors of the breast, bladder, colon, kidney, and lung (Kandoth et al. 2013).
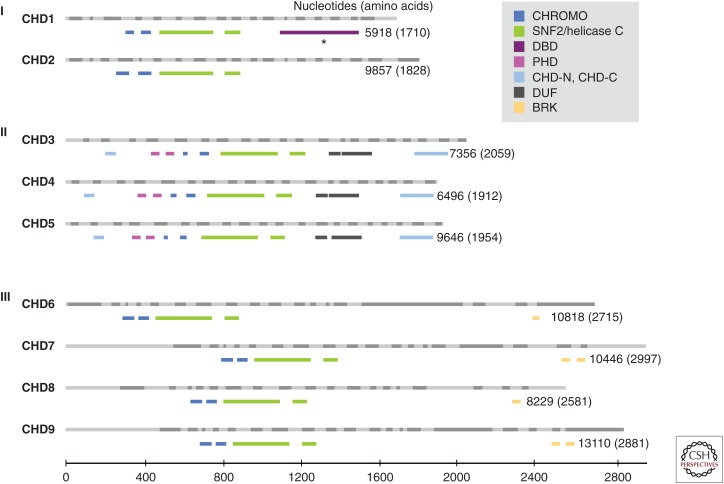
The chromodomain helicase DNA-binding (CHD) family of chromatin remodelers is one type of chromatin regulator that is frequently lost or inactivated in a diverse array of human cancers. The CHD family consists of nine members, CHD1–9 (Fig. 1). CHD5 was the first CHD protein shown to have a functional role in cancer (Bagchi et al. 2007). CHD family members share chromatin organizing (CHROMO) domains that bind specifically modified histones and an SNF2-like ATP-dependent helicase domain that facilitates nucleosome mobilization (Marfella and Imbalzano 2007). The family name reflects the fact that CHD1, the original CHD protein identified, tends to interact with AT-rich regions of DNA, implying that it has a DNA-binding domain (Delmas et al. 1993). In addition to core motifs characteristic of the family as a whole, individual CHD proteins have other domains that classify them into three subfamilies. Subfamily I, which includes human CHD1 and CHD2, is classified based on members having SNF2 domains homologous to CHD1 proteins of other organisms, with mouse Chd1 having an AT-rich DNA-binding domain (Delmas et al. 1993). Subfamily II, which includes CHD3, CHD4, and CHD5, has dual plant homeodomains (PHDs). Subfamily III, which includes CHD6, CHD7, CHD8, and CHD9, has Brahma and Kismet domains. CHD proteins affect chromatin compaction and therefore the cellular machinery’s access to DNA; thus, these enzymes control fundamental biological processes, including transcription, cellular proliferation, and DNA damage repair. Given their key role in these crucial cellular processes, it is perhaps not surprising that loss or inactivation of CHD proteins is pivotal in a range of developmental syndromes and cancers. This review focuses on the mechanisms by which perturbation of CHD-mediated chromatin dynamics regulates tumorigenesis. I discuss what is known about the biological roles of these chromatin remodelers, highlight recent evidence for the genetic and epigenetic factors that are upstream, in parallel, and downstream from CHD proteins, and discuss the current view on how perturbation of different members of each CHD subfamily modulates the tumorigenic process and affects cancer patient survival.
Figure 1.
The chromodomain helicase DNA-binding (CHD) family of chromatin remodelers. CHD proteins are classified into three subfamilies (Roman numerals) based on their functional motifs (see legend). The human CHD family based on Ensembl is drawn to scale, with light and dark gray bars depicting alternating exons (above) and the functional motifs from PFAM (a database of protein families of multiple sequence alignments generated using hidden Markov models) shown in color (below) for each CHD member. The number of nucleotides and amino acid residues for the CHD transcript and protein, respectively, are shown. BRK, Brahma and Kismet domains; CHD, chromodomain helicase DNA binding; CHROMO, chromodomain; CHD-N, CHD-C: CHD_N and CHD_C are shown in upstream and downstream regions, respectively; DBD, DNA-binding domain (based on Delmas et al. 1993) rather than PFAM; DUF, domain of unknown function; PHD, plant homeodomain; SNF2/Helicase C, SNF2_N, and Helicase_C are shown in upstream and downstream regions, respectively. (From Li and Mills 2014; reproduced and modified, with express permission, from Future Medicine, 2014.)
SUBFAMILY I: CHD1 and CHD2
CHD1—the founding member of the CHD family—was initially discovered as a DNA-binding protein that, based on its functional motifs, was proposed to regulate chromatin structure and gene expression (Delmas et al. 1993; Stokes and Perry 1995). Subfamily I CHD proteins have been implicated in transcriptional regulation, and this has recently been shown to impact proliferation, repair of DNA damage, and pluripotency. The first demonstration for any CHD protein playing a functional role in cancer was when the subfamily II member CHD5 was identified as a tumor suppressor mapping to human 1p36—a genomic interval frequently deleted in a variety of cancers (Bagchi et al. 2007; Bagchi and Mills 2008) (discussed below). But more recently, it has become apparent that like CHD5, subfamily I CHD proteins are also lost or inactivated in several cancers. However, their gain of function has also been shown to promote cancer. Deregulation of subfamily I proteins has a profound effect on invasion, metastasis, and patient survival. CHD1 is the CHD subfamily I member with the tightest link to cancer, although CHD2 has also been implicated in tumor suppression.
Factors upstream of CHD subfamily I proteins include environmental factors, hormones, chromatin regulators, and signaling pathways. Exposure to cigarette smoke correlates with hypermethylation of the CHD1 promoter (Lyn-Cook et al. 2014). Promoter methylation is associated with RNA polymerase II (Pol II) stalling and compromised transcriptional activation, whereas demethylating agents abrogate this effect by inducing Pol II phosphorylation at serine 2 to promote transcriptional elongation (Tao et al. 2011). Estrogen signaling plays a key role in promoting proliferation of estrogen receptor (ER)-positive breast cancer. Estrogen inhibits expression of miR26a and miR26b, microRNAs that target and degrade CHD1 transcript and promote proliferation of breast cancer cells (Tan et al. 2014). c-MYC is required for both the inhibition of miR26 and the increase in CHD1 expression in response to estrogen. The investigators show that depletion of CHD1 abrogates the pro-proliferative effect of estrogen, indicating that CHD1 potentiates oncogenesis, at least in the context of ERα-positive breast cancer. Factors interacting with components of the transcriptional machinery and histone modifiers also converge upstream of CHD1 to modulate its expression. For example, the Pol II–associated factor hPAF2/PD2 mediates MLL-mediated deposition of the H3K4me2/3 covalent modifications characteristic of transcriptionally active genes and facilitates CHD1 expression in pancreatic cancer cells (Dey et al. 2011). Another example is the protein arginine methyltransferase Prmt6, which inhibits expression of Chd1 (Lee et al. 2012). Prmt6 evokes H3R2me2—a covalent modification antagonistic to the H3K4me3 mark associated with transcriptional activation. CHD1 is also modified by SUMOylation in KRAS mutant colorectal cancer cells by the SUMO E2 ligase UBC9 (Yu et al. 2015a). While this study indicates that activation of the RAS/RAF pathway covalently modifies CHD1 and that UBC9 is required for KRAS-mediated transformation, the finding that CHD1 depletion inhibits the transformed phenotype suggests that SUMOylation endows CHD1 with pro-oncogenic activity, rather than it inhibiting its tumor-suppressive activity.
Factors working in parallel with CHD1 include hPAF1/PD2 that binds and facilitates nuclear import of CHD1 where it is then positioned to bind H3K4me2/3 via its dual CHROMO domains (Flanagan et al. 2005), promote nucleosome destabilization, and modulate transcription in pancreatic cancer cells (Dey et al. 2011). Because hPAF1/PD2 is proposed to have an oncogenic function and is aberrantly overexpressed in pancreatic cancer, and hPAF/PD2 facilitates CHD1 activity, this again suggests that CHD1 plays a pro-oncogenic role. There is some indication that CHD1 functionally interacts with MAP3K7, as co-deletion of CHD1 and MAP3K7 occurs in prostate cancer and co-suppression of Chd1 and Map3k7 in mouse prostate epithelial stem/progenitor cells inhibits differentiation and causes aggressive prostate tumors (Rodrigues et al. 2015). CHD1 also works in concert with the androgen receptor (AR), as it is required for AR-dependent transcriptional activation of androgen-responsive genes in prostate cancer (Burkhardt et al. 2013).
Factors downstream from CHD1 include the AR-responsive tumor suppressor genes NKX3-1, FOXO1, and PPARγ, consistent with CHD1 being a coactivator of AR-mediated transcriptional activation (Burkhardt et al. 2013). In mouse embryonic stem (ES) cells, Chd1 facilitates expression of the pluripotency genes Oct4 and Nanog (Lee et al. 2012). The mechanism proposed is that increased expression of the histone arginine methyltransferase Prmt6 occurs upon differentiation thereby evoking H3R2me2, a covalent histone modification that counteracts the transcriptional activation mark H3K4me3. Simultaneously, Chd1 expression is compromised, and there is also less Chd1 bound at the promoters of Oct4 and Nanog because these regions have less H3K4me3 compared with undifferentiated ES cells, and Chd1 is known to bind H3K4me3 via its CHROMO domains. A separate study found that H3K4me3 is not sufficient for recruiting CHD1 to promoters; instead, CHD1 is recruited to target loci through its interaction with components of the transcriptional machinery in an activation-dependent manner where it regulates H3/H3.3 occupancy and chromatin accessibility at transcriptional start sites of its target genes (Siggens et al. 2015).
Inactivating lesions affecting CHD1 include promoter methylation in breast and other cancers (Lyn-Cook et al. 2014), and mutation in colorectal cancers with high levels of microsatellite instability (Kim et al. 2011). But the most striking evidence for CHD1 inactivation is in prostate cancer, where it is deleted or mutated (Grasso et al. 2012; Huang et al. 2012; Liu et al. 2012; Burkhardt et al. 2013; Martin et al. 2013; Blattner et al. 2014; Gao et al. 2014; Scott et al. 2014; Tereshchenko et al. 2014; Attard et al. 2015; Fisher et al. 2015; Sowalsky et al. 2015). Indeed, homozygous deletion of CHD1 is the second most common genetic event in prostate cancer after PTEN deletion (Liu et al. 2012). Chromosome rearrangements that cause overexpression of ETS family members, most commonly translocations between the androgen-regulated gene transmembrane protease serine 2 (TMPRSS2) and the ERG gene, are frequent in some types of prostate cancer (Clark and Cooper 2009). CHD1 lesions occur in ETS fusion-negative prostate cancer (Grasso et al. 2012; Martin et al. 2013; Tereshchenko et al. 2014), indicating that the CHD1 status defines a unique prostate cancer subtype (Attard et al. 2015; Fisher et al. 2015). Whereas CHD1 mutation and ETS fusions are mutually exclusive, CHD1 inactivation co-occurs with speckle-type PTB/POZ protein mutations (Blattner et al. 2014) and MAP3K7 deletion (Rodrigues et al. 2015), suggesting that these lesions cooperate with CHD1 loss to drive tumorigenesis in the prostate. Inactivation of CHD1 has also been correlated with anchorage-independent growth (Yu et al. 2015a), enhanced invasiveness (Huang et al. 2012; Liu et al. 2012), and compromised differentiation and increased stemness in mouse prostate epithelial stem/progenitor cells (Rodrigues et al. 2015). These findings from human studies are in agreement with the observations made from work in the mouse. For example, Prmt6 inhibits Chd1 occupancy at the pluropotency genes Oct4 and Nanog, and Chd1 inactivation augments expression of Oct4 and Nanog and enhances stemness (Lee et al. 2012). Furthermore, Chd1 is essential for the open chromatin state and pluripotency of ES cells and is required for the reprogramming of somatic cells (Gaspar-Maia et al. 2009).
There is also some evidence that CHD2 plays a role in cancer, although this view is not nearly as clear as it is for CHD1. In this regard, there are some similarities between CHD1 and CHD2. For example, CHD2 may also be hormone responsive, as human chorionic gonadotropin that is released systemically during pregnancy causes transcriptional induction of CHD2, which has been proposed, along with other chromatin regulators, to prevent breast cancer (Russo and Russo 2012). CHD2 also regulates H3/H3.3 occupancy (Siggens et al. 2015). As is the case for CHD1 (and other CHDs, see below), colorectal tumors with high microsatellite instability have CHD2 mutations (Kim et al. 2011). There is some, although scant, evidence that CHD2 is inactivated in human cancers, including chronic lymphoblastic leukemia (CLL) and monoclonal B lymphocytosis, a B-cell expansion syndrome that can progress to CLL (Rodriguez et al. 2015). These CHD2 mutations in CLL are associated with mutations in genes encoding immunoglobulin heavy chain variable regions. CHD2 is also down-regulated in colorectal cancer (Bandres et al. 2007). CHD2 has also been implicated in neurodevelopment, as haploinsufficiency of CHD2 is associated with neurological deficits, including developmental delay, intellectual disability, epilepsy, and behavioral anomalies (Chenier et al. 2014). Neurological symptoms are a characteristic feature of deregulation of CHD subfamily III (and to a lesser extent to subfamily II) members (discussed below).
Mouse models echo the theme that Chd2 functions more in development than in tumorigenesis, as Chd2−/− mice have compromised viability, growth delay (Marfella et al. 2006), and lordokyphosis (Kulkarni et al. 2008). Furthermore, a congenic mouse backcross study identified Chd2 as a candidate obesity gene (Sarahan et al. 2011). Perhaps in line with the idea that Chd2 has some cancer-specific roles, genetic linkage analysis identified Chd2 as one of three candidate genes for a genetic modifier of breast cancer, suggesting an explanation for why p53 heterozygous mutant mice are uniquely susceptible to developing mammary gland tumors when established in the BALB/c genetic background (Koch et al. 2007). Heterozygosity for mutant Chd2 alleles is associated with extramedullary hematopoiesis and susceptibility to lymphoma (Nagarajan et al. 2009). In a follow-up study, the investigators found that Chd2-deficient cells are sensitive to DNA-damaging agents and do not efficiently repair DNA damage induced by ultraviolet or ionizing radiation, leading them to conclude that Chd2 (like other Chd proteins, discussed below) facilitates DNA repair and maintains genomic stability (Rajagopalan et al. 2012). Yet reports using a different Chd2 compromised mouse model concluded that heterozygotes succumb to non-neoplastic lesions in a number of organs, but are not susceptible to frank cancer (Marfella et al. 2006). Thus, while there is ample evidence that CHD1 functions as a tumor suppressor particularly in the prostate, and that in some cases it promotes oncogenesis, there is currently only tangential evidence that CHD2 shares these cancer-associated roles with its closest sibling.
SUBFAMILY II: CHD3, CHD4, and CHD5
Several of the biological processes ascribed to CHD subfamily II proteins are similar to those of subfamily I; for example, essentially all subfamily II proteins control transcription (Zhang et al. 1998; Srinivasan et al. 2006; Denslow and Wade 2007; Lee and Das 2010) and DNA damage repair (Stanley et al. 2013; Hall et al. 2014). But whereas subfamily I members function to maintain the pluripotent state in ES cells (Gaspar-Maia et al. 2009; Lee et al. 2012), the CHD II subfamily of proteins includes potent modulators of cellular proliferation, senescence, and apoptosis (Bagchi et al. 2007). The prototypical member of this family, CHD5, is a tumor suppressor whose inactivation is a predominant theme in a variety of human cancers. Lesions in CHD4, and to a lesser extent in CHD3, also occur in human cancer, with loss or inactivation of CHD subfamily II members being associated with chemoresistance, epithelial–mesenchymal transition (EMT), metastasis (Wang et al. 2011; Wu et al. 2012), and poor overall patient survival (Garcia et al. 2010; Wong et al. 2011; Wu et al. 2012; Du et al. 2013; Wang et al. 2013; Hall et al. 2014; Xie et al. 2015).
The best body of evidence for a CHD II subclass protein having a functional role in cancer exists for CHD5, likely due at least in part to the fact that CHD5 was the earliest CHD member defined to have a tumor-suppressive role (Bagchi et al. 2007). CHD5 maps to 1p36—a region of the genome frequently deleted in human cancers; modeling these deletions in the mouse using chromosome engineering pinpointed a 4.3-Mb genomic interval that encodes a product with potent tumor-suppressive activity. Although heterozygous loss of this interval leads to immortalization, oncogenic transformation, and spontaneous tumorigenesis, gain of dosage of this interval induces cellular senescence, excessive apoptosis, and perinatal lethality. Because the gain of dosage phenotypes of compromised proliferation and enhanced senescence can be rescued by depleting Chd5, we conclude that Chd5 (and not any of the other genes in the duplicated region that were tested) is responsible for these phenotypes. Thus, Chd5 is a highly dosage-dependent tumor suppressor that must be diploid: having only one copy predisposes to cancer; having three copies causes death.
Factors upstream of CHD subfamily II members include environmental factors, DNA methylation, miRNAs, DNA tumor virus-encoded oncogenes, chromatin regulators, transcription factors, and signaling pathways. CHD5 responds to environmental cues; for example, genestein—a compound present in soybeans—enhances CHD5 expression (Li et al. 2012). In neuroblastoma cells in which CHD5 is silenced by promoter methylation, genestein inhibits expression of the DNA methyltransferase DNMT3B, thereby reducing CHD5 methylation and activating its transcription. MicroRNAs (miRNAs), miR211 and miR454, target and degrade CHD5 mRNA in colorectal cancer (Cai et al. 2012) and hepatocellular carcinoma (Yu et al. 2015b), respectively. The chromatin regulator JMJD2A (also known as KDM4A, a member of the jumonji domain containing two families of lysine demethylases) inhibits RAS-mediated senescence to drive transformation (Mallette 2012). Importantly, JMJD2A was shown to inhibit the ability of RAS to induce CHD5 expression (Mallette 2012), thereby compromising p53-mediated pathways that are downstream (Bagchi 2007). The promoter of CHD5 has binding sites for transcription factors, including LEF1/TCF, SP1, and AP2, suggesting that CHD5 is transcriptionally regulated by components of the WNT/β-catenin pathway (Fatemi et al. 2014). Aberrant insulin-like growth factor 1 (IGF-1) signaling promotes CHD5 promoter hypermethylation, thereby inhibiting CHD5 expression to drive hepatocellular carcinoma development (Fang et al. 2015). RAS normally induces CHD5 expression, but aberrant up-regulation of JMJD2A inhibits this activation (Mallette and Richard 2012). This finding is consistent with Chd5-compromised cells being exquisitely sensitive to oncogenic transformation (Bagchi et al. 2007).
CHD5 is a component of the nucleosome remodeling and deacetylase (NURD) complex (Quan and Yusufzai 2014; Quan et al. 2014; Kolla et al. 2015). Therefore, factors functioning in parallel with CHD5 include NURD complex components such as MTA, GATAD2A, HDAC1/2, RBBP4/7, MDB2/3. Like CHD1, CHD5 is a nucleosome remodeler—an ATP-dependent enzyme that repositions or exchanges nucleosomes (Quan and Yusufzai 2014). A unique nucleosome “unwrapping” activity was discovered for CHD5 that at least in vitro, appeared to be distinct from the subfamily II member CHD4; perhaps this capability provides CHD5 with specific roles. In addition, the carboxyl terminus of CHD5 is distinct from CHD3 and CHD4, which may equip it with exclusive function distinct from its closest subfamily members.
Factors downstream from CHD5 include its transcriptional targets. Chd5 inhibits proliferation by transcriptionally activating Cdkn2a, a locus that encodes multiple tumor suppressors, including p16Ink4a and p19Arf (Bagchi et al. 2007; Bagchi and Mills 2008). Decreased Chd5 dosage cripples p16Ink4a/Rb and p19Arf/p53-tumor-suppressive pathways, setting the stage for cancer; on the other hand, enhanced dosage of the interval encoding Chd5 exacerbates these tumor-suppressive pathways, causing over exuberant apoptosis that depletes stem cells and is incompatible with life. What distinguishes subfamily II from other CHD proteins is the presence of tandem PHD zinc-finger motifs (see Fig. 1). We and others reported that the PHDs of CHD5 bind the amino-terminal tail of unmodified histone H3 (Oliver et al. 2012; Paul et al. 2013). The dual nature of the juxtaposed PHDs may have functional importance, as PHD1 and PHD2 of CHD5 simultaneously bind two H3 amino termini, which together enhance binding affinity four- to 11-fold (Oliver et al. 2012). PHD-mediated H3 binding is crucial for Chd5’s ability to regulate transcription, inhibit proliferation, and function as a tumor suppressor (Paul et al. 2013). In addition to Chd5-inducing Cdkn2a expression, it regulates gene expression globally. We identified Chd5-bound loci across the genome and found that Chd5 regulates a cascade of cancer pathways and chromatin regulators such as the polycomb repressive group complex (PcG) oncoprotein Bmi1 (Paul et al. 2013). CHD5 is also linked to other PcG proteins, as CHD5 and EZH2 transcriptionally inhibit each other’s expression (Xie et al. 2015). CHD5 can also induce expression of WEE1 to engage cell-cycle arrest at the G2/M checkpoint (Quan et al. 2014).
Mouse models were key for revealing that Chd5 plays a critical role in maintaining genomic integrity. Chd5 plays a dynamic role in remodeling the genome during maturation of the male germline (Li et al. 2014; Zhuang et al. 2014). The process of sperm maturation or “spermiogenesis,” an intricate process that occurs in haploid spermatids following meiosis, is one of the most extensive examples of chromatin remodeling known. A consequence of Chd5 deficiency in the male germline is alterations in chromatin compaction because of inefficient removal of canonical histones, deregulated incorporation of transition proteins and protamines, unbridled DNA damage, and compromised fertility (Li and Mills 2014; Li et al. 2014). These findings are in agreement with the low CHD5 expression levels found in testes of infertile men. CHD5’s role in unpackaging the genome to remove canonical histones and repackaging it, first with transition proteins and ultimately with protamines in mature sperm, and by doing so to maintain genomic integrity, may be unique to this particular subfamily member, as to date neither CHD3 nor CHD4 have been implicated in infertility. Perhaps Chd5’s role in the male germline is a result of its unique nucleosome unwrapping activity (Quan and Yusufzai 2014) or its distinctive carboxy-terminal region. Another possibility is that the three Chd II family members are expressed differently in testes. CHD5 also functions to maintain the genome in somatic cells, as compromised CHD5 enhances the DNA damage response in pancreatic adenocarcinoma cells, a finding that correlates with decreased patient survival (Hall et al. 2014).
CHD5 is frequently lost or inactivated in diverse human cancers. Loss-of-function CHD5 lesions, including compromised expression, promoter hypermethylation, deletion, and/or mutation, have been reported in glioma (Bagchi et al. 2007; Mulero-Navarro and Esteller 2008; Wang et al. 2013), neuroblastoma (Fujita et al. 2008; Garcia et al. 2010; Koyama et al. 2012; Li et al. 2012), lung cancer (Zhao et al. 2012), prostate cancer (Robbins et al. 2011), breast cancer (Mulero-Navarro and Esteller 2008; Wu et al. 2012), pancreatic adenocarcinoma (Hall et al. 2014), gastric cancer (Wang et al. 2009; Qu et al. 2013), bladder cancer (Wu et al. 2015), ovarian cancer (Gorringe et al. 2008; Wong et al. 2011), gallbladder carcinoma (Du et al. 2013), colorectal cancer (Mulero-Navarro and Esteller 2008; Mokarram et al. 2009; Cai et al. 2012; Fatemi et al. 2014), hepatocellular carcinoma (Zhao et al. 2014; Fang et al. 2015; Xie et al. 2015), melanoma (Lang et al. 2011), leukemia (Zhao et al. 2014), and laryngeal squamous cell carcinoma (Wang et al. 2011).
CHD5 expression correlates directly with overall patient survival for several cancers, including glioma (Wang et al. 2013), neuroblastoma (Garcia et al. 2010), as well as for cancers of the ovary (Wong et al. 2011), breast (Wu et al. 2012), gallbladder (Du et al. 2013), pancreas (Hall et al. 2014), and liver (Xie et al. 2015). The fact that CHD5 lesions tend to be heterozygous (Henrich et al. 2012) suggests that reactivation of the wild-type locus may be effective as a therapeutic strategy. Indeed, CHD5 induction using demethylating agents and transcriptional up-regulation of CHD5 decreases proliferation and compromises invasion (Fatemi et al. 2014). Thus, there is ample experimental and clinical evidence for CHD5’s potent tumor-suppressive role, suggesting that strategies to induce it could provide new avenues for treating diverse types of cancer.
The subfamily II CHD protein CHD4 has a number of similarities with CHD5. CHD4 is also a component of the NURD and is also a chromatin remodeler that regulates transcription, proliferation, and DNA damage repair. Like CHD5, CHD4 has strong ATPase activity (Quan and Yusufzai 2014). However, there are important differences, for instance, assays performed in vitro suggest that CHD4 does not unwrap nucleosomes nearly as efficiently as CHD5. The distinction between the different CHD II subfamily members in vivo, however, is not at present clear.
As found for CHD5, factors upstream of CHD4 include environmental insults such as tobacco smoke (Yamada et al. 2015). MYC enhances CHD4’s interaction with MTA and NURD during transformation (Zhang et al. 2005). The HPV16 oncoprotein E7 binds CHD4, evokes histone deacetylase activity, and enhances proliferation (Brehm et al. 1999).
Factors working in parallel with CHD4 include the NURD components (Quan and Yusufzai 2014; Kolla et al. 2015). Interestingly, it has been proposed that NURD complexes containing CHD4 are mutually exclusive with those containing CHD5 (Quan and Yusufzai 2014). CHD4 interacts with BRD4/NSD3 (Rahman et al. 2011), ZFHX4 (Chudnovsky et al. 2014), p300 acetyltransferase (Qi et al. 2015), p300/GATA4 (Hosokawa et al. 2013), TWIST (Fu et al. 2011), and HSF (Khaleque et al. 2008).
Factors downstream from CHD4 include its transcriptional targets. CHD4 has been shown to couple histone deacetylase activity to promoter hypermethylation in colorectal cancer (Cai et al. 2014). CHD4 interacts with NAB2 to regulate expression of genes encoding early growth response (EGR) transactivators (Srinivasan et al. 2006). CHD4 is recruited to MBD2/p66α-bound methylated DNA, an interaction abrogated in breast cancer (Desai et al. 2015). CHD4 inhibits E-cadherin, thereby inhibiting EMT and metastasis of lung cancer cells (Fu et al. 2011).
CHD4 lesions include its mutation in endometrial cancer (Le Gallo et al. 2012) and uterine serous carcinoma (Zhao et al. 2013), perhaps analogous to the single-nucleotide polymorphisms in CHD5 that are associated with endometriosis (Falconer et al. 2012). CHD4 maintains tumor-initiating cells in glioblastoma (Chudnovsky et al. 2014). As is the case for CHD5, CHD4 functions in the DNA damage response (Stanley et al. 2013; Qi et al. 2015). CHD4 deficiency has been reported to contribute to chemoresistance in BRCA mutant cells (Guillemette et al. 2015), and targeting CHD4 is able to deplete EpCam+ liver cancer cells (Nio et al. 2015). Thus, while there are some indications that CHD4 functions as a tumor suppressor, targeting it therapeutically has been proposed as a way to overcome chemoresistance (Nio et al. 2015), a finding that has also been shown for CHD subfamily I and III members (discussed below).
There is scant evidence for a role for CHD3 in cancer. Like its closest siblings, CHD3 is part of the NURD complex (Kolla et al. 2015), and the crystal and NMR structures were recently solved (Torchy et al. 2015). CHD3, like CHD4, interacts with the transcriptional corepressors NAB2 to inhibit expression of EGR target genes (Srinivasan et al. 2006). CHD3 appears to take on the family job of inducing the DNA damage repair response (Stanley et al. 2013; Klement et al. 2014). CHD3 regulates heterochromatin formation, thereby stimulating ATM-induced double-strand break repair, KAP-1 phosphorylation, and recruitment of ACF/SNF2 to sites of DNA damage (Klement et al. 2014). Thus, while there is substantial evidence that CHD5 functions as a tumor suppressor, and there is accumulating support for CHD4 playing a somewhat similar role, it is early days for CHD3. Time will tell whether this trait is conserved throughout the subfamily.
SUBFAMILY III: CHD6, CHD7, CHD8, and CHD9
Like the other subfamilies, subfamily III CHD proteins have been implicated in transcriptional regulation, cellular proliferation, and repair of DNA damage, and therefore have also been shown to be deregulated in cancer and to affect overall patient survival. But mutations in members of this subfamily have also been heavily implicated in developmental and neurological syndromes that are not associated with frank malignancy (Ronan et al. 2013), including coloboma of the eye, heart defects, atresia of the choanae, retardation of growth and/or development, genital and/or urinary abnormalities, and ear abnormalities and deafness (CHARGE) syndrome, schizophrenia, and autism (Layman et al. 2010). Furthermore, similar to the subfamily I CHD proteins, loss or inactivation of members of subfamily III CHD proteins sometimes correlate with enhanced patient survival, suggesting that members of this subfamily have both oncogenic and tumor-suppressive functions. At present, CHD8 appears to have the strongest link with cancer, although CHD7 has also been reported to modulate cancer-related pathways and to impact patient survival.
Factors upstream of CHD subfamily III proteins include hormones, environmental factors, and DNA methylation. Estrogen’s pro-proliferative effect in breast cancer cells effect occurs via cyclin D1-mediated activation of cyclin E2/CDK2, and CHD8 is required for efficient E2F1 recruitment to the promoter of cyclin E2 and its transcriptional activation (Caldon et al. 2009). In gastric cancers, CHD8 mutations are associated with the presence of Fusibacterium, a pathogen that is part of the gut microbiome (Tahara et al. 2014a), and CHD8 is silenced by promoter methylation in prostate cancer (Damaschke et al. 2014). At the parallel level, CHD8 interacts with c-MYC (Dingar et al. 2015). CHD8 also interacts with CCCTC-binding factor thereby affecting transcriptional output through modulation of chromatin insulation, DNA methylation, and histone acetylation (Ishihara et al. 2006). Indeed, CHD8 protein expression is reduced in prostate cancer and demethylating agents such as 5-aza-2′-deoxycytidine induces CHD8 expression at the transcriptional level (Damaschke et al. 2014). CHD8 is a coregulator of AR, and transcriptional activation the AR-responsive genes, such as TMPRSS2, requires CHD8 (Menon et al. 2010). CHD8 transcriptionally regulates genes encoding components of the WNT/β-catenin pathway and cell-cycle regulators (Sawada et al. 2013) and also has an effect on expression of genes implicated in cancer and neurogenesis (Sugathan et al. 2014). CHD8 inhibits β-catenin signaling by recruiting histone H1 to promoters of WNT target genes (Nishiyama et al. 2012). CHD8 inhibits p53-mediated apoptosis by its ability to recruit histone H1, a process that occurs during embryogenesis (Nishiyama et al. 2009) but apparently not in the context of malignancy—at least in the setting that was analyzed (Sawada et al. 2013).
CHD8 is mutated in breast cancer (Pongor et al. 2015), deleted in 36% of gastric cancers and 29% of colorectal cancers (Kim et al. 2011), and silenced by promoter methylation in prostate cancer (Damaschke et al. 2014). CHD8 is mutated in CpG island methylator phenotype 1 (CIMP1)-positive colorectal cancers (Tahara et al. 2014a) and CHD8 mutations correlate with Fusibacterium status, CIMP1 positivity, microsatellite instability, as well as mutations in BRAF, KRAS, and P53 (Tahara et al. 2014b). A mouse model of BCR-Abl-driven acute lymphoblastic leukemia found that Chd8 knockdown causes apoptosis, suggesting that targeting CHD8 is an effective therapy for patients with B-lymphoid malignancies (Shingleton and Hemann 2015). A genetic screen in a mouse model of acute myelogenous leukemia (AML) revealed that CHD8 is required for the ability of BRD4 to maintain AML through the H3K36-specific methyltransferase NSD3-short (Shen et al. 2015). Seemingly at odds with these mouse studies, high CHD8 expression in gastric cancers correlates with favorable patient survival (Sawada et al. 2013). In fact, enhanced nuclear expression of CHD8 has been shown to correlate with decreased survival and increased metastasis in patients with prostate cancer (Damaschke et al. 2014). Thus, while some reports suggest that CHD8 has tumor-suppressive functions, others clearly indicate that it is endowed with more nefarious pro-oncogenic capabilities. This apparent dichotomy warrants further clarification.
Some of the factors upstream of CHD7 are similar to those that regulate CHD8, such as environmental insults by Fusibacterium and the correlation between CHD7 mutation, CIMP1, and genomic instability (Kim et al. 2011; Tahara et al. 2014a,b). CHD7 is also mutated in response to tobacco smoke in small-cell lung cancer, having an in-frame duplication of exons 3–7, or being expressed as a fusion with PVT-1 (Pleasance et al. 2010). The factors that work in parallel to CHD7 are not well understood, although one example is that studies in mouse show that Chd7 is recruited by Smad1, Smad5, and Smad8 to promoters of cardiogenic genes (Liu et al. 2014). Pathways downstream from Chd7 include induction of Bmp signaling in mice (Jiang et al. 2012; Liu et al. 2014) and inhibition of CHK1 phosphorylation-dependent DNA damage repair in response to gemcitabine in human pancreatic cancer cells (Colbert et al. 2014). CHD7 and ES cell genes are aberrantly up-regulated in cutaneous T-cell lymphoma, leading to stem-cell-like features (Litvinov et al. 2014). Consistent with these findings made using human cells, conditional deletion of Chd7 in mice reveals that Chd7 maintains quiescence, thereby preventing premature depletion of neural stem cells (Jones et al. 2015).
A number of mouse models show Chd7’s pleiotropic roles in development. For example, the spontaneous heterozygous Chd7 mutations in “looper” and “whirligig” mice lead to CHARGE syndrome-like features (Ogier et al. 2014) and olfaction and reproductive defects (Bergman et al. 2010), respectively. Heterozygous disruption of Chd7 causes hearing loss (Hurd et al. 2011), ear defects (Adams et al. 2007; Tian et al. 2012), and defects in puberty and reproduction (Layman et al. 2011), whereas homozygous disruption of Chd7 causes embryonic lethality at 11 d of gestation (Sperry et al. 2014). While these studies highlight the critical role of Chd7 in development (reviewed in Layman et al. 2010), it is also clear that Chd7 modulates pathways central in tumorigenesis. Indeed, the CHARGE syndrome phenotypes of Chd7-compromised mice are at least partially a result of enhanced p53 activity (Van Nostrand and Attardi 2014). This is reminiscent of the finding that gain of Chd5 dosage enhances p53 activity, leading to developmental abnormalities and neonatal lethality caused by over exuberant apoptosis (Bagchi et al. 2007). But in contrast to Chd5, which promotes p53-mediated apoptosis, Chd7 appears to inhibit it (Bagchi et al. 2007).
Whereas there are loss-of-function mutations in CHD7 (like CHD8) in colorectal cancer, and these lesions correlate with mutations in BRAF, P53, and KRAS (Tahara et al. 2014b), there is enhanced expression of CHD7 in cutaneous T-cell lymphoma (Litvinov et al. 2014). Consistent with the concept that CHD7 promotes oncogenesis, low-level CHD7 protein expression correlates with enhanced survival of patients with pancreatic cancer (Colbert et al. 2014). This study showed that CHD7 depletion enhances the sensitivity of pancreatic cancer cells to gemcitabine by triggering DNA damage via ATR-mediated phosphorylation of CHK1. As has been suggested for CHD8, CHD7 depletion may render current therapies more effective by enhancing cell death, at least in the case of pancreatic cancer.
The roles of the remaining subfamily III proteins, CHD6 and CHD9, in cancer are at present much more obscure. CHD6 has been reported to map within a minimally common region of amplification in colorectal cancer (Ali Hassan et al. 2014), and CHD6 is mutated in both colorectal tumors (Mouradov et al. 2014) and transitional cell carcinoma of the bladder (Gui et al. 2011). In addition, it has been suggested that CHD6, like many other CHD proteins, regulates DNA damage repair (Stanley et al. 2013). Evidence for CHD9 playing a role in cancer is even less compelling, but CHD9 mutations have been reported in gastric and colorectal cancers (Kim et al. 2011). Whether these mutations are bone fide drivers or merely passengers of tumorigenesis remains to be evaluated. Thus, there is clear evidence that CHD subfamily III members, in particular CHD8 and to some extent CHD7, are critical cancer genes. However, in stark contrast to the tumor-suppressive roles ascribed to members of subfamily II, subfamily III members also have pro-oncogenic roles in some settings and their inhibition is proving to be an effective therapeutic strategy.
CONCLUDING REMARKS
The CHD family shares core motifs, with unique features equipping different members with highly variable functions. While subfamily II members have been defined as potent tumor suppressors, members of subfamily I and subfamily III have tumor-suppressive capabilities in some contexts but oncogenic capabilities in others. Lesions in members of each subfamily can define tumor subtype and predict patient survival. Whereas activation of subfamily II CHD members may hold promise as an effective therapeutic strategy, inactivation of subfamily I and III CHD members reveal vulnerabilities that conquer chemoresistance, which may be exploited in the oncology clinic.
ACKNOWLEDGMENTS
I thank members of my laboratory for helpful discussions. While every effort has been made to include a thorough and comprehensive review of the current literature, relevant publications may have been inadvertently omitted. This work was supported by the National Institutes of Health (R01CA190997 and R21OD018332) and the Stanley Family Foundation.
Footnotes
Editors: Scott A. Armstrong, Steven Henikoff, and Christopher R. Vakoc
Additional Perspectives on Chromatin Deregulation in Cancer available at www.perspectivesinmedicine.org
REFERENCES
- Adams ME, Hurd EA, Beyer LA, Swiderski DL, Raphael Y, Martin DM. 2007. Defects in vestibular sensory epithelia and innervation in mice with loss of Chd7 function: Implications for human CHARGE syndrome. J Comp Neurol 504: 519–532. [DOI] [PubMed] [Google Scholar]
- Ali Hassan NZ, Mokhtar NM, Kok Sin T, Mohamed Rose I, Sagap I, Harun R, Jamal R. 2014. Integrated analysis of copy number variation and genome-wide expression profiling in colorectal cancer tissues. PLoS ONE 9: e92553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attard G, Parker C, Eeles RA, Schroder F, Tomlins SA, Tannock I, Drake CG, de Bono JS. 2016. Prostate cancer. Lancet 387: 70–82. [DOI] [PubMed] [Google Scholar]
- Bagchi A, Mills AA. 2008. The quest for the 1p36 tumor suppressor. Cancer Res 68: 2551–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagchi A, Papazoglu C, Wu Y, Capurso D, Brodt M, Francis D, Bredel M, Vogel H, Mills AA. 2007. CHD5 is a tumor suppressor at human 1p36. Cell 128: 459–475. [DOI] [PubMed] [Google Scholar]
- Bandres E, Malumbres R, Cubedo E, Honorato B, Zarate R, Labarga A, Gabisu U, Sola JJ, Garcia-Foncillas J. 2007. A gene signature of 8 genes could identify the risk of recurrence and progression in Dukes’ B colon cancer patients. Oncol Rep 17: 1089–1094. [PubMed] [Google Scholar]
- Bergman JE, Bosman EA, van Ravenswaaij-Arts CM, Steel KP. 2010. Study of smell and reproductive organs in a mouse model for CHARGE syndrome. Eur J Hum Genet 18: 171–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner M, Lee DJ, O’Reilly C, Park K, MacDonald TY, Khani F, Turner KR, Chiu YL, Wild PJ, Dolgalev I, et al. 2014. SPOP mutations in prostate cancer across demographically diverse patient cohorts. Neoplasia 16: 14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehm A, Nielsen SJ, Miska EA, McCance DJ, Reid JL, Bannister AJ, Kouzarides T. 1999. The E7 oncoprotein associates with Mi2 and histone deacetylase activity to promote cell growth. EMBO J 18: 2449–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhardt L, Fuchs S, Krohn A, Masser S, Mader M, Kluth M, Bachmann F, Huland H, Steuber T, Graefen M, et al. 2013. CHD1 is a 5q21 tumor suppressor required for ERG rearrangement in prostate cancer. Cancer Res 73: 2795–2805. [DOI] [PubMed] [Google Scholar]
- Cai C, Ashktorab H, Pang X, Zhao Y, Sha W, Liu Y, Gu X. 2012. MicroRNA-211 expression promotes colorectal cancer cell growth in vitro and in vivo by targeting tumor suppressor CHD5. PLoS ONE 7: e29750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Geutjes EJ, de Lint K, Roepman P, Bruurs L, Yu LR, Wang W, van Blijswijk J, Mohammad H, de Rink I, et al. 2014. The NuRD complex cooperates with DNMTs to maintain silencing of key colorectal tumor suppressor genes. Oncogene 33: 2157–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldon CE, Sergio CM, Schutte J, Boersma MN, Sutherland RL, Carroll JS, Musgrove EA. 2009. Estrogen regulation of cyclin E2 requires cyclin D1 but not c-Myc. Mol Cell Biol 29: 4623–4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenier S, Yoon G, Argiropoulos B, Lauzon J, Laframboise R, Ahn JW, Ogilvie CM, Lionel AC, Marshall CR, Vaags AK, et al. 2014. CHD2 haploinsufficiency is associated with developmental delay, intellectual disability, epilepsy and neurobehavioural problems. J Neurodev Disord 6: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chudnovsky Y, Kim D, Zheng S, Whyte WA, Bansal M, Bray MA, Gopal S, Theisen MA, Bilodeau S, Thiru P, et al. 2014. ZFHX4 interacts with the NuRD core member CHD4 and regulates the glioblastoma tumor-initiating cell state. Cell Rep 6: 313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark JP, Cooper CS. 2009. ETS gene fusions in prostate cancer. Nat Rev Urol 6: 429–439. [DOI] [PubMed] [Google Scholar]
- Colbert LE, Petrova AV, Fisher SB, Pantazides BG, Madden MZ, Hardy CW, Warren MD, Pan Y, Nagaraju GP, Liu EA, et al. 2014. CHD7 expression predicts survival outcomes in patients with resected pancreatic cancer. Cancer Res 74: 2677–2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damaschke NA, Yang B, Blute ML Jr, Lin CP, Huang W, Jarrard DF. 2014. Frequent disruption of chromodomain helicase DNA-binding protein 8 (CHD8) and functionally associated chromatin regulators in prostate cancer. Neoplasia 16: 1018–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas V, Stokes DG, Perry RP. 1993. A mammalian DNA-binding protein that contains a chromodomain and an SNF2/SWI2-like helicase domain. Proc Natl Acad Sci 90: 2414–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denslow SA, Wade PA. 2007. The human Mi-2/NuRD complex and gene regulation. Oncogene 26: 5433–5438. [DOI] [PubMed] [Google Scholar]
- Desai MA, Webb HD, Sinanan LM, Scarsdale JN, Walavalkar NM, Ginder GD, Williams DC Jr. 2015. An intrinsically disordered region of methyl-CpG binding domain protein 2 (MBD2) recruits the histone deacetylase core of the NuRD complex. Nucleic Acids Res 43: 3100–3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey P, Ponnusamy MP, Deb S, Batra SK. 2011. Human RNA polymerase II–association factor 1 (hPaf1/PD2) regulates histone methylation and chromatin remodeling in pancreatic cancer. PLoS ONE 6: e26926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingar D, Kalkat M, Chan PK, Srikumar T, Bailey SD, Tu WB, Coyaud E, Ponzielli R, Kolyar M, Jurisica I, et al. 2015. BioID identifies novel c-MYC interacting partners in cultured cells and xenograft tumors. J Proteomics 118: 95–111. [DOI] [PubMed] [Google Scholar]
- Du X, Wu T, Lu J, Zang L, Song N, Yang T, Zhao H, Wang S. 2013. Decreased expression of chromodomain helicase DNA-binding protein 5 is an unfavorable prognostic marker in patients with primary gallbladder carcinoma. Clin Transl Oncol 15: 198–204. [DOI] [PubMed] [Google Scholar]
- Falconer H, Sundqvist J, Xu H, Vodolazkaia A, Fassbender A, Kyama C, Bokor A, D’Hooghe TM. 2012. Analysis of common variations in tumor-suppressor genes on chr1p36 among Caucasian women with endometriosis. Gynecol Oncol 127: 398–402. [DOI] [PubMed] [Google Scholar]
- Fang QL, Yin YR, Xie CR, Zhang S, Zhao WX, Pan C, Wang XM, Yin ZY. 2015. Mechanistic and biological significance of DNA methyltransferase 1 upregulated by growth factors in human hepatocellular carcinoma. Int J Oncol 46: 782–790. [DOI] [PubMed] [Google Scholar]
- Fatemi M, Paul TA, Brodeur GM, Shokrani B, Brim H, Ashktorab H. 2014. Epigenetic silencing of CHD5, a novel tumor-suppressor gene, occurs in early colorectal cancer stages. Cancer 120: 172–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher KW, Montironi R, Lopez Beltran A, Moch H, Wang L, Scarpelli M, Williamson SR, Koch MO, Cheng L. 2015. Molecular foundations for personalized therapy in prostate cancer. Curr Drug Target 16: 103–114. [DOI] [PubMed] [Google Scholar]
- Flanagan JF, Mi LZ, Chruszcz M, Cymborowski M, Clines KL, Kim Y, Minor W, Rastinejad F, Khorasanizadeh S. 2005. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature 438: 1181–1185. [DOI] [PubMed] [Google Scholar]
- Fu J, Qin L, He T, Qin J, Hong J, Wong J, Liao L, Xu J. 2011. The TWIST/Mi2/NuRD protein complex and its essential role in cancer metastasis. Cell Res 21: 275–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita T, Igarashi J, Okawa ER, Gotoh T, Manne J, Kolla V, Kim J, Zhao H, Pawel BR, London WB, et al. 2008. CHD5, a tumor suppressor gene deleted from 1p36.31 in neuroblastomas. J Natl Cancer Inst 100: 940–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D, Vela I, Sboner A, Iaquinta PJ, Karthaus WR, Gopalan A, Dowling C, Wanjala JN, Undvall EA, Arora VK, et al. 2014. Organoid cultures derived from patients with advanced prostate cancer. Cell 159: 176–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia I, Mayol G, Rodriguez E, Sunol M, Gershon TR, Rios J, Cheung NK, Kieran MW, George RE, Perez-Atayde AR, et al. 2010. Expression of the neuron-specific protein CHD5 is an independent marker of outcome in neuroblastoma. Mol Cancer 9: 277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar-Maia A, Alajem A, Polesso F, Sridharan R, Mason MJ, Heidersbach A, Ramalho-Santos J, McManus MT, Plath K, Meshorer E, et al. 2009. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature 460: 863–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorringe KL, Choong DY, Williams LH, Ramakrishna M, Sridhar A, Qiu W, Bearfoot JL, Campbell IG. 2008. Mutation and methylation analysis of the chromodomain-helicase-DNA binding 5 gene in ovarian cancer. Neoplasia 10: 1253–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC, et al. 2012. The mutational landscape of lethal castration-resistant prostate cancer. Nature 487: 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui Y, Guo G, Huang Y, Hu X, Tang A, Gao S, Wu R, Chen C, Li X, Zhou L, et al. 2011. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet 43: 875–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemette S, Serra RW, Peng M, Hayes JA, Konstantinopoulos PA, Green MR, Cantor SB. 2015. Resistance to therapy in BRCA2 mutant cells due to loss of the nucleosome remodeling factor CHD4. Genes Dev 29: 489–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall WA, Petrova AV, Colbert LE, Hardy CW, Fisher SB, Saka B, Shelton JW, Warren MD, Pantazides BG, Gandhi K, et al. 2014. Low CHD5 expression activates the DNA damage response and predicts poor outcome in patients undergoing adjuvant therapy for resected pancreatic cancer. Oncogene 33: 5450–5456. [DOI] [PubMed] [Google Scholar]
- Henrich KO, Schwab M, Westermann F. 2012. 1p36 tumor suppression—A matter of dosage? Cancer Res 72: 6079–6088. [DOI] [PubMed] [Google Scholar]
- Hosokawa H, Tanaka T, Suzuki Y, Iwamura C, Ohkubo S, Endoh K, Kato M, Endo Y, Onodera A, Tumes DJ, et al. 2013. Functionally distinct Gata3/Chd4 complexes coordinately establish T helper 2 (Th2) cell identity. Proc Natl Acad Sci 110: 4691–4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Gulzar ZG, Salari K, Lapointe J, Brooks JD, Pollack JR. 2012. Recurrent deletion of CHD1 in prostate cancer with relevance to cell invasiveness. Oncogene 31: 4164–4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd EA, Adams ME, Layman WS, Swiderski DL, Beyer LA, Halsey KE, Benson JM, Gong TW, Dolan DF, Raphael Y, et al. 2011. Mature middle and inner ears express Chd7 and exhibit distinctive pathologies in a mouse model of CHARGE syndrome. Hear Res 282: 184–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara K, Oshimura M, Nakao M. 2006. CTCF-dependent chromatin insulator is linked to epigenetic remodeling. Mol Cell 23: 733–742. [DOI] [PubMed] [Google Scholar]
- Jiang X, Zhou Y, Xian L, Chen W, Wu H, Gao X. 2012. The mutation in Chd7 causes misexpression of Bmp4 and developmental defects in telencephalic midline. Am J Pathol 181: 626–641. [DOI] [PubMed] [Google Scholar]
- Jones KM, Saric N, Russell JP, Andoniadou CL, Scambler PJ, Basson MA. 2015. CHD7 maintains neural stem cell quiescence and prevents premature stem cell depletion in the adult hippocampus. Stem Cells 33: 196–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, et al. 2013. Mutational landscape and significance across 12 major cancer types. Nature 502: 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaleque MA, Bharti A, Gong J, Gray PJ, Sachdev V, Ciocca DR, Stati A, Fanelli M, Calderwood SK. 2008. Heat shock factor 1 represses estrogen-dependent transcription through association with MTA1. Oncogene 27: 1886–1893. [DOI] [PubMed] [Google Scholar]
- Kim MS, Chung NG, Kang MR, Yoo NJ, Lee SH. 2011. Genetic and expressional alterations of CHD genes in gastric and colorectal cancers. Histopathology 58: 660–668. [DOI] [PubMed] [Google Scholar]
- Klement K, Luijsterburg MS, Pinder JB, Cena CS, Del Nero V, Wintersinger CM, Dellaire G, van Attikum H, Goodarzi AA. 2014. Opposing ISWI- and CHD-class chromatin remodeling activities orchestrate heterochromatic DNA repair. J Cell Biol 207: 717–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch JG, Gu X, Han Y, El-Naggar AK, Olson MV, Medina D, Jerry DJ, Blackburn AC, Peltz G, Amos CI, et al. 2007. Mammary tumor modifiers in BALB/cJ mice heterozygous for p53. Mamm Genome 18: 300–309. [DOI] [PubMed] [Google Scholar]
- Kolla V, Naraparaju K, Zhuang T, Higashi M, Kolla S, Blobel GA, Brodeur GM. 2015. The tumour suppressor CHD5 forms a NuRD-type chromatin remodelling complex. Biochem J 468: 345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama H, Zhuang T, Light JE, Kolla V, Higashi M, McGrady PW, London WB, Brodeur GM. 2012. Mechanisms of CHD5 inactivation in neuroblastomas. Clin Cancer Res 18: 1588–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni S, Nagarajan P, Wall J, Donovan DJ, Donell RL, Ligon AH, Venkatachalam S, Quade BJ. 2008. Disruption of chromodomain helicase DNA binding protein 2 (CHD2) causes scoliosis. Am J Med Genet A 146A: 1117–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang J, Tobias ES, Mackie R. 2011. Preliminary evidence for involvement of the tumour suppressor gene CHD5 in a family with cutaneous melanoma. Br J Dermatol 164: 1010–1016. [DOI] [PubMed] [Google Scholar]
- Layman WS, Hurd EA. 2011. Reproductive dysfunction and decreased GnRH neurogenesis in a mouse model of CHARGE syndrome. Hum Mol Genet 20: 3138–3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layman WS, Hurd EA, Martin DM. 2010. Chromodomain proteins in development: Lessons from CHARGE syndrome. Clin Genet 78: 11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Das HK. 2010. Transcriptional regulation of the presenilin-1 gene controls γ-secretase activity. Front Biosci 2: 22–35. [DOI] [PubMed] [Google Scholar]
- Lee YH, Ma H, Tan TZ, Ng SS, Soong R, Mori S, Fu XY, Zernicka-Goetz M, Wu Q. 2012. Protein arginine methyltransferase 6 regulates embryonic stem cell identity. Stem Cells Dev 21: 2613–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gallo M, O’Hara AJ, Rudd ML, Urick ME, Hansen NF, O’Neil NJ, Price JC, Zhang S, England BM, Godwin AK, et al. 2012. Exome sequencing of serous endometrial tumors identifies recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes. Nat Genet 44: 1310–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Mills AA. 2014. Packing for the journey: CHD5 remodels the genome. Cell Cycle 13: 1833–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Xu W, Huang Y, Huang X, Xu L, Lv Z. 2012. Genistein demethylates the promoter of CHD5 and inhibits neuroblastoma growth in vivo. Int J Mol Med 30: 1081–1086. [DOI] [PubMed] [Google Scholar]
- Li W, Wu J, Kim SY, Zhao M, Hearn SA, Zhang MQ, Meistrich ML, Mills AA. 2014. Chd5 orchestrates chromatin remodelling during sperm development. Nat Commun 5: 3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvinov IV, Netchiporouk E, Cordeiro B, Zargham H, Pehr K, Gilbert M, Zhou Y, Moreau L, Woetmann A, Odum N, et al. 2014. Ectopic expression of embryonic stem cell and other developmental genes in cutaneous T-cell lymphoma. Oncoimmunology 3: e970025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Lindberg J, Sui G, Luo J, Egevad L, Li T, Xie C, Wan M, Kim ST, Wang Z, et al. 2012. Identification of novel CHD1-associated collaborative alterations of genomic structure and functional assessment of CHD1 in prostate cancer. Oncogene 31: 3939–3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Harmelink C, Peng Y, Chen Y, Wang Q, Jiao K. 2014. CHD7 interacts with BMP R-SMADs to epigenetically regulate cardiogenesis in mice. Hum Mol Genet 23: 2145–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyn-Cook L, Word B, George N, Lyn-Cook B, Hammons G. 2014. Effect of cigarette smoke condensate on gene promoter methylation in human lung cells. Tob Induc Dis 12: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallette FA, Richard S. 2012. JMJD2A promotes cellular transformation by blocking cellular senescence through transcriptional repression of the tumor suppressor CHD5. Cell Rep 2: 1233–1243. [DOI] [PubMed] [Google Scholar]
- Marfella CG, Imbalzano AN. 2007. The Chd family of chromatin remodelers. Mutat Res 618: 30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marfella CG, Ohkawa Y, Coles AH, Garlick DS, Jones SN, Imbalzano AN. 2006. Mutation of the SNF2 family member Chd2 affects mouse development and survival. J Cell Physiol 209: 162–171. [DOI] [PubMed] [Google Scholar]
- Martin TJ, Peer CJ, Figg WD. 2013. Uncovering the genetic landscape driving castration-resistant prostate cancer. Cancer Biol Ther 14: 399–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon T, Yates JA, Bochar DA. 2010. Regulation of androgen-responsive transcription by the chromatin remodeling factor CHD8. Mol Endocrinol 24: 1165–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokarram P, Kumar K, Brim H, Naghibalhossaini F, Saberi-firoozi M, Nouraie M, Green R, Lee E, Smoot DT, Ashktorab H. 2009. Distinct high-profile methylated genes in colorectal cancer. PLoS ONE 4: e7012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouradov D, Sloggett C, Jorissen RN, Love CG, Li S, Burgess AW, Arango D, Strausberg RL, Buchanan D, Wormald S, et al. 2014. Colorectal cancer cell lines are representative models of the main molecular subtypes of primary cancer. Cancer Res 74: 3238–3247. [DOI] [PubMed] [Google Scholar]
- Mulero-Navarro S, Esteller M. 2008. Chromatin remodeling factor CHD5 is silenced by promoter CpG island hypermethylation in human cancer. Epigenetics 3: 210–215. [DOI] [PubMed] [Google Scholar]
- Nagarajan P, Onami TM, Rajagopalan S, Kania S, Donnell R, Venkatachalam S. 2009. Role of chromodomain helicase DNA-binding protein 2 in DNA damage response signaling and tumorigenesis. Oncogene 28: 1053–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nio K, Yamashita T, Okada H, Kondo M, Hayashi T, Hara Y, Nomura Y, Zeng SS, Yoshida M, Hayashi T, et al. 2015. Defeating EpCAM+ liver cancer stem cells by targeting chromatin remodeling enzyme CHD4 in human hepatocellular carcinoma. J Hepatol 63: 1164–1172. [DOI] [PubMed] [Google Scholar]
- Nishiyama M, Oshikawa K, Tsukada Y, Nakagawa T, Iemura S, Natsume T, Fan Y, Kikuchi A, Skoultchi AI, Nakayama KI. 2009. CHD8 suppresses p53-mediated apoptosis through histone H1 recruitment during early embryogenesis. Nat Cell Biol 11: 172–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama M, Skoultchi AI, Nakayama KI. 2012. Histone H1 recruitment by CHD8 is essential for suppression of the Wnt-β-catenin signaling pathway. Mol Cell Biol 32: 501–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogier JM, Carpinelli MR, Arhatari BD, Symons RC, Kile BT, Burt RA. 2014. CHD7 deficiency in “Looper,” a new mouse model of CHARGE syndrome, results in ossicle malformation, otosclerosis and hearing impairment. PLoS ONE 9: e97559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver SS, Musselman CA, Srinivasan R, Svaren JP, Kutateladze TG, Denu JM. 2012. Multivalent recognition of histone tails by the PHD fingers of CHD5. Biochemistry 51: 6534–6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Kuo A, Schalch T, Vogel H, Joshua-Tor L, McCombie WR, Gozani O, Hammell M, Mills AA. 2013. Chd5 requires PHD-mediated histone 3 binding for tumor suppression. Cell Rep 3: 92–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance ED, Stephens PJ, O’Meara S, McBride DJ, Meynert A, Jones D, Lin ML, Beare D, Lau KW, Greenman C, et al. 2010. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature 463: 184–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pongor L, Kormos M, Hatzis C, Pusztai L, Szabo A, Gyorffy B. 2015. A genome-wide approach to link genotype to clinical outcome by utilizing next generation sequencing and gene chip data of 6,697 breast cancer patients. Genome Med 7: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W, Chen H, Xiao T, Wang R, Li T, Han L, Zeng X. 2015. Acetyltransferase p300 collaborates with chromodomain helicase DNA-binding protein 4 (CHD4) to facilitate DNA double-strand break repair. Mutagenesis 31: 193–203. [DOI] [PubMed] [Google Scholar]
- Qu Y, Dang S, Hou P. 2013. Gene methylation in gastric cancer. Int J Clin Chem 424: 53–65. [DOI] [PubMed] [Google Scholar]
- Quan J, Yusufzai T. 2014. The tumor suppressor chromodomain helicase DNA-binding protein 5 (CHD5) remodels nucleosomes by unwrapping. J Biol Chem 289: 20717–20726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan J, Adelmant G, Marto JA, Look AT, Yusufzai T. 2014. The chromatin remodeling factor CHD5 is a transcriptional repressor of WEE1. PLoS ONE 9: e108066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S, Sowa ME, Ottinger M, Smith JA, Shi Y, Harper JW, Howley PM. 2011. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol Cell Biol 31: 2641–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan S, Nepa J, Venkatachalam S. 2012. Chromodomain helicase DNA-binding protein 2 affects the repair of X-ray and UV-induced DNA damage. Environ Mol Mutagen 53: 44–50. [DOI] [PubMed] [Google Scholar]
- Robbins CM, Tembe WA, Baker A, Sinari S, Moses TY, Beckstrom-Sternberg S, Beckstrom-Sternberg J, Barrett M, Long J, Chinnaiyan A, et al. 2011. Copy number and targeted mutational analysis reveals novel somatic events in metastatic prostate tumors. Genome Res 21: 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues LU, Rider L, Nieto C, Romero L, Karimpour-Fard A, Loda M, Lucia MS, Wu M, Shi L, Cimic A, et al. 2015. Coordinate loss of MAP3K7 and CHD1 promotes aggressive prostate cancer. Cancer Res 75: 1021–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez D, Bretones G, Quesada V, Villamor N, Arango JR, Lopez-Guillermo A, Ramsay AJ, Baumann T, Quiros PM, Navarro A, et al. 2015. Mutations in CHD2 cause defective association with active chromatin in chronic lymphocytic leukemia. Blood 126: 195–202. [DOI] [PubMed] [Google Scholar]
- Ronan JL, Wu W, Crabtree GR. 2013. From neural development to cognition: Unexpected roles for chromatin. Nat Rev Genet 14: 347–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo J, Russo IH. 2012. Molecular basis of pregnancy-induced breast cancer prevention. Horm Mol Biol Clin Investig 9: 3–10. [DOI] [PubMed] [Google Scholar]
- Sarahan KA, Fisler JS, Warden CH. 2011. Four out of eight genes in a mouse chromosome 7 congenic donor region are candidate obesity genes. Physiol Genomics 43: 1049–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada G, Ueo H, Matsumura T, Uchi R, Ishibashi M, Mima K, Kurashige J, Takahashi Y, Akiyoshi S, Sudo T, et al. 2013. CHD8 is an independent prognostic indicator that regulates Wnt/β-catenin signaling and the cell cycle in gastric cancer. Oncol Rep 30: 1137–1142. [DOI] [PubMed] [Google Scholar]
- Scott AF, Mohr DW, Ling H, Scharpf RB, Zhang P, Liptak GS. 2014. Characterization of the genomic architecture and mutational spectrum of a small cell prostate carcinoma. Genes 5: 366–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C, Ipsaro JJ, Shi J, Milazzo JP, Wang E, Roe JS, Suzuki Y, Pappin DJ, Joshua-Tor L, Vakoc CR. 2015. NSD3-short is an adaptor protein that couples BRD4 to the CHD8 chromatin remodeler. Mol Cell 60: 847–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingleton JR, Hemann MT. 2015. The chromatin regulator CHD8 is a context-dependent mediator of cell survival in murine hematopoietic malignancies. PloS ONE 10: e0143275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siggens L, Cordeddu L, Ronnerblad M, Lennartsson A, Ekwall K. 2015. Transcription-coupled recruitment of human CHD1 and CHD2 influences chromatin accessibility and histone H3 and H3.3 occupancy at active chromatin regions. Epigenetics Chromatin 8: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowalsky AG, Xia Z, Wang L, Zhao H, Chen S, Bubley GJ, Balk SP, Li W. 2015. Whole transcriptome sequencing reveals extensive unspliced mRNA in metastatic castration-resistant prostate cancer. Mol Cancer Res 13: 98–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperry ED, Hurd EA, Durham MA, Reamer EN, Stein AB, Martin DM. 2014. The chromatin remodeling protein CHD7, mutated in CHARGE syndrome, is necessary for proper craniofacial and tracheal development. Dev Dyn 243: 1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan R, Mager GM, Ward RM, Mayer J, Svaren J. 2006. NAB2 represses transcription by interacting with the CHD4 subunit of the nucleosome remodeling and deacetylase (NuRD) complex. J Biol Chem 281: 15129–15137. [DOI] [PubMed] [Google Scholar]
- Stanley FK, Moore S, Goodarzi AA. 2013. CHD chromatin remodelling enzymes and the DNA damage response. Mutat Res 750: 31–44. [DOI] [PubMed] [Google Scholar]
- Stokes DG, Perry RP. 1995. DNA-binding and chromatin localization properties of CHD1. Mol Cell Biol 15: 2745–2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugathan A, Biagioli M, Golzio C, Erdin S, Blumenthal I, Manavalan P, Ragavendran A, Brand H, Lucente D, Miles J, et al. 2014. CHD8 regulates neurodevelopmental pathways associated with autism spectrum disorder in neural progenitors. Proc Natl Acad Sci 111: E4468–E4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahara T, Yamamoto E, Madireddi P, Suzuki H, Maruyama R, Chung W, Garriga J, Jelinek J, Yamano HO, Sugai T, et al. 2014a. Colorectal carcinomas with CpG island methylator phenotype 1 frequently contain mutations in chromatin regulators. Gastroenterology 146: 530–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahara T, Yamamoto E, Suzuki H, Maruyama R, Chung W, Garriga J, Jelinek J, Yamano HO, Sugai T, An B, et al. 2014b. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res 74: 1311–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S, Ding K, Li R, Zhang W, Li G, Kong X, Qian P, Lobie PE, Zhu T. 2014. Identification of miR-26 as a key mediator of estrogen stimulated cell proliferation by targeting CHD1, GREB1 and KPNA2. Breast Cancer Res 16: R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Liu S, Briones V, Geiman TM, Muegge K. 2011. Treatment of breast cancer cells with DNA demethylating agents leads to a release of Pol II stalling at genes with DNA-hypermethylated regions upstream of TSS. Nucleic Acids Res 39: 9508–9520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tereshchenko IV, Zhong H, Chekmareva MA, Kane-Goldsmith N, Santanam U, Petrosky W, Stein MN, Ganesan S, Singer EA, Moore D, et al. 2014. ERG and CHD1 heterogeneity in prostate cancer: Use of confocal microscopy in assessment of microscopic foci. Prostate 74: 1551–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C, Yu H, Yang B, Han F, Zheng Y, Bartels CF, Schelling D, Arnold JE, Scacheri PC, Zheng QY. 2012. Otitis media in a new mouse model for CHARGE syndrome with a deletion in the Chd7 gene. PLoS ONE 7: e34944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torchy MP, Hamiche A, Klaholz BP. 2015. Structure and function insights into the NuRD chromatin remodeling complex. Cell Mol Life Sci 72: 2491–2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand JL, Attardi LD. 2014. Guilty as CHARGED: p53’s expanding role in disease. Cell Cycle 13: 3798–3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lau KK, So LK, Lam YW. 2009. CHD5 is down-regulated through promoter hypermethylation in gastric cancer. J Biomed Sci 16: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Chen H, Fu S, Xu ZM, Sun KL, Fu WN. 2011. The involvement of CHD5 hypermethylation in laryngeal squamous cell carcinoma. Oral Oncol 47: 601–608. [DOI] [PubMed] [Google Scholar]
- Wang L, He S, Tu Y, Ji P, Zong J, Zhang J, Feng F, Zhao J, Gao G, Zhang Y. 2013. Downregulation of chromatin remodeling factor CHD5 is associated with a poor prognosis in human glioma. J Clin Neurosci 20: 958–963. [DOI] [PubMed] [Google Scholar]
- Wong RR, Chan LK, Tsang TP, Lee CW, Cheung TH, Yim SF, Siu NS, Lee SN, Yu MY, Chim SS, et al. 2011. CHD5 downregulation associated with poor prognosis in epithelial ovarian cancer. Gynecol Obstet Invest 72: 203–207. [DOI] [PubMed] [Google Scholar]
- Wu X, Zhu Z, Li W, Fu X, Su D, Fu L, Zhang Z, Luo A, Sun X, Fu L, et al. 2012. Chromodomain helicase DNA binding protein 5 plays a tumor suppressor role in human breast cancer. Breast Cancer Res 14: R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Yang Z, Ye R, An D, Li C, Wang Y, Wang Y, Huang Y, Liu H, Li F, et al. 2015. Novel variants in MLL confer to bladder cancer recurrence identified by whole-exome sequencing. Oncotarget 7: 2629–2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie CR, Li Z, Sun HG, Wang FQ, Sun Y, Zhao WX, Zhang S, Zhao WX, Wang XM, Yin ZY. 2015. Mutual regulation between CHD5 and EZH2 in hepatocellular carcinoma. Oncotarget 6: 40940–40952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Sato N, Ikeda S, Arai T, Sawabe M, Mori S, Yamada Y, Muramatsu M, Tanaka M. 2015. Association of the chromodomain helicase DNA-binding protein 4 (CHD4) missense variation p.D140E with cancer: Potential interaction with smoking. Genes Chromosomes Cancer 54: 122–128. [DOI] [PubMed] [Google Scholar]
- Yu B, Swatkoski S, Holly A, Lee LC, Giroux V, Lee CS, Hsu D, Smith JL, Yuen G, Yue J, et al. 2015a. Oncogenesis driven by the Ras/Raf pathway requires the SUMO E2 ligase Ubc9. Proc Natl Acad Sci 112: E1724–E1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Gong X, Sun L, Yao H, Lu B, Zhu L. 2015b. miR-454 functions as an oncogene by inhibiting CHD5 in hepatocellular carcinoma. Oncotarget 6: 39225–39234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. 1998. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95: 279–289. [DOI] [PubMed] [Google Scholar]
- Zhang XY, DeSalle LM, Patel JH, Capobianco AJ, Yu D, Thomas-Tikhonenko A, McMahon SB. 2005. Metastasis-associated protein 1 (MTA1) is an essential downstream effector of the c-MYC oncoprotein. Proc Natl Acad Sci 102: 13968–13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Yan Q, Lv J, Huang H, Zheng W, Zhang B, Ma W. 2012. CHD5, a tumor suppressor that is epigenetically silenced in lung cancer. Lung cancer 76: 324–331. [DOI] [PubMed] [Google Scholar]
- Zhao S, Choi M, Overton JD, Bellone S, Roque DM, Cocco E, Guzzo F, English DP, Varughese J, Gasparrini S, et al. 2013. Landscape of somatic single-nucleotide and copy-number mutations in uterine serous carcinoma. Proc Natl Acad Sci 110: 2916–2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Meng F, Wang N, Ma W, Yan Q. 2014. Silencing of CHD5 gene by promoter methylation in leukemia. PLoS ONE 9: e85172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang T, Hess RA, Kolla V, Higashi M, Raabe TD, Brodeur GM. 2014. CHD5 is required for spermiogenesis and chromatin condensation. Mech Dev 131: 35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]