Abstract
Design automation refers to a category of software tools for designing systems that work together in a workflow for designing, building, testing, and analyzing systems with a target behavior. In synthetic biology, these tools are called bio-design automation (BDA) tools. In this review, we discuss the BDA tools areas—specify, design, build, test, and learn—and introduce the existing software tools designed to solve problems in these areas. We then detail the functionality of some of these tools and show how they can be used together to create the desired behavior of two types of modern synthetic genetic regulatory networks.
Engineering biological systems is a complex undertaking and requires computational approaches. Bio-design automation tools in five areas—specification, design, building, testing, and learning—will accelerate progress.
Synthetic biology as a discipline has concerned itself with “forward engineering” living systems. This is a purposefully grand goal and is faced with numerous scientific, engineering, political, and ethical challenges. These engineering challenges include the storage of biological information, the design of complex, interacting biological systems using that information, the physical creation of these systems, and the dissemination of foundational principles as abstractions on which the next technical advances can be made. What should be clear to even the most casual observer is that computers, and the computational capabilities they bring with them, are going to be required if the field is going to succeed going forward. Debates remain regarding the framing of the computational approaches (Andrianantoandro et al. 2006; Lux et al. 2012; Densmore and Bhatia 2014), but there are few who debate the important and increasingly prominent role of computation in the field.
Synthetic biology and its associated promise have attracted a wide variety of participants. In particular, computer engineers and computer scientists began developing computational tools for engineering these genetic systems using techniques from their disciplines. Electronic design automation (EDA) showed a clear process by which design formalisms could be built, software tools created, and a larger, separate industry developed (see latticeautomation.com; teselagen.com; deskgen.com; benchling.com). This pursuit in the synthetic biology field has recently been coined bio-design automation (BDA) (Densmore 2012). BDA often uses a “divide and conquer” approach for solving small parts of a larger problem one piece at a time and allows for specialists to tackle specific narrow problems in which they have considerable expertise. After all necessary pieces are defined and solved, solutions for each subproblem can be automated, connected, and reused to solve larger problems. The central conjecture of this approach to synthetic biology is that the application of these core engineering concepts will accelerate synthetic biology workflows, increase abstraction and reuse, and greatly scale the size and complexity of systems that can be successfully created.
A key contemporary challenge in BDA is that there are few standards and little documentation for repeatedly engineering these systems. In response, multiple efforts have begun working toward community standards (Endy 2005; Arkin 2008; Canton et al. 2008; Kelly et al. 2009; Smolke 2009; Galdzicki et al. 2011) for data models and data exchange. This effort has recently gained more momentum (Galdzicki et al. 2014) and seen support and involvement from larger government organizations (Hayden 2015). Currently, some of the core requirements for representing sequence information have been discussed at length and interest has shifted to establishing standards for additional related subproblems.
The current BDA landscape can be divided into five main areas: specification, design, build, test, and learn. “Specification” concerns a precise, formal definition of the desired function and design of a target genetic system. “Design” concerns the set of decisions needed to determine the DNA constructs to be used and any needed modification to those constructions. “Build” refers to the processes and decisions involved in creating a plan for composing the DNA constructs from their elements using specific cloning methodologies and physically implementing a DNA-assembly plan (e.g., many approaches integrate robotics). “Test” concerns the design and physical implementation of experiments for characterizing engineered systems and the accompanying analysis and interpretation of acquired data. “Learn” includes machine-learning approaches to allow for the automated revision of designs based on processed experimental outcomes.
This review provides a survey of BDA approaches in these areas; it reflects the investigators’ experience in the field since 2007 and attempts to complement other similar surveys (Kahl and Endy 2013) by both updating these offerings to reflect the state-of-the-art in 2016 as well as providing examples of some core workflows.
SPECIFICATION AND DESIGN OF GENETIC SYSTEMS
The first step in engineering a genetic system is to specify its intended structure and/or function. Such a specification may take many different forms, ranging from a mathematical description of system behavior to a rule-based description of system architecture. Once a specification has been made, it must be mapped to a set of genetic parts and/or larger devices with defined genetic sequences that can conceivably satisfy its requirements. This mapping process constitutes the problem of genetic design, which can be solved via many alternate approaches. In this section, we will discuss BDA tools for specification and design and classify them as supporting a rational or combinatorial approach to genetic design. We will also highlight the use of several exemplar tools to specify and design a genetic AND gate and dual-feedback oscillator similar to those originally designed by Nielsen et al. (2016) and Stricker et al. (2008), respectively.
Rational Specification and Design
In general, rational approaches to genetic design are those in which design decisions are based on conclusions drawn from a biophysical or biochemical model. For example, a designer might decide to use one genetic part over another based on its impact on gene expression as predicted by a model. Roughly, two-thirds of all the specification and design tools listed in Table 1 can be classified as supporting a rational approach to genetic design. The specification tools belonging to this classification can be further divided into two non-mutually exclusive categories: languages and tools for directly building biophysical/biochemical models of genetic systems (Antimony [Smith et al. 2009], iBioSim [Myers et al. 2009], and ProMot [Mirschel et al. 2009]) and languages and tools for composing less-detailed descriptions of system behavior and/or architecture that are compiled into more detailed models (Cello [Nielsen et al. 2016], GEC [Pedersen and Phillips 2009], GenoCAD [Czar et al. 2009], iBioSim [Myers et al. 2009], Kera [Dhar 2010], the Proto Biocompiler [Beal et al. 2011], and TinkerCell [Chandran et al. 2009]). Specification tools belonging to the first class tend to give the user greater control over the form of model built with them, whereas specification tools belonging to the second class tend to remove this control from the user to automate specification and reduce the expert knowledge required during this process.
Table 1.
A comprehensive list of software tools available for solving bio-design automation tasks
| Realm | Tool name | Description |
|---|---|---|
| Specification | Eugene (Oberortner and Densmore 2014) | A rule-based specification language, which is able to incorporate abstraction layers and automatically generate combinatorial biological devices from components (eugenecad.org) |
| Pigeon (Bhatia and Densmore 2013) | Translates textual representations of nucleic acids into standardized visual representations (pigeoncad.org) | |
| GEC (Pedersen and Phillips 2009) | A language for modular specification of proteins and genes and their interactions; such specifications can then be compiled into standard biological sequences (biology.azurewebsites.net/gec) | |
| Phoenix | A tool that guides users through an automated, iterative design–build–test–learn cycle; with an ability to learn from empirical data, it can complete the closed-loop automation | |
| Proto/Biocompiler (Beal et al. 2011) | Generates optimized genetic regulatory network designs from specifications written in a high-level programming language (synbiotools.bbn.com) | |
| GSL (Wilson et al. 2016) | A language that facilitates design of large and complex DNA constructs used to engineer genomes; it also incorporates a set of low-level DNA manipulation primitives | |
| Kera (Dhar 2010) | An object-oriented programming language for synthetic biology, which is tempered by the biopart rule library, Samhita; Kera captures the knowledge regarding the interaction of genome components and catalytic molecules | |
| Antimony (Smith et al. 2009) | Text-based modular human-readable/writable model definition language with capabilities of easy translation into SBML | |
| ProMot (Mirschel et al. 2009) | A tool for construction and manipulation of biological dynamic models based on differential-algebraic equations ProMot is matched with the simulation environment Diana (www2.mpi-magdeburg.mpg.de/projects/promot) | |
| Molecula Maxima | Contains a programming IDE called Cytostudio for a synthetic biology programming language called Synthetic; Synthetic is a bio-hardware description language (moleculamaxima.com) | |
| MoSeC (Misirli et al. 2011) | Java application capable of generating DNA sequences. MoSeC has the ability to generate FASTA, GenBank, EMBL, and SBOL formats from SBML and CellML models (ico2s.org/software/mosec.html) | |
| Asmparts (Rodrigo et al. 2007) | A deprecated tool for modular specification of biological devices; Asmparts takes advantage of standardization by considering a model for each part in the design | |
| Design | Cello (Nielsen et al. 2016) | Compiles Verilog code specifying a logic circuit to a DNA sequence, with prediction of the design correctness; outputs are Eugene and SBOL compatible (cellocad.org) |
| Merlin (Quintin et al. 2016) | Genomic reprogramming tool exploiting Multiplex Automated Genome Engineering (MAGE), resulting in simultaneous multiregion genome modification (merlincad.org) | |
| Double Dutch (Roehner et al. 2016a ) | Web application for designing libraries of biosynthetic pathway variants based on design of experiments (DOE) matrices (clothocad.org/doubleDutch) | |
| RBS Calculator (Salis et al. 2009; Borujeni et al. 2013) | Allows for predicting and controlling translation initiation and protein expression in bacteria; can also be used for optimizing synthetic RBS sequences to achieve a targeted translation initiation rate (denovodna.com/software) | |
| GenoCAD (Czar et al. 2009) | A gene design and simulation tool with its own repository of biological parts; it incorporates various grammars/libraries and supports GenBank format (genocad.com) | |
| TeselaGen | A cloud-based SBOL compliant solution for sequence editing, combinatorial gene design, and assembly automation (teselagen.com) | |
| BioJADE (Goler 2004) | A java-based design and simulation tool for synthetic biological systems with interactive use of BioBrick repositories (web.mit.edu/jagoler/www/biojade) | |
| DeviceEditor | Maps the DNA sequence and feature annotations underlying each part to a graphic icon; icons can then be drag-and-dropped about a visual design canvas (j5.jbei.org/bin/deviceeditor.pl) | |
| SBROME (Huynh et al. 2013) | Given genetic circuit topology and dynamics, designs circuit with parts that minimize cross talk and incompatibility (tagkopouloslab.ucdavis.edu/software.html) | |
| Salis Lab calculators (Borujeni and Salis 2016) | A number of mathematical tools for synthetic biology design process (salislab.net/software) | |
| DeskGen | A genome-editing platform that combines the use of a CRISPR/Cas9 tool with the convenience of a genome browser (deskgen.com) | |
| Genome Compiler | A software platform incorporating advanced design tools, a variety of plasmids and parts repositories, direct connections to an array of DNA synthesis providers and laboratory automation services (genomecompiler.com) | |
| MatchMaker (Yaman et al. 2012) | Automates three steps: feature matching for regulatory relationship satisfaction, signal matching for expression level compatibility, and part matching to find physical DNA sequence in a database | |
| Mfold (Zuker 2003) | Analysis of DNA/RNA folding as well as free energy determination and structure display (unafold.rna.albany.edu) | |
| Rosetta (Leaver-Fay et al. 2011) | Computes the expected structure of natural proteins and design of new proteins (boinc.bakerlab.org/rosetta) | |
| GLAMM (Bates et al. 2011) | Provides maps for metabolic pathways, biosynthesis of secondary metabolites, and macrobial metabolism (glamm.lbl.gov) | |
| OptCircuit (Dasika and Maranas 2008) | An optimization-based framework that automatically identifies the circuit components from a list and connectivity that brings about the desired functionality. | |
| AutoBioCAD (Rodrigo and Jaramillo 2013) | Fully automated computational design of regulatory circuits in Escherichia coli; is able to compile the design into a nucleotide sequence | |
| Parts & Pools (Marchisio 2014) | A framework for modular design of bacterial synthetic genetic circuits from standard biological parts and pools of molecules as signal carriers | |
| Build/assembly | Puppeteer (Vasilev et al. 2011) | Liquid-handling automation tool capable of handling various protocols and laboratory resources; output is Common Human Robot Instruction Set (CHRIS); a compiler then translates CHRIS into a human/machine executable language |
| Raven (Appleton et al. 2014a) | DNA assembly planning tool for high throughput cloning; supports modern assembly methods such as BioBrick, MoClo, and Gibson Assembly; can output computer-readable or human-readable assembly instructions for liquid-handling automation (ravencad.org) | |
| DNALD (Blakes et al. 2014) | DNA assembly planning tool for binary assembly; improves on algorithm performance of prior binary algorithms (dnald.org/planner/index.html) | |
| Fluigi (Huang and Densmore 2014) | Automates the design of microfluidic devices used in synthetic biology by optimizing their layout; specifications are in the text-based MINT language | |
| 3DuF | Visual CAD tool for the design of microfluidic devices; outputs STL for CNC milling and SVG for lithography, as well as a textual JSON representation (cidarlab.github.io/3DuF) | |
| j5 (Hillson et al. 2011) | Web-based DNA assembly automation tool with ability to design assemblies and oligonucleotides for SLIC/Gibson/CPEC/SLiCE and Golden Gate; also performs cost analysis for choices of full synthesis, PCR/SOE, or oligo embedding (j5.jbei.org/bin/j5_entry_form.pl) | |
| PR-PR (Linshiz et al. 2014) | A cross-platform high-level biologist-friendly standard language used for management of liquid-handling robotics and microfluidic automated platforms. | |
| Aquarium | An assembly design tool for generating scalable, reproducible, and transferable molecular biology workflows (aquarium.bio) | |
| Primer3 (Koressaar and Remm 2007; Untergasser et al. 2012) | A tool used to design and analyze primers for PCR reactions with the ability to select primers for sequencing reactions and hybridization probes (bioinfo.ut.ee/primer3) | |
| Test/analysis | TASBE tools | A variety of synthetic biology automation software tool for tasks from high-level specification to part assignment and assembly (synthetic-biology.bbn.com/tasbe.html) |
| Bioconductor (Huber et al. 2015) | Analysis and comprehension tool that uses the open-source statistical language, R; AMI and Docker images are available (bioconductor.org) | |
| FlowCal (Castillo-Hair et al. 2016) | A library for reading, analyzing, and calibrating flow cytometry data; it accepts FCS files as input and is compatible with different calibration particles, fluorescent probes, and cell types | |
| FlowJo | A software application with an integrated environment for viewing and analyzing flow cytometry data; the user interface (UI) is presented as the workspace of experimental data, statistics, gates, as well as tabular and graphical layouts (flowjo.com) | |
| Data | ICE (Ham et al. 2012) | A registry platform for robust data storage for DNA components, integrated tools for part characterization, and secure access and information sharing (public-registry.jbei.org) |
| SBOL Stack (Madsen et al. 2016) | An RDF-based repository of SBOL represented synthetic biology parts and devices with an ability to execute search queries (sbolstack.org) | |
| GenBank (Bilofsky and Christian 1988) | A genetic sequence database containing a complete repository of public DNA sequences with corresponding annotations; GenBank also has a widely used file format for representing such sequences (ncbi.nlm.nih.gov/genbank) | |
| Registry of Standard Biological Parts | The iGEM repository including parts that are used in BioBrick assembly; this repository has more than 20,000 documented parts (parts.igem.org) | |
| Clotho (Densmore et al. 2009) | A database for storing synthetic biological parts data as well as a framework for applications to engineer them (clothocad.org) | |
| Owl (Appleton et al. 2014b) | A tool for creating datasheets for biological entities with an ability to perform data analytics services (cidar.bu.edu/owl) | |
| Phagebook | A Clotho 3.0 app that serves as a social network that also integrates laboratory inventory management; publication announcements are also supported | |
| Viz-a-Brick | A search and navigation tool for the iGEM registry of standard biological parts with quick and easy access to global and local relationships between parts in the registry (gcat.davidson.edu/vizabrick) | |
| BioModels database (Chelliah et al. 2013) | A reference repository hosting mathematical models that describe the dynamic interactions of biological components at various scales; most model components are cross-linked with external resources to facilitate interoperability (ebi.ac.uk/biomodels-main) | |
| Simulation | iBioSim (Myers et al. 2009) | A simulation and design tool focused on genetic circuits specified in SBML or SBOL format; has the ability to analyze metabolic networks and cell-signaling pathways, as well as modeling and visualization of multicellular and spatial models (async.ece.utah.edu/iBioSim) |
| COPASI (Hoops et al. 2006) | Simulation and analysis tool for biochemical networks and their dynamics; accepts models in SBML format and analyzes them using ODE as well as stochastic algorithms (copasi.org) | |
| SynBioSS (Hill et al. 2008) | A suite of software for the modeling and simulation of synthetic genetic constructs; it uses the registry of standard biological parts, a database of kinetic parameters, and both graphical and command-line interfaces to multiscale simulation algorithms (synbioss.sourceforge.net) | |
| TinkerCell (Chandran et al. 2009) | Incorporates a diagram that is detailed enough so that it can be mapped to models or experimental results; various mathematical analyses can then be run on those models (tinkercell.com) | |
| Gro (Jang et al. 2012) | A language for programming, modeling, specifying, and simulating the behavior of cells in growing microcolonies of microorganisms; Gro can simulate cell growth, division, intrinsic and extrinsic noise, as well as diffusing molecular signals (depts.washington.edu/soslab/gro) | |
| Morpheus (Starruß et al. 2014) | A modeling environment for simulating cell-based models with ordinary differential equations and reaction–diffusion systems; multiscale biological models can be defined in biological terms and mathematical expressions—it supports model construction, simulation, visualization, and batch processing (imc.zih.tu-dresden.de/wiki/morpheus/doku.php) | |
| PySCeS (Olivier et al. 2004) | Provides a variety of tools for the analysis of cellular systems; these include a human readable model description language, a structural, and a bifurcation analysis module—PySCeS supports SBML and SED-ML (pysces.sourceforge.net) | |
| CellDesigner (Funahashi et al. 2008) | A structured diagram editor for drawing gene-regulatory and biochemical networks, with integrated SBML ODE solver, SBML simulation core, and COPASI (celldesigner.org) | |
| Jarnac (Sauro 2000) | A Systems Biology Workbench (SBW) reaction simulator that includes both deterministic and stochastic time course simulation, steady-state analysis, basic structural properties of networks, and more (sbw.kgi.edu/software/jarnac.htm) | |
| JDesigner | An SBW network design tool with ability to export SBML file format; JDesigner is capable of connecting to Jarnac to perform simulation tasks (sbw.kgi.edu/software/jdesigner.htm) | |
| Mathematica | Incorporates an exclusive solution for synthetic biology; it is capable of performing the complete workflow from data import to analysis | |
| RoVerGeNe | A tool for analysis of dynamical properties regarding genetic regulatory networks; RoVerGeNe is capable of robustness analysis and parameter constraint synthesis (sites.bu.edu/hyness/rovergene) | |
| BioPSy (Madsen et al. 2015) | Performs guaranteed parameter set synthesis for ODE biological models expressed in SBML given a desired behavior expressed by time-series data (github.com/dreal/biology) | |
| D-VASim | A genetic network simulator that is used to perform the timing and propagation delay analysis on genetic logic circuits; D-VASim supports SBML format (bda.compute.dtu.dk/tools-2/d-vasim) | |
| RoadRunner | A portable simulation engine for systems and synthetic biology models in SBML format; RoadRunner is written in C# and incorporates C, C++, and Python APIs (libroadrunner.org) | |
| Sequence editing | Benchling | Sequence repository capable of storing and showing annotations and searching; Benchling is also a cloud-based framework for life sciences applications (benchling.com) |
| ApE | A sequence editor supporting FASTA, GenBank, DNA Strider, EMBL, and ABI formats; it also supports direct connection to BLAST (biologylabs.utah.edu/jorgensen/wayned/ape) | |
| VectorEditor | Web-based DNA sequence-editing and analysis tool with restriction enzyme manager, amino acid translation, and gel digest prediction (public-registry.jbei.org/vectoreditor) | |
| GeneDesigner | Encompasses a set of in silico sequence design tools and algorithms, such as cloning, codon optimization, back translation and primer design; supports GenBank and FASTA formats (dna20.com/resources/genedesigner) | |
| VectorNTI/Express | Sequence analysis and design tool with an integrated graphical environment that provides the ability to manage, view, analyze, transform, share, and publish molecular biology data | |
| Geneious (Drummond et al. 2010) | Sequence editing and alignment tool with support for annotations, incorporating multiple alignment algorithms and easy switch between them (geneious.com/features/genome-alignment) | |
| GeneDesign | Web-based synthetic gene designer including various modules such as reverse translator, codon juggler, etc. for sequence manipulation (54.235.254.95/gd) | |
| Sequence REFINER (Chakrabarti et al. 2006) | Sequence alignment tool that refines a multiple sequence alignment by iterative realignment of its individual sequences with the predetermined conserved core model of a protein family (ftp.ncbi.nih.gov/pub/REFINER) | |
| SnapGene Editor | A sequence-editing software capable of creating, browsing, and sharing richly annotated DNA sequence files up to 1 Gb in length (snapgene.com) | |
| Synthetic Gene Designer (Wu et al. 2006) | Generates optimal gene sequences taking codon bias, like GC content, into consideration (userpages.umbc.edu/wug1/codon/sgd) | |
| SBOL Designer | CAD tool for creating and manipulating genetic construct sequences using the SBOL data model (async.ece.utah.edu/SBOLDesigner) |
While rational specification tools are used to specify the intended function of a genetic system, rational design tools are used to map from such a specification to an implementing set of genetic parts and/or devices with defined sequences. A majority of the rational design tools in Table 1 (AutoBioCAD [Rodrigo and Jaramillo 2013], BioJade [Goler 2004], Cello [Nielsen et al. 2016], GEC [Pedersen and Phillips 2009], GenoCAD [Czar et al. 2009], iBioSim [Myers et al. 2009], Kera [Dhar 2010], Matchmaker [Yaman et al. 2012], MoSeC [Misirli et al. 2011], OptCircuit [Dasika and Maranas 2008], Parts&Pools [Marchisio 2014], and SBROME [Huynh et al. 2013; Huynh and Tagkopoulos 2014]) feature dedicated support for designing genetic regulatory networks/circuits, but none feature the same level of support for designing biosynthetic/metabolic pathways. This omission is partly a reflection of the current membership of the BDA community, many of whom work at the intersection between synthetic biology, systems biology, and electrical and computer engineering, thus favoring the development of tools for genetic circuit design. However, this omission also highlights that existing tools for rational pathway design, such as Cobra 2.0 (Schellenberger et al. 2011) and OptFlux (Rocha et al. 2010), have been developed based on traditional approaches to pathway engineering as opposed to newer approaches from synthetic biology. The former approaches tend to focus on knocking out genes from a known pathway to optimize its performance (Kim et al. 2015), although the latter approaches emphasize the optimal design of a synthetic pathway under the control of standardized parts (Smanski et al. 2014). Of the remaining rational design tools in Table 1, Rosetta (Leaver-Fay et al. 2011) and the RBS Calculator (Salis et al. 2009) have perhaps the broadest application in the sense that they are based on principles from biophysics and can be used to design some of the primitive genetic parts (proteins and RBSs, respectively) found in most genetic systems.
The example at the end of this section features Cello, GEC, and the RBS Calculator as effective BDA tools for rational specification and design. Cello was selected as it represents one of the most dedicated and experimentally vetted tools for genetic circuit design. Cello is also notable for basing its designs on an empirical model of circuit behavior in which the transfer curves for the individual logic gates that make up a circuit are multiplied to obtain a steady-state relationship between its input and output. This is in stark contrast with the mechanistic models of genetic regulatory networks leveraged by most other circuit design tools. In this way, Cello obviates the need to directly measure or estimate chemical kinetic parameters, but at the cost of being unable to predict the transient, non-steady-state behavior of its designed circuits. Consequently, for our example, Cello is best suited to the design of the AND gate as opposed to the dual-feedback oscillator. For the design of the latter circuit, GEC was selected by reason of its highly developed language for abstractly specifying genetic function and its capability to export biochemical models written in the Systems Biology Markup Language (SBML) (Hucka et al. 2003), a standard leveraged by many other BDA tools, including simulation tools featured in a later section (see Data Analysis and Simulation) (iBioSim [Myers et al. 2009] and COPASI [Hoops et al. 2006]). Last, the RBS calculator was chosen for its immediate accessibility compared with other biophysics-based tools, its high potential for complementing other BDA tools, and its widespread use among synthetic biologists outside of the BDA community.
Combinatorial Specification and Design
Combinatorial approaches to genetic design typically involve designing many different variants of a system, such that these variants can be efficiently screened and/or undergo selection to obtain those that optimize system function. For example, an engineer might design a library of biosynthetic pathway variants in which the genetic parts controlling expression are varied to enable screening for the set of parts that optimize pathway yield (Smanski et al. 2014). Alternatively, one might design a library of oligonucleotides to make a large number of targeted edits to the genome of a host organism and then select for the most productive strain (Wang et al. 2009). One-third of the specification and design tools listed in Table 1 can be classified as supporting a combinatorial approach to genetic design. At least four of these tools are dedicated to combinatorial specification, with three tools providing languages for specifying library architectures and/or target sequences for genome engineering (Eugene [Bilitchenko et al. 2011], GSL [Wilson et al. 2016], and Molecula Maxima) and one tool providing the means to generate experimental design matrices that can be interpreted as specifications of library function (Double Dutch [Roehner et al. 2016a]).
While combinatorial specification tools are used to specify the intended structure and/or function of a genetic library, combinatorial design tools are used to map from such a specification to an implementing set of genetic parts and/or devices with defined sequences. The combinatorial design tools in Table 1 are a superset of the combinatorial specification tools in this table and can be broadly divided into three groups: tools focused on genome engineering (MAGE [Wang et al. 2009] with Merlin [Quintin et al. 2016] and CRISPR [Cong et al. 2013] with DeskGen [see deskgen.com]), tools for general purpose design and/or assembly of combinatorial libraries (Device Editor [Hillson 2014], Eugene [Oberortner et al. 2014], Genome Compiler, GSL [Wilson et al. 2016], j5 [Hillson et al. 2011], and Teselagen [see teselagen.com]), and tools focused on designing libraries that adhere to experimental designs (Double Dutch [Roehner et al. 2016a]). These three groups can be viewed as a spectrum in which tools from the first and second groups are well suited to combinatorial approaches that rely exclusively on screening and/or selection, whereas tools from the second and third groups are well suited to semirational approaches that supplement screening with statistical methods such as regression analysis to guide the search for optimal variants. The majority of the combinatorial design tools in Table 1 belong to the first half of this spectrum rather than the second, which suggests that the BDA community has thus far focused its attention on combinatorial design and assembly as opposed to semirational design. This gap in tooling may also reflect a difference in research focus between academia and industry, given that the academic tools in Table 1 are overrepresented in the category of rational design, whereas the industrial tools are overrepresented in the category of combinatorial design but underrepresented in the subcategory of semirational design.
The example at the end of this section features Eugene and Double Dutch as effective BDA tools for combinatorial specification and design. Eugene was chosen for its flexibility as a domain-specific programming language, its incorporation into both academic and industrial tools (Cello [Nielsen et al. 2016], Device Editor [Hillson 2014], j5 [Hillson et al. 2011], and Teselagen [see teselagen.com]), and its capacity to export the Synthetic Biology Open Language (SBOL) (Galdzicki et al. 2011, 2014). SBOL is a prominent design standard that has been developed by a subset of the BDA community and has growing support among BDA tools (see section on Data Exchange and Standardization). Last, Double Dutch was chosen as the sole combinatorial design tool in Table 1 with explicit support for semirational design of biological systems.
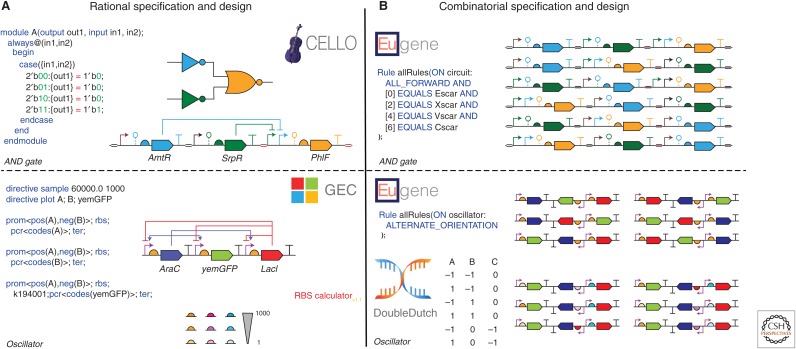
Example
In this example, we will discuss how Cello (Nielsen et al. 2016) and Eugene (Oberortner et al. 2014) can be used to design a genetic AND gate, and how GEC (Pedersen and Phillips 2009), the RBS Calculator (Salis et al. 2009), Eugene, and Double Dutch (Roehner et al. 2016a) can be used to design a dual-feedback oscillator (Fig. 1). The first step in the design of the genetic AND gate is to submit a Verilog (Thomas and Moorby 1995) case statement for the appropriate Boolean logic as input to Cello. Cello then compiles this Verilog specification to a gate netlist and optimizes the assignment of genetic logic gates to this netlist (colors the netlist) based on predicted circuit behavior. Because the correct behavior of a genetic circuit in vivo is the result of many factors besides just gate choice, oftentimes an additional combinatorial design step is performed to increase the likelihood of success. Among the outputs of Cello is a Eugene file that encodes the theoretical best circuit design. This file can be modified through the addition or subtraction of Eugene rules to customize the design and potentially specify a library of design variants. In this case, the rules specifying gene order are removed, whereas the rules specifying scar positions are kept. When the modified Eugene file is executed, the result is a library of six AND gates, one for every possible gene order. Following testing, we can attempt to analyze the effect of gene order on circuit performance and use the circuit variant that best satisfies the original Verilog specification made using Cello.
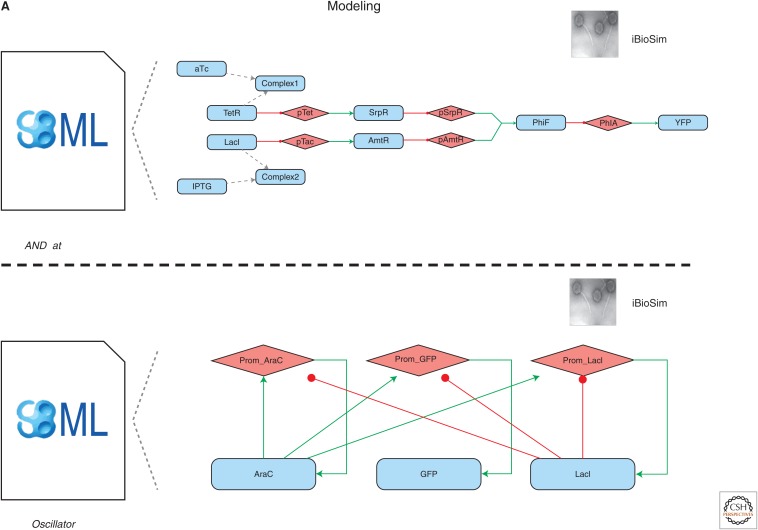
Figure 1.
Specification and design of a genetic AND gate and dual-feedback oscillator. All genetic part symbols are defined by the SBOL Visual standard (Quinn et al. 2015). Parts of the same symbol but different colors have different sequences, whereas parts of different symbols but the same color belong to same genetic logic gate. (A) Cello (Nielsen et al. 2016) takes a Verilog (Thomas and Moorby 1995) specification for an AND gate as input, compiles and colors a gate netlist, and outputs a genetic circuit design (top). GEC (Pedersen and Phillips 2009) takes a GEC specification for a dual-feedback oscillator as input and outputs a genetic circuit design (bottom). The RBS calculator (Salis et al. 2009) outputs a library of RBSs with a range of translation initiation rates (bottom). (B) Eugene (Oberortner et al. 2014) takes Eugene specifications as input and outputs a library of AND gate designs that vary in gene order (top) and a library of oscillator designs that vary in gene order and orientation (bottom). Double Dutch (Roehner et al. 2016a) takes a design of experiments (DOE) matrix as input and outputs a commensurate library of oscillator designs with varying RBSs designed by the RBS calculator (bottom).
Next, the design of the dual-feedback oscillator begins with writing a specification in the GEC language for its genetic architecture (number and types of genetic parts) and regulatory relationships. GEC then compiles this specification to all genetic circuit designs that satisfy the specified constraints. In this example, only one design is generated because the default part database provided with GEC is relatively small and must be extended with a dual-regulatory promoter from the iGEM (Smolke 2009; Vilanova and Porcar 2014) Registry (BioBrick K094120) to obtain any solutions. To predict which design will have optimal performance, one must use GEC to compile each design to a biochemical model and simulate each model using GEC or another tool. In practice, however, it can be difficult to obtain predictive biochemical models for this step, as many part databases lack the chemical kinetic parameters necessary to inform such models. Hence, in this example, the RBS calculator is used to design a library of nine RBSs with translation initiation rates spanning three orders of magnitude. This set of parts is used later as part of a semirational design strategy with Double Dutch. Unlike Cello, GEC does not produce a Eugene file that can be used for combinatorial design. Instead, this file must be written by the user. In this case, the Eugene file for the oscillator varies not only gene order, but also gene orientation, constraining the latter with a rule that requires genes to have alternating orientations (e.g., forward, reverse, forward). This rule can be useful for reducing read-through transcription of genes. Once the most promising of the six rule-conformant architectures has been identified through testing, a library of oscillators that share this architecture but have different RBSs is designed using Double Dutch. Double Dutch bases this library design on a design of experiments (DOE) matrix so that a user can perform regression analysis following testing and correlate RBS strength with oscillator performance. Consequently, further design decisions can be based on a regression model, and the oscillator can be fine-tuned when a biochemical model like those used in GEC is unavailable or not predictive.
BUILDING AND EDITING DNA SEQUENCES
Once a specification has been mapped to a set of target DNA constructs, they must be assembled. This problem, called the DNA assembly problem, has both design optimization and physical implementation aspects and can be broken into at least three subproblems: (1) vector design; (2) assembly planning; and (3) liquid handling. Traditionally, many of these problems have been solved manually and simultaneously in a process that became known as molecular cloning. In the early days of molecular cloning, these methods were all ad hoc; for each target clone, a human would have to study the fragment and vector of interest and select an intelligent set of restriction enzymes to cut the vector and the amplified template and then perform a separate ligation reaction to create the final clone. Some of the challenges of this approach include selecting compatible sets of restriction enzymes and compatible sets of sticky ends that were likely to result in a successful ligation. This method of cloning is called “traditional cloning.”
As our knowledge of cloning enzymes and DNA synthesis technologies improved, some groups started to gain interest in high-throughput DNA assembly where many fragments could be assembled at once in parallel. For these technologies, we could not require humans to make each assembly decision in such a large set and so some groups invented standard cloning frameworks called “DNA assembly methods,” which asserted standard conventions for the design of flanking ends of DNA fragments and the enzymes used in cloning reactions. In recent years, numerous methods have been developed with variations in these categories (Li and Elledge 2007; Engler et al. 2008; Shetty et al. 2008; Gibson et al. 2009; Quan and Tian 2009; Weber et al. 2010; Sarrion-Perdigones et al. 2011; Iverson et al. 2016), but all have the goal of scaling and modularizing DNA assembly capabilities. Some methods place a heavy emphasis on modularity (Weber et al. 2010; Iverson et al. 2016) and standardization (Shetty et al. 2008), whereas others place a heavier emphasis on the scale of fragments that can be reliably assembled (Gibson et al. 2009). In addition to these biochemical technology developments, software was developed to assist problem solving in each of the three DNA assembly problem areas.
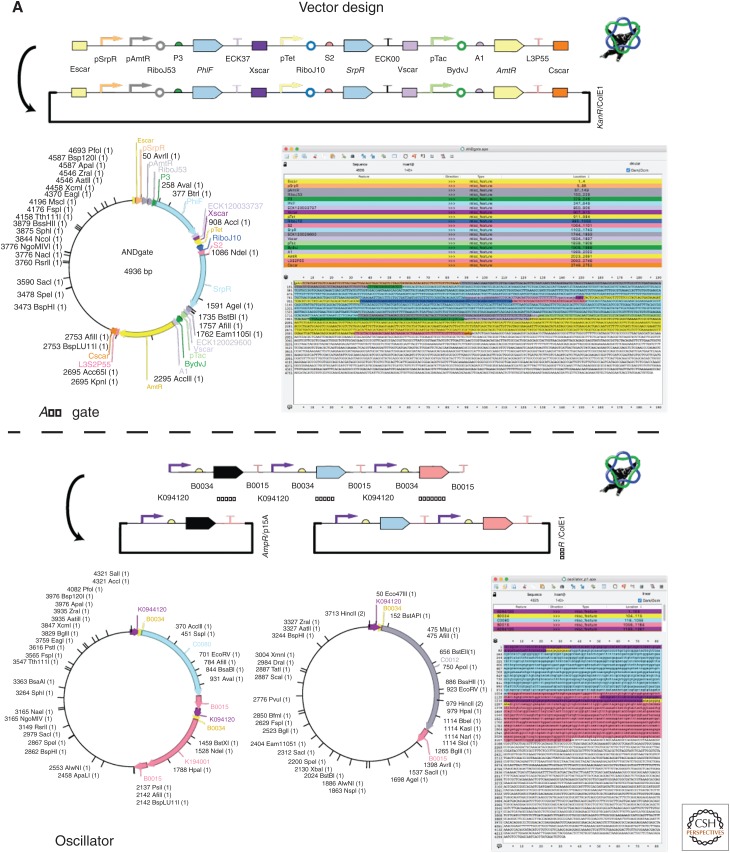
Vector Design
The first category of problems to solve in DNA assembly is the conversion of designs into replicating vectors or sets of replicating vectors. This problem, called vector design, is a commonly overlooked subproblem in the current software landscape. Presently, there are no known algorithmic software tools dedicated exclusively to this task and this problem is generally solved manually within sequence-editing software. To perform this task, a user copy–pastes their sequences into the editor and highlights important features and candidate restriction sites (Fig. 2A). There are a variety of popular proprietary (VectorNTI, Benchling, Geneious, Vector Express, SnapGene) and open-source (ApE, GeneDesign, Sequence Refiner, Vector Editor, Synthetic Gene Designer) sequence-editing tools available for solving this problem. These tools often include many additional features for curating sequence libraries and data management and have become the standard technology for storing sequence information in most laboratories.
Figure 2.
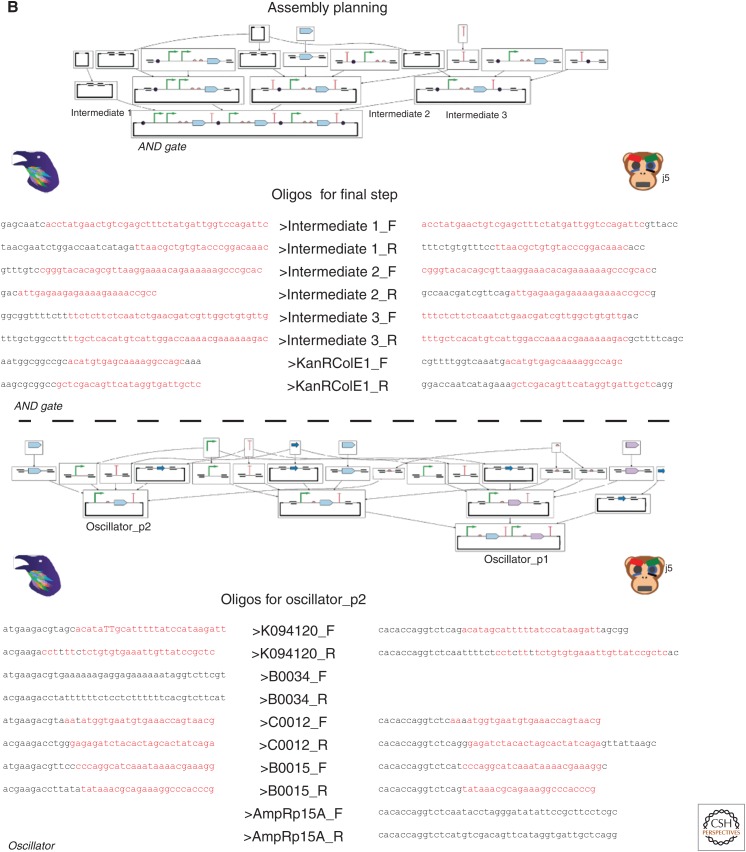
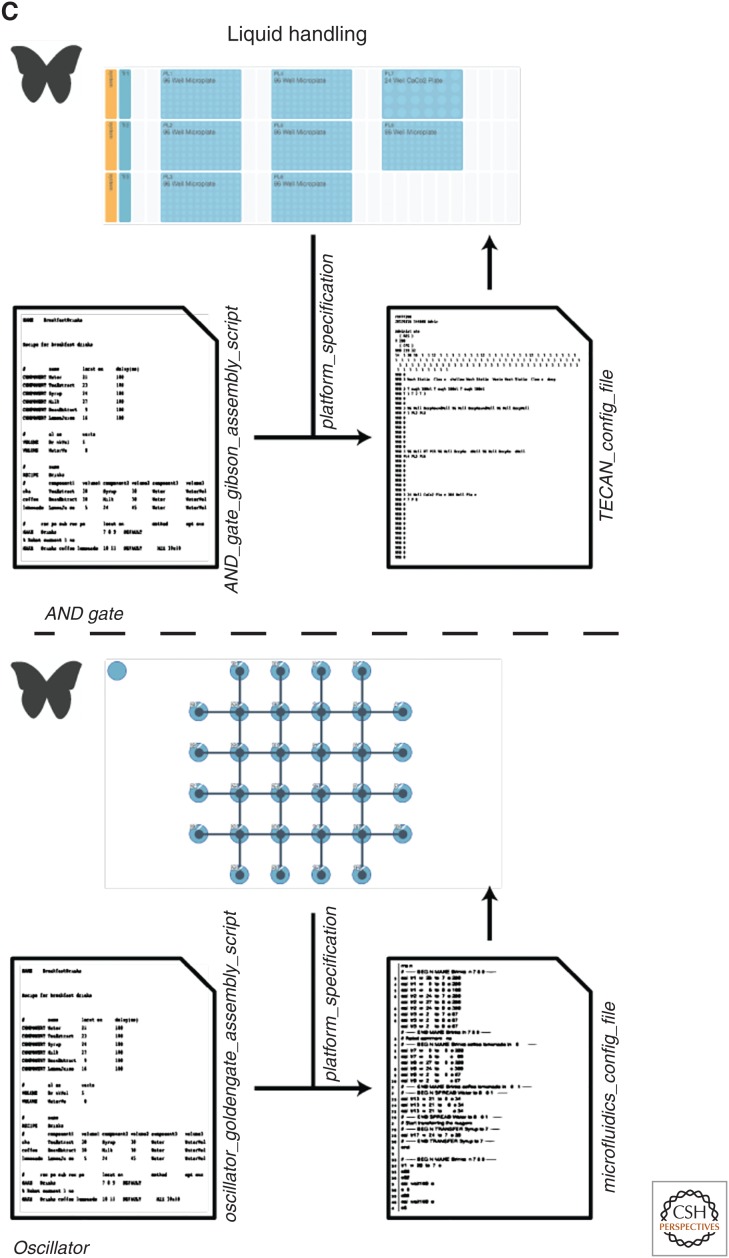
Construction of a complex logic gate and oscillator. (A) Using ApE, one can take a sequence of parts from a design, assign to one or more vectors, and edit any parts or junctions at the DNA sequence level. (B) Raven is used to determine a hierarchical assembly plan for each construct using a specific assembly method. Assembling an AND gate with Gibson requires two stages, four steps, four synthesis steps, and six polymerase chain reaction (PCR) steps, whereas assembling the oscillator constructs with GoldenGate requires two stages, four steps, and 16 PCR steps. Raven generates oligonucleotides to perform these assemblies, but j5 can be used for more refined primer designs for specific cloning steps. The common sequences are highlighted in red. (C) Once a complete assembly plan is determined, PR-PR can be used to create instruction files for liquid-handling robotics or microfluidics platforms to distribute liquids.
Assembly Planning
Hierarchical assembly planning refers to the selection of which cloning reactions to perform in parallel and in which series to ultimately create all final clones. For large assemblies or large sets of assemblies, it is desirable to maximize fragment reuse and minimize cloning steps to minimize time and material expense. Some algorithms have been developed to automate this design process (Densmore et al. 2010; Appleton et al. 2014a; Blakes et al. 2014) and have resulted in two web-based open-source software tools: DNALD (see dnald.org/planner) and Raven (see cidar.bu.edu/ravencad). DNALD inputs sets of target parts made of combinations of “basic part” primitives and determines an optimized hierarchical assembly plan for building all target parts assuming that only two fragments can ever be joined in one cloning step (called “binary assembly”). Raven also calculates hierarchical assembly plans for sets of target parts composed of “basic part” primitives, but offers several additional features. First, it partially addresses the vector design problem by allowing for the association of parts with the vectors they originate from and the vectors the final clone must be in. Second, it has algorithms for “n-ary” assembly in which more than two fragments may be connected in one cloning reaction, which is a common trait of modern “one-pot” cloning techniques categories (Fig. 2B) (Li and Elledge 2007; Engler et al. 2008; Shetty et al. 2008; Gibson et al. 2009; Quan and Tian 2009; Weber et al. 2010; Sarrion-Perdigones et al. 2011; Iverson et al. 2016).
After the hierarchical assembly plan is determined, flanking sequences for each of these fragments must be determined and primers must be designed for polymerase chain reaction (PCR) such that the correct flanking regions are added to each fragment. Raven provides algorithms for optimizing assembly junction sequence reuse, which is important when cloning libraries for combinatorial assembly (Engler et al. 2008; Weber et al. 2010; Sarrion-Perdigones et al. 2011; Iverson et al. 2016), and provides basic primer designs but does optimize them for assembly-method-specific cloning reaction chemistry concerns. However, the j5 tool addresses this problem in great detail for many contemporary cloning methods (Hillson et al. 2011) and produces highly optimized primers with a high likelihood of improving the success of each individual cloning reaction (Fig. 2B). Most sequence-editing software also has integrated tools for informing these decisions, but do not optimize primers as well as j5. Other tools like Primer3 can optimize primers for sequencing or other assembly-related tasks such as genome editing.
Liquid Handling
After assembly planning, the selection of chemical protocols and plans for dispensing liquid to carry out these protocols must be determined. Some of these protocols are standardized by the chemical manufacturer, but many cloning reaction reagents are not available in the form of a standard master mix, so they must be designed by a biologist. After a set of protocols is determined, the problem of how to execute all necessary protocols efficiently is called the liquid-handling problem. Some open-source tools (PR-PR [Linshiz et al. 2014] and Aquarium) have been created to solve these problems and subsequently produce step-by-step instructions for executing these tasks in an effort to reduce human cost and improve protocol efficiency plus reproducibility. One such tool for this task, PR-PR, inputs a formatted file to describe all of the reactions that must be performed and a file to describe the configuration of a supported platform before generating liquid-handling instructions for a liquid-handling robot, microfluidic platform, or human user (Fig. 2C).
Example
To provide an example of how to use these tools, we will examine the construction of the AND gate and oscillator designs from the previous section (Fig. 2). To solve the vector design problem, one can import the sequences from each part of the overall design into ApE and paste them into a single (as per the AND gate) or multiple (as per the oscillator) replicating vector(s) (Fig. 2A). Next, we can use Raven to determine hierarchical assembly plans using Gibson assembly for the AND gate and GoldenGate for oscillator. Raven generates primers and synthesis fragments for these plans, but j5 can offer more refined primers for each specific cloning step that can improve the likelihood that the cloning step will work on the first try (Fig. 2B) (in this case, we determine primers for the last cloning step of the AND gate and for the smaller plasmid from the oscillator). Finally, this assembly plan can be written up in a standard format and converted to protocol planning files that can be compiled by the PR-PR software to produce assembly instructions that can be interpreted by specific liquid-handling robotics platforms, microfluidic rigs, or a human with a pipettor (Fig. 2C).
DATA ANALYSIS AND SIMULATION
Data generated in the previous steps can be simulated and analyzed to evaluate and validate the biological system. The first step in this analysis process typically involves the application of a computational model. This model can either be simulated to produce traces representing the behavior of the system or analyzed to validate properties of the system. Depending on what types of properties a researcher wants to check, there are different modeling techniques as well as a variety of simulation and analysis methods that can be used.
Modeling
To reason about biological data, researchers must construct models. There are a plethora of different modeling techniques that can be used to accomplish this goal, which mainly fall into two categories: qualitative and quantitative (Jong 2002). Qualitative approaches typically abstract many details of the system, reducing it to a collection of states and transitions. An example of one of these types of models is a Boolean network that views each species in the system as being either in the “on” or “off” state and each reaction as a logical transition of species from “on” to “off” and vice versa. If more “levels” are desired (e.g., “high,” “low,” and “medium”), the model can be extended into a qualitative network. These types of models are often used in multicellular simulators such as Gro or Morpheus, in which simplifying the logic for each cell aids in reducing the complexity of the entire multicellular system.
Another type of qualitative model for describing chemical processes is a Petri net (Petri 1966). Petri nets are useful for modeling systems with a lot of concurrency and are comprised of bipartite graphs made up of a collection of transitions (i.e., events or reactions) and places (i.e., states or conditions). Each place can have a number of tokens representing anything such as the concentration of a species or the number of times a reaction has occurred. Tokens are moved from one place to another by firing a transition between them. This firing can represent many things such as an interaction of two species or the advancement of time. Among its modeling options, iBioSim supports the creation of Petri net models of biological systems.
Although qualitative models have been used to successfully predict different behaviors in biological systems, they often fall short in producing quantitative results. Quantitative models, on the other hand, strive to be as real-to-life as possible and often include very complex interactions that are described using reaction rate equations. In these types of models, the modeler will often define the quantity of each chemical species, a set of chemical reactions between species, and a collection of parameters used in the rate equations. These models can then be either translated into a collection of ordinary differential equations (ODEs) using classical chemical kinetics (Waage and Guldberg 1864) or into a collection of propensity functions using stochastic chemical kinetics (Gillespie 1992), depending on which types of simulation or analysis a user wishes to perform. Quantitative models are the most widely used in synthetic biology as they can be built and tuned to give the most predictive power. Nearly all of the simulation tools presented in this review are capable of producing and/or processing quantitative models.
Simulation
Simulation largely falls into two categories: deterministic and stochastic (or probabilistic) (Lecca et al. 2013). Deterministic simulation is often performed after applying classical chemical kinetics to a model to produce a set of ODEs. These ODEs are then solved using a standard method such as the Euler or Runge-Kutta methods to produce time series data for each species in the system. ODE simulation is deterministic because the solution to the ODEs is always the same given the same initial conditions on the model. Hence, the system only needs to be simulated once to produce a trace representing its behavior.
Classical chemical kinetics assume that concentrations of species in a system are large and that changes in these concentrations occur continuously and deterministically. This assumption does not always hold for biological systems, particularly genetic regulatory circuits where molecule counts are small and reaction firings cause large fluctuations in species counts (McAdams and Arkin 1999). For these systems, stochastic simulation is necessary to produce accurate results.
To prepare the model for stochastic simulation, the propensity of each reaction in the system must be calculated. Each propensity represents the probability that its associated reaction will fire within the next infinitesimal time interval, and they are computed by multiplying the reactants in the reaction by the reaction rate constants. With these propensities, the simulator can use a method such as Gillespie’s stochastic simulation algorithm (SSA) (Gillespie 1976, 1977). This algorithm roughly works by determining the next reaction to fire based on propensities, probabilistically determining a time for the reaction to fire, stepping over all of the uneventful time to this firing time, firing the reaction to update the state, and then repeating this process until the desired time limit has been reached. Traces produced by this algorithm will vary each time it is run because the next reaction and the next firing times are always selected probabilistically. This fact means that a system may need to be simulated many times to determine its average behavior or to observe a rare event. Additionally, some systems have many fast reactions (reactions with large propensities), which cause the simulator to spend an exorbitant amount of simulation time firing these reactions while not advancing time very much. For these systems, improvements to the SSA such as tau-leaping (Gillespie and Petzold 2003) (selecting a larger time step and firing several reactions instead of one) and slow-scale SSA (Cao et al. 2005) (partitioning the system into fast and slow reactions and simulating them on different time scales with the help of steady-state approximations) have aided in their simulation.
Almost all of the tools mentioned in this review with simulation capabilities support both deterministic and stochastic simulation. Some of them (e.g., TinkerCell, CellDesigner, JDesigner), however, connect to another simulation tool (e.g., COPASI, Jarnac) for this support.
Analysis
Beyond simulation, there exist many types of analysis that can be performed on a model. These include model checking over steady-state or transient properties of the system, flux balance analysis, parameter estimation, and sensitivity analysis. Model checking approaches, especially those with a probabilistic component, can either be analyzed with statistical or numerical approaches (Younes et al. 2006; Kwiatkowska et al. 2007). Statistical approaches typically involve stochastically simulating the system a large number of times and then computing statistics over the resulting traces, whereas numerical analysis usually involves finding the state space of the model, often represented with a matrix, and performing computations directly on it. Without translating the model into the appropriate format and exporting it to an external model-checking tool like PRISM (Hinton et al. 2006), the only biological simulation tool capable of performing model checking is iBioSim.
Flux balance analysis is used to calculate the steady-state metabolic fluxes of biological systems by assuming that the input to the system roughly equals the output plus or minus one or a few factors (Raman and Chandra 2009). It is fairly easy to implement and is included as a feature in iBioSim, COPASI, PySCeS, and Jarnac.
Parameter estimation methods attempt to find values for parameters based on some experimental data (Lynch and Walsh 1998; Thompson et al. 2005). Most of these methods involve choosing some values for the parameters and then simulating the system to see how closely the resulting trace matches the data. If the trace matches well enough, the parameters are returned as acceptable; otherwise, the parameters are perturbed and the process repeats. This perturbation can be performed using a variety of methods, from genetic algorithms to particle swarms to hill-climbing techniques. iBioSim has newly added support for parameter estimation, but COPASI has the largest number of implemented parameter estimation methods. For those looking to take parameter estimation a step further and instead try to solve the parameter synthesis problem to determine ranges of acceptable parameters, BioPSy can be used.
Sensitivity analysis involves determining how much different parameters affect the dynamics of the system (Nestorov 1999). Like model checking, sensitivity analysis can either be performed through simulations of the model or by analyzing the model numerically. Many tools including COPASI, PySCeS, and Jarnac are capable of performing this type of analysis.
Example
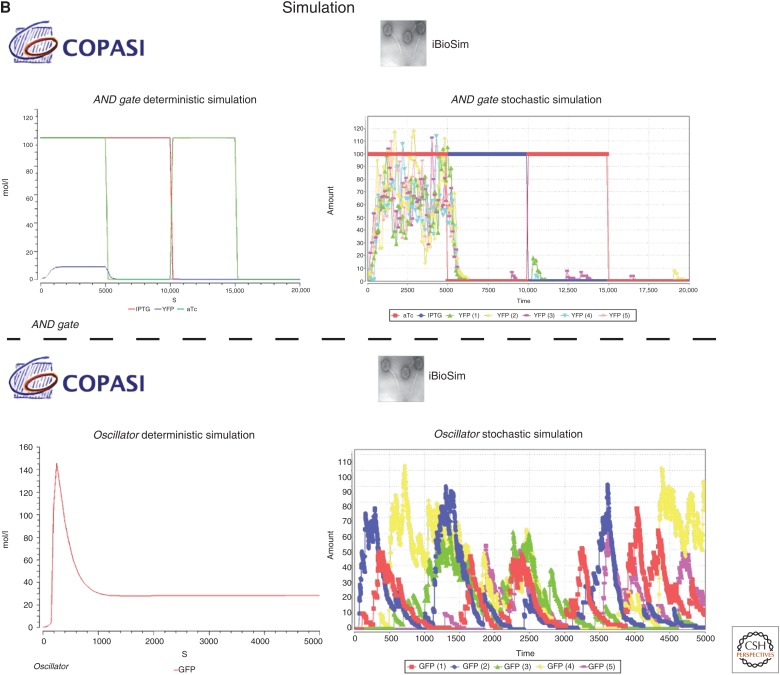
In this example, we apply modeling and simulation to the genetic AND gate and the dual-feedback genetic oscillator using iBioSim and COPASI (Fig. 3). The first step is generating a mathematical model for each of these systems that we can simulate. For the genetic AND gate, the SBOL file generated by Cello is imported into iBioSim and converted into SBML. Although the converted model can be simulated outright, it is missing tunable inputs (i.e., the small molecules aTc and IPTG) and an output (i.e., the florescent protein YFP). Using the model editor in iBioSim, aTc, and IPTG are added to the model in addition to their complex formation reactions with TetR and LacI, respectively. Additionally, YFP is added, a repression arc is drawn between PhlF and YFP, and the final model is shown (Fig. 3A). This model is then exported as an SBML model that is passed to COPASI and simulated using ODEs while varying the values of aTc and IPTG (Fig. 3B shows simulation plots for this model). The ODE simulation shows that the value of YFP is highest when both aTc and IPTG are present and lowest when only one or neither are present. However, the high value of YFP is still fairly low as the ODE simulation treats reactions as continuous, deterministic events. Using iBioSim, we can perform a few stochastic simulations while still varying the values of aTc and IPTG. These simulations show that again the value of YFP is highest when both aTc and IPTG are present and lowest when only one or neither are present. Because of stochastic noise, this simulation reveals that there is occasionally sporadic production of YFP, but YFP quickly degrades away because of the lack of one or both inputs.
Figure 3.
Modeling and simulation results of a genetic AND gate and dual-feedback oscillator. (A) SBML models are created in iBioSim for the AND gate and oscillator. The AND gate’s model is automatically generated by converting the SBOL file produced by Cello to SBML. This model is then extended to include the regulation by the proteins LacI and TetR, the complex formation of LacI and TetR with the small molecules IPTG and aTc, respectively, and the regulation of the florescent protein YFP. The oscillator model is reconstructed using information (e.g., parameter values, rate constants) from the SBML generated by GEC. (B) Simulations are performed using both COPASI and iBioSim. Deterministic simulations are performed using the LSODA ODE solver in COPASI. Stochastic simulations are performed in iBioSim using an implementation of Gillespie’s SSA.
The SBML model for the dual-feedback genetic oscillator produced by GEC was not able to be simulated directly using iBioSim because of some differences in support of SBML packages; however, this model is recreated in iBioSim (Fig. 3A). As with the genetic AND gate, the oscillator’s model is exported to SBML that is fed into COPASI and simulated using deterministic methods (Fig. 3B). In this simulation, the value of GFP initially increases to a peak value and then declines until it levels off at a steady-state value. Needless to say, this is not the behavior we would expect from an oscillator, and it is obvious that the ODE simulation is failing to capture some of the noisy, stochastic events that lead to oscillations. Using iBioSim, on the other hand, several stochastic simulations are performed, which show this oscillatory behavior.
DATA EXCHANGE AND STANDARDIZATION
At all stages of the design cycle described in the introductory section, there exist opportunities to store and exchange data on genetic designs, including their structure (genetic sequences, X-ray crystal structures, etc.), function (regulatory relationships, chemical kinetic models, etc.), and measurements (flow cytometry histograms, microscope images, etc.), among other types of data. Table 1 contains a list of BDA databases and tools for storing and exchanging these data types. In this section, we will briefly describe two of the standards most widely used by these and other BDA tools, and discuss how these standards facilitate data exchange in the examples presented in this review.
The Synthetic Biology Open Language (SBOL)
SBOL (Galdzicki et al. 2014; Roehner et al. 2016b) is an emerging data exchange standard for synthetic biology with growing support among BDA tools, including database tools such as ICE (Ham et al. 2012) and the SBOL Stack (Madsen et al. 2016). SBOL has been developed by members of the BDA and greater synthetic biology communities to document genetic components (DNA, RNA, protein, etc.) for the purpose of engineering design. Unlike existing standards that were originally conceived for documenting naturally occurring genetic sequences, such as FASTA (Pearson and Lipman 1988) and GenBank (Bilofsky and Christian 1988), SBOL can be used to document partial genetic designs and recursively annotate the sequences of genetic components with other components in a hierarchical fashion. These capabilities of SBOL address the iterative, modular character of engineering designs in a way that current standards for genetic sequences neglect. Furthermore, SBOL is an extensible standard that can be adapted to meet the evolving needs of the synthetic biology community, such as the need to document the context-dependence of functioning genetic systems.
In the examples presented in this review, SBOL facilitates the exchange of design data for the genetic AND gate. In particular, Cello exports an SBOL file for the AND gate that is imported and compiled by iBioSim into an SBML model (Roehner and Myers 2014). This model is then extended and simulated as described above (see sections Data Analysis and Simulation and Example). In this way, the SBOL-documented structure and function of the AND gate directly inform the creation of a quantitative model of its chemical kinetics.
The Systems Biology Markup Language (SBML)
SBML (Hucka et al. 2003) is a biological modeling standard that has been developed by the systems biology community and is currently supported by more than 250 different software tools. There also exists a public database for storing and accessing SBML models known as the BioModels database (Chelliah et al. 2013). The primary goal of SBML is to enable intersoftware exchange of most essential features of models for biological systems, especially biochemical models for describing cell signaling, metabolism, and genetic regulation, among other phenomena. Accordingly, SBML is a machine-readable Extensible Markup Language (XML) that is independent of any proprietary software language or particular analytical framework (e.g., ODE or stochastic analysis).
In the examples presented in this review, SBML enables models of the genetic AND gate and dual-feedback oscillator to be simulated across tools. In particular, models of the AND gate and oscillator exported from iBioSim (Myers et al. 2009) can be imported and simulated by COPASI (Hoops et al. 2006). This allows users to take advantage of the analysis methods that are unique to each tool without having to translate their models between formats and risk losing information. Even in the case when tools adhering to a standard are unable to directly exchange models because of implementation errors or differences in support, the standard can facilitate the recreation of models across tools. This is currently the case when exchanging SBML models between GEC (Pedersen and Phillips 2009) and iBioSim. Even though the oscillator model exported by GEC cannot be directly imported into iBioSim, its SBML encoding permits a researcher to more easily interpret and recreate this model in iBioSim.
CONCLUSION
As shown in this overview, great progress has been made in design automation in synthetic biology in a relatively short period of time (<9 years at the time of writing as measured by the first iGEM software competition). These efforts have validated many approaches (Beal et al. 2012), created interwoven design flows (Galdzicki et al. 2014), and began to foster a self-sustaining industrial development ecosystem and associated academic discipline. However, much more work needs to be performed. Examples include:
Specification—the specification of the desired performance (e.g., timing, yield, expression level) of a biological system separated from the behavior of that system remains largely unaddressed. Formal languages for describing the diverse domains in synthetic biology (e.g., metabolic networks, genetic circuits, engineered organisms) will need to be developed and related to one another when appropriate. Abstraction terminology should also work toward standardization.
Design—rules and constraints need to be formally captured and shared more broadly. These constraints need to be validated experimentally and parameterized for specific contexts (e.g., host organisms, environmental conditions).
Build—microfluidics and liquid-handling robot platforms need to be more fully integrated across the entire assembly process (e.g., device design to organism growth). Interfaces for these devices need to be standardized and widely understood. Support for “cloud labs” (emeraldcloudlab.com; transcriptic.com) should increase and be part of the software toolchain.
Test—standardized assays amendable to BDA tools should be encouraged and expected of experimentalists in the field. Measurement standards and metrics (programmingbiology.org; Beal 2015) should be developed and promoted.
Learn—large-scale data mining and storage need to be available to communities outside of synthetic biology so that experts in these fields can apply the latest analytical methods.
Recently SBOL has been adopted by the ACS Synthetic Biology journal (Hillson et al. 2016) as an acceptable format to submit along with research manuscripts. This represents an important milestone in the promotion of BDA principles.
In addition, curated open-source software efforts related to BDA may prove valuable for academics as a means to sustain their software over long periods of time and unpredictable funding climates. These may also be useful for industry looking to benefit from early technical advances with little upfront development costs. The Nona Research Foundation (see nonasoftware.org) is a U.S.-based nonprofit that looks to provide open-source software to the entire community (academic and industrial). This organization and others like the BioBricks Foundation and BioBuilder can also serve as educational hubs to communicate the importance of BDA.
Footnotes
Editors: Daniel G. Gibson, Clyde A. Hutchison III, Hamilton O. Smith, and J. Craig Venter
Additional Perspectives on Synthetic Biology available at www.cshperspectives.org
REFERENCES
- Andrianantoandro E, Basu S, Karig DK, Weiss R. 2006. Synthetic biology: New engineering rules for an emerging discipline. Mol Syst Biol 2: 2006.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appleton E, Tao J, Haddock T, Densmore D. 2014a. Interactive assembly algorithms for molecular cloning. Nat Methods 11: 657–662. [DOI] [PubMed] [Google Scholar]
- Appleton E, Tao J, Wheatley FC, Desai DH, Lozanoski TM, Shah PD, Awtry JA, Jin SS, Haddock TL, Densmore DM. 2014b. Owl: Electronic datasheet generator. ACS Synth Biol 3: 966–968. [DOI] [PubMed] [Google Scholar]
- Arkin A. 2008. Setting the standard in synthetic biology. Nat Biotechnol 26: 771–774. [DOI] [PubMed] [Google Scholar]
- Bates JT, Chivian D, Arkin AP. 2011. GLAMM: Genome-linked application for metabolic maps. Nucleic Acids Res 39: W400–W405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal J. 2015. Signal-to-noise ratio measures efficacy of biological computing devices and circuits. Front Bioeng Biotechnol 3: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal J, Lu T, Weiss R. 2011. Automatic compilation from high-level biologically oriented programming language to genetic regulatory networks. PLoS ONE 6: e22490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal J, Weiss R, Densmore D, Adler A, Appleton E, Babb J, Bhatia S, Davidsohn N, Haddock T, Loyall J, et al. 2012. An end-to-end workflow for engineering of biological networks from high-level specifications. ACS Synth Biol 1: 317–331. [DOI] [PubMed] [Google Scholar]
- Bhatia S, Densmore D. 2013. Pigeon: A design visualizer for synthetic biology. ACS Synth Biol 2: 348–350. [DOI] [PubMed] [Google Scholar]
- Bilitchenko L, Liu A, Cheung S, Weeding E, Xia B, Leguia M, Anderson JC, Densmore D. 2011. Eugene: A domain specific language for specifying and constraining synthetic biological parts, devices, and systems. PLoS ONE 6: e18882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilofsky HS, Christian B. 1988. The GenBank genetic sequence data bank. Nucleic Acids Res 16: 1861–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakes J, Raz O, Feige U, Bacardit J, Widera P, Ben-Yehezkel T, Shapiro E, Krasnogor N. 2014. Heuristic for maximizing DNA reuse in synthetic DNA library assembly. ACS Synth Biol 3: 529–542. [DOI] [PubMed] [Google Scholar]
- Borujeni AE, Salis HM. 2016. Translation initiation is controlled by RNA folding kinetics via a ribosome drafting mechanism. J Am Chem Soc 138: 7016–7023. [DOI] [PubMed] [Google Scholar]
- Borujeni AE, Channarasappa AS, Salis HM. 2013. Translation rate is controlled by coupled trade-offs between site accessibility, selective RNA unfolding and sliding at upstream standby sites. Nucleic Acids Res 42: 2646–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canton B, Labno A, Endy D. 2008. Refinement and standardization of synthetic biological parts and devices. Nat Biotechnol 26: 787–793. [DOI] [PubMed] [Google Scholar]
- Cao Y, Gillespie DT, Petzold LR. 2005. The slow-scale stochastic simulation algorithm. J Chem Phys 10.1063/1.1824902. [DOI] [PubMed] [Google Scholar]
- Castillo-Hair SM, Sexton JT, Landry BP, Olson EJ, Igoshin OA, Tabor JJ. 2016. FlowCal: A user-friendly, open source software tool for automatically converting flow cytometry data from arbitrary to calibrated units. ACS Synth Biol 5: 774–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S, Lanczycki CJ, Panchenko AR, Przytycka TM, Thiessen PA, Bryant SH. 2006. Refining multiple sequence alignments with conserved core regions. Nucleic Acids Res 34: 2598–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran D, Bergmann F, Sauro H. 2009. TinkerCell: Modular CAD tool for synthetic biology. J Biol Eng 3: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelliah V, Laibe C, Novere NL. 2013. BioModels database: A repository of mathematical models of biological processes. Methods Mol Biol 1021: 189–199. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czar MJ, Cai Y, Peccoud J. 2009. Writing DNA with GenoCAD. Nucleic Acids Res 37: W40–W47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasika MS, Maranas CD. 2008. OptCircuit: An optimization based method for computational design of genetic circuits. BMC Syst Biol 2: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Densmore D. 2012. Bio-design automation: Nobody said it would be easy. ACS Synth Biol 1: 296–296. [DOI] [PubMed] [Google Scholar]
- Densmore DM, Bhatia S. 2014. Bio-design automation: Software + biology + robots. Trends Biotechnol 32: 111–113. [DOI] [PubMed] [Google Scholar]
- Densmore D, Van Devender A, Johnson M, Sritanyaratana N. 2009. A platform-based design environment for synthetic biological systems. In The Fifth Richard Tapia Celebration of Diversity in Computing Conference: Intellect, Initiatives, Insight, and Innovations, pp. 24–29. Portland, OR. [Google Scholar]
- Densmore D, Hsiau TH, Kittleson JT, DeLoache W, Batten C, Anderson JC. 2010. Algorithms for automated DNA assembly. Nucleic Acids Res 38: 2607–2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar PK. 2010. The language of life. Nat India 10.1038/nindia.2010.145. [DOI] [Google Scholar]
- Drummond A, et al. 2010. Geneious v5.5. http://www.geneious.com. [Google Scholar]
- Endy D. 2005. Foundations for engineering biology. Nature 438: 449–453. [DOI] [PubMed] [Google Scholar]
- Engler C, Kandzia R, Marillonnet S. 2008. A one pot, one step, precision cloning method with high throughput capability. PLoS ONE 3: e3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funahashi A, Matsuoka Y, Jouraku A, Morohashi M, Kikuchi N, Kitano H. 2008. CellDesigner 3.5: A versatile modeling tool for biochemical networks. Proc IEEE 96: 1254–1265. [Google Scholar]
- Galdzicki M, Wilson ML, Rodriguez CA, Adam L, Adler A, Anderson JC, Beal J, Chandran D, Densmore D, Drory OA, et al. 2011. Synthetic Biology Open Language (SBOL) Version 1.0.0. RFC 85. doi: 1721.1/66172. [Google Scholar]
- Galdzicki M, Clancy KP, Oberortner E, Pocock M, Quinn JY, Rodriguez CA, Roehner N, Wilson ML, Adam L, Anderson JC, et al. 2014. The Synthetic Biology Open Language (SBOL) provides a community standard for communicating designs in synthetic biology. Nat Biotechnol 32: 545–550. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6: 343–345. [DOI] [PubMed] [Google Scholar]
- Gillespie DT. 1976. A general method for numerically simulating the stochastic time evolution of coupled chemical reactions. J Comp Phys 22: 403–434. [Google Scholar]
- Gillespie DT. 1977. Exact stochastic simulation of coupled chemical reactions. J Phys Chem 81: 2340–2361. [Google Scholar]
- Gillespie DT. 1992. Markov processes: An introduction for physical scientists. Academic, New York. [Google Scholar]
- Gillespie DT, Petzold LR. 2003. Tau leaping. J Chem Phys 119: 8229–8234. [Google Scholar]
- Goler J. 2004. “BioJADE: A design and simulation tool for synthetic biological systems.” Master's thesis, MIT. [Google Scholar]
- Ham TS, Dmytriv Z, Plahar H, Chen J, Hillson NJ, Keasling JD. 2012. Design, implementation and practice of JBEI-ICE: An open source biological part registry platform and tools. Nucleic Acids Res 40: e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden EC. 2015. Synthetic biologists seek standards for nascent field. Nature 520: 141–142.25855435 [Google Scholar]
- Hill AD, Tomshine JR, Weeding EMB, Sotiropoulos V, Kaznessis YN. 2008. SynBioSS: The synthetic biology modeling suite. Bioinformatics 24: 2551–2553. [DOI] [PubMed] [Google Scholar]
- Hillson NJ. 2014. j5 DNA assembly design automation, pp. 245–269. Humana, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- Hillson N, Rosengarten RD, Keasling J. 2011. j5 DNA assembly design automation software. ACS Synth Biol 1: 14–21. [DOI] [PubMed] [Google Scholar]
- Hillson NJ, Plahar HA, Beal J, Prithviraj R. 2016. Improving synthetic biology communication: Recommended practices for visual depiction and digital submission of genetic designs. ACS Synth Biol 5: 449–451. [DOI] [PubMed] [Google Scholar]
- Hinton A, Kwiatkowska M, Norman G, Parker D. 2006. 12th International Conference on Tools and Algorithms for the Construction and Analysis of Systems (TACAS’06) (ed. Hermanns H, Palsberg J), pp. 441–444. Springer, Berlin. [Google Scholar]
- Hoops S, Sahle S, Gauges R, Lee C, Pahle J, Simus N, Singhal M, Xu L, Mendes P, Kummer U. 2006. COPASI—A COmplex PAthway SImulator. Bioinformatics 22: 3067–3074. [DOI] [PubMed] [Google Scholar]
- Huang H, Densmore D. 2014. Fluigi: Microfluidic device synthesis for synthetic biology. ACM J Emerg Technol Comput Syst 11: 1–19. [Google Scholar]
- Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, et al. 2015. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods 12: 115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, Arkin AP, Bornstein BJ, Bray D, Cornish-Bowden A, et al. 2003. The systems biology markup language (SBML): A medium for representation and exchange of biochemical network models. Bioinformatics 19: 524–531. [DOI] [PubMed] [Google Scholar]
- Huynh L, Tagkopoulos I. 2014. Optimal part and module selection for synthetic gene circuit design automation. ACS Synth Biol 3: 556–564. [DOI] [PubMed] [Google Scholar]
- Huynh L, Tsoukalas A, Köppe M, Tagkopoulos I. 2013. SBROME: A scalable optimization and module matching framework for automated biosystems design. ACS Synth Biol 2: 263–273. [DOI] [PubMed] [Google Scholar]
- Iverson SV, Haddock TL, Beal J, Densmore DM. 2016. CIDAR MoClo: Improved MoClo assembly standard and new E. coli part library enable rapid combinatorial design for synthetic and traditional biology. ACS Synth Biol 5: 99–103. [DOI] [PubMed] [Google Scholar]
- Jang SS, Oishi KT, Egbert RG, Klavins E. 2012. Specification and simulation of synthetic multicelled behaviors. ACS Synth Biol 1: 365–374. [DOI] [PubMed] [Google Scholar]
- Jong HD. 2002. Modeling and simulation of genetic regulatory systems: A literature review. J Comp Biol 9: 67–103. [DOI] [PubMed] [Google Scholar]
- Kahl LJ, Endy D. 2013. A survey of enabling technologies in synthetic biology. J Biol Eng 7: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly JR, Rubin AJ, Davis JH, Ajo-Franklin CM, Cumbers J, Czar MJ, de Mora K, Glieberman AL, Monie DD, Endy D. 2009. Measuring the activity of BioBrick promoters using an in vivo reference standard. J Biol Eng 3: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TY, Park JM, Kim HU, Cho KM, Lee SY. 2015. Design of homo-organic acid producing strains using multi-objective optimization. Metab Eng 28: 63–73. [DOI] [PubMed] [Google Scholar]
- Koressaar T, Remm M. 2007. Enhancements and modifications of primer design program Primer3. Bioinformatics 23: 1289–1291. [DOI] [PubMed] [Google Scholar]
- Kwiatkowska M, Norman G, Parker D. 2007. Stochastic model checking. In Formal Methods for the Design of Computer, Communication and Software Systems: Performance Evaluation (SFM’07), Vol. 4486 (ed. Bernardo M, Hillston J), pp. 220–270. Springer, Berlin. [Google Scholar]
- Leaver-Fay A, Tyka M, Lewis SM, Lange OF, Thompson J, Jacak R, Kaufman K, Renfrew PD, Smith CA, Sheffler W, et al. 2011. ROSETTA3: An object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol 487: 545–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecca P, Laurenzi I, Jordan F. 2013. Deterministic versus stochastic modelling in biochemistry and systems biology. Woodhead, Oxford. [Google Scholar]
- Li MZ, Elledge SJ. 2007. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat Methods 4: 251–256. [DOI] [PubMed] [Google Scholar]
- Linshiz G, Stawski N, Goyal G, Bi C, Poust S, Sharma M, Mutalik V, Keasling JD, Hillson NJ. 2014. PR-PR: Cross-platform laboratory automation system. ACS Synth Biol 3: 515–524. [DOI] [PubMed] [Google Scholar]
- Lux M, Bramlett BW, Ball DA, Peccoud J. 2012. Genetic design automation: Engineering fantasy or scientific renewal? Trends Biotechnol 30: 120–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Walsh B. 1998. Genetics and analysis of quantitative traits. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Madsen C, Shmarov F, Zuliani P. 2015. Computational methods in systems biology, pp. 182–194. Springer, New York. [Google Scholar]
- Madsen C, McLaughlin JA, Misirli G, Pocock M, Flanagan K, Hallinan J, Wipat A. 2016. The SBOL Stack: A platform for storing, publishing, and sharing synthetic biology designs ACS Synth Biol 5: 487–497. [DOI] [PubMed] [Google Scholar]
- Marchisio MA. 2014. Parts & pools: A framework for modular design of synthetic gene circuits. Front Bioeng Biotechnol 2: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdams HH, Arkin A. 1999. Genetic regulation at the nanomolar scale: It’s a noisy business. Trends Genet 15: 65–69. [DOI] [PubMed] [Google Scholar]
- Mirschel S, Steinmetz K, Rempel M, Ginkel M, Gilles ED. 2009. PROMOT: Modular modeling for systems biology. Bioinformatics 25: 687–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misirli G, Hallinan JS, Yu T, Lawson JR, Wimalaratne SM, Cooling MT, Wipat A. 2011. Model annotation for synthetic biology: Automating model to nucleotide sequence conversion. Bioinformatics 27: 973–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers CJ, Barker N, Jones K, Kuwahara H, Madsen C, Nguyen NP. 2009. iBioSim: A tool for the analysis and design of genetic circuits. Bioinformatics 25: 2848–2849. [DOI] [PubMed] [Google Scholar]
- Nestorov IA. 1999. Sensitivity analysis of pharmacokinetic and pharmacodynamic systems. I: A structural approach to sensitivity analysis of physiologically based pharmacokinetic models. J Pharmacokinet Biopharm 27: 577–596. [DOI] [PubMed] [Google Scholar]
- Nielsen AA, Der BS, Shin J, Vaidyanathan P, Paralanov V, Strychalski EA, Ross D, Densmore D, Voigt CA. 2016. Genetic circuit design automation. Science 352: aac7341. [DOI] [PubMed] [Google Scholar]
- Oberortner E, Densmore D. 2014. Web-based software tool for constraint-based design specification of synthetic biological systems. ACS Synth Biol 4: 757–760. [DOI] [PubMed] [Google Scholar]
- Oberortner E, Bhatia S, Lindgren E, Densmore D. 2014. A rule-based design specification language for synthetic biology. ACM J Emerg Technol Comput Syst 11z: 25. [Google Scholar]
- Olivier BG, Rohwer JM, Hofmeyr JHS. 2004. Modelling cellular systems with PySCeS. Bioinformatics 21: 560–561. [DOI] [PubMed] [Google Scholar]
- Pearson WR, Lipman DJ. 1988. Improved tools for biological sequence comparison. Proc Natl Acad Sci 85: 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen M, Phillips A. 2009. Towards programming languages for genetic engineering of living cells. J R Soc Interface 6: S437–S450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petri CA. 1966. “Communication with automata.” PhD thesis, Universität Hamburg, Hamburg, Germany. [Google Scholar]
- Quan J, Tian J. 2009. Circular polymerase extension cloning of complex gene libraries and pathways. PLoS ONE 4: e6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn JY, Cox RS III, Adler A, Beal J, Bhatia S, Cai Y, Chen J, Clancy K, Galdzicki M, Hillson NJ, et al. 2015. SBOL Visual: A graphical language for genetic designs. PLoS Biol 13: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintin M, Ma NJ, Ahmed S, Bhatia S, Lewis A, Isaacs FJ, Densmore D. 2016. Merlin: Computer-aided oligonucleotide design for large scale genome engineering with MAGE. ACS Synth Biol 5: 452–458. [DOI] [PubMed] [Google Scholar]
- Raman K, Chandra N. 2009. Flux balance analysis of biological systems: Applications and challenges. Brief Bioinform 10: 435–449. [DOI] [PubMed] [Google Scholar]
- Rocha I, Maia P, Evangelista P, Vilaça P, Soares S, Pinto JP, Nielsen J, Patil KR, Ferreira EC, Rocha M. 2010. OptFlux: An open-source software platform for in silico metabolic engineering. BMC Syst Biol 4: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo G, Jaramillo A. 2013. AutoBioCAD: Full biodesign automation of genetic circuits. ACS Synth Biol 2: 230–236. [DOI] [PubMed] [Google Scholar]
- Rodrigo G, Carrera J, Jaramillo A. 2007. Asmparts: Assembly of biological model parts. Syst Synth Biol 1: 167–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roehner N, Myers CJ. 2014. A methodology to annotate systems biology markup language models with the synthetic biology open language. ACS Synth Biol 3: 57–66. [DOI] [PubMed] [Google Scholar]
- Roehner N, Young EM, Voigt CA, Gordon DB, Densmore D. 2016a. Double Dutch: A tool for designing combinatorial libraries of biological systems. ACS Synth Biol 5: 507–517. [DOI] [PubMed] [Google Scholar]
- Roehner N, Beal J, Clancy K, Bartley B, Misirli G, Grünberg R, Oberortner E, Pocock M, Bissell M, Madsen C, et al. 2016b. Sharing structure and function in biological design with SBOL 2.0. ACS Synth Biol 5: 498–506. [DOI] [PubMed] [Google Scholar]
- Salis HM, Mirsky EA, Voigt CA. 2009. Automated design of synthetic ribosome binding sites to control protein expression. Nat Biotechnol 27: 946–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrion-Perdigones A, Falconi EE, Zandalinas SI, Juárez P, Fernández-del-Carmen A, Granell A, Orzaez D. 2011. GoldenBraid: An iterative cloning system for standardized assembly of reusable genetic modules. PLoS ONE 6: e21622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauro H. 2000. Jarnac: An interactive metabolic systems language in computation in cells. In Proceedings of an EPSRC Emerging Computing Paradigms Workshop (ed. Bolouri H, Paton R). University of Hertfordshire, Hertfordshire, UK. [Google Scholar]
- Schellenberger J, Que R, Fleming RM, Thiele I, Orth JD, Feist AM, Zielinski DC, Bordbar A, Lewis NE, Rahmanian S, et al. 2011. Quantitative prediction of cellular metabolism with constraint-based models: The COBRA Toolbox v2.0. Nat Protoc 1: 1290–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty RP, Endy D, Knight TF. 2008. Engineering BioBrick vectors from BioBrick parts. J Biol Eng 2: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smanski MJ, Bhatia S, Zhao D, Park Y, B A Woodruff L, Giannoukos G, Ciulla D, Busby M, Calderon J, Nicol R, et al. 2014. Functional optimization of gene clusters by combinatorial design and assembly. Nat Biotechnol 32: 1241–1249. [DOI] [PubMed] [Google Scholar]
- Smith LP, Bergmann FT, Chandran D, Sauro HM. 2009. Antimony: A modular model definition language. Bioinformatics 25: 2452–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolke CD. 2009. Building outside of the box: iGEM and the BioBricks Foundation. Nat Biotechnol 27: 1099–1102. [DOI] [PubMed] [Google Scholar]
- Starruß Jo, de Back W, Brusch L, Deutsch A. 2014. Morpheus: A user-friendly modeling environment for multiscale and multicellular systems biology. Bioinformatics 30: 1331–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stricker J, Cookson S, Bennett MR, Mather WH, Tsimring LS, Hasty J. 2008. A fast, robust and tunable synthetic gene oscillator. Nature 456: 516–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DE, Moorby P. 1995. The Verilog hardware description language (2nd ed.). Kluwer, New York. [Google Scholar]
- Thompson R, Brotherstone S, White IMS. 2005. Estimation of quantitative genetic parameters. Philos Trans R Soc B Biol Sci 360: 1469–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. 2012. Primer3—New capabilities and interfaces. Nucleic Acids Res 40: e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilev V, Liu C, Haddock T, Bhatia S, Adler A, Yaman F, Beal J, Babb J, Weiss R, Densmore D. et al. 2011. A software stack for specification and robotic execution of protocols for synthetic biological engineering. In 3rd International Workshop on Bio-Design Automation (IWDBDA), pp. 24–25. San Diego, CA. [Google Scholar]
- Vilanova C, Porcar M. 2014. iGEM 2.0—Refoundations for engineering biology. Nat Biotechnol 32: 420–424. [DOI] [PubMed] [Google Scholar]
- Waage P, Guldberg CM. 1864. Studies concerning affinity. J Chem Educ 63: 1044. [Google Scholar]
- Wang HH, Isaacs FJ, Carr PA, Sun ZZ, Xu G, Forest CR, Church GM. 2009. Programming cells by multiplex genome engineering and accelerated evolution. Nature 460: 894–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber E, Engler C, Gruetzner R, Werner S, Marillonet S. 2010. A modular cloning system for standardized assembly of multigene constructs. PLoS ONE 10.1371/journal.pone.0016765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson EH, Sagawa S, Weis JW, Schubert MG, Bissell M, Hawthorne B, Reeves CD, Dean J, Platt D. 2016. Genotype specification language. ACS Synth Biol 5: 471–478. [DOI] [PubMed] [Google Scholar]
- Wu G, Bashir-Bello N, Freeland SJ. 2006. The synthetic gene designer: A flexible web platform to explore sequence manipulation for heterologous expression. Prot Expres Purif 47: 441–445. [DOI] [PubMed] [Google Scholar]
- Yaman F, Bhatia S, Adler A, Densmore D, Beal J. 2012. Automated selection of parts for genetic regulatory networks. ACS Synth Biol 1: 332–344. [DOI] [PubMed] [Google Scholar]
- Younes H, Kwiatkowska M, Norman G, Parker D. 2006. Numerical vs. statistical probabilistic model checking. Int J Softw Tools Technol Transf 8: 216–228. [Google Scholar]
- Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]