Abstract
Cancers evolve as a consequence of multiple somatic lesions, with competition between subclones and sequential subclonal evolution. Some driver mutations arise either early or late in the evolution of different individual tumors, suggesting that the final malignant properties of a subclone reflect the sum of mutations acquired rather than the order in which they arose. However, very little is known about the cellular consequences of altering the order in which mutations are acquired. Recent studies of human myeloproliferative neoplasms show that the order in which individual mutations are acquired has a dramatic impact on the cell biological and molecular properties of tumor-initiating cells. Differences in clinical presentation, complications, and response to targeted therapy were all observed and implicate mutation order as an important player in cancer biology. These observations represent the first demonstration that the order of mutation acquisition influences stem and progenitor cell behavior and clonal evolution in any cancer. Thus far, the impact of different mutation orders has only been studied in hematological malignancies, and analogous studies of solid cancers are now required.
Cancers are driven by somatic mutations and their functional consequences. Recent studies have shown that the order in which mutations are acquired may dramatically impact cancer biology, clinical presentation, and therapy.
From a cell biological point of view, cancers arise as a result of a clone of cells outcompeting their neighbors, thereby causing an accumulation of immature cells that can no longer carry out the required tasks of cells in that tissue. The mechanistic underpinnings of this increased competitive ability have been the focus of many research efforts and have identified key properties of tumors that facilitate this process (reviewed in Knudson 2001; Hanahan and Weinberg 2011). These properties range from avoiding immune cell destruction through to increased genome instability, but in some way or another they help promote the expansion of the cancer cells relative to their noncancerous counterparts (reviewed in Hahn and Weinberg 2002; Hanahan and Weinberg 2011). Traditionally, these abnormal cellular attributes have been linked to individual somatic genetic mutations, each conferring an oncogenic property, and they have been loosely divided into loss-of-function mutations (a tumor suppressor) and gain-of-function mutations (an oncogene) (Balmain 2001; Futreal et al. 2004).
The field of cancer genetics was heavily informed by the discovery of dominant oncogenes and tumor suppressors that strongly associated with cancer (e.g., Ras [Fearon and Vogelstein 1990], p53 [Finlay et al. 1989], BRCA2 [Wooster et al. 1994], Rb [Friend et al. 1986]). These studies laid the foundation for cancer being considered a disease of genetic mutations and substantial resources have subsequently been invested in characterizing the complete genetic architecture of individual cancers. Initially, genetic changes were screened for by comparative genome hybridization efforts that identified large genetic changes, such as copy number alterations, amplifications, or deletions that correspond to over- or underactivation of a particular gene (reviewed in Emanuel and Saitta 2007). However, these studies were unable to identify single activating or inactivating point mutations in individual cancers. This changed with the introduction of high throughput sequencing methods that evolved from efforts to sequence the human genome in the 1990s. Mutations in driver genes associated with various cancers were identified and databases for such mutations now exist that catalog thousands of cancer-related mutations in hundreds of genes (e.g., COSMIC [Forbes et al. 2015], CBioPortal [Cerami et al. 2012]). These include a number of hereditary risk alleles that have been identified across cancer subtypes (e.g., BRCA2, APC) and impose an increased risk of developing disease. Subsequent large-scale genome-wide association studies (GWASs) also identified constitutional single nucleotide polymorphisms (SNPs) that were enriched (or not) in particular cohorts of patients (reviewed in Gibson 2011). These SNPs correlated to areas in the genome where previously identified mutations were located but also identified a number of additional loci of interest that are still under scrutiny, somewhat obfuscated by the challenges associated with population-wide SNP analyses (reviewed in Evangelou and Ioannidis 2013).
Overall, this patchwork of mutations and genomic variants and their associated gain or loss of functional properties drives a cancer. Indeed, the last two decades have resulted in a near complete set of coding mutations that associate with individual cancers. Larger projects such as those in breast cancer (Curtis et al. 2012) and acute myeloid leukemia (Cancer Genome Atlas Research Network 2013) make a convincing case that the scientific community has exhausted the search for novel driver mutations in coding genes, yet the evolution of individual cancers remains a complex puzzle with little to no treatment progress in many cancers (Coombs et al. 2015; Dan et al. 2015; Bible and Ryder 2016).
Several studies have already underscored this complexity in hematological malignancies in which tumors often undergo branching evolution with respect to additional mutations after their initial clonal advantage (Anderson et al. 2011; Notta et al. 2011). Whereas early mutations are present throughout the entire malignant clone (e.g., at the base of a branching tree), later mutations may or may not represent a significant proportion of the malignant clone. Instead of being a homogeneous mass of genetically identical cells, a tumor rather becomes an entire family of clonally diverse cells that have their origins in a common ancestral clone that may or may not still exist (see Curtius et al. 2016; Enver 2016). This has significant clinical implications because this pool of genetically diverse clones is a common source of therapy-resistant clones (see Gatenby 2016; Purroy and Wu 2016; Quezada and Peggs 2016; Taylor and Costello 2016).
Moreover, there are significant examples of contradictory functions of genes associated with cancer (Pallante et al. 2015; Koppens and van Lohuizen 2016). For example, increased expression of Ezh2 commonly associates with multiple cancers, but loss-of-function mutations in Ezh2 have been shown to be sufficient to drive T-ALL in mouse models (Hock 2012; Simon et al. 2012; Kim and Roberts 2016). Further complicating the picture is the clear evidence of cancer genes interacting to drive a phenotype (e.g., Ras and p53) in which mutations in the same genes are more commonly found together in particular cancers. Such gene interactions are starting to form the basis for risk stratification and targeted therapy (Flaherty et al. 2012; Nangalia et al. 2013; Papaemmanuil et al. 2013).
The idea of targeting two individual genes therapeutically has thus far implied a static state of cancer that can be targeted (e.g., the cancer has mutation A and B, treat with inhibitors to A and B) (Basheer and Huntly 2015; Melero et al. 2015; Epner et al. 2016). However, this sort of approach fails to consider whether the historical path to a cancer could give insight into its growth, its response to therapy, and its subsequent evolution. Previous studies using multiregion sequencing analyses of cancer samples has formally showed the temporal sequence of mutations in many cases (e.g., kidney [Gerlinger et al. 2014], breast [Yates et al. 2015]); however, little was known about whether or not the temporal sequence of individual mutations mattered. The remainder of this review will focus on the mechanistic and therapeutic implications of the recent discovery that the order of genetic mutations impacts disease evolution.
MUTATION ORDER MATTERS
As described above, cancer is often considered to be the sum of the phenotypes that individual mutations confer. The traditional hallmarks of cancer are almost always discussed as isolated units in which one mutation drives a particular phenotype and a second mutation drives another complementary phenotype and so on. The first wave of combinatorial studies therefore involved studying individual mutations and how the subsequent mutant gene interactions drive the malignant properties of a clone. Indeed it turns out that specific mutations have preferential partner mutations in particular cancers and numerous such studies have been published (Cancer Genome Atlas Research Network 2012, 2013; Curtis et al. 2012; Nangalia et al. 2013; Papaemmanuil et al. 2013; Skoulidis et al. 2015). This also prompted the classification of cancer genes into categories that defined types of activities (e.g., differentiation block + proliferation advantage = cancer) (reviewed in Tenen 2003). However, this view does not take into account the dynamic nature of cancer evolution nor does it account for the possibility that one mutation may need to precede the other for the cancer to develop.
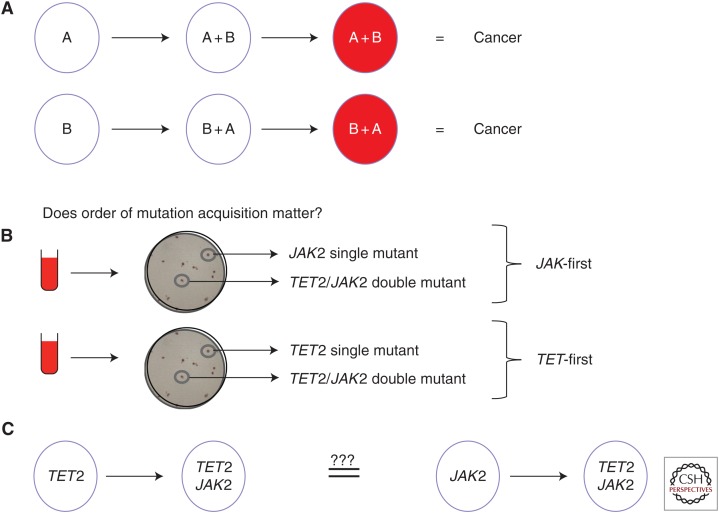
The first suggestion that the temporal sequence of two mutations might impact tumorigensis was in a mouse model of adrenocortical tumors (Herbet et al. 2012). When oncogenic Ras was overexpressed before loss of p53, highly malignant tumors with metastatic properties were formed in contrast to the benign tumors that resulted from the reverse mutational order. These experiments were in mice and involved overexpression of Ras at nonphysiological levels, so it remained unclear whether or not the order of endogenous mutations had any impact on human cancers. It did, however, suggest that the same two genetic mutations might not always lead to the same phenotypic outcome. Our group therefore recently undertook a study that was aimed at determining whether or not the order of mutation matters for human tumor evolution (Fig. 1).
Figure 1.
Determining the mutation order in hematological malignancies. (A) Different scenarios for how two mutations could lead to cancer with the only difference being the order in which mutations are acquired. (B) How the colony assay can be used to determine mutation order. Briefly, single colonies are grown from peripheral blood or bone marrow samples and individually picked and sequenced for JAK2 and TET2 mutations. The presence of a single-mutant clone for one of these genes allows determination of mutation order as displayed.
Such a question was most easily addressed in malignancies that are not genetically complex, and we therefore elected to study the chronic myeloprolifertive neoplasms (MPNs). MPNs are clonal myeloid malignancies that harbor an average of five to 10 somatic mutations, only a minority of which have been determined to be driver mutations, making them a tractable disease for studying the impact of mutation acquisition order (Klampfl et al. 2013; Nangalia et al. 2013). In particular, MPNs readily permit clonal analysis and are chronic diseases, thereby facilitating the dissection of disease evolution and intratumor clonal architecture. Moreover, MPNs are not associated with a differentiation block, allowing progenitors to be assessed for differences in downstream blood cell production. In 2005, several groups identified a single gain-of-function point mutation (JAK2 V617F) that was present in the majority of MPNs (reviewed in Levine and Gilliland 2007) and subsequent studies have identified collaborating driver mutations in genes that were either commonly co-mutated (e.g., TET2 [Delhommeau et al. 2009]) or independently mutated (e.g., CALR [Klampfl et al. 2013; Nangalia et al. 2013]).
Because TET2 was the most commonly co-mutated gene alongside the JAK2 V617F driver mutation, our studies focused on a cohort of patients who had both mutations. Colony assays of peripheral blood cells were used to unambiguously determine mutation order (Fig. 1B) and it was determined that about half of the patients acquired JAK2 V617F before acquisition of a TET2 mutation (JAK2-first patients) (Ortmann et al. 2015). In some patients, mutation order could not be determined because single-mutant clones could not be isolated. This latter point poses a significant problem for studies in other, more advanced, cancers in which the single-mutant parent clone may not always be identifiable, thereby precluding straightforward studies of mutation order.
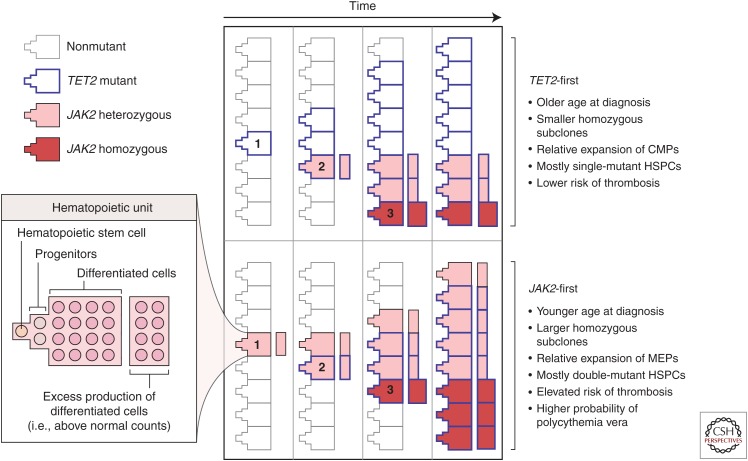
Once stratified into JAK2-first and TET2-first subcohorts, it became obvious that there were different disease characteristics associated with the different mutation order (Fig. 2). Most strikingly, JAK2-first patients presented in clinic more than a decade before TET2-first patients, suggesting that disease symptoms developed at a different rate depending on the order of mutation acquisition. Differences were also observed in disease subtype, thrombosis free-survival, and progenitor composition (Ortmann et al. 2015). Thus, JAK2-first patients were more likely to present with polycythemia vera (PV), to develop thromboses, and they also harbored more megakaryocyte and erythroid progenitors.
Figure 2.
Order of mutation acquisition influences the evolution of disease. This model depicts the manner in which single hematopoietic units (left), consisting of stem cells, progenitors, and differentiated cells, acquire mutations over time. Some units are hyperproliferative and produce excess differentiated cells that contribute to the disease phenotype. The numbers represent the acquisition of the first mutation (1), second mutation (2), and JAK2 V617F homozygosity (3). Patients who acquire a TET2 mutation first gain a self-renewal advantage but do not overproduce downstream progeny. The expansion of the TET2-alone clone (bold borders) without excess differentiated cells leads to clonal expansion without immediate clinical presentation. Hematopoietic stem cells that acquire a secondary JAK2 mutation (pink fill) compete with the TET2-alone clone, and their increased proliferation at the progenitor level drives an overproduction of terminal cells. When homozygosity is acquired as a third event (red fill), this clone has limited space to expand because of the high self-renewal activity of TET2-alone and TET2–JAK2–heterozygous clones. Patients who acquire a JAK2 mutation first (pink fill, lower panel) produce excess differentiated cells in the absence of a distinct self-renewal advantage in the hematopoietic stem cells. When a secondary TET2 mutation is acquired, hematopoietic stem cells obtain a self-renewal advantage and JAK2–TET2–mutant cells expand at the stem-cell level. Hematopoietic stem cells with loss of heterozygosity of JAK2 V617F (acquired before or after the TET2 mutation) (red fill) also have space to expand and result in a more pronounced excess of differentiated cells. This excess production would explain both the presentation as a polycythemia vera and the elevated risk of thrombotic events in JAK2-first patients. (Both the figure and legend are from Ortmann et al. 2015, Massachusetts Medical Society; reprinted, with permission, © 2015.)
Another key observation of this study was that the most evolved clone (the clone with the most number of mutations) was not the dominant clone within the stem and progenitor cell compartment of TET2-first patients. This runs counter to the commonly held belief that the clone with the most number of driver mutations is, or at least will become, the most dominant. Sequential follow-up of TET2-first patients showed that the clonal architecture was typically stable over months to years with the most evolved clone continuing to represent a minor subclone (Ortmann et al. 2015).
SAME TWO MUTATIONS, DIFFERENT BIOLOGICAL FUNCTION, AND MOLECULAR PROFILES
Single-cell assays of blood stem and progenitor cells showed that genetically similar cells (i.e., those harboring both a JAK2 and a TET2 mutation) possessed distinct functional properties: the number of progenitor cells created by a double-mutant HSC was increased in JAK2-first patients and decreased in TET2-first patients. This implied the existence of an intrinsic epigenetic mechanism in which acquiring one mutation before the other altered the HSCs ability to produce progenitor cells despite carrying the exact same two genetic lesions. Indeed expression profiling studies showed that the transcriptional consequences of a JAK2 V617F mutation were altered by the presence of a prior TET2 mutation. Together, these data show that single cells and their clonal progeny can have the same two mutations but have distinct biological and molecular properties (Ortmann et al. 2015).
POTENTIAL MECHANISMS FOR HOW MUTATION ORDER ALTERS DISEASE EVOLUTION
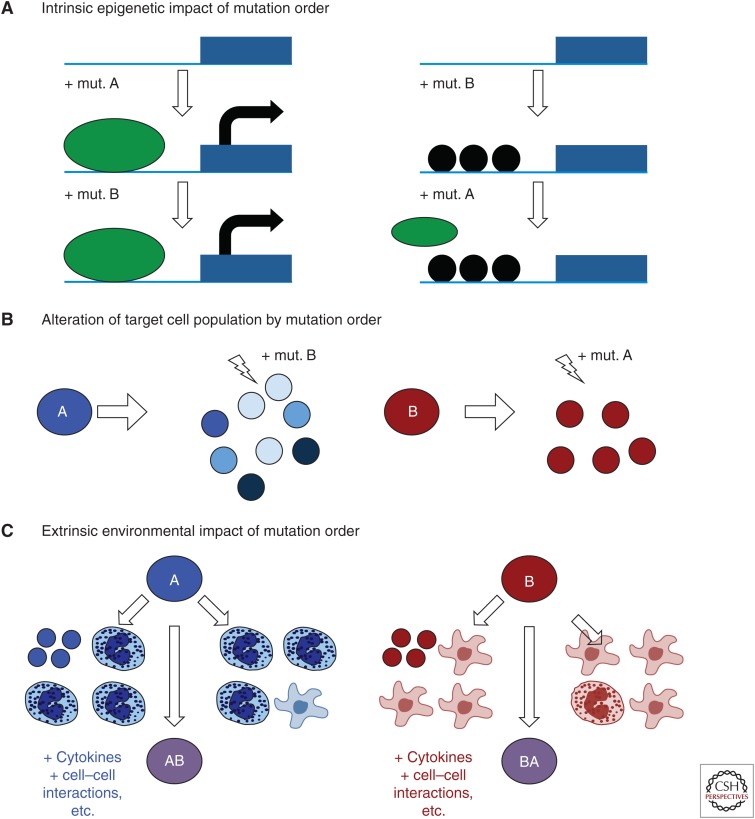
The mechanism by which mutation order influences disease evolution is currently unknown. At least three mechanisms (not mutually exclusive) may explain the effects of different mutation order and these are shown in Figure 3: (1) The first mutation permanently alters the gene expression of clonal progeny by an intrinsic epigenetic mechanism, (2) the first mutation creates a heterogeneous and unique pool of potential target cells, and (3) the first mutation drives a clonal expansion of progeny, thereby creating a cellular microenvironment that exerts an extrinsic effect on the newly minted double-mutant clone. These are described in more detail below.
Figure 3.
Potential mechanisms for how mutation order alters disease evolution. (A) The intrinsic epigenetic impact of mutation order. Here, the first mutation impacts the accessibility of particular genomic loci such that the second mutation cannot make the changes it would normally make on a wild-type (WT) background. In this example, black circles represent chromatin compaction by histones induced by mutation B that block the accessibility of a promoter region for a particular transcription factor (green oval) that is stimulated by mutation A. The gene would be turned ON if mutation A came first and OFF if B came first. (B) How a single mutation could create a different collection of target cells. In this case, mutation A induces rapid cell growth and differentiation from the initial cell, creating many distinct target cell types, whereas mutation B creates a slower growing, more undifferentiated clone. The second mutation could therefore occur in a different target cell depending on which mutation comes first. (C) The extrinsic environmental impact of mutation order. Here the first mutation (A or B) gives rise to different numbers and types of mature cells, thereby altering the cellular environment that the double-mutant clone (AB) finds itself when it is first created. These distinct cellular environments may contribute to distinct disease evolution.
Intrinsic Epigenetic Impact of Mutation Order
The gene expression studies described in Ortmann et al. (2015) suggest that certain genes are differently regulated depending on which mutation comes first. One possible explanation for this would be an epigenetic mechanism in which the first mutation alters the accessibility of particular genomic regions (e.g., induces a local chromatin compaction) and the second mutation can no longer activate/repress that region as it would have done on a nonmutant background. An illustration of how this might occur is presented in Figure 3A. In the case of JAK2 V617F and TET2 double-mutant patients, this seems especially plausible because both TET2 (Ito et al. 2010; Shih et al. 2012) and JAK2 (Dawson et al. 2009) have been shown to play a role in epigenetic regulation and chromatin alteration. Moreover, many cancers have had mutations in epigenetic modifiers described (reviewed in Baylin and Jones 2011; Azad et al. 2013; Eriksson et al. 2015) making this mode of action potentially applicable to numerous other malignancies.
Production of a Different Target Cell Pool
The dynamics of early disease evolution are very poorly understood and are often ignored by consequence. The first mutation almost certainly creates multiple immature progeny, with each cell having a high proliferative index and therefore representing a potential target cell for the next mutation. For example, cells resulting from one mutation, as opposed to the other, could exist in a different differentiation status and may even drive clonal exhaustion before developing into disease. Moreover, it has been speculated previously that early clonal heterogeneity might even drive distinct disease subtypes (Prick et al. 2014).
Extrinsic Cellular Environmental Impact of Mutation Order
Mutation order may also alter the cellular context within which the mutant clone evolves. Numerous studies have shown that the tumor microenvironment plays a role in shaping the clonal expansion that results in cancer (Alderton 2015; Hirata and Sahai 2016; Martin et al. 2016; Yuan 2016). Following the first mutation, single-mutant progeny may expand and create a distinct microenvironment as illustrated in Figure 3B, especially in cases in which the first mutation has been present for a number of years or decades as in chronic premalignant conditions (Campbell and Green 2006). Consistent with this concept, some mature hematopoietic cell types (e.g., monocytes and megakaryocytes) have been shown to exert effects on the stem and progenitor cell compartment by secreting key molecules or activating key pathways that influence cell fate choice. Indeed, it has been shown that sequential removal of mature cells and the molecules they secrete from HSC expansion cultures markedly increases HSC self-renewal capacity (Csaszar et al. 2012; Fares et al. 2014). Therefore, if the two mutations are different in the way they alter the differentiation and proliferation of the mutant clone, then the double-mutant clone will find itself in a cellular environment that depends on the nature (and perhaps length of existence) of the first mutation. This in turn could drive different signaling responses and alter the evolution and/or expansion of the double-mutant clone.
CLINICAL SIGNIFICANCE OF MUTATION ORDER—A CHALLENGE FOR SOLID CANCERS
There are at least two areas of clinical practice that could be impacted by identification of distinct mutation order in disease. The first is in disease management in which one mutation order may lead to complications that are not as frequently observed when the other mutation occurs first. In the MPNs, the evidence for this is already clear—JAK2-first patients have an increased likelihood of thrombotic events. The second is the prediction that patients with one mutation order would respond to therapy and patients with the other order would not. It has been suggested based on in vitro data that JAK2 inhibition would be more effective in JAK2-first patients compared to TET2-first patients, but a prospective clinical trial would be needed to validate these findings. In any event, it would appear prudent to incorporate mutation order into future clinical trial design to assess whether distinct responses are because of different mutation orders.
For other cancers, however, there are significant issues that must be addressed before these sorts of studies can be undertaken. First and foremost, it is easier to determine mutation order in MPNs for two reasons. First, there are well-defined clonal assays (e.g., colony-forming cell, long-term culture-initiating cell, and competitive repopulation assays) (Eaves 2015) and second, they represent a very early stage of malignancy. This makes studies of mutation order in other more advanced cancers quite challenging. The first issue will almost certainly be addressed by developments in single-cell genomics, which are allowing the characterization of single cells from tumors. The issue of tumor stage, however, may not be so easily resolved.
Samples from vast numbers of cancers have been subjected to large-scale sequencing projects, and bioinformatic analysis of exome sequencing data has been shown to be able to determine mutation order accurately in a proportion of patients (Ortmann et al. 2015). However, by the time of their diagnosis, the clonal architecture of solid tumors may be dominated by a particular subclone, although in some cases multisite analysis can address this problem. Nevertheless, if ancestral clones are very rare it may be difficult to ascertain mutation order. Either way, such analyses will be challenging in some patients; the mutation burden is not sufficiently high to conclusively prove that the mutations coexist within the same clone (e.g., mutation A is in 20% of cells, mutation B is present in 10%—these mutations could be from independent clones), although novel approaches using deep targeted sequencing and copy number alteration information have proved effective in several cases (Roth et al. 2014). In other patients, the variant allele frequency may be too close together for two mutations to conclude that one preceded the other. In still other patients, loss of heterozygosity at some loci may have occurred in some cells and resulted in an overestimate of the number of cells that bear the mutation, although this issue could potentially be addressed by subsequent allele specific analyses.
A further complication is that when only one mutation order is observed (e.g., mutation B is always preceded by mutation A) and the remainder of patients are exclusively double mutant, it remains impossible to decipher whether or not the other mutation order (B then A) existed. It may be that one mutation order differentiates the clone to exhaustion (and consequently no tumor ever emerges) and the other mutation order drives an expansion that results in a tumor. In this case, all patients who have tumors would have had to acquire one mutation before the other. Alternatively, it may be that the double-mutant clone has such a strong clonal advantage that the single-mutant clone is quickly outcompeted and no longer observable. In this case, patients would have different mutation orders with potentially different disease management strategies.
In both cases, however, the result would be the same: no observable single-mutant clones of one order. Indeed, an example has already been described in the MPNs in which mutations in Dnmt3a appear to occur first more frequently than JAK2 V617F or MPL mutations (Nangalia et al. 2015). On closer inspection using earlier and/or diagnostic clinical samples, it turned out that some patients in whom order could not be determined were JAK2/MPL-first and the single-mutant clone had been completely overtaken by the more dominant double-mutant clone (Nangalia et al. 2015). It remains unclear whether this is a general rule across cancers or an anomaly of the blood system, perhaps as a consequence of high tissue turnover.
The most immediate clinical impact of mutation order is for JAK2/TET2 double-mutant MPN patients in which JAK2-first patients are much more likely to develop PV and have a significantly increased risk of a thrombotic event. Monitoring of patients from other centers is required to validate the observations from this study and significant follow-up studies are required to determine whether or not disease transformation to acute myeloid leukemia and differential response to JAK2 inhibitors are mutation order dependent in patients.
The in vitro data suggest that JAK2-first patients would be more likely to respond to JAK2 inhibitors and if a different therapeutic response based on mutation order is borne out in a prospective clinical trial, there will be two important clinical consequences. The first is that previous clinical trials in which no significant differences in treatment groups were observed could be reevaluated if genomic data from those patients were available. It may be that patients of one mutation order respond perfectly well to treatment and could be useful in this subset of patients. The second is that mutation order could be factored into disease management in which patients are treated with a targeted therapy if they acquire mutations in one order but not the other (e.g., in the case of MPNs, this would mean prescribing targeted JAK2 inhibitors for JAK2-first patients but not for TET2-first patients). Moreover, if it turns out that different mutation orders induce different processes of disease evolution and maintenance, then it will become important to identify the mechanism of action for each individual set of mutations to identify novel therapeutic targets.
CONCLUDING REMARKS
Given the high frequency of mutations in epigenetic modifiers that have been identified in hematological and nonhematological cancers, it seems reasonable to suggest that a second mutation may not be able to induce the same genetic changes that it would have done in the absence of a mutant epigenetic modifier. Whether or not such changes directly determine disease evolution and/or therapeutic response in solid cancers remains unclear but we suspect this will be the subject of many future biological and clinical investigations.
What may not be studied as much, but could be just as important, is the cellular context in which a newly mutated progenitor finds itself. Differences in cell–cell interactions with distinct cell types, cytokine concentrations, and feedback loops from single-mutant cells that have preexisted for several months or years could all play a role in shaping the eventual disease. Mutation order may well determine this cellular context with each single mutation causing the development of a different milieu of target cells for the second mutation, mature cell types, secreted molecules, and clonal fitness. However, the experimental systems are not as well defined for such questions of cellular environment or early clonal propagation, representing a major challenge for the field moving forward that will likely require the introduction of longitudinal microenvironmental monitoring of serum cytokines, use of mouse models of single- and double-mutant cell populations, and comprehensive phenotyping of the cell types created by initiating mutations.
With a reasonable understanding of the mutational spectra involved in different cancer types now in place, research questions must evolve to consider cancer development in the context of tumor heterogeneity, clonal competition, and disease evolution. This next stage of multilevel dissection of a dynamic growth process will be increasingly complex but should allow scientists and clinicians to make better predictions of where a cancer is going by understanding where it has come from.
ACKNOWLEDGMENTS
Work in the Kent laboratory is supported by a Bloodwise Bennett Fellowship (15008) and a European Hematology Association Non-Clinical Advanced Research Fellowship. Work in the Green laboratory is supported by Bloodwise, Cancer Research UK, the Kay Kendall Leukaemia Fund, the National Institute for Health Research Cambridge Biomedical Research Centre, the Cambridge Experimental Cancer Medicine Centre, and the Leukemia and Lymphoma Society of America. D.G.K and A.R.G. are supported by a core support grant from the Wellcome Trust and MRC to the Wellcome Trust—Medical Research Council Cambridge Stem Cell Institute.
Footnotes
Editors: Charles Swanton, Alberto Bardelli, Kornelia Polyak, Sohrab Shah, and Trevor A. Graham
Additional Perspectives on Cancer Evolution available at www.perspectivesinmedicine.org
REFERENCES
*Reference is also in this collection.
- Alderton GK. 2015. Tumour microenvironment: Driving relapse. Nat Rev Cancer 15: 195. [DOI] [PubMed] [Google Scholar]
- Anderson K, Lutz C, van Delft FW, Bateman CM, Guo Y, Colman SM, Kempski H, Moorman AV, Titley I, Swansbury J, et al. 2011. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature 469: 356–361. [DOI] [PubMed] [Google Scholar]
- Azad N, Zahnow CA, Rudin CM, Baylin SB. 2013. The future of epigenetic therapy in solid tumours—lessons from the past. Nat Rev Clin Oncol 10: 256–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmain A. 2001. Cancer genetics: From Boveri and Mendel to microarrays. Nat Rev Cancer 1: 77–82. [DOI] [PubMed] [Google Scholar]
- Basheer F, Huntly BJ. 2015. BET bromodomain inhibitors in leukemia. Exp Hematol 43: 718–731. [DOI] [PubMed] [Google Scholar]
- Baylin SB, Jones PA. 2011. A decade of exploring the cancer epigenome–biological and translational implications. Nat Rev Cancer 11: 726–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bible KC, Ryder M. 2016. Evolving molecularly targeted therapies for advanced-stage thyroid cancers. Nat Rev Clin Oncol 13: 403–416. [DOI] [PubMed] [Google Scholar]
- Campbell PJ, Green AR. 2006. The myeloproliferative disorders. N Engl J Med 355: 2452–2466. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. 2012. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487: 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. 2013. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 368: 2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. 2012. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov 2: 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombs CC, Tallman MS, Levine RL. 2015. Molecular therapy for acute myeloid leukaemia. Nat Rev Clin Oncol 13: 305–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csaszar E, Kirouac DC, Yu M, Wang W, Qiao W, Cooke MP, Boitano AE, Ito C, Zandstra PW. 2012. Rapid expansion of human hematopoietic stem cells by automated control of inhibitory feedback signaling. Cell Stem Cell 10: 218–229. [DOI] [PubMed] [Google Scholar]
- Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. 2012. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486: 346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Curtius K, Wright NA, Graham TA. 2016. Evolution of premalignant disease. Cold Spring Harb Perspect Med 10.1101/cshperspect.a026542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan C, Chi J, Wang L. 2015. Molecular mechanisms of the progression of myelodysplastic syndrome to secondary acute myeloid leukaemia and implication for therapy. Ann Med 47: 209–217. [DOI] [PubMed] [Google Scholar]
- Dawson MA, Bannister AJ, Gottgens B, Foster SD, Bartke T, Green AR, Kouzarides T. 2009. JAK2 phosphorylates histone H3Y41 and excludes HP1α from chromatin. Nature 461: 819–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A, Kosmider O, Le Couedic JP, Robert F, Alberdi A, et al. 2009. Mutation in TET2 in myeloid cancers. N Engl J Med 360: 2289–2301. [DOI] [PubMed] [Google Scholar]
- Eaves CJ. 2015. Hematopoietic stem cells: Concepts, definitions, and the new reality. Blood 125: 2605–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuel BS, Saitta SC. 2007. From microscopes to microarrays: Dissecting recurrent chromosomal rearrangements. Nat Rev Genet 8: 869–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Enver T. 2016. Cancer stem cells and clonal evolution. Cold Spring Harb Perspect Med 10.1101/cshperspect.a026559. [DOI] [Google Scholar]
- Epner EM, Saroya BS, Hasanali ZS, Loughran TP Jr. 2016. Combination epigenetic and immunotherapy overcomes resistance to monoclonal antibodies in hematologic malignancies: A new therapeutic approach. Exp Hematol 44: 157–160. [DOI] [PubMed] [Google Scholar]
- Eriksson A, Lennartsson A, Lehmann S. 2015. Epigenetic aberrations in acute myeloid leukemia: Early key events during leukemogenesis. Exp Hematol 43: 609–624. [DOI] [PubMed] [Google Scholar]
- Evangelou E, Ioannidis JP. 2013. Meta-analysis methods for genome-wide association studies and beyond. Nat Rev Genet 14: 379–389. [DOI] [PubMed] [Google Scholar]
- Fares I, Chagraoui J, Gareau Y, Gingras S, Ruel R, Mayotte N, Csaszar E, Knapp DJ, Miller P, Ngom M, et al. 2014. Cord blood expansion. Pyrimidoindole derivatives are agonists of human hematopoietic stem cell self-renewal. Science 345: 1509–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon ER, Vogelstein B. 1990. A genetic model for colorectal tumorigenesis. Cell 61: 759–767. [DOI] [PubMed] [Google Scholar]
- Finlay CA, Hinds PW, Levine AJ. 1989. The p53 proto-oncogene can act as a suppressor of transformation. Cell 57: 1083–1093. [DOI] [PubMed] [Google Scholar]
- Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, Hamid O, Schuchter L, Cebon J, Ibrahim N, et al. 2012. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med 367: 1694–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, Ding M, Bamford S, Cole C, Ward S, et al. 2015. COSMIC: Exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res 43: D805–D811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friend SH, Bernards R, Rogelj S, Weinberg RA, Rapaport JM, Albert DM, Dryja TP. 1986. A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature 323: 643–646. [DOI] [PubMed] [Google Scholar]
- Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. 2004. A census of human cancer genes. Nat Rev Cancer 4: 177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Gatenby RA. 2016. Cancer drug resistance in light of clonal evolution: Novel trial designs. Cold Spring Harb Perspect Med 10.1101/cshperspect.a027078. [DOI] [Google Scholar]
- Gerlinger M, Horswell S, Larkin J, Rowan AJ, Salm MP, Varela I, Fisher R, McGranahan N, Matthews N, Santos CR, et al. 2014. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet 46: 225–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G. 2011. Rare and common variants: Twenty arguments. Nat Rev Genet 13: 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn WC, Weinberg RA. 2002. Modelling the molecular circuitry of cancer. Nat Rev Cancer 2: 331–341. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. 2011. Hallmarks of cancer: The next generation. Cell 144: 646–674. [DOI] [PubMed] [Google Scholar]
- Herbet M, Salomon A, Feige JJ, Thomas M. 2012. Acquisition order of Ras and p53 gene alterations defines distinct adrenocortical tumor phenotypes. PLoS Genet 8: e1002700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Hirata E, Sahai E. 2016. Tumor microenvironment and differential responses to therapy. Cold Spring Harb Perspect Med 10.1101/cshperspect.a026781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock H. 2012. A complex polycomb issue: The two faces of EZH2 in cancer. Genes Dev 26: 751–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. 2010. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466: 1129–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH, Roberts CW. 2016. Targeting EZH2 in cancer. Nat Med 22: 128–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klampfl T, Gisslinger H, Harutyunyan AS, Nivarthi H, Rumi E, Milosevic JD, Them NC, Berg T, Gisslinger B, Pietra D, et al. 2013. Somatic mutations of calreticulin in myeloproliferative neoplasms. N Engl J Med 369: 2379–2390. [DOI] [PubMed] [Google Scholar]
- Knudson AG. 2001. Two genetic hits (more or less) to cancer. Nat Rev Cancer 1: 157–162. [DOI] [PubMed] [Google Scholar]
- Koppens M, van Lohuizen M. 2016. Context-dependent actions of Polycomb repressors in cancer. Oncogene 35: 1341–1352. [DOI] [PubMed] [Google Scholar]
- Levine RL, Gilliland DG. 2007. JAK-2 mutations and their relevance to myeloproliferative disease. Curr Opin Hematol 14: 43–47. [DOI] [PubMed] [Google Scholar]
- *.Martin JD, Fukumura D, Duda DG, Boucher Y, Jain RK. 2016. Reengineering the tumor microenvironment to alleviate hypoxia and overcome cancer heterogeneity. Cold Spring Harb Perspect Med 10.1101/cshperspect.a027094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melero I, Berman DM, Aznar MA, Korman AJ, Perez Gracia JL, Haanen J. 2015. Evolving synergistic combinations of targeted immunotherapies to combat cancer. Nat Rev Cancer 15: 457–472. [DOI] [PubMed] [Google Scholar]
- Nangalia J, Massie CE, Baxter EJ, Nice FL, Gundem G, Wedge DC, Avezov E, Li J, Kollmann K, Kent DG, et al. 2013. Somatic CALR mutations in myeloproliferative neoplasms with nonmutated JAK2. N Engl J Med 369: 2391–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nangalia J, Nice FL, Wedge DC, Godfrey AL, Grinfeld J, Thakkerm C, Massie CE, Baxter J, Sewell D, Silber Y, et al. 2015. DNMT3A mutations occur early or late in patients with myeloproliferative neoplasms and mutation order influences phenotype. Haematologica 100: e438–e442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notta F, Mullighan CG, Wang JC, Poeppl A, Doulatov S, Phillips LA, Ma J, Minden MD, Downing JR, Dick JE. 2011. Evolution of human BCR-ABL1 lymphoblastic leukaemia-initiating cells. Nature 469: 362–367. [DOI] [PubMed] [Google Scholar]
- Ortmann CA, Kent DG, Nangalia J, Silber Y, Wedge DC, Grinfeld J, Baxter EJ, Massie CE, Papaemmanuil E, Menon S, et al. 2015. Effect of mutation order on myeloproliferative neoplasms. N Engl J Med 372: 601–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallante P, Forzati F, Federico A, Arra C, Fusco A. 2015. Polycomb protein family member CBX7 plays a critical role in cancer progression. Am J Cancer Res 5: 1594–1601. [PMC free article] [PubMed] [Google Scholar]
- Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, Van Loo P, Yoon CJ, Ellis P, Wedge DC, Pellagatti A, et al. 2013. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood 122: 3616–3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prick J, de Haan G, Green AR, Kent DG. 2014. Clonal heterogeneity as a driver of disease variability in the evolution of myeloproliferative neoplasms. Exp Hematol 42: 841–851. [DOI] [PubMed] [Google Scholar]
- *.Purroy N, Wu CJ. 2016. Co-evolution of leukemia and host immune cells in chronic lymphocytic leukemia (CLL). Cold Spring Harb Perspect Med 10.1101/cshperspect.a026740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Quezada S, Peggs K. 2016. Tumor heterogeneity—The Achilles heel for cancer immunotherapy. Cold Spring Harb Perspect Med 10.1101/cshperspect.a027086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A, Khattra J, Yap D, Wan A, Laks E, Biele J, Ha G, Aparicio S, Bouchard-Cote A, Shah SP. 2014. PyClone: Statistical inference of clonal population structure in cancer. Nat Methods 11: 396–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih AH, Abdel-Wahab O, Patel JP, Levine RL. 2012. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer 12: 599–612. [DOI] [PubMed] [Google Scholar]
- Simon C, Chagraoui J, Krosl J, Gendron P, Wilhelm B, Lemieux S, Boucher G, Chagnon P, Drouin S, Lambert R, et al. 2012. A key role for EZH2 and associated genes in mouse and human adult T-cell acute leukemia. Genes Dev 26: 651–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoulidis F, Byers LA, Diao L, Papadimitrakopoulou VA, Tong P, Izzo J, Behrens C, Kadara H, Parra ER, Canales JR, et al. 2015. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov 5: 860–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Taylor B, Costello J. 2016. Therapy-induced cancer evolution. Cold Spring Harb Perspect Med 10.1101/cshperspect.a026617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenen DG. 2003. Disruption of differentiation in human cancer: AML shows the way. Nat Rev Cancer 3: 89–101. [DOI] [PubMed] [Google Scholar]
- Wooster R, Neuhausen SL, Mangion J, Quirk Y, Ford D, Collins N, Nguyen K, Seal S, Tran T, Averill D, et al. 1994. Localization of a breast cancer susceptibility gene, BRCA2, to chromosome 13q12-13. Science 265: 2088–2090. [DOI] [PubMed] [Google Scholar]
- Yates LR, Gerstung M, Knappskog S, Desmedt C, Gundem G, Van Loo P, Aas T, Alexandrov LB, Larsimont D, Davies H, et al. 2015. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat Med 21: 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Yuan Y. 2016. Spatial heterogeneity in the tumor microenvironment. Cold Spring Harb Perspect Med 10.1101/cshperspect.a026583. [DOI] [PMC free article] [PubMed] [Google Scholar]