Abstract
Neurodegenerative dementias are clinically heterogeneous, progressive diseases with frequently overlapping symptoms, such as cognitive impairments and behavior and movement deficits. Although a majority of cases appear to be sporadic, there is a large genetic component that has yet to be fully explained. Here, we review the recent genetic and genomic findings pertaining to Alzheimer’s disease, frontotemporal dementia, Lewy body dementia, and prion dementia. In this review, we describe causal and susceptibility genes identified for these dementias and discuss recent research pertaining to the molecular function of these genes. Of particular interest, there is a large overlap in clinical phenotypes, genes, and/or aggregating protein products involved in these diseases, as well as frequent comorbid presentation, indicating that these dementias may represent a continuum of syndromes rather than individual diseases.
Many gene variants associated with neurodegenerative dementias have been identified in the past decade. Some genes (e.g., SNCA and MAPT) and their protein products appear to be involved in several dementias.
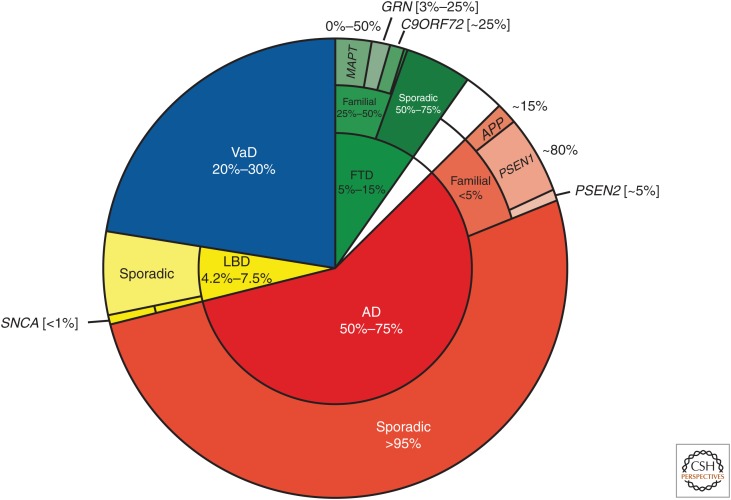
Neurodegenerative dementias are clinically heterogeneous, progressive diseases with frequently overlapping symptoms, such as cognitive impairments and behavior and movement deficits. This spectrum of disease includes vascular dementia (VaD), Lewy body dementia (LBD), Alzheimer’s disease (AD), frontotemporal dementia (FTD), and prion dementias (Fig. 1). Recent estimates indicate that 5.9%–9.4% of people >65 years of age suffer from dementia, with approximately 44 million affected worldwide (Prince et al. 2014). Moreover, as a result of our aging population, the number of people affected with dementia is expected to double roughly every 20 years. Of these dementias, AD is by far the most frequent, comprising 50%–75% of all cases (Fig. 1) (Prince et al. 2014).
Figure 1.
Frequency of specific neurodegenerative dementias. The inner circle shows the frequency of specific neurodegenerative dementias (indicated by different colors) as an approximate percentage of all cases of disease. The middle circle shows the percentage of patients that show familial or sporadic patterns of inheritance for each disease. The outer circle depicts the approximate percent of patients with familial inheritance patterns that harbor mutations in specific known causal genes.
Over the last few decades, substantial progress has been made in understanding the molecular genetics of neurodegenerative dementias and identifying the pathologically aggregating proteins involved. In large part, this can be attributed to advances in sequencing techniques and bioinformatic analysis approaches. Beginning in the early 1990s with the identification of microsatellites (also known as short tandem repeats) and single nucleotide polymorphisms (SNPs), researchers began to apply an approach termed linkage analysis. This technique makes use of these genomic markers to identify chromosomal regions shared by affected family members in large families with apparent dominant transmission. The assumption is that the pathogenic mutation lies within this shared chromosomal region, which carries disease risk, as it segregates with the disease in the family. Linkage analysis led to the rapid identification of a number of highly penetrant disease-causing mutations (Kerem et al. 1989; MacDonald et al. 1993; Hästbacka et al. 1994; Feder et al. 1996; Poorkaj et al. 1998; Amir et al. 1999).
However, only a small subset (∼5%) of neurodegenerative dementia cases show a pattern of autosomal dominant inheritance, whereas most cases appear to be sporadic. Early on, using genetic approaches such as linkage analysis, the majority of disease genes involved in these familial, often early-onset cases were discovered. More recently, genome-wide association studies (GWAS) have been applied to identify a multitude of risk variants (mutations with low penetrance) in idiopathic dementia. This new approach was enabled by the development of comprehensive genome-wide arrays, allowing for the simultaneous evaluation of millions of SNPs in thousands of samples in a cost-effective manner. GWAS, by comparing the SNPs of millions of patients to SNPs from millions of unaffected controls, are used to identify genetic variants that are common in the population but modulate the risk of disease. Recently, GWAS and, subsequently, meta-analyses of GWAS, which pool samples from multiple studies to increase statistical power, have led to the identification of hundreds of mutations in so-called susceptibility genes, most with very small effect sizes (odds ratios usually <2; for review, see Visscher et al. 2012). Although the effect size of these variants is not large enough to inform disease prediction, these loci may shed light on novel genes and molecular pathways dysregulated in disease. However, it is important to keep in mind that GWAS use SNPs as proxies for tagging chromosomal regions. Thus, this type of analysis can determine that a specific SNP or genomic locus is associated with disease risk, but does not necessarily mean that a particular SNP functionally contributes to disease risk or which gene in a genomic locus may be associated with the pathology.
In the past few years, with the cost of sequencing dropping and bioinformatics technology advancing, next-generation sequencing (NGS) approaches are becoming more commonly used. Rather than limiting sequencing to a gene or a small region of the chromosome, it is now possible to sequence the entire genome (whole-genome sequencing, WGS) or only the gene-coding regions, the exons (whole-exome sequencing, WES) (Ng et al. 2009). WES, as a result of focusing on only ∼2% of the genome, is still considerably cheaper, and the analytical processing power, as well as the storage space required to handle the data produced, is more feasible to acquire for individual laboratories (Singleton 2011; Wang et al. 2013). Importantly, these NGS approaches allow for the direct identification of rare variants in disease, without prior linkage or GWAS. As compared to WES, WGS has the advantage of better coverage of the exome, owing in part to longer reads and more consistent read depth. Furthermore, WGS can be used to investigate regulatory variants, which are increasingly thought to play a significant role in the cause of and risk for neurodegenerative disease (Wang et al. 2013).
Despite these advances, much of the genetic contribution to the heritable liability of most sporadic forms of dementia has yet to be explained. Here, we review the recent genetic and genomic findings in studies of neurodegenerative dementias that involve abnormal protein aggregation and discuss the idea that these diseases may represent a continuum of disorders rather than individual diseases.
ALZHEIMER’S DISEASE
By far the most common form of age-related dementia, AD is clinically heterogeneous and slowly progressive, characterized by a gradual decline in memory and other cognitive functions (such as anomia, agnosia, and apraxia), depression, and apathy (www.alz.org/research/diagnostic_criteria). The brain regions most affected by neurodegeneration are the cortex, hippocampus, amygdala, basal forebrain, and brainstem (Rademakers and Rovelet-Lecrux 2009). Progressive atrophy in these regions is preceded by the accumulation of extracellular β-amyloid plaques formed by cleavage products of amyloid precursor protein (APP) and intracellular neurofibrillary tangles (NFTs) composed of hyperphosphorylated microtubule binding protein tau (MAPT). Plaques and tangles have been shown to interfere with calcium signaling and synaptic transmission, to induce a persistent inflammatory response, and to lead to synapse loss and ultimately neuronal degeneration. The presence of NFTs is strongly correlated with neuronal dysfunction and disease progression (for reviews of AD pathology, see Braak and Braak 1997, 1998; Serrano-Pozo et al. 2011).
AD is classified as early onset (before 65 years of age) versus late onset (after 65). Less than 10% of cases are early onset, and these usually follow an autosomal dominant inheritance pattern in which mutations in a single gene can cause the disease. Late-onset AD is much more common and far more genetically complex, possibly involving concurrent mutations in multiple genes and interactions among these susceptibility genes with each other, as well as unknown environmental factors. Although late-onset AD is largely idiopathic, there is still a significant genetic contribution to susceptibility, with twin studies predicting heritability of 60%–80% (Gatz et al. 2006).
Genes in Which Disease-Causing Mutations Have Been Identified
In early-onset AD, three different causal genes have been identified via linkage analysis (Table 1). The amyloid precursor protein gene (APP), located at chromosome 21q21, encodes the ubiquitously expressed transmembrane protein APP. APP is cleaved via the subsequent action of two proteases, β-secretase and γ-secretase, to release Aβ-peptides (Thinakaran and Koo 2008). Depending on the specific site of cleavage, peptides of either 40 (Aβ40) or 42 (Aβ42) amino acids are produced. Aβ42 has been shown to be more prone to aggregate into pathogenic amyloid plaques than Aβ40. More than 30 different mutations have been described in APP (www.molgen.vib-ua.be/ADMutations). Most of these mutations are heterozygous missense mutations in or near exons 16 and 17 (the site of proteolytic processing by the secretases), whole-gene duplications, rare recessive small deletions, and recessive missense mutations (reviewed in Guerreiro et al. 2012; Karch et al. 2014). These mutations most frequently result in either altered Aβ production, changes in the ratios of Aβ42 to Aβ40, and/or increased NFT formation. Interestingly, a protective variant (A676T) in APP has recently been identified for AD in the Icelandic population (Jonsson et al. 2012). This variant is located adjacent to the β-secretase cleavage site and has been shown to reduce β-secretase-1-mediated formation of Aβ peptides. This reduction in Aβ production is thought to modulate neurodegeneration in A676T carriers and is proposed to mediate the protective effect.
Table 1.
Alzheimer’s disease—known causal genes
| Gene | Protein | Chromosome | Mutation frequency in early onset | Types of mutations reported | Known function | Pathway |
|---|---|---|---|---|---|---|
| APP | Amyloid precursor protein | 21 | 15% | Missense, duplication, deletions | Neurite outgrowth, adhesion, and axonogenesis | APP processing |
| PSEN1 | Presenilin-1 | 14 | 80% | Missense, duplication, deletions | Component of γ-secretase complex; proteolytic cleavage of membrane proteins, including APP | APP processing |
| PSEN2 | Presenilin-2 | 1 | 5% | Missense | Component of γ-secretase complex; proteolytic cleavage of membrane proteins, including APP | APP processing |
The majority of mutations causing early-onset AD are found in the presenilin 1 (PSEN1) gene, located at chromosome 14q24.3, and in its homolog presenilin 2 (PSEN2), located at chromosome 1q31. More than 180 dominant, pathogenic mutations have been identified in PSEN1, whereas approximately 13 dominant, pathogenic mutations have been identified in PSEN2 (www.molgen.vib-ua.be/ADMutations). These mutations are distributed throughout the two proteins, but the mutations have a tendency to cluster in the transmembrane portions of both proteins. Interestingly, PSEN1 and PSEN2 encode structurally similar, integral membrane proteins that are essential components of the γ-secretase complex, which cleaves APP into Aβ peptides (Wilquet and De Strooper 2004). The finding that all three causative genes are involved in the production of Aβ peptides led to the “amyloid cascase hypothesis” (Hardy and Higgins 1992; reviewed in Karran et al. 2011). This theory states that changes in APP homeostasis or cleavage lead to aggregation of Aβ and its deposition in plaques, which is sufficient to initiate the cascade of neuropathological changes, including the aggregation of tau, and results in neuronal atrophy. Although familial, early-onset AD only accounts for a small number of total AD cases, identification of these genes highlights molecular pathways involved in disease and may therefore enable identification of potential therapeutic targets in both familial and idiopathic AD.
Risk Variants
The apolipoprotein E gene (APOE) was the first late-onset AD risk factor identified and, thus far, remains the risk factor carrying the greatest proportion of the population variance in liability to the disease (for review, see Ashford 2004). Located at chromosome 19q13.2, APOE encodes a multifunctional glycoprotein, involved in mobilization and redistribution of cholesterol (Mahley 1988). APOE has been shown to bind to Aβ and influence its metabolism, both in terms of clearance of soluble Aβ and Aβ aggregation (Kim et al. 2009; Castellano et al. 2011; Liu et al. 2013). There are three major alleles of APOE (ɛ2, ɛ3, and ɛ4) that result from a single amino-acid substitution between each pair of isoforms on the protein level. The presence of a single ɛ4 allele increases the risk for AD about threefold, whereas individuals homozygous for this allele have ∼15-fold increased risk compared to the most common genotype (ɛ3 homozygous) (Corder et al. 1993; Strittmatter et al. 1993). APOEɛ4 is usually thought of as a genetic risk factor, which is defined as making only a small contribution to disease and is neither necessary nor sufficient to cause disease. However, the very high-risk estimates for APOEɛ4 carriers have led some to assign APOEɛ4 the status of a “moderately penetrant gene with semidominant inheritance.” This status acknowledges that not all ɛ4 carriers develop the disease and that heterozygous ɛ4 carriers have intermediate risk compared with homozygous carriers (Genin et al. 2011). Interestingly, the APOEɛ2 allele appears to be protective and has been shown to decrease the risk of AD as well as to increase longevity (Corder et al. 1994).
Numerous SNP-based GWAS, sequence-based studies, and meta-analysis have recently uncovered hundreds of additional risk factors for AD with varying levels of support (Lambert et al. 2009, 2013; Harold et al. 2009; Seshadri et al. 2010; Naj et al. 2011; Hollingworth et al. 2011; Jonsson et al. 2013; Reitz et al. 2013; Cruchaga et al. 2013; Miyashita et al. 2013; Beecham et al. 2014; Chen et al. 2015). More than 20 of these novel common risk factors are now well established (Table 2) at the stringent level of genome-wide significance (p-value ≤ 5 × 108) in GWAS meta-analysis and have been replicated in independent studies. Most confer much smaller risk (odds ratio of ∼0.8–1.25) than APOEɛ4, and many of the specific pathogenic variants in or near these susceptibility genes remain to be characterized. More recently, as NGS technologies become available, a handful of low-frequency (rare) risk variants have been uncovered (Table 3), many of which have an odds ratio >2 (for review, see Lord et al. 2014). Together, these variants may be essential in suggesting additional, novel regulatory pathways contributing to disease pathogenesis, apart from Aβ metabolism and clearance. Areas of molecular dysfunction implicated by gene ontology analyses of these novel risk factors include the neural-immune system (CLU, CR1, ABCA7, EPHA1, CD33, INPP5D, TREM2, HLA complex), synaptic function (PICALM, BIN1, EPHA1, CD2AP, MEF2C, PTK2B, AKAP9), endocytosis (PICALM, BIN1, EPHA1, CD33, CD2AP, SORL1, RIN3), and lipid metabolism (APOE, CLU, ABCA7, PLD3) (for review, see Rosenthal and Kamboh 2014; Karch and Goate 2015). Many studies are actively underway to identify additional risk variants in AD, and with the rise of NGS techniques, these may soon include regulatory variants in noncoding regions, such as promoters, enhancers, and noncoding RNA.
Table 2.
Alzheimer's disease—known common susceptibility variants
| Gene | Protein | Chromosome | Pathogenic/protective | Known function | Pathway |
|---|---|---|---|---|---|
| APOE | Apolipoprotein E | 19 | APOEɛ2, protective; APOEɛ4, pathogenic | Lipoprotein metabolism and homeostasis | Lipid metabolism |
| CR1 | Complement component (3b/4b) receptor 1 | 1 | Pathogenic | Regulates complement activation | Immune response |
| BIN1 | Bridging integrator 1 | 2 | Pathogenic | Regulates endocytosis; interacts with clathrin and AP2; binds to lipid membranes and induces membrane curvature | Endocytosis; synaptic function |
| INPP5D | Inositol polyphosphate-5-phosphatase D | 2 | Pathogenic | Inflammatory response; regulation of cytokine signaling | Immune response |
| MEF2C | Myocyte enhancer factor 2C | 5 | Pathogenic | Transcription enhancer involved in synaptic plasticity | Synaptic function |
| CD2AP | CD2-associated protein | 6 | Pathogenic | Scaffolding molecule that regulates the actin cytoskeleton; required for synapse formation | Endocytosis; synaptic function |
| HLA-DRB1/HLA-DRB5 | Major histocompatibility complex class II subunits | 6 | Pathogenic | Member of the major histocompatibility complex | Immune response |
| NME8 | NME/NM23 family member 8 | 7 | Pathogenic | Ciliary functions | — |
| ZCWPW1 | Zinc finger, CW domain with PWWP domain 1 | 7 | Pathogenic | Contains domains identified in a number of other proteins responsible for epigenetic regulation | — |
| EPHA1 | EPH receptor A1 | 7 | Pathogenic | Member of tyrosine kinase receptor family involved in intercellular signaling, synapse formation and plasticity, axonal guidance | Immune response; endocytosis; synaptic function |
| PTK2B | Protein tyrosine kinase 2β | 8 | Pathogenic | Calcium-induced regulation of ion channels and activation of the map kinase signaling pathway; synaptic LTP | Synaptic signaling |
| CLU | Clusterin | 8 | Pathogenic | Secreted, stress-activated chaperone involved in apoptosis, lipid transport, and Aβ clearance | Lipid metabolism; immune response |
| CELF1 | CUGBP, Elav-like family member 1 | 11 | Pathogenic | Regulates splicing, mRNA editing, and translation | — |
| MS4A4A, MS4A6E | Membrane spanning four domains A4A / A6E | 11 | Pathogenic | Little known; potentially involved in inflammatory response | Immune response |
| PICALM | Phosphatidylinositol-binding clathrin assembly protein | 11 | Pathogenic | Clathrin-mediated endocytosis; synaptic vesicle fusion to the presynaptic membrane via VAMP2 trafficking | Endocytosis; synaptic function |
| SORL1 | Sortilin-related receptor | 11 | Pathogenic | Vesicle trafficking from the cell surface to the Golgi–endoplasmic reticulum | Endocytosis |
| FERMT2 | Fermitin family member 2 | 14 | Pathogenic | Actin assembly and cell shape | — |
| SLC24A/RIN3 | Ras and Rab interactor 3 | 14 | Pathogenic | Membrane budding and trafficking | Endocytosis |
| ABCA7 | ATP-binding cassette A7 | 19 | Pathogenic | Intra- and extracellular transmembrane transport | Lipid metabolism; immune response |
| CD33 | CD33 molecule | 19 | Pathogenic | Triggers immune cell–cell interactions via clathrin-independent endocytosis | Endocytosis; immune response |
| CASS4 | Cas scaffolding protein family member 4 | 20 | Pathogenic | Possibly involved in cell adhesion and cytoskeletal regulation | — |
Table 3.
Alzheimer's disease—known rare susceptibility variants
| Gene | Protein | Chromosome | Pathogenic/protective | Known function | Pathway |
|---|---|---|---|---|---|
| UNC5C | Unc-5 homolog C | 4 | T835M, pathogenic | Possible role in apoptosis in neurons | [Apoptosis] |
| TREM2 | Triggering receptor expressed on myeloid cells 2 | 6 | R47H/R62H, pathogenic | Regulation of phagocytosis; suppression of inflammation | Immune response |
| AKAP9 | A-kinase anchor protein 9 | 7 | Pathogenic | Scaffold protein attaching kinases to NMDA receptor | Synaptic function |
| ADAM10 | A disintegrin and metalloproteinase domain-containing protein 10 | 15 | Q170H/R181G, pathogenic | Membrane protein cleavage | APP processing |
| MAPT | Microtubule-binding protein tau | 17 | A152T, pathogenic | Assembly and stabilization of tubulin microtubules | Axonal transport; synaptic function |
| APOE | Apolipoprotein E | 19 | V236E, protective | Lipoprotein metabolism and homeostasis | Lipid metabolism |
| PLD3 | Phospholipase D family, member 3 | 19 | V232M, pathogenic | Hydrolysis of membrane phospholipids | Lipid metabolism |
| APP | Amyloid precursor protein | 21 | A637T, protective | Neurite outgrowth, adhesion, and axonogenesis | APP processing |
FRONTOTEMPORAL DEMENTIA
FTD can be divided into the following three major clinical subtypes: behavioral variant (bvFTD), semantic variant primary progressive aphasia, and nonfluent variant primary progressive aphasia (Sieben et al. 2012; Bang et al. 2015). bvFTD is the most common and is associated with frontal and anterior cortex atrophy, which leads to progressive personality changes, including disinhibition, apathy, loss of sympathy, and stereotyped behaviors. As the disease progresses cognitive deficits appear, but memory and visuospatial functions remain relatively spared (Riedl et al. 2014). FTD is a genetically and pathologically heterogenous disorder, typically with an earlier age of onset (between 50 and 65 years of age) and a higher incidence of familial cases than AD (Rademakers et al. 2012). Almost 50% of individuals with FTD have a positive family history, and an autosomal-dominant mode of transmission can be identified in ∼10%–20% of patients (Chow et al. 1999; Seelaar et al. 2008; Rohrer et al. 2009). However, many of the causative genes identified have variable penetrance, and most result in a spectrum of phenotypes, extending from amyotrophic lateral sclerosis (ALS) through classic FTD variants to corticobasal syndrome and progressive supranuclear palsy (PSP) (Ng et al. 2015).
Genes in Which Disease-Causing Mutations Have Been Identified
Mutations in six genes have been implicated in FTD (Table 4). Using linkage analysis, the first causal gene for FTD to be identified was MAPT (coding for microtubule associated protein tau). MAPT is located on chromosome 17q21.1, and since its initial identification as a causal gene for FTD (Hutton et al. 1998; Poorkaj et al. 1998; Spillantini et al. 1998), >40 different pathogenic mutations (mainly missense and splicing mutations) have been described (www.molgen.vib-ua.be/FTDMutations). These mutations can account for up to 50% of familial frontotemporal lobar dementia (FTLD) cases in certain populations (Morris et al. 2001; Sieben et al. 2012). MAPT promotes the assembly and modulates the stability of tubulin microtubules. Most of the mutations identified in MAPT are found in exons 9–13, which code for the microtubule binding domains that mediate protein function. These mutations can alter the expression ratio of different isoforms of tau (between those containing four microtubule binding domains [4R] and those that contain only three [3R]), as well as modulate tau phosphorylation (Spillantini and Goedert 2013). This leads to abnormal accumulation of MAPT, which results in the formation of pathological NFTs within neurons and glia and impairs neuronal axon transport, among other cellular functions (for review, see Rademakers et al. 2004; Brandt et al. 2005; Ghetti et al. 2015). The discovery that MAPT mutations cause dementia runs counter to the “amyloid cascade hypothesis,” which states that abnormalities in APP homeostasis are the most significant molecular event necessary to initiate the cascade of neuropathological changes that cause neurodegeneration (Hardy and Higgins 1992). The amyloid cascade hypothesis usually asserts that aggregation of tau is only a secondary event. Instead, these MAPT mutations show that abnormalities in tau are sufficient to induce atrophy and indicate that tau pathology may also play an important role in AD pathogenesis. Interestingly, a variant in MAPT, previously identified in PSP, was recently discovered in AD patients (Coppola et al. 2012), further eroding support for a dogmatic view of the “amyloid cascade hypothesis.”
Table 4.
Frontotemporal dementia—known causal genes
| Gene | Protein | Chromosome | Mutation frequency in early onset | Types of mutations reported | Known function | Pathway |
|---|---|---|---|---|---|---|
| MAPT | Microtubule binding protein tau | 17 | 0%–50% | Missense, deletions | Assembly and stabilization of tubulin microtubules | Axonal transport; synaptic function |
| GRN | Progranulin | 17 | 3%–26% | Missense, deletions, insertions | Multifunctional protein suggested to be both aneurotrophic and anti-inflammatory factor | Growth factor; anti-inflammatory |
| C9orf72 | Chromosome 9 open reading frame 72 | 9 | 25% | Hexanucleotide repeat in regulatory region | Unknown | — |
| VCP | Valosin-containing protein | 9 | <1% | Missense | Multifunctional ATPase | DNA damage response; authophagy and protein degradation |
| TARDBP | TAR DNA-binding protein [TDP-43] | 1 | <1% | Missense | Transcriptional regulator involved in RNA splicing and stability | Transcriptional regulation |
GRN, a disease-causing gene first identified in 2006 (Baker et al. 2006; Cruts et al. 2006), is located only 1.7 Mb away from MAPT on chromosome 17q21. Its proximity to MAPT originally made it difficult to identify through linkage mapping studies. GRN codes for progranulin, a cysteine-rich secreted glycoprotein, which is cleaved by enzymes such as elastase into small peptides (granulins) (Toh et al. 2011). The exact function of progranulin and granulins is still unknown in the central nervous system, but some hypothesize that these proteins have opposing cellular functions. Progranulin has been implicated in neurotrophic and anti-inflammatory pathways, as well as in modulation of Wnt signaling (Van Damme et al. 2008; Ryan et al. 2009; Laird et al. 2010; Rosen et al. 2011; for review, see Petkau and Leavitt 2014). A range of mutations, >65 of which are thought to be pathogenic, have been identified in GRN (Yu et al. 2010; www.molgen.vib-ua.be/FTDMutations). Mutations are found in almost all GRN exons (the exception being exon 13 located at the very 3′ end of the coding sequence) and include nonsense and splice-site mutations, as well as insertions and deletions leading to a frameshift in the transcript. The majority of these mutations result in the loss of function of the mutated allele, and it is now generally accepted that GRN mutations cause disease through haploinsufficiency (Kleinberger et al. 2013), which is supported by downregulation of progranulin in the blood of mutation carriers (Coppola et al. 2008). Aggregation of ubiquitin and TDP-43 are characteristic of all GRN mutation carriers (Eriksen and Mackenzie 2007).
GRN mutations account for up to 26% of familial FTD cases in some populations and 1%–5% of sporadic cases. However, much variability is observed in the clinical phenotype associated with GRN mutations, and penetrance is incomplete (reviewed in Cruts and Van Broeckhoven 2008; Ng et al. 2015). Therefore, genetic variability—both on the wild-type GRN allele and in other genes—is thought to contribute to disease pathogenesis in individuals with GRN mutations. One such factor is variance in the transmembrane protein 106B gene (TMEM106B), which has been shown to associate with progranulin in endolysosomes (Lang et al. 2012). Some genetic variants in or near TMEM106B appear to protect GRN mutation carriers from FTD or delay its onset, whereas others seem to increase the risk for FTD (Finch et al. 2011; van der Zee et al. 2011; Lattante et al. 2014). Both types of variants may possibly assert their effect by modulating the levels of progranulin. Furthermore, a number of miRNAs (specifically, miR29b and miR107) may regulate GRN expression and thereby contribute to disease penetrance and age of onset (Jiao et al. 2010; Wang et al. 2010).
In 2011, the abnormal expansion of a GGGGCC hexanucleotide repeat in the noncoding portion (first intron) of C9ORF72 was found to account for up to ∼25% of familial FTD cases and 40% of familial ALS cases (Renton et al. 2011; DeJesus-Hernandez et al. 2011; Majounie et al. 2012a). Although C9ORF72 codes for a protein of unknown function, structural comparison studies indicate that it is a potential DENN-type guanine nucleotide exchange factor, regulating Rab GTPases (Levine et al. 2013). Unaffected individuals carry less than 30 repeats, whereas affected mutation carriers have been identified with an excess of 4000 repeats. Recently, Gijselinck and colleagues (2015) showed that repeat size correlates with the disease age of onset as well as methylation of CpGs in the C9orf72 promoter in a cohort of 72 Belgian patients with FTD, FTD-ALS, or ALS. Furthermore, the researchers were able to provide some evidence for genetic anticipation, as several parent–child pairs showed decreased age of onset, increased expansion size, and/or increased promoter methylation with transmission to the next generation.
A variety of pathological mechanisms by which C9ORF72 repeat expansions result in neurodegeneration have been proposed. Haploinsufficiency, triggered by epigenetic changes to the expanded repeat region, has been suggested, and recent studies have shown reduced C9ORF72 protein levels in the frontal cortex of expanded repeat carriers (Waite et al. 2014). Furthermore, RNA foci have been identified in neurons of C9ORF72 expansion cases (Lee et al. 2013; Haeusler et al. 2014). As in other repeat-expansion disorders (such as fragile-X syndrome), C9ORF72 repeat-expansion transcripts may aggregate into foci, which in turn are thought to sequester RNA-binding proteins and result in major RNA-processing alterations (Wojciechowska and Krzyzosiak 2011; Cooper-Knock et al. 2015; Prudencio et al. 2015). However, no correlation between RNA focus burden and neurodegeneration has been reported for C9ORF72 expansion carriers thus far, and, in fact, some propose that RNA foci may be neutral intermediates or possibly even neuroprotective (Tran et al. 2015), calling this proposed mode of pathogenicity into question. Furthermore, unconventional, repeat-associated, non-ATG dependent translation (RAN translation) has been shown to cause expression of a variety of dipeptide mutant proteins from the expansion (Ash et al. 2013; Mori et al. 2013). Protein-labeling studies with antibodies designed against each of the six possible dipeptides show that these dipeptides cluster into insoluble inclusions in neurons in the cortex. Most recently, in large-scale genetic screens in yeast and Drosophila, several groups have identified that this unconventional translation of dipeptides may lead to nucleocytoplasmic transport defects and neurodegeneration (Freibaum et al. 2015; Jovičić et al. 2015; Zhang et al. 2015; reviewed in van Blitterswijk and Rademakers 2015). Finally, there is emerging evidence that like PGRN, C9ORF72 mutation carriers have elevated prevalence of specific classes of autoimmune disorders, further implicating inflammatory mechanisms in disease pathophysiology (Miller et al. 2016). Most likely, many if not all of these mechanisms contribute to neurodegeneration in C9ORF72 expansion carriers to some extent, possibly with synergistic effects.
Together, mutations in MAPT, GRN, and C9ORF72 repeat expansions are the most common causes of familial and sporadic FTD and usually result in bvFTD. However, clinical presentation is considerably variable; mutations in GRN alone have been reported among individuals diagnosed with FTD, AD, corticobasal degeneration, and mild cognitive impairments. Furthermore, the age of onset varies widely between carriers of these causative mutations, even within families (Ng et al. 2015).
Four additional genes have been implicated in FTD. Very rare, dominant-negative mutations have been identified in charged multivesicular body protein 2B (CHMP2B) (Skibinski et al. 2005), which is involved in the endosomal sorting complex, required for the formation of the multivesicular body, an early precursor to the lysosome (Tanikawa et al. 2012). The discovery of mutations in CHMP2B further underscores the importance of membrane dynamics and autophagy in neurodegenerative disease.
TARDBP, located on chromosome 1q36, codes for TAR DNA-binding protein (TDP-43), a ribonucleoprotein functioning as a transcriptional regulator involved in RNA splicing and stability (Janssens and Van Broeckhoven 2013). TDP-43 aggregates are found in the majority of tau-negative, ubiquitin-positive inclusions in FTD (Rademakers et al. 2012). Although missense and nonsense mutations in TARDBP have been identified in FTD cases, these are exceedingly rare (<1%) (Sieben et al. 2012). However, TARDBP is an important causal factor in the related motor neuron diseases without dementia—recent studies have found mutations in TARDBP in up to ∼3% of ALS patients (Kabashi et al. 2008; Rutherford et al. 2008; Sreedharan et al. 2008; Mentula et al. 2012; reviewed in Valdmanis et al. 2009).
FUS is located at chromosome 16q11 and codes for fused in sarcoma (FUS), which has been shown to co-localize with ubiquitin-immunoreactive inclusions in FTD patients (Rademakers et al. 2012). However, whereas mutations in FUS have been identified in both familial and sporadic ALS, none have been associated with FTD. When located in the nucleus, FUS is thought to be involved in regulating transcription and pre-mRNA splicing, whereas cytoplasmic FUS is implicated in mRNA transport and local protein synthesis at the synapse (Colombrita et al. 2012). The discovery of mutations in FUS and TARDBP, as well as the suggestion that C9ORF72 repeat expansions may lead to RNA-processing alterations, highlights the importance of RNA metabolism in both FTD and ALS.
VCP, located at chromosome 9q13, codes for valosin-containing protein, a highly abundant, multifunctional ATPase involved in a variety of cellular pathways (Yamanaka et al. 2012). Deficiency in VCP in neurons leads to mitochondrial uncoupling and significant reduction of cellular ATP production (Bartolome et al. 2013). Mutations in VCP cause a pleiotropic degenerative disorder called multisystem proteinopathy, which can manifest clinically as a variety of neurodegenerative diseases including classical ALS, FTD, inclusion body myopathy, Paget’s disease of bone, or as a combination of these disorders (Watts et al. 2004).
Common, non-Mendelian risk variants for FTLD remain to be explored for the most part. As described previously, variants in TMEM106B are thought to modulate levels of granulin secretion and may therefore influence GRN-mutation-carrier pathological phenotype (Lattante et al. 2014). Furthermore, Ferrari and colleagues (2014) recently identified two new FTD-associated risk variants in a GWAS with more than 3500 cases and more than 9400 controls (Ferrari et al. 2014). First, the HLA locus at 6p21.3, coding for key molecular components of the immune system, showed significant genome-wide association. Second, in a separate association analysis for each of the different subtypes of FTD, the RAB38/CTSC locus, involved in lysosomal processing (Bultema et al. 2012), was significantly associated with bvFTD cases specifically.
With the discovery of the hexanucleotide repeat expansion in C9ORF72, and mutations in TARDBP and VCP, which result in FTD, ALS, and concomitant FTD-ALS, it has become clear that cases of FTD-ALS are not the result of simple coincidence, but most likely represent a continuum of disease between classical FTD and motor neuron disease (Ng et al. 2015). This is further supported by the clustering of neurodegenerative diseases in relatives of patients with ALS (Al-Chalabi et al. 2012). As has been recently described, up to 50% of ALS cases show some functional loss in frontal lobe tests, whereas in ∼15% of cases, frontal lobe functional loss was enough to justify diagnosis of FTD. Similarly, ∼40% of FTD cases show measurable motor dysfunction, whereas up to 15% of FTD cases are eventually diagnosed with concomitant ALS (for review, see Callister and Pickering-Brown 2014; Ng et al. 2015). From the progress that has been made in our understanding of the molecular genetics of FTD and ALS over the last decade, it is evident that these diseases form part of a spectrum, with the same genes often implicated in both.
How mutations in the same genes and/or aggregation of their protein products cause these divergent phenotypes is a key question that we now have the genetic tools to answer. Using next-generation sequencing approaches, we can identify rare variants that contribute to a slew of what were previously thought of as separate dementias and motor neuron diseases. One example is the MAPT A152T variant, which was originally found in a patient with PSP but has been recently shown to be associated with FTD, AD, and LBD, as well as other atypical tauopathies (Coppola et al. 2012; Kara et al. 2012; Labbé et al. 2015). In the future, careful application of such techniques will be useful in mapping the genetic and molecular commonalities and differences of diseases on this spectrum.
LEWY BODY DEMENTIA
LBD is a complex neurodegenerative disease characterized by progressive cognitive decline, motor symptoms, visual hallucination, and fluctuating levels of attention and alertness (Molano 2013). It is considered to be a common cause of neurodegenerative dementia, with a prevalence of up to 7.5% (Vann Jones and O’Brien 2014). Being a member of the family of synucleinopathies, which also includes Parkinson’s disease (PD), a key feature of LBD pathology is interneuronal α-synuclein aggregation into Lewy bodies. However, most LBD-affected brains also show comorbid AD pathology in the form of β-amyloid plaques and NFTs (Colom-Cadena et al. 2013).
Although a number of families have been identified in which a mixed phenotype of dementia and parkinsonism is inherited in a Mendelian manner (Meeus et al. 2012), most LBD is late onset and sporadic, with twin studies not lending much support for a major genetic etiology (Wang et al. 2009). Linkage analysis in a large Belgian family with prominent dementia and parkinsonism showed significant genetic linkage to a novel locus on the long arm of chromosome 2. However, extensive sequence analysis of five candidate genes within this 2q35-q36 region did not reveal a single nucleotide or structural chromosomal variant (gene dosage) mutation that cosegregated with LBD (Bogaerts et al. 2007).
In cohorts of unrelated and largely sporadic LBD patients, molecular genetic investigations have revealed mutations in genes that have previously been implicated both with Parkinson’s disease (SNCA) (Yamaguchi et al. 2005) and with AD (APP and PSEN1) (Ishikawa et al. 2005; Guyant-Marechal et al. 2008), indicating that these disorders may be members of the same disease continuum. The most strongly implicated LBD-causative gene is SNCA, in which a number of rare mutations have been identified associated with LBD. SNCA, located at chromosome 4q21, codes for α-synuclein, a protein thought to be involved in neurotransmitter release and vesicle turnover at presynaptic terminals (Bendor et al. 2013). Of note, different mutations in SNCA appear to be loosely correlated with different clinical presentations (Bonifati 2008). Although duplications are more often associated with classic PD (Ibáñez et al. 2004; Chartier-Harlin et al. 2004), triplications result in PD with dementia and LBD (Singleton et al. 2003; Fuchs et al. 2007). Furthermore, the missense mutation A30P is rarely associated with dementia. Instead, patients with this mutation present with typical PD (Krüger et al. 2001). In contrast, the missense mutations E46K and A53T, which have been shown to promote α-synuclein polymerization into amyloid in vitro, are associated with a mixed presentation of PD, PD with dementia, and LBD (Zarranz et al. 2004; Yamaguchi et al. 2005). Although mutations in β-synuclein have not been associated with PD, two missense mutations (V70M and P123H) were identified in one LBD patient each (Ohtake et al. 2004). Further genetic studies are needed to verify pathogenicity.
Carriers of mutations in genes usually associated with AD, including APP, PSEN1, and PSEN2, often show Lewy body pathology as well. Increased expression of APP and point mutations near the β-secretase cleavage sites (V717I) can result in Lewy body formation (Meeus et al. 2012). However, the process by which these mutations cause Lewy body pathology in some patients but not others is still unresolved.
Recently, mutations in glucocerebrosidase (GBA) have been identified as potential risk variants for both PD and LBD. Goker-Alpan and colleagues found a higher incidence of heterozygous GBA variant carriers among patients with LBD than healthy controls (Goker-Alpan et al. 2006). This study was expanded on by Tsuang and colleagues, who screened the entire coding sequence of GBA in 79 pure LBD cases and almost 400 controls to show that LBD patients had an increased odds ratio of 7.6 (Tsuang et al. 2012). Interestingly, GBA is an important enzyme involved in lysosomal storage, which has been recently implicated in α-synuclein processing and homeostasis (Cullen et al. 2011). A second main risk factor for LBD is APOEɛ4. Investigations of clinically confirmed LBD cohorts show that APOEɛ4 is overrepresented in LBD cases compared with controls, and individuals with ɛ4 alleles have a threefold increased risk for LBD, which is similar to the increased risk associated with APOEɛ4 for AD (Tsuang et al. 2013). As most LBD-affected brains show comorbid AD pathology in the form of β-amyloid plaques, and APOEɛ4 is thought to modulate Aβ homeostasis and clearance, it is not altogether surprising that APOEɛ4 is also a prominent risk factor for LBD. However, these genes explain only a minority of cases, and identification of novel risk loci, including possible pathogenic regulatory variants, remains an important goal.
PRION DISEASES
Human prion diseases, a rare group of neurodegenerative disorders, are characterized by progressive cognitive decline (most commonly presenting as memory impairment and executive and/or language dysfunction), parkinsonism, and behavioral symptoms (Takada and Geschwind 2013). Prion diseases can be classified into three groups: sporadic (most common), genetic, and acquired (Brown and Mastrianni 2010). In all three forms, neurodegeneration is caused by the propagation of conformational remodeling of properly folded, cellular prion-related protein (PrPC; predominantly α-helical) into the aberrant, aggregating PrPSc (β-sheet enriched, in which the superscript “Sc” refers to “scrapie”). Importantly, it is the exposure of PrPC to PrPSc that induces its pathogenic conformational change. PrPSc can aggregate to form oligomers and subsequent intra- and extracellular depositions of amyloid fibrils. PrPC is a membrane-bound protein that is most likely involved in synapse formation during neuronal development. Whether loss of function of PrPC, either via conformational change or oligomerization, leads to neuronal impairment and cell death or whether PrPSc oligomers are neurotoxic is unclear.
PrP is encoded by the prion-related protein gene (PRNP) located at chromosome 20q13. Genetic prion disease is caused by dominantly inherited mutations in PRNP, which are found in ∼10% of all cases (Takada and Geschwind 2013). Interestingly, not all of these genetic prion disease cases have a positive family history of prion disease. This may be caused by a number of factors, including misdiagnosis of family members, late onset of disease symptoms, rare occurrence of de novo mutations in PRNP, and incomplete penetrance of some of the disease-causing mutations in PRNP (Kovács et al. 2005). More than 30 pathogenic mutations in PRNP have been identified, including missense mutations, insertions (specifically an octapeptide-repeat insertion), and deletions, though some of these mutations are very rare, and occurrence is restricted to specific geographic regions (Mastrianni 2010). Penetrance is high for most PRNP mutations but is strongly age-dependent, with disease onset usually between 40 and 60 years of age. Although specific clinical features are often associated with specific PRNP mutations, a single PRNP mutation can be associated with variable phenotypes even within a family (Kovács et al. 2005).
Polymorphisms in PRNP are also the most common risk factors identified in sporadic prion disease to date. Specifically, individuals who are heterozygous for PRNP codon 129 (which can code for either methionine or valine) have reduced risk for developing both sporadic and acquired prion disease, whereas homozygous individuals (either MM or VV) are overrepresented in prion disease affected populations (Parchi et al. 1999). Homozygosity at this codon can decrease the age of onset in genetic prion disease cases, and whether methionine or valine is present in the mutant PrP protein can influence clinical presentation (Mead et al. 2009a). Furthermore, two protective variants in PRNP have been described. The E219K variant has been reported in roughly 6% of the Japanese population but is absent in prion disease cases (Shibuya et al. 1998), whereas the G127V polymorphism has been identified to reduce the risk of developing kuru, a prion disease transmitted by cannibalism (Mead et al. 2009b). Although variants in genes other than PRNP are likely to contribute to disease risk, the low incidence of prion disease makes statistically significant identification of these risk factors of modest effect via SNP-based GWAS exceedingly difficult.
DISCUSSION
Independent Diseases or Spectrum Disorder?
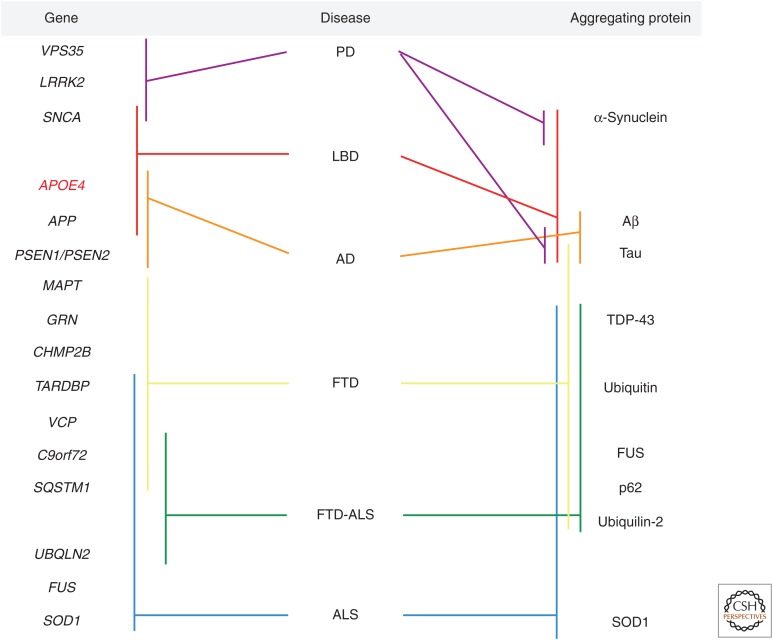
A number of the same genes and their pathologically aggregating protein products are involved in several clinically and pathologically distinct neurodegenerative disorders (Fig. 2). Mutations in SNCA and aggregation of its protein product α-synuclein are characteristic of both PD and LBD, whereas APP mutations and pathological aggregations of the resulting amyloid protein cleavage product Aβ are found in both LBD and AD. Although mutations in MAPT are commonly associated only with FTD, aggregation of hyperphosphorylated tau into NFTs is common to LBD, AD, and FTD. More recently, it has become clear that the hexanucleotide repeat expansion in C9ORF72 and mutations in TARDBP and VCP are common to FTD, ALS, and concomitant FTD-ALS (Callister and Pickering-Brown 2014).
Figure 2.
Neurodegenerative dementias as a spectrum disorder. Genes (left) and their pathologically aggregating protein products (right) and the neurodegenerative disorders (center) in which they have been implicated are shown.
Furthermore, a number of rare cases have been described in which a gene previously thought to be associated with only one specific form of dementia was found to be mutated in patients whose clinical phenotype and neuropathology matched a different neurodegenerative disorder. Examples of such cases include, but are not limited to, the discovery of MAPT mutations (R406T, A406W, and A152T) in AD (Rademakers et al. 2003; Coppola et al. 2012; Wojtas et al, 2012); PSEN1 G183V and M146V missense mutations in Pick’s disease (Dermaut et al. 2004) and FTD with amyloid plaques, NFTs, and Pick’s bodies (Riudavets et al. 2013), respectively; and a number of patients with C9ORF72 repeat expansions that presented with AD, PD, and LBD (Majounie et al. 2012b; Wojtas et al. 2012; Kohli et al. 2013; Robinson et al. 2014; for review, see Liu et al. 2014). Moreover, mutations in GRN have been identified in both AD and PD cases (Brouwers et al. 2007, 2008; Wojtas et al. 2012), and there is mounting evidence that GRN may act as a susceptibility gene for AD (Sheng et al. 2014).
As the clinical phenotype of these disorders is often highly variable, it is possible that some of these cases are examples of an initial misdiagnosis of the inherent condition. However, the overlap in clinical phenotype of these diseases and frequent comorbid presentation, along with the phenotypic variability associated with genetic variants described above, speaks to the fact that these neurodegenerative dementias may be better considered as part of a disease spectrum, ranging from PD to LBD, AD, FTD, and, on the far end, to ALS, than distinct disorders. Although neurodegeneration is typically targeted to distinct brain regions, which in turn affects clinical phenotypes, the underlying genetic variants, molecular pathology, and general cellular pathways involved in disease progression may show remarkable overlap throughout this disease continuum.
Prion-Like Behavior of Aggregation-Prone Pathogenic Proteins Underlying Neurodegenerative Dementias
It has been proposed recently that a common pathogenic mechanism, the prion-like behavior of aggregation-prone protein species, may underlie most neurodegenerative diseases (Prusiner 2012, 2013; Peggion et al. 2014). “Prion-like” refers to the capacity of an abnormally folded peptide or protein to induce the same abnormal conformation in a regularly folded protein of the same kind, thereby initiating a self-amplifying cascade. It is postulated that aging—and with it impairment of misfolded protein clearance mechanisms—is necessary as a pathogenic event. Only when misfolded prion-like proteins reach a certain threshold does self-propagation, which leads to neuronal dysfunction, occur. Some evidence that such abnormal conformational changes occur in proteins associated with neurodegenerative diseases and result in atrophy of affected tissue has been provided for Aβ (Stöhr et al. 2014; Watts et al. 2014; reviewed in Prusiner 2013), tau (Frost et al. 2009; Guo and Lee 2011; Holmes et al. 2014; Sanders et al. 2014), α-synuclein (Prusiner et al. 2015; reviewed in Oueslati et al. 2014), SOD-1 (Grad et al. 2015), and TDP-43 (Smethurst et al. 2015), but these findings are mainly restricted to mouse models thus far (for review, see Peggion et al. 2014).
However, this hypothesis is attractive, as it would explain the sporadic nature and late onset of most neurodegenerative diseases as well as the progressive spread of atrophy to different areas of the brain. Furthermore, different conformations of aggregating protein could act as different “strains” of the same disease, explaining different cellular pathology and clinical symptoms seen in diseases with the same underlying mutation or aggregating protein. Lastly, some disease-causing mutations and risk variants may exert their pathogenic effect by modulating specifically the “prion-like” properties of aggregation-prone proteins.
These findings suggest that it may be helpful to consider neurodegenerative dementias as a spectrum disorder with shared pathogenic mechanisms.
CONCLUSION
Over the last decade, a multitude of disease variants implicated in neurodegenerative dementias has been identified using linkage-based analysis, SNP-mediated GWAS techniques, and, more recently, next-generation sequencing approaches. Genetic screening for a number of these disease-causing mutations and risk variants is now commonly available. This not only will improve patient care but also may enable the identification of case and control cohorts for drug trials that aim to test the efficacy of new treatments before onset of widespread neuronal atrophy and associated clinical phenotypes. Furthermore, the function of proteins encoded by disease-causing and susceptibility genes and the molecular pathways these act as part of will inform and direct future investigations of the underlying cellular pathology. Forty-four million people currently suffer from neurodegenerative dementias, and the associated health-care costs for AD in the United States in 2013 alone exceeded $200 billion (alz.org), underscoring the severe need to develop currently lacking treatment options. Hopefully, in the future, a multidisciplinary approach that combines genetic, transcriptomic, proteomic, and epigenetic information will provide a more complete picture of underlying disease mechanisms and point the way toward improved therapeutic options.
ACKNOWLEDGMENTS
We thank the members of the Geschwind laboratory for their helpful discussions and critical reading of the manuscript.
Footnotes
Editor: Stanley B. Prusiner
Additional Perspectives on Prion Biology available at www.cshperspectives.org
REFERENCES
- Al-Chalabi A, Jones A, Troakes C, King A, Al-Sarraj S, van den Berg LH. 2012. The genetics and neuropathology of amyotrophic lateral sclerosis. Acta Neuropathol 124: 339–352. [DOI] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. 1999. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet 23: 185–188. [DOI] [PubMed] [Google Scholar]
- Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, DeJesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW III, Rademakers R, et al. 2013. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77: 639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashford JW. 2004. APOE genotype effects on Alzheimer’s disease onset and epidemiology. J Mol Neurosci 23: 157–165. [DOI] [PubMed] [Google Scholar]
- Baker M, Mackenzie IR, Pickering-Brown SM, Gass J, Rademakers R, Lindholm C, Snowden J, Adamson J, Sadovnick AD, Rollinson S, et al. 2006. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature 442: 916–919. [DOI] [PubMed] [Google Scholar]
- Bang J, Spina S, Miller BL. 2015. Frontotemporal dementia. Lancet 386: 1672–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolome F, Wu HC, Burchell VS, Preza E, Wray S, Mahoney CJ, Fox NC, Calvo A, Canosa A, Moglia C, et al. 2013. Pathogenic VCP mutations induce mitochondrial uncoupling and reduced ATP levels. Neuron 78: 57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beecham GW, Hamilton K, Naj AC, Martin ER, Huentelman M, Myers AJ, Corneveaux JJ, Hardy J, Vonsattel JP, Younkin SG, et al. 2014. Genome-wide association meta-analysis of neuropathologic features of Alzheimer’s disease and related dementias. PLoS Genet 10: e1004606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendor JT, Logan TP, Edwards RH. 2013. The function of α-synuclein. Neuron 79: 1044–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennion Callister J, Pickering-Brown SM. 2014. Pathogenesis/genetics of frontotemporal dementia and how it relates to ALS. Exp Neurol 262: 84–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogaerts V, Engelborghs S, Kumar-Singh S, Goossens D, Pickut B, van der Zee J, Sleegers K, Peeters K, Martin JJ, Del-Favero J, et al. 2007. A novel locus for dementia with Lewy bodies: A clinically and genetically heterogeneous disorder. Brain 130: 2277–2291. [DOI] [PubMed] [Google Scholar]
- Bonifati V. 2008. Recent advances in the genetics of dementia with lewy bodies. Curr Neurol Neurosci Rep 8: 187–189. [DOI] [PubMed] [Google Scholar]
- Braak H, Braak E. 1997. Diagnostic criteria for neuropathologic assessment of Alzheimer’s disease. Neurobiol Aging 18: S85–S88. [DOI] [PubMed] [Google Scholar]
- Braak H, Braak E. 1998. Evolution of neuronal changes in the course of Alzheimer’s disease. J Neural Trans Supplementum 53: 127–140. [DOI] [PubMed] [Google Scholar]
- Brandt R, Hundelt M, Shahani N. 2005. Tau alteration and neuronal degeneration in tauopathies: Mechanisms and models. Biochim Biophys Acta 1739: 331–354. [DOI] [PubMed] [Google Scholar]
- Brouwers N, Nuytemans K, van der Zee J, Gijselinck I, Engelborghs S, Theuns J, Kumar-Singh S, Pickut BA, Pals P, Dermaut B, et al. 2007. Alzheimer and Parkinson diagnoses in progranulin null mutation carriers in an extended founder family. Arch Neurol 64: 1436–1446. [DOI] [PubMed] [Google Scholar]
- Brouwers N, Sleegers K, Engelborghs S, Maurer-Stroh S, Gijselinck I, van der Zee J, Pickut BA, Van den Broeck M, Mattheijssens M, Peetrs K, et al. 2008. Genetic variability in progranulin contributes to risk for clinically diagnosed Alzheimer disease. Neurology 71: 656–664. [DOI] [PubMed] [Google Scholar]
- Brown K, Mastrianni JA. 2010. The prion diseases. J Geriatr Psychiatry Neurol 23: 277–298. [DOI] [PubMed] [Google Scholar]
- Bultema JJ, Ambrosio AL, Burek CL, Di Pietro SM. 2012. BLOC-2, AP-3, and AP-1 proteins function in concert with Rab38 and Rab32 proteins to mediate protein trafficking to lysosome-related organelles. J Biol Chem 287: 19550–19563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano JM, Kim J, Stewart FR, Jiang H, DeMattos RB, Patterson BW, Fagan AM, Morris JC, Mawuenyega KG, Cruchaga C, et al. 2011. Human apoE isoforms differentially regulate brain amyloid-β peptide clearance. Sci Transl Med 3: 89ra57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartier-Harlin MC, Kachergus J, Roumier C, Mouroux V, Douay X, Lincoln S, Levecque C, Larvor L, Andrieux J, Hulihan M, et al. 2004. α-Synuclein locus duplication as a cause of familial Parkinson’s disease. Lancet 364: 1167–1169. [DOI] [PubMed] [Google Scholar]
- Chen JA, Wang Q, Davis-Turak J, Li Y, Karydas AM, Hsu SC, Sears RL, Chatzopoulou D, Huang AY, Wojta KJ, et al. 2015. A multiancestral genome-wide exome array study of Alzheimer disease, frontotemporal dementia, and progressive supranuclear palsy. JAMA Neurol 72: 414–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow TW, Miller BL, Hayashi VN, Geschwind DH. 1999. Inheritance of frontotemporal dementia. Arch Neurol 7: 817–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombrita C, Onesto E, Megiorni F, Pizzuti A, Baralle FE, Buratti E, Silani V, Ratti A. 2012. TDP-43 and FUS RNA-binding proteins bind distinct sets of cytoplasmic messenger RNAs and differently regulate their post-transcriptional fate in motoneuron-like cells. J Biol Chem 287: 15635–15647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colom-Cadena M, Gelpi E, Charif S, Belbin O, Blesa R, Martí MJ, Clarimón J, Lleó A. 2013. Confluence of α-synuclein, tau, and β-amyloid pathologies in dementia with Lewy bodies. J Neuropathol Exp Neurol 72: 1203–1212. [DOI] [PubMed] [Google Scholar]
- Cooper-Knock J, Bury JJ, Heath PR, Wyles M, Higginbottom A, Gelsthorpe C, Highley JR, Hautbergue G, Rattray M, Kirby J, et al. 2015. C9ORF72 GGGGCC expanded repeats produce splicing dysregulation which correlates with disease severity in amyotrophic lateral sclerosis. PLoS ONE 10: e0127376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppola G, Karydas A, Rademakers R, Wang Q, Baker M, Hutton M, Miller BL, Geschwind DH. 2008. Gene expression study on peripheral blood identifies progranulin mutations. Ann Neurol 64: 92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppola G, Chinnathambi S, Lee JJ, Dombroski BA, Baker MC, Soto-Ortolaza AI, Lee SE, Klein E, Huang AY, Sears R, et al. 2012. Evidence for a role of the rare p.A152T variant in MAPT in increasing the risk for FTD-spectrum and Alzheimer’s diseases. Hum Mol Genet 21: 3500–3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. 1993. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science 261: 921–923. [DOI] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Risch NJ, Strittmatter WJ, Schmechel DE, Gaskell PC Jr, RImmler JB, Locke PA, Conneally PM, Schmader KE, et al. 1994. Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat Genet 7: 180–184. [DOI] [PubMed] [Google Scholar]
- Cruchaga C, Kauwe JSK, Harari O, Jin SC, Cai Y, Karch CM, Benitez BA, Jeng AT, Skorupa T, Carrell D, et al. 2013. GWAS of cerebrospinal fluid tau levels identifies risk variants for Alzheimer’s disease. Neuron 78: 256–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruts M, Van Broeckhoven C. 2008. Loss of progranulin function in frontotemporal lobar degeneration. Trends Genet 4: 186–194. [DOI] [PubMed] [Google Scholar]
- Cruts M, Gijselinck I, van der Zee J, Engelborghs S, Wils H, Pirici D, Rademakers R, Vandenberghe R, Dermaut B, Martin JJ, et al. 2006. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature 442: 920–924. [DOI] [PubMed] [Google Scholar]
- Cullen V, Sardi SP, Ng J, Xu YH, Sun Y, Tomlinson JJ, Kolodziej P, Kahn I, Saftig P, Woulfe J, et al. 2011. Acid β-glucosidase mutants linked to Gaucher disease, Parkinson disease, and Lewy body dementia alter α-synuclein processing. Ann Neurol 69: 940–953. [DOI] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, et al. 2011. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72: 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermaut B, Kumar-Singh S, Engelborghs S, Theuns J, Rademakers R, Saerens J, Pickut BA, Peeters K, van den Broeck M, Vennekens K, et al. 2004. A novel presenilin 1 mutation associated with Pick’s disease but not β-amyloid plaques. Ann Neurol 55: 617–626. [DOI] [PubMed] [Google Scholar]
- Eriksen JL, Mackenzie IRA. 2007. Progranulin: Normal function and role in neurodegeneration. J Neurochem 104: 287–297. [DOI] [PubMed] [Google Scholar]
- Feder JN, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy DA, Basava A, Dormishian F, Domingo R Jr, Ellis MC, Fullan A, et al. 1996. A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet 13: 399–408. [DOI] [PubMed] [Google Scholar]
- Ferrari R, Hernandez DG, Nalls MA, Rohrer JD, Ramasamy A, Kwok JBJ, Dobson-Stone C, Brooks WS, Schofield PR, Halliday GM, et al. 2014. Frontotemporal dementia and its subtypes: A genome-wide association study. Lancet Neurol 13: 686–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch N, Carrasquillo MM, Baker M, Rutherford NJ, Coppola G, Dejesus-Hernandez M, Crook R, Hunter T, Ghidoni R, Benussi L, et al. 2011. TMEM106B regulates progranulin levels and the penetrance of FTLD in GRN mutation carriers. Neurology 76: 467–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freibaum BD, Lu Y, Lopez-Gonzalez R, Kim NC, Almeida S, Lee KH, Badders N, Valentine M, Miller BL, Wong PC, et al. 2015. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature 525: 129–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost B, Jacks RL, Diamond MI. 2009. Propagation of tau misfolding from the outside to the inside of a cell. J Biol Chem 284: 12845–12852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs J, Nilsson C, Kachergus J, Munz M, Larsson EM, Schüle B, Langston JW, Middleton FA, Ross OA, Hulihan M, et al. 2007. Phenotypic variation in a large Swedish pedigree due to SNCA duplication and triplication. Neurology 68: 916–922. [DOI] [PubMed] [Google Scholar]
- Gatz M, Mortimer JA, Fratiglioni L, Johansson B, Berg S, Reynolds CA, Pedersen NL. 2006. Potentially modifiable risk factors for dementia in identical twins. Alzheimer's Dement 2: 110–117. [DOI] [PubMed] [Google Scholar]
- Genin E, Hannequin D, Wallon D, Sleegers K, Hiltunen M, Combarros O, Bullido MJ, Engelborghs S, De Deyn P, Berr C, et al. 2011. APOE and Alzheimer disease: A major gene with semi-dominant inheritance. Mol Psychiatry 16: 903–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghetti B, Oblak AL, Boeve BF, Johnson KA, Dickerson BC, Goedert M. 2015. Invited review: Frontotemporal dementia caused by microtubule-associated protein tau gene (MAPT) mutations: A chameleon for neuropathology and neuroimaging. Neuropathol Appl Neurobiol 41: 24–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gijselinck I, Van Mossevelde S, van der Zee J, Sieben A, Engelborghs S, De Bleecker J, Ivanoiu A, Deryck O, Edbauer D, Zhang M, et al. 2015. The C9orf72 repeat size correlates with onset age of disease, DNA methylation and transcriptional downregulation of the promoter. Mol Psychiatry 21: 1112–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert M. 2015. Neurodegeneration. Alzheimer’s and Parkinson’s diseases: The prion concept in relation to assembled Aβ, tau, and α-synuclein. Science 349: 1255555. [DOI] [PubMed] [Google Scholar]
- Goker-Alpan O, Giasson BI, Eblan MJ, Nguyen J, Hurtig HI, Lee VM, Trojanowski JQ, Sidransky E. 2006. Glucocerebrosidase mutations are an important risk factor for Lewy body disorders. Neurology 67: 908–910. [DOI] [PubMed] [Google Scholar]
- Grad LI, Fernando SM, Cashman NR. 2015. From molecule to molecule and cell to cell: Prion-like mechanisms in amyotrophic lateral sclerosis. Neurobiol Dis 77: 257–265. [DOI] [PubMed] [Google Scholar]
- Guerreiro RJ, Gustafson DR, Hardy J. 2012. The genetic architecture of Alzheimer’s disease: Beyond APP, PSENs and APOE. Neurobiol Aging 33: 437–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JL, Lee VM. 2011. Seeding of normal tau by pathological tau conformers drives pathogenesis of Alzheimer-like tangles. J Biol Chem 286: 15317–15331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyant-Marechal I, Berger E, Laquerrière A, Rovelet-Lecrux A, Viennet G, Frebourg T, Rumbach L, Campion D, Hannequin D. 2008. Intrafamilial diversity of phenotype associated with app duplication. Neurology 71: 1925–1926. [DOI] [PubMed] [Google Scholar]
- Haeusler AR, Donnelly CJ, Periz G, Simko EAJ, Shaw PG, Kim MS, Maragakis NJ, Troncoso JC, Pandey A, Sattler R, et al. 2014. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature 507: 195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy JA, Higgins GA. 1992. Alzheimer’s disease: The amyloid cascade hypothesis. Science 256: 184–185. [DOI] [PubMed] [Google Scholar]
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, et al. 2009. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet 41: 1088–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hästbacka J, de la Chapelle A, Mahtani MM, Clines G, Reeve-Daly MP, Daly M, Hamilton BA, Kusumi K, Trivedi B, Weaver A, et al. 1994. The diastrophic dysplasia gene encodes a novel sulfate transporter: Positional cloning by fine-structure linkage disequilibrium mapping. Cell 78: 1073–1087. [DOI] [PubMed] [Google Scholar]
- Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, Abraham R, Hamshere ML, Pahwa JS, Moskvina V, et al. 2011. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet 43: 429–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes BB, Furman JL, Mahan TE, Yamasaki TR, Mirbaha H, Eades WC, Belaygorod L, Cairns NJ, Holtzman DM, Diamond MI. 2014. Proteopathic tau seeding predicts tauopathy in vivo. Proc Natl Acad Sci 111: E4376–E4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H, Pickering-Brown S, Chakraverty S, Isaacs A, Grover A, et al. 1998. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature 393: 702–705. [DOI] [PubMed] [Google Scholar]
- Ibáñez P, Bonnet AM, Débarges B, Lohmann E, Tison F, Pollak P, Agid Y, Dürr A, Brice A. 2004. Causal relation between α-synuclein gene duplication and familial Parkinson’s disease. Lancet 364: 1169–1171. [DOI] [PubMed] [Google Scholar]
- Ishikawa A, Piao YS, Miyashita A, Kuwano R, Onodera O, Ohtake H, Suzuki M, Nishizawa M, Takahashi H. 2005. A mutant PSEN1 causes dementia with Lewy bodies and variant Alzheimer’s disease. Ann Neurol 57: 429–434. [DOI] [PubMed] [Google Scholar]
- Janssens J, Van Broeckhoven C. 2013. Pathological mechanisms underlying TDP-43 driven neurodegeneration in FTLD-ALS spectrum disorders. Hum Mol Genet 22: R77–R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao J, Herl LD, Farese RV, Gao FB. 2010. MicroRNA-29b regulates the expression level of human progranulin, a secreted glycoprotein implicated in frontotemporal dementia. PLoS ONE 5: e10551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson T, Atwal JK, Steinberg S, Snaedal J, Jonsson PV, Bjornsson S, Stefansson H, Sulem P, Gudbjartsson D, Maloney J, et al. 2012. A mutation in APP protects against Alzheimer’s disease and age-related cognitive decline. Nature 488: 96–99. [DOI] [PubMed] [Google Scholar]
- Jonsson T, Stefansson H, Steinberg S, Jonsdottir I, Jonsson PV, Snaedal J, Bjornsson S, Huttenlocher J, Levey AI, Lah JJ, et al. 2013. Variant of TREM2 associated with the risk of Alzheimer’s disease. N Engl J Med 368: 107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovičić A, Mertens J, Boeynaems S, Bogaert E, Chai N, Yamada SB, Paul JW III, Sun S, Herdy JR, Bieri G, et al. 2015. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat Neurosci 18: 1226–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashi E, Valdmanis PN, Dion P, Spiegelman D, McConkey BJ, Vande Velde C, Bouchard JP, Lacomblez L, Pochigaeva K, Salachas F, et al. 2008. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet 40: 572–574. [DOI] [PubMed] [Google Scholar]
- Kara E, Ling H, Pittman AM, Shaw K, de Silva R, Simone R, Holton JL, Warren JD, Rohrer JD, Xiromerisiou G, et al. 2012. The MAPT p.A152T variant is a risk factor associated with tauopathies with atypical clinical and neuropathological features. Neurobiol Aging 33: 2231.e7–2231.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karch CM, Goate AM. 2015. Alzheimer’s disease risk genes and mechanisms of disease pathogenesis. Biol Psychiatry 77: 43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karch CM, Cruchaga C, Goate AM. 2014. Alzheimer’s disease genetics: From the bench to the clinic. Neuron 83: 11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karran E, Mercken M, De Strooper B. 2011. The amyloid cascade hypothesis for Alzheimer’s disease: An appraisal for the development of therapeutics. Nat Rev Drug Discov 10: 698–712. [DOI] [PubMed] [Google Scholar]
- Kerem B, Rommens JM, Buchanan JA, Markiewicz D, Cox TK, Chakravarti A, Buchwald M, Tsui LC. 1989. Identification of the cystic fibrosis gene: Genetic analysis. Science 245: 1073–1080. [DOI] [PubMed] [Google Scholar]
- Kim J, Basak JM, Holtzman DM. 2009. The role of apolipoprotein E in Alzheimer’s disease. Neuron 63: 287–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinberger G, Capell A, Haass C, Van Broeckhoven C. 2013. Mechanisms of granulin deficiency: Lessons from cellular and animal models. Mol Neurobiol 47: 337–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli MA, John-Williams K, Rajbhandary R, Naj A, Whitehead P, Hamilton K, Carney RM, Wright C, Crocco E, Gwirtzman HE, et al. 2013. Repeat expansions in the C9ORF72 gene contribute to Alzheimer’s disease in Caucasians. Neurobiol Aging 34: 1519.e5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovács GG, Puopolo M, Ladogana A, Pocchiari M, Budka H, van Duijn C, Collins SJ, Boyd A, Giulivi A, Coulthart M, et al. 2005. Genetic prion disease: The EUROCJD experience. Hum Genet 118: 166–174. [DOI] [PubMed] [Google Scholar]
- Krüger R, Kuhn W, Leenders KL, Sprengelmeyer R, Müller T, Woitalla D, Portman AT, Maguire RP, Veenma L, Schröder U, et al. 2001. Familial parkinsonism with synuclein pathology: Clinical and PET studies of A30P mutation carriers. Neurology 56: 1355–1362. [DOI] [PubMed] [Google Scholar]
- Labbé C, Ogaki K, Lorenzo-Betancor O, Soto-Ortolaza AI, Walton RL, Rayaprolu S, Fujioka S, Murray ME, Heckman MG, Puschmann A, et al. 2015. Role for the microtubule-associated protein tau variant p.A152T in risk of α-synucleinopathies. Neurology 85: 1680–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird AS, Van Hoecke A, De Muynck L, Timmers M, Van den Bosch L, Van Damme P, Robberecht W. 2010. Progranulin is neurotrophic in vivo and protects against a mutant TDP-43 induced axonopathy. PLoS ONE 5: e13368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, et al. 2009. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet 41: 1094–1099. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, DeStafano AL, Bis JC, Beecham GW, Grenier-Boley B, et al. 2013. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet 45: 1452–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang CM, Fellerer K, Schwenk BM, Kuhn PH, Kremmer E, Edbauer D, Capell A, Haass C. 2012. Membrane orientation and subcellular localization of transmembrane protein 106B (TMEM106B), a major risk factor for frontotemporal lobar degeneration. J Biol Chem 287: 19355–19365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lattante S, Le Ber I, Galimberti D, Serpente M, Rivaud-Péchoux S, Camuzat A, Clot F, Fenoglio C; French research network on FTD and FTD-ALS, Scarpini E, et al. 2014. Defining the association of TMEM106B variants among frontotemporal lobar degeneration patients with GRN mutations and C9orf72 repeat expansions. Neurobiol Aging 35: 2658.e1–2658.e5. [DOI] [PubMed] [Google Scholar]
- Lee YB, Chen HJ, Peres JN, Gomez-Deza J, Attig J, Stalekar M, Troakes C, Nishimura AL, Scotter EL, Vance C, et al. 2013. Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep 5: 1178–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine TP, Daniels RD, Gatta AT, Wong LH, Hayes MJ. 2013. The product of C9orf72, a gene strongly implicated in neurodegeneration, is structurally related to DENN Rab-GEFs. Bioinformatics 29: 499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CC, Liu CC, Kanekiyo T, Xu H, Bu G. 2013. Apolipoprotein E and Alzheimer disease: Risk, mechanisms and therapy. Nat Rev Neurol 9: 106–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Yu JT, Zong Y, Zhou J, Tan L. 2014. C9ORF72 mutations in neurodegenerative diseases. Mol Neurobiol 49: 386–398. [DOI] [PubMed] [Google Scholar]
- Lord J, Lu AJ, Cruchaga C. 2014. Identification of rare variants in Alzheimer’s disease. Front Genet 5: 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald ME, Ambrose CM, Duyao MP, Myers RH, Lin C, Srinidhi L, et al. 1993. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 72: 971–983. [DOI] [PubMed] [Google Scholar]
- Mahley RW. 1988. Apolipoprotein E: Cholesterol transport protein with expanding role in cell biology. Science 240: 622–630. [DOI] [PubMed] [Google Scholar]
- Majounie E, Abramzon Y, Renton AE, Perry R, Bassett SS, Pletnikova O, Troncoso JC, Hardy J, Singleton AB, Traynor BJ. 2012a. Repeat expansion in C9ORF72 in Alzheimer’s disease. N Engl J Med 366: 283–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majounie E, Renton AE, Mok K, Dopper EG, Waite A, Rollinson S, Chiò A, Restagno G, Nicolaou N, Simon-Sanchez J, et al. 2012b. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: A cross-sectional study. Lancet Neurol 11: 323–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrianni JA. 2010. The genetics of prion diseases. Gen Med 12: 187–195. [DOI] [PubMed] [Google Scholar]
- Mead S, Poulter M, Uphill J, Beck J, Whitfield J, Webb TE, Campbell T, Adamson G, Deriziotis P, Tabrizi SJ, et al. 2009a. Genetic risk factors for variant Creutzfeldt–Jakob disease: A genome-wide association study. Lancet Neurol 8: 57–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead S, Whitfield J, Poulter M, Shah P, Uphill J, Campbell T, Al-Dujaily H, Hummerich H, Beck J, Mein CA, et al. 2009b. A novel protective prion protein variant that colocalizes with kuru exposure. N Engl J Med 361: 2056–2065. [DOI] [PubMed] [Google Scholar]
- Meeus B, Theuns J, Van Broeckhoven C. 2012. The genetics of dementia with Lewy bodies: What are we missing? Arch Neurol 69: 1113–1118. [DOI] [PubMed] [Google Scholar]
- Mentula HK, Tuovinen L, Penttilä S, Suominen T, Udd B, Palmio J. 2012. TARDBP mutations are not a frequent cause of ALS in Finnish patients. Acta Myologica 31: 134–138. [PMC free article] [PubMed] [Google Scholar]
- Miller ZA, Sturm VE, Camsari GB, Karydas A, Yokoyama JS, Grinberg LT, Boxer AL, Rosen HJ, Rankin KP, Gorno-Tempini ML, et al. 2016. Increased prevalence of autoimmune disease within C9 and FTD/MND cohorts: Completing the picture. Neurol Neuroimmunol Neuroinflamm 6: 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita A, Koike A, Jun G, Wang LS, Takahashi S, Matsubara E, Kawarabayashi T, Shoji M, Tomita N, Arai H, et al. 2013. SORL1 is genetically associated with late-onset Alzheimer’s disease in Japanese, Koreans and Caucasians. PLoS ONE 8: e58618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molano JRV. 2013. Dementia with Lewy bodies. Semin Neurol 33: 330–335. [DOI] [PubMed] [Google Scholar]
- Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C, et al. 2013. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 339: 1335–1338. [DOI] [PubMed] [Google Scholar]
- Morris HR, Khan MN, Janssen JC, Brown JM, Perez-Tur J, Baker M, Ozansoy M, Hardy J, Hutton M, Wood NW, et al. 2001. The genetic and pathological classification of familial frontotemporal dementia. Arch Neurol 58: 1813–1816. [DOI] [PubMed] [Google Scholar]
- Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK, et al. 2011. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet 43: 436–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, Shaffer T, Wong M, Bhattacharjee A, Eichler EE, et al. 2009. Targeted capture and massively parallel sequencing of 12 human exomes. Nature 461: 272–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng ASL, Rademakers R, Miller BL. 2015. Frontotemporal dementia: A bridge between dementia and neuromuscular disease. Ann NY Acad Sci 1338: 71–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtake H, Limprasert P, Fan Y, Onodera O, Kakita A, Takahashi H, Bonner LT, Tsuang DW, Murray IV, Lee VM, et al. 2004. β-synuclein gene alterations in dementia with Lewy bodies. Neurology 63: 805–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oueslati A, Ximerakis M, Vekrellis K. 2014. Protein transmission, seeding and degradation: Key steps for α-synuclein prion-like propagation. Exp Neurobiol 23: 324–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parchi P, Giese A, Capellari S, Brown P, Schulz-Schaeffer W, Windl O, Zerr I, Budka H, Kopp N, Piccardo P, et al. 1999. Classification of sporadic Creutzfeldt–Jakob disease based on molecular and phenotypic analysis of 300 subjects. Ann Neurol 46: 224–233. [PubMed] [Google Scholar]
- Peggion C, Sorgato MC, Bertoli A. 2014. Prions and prion-like pathogens in neurodegenerative disorders. Pathogens 3: 149–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkau TL, Leavitt BR. 2014. Progranulin in neurodegenerative disease. Trends Neurosci 37: 388–398. [DOI] [PubMed] [Google Scholar]
- Poorkaj P, Bird TD, Wijsman E, Nemens E, Garruto RM, Anderson L, Andreadis A, Wiederholt WC, Raskind M, Schellenberg GD. 1998. Tau is a candidate gene for chromosome 17 frontotemporal dementia. Ann Neurol 43: 815–825. [DOI] [PubMed] [Google Scholar]
- Prince M, Albanese E, Guerchet M, Prina M. 2014. World Alzheimer report 2014. Dementia and risk reduction: An analysis of protective and modifiable factors. Alzheimer's Disease International, London: https://www.alz.co.uk/research/WorldAlzheimerReport2014.pdf. [Google Scholar]
- Prudencio M, Belzil VV, Batra R, Ross CA, Gendron TF, Pregent LJ, Murray ME, Overstreet KK, Piazza-Johnston AE, Desaro P, et al. 2015. Distinct brain transcriptome profiles in C9orf72-associated and sporadic ALS. Nat Neurosci 18: 1175–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB. 2012. Cell biology. A unifying role for prions in neurodegenerative diseases. Science 336: 1511–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB. 2013. Biology and genetics of prions causing neurodegeneration. Ann Rev Genet 47: 601–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB, Woerman AL, Mordes DA, Watts JC, Rampersaud R, Berry DB, Patel S, Oehler A, Lowe JK, Kravitz SN, et al. 2015. Evidence for -synuclein prions causing multiple systems atrophy in humans with parkinsonism. Prot Natl Acad Sci 38: E5308–E5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademakers R, Rovelet-Lecrux A. 2009. Recent insights into the molecular genetics of dementia. Trends Neurosci 32: 451–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademakers R, Dermaut B, Peeters K, Cruts M, Heutink P, Goate A, van Broeckhoven C. 2003. Tau (MAPT) mutation Arg406Trp presenting clinically with Alzheimer disease does not share a common founder in Western Europe. Hum Mutat 22: 409–411. [DOI] [PubMed] [Google Scholar]
- Rademakers R, Cruts M, van Broeckhoven C. 2004. The role of tau (MAPT) in frontotemporal dementia and related tauopathies. Hum Mutat 24: 277–295. [DOI] [PubMed] [Google Scholar]
- Rademakers R, Neumann M, Mackenzie IR. 2012. Advances in understanding the molecular basis of frontotemporal dementia. Nat Rev Neurol 8: 423–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitz C, Jun G, Naj A, Rajbhandary R, Vardarajan BN, Wang LS, Valladares O, Lin CF, Larson EB, Graff-Radford NR, et al. 2013. Variants in the ATP-binding cassette transporter (ABCA7), apolipoprotein E ɛ4,and the risk of late-onset Alzheimer disease in African Americans. JAMA 309: 1483–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simón-Sánchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, et al. (2011). A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72: 257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl L, Mackenzie IR, Förstl H, Kurz A, Diehl-Schmid J. 2014. Frontotemporal lobar degeneration: Current perspectives. Neuropsychiatr Dis Treat 10: 297–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riudavets MA, Bartoloni L, Troncoso JC, Pletnikova O, St George-Hyslop P, Schultz M, Sevlever G, Allegri RF. 2013. Familial dementia with frontotemporal features associated with M146V presenilin-1 mutation. Brain Pathol 23: 595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson A, Davidson Y, Snowden JS, Mann DMA. 2014. C9ORF72 in dementia with Lewy bodies. J Neurol Neurosurg Psychiatry 85: 1435–1436. [DOI] [PubMed] [Google Scholar]
- Rohrer JD, Guerreiro R, Vandrovcova J, Uphill J, Reiman D, Beck J, Isaacs AM, Authier A, Ferrari R, Fox NC, et al. 2009. The heritability and genetics of frontotemporal lobar degeneration. Neurology 73: 1451–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen EY, Wexler EM, Versano R, Coppola G, Gao F, Winden KD, Oldham MC, Martens LH, Zhou P, Farese RV Jr, et al. 2011. Functional genomic analyses identify pathways dysregulated by progranulin deficiency, implicating Wnt signaling. Neuron 71: 1030–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal SL, Kamboh MI. 2014. Late-onset Alzheimer’s disease genes and the potentially implicated pathways. Curr Genet Med Rep 2: 85–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford NJ, Zhang YJ, Baker M, Gass JM, Finch NA, Xu YF, Stewart H, Kelley BJ, Kuntz K, Crook RJ, et al. 2008. Novel mutations in TARDBP (TDP-43) in patients with familial amyotrophic lateral sclerosis. PLoS Genet 4: e1000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan CL, Baranowski DC, Chitramuthu BP, Malik S, Li Z, Cao M, Minotti S, Durham HD, Kay DG, Shaw CA, et al. 2009. Progranulin is expressed within motor neurons and promotes neuronal cell survival. BMC Neurosci 10: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders DW, Kaufman SK, DeVos SL, Sharma AM, Mirbaha H, Li A, Barker SJ, Foley AC, Thorpe JR, et al. 2014. Distinct tau prion strains propagate in cells and mice and define different tauopathies. Neuron 82: 1271–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelaar H, Kamphorst W, Rosso SM, Azmani A, Masdjedi R, de Koning I, Maat-Kievit JA, Anar B, Donker Kaat L, Breedveld GJ, et al. 2008. Distinct genetic forms of frontotemporal dementia. Neurology 71: 1220–1226. [DOI] [PubMed] [Google Scholar]
- Serrano-Pozo A, Frosch MP, Masliah E, Hyman BT. 2011. Neuropathological alterations in Alzheimer disease. Cold Spring Harb Perspect Med. 1: a006189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, Bis JC, Smith AV, Carassquillo MM, Lambert JC, et al. 2010. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA 303: 1832–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng J, Su L, Xu Z, Chen G. 2014. Progranulin polymorphism rs5848 is associated with increased risk of Alzheimer’s disease. Gene 542: 141–145 [DOI] [PubMed] [Google Scholar]
- Shibuya S, Higuchi J, Shin RW, Tateishi J, Kitamoto T. 1998. Protective prion protein polymorphisms against sporadic Creutzfeldt–Jakob disease. Lancet 351: 419. [DOI] [PubMed] [Google Scholar]
- Sieben A, Van Langenhove T, Engelborghs S, Martin JJ, Boon P, Cras P, De Deyn PP, Santens P, Van Broeckhoven C, Cruts M. 2012. The genetics and neuropathology of frontotemporal lobar degeneration. Acta Neuropathol 124: 353–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton AB. 2011. Exome sequencing: A transformative technology. Lancet Neurol 10: 942–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, Hulihan M, Peuralinna T, Dutra A, Nussbaum R, et al. 2003. α-Synuclein locus triplication causes Parkinson’s disease. Science 302: 841. [DOI] [PubMed] [Google Scholar]
- Skibinski G, Parkinson NJ, Brown JM, Chakrabarti L, Lloyd SL, Hummerich H, Nielsen JE, Hodges JR, Spillantini MG, Thusgaard T, et al. 2005. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat Genet 37: 806–808. [DOI] [PubMed] [Google Scholar]
- Smethurst P, Sidle KCL, Hardy J. 2015. Review: Prion-like mechanisms of transactive response DNA binding protein of 43 kDa (TDP-43) in amyotrophic lateral sclerosis (ALS). Neuropathol Appl Neurobiol 41: 578–597. [DOI] [PubMed] [Google Scholar]
- Spillantini MG, Goedert M. 2013. Tau pathology and neurodegeneration. Lancet Neurol 12: 609–622. [DOI] [PubMed] [Google Scholar]
- Spillantini MG, Murrell JR, Goedert M, Farlow MR, Klug A, Ghetti B. 1998. Mutation in the tau gene in familial multiple system tauopathy with presenile dementia. Proc Natl Acad Sci 95: 7737–7741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, Ackerley S, Durnall JC, Williams KL, Buratti E, et al. 2008. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 319: 1668–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöhr J, Condello C, Watts JC, Bloch L, Oehler A, Nick M, DeArmond SJ, Giles K, DeGrado WF, Prusiner SB. 2014. Distinct synthetic Aβ prion strains producing different amyloid deposits in bigenic mice. Proc Natl Acad Sci 111: 10329–10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strittmatter WJ, Weisgraber KH, Huang DY, Dong LM, Salvesen GS, Pericak-Vance M, Schmechel D, Saunders AM, Goldgaber D, Roses AD. 1993. Binding of human apolipoprotein E to synthetic amyloid β peptide: Isoform-specific effects and implications for late-onset Alzheimer disease. Proc Natl Acad Sci 90: 8098–8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada LT, Geschwind MD. 2013. Prion diseases. Semin Neurol 33: 348–356. [DOI] [PubMed] [Google Scholar]
- Tanikawa S, Mori F, Tanji K, Kakita A, Takahashi H, Wakabayashi K. 2012. Endosomal sorting related protein CHMP2B is localized in Lewy bodies and glial cytoplasmic inclusions in α-synucleinopathy. Neurosci Lett 527: 16–21. [DOI] [PubMed] [Google Scholar]
- Thinakaran G, Koo EH. 2008. Amyloid precursor protein trafficking, processing, and function. J Biol Chem 283: 29615–29619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh H, Chitramuthu BP, Bennett HPJ, Bateman A. 2011. Structure, function, and mechanism of progranulin; the brain and beyond. J Mol Neurosci 45: 538–548. [DOI] [PubMed] [Google Scholar]
- Tran H, Almeida S, Moore J, Gendron TF, Chalasani U, Lu Y, Du X, Nickerson JA, Petrucelli L, Weng Z, et al. 2015. Differential toxicity of nuclear RNA foci versus dipeptide repeat proteins in a Drosophila model of C9ORF72 FTD/ALS. Neuron 87: 1207–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuang D, Leverenz JB, Lopez OL, Hamilton RL, Bennett DA, Schneider JA, Buchman AS, Larson EB, Crane PK, Kaye JA, et al. 2012. GBA mutations increase risk for Lewy body disease with and without Alzheimer disease pathology. Neurology 79: 1944–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuang D, Leverenz JB, Lopez OL, Hamilton RL, Bennett DA, Schneider JA, Buchman AS, Larson EB, Crane PK, Kaye JA, et al. 2013. APOE ɛ4 increases risk for dementia in pure synucleinopathies. JAMA Neurol 70: 223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdmanis PN, Daoud H, Dion PA, Rouleau GA. 2009. Recent advances in the genetics of amyotrophic lateral sclerosis. Curr Neurol Neurosci Rep 9: 198–205. [DOI] [PubMed] [Google Scholar]
- van Blitterswijk M, Rademakers R. 2015. Neurodegenerative disease: C9orf72 repeats compromise nucleocytoplasmic transport. Nat Rev Neurol 11: 670–672. [DOI] [PubMed] [Google Scholar]
- Van Damme P, Van Hoecke A, Lambrechts D, Vanacker P, Bogaert E, van Swieten J, Carmeliet P, Van Den Bosch L, Robberecht W. 2008. Progranulin functions as a neurotrophic factor to regulate neurite outgrowth and enhance neuronal survival. J Cell Biol 181: 37–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Zee J, Van Langenhove T, Kleinberger G, Sleegers K, Engelborghs S, Vandenberghe R, Santens P, Van den Broeck M, Joris G, Brys J, et al. 2011. TMEM106B is associated with frontotemporal lobar degeneration in a clinically diagnosed patient cohort. Brain 134: 808–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vann Jones SA, O’Brien JT. 2014. The prevalence and incidence of dementia with Lewy bodies: A systematic review of population and clinical studies. Psychol Med 44: 673–683. [DOI] [PubMed] [Google Scholar]
- Visscher PM, Brown MA, McCarthy MI, Yang J. 2012. Five years of GWAS discovery. Am J Hum Genet 90: 7–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waite AJ, Bäumer D, East S, Neal J, Morris HR, Ansorge O, Blake DJ. 2014. Reduced C9orf72 protein levels in frontal cortex of amyotrophic lateral sclerosis and frontotemporal degeneration brain with the C9ORF72 hexanucleotide repeat expansion. Neurobiol Aging 35: 1779.e5–1779.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CSM, Burke JR, Steffens DC, Hulette CM, Breitner JCS, Plassman BL. 2009. Twin pairs discordant for neuropathologically confirmed Lewy body dementia. J Neurol Neurosurg Psychiatry 80: 562–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WX, Wilfred BR, Madathil SK, Tang G, Hu Y, Dimayuga J, Stromberg AJ, Huang Q, Saatman KE, Nelson PT. 2010. miR-107 regulates granulin/progranulin with implications for traumatic brain injury and neurodegenerative disease. Am J Pathol 177: 334–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Liu X, Yang BZ, Gelernter J. 2013. The role and challenges of exome sequencing in studies of human diseases. Front Genet 4: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts GD, Wymer J, Kovach MJ, Mehta SG, Mumm S, Darvish D, Pestronk A, Whyte MP, Kimonis VE. 2004. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet 36: 377–381. [DOI] [PubMed] [Google Scholar]
- Watts JC, Condello C, Stöhr J, Oehler A, Lee J, DeArmond SJ, Lannfelt L, Ingelsson M, Giles K, Prusiner SB. 2014. Serial propagation of distinct strains of Aβ prions from Alzheimer’s disease patients. Proc Natl Acad Sci 111: 10323–10328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilquet V, De Strooper B. 2004. Amyloid-β precursor protein processing in neurodegeneration. Curr Opin Neurobiol 14: 582–588. [DOI] [PubMed] [Google Scholar]
- Wojciechowska M, Krzyzosiak WJ. 2011. Cellular toxicity of expanded RNA repeats: Focus on RNA foci. Hum Mol Genet 20: 3811–3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtas A, Heggeli KA, Finch N, Baker M, DeJesus-Hernandez M, Younkin SG, Dickson DW, Graff-Radford NR, Rademakers R. 2012. C9ORF72 repeat expansions and other FTD gene mutations in a clinical AD patient series from Mayo Clinic. Am J Neurodegener Dis 1: 107–118. [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K, Cochran EJ, Murrell JR, Polymeropoulos MH, Shannon KM, Crowther RA, Goedert M, Ghetti B. 2005. Abundant neuritic inclusions and microvacuolar changes in a case of diffuse Lewy body disease with the A53T mutation in the α-synuclein gene. Acta Neuropathol 110: 298–305. [DOI] [PubMed] [Google Scholar]
- Yamanaka K, Sasagawa Y, Ogura T. 2012. Recent advances in pp97/VCP/Cdc48 cellular functions. Biochim Biophys Acta 1823: 130–137. [DOI] [PubMed] [Google Scholar]
- Yu CE, Bird TD, Bekris LM, Montine TJ, Leverenz JB, Steinbart E, Galloway NM, Feldman H, Woltjer R, Miller CA, et al. 2010. The spectrum of mutations in progranulin: A collaborative study screening 545 cases of neurodegeneration. Arch Neurol 67: 161–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarranz JJ, Alegre J, Gómez-Esteban JC, Lezcano E, Ros R, Ampuero I, Vidal L, Hoenicka J, Rodriguez O, Atarés B, et al. 2004. The new mutation, E46K, of α-synuclein causes Parkinson and Lewy body dementia. Ann Neurol 55: 164–173. [DOI] [PubMed] [Google Scholar]
- Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P, Daley EL, Miller SJ, Cunningham KM, Vidensky S, et al. 2015. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature 525: 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]