Abstract
Important gaps remain in our understanding of the spread of farming into Europe, due partly to apparent contradictions between studies of contemporary genetic variation and ancient DNA. It seems clear that farming was introduced into central, northern, and eastern Europe from the south by pioneer colonization. It is often argued that these dispersals originated in the Near East, where the potential source genetic pool resembles that of the early European farmers, but clear ancient DNA evidence from Mediterranean Europe is lacking, and there are suggestions that Mediterranean Europe may have resembled the Near East more than the rest of Europe in the Mesolithic. Here, we test this proposal by dating mitogenome founder lineages from the Near East in different regions of Europe. We find that whereas the lineages date mainly to the Neolithic in central Europe and Iberia, they largely date to the Late Glacial period in central/eastern Mediterranean Europe. This supports a scenario in which the genetic pool of Mediterranean Europe was partly a result of Late Glacial expansions from a Near Eastern refuge, and that this formed an important source pool for subsequent Neolithic expansions into the rest of Europe.
Keywords: phylogeography, Neolithic, Late Glacial, European origins, mitogenomes, haplogroups
1. Background
Radiocarbon evidence for the spread of agriculture provides a detailed, fine-structure chronology for the expansion of Early Neolithic material culture from a Near Eastern source into and across Europe, beginning in Greece approximately 8.5 thousand years ago (ka) [1–4], but the extent to which these dates portray the movement of human beings can only be settled by genetics.
This was for many years restricted to inferences from modern genetic distributions, but has been augmented more recently by a substantial and highly influential body of ancient DNA (aDNA) research. This points to a major break in genetic continuity in central, northern, and eastern Europe with the arrival of the Early Neolithic from about 7.5 ka, evident in signatures of mitochondrial DNA (mtDNA) [5–10], Y-chromosome [11–14], and genome-wide variation. In the latter, a novel ancestral component, present in most modern Europeans, but not in any pre-Neolithic European individuals so far tested, has been inferred at high levels in all Early Neolithic Europeans sampled [14–16]. This discontinuity can be traced back to at least the Starčevo culture of Romania/Croatia [17], with some analyses suggesting a greater affinity of the Neolithic newcomers to modern Near Easterners than to Europeans [6,16]. This has led to the conclusion that there was a wide-scale replacement across Europe from the Near East in the Early Neolithic [18,19].
A Near Eastern source has recently been bolstered by direct evidence from Anatolia and the Levant. Mathieson et al. [20] analysed both mitogenomes and genome-wide markers from 28 individuals from the Early Neolithic in Anatolia, showing some mtDNA (such as N1a1a, K1a, T2b, J1c, U3, and X2) and Y-chromosome haplogroups (especially G2a) shared with central and west European Early Neolithic populations and the presence of the European Neolithic genome-wide component at more than 90%. Omrak et al. [21] have also inferred an Anatolian source, on similar grounds, and Lazaridis et al. [22] have extended the genome-wide analysis to the Levant, showing that the European Neolithic component reaches approximately 100% there.
Nevertheless, there are significant gaps in the aDNA evidence. There are few Mesolithic data from Mediterranean Europe, owing to sample availability and poor preservation. Hofmanová et al. [23] have recovered mtDNA from two Mesolithic samples from Greece—but they have found them to belong to haplogroup K1c, which seems to place them closer to Neolithic populations in Europe and Anatolia than to Mesolithic central and northern Europeans. Therefore, they point out, the precise source for the early central European farmers remains in doubt—it could have been Anatolia, but it could equally have been Greece or the Balkans.
There is, in fact, some evidence for a Mediterranean European source from analyses of contemporary populations. Many modern European mtDNA haplogroup J, T, I, and W lineages, as well as some from R0a (and potentially others, including from HV), have been estimated to have arrived in Europe in the Late Glacial and immediate postglacial period, before the onset of the Neolithic, from one or more Near Eastern glacial refugia [24–26].
In the absence of sufficient aDNA evidence from the Mediterranean, we have conducted instead a more detailed analysis of the major component of these lineages, those belonging to haplogroups J and T, in order to estimate their arrival times in different parts of Europe. We find that while they largely spread into central Europe and Iberia with the Neolithic, they reached eastern/central Mediterranean Europe earlier, a conclusion that is consistent with recent statistical analyses of autosomal aDNA [27].
2. Methods
We sequenced 203 haplogroup JT mitogenomes, using Sanger sequencing, and analysed them alongside 1 476 published JT mitogenomes. We estimated trees using reduced-median network analysis together with PhyloTree (electronic supplementary material, data S1). For time estimates (electronic supplementary material, table S1, data S2), we used rho (ρ) and maximum-likelihood (ML), with a mutation rate of one substitution every 3 624 years, corrected for purifying selection, and a synonymous rate of one substitution every 7 884 years [28]. We estimated migration times using founder analysis [29–31] (electronic supplementary material, data S2) in three ways: (i) from the Near East (including Anatolia) to Mediterranean Europe (excluding Iberia), (ii) from the Near East and eastern/central Mediterranean Europe to Iberia, and (iii) from the Near East and Mediterranean Europe to central/northern Europe. We also performed the founder analysis considering Mediterranean Europe and the Near East as independent sources to Iberia as well as to central and northern Europe. In addition, we performed a reciprocal founder analysis, reversing the migration direction of all the models described. We obtained Bayesian skyline plots using BEAST. For full details, see electronic supplementary material, data S3.
3. Results
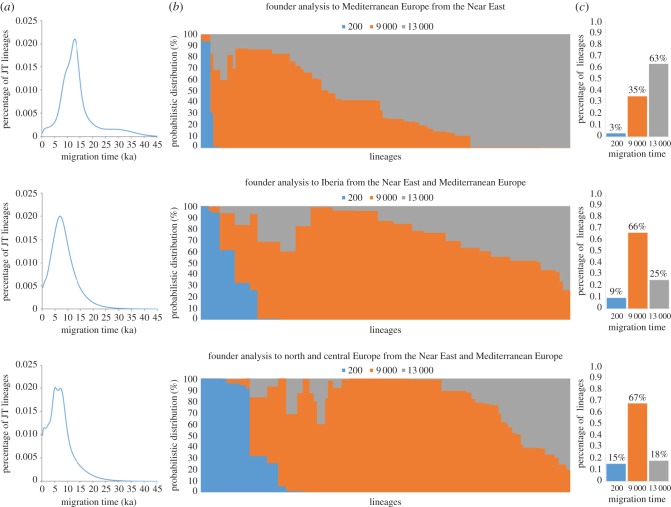
To further assess our earlier inferences [24], we first carried out a founder analysis for haplogroups J and T in Mediterranean Europe, excluding Iberia, using Anatolia and the Near East as the source pool (figure 1, top panel). The migration scan, which plots the fraction of arriving lineages against time, showed a single main peak at 12.8 ka, but was slightly humped suggesting a possible second, masked peak approximately 9 ka (figure 1a, top panel). Thus, the analysis points to a major dispersal from the Near East into Mediterranean Europe in the Late Glacial, approximately 13 ka, with a smaller-scale postglacial or Neolithic dispersal at approximately 9 ka.
Figure 1.
Founder analysis results for whole mitogenomes. Top panel: from a Near East source to Mediterranean Europe (excluding Iberia); middle panel: from the Near East and Mediterranean Europe to Iberia; bottom panel: from the Near East and Mediterranean Europe to north and central Europe. (a) Probabilistic distribution across migration times scanned at 200 year intervals from 0 to 70 ka; (b) probabilistic distribution across migration times scanned at 200 year intervals from 0 to 70 ka; (c) proportion of Late Glacial, Neolithic and recent founder lineages in a heuristic three-migration model.
The full set of dated founders is shown in electronic supplementary material, table S2. Aside from a possible pre-glacial presence of J2a, the major Late Glacial founders (approx. 13 ka) were J1c, J2b1, T2b, and T1a1c. The main postglacial founders (11.5–9 ka) were T2e, J2b1, and J1c2, but the identification of J1c2 as a distinct founder, dating to approximately 9 ka, may be incorrect (owing to back-migration to the Near East), given the evidence from the phylogeny. T1a1a1 dates to the Early Neolithic for this region, at approximately 8.6 ka. Overall, we estimated that Late Glacial dispersals accounted for around two-thirds of the JT lineages, and postglacial/Neolithic dispersals for the remaining one-third (figure 1b,c, top panel).
We next assumed a combined Near Eastern and central/eastern European Mediterranean source, and estimated dispersals into Iberia (figure 1, middle panel; electronic supplementary material, table S3 in supplementary data S2), where the Neolithic began approximately 7.5 ka [32,33]. Here there was a clear, but quite distinct, pattern, with a single major peak dating to 7.0 ka. Two of the major founders (T2b, T1a1a1) dated to the Early Neolithic; of these, the former has a Late Glacial ancestry in central Mediterranean Europe, whereas the latter dispersed throughout the Mediterranean from the Near East with the Neolithic. Two others (J1c5c, T2b3) date to 8.5–9.5 ka, but with the reduced Iberian dataset this may reflect imprecision in the founder estimates rather than genuinely Mesolithic arrivals. In any case, both evidently have Late Glacial ancestry in the central Mediterranean. Unusually for Iberia, T2a1a dates to the Early Bronze Age, approximately 4.0 ka, while having an early postglacial ancestry further to the east.
Overall, the situation in Iberia is reversed by comparison with the region further east: two-thirds of the lineages arrived in the Neolithic, with only a quarter dating to the Late Glacial. We obtained a similar picture when using the Near East and Mediterranean Europe separately as sources, with the main Neolithic founder (T2b) shared across these analyses, albeit with a greater fraction of Late Glacial founders (e.g. J1c1) with the Near East as the source, as expected considering that Mediterranean Europe is likely the direct source for Iberia (electronic supplementary material, figure S1 in supplementary data S3, tables S4 and S5 in supplementary data S2).
The picture is slightly more complex for central/northern Europe (figure 1, bottom panel and electronic supplementary material, table S6 in supplementary data S2). When we assumed a solely Near Eastern source and estimated dispersals to central/northern Europe, we obtained a primarily Late Glacial ancestry, with more than half of the lineages dating to approximately 12 ka, and the remainder partitioned roughly between the Early Neolithic and the Late Neolithic (electronic supplementary material, figure S2 in supplementary data S3, table S7 in supplementary data S2). But when we assumed a solely European Mediterranean source, most of the lineages fell into the Neolithic (electronic supplementary material, figure S2 in supplementary data S3, table S8 in supplementary data S2), implying once again that the Near East alone was not adequately representing the source region and that important founders were present in the European Mediterranean.
When we therefore assumed a combined Near Eastern/European Mediterranean source and estimated dispersals to central/northern Europe, the result was similar, with two major, roughly equal peaks at 7.2 and 5.2 ka (figure 1a, bottom panel), accounting for two-thirds of the lineages between them. The earlier peak corresponds well with the arrival of the Early Neolithic in central Europe, with the linear pottery culture (LBK), which spread rapidly approximately 7.5 ka [2]. The clearest Early Neolithic founder lineages, dating to approximately 7.2–8.3 ka, are J1c2 and T2b, which likely have Late Glacial ancestry in Mediterranean Europe. There is a minor Late Glacial/early postglacial signal, in particular from J1c7a1 at approximately 12.6 ka, but amounting to less than a fifth of the overall total.
Most of the more recent peak in central and northern Europe is due to the dramatic expansion across Europe of T1a1a1 in the Late Neolithic, approximately 5.0 ka. Although T1a1a1 is very occasionally seen in aDNA samples before 5 ka, most are from eastern Europe, and it does not rise to appreciable levels (approx. 4%) in central Europe until the Late Neolithic, possibly spreading with the Corded Ware from further east [8,9].
Finally, in order to check that we assigned source and sink regions correctly, we performed reverse founder analyses for every combination of source and sink (electronic supplementary material, figure S3 in supplementary data S3, tables S9–S15 in supplementary data S2). In every case, the reverse analysis yielded higher age estimates, mostly in the Late Glacial, reflecting the higher proportion of private variants in the source regions, especially in the Near East, but also to a lesser extent in Mediterranean Europe.
This result mirrors similar source/sink dynamics in other parts of the world such as Island Southeast Asia/Taiwan [34] and North Africa/Iberia [35]. As well as confirming that the Near East and Mediterranean Europe are both acting as source regions for Iberia and central/northern Europe, these reciprocal analyses also show that, although Mediterranean Europe is evidently acting as a reservoir for some Late Glacial Near Eastern diversity, it is nevertheless a clearly distinct genetic pool from the Near East: the two regions are unlikely to simply form a single continuous population resulting from high levels of recurrent Holocene gene flow.
The patterns revealed by the founder analyses are closely reflected in a very different kind of analysis: skyline plots of effective population size (electronic supplementary material, figure S4 in supplementary data S3). For haplogroup J (left panel), there is a dramatic Late Glacial expansion in the Near East/Arabia at approximately 15 ka, and a little later approximately 13 ka in South Caucasus/Anatolia, levelling off in the Holocene. In Mediterranean Europe, we see this expansion again, but with a second increment superimposed after approximately 7 ka. Iberia and central Europe, by contrast, show a single main burst of expansion with the Early Neolithic, after approximately 8 ka. The pattern is broadly similar for haplogroup T (right panel), but here northern/central Europe is distinct: the main expansion is later, after 5 ka, reflecting the fact that it is largely haplogroup T lineages (especially T1a1a1) in central/northern Europe that show signs of major Late Neolithic dispersal (matching the additional, more recent peak in the founder analysis for this region).
4. Discussion
The earliest studies on classical genetic markers led to the proposal for a large-scale demic diffusion of food-producing communities from a source in the Near East [36]. This was eventually questioned by analyses of molecular data from the maternally inherited mtDNA, which estimated a minor Neolithic Near Eastern contribution, leading to the suggestion of pioneer colonization, followed by assimilation of indigenous European lineages [29,37,38]. This overall picture received support from the emerging genome-wide studies, which indicated a majority autosomal genetic ancestry among Europeans of a component that is minor in the Near East and most frequent in Saami, who are assumed to lack Neolithic ancestry [39,40].
However, this interpretation has, in turn, been cast into doubt by analyses of aDNA [9,18]. European Upper Palaeolithic individuals carry mainly the ancient lineages U8 (excluding its major subclade, haplogroup K), U5, and U2, with several episodes of apparent turnover, especially immediately following the Last Glacial Maximum and in the Late Glacial [27,41,42]. These lineages amount to only approximately 15% of modern European mtDNAs. Central, northern, and eastern Mesolithic samples tested to date carry mainly U5a, U5b, and U4 [7,43]. In stark contrast, Early Neolithic individuals from the same areas have minimal fractions of these lineages, carrying instead mainly haplogroups H, HV*, K1, N1a1a, T2, and J [5,6,8–10]. A similar pattern of apparent discontinuity has been seen for Y-chromosome lineages [11,12,44]. Moreover, Early Neolithic individuals in these regions, and also Iberia, carry a majority genome-wide ancestry component distinct from the earlier hunter–gatherers, common throughout the Near East and Mediterranean today, but which reaches its highest levels in modern Sardinians (more than 50%) [45,46], Neolithic Anatolians (roughly 90%) [20], and Neolithic and Epipalaeolithic Levantine populations (around 100%) [22].
This has often been regarded as a decisive argument in favour of large-scale dispersal from Anatolia into Europe at the onset of the Neolithic, with rather little assimilation of native European groups [16]. If this were the case, however, how do we explain the results from both the extant mtDNA and genome-wide data, which indicate a minority Near Eastern Neolithic contribution?
One obvious answer is that we are indeed seeing pioneer-colonists with a new genetic make-up in the Early European Neolithic, and that assimilation of indigenous lineages took place later, in the Middle Neolithic/Chalcolithic. However, this is only a small part of the explanation—the Neolithic fraction remains more than 50% even by the Chalcolithic, for prehistoric Europeans sampled so far. A bigger part of the picture concerns fresh immigration in the Late Neolithic from parts of eastern Europe that the Neolithic fraction had not previously reached significantly. aDNA studies show that a third ancestral pool spread across Europe from the east after about 5.0 ka. This ‘steppe’ or ‘Caucasus hunter–gatherer’ component most likely had its source in the Mesolithic of the Caucasus region/eastern Fertile Crescent, where it approaches 100% in pre-Neolithic individuals and remains at high levels today [47], and likely spread from the Pontic/Caspian region across Europe in the Late Neolithic/Early Bronze Age [14,48], as well as eastwards into Central Asia and south into South Asia. It was likely accompanied by a similar fraction of the Mesolithic European component, so that a substantial element of indigenous eastern European lineages may have spread across central and western Europe at this time [8]. This eastern component was also present at high levels in the Mesolithic and Neolithic of Iran, and spread across the Near East into Anatolia from either there or the Caucasus by the Late Neolithic/Bronze Age [22]. Thus, populations have been substantially reshaped genetically across West Eurasia in the last 5 ka, and both the eastern European part of the Mesolithic European ancestral pool and the ‘steppe’ component carried deep-rooting autochthonous European mtDNA lineages [49].
However, there is also a third possible explanation for the low estimation of Near Eastern Neolithic impact on the present-day European population. Previous analyses of mtDNA haplogroups J and T in extant populations have suggested that many of them entered Europe from a Near Eastern glacial refuge before the Neolithic, approximately 10–20 ka [24]. If this were the case, they may have been restricted to the Mediterranean region in the postglacial period, and as a consequence they may even have contributed, perhaps substantially, to the ancestry of the Early Neolithic Europeans that dispersed from south to north. Given that Neolithic and Mesolithic/Epipalaeolithic Anatolian and Levantine populations mainly carry the ‘Mediterranean’ genome-wide ancestry component, populations dispersing from there in the Late Glacial/early postglacial would likely also have carried this component.
This possibility has not yet been directly tested using aDNA, because few pre-Neolithic genomes have been retrieved from Mediterranean European samples to date. However, two Mesolithic samples from Greece, although untested for genome-wide markers, have been found to carry mtDNA haplogroup K1c. K1 has so far been associated with the Neolithic dispersal from the Near East and not seen in Mesolithic Europeans, although K nests within haplogroup U8, which is known to have a European Palaeolithic ancestry [41,50]. Moreover, recent statistical analyses of genome-wide data from several dozen samples from the Upper Palaeolithic have suggested a major Near Eastern component [27].
This conclusion is supported qualitatively by the appearance of the Y-chromosome haplogroup R1b1 in the sole Late Glacial sample from Villabruna in Italy [27], and a number of specific derived autosomal alleles. The interpretation is not straightforward, because the same Villabruna sample, which is the ‘type specimen’ for this Late Glacial cluster, carries a U5b mtDNA lineage, rather than J or T, and indeed, the other members of this Late Glacial cluster have been sampled from further north and west in Europe, and also mainly carry haplogroup U5 lineages. However, we should not assume that each genetic marker system tells exactly the same story, and it is likely that any Late Glacial arrivals from the east in Italy melded with local forager populations, assimilating local mtDNA, whereas some of their genome-wide variation diffused further afield into indigenous Late Glacial groups.
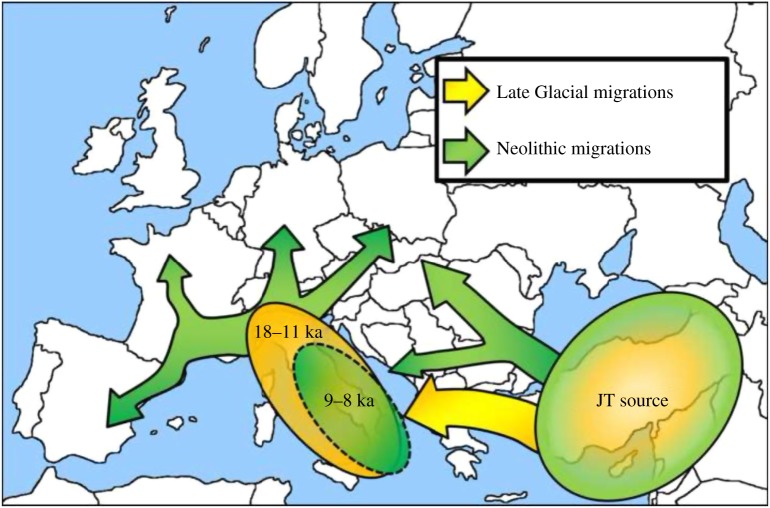
We tested this hypothesis by investigating dispersals into Europe of Near Eastern J and T lineages using regional founder analyses to date their arrival times in distinct regions of Europe. There was a very clear result: the majority of J and T lineages indeed arrived in eastern/central Mediterranean Europe before the advent of the Neolithic, but their further dispersals not only into continental Europe, but also into Iberia, largely did not take place until the Early Neolithic (figure 2). Thus, eastern/central Mediterranean Europe is a potential source pool for the Neolithic of the rest of Europe, reconciling the evidence of contemporary and aDNA.
Figure 2.
Late Glacial/early postglacial and Neolithic migrations from the Near East into Europe and the subsequent dispersals within Europe triggered by the arrival of the first farmers, involving both new Near Eastern genetic input and indigenous European lineages.
We should emphasize that haplogroups J and T make up only approximately 20% of European mtDNAs, but our focus reflects their previously proposed roles in both the Neolithic and the Levantine refuge. As noted above, much of haplogroup U is indigenous to pre-Neolithic Europe, whereas the source for R0 may be in the Near East [26], the origins of haplogroup H remain uncertain, although different major subclades seem likely to have arisen in Pleistocene Iberia [9,51–55] and southern Russia/Caucasus [14,47]. Thus, our evidence should not be taken as implying only a minor role for Mesolithic Iberians in subsequent times, for example, owing to the ‘8.2 ka event’ cold-snap as sometimes suggested [32,56], although they clearly highlight an important role for pioneer colonization in the west [57].
These analyses confirm that the Near East was an important glacial refuge area for Europe [24–26] and that Late Glacial expansions took place not only within Europe, but also from a source in the Near East, likely the Levant—a view now indirectly supported by genome-wide aDNA analyses [27]. They do not deny a role for Anatolia as a source pool for the European Neolithic or that some dispersals took place from there to Europe approximately 8.5–9 ka, but they do imply that the Neolithic of Europe also has some deeper genetic roots than this model implies, and that acculturation or assimilation were likely especially important around the Mediterranean.
The archaeological evidence provides support for such a scenario [58], especially in northern Italy. The rich Mesolithic Sauveterrean tradition flourished there for several thousand years prior to the arrival of farming, resulting in refugia for Mesolithic practices that survived for several hundred years as farming expanded around them, alongside evidence for the emergence of hybrid hunter–farmer communities [58–60]. Some Mesolithic communities came to adopt a fairly sedentary marine-based lifestyle on the coasts, whereas others evidently developed seafaring, judging from the evidence for obsidian trade and deep-sea fishing [58,61], in some cases becoming more mobile as a result, leading to the concept of Early Holocene ‘foraging seascapes’ [62].
Moreover, a scenario has been suggested in which a fusion of forager maritime mobility with terrestrial farming in Cyprus and the Aegean led to leapfrogging Neolithic pioneer colonization along the Mediterranean to Iberia [58,61,63]. The increased pace of westwards expansion from Italy has suggested to some that dispersal and uptake was through the indigenous forager communities, but the renewed maritime emphasis, in a region where seafaring had been less pronounced in the Mesolithic, rather implies again an important role for leapfrog migration, as also suggested by our results [57,58]. Especially towards the west, many Mesolithic populations may have been hit hard by the ‘8.2 ka’ climatic downturn, whereas northern Italy acted as a refugium allowing longer survival for Mesolithic groups and a greater likelihood of ultimate assimilation into the Neolithic communities [56,58].
Our results reconcile the apparent conflict in the patterns seen in contemporary and aDNA studies, and suggest that the spread of the Neolithic into Europe may have been a much more complex process than sometimes envisioned in aDNA research. Indeed, the strong, hitherto unsuspected patterning in our results for the different regions of Europe, and the congruence with existing aDNA analyses for central/northern Europe, lends credence to our analytical approach.
The results also have implications for Iberia, where aDNA data for the onset of the Neolithic do exist but are less clear owing to the lack of decisive Mesolithic evidence across most of the peninsula (mainly based on the mtDNA control region, and with some potential contamination issues [64]). The mtDNA patterns appear to be distinctive [55,65,66], but Y-chromosome variation may point to immigration from further east [11,12], and there are very close similarities in the genome-wide signal for the central European LBK and the Iberian Early Neolithic [20]—although Chalcolithic Basque samples seem to carry a higher fraction of Mesolithic European ancestry [67].
For at least 15 years, the dominant paradigm for the arrival of the Neolithic in Iberia has been the ‘maritime pioneer colonization’ model [68], involving the immigration of new groups carrying the ‘Neolithic package’ and later acculturation or assimilation of Mesolithic groups to the new economy. Archaeological evidence for a dispersal of the Neolithic from Italy includes the spread of Cardial impressed ware, which appears to be several hundred years older in Italy (almost 8 ka) compared with Iberia (approx. 7.5 ka) [32,68]. It has more recently been argued that an earlier Impressa ware may be the direct sign of colonization from Liguria, with the Cardial ceramics marking the coalescence of incoming Neolithic and local Mesolithic groups, but there is little clear evidence for the precedence of the few Impressa ware deposits at present [32]. Recent archaeological assessments have moved away from the colonization model to an emphasis on exchange [32], but whatever the deficiencies of the radiocarbon record, our genetic evidence points to an important role for immigration to Iberia from central/eastern Mediterranean with the onset of the Neolithic. In a sense, our analysis of central Europe acts as a control for this conclusion, because both the aDNA and archaeological evidence point strongly to pioneer colonization for the LBK.
5. Conclusion
Gamble distinguished two very different zones within Europe—the ‘biotidal’ zone north of the continental divide, and the ‘refugium’ zone in the south, including the major glacial refuge areas of Iberia/southern France, Italy, and the Balkans and the east European plain [69]. He argued that human populations have washed through the biotidal zone repeatedly throughout prehistoric times, from sources in the continuously fertile refugium zone around the Mediterranean and Black Seas, where continuity and stability rather than rupture and replacement were the norm. This provides a richer framework for understanding European population history than simple dichotomies between Europe and the Near East. We argue that the genetic evidence now strongly favours such a model, while noting that Iberia appears to be a mosaic of the two zones, acting as both refuge and sink at different times in its history.
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank Dan Bradley, Anne Cambon–Thomsen, CEPH, Pierre-Marie Danze, Dimitar Dimitrov, Mukaddes Gölge, Oksana Naumova, CEPH, Steve Jones, Ariella Oppenheim, Surinder Papiha, Mark Thomas, and the donors for providing DNA samples.
Ethics
All individuals provided informed consent and the work was approved by the University of Leeds, Faculty of Biological Sciences Ethics Committee and that of the School of Applied Sciences, University of Huddersfield.
Data accessibility
The datasets supporting this article have been uploaded as part of the supplementary material, supplementary data S1, supplementary data S2, and supplementary data S3. The sequences data are deposited in GenBank under accessions numbers KX440187-KX440389.
Authors' contributions
M.B.R., L.P., and P.S. devised and supervised the project, J.B.P., M.D.C., and L.S. performed laboratory work, J.B.P., D.V., M.P., and P.S. performed data analyses, J.B.P. and M.B.R. wrote the manuscript, N.H., L.C., F.A., J.H., S.R., G.S., T.K., and A.T. discussed the results and helped revise the manuscript. All authors gave final approval for publication.
Competing interests
We have no competing interests.
Funding
FCT, the Portuguese Foundation for Science and Technology, supported this work through the research project PTDC/CS–ANT/113832/2009 and personal grants to J.B.P. (SFRH/BD/45657/2008) and M.D.C. (SFRH/BD/48372/2008). P.S. is supported by FCT, ESF, POPH, and the FCT Investigator Programme (IF/01641/2013) and acknowledges FCT I.P. and ERDF (through COMPETE2020-POCI) for CBMA's strategic programme UID/BIA/04050/2013 (POCI-01-0145-FEDER-007569). AT received support by the University of Pavia strategic theme ‘Towards a governance model for international migration: an interdisciplinary and diachronic perspective (MIGRAT.IN.G)’ and the Italian Ministry of Education, University and Research: Progetti Ricerca Interesse Nazionale 2012. M.B.R. received support from The Leverhulme Trust (research project grant no. F/10 105/D) and M.P. from the Newton International Fellowship scheme. This work was financed by FEDER-Fundo Europeu de Desenvolvimento Regional funds through COMPETE 2020-Operational Programme for Competitiveness and Internationalization (POCI), Portugal 2020, and by Portuguese funds through FCT/Ministério da Ciência, Tecnologia e Inovação in the framework of the project ‘Institute for Research and Innovation in Health Sciences’ (POCI-01-0145-FEDER-007274).
References
- 1.Reingruber A, Thissen L. 2009. Depending on 14C data: chronological frameworks in the Neolithic and Chalcolithic of Southeastern Europe. Radiocarbon 51, 751–770. ( 10.2458/azu_js_rc.51.3531) [DOI] [Google Scholar]
- 2.Manning K, Timpson A, Colledge S, Crema E, Edinborough K, Kerig T, Shennan S. 2014. The chronology of culture: a comparative assessment of European Neolithic dating approaches. Antiquity 88, 1065–1080. ( 10.1017/S0003598X00115327) [DOI] [Google Scholar]
- 3.Brami MN. 2015. A graphical simulation of the 2,000-year lag in Neolithic occupation between Central Anatolia and the Aegean basin. Archaeol. Anthropol. Sci. 7, 319–327. ( 10.1007/s12520-014-0193-4) [DOI] [Google Scholar]
- 4.Shennan S, Edinborough K. 2007. Prehistoric population history: from the Late Glacial to the Late Neolithic in Central and Northern Europe. J. Archaeol. Sci. 34, 1339–1345. ( 10.1016/j.jas.2006.10.031) [DOI] [Google Scholar]
- 5.Haak W, et al. 2005. Ancient DNA from the first European farmers in 7500-year-old Neolithic sites. Science (New York, NY) 310, 1016–1018. ( 10.1126/science.1118725) [DOI] [PubMed] [Google Scholar]
- 6.Haak W, et al. 2010. Ancient DNA from European Early Neolithic farmers reveals their Near Eastern affinities. PLoS Biol. 8, e1000536 ( 10.1371/journal.pbio.1000536) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bramanti B, et al. 2009. Genetic discontinuity between local hunter-gatherers and central Europe's first farmers. Science (New York, NY) 326, 137–140. ( 10.1126/science.1176869) [DOI] [PubMed] [Google Scholar]
- 8.Brandt G, et al. 2013. Ancient DNA reveals key stages in the formation of central European mitochondrial genetic diversity. Science (New York, NY) 342, 257–261. ( 10.1126/science.1241844) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brandt G, Szecsenyi-Nagy A, Roth C, Alt KW, Haak W. 2015. Human paleogenetics of Europe--the known knowns and the known unknowns. J. Hum. Evol. 79, 73–92. ( 10.1016/j.jhevol.2014.06.017) [DOI] [PubMed] [Google Scholar]
- 10.Deguilloux MF, Leahy R, Pemonge MH, Rottier S. 2012. European neolithization and ancient DNA: an assessment. Evol. Anthropol. 21, 24–37. ( 10.1002/evan.20341) [DOI] [PubMed] [Google Scholar]
- 11.Lacan M, Keyser C, Ricaut FX, Brucato N, Duranthon F, Guilaine J, Crubezy E, Ludes B. 2011. Ancient DNA reveals male diffusion through the Neolithic Mediterranean route. Proc. Natl Acad. Sci. USA 108, 9788–9791. ( 10.1073/pnas.1100723108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lacan M, Keyser C, Ricaut FX, Brucato N, Tarrus J, Bosch A, Guilaine J, Crubezy E, Ludes B. 2011. Ancient DNA suggests the leading role played by men in the Neolithic dissemination. Proc. Natl Acad. Sci. USA 108, 18 255–18 259. ( 10.1073/pnas.1113061108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gamba C, et al. 2014. Genome flux and stasis in a five millennium transect of European prehistory. Nat. Commun. 5, 5257 ( 10.1038/ncomms6257) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haak W, et al. 2015. Massive migration from the steppe is a source for Indo-European languages in Europe. Nature 522, 207–211. ( 10.1038/nature14317) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skoglund P, Malmstrom H, Raghavan M, Stora J, Hall P, Willerslev E, Gilbert MT, Gotherstrom A, Jakobsson M. 2012. Origins and genetic legacy of Neolithic farmers and hunter-gatherers in Europe. Science (New York, NY) 336, 466–469. ( 10.1126/science.1216304) [DOI] [PubMed] [Google Scholar]
- 16.Lazaridis I, et al. 2014. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature 513, 409–413. ( 10.1038/nature13673) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szecsenyi-Nagy A, et al. 2015. Tracing the genetic origin of Europe's first farmers reveals insights into their social organization. Proc. R. Soc. B 282, 20150339 ( 10.1098/rspb.2015.0339) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pinhasi R, Thomas MG, Hofreiter M, Currat M, Burger J. 2012. The genetic history of Europeans. Trends Genet. 28, 496–505. ( 10.1016/j.tig.2012.06.006) [DOI] [PubMed] [Google Scholar]
- 19.Sanchez-Quinto F, et al. 2012. Genomic affinities of two 7,000-year-old Iberian hunter-gatherers. Curr. Biol. 22, 1494–1499. ( 10.1016/j.cub.2012.06.005) [DOI] [PubMed] [Google Scholar]
- 20.Mathieson I, et al. 2015. Genome-wide patterns of selection in 230 ancient Eurasians. Nature 528, 499–503. ( 10.1038/nature16152) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Omrak A, et al. 2016. Genomic evidence establishes Anatolia as the source of the European Neolithic gene pool. Curr. Biol. 26, 270–275. ( 10.1016/j.cub.2015.12.019) [DOI] [PubMed] [Google Scholar]
- 22.Lazaridis I, et al. 2016. Genomic insights into the origin of farming in the ancient Near East. Nature 536, 419–424. ( 10.1038/nature19310) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hofmanová Z, et al. 2016. Early farmers from across Europe directly descended from Neolithic Aegeans. Proc. Natl Acad. Sci. USA 113, 6886–6891. ( 10.1073/pnas.1523951113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pala M, et al. 2012. Mitochondrial DNA signals of Late Glacial recolonization of Europe from Near Eastern refugia. Am. J. Hum. Genet. 90, 915–924. ( 10.1016/j.ajhg.2012.04.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olivieri A, et al. 2013. Mitogenomes from two uncommon haplogroups mark Late Glacial/postglacial expansions from the Near East and Neolithic dispersals within Europe. PLoS ONE 8, e70492 ( 10.1371/journal.pone.0070492) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gandini F, et al. 2016. Mapping human dispersals into the Horn of Africa from Arabian Ice Age refugia using mitogenomes. Sci. Rep. 6, 25472 ( 10.1038/srep25472) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu Q, et al. 2016. The genetic history of Ice Age Europe. Nature 534, 200–205. ( 10.1038/nature17993) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soares P, et al. 2009. Correcting for purifying selection: an improved human mitochondrial molecular clock. Am. J. Hum. Genet. 84, 740–759. ( 10.1016/j.ajhg.2009.05.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richards M, et al. 2000. Tracing European founder lineages in the Near Eastern mtDNA pool. Am. J. Hum. Genet. 67, 1251–1276. ( 10.1016/S0002-9297(07)62954-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soares PA, et al. 2016. Resolving the ancestry of Austronesian-speaking populations. Hum. Genet. 135, 309–326. ( 10.1007/s00439-015-1620-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rito T, Richards MB, Fernandes V, Alshamali F, Cerny V, Pereira L, Soares P. 2013. The first modern human dispersals across Africa. PLoS ONE 8, e80031 ( 10.1371/journal.pone.0080031.eCollection2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berrocal MC. 2012. The Early Neolithic in the Iberian Peninsula and the Western Mediterranean: a review of the evidence on migration. J. World Prehist. 25, 123–156. ( 10.1007/s10963-012-9059-9) [DOI] [Google Scholar]
- 33.Martins H, Oms FX, Pereira L, Pike AWG, Rowsell K, Zilhão J. 2015. Radiocarbon dating the beginning of the Neolithic in Iberia: new results, new problems. J. Mediterranean Archaeol. 28, 105–131. ( 10.1558/jmea.v28i1.27503) [DOI] [Google Scholar]
- 34.Soares P, et al. 2008. Climate change and postglacial human dispersals in southeast Asia. Mol. Biol. Evol. 25, 1209–1218. ( 10.1093/molbev/msn068) [DOI] [PubMed] [Google Scholar]
- 35.Hernandez CL, et al. 2015. Early Holocenic and historic mtDNA African signatures in the Iberian Peninsula: the Andalusian region as a paradigm. PLoS ONE 10, e0139784 ( 10.1371/journal.pone.0139784) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cavalli-Sforza LL, Menozzi P, Piazza A. 1993. Demic expansions and human evolution. Science 259, 639–646. ( 10.1126/science.8430313) [DOI] [PubMed] [Google Scholar]
- 37.Richards M, et al. 1996. Paleolithic and Neolithic lineages in the European mitochondrial gene pool. Am. J. Hum. Genet. 59, 185–203. [PMC free article] [PubMed] [Google Scholar]
- 38.Soares P, Achilli A, Semino O, Davies W, Macaulay V, Bandelt H-J, Torroni A, Richards MB. 2010. The archaeogenetics of Europe. Curr. Biol. 20, R174–R183. ( 10.1016/j.cub.2009.11.054) [DOI] [PubMed] [Google Scholar]
- 39.Huyghe JR, et al. 2011. A genome-wide analysis of population structure in the Finnish Saami with implications for genetic association studies. Eur. J. Hum. Genet. 19, 347–352. ( 10.1038/ejhg.2010.179) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pala M, Chaubey G, Soares P, Richards MB. 2014. The archaeogenetics of European ancestry. In Encyclopedia of life sciences (ELS), Chichester, UK: John Wiley and Sons. [Google Scholar]
- 41.Posth C, et al. 2016. Pleistocene mitochondrial genomes suggest a single major dispersal of non-Africans and a Late Glacial population turnover in Europe. Curr. Biol. 26, 827–833. ( 10.1016/j.cub.2016.01.037) [DOI] [PubMed] [Google Scholar]
- 42.Richards MB, Soares P, Torroni A. 2016. Palaeogenomics: mitogenomes and migrations in Europe's past. Curr. Biol. 26, R243–R246. ( 10.1016/j.cub.2016.01.044) [DOI] [PubMed] [Google Scholar]
- 43.Malmstrom H, et al. 2009. Ancient DNA reveals lack of continuity between Neolithic hunter-gatherers and contemporary Scandinavians. Curr. Biol. 19, 1758–1762. ( 10.1016/j.cub.2009.09.017) [DOI] [PubMed] [Google Scholar]
- 44.Lacan M, Keyser C, Crubezy E, Ludes B. 2013. Ancestry of modern Europeans: contributions of ancient DNA. Cell. Mol. Life Sci. 70, 2473–2487. ( 10.1007/s00018-012-1180-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keller A, et al. 2012. New insights into the Tyrolean Iceman's origin and phenotype as inferred by whole-genome sequencing. Nat. Commun. 3, 698 ( 10.1038/ncomms1701) [DOI] [PubMed] [Google Scholar]
- 46.Sikora M, et al. 2014. Population genomic analysis of ancient and modern genomes yields new insights into the genetic ancestry of the Tyrolean Iceman and the genetic structure of Europe. PLoS Genet. 10, e1004353 ( 10.1371/journal.pgen.1004353) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones ER, et al. 2015. Upper Palaeolithic genomes reveal deep roots of modern Eurasians. Nat. Commun. 6, 8912 ( 10.1038/ncomms9912) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Allentoft ME, et al. 2015. Population genomics of Bronze Age Eurasia. Nature 522, 167–172. ( 10.1038/nature14507) [DOI] [PubMed] [Google Scholar]
- 49.Pala M, Soares P, Richards MB. 2016. Archaeogenetic and palaeogenetic evidence for Metal Age mobility in Europe. In Celtic from the West 3: Atlantic Europe in the Metal Ages: questions of shared language (eds Koch JT, Cunliffe B, Cleary K, Gibson CD), pp. 351–384. Oxford, UK: Oxbow Books. [Google Scholar]
- 50.Costa MD, et al. 2013. A substantial prehistoric European ancestry amongst Ashkenazi maternal lineages. Nat. Commun. 4, 2543 ( 10.1038/ncomms3543) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Torroni A, et al. 1998. mtDNA analysis reveals a major late Paleolithic population expansion from southwestern to northeastern Europe. Am. J. Hum. Genet. 62, 1137–1152. ( 10.1086/301822) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Torroni A, et al. 2001. A signal, from human mtDNA, of postglacial recolonization in Europe. Am. J. Hum. Genet. 69, 844–852. ( 10.1086/323485) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pereira L, et al. 2005. High-resolution mtDNA evidence for the late-glacial resettlement of Europe from an Iberian refugium. Genome Res. 15, 19–24. ( 10.1101/gr.3182305) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Achilli A, et al. 2004. The molecular dissection of mtDNA haplogroup H confirms that the Franco-Cantabrian glacial refuge was a major source for the European gene pool. Am. J. Hum. Genet. 75, 910–918. ( 10.1086/425590) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hervella M, Izagirre N, Alonso S, Fregel R, Alonso A, Cabrera VM, de la Rua C. 2012. Ancient DNA from hunter–gatherer and farmer groups from Northern Spain supports a random dispersion model for the Neolithic expansion into Europe. PLoS ONE 7, e34417 ( 10.1371/journal.pone.0034417) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Berger JF, Guilaine J. 2009. The 8200 cal BP abrupt environmental change and the Neolithic transition: a Mediterranean perspective. Quat. Int. 200, 31–49. ( 10.1016/j.quaint.2008.05.013) [DOI] [Google Scholar]
- 57.Zilhão J. 2000. From the Mesolithic to the Neolithic in the Iberian Peninsula. In Europe’s first farmers (ed. Price TD.), pp. 144–182. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 58.Broodbank C. 2013. The making of the middle sea: a history of the Mediterranean from the beginning to the emergence of the classical world. London, UK: Thames & Hudson. [Google Scholar]
- 59.Binder D. 2000. Mesolithic and Neolithic interaction in southern France and northern Italy: new data and current hypotheses. In Europe's first farmers (ed. Price TD.), pp. 117–143. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 60.Rowley-Conwy PA, Gourichon L, Helmer D, Vigne J-D. 2013. Early domestic animals in Italy, Istria, the Tyrrhenian Islands, and Southern France. In The Origins and Spread of Domestic Animals in Southwest Asia and Europe (eds Colledge S, Conolly J, Dobney K, Manning K, Shennan S), pp. 161–194. Walnut Creek, CA: Left Coast Press. [Google Scholar]
- 61.Perlès C. 2001. The Early Neolithic in Greece. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 62.Barker G. 2005. Agriculture, pastoralism and Mediterranean landscapes in prehistory. In The archaeology of Mediterranean prehistory (eds Blake E, Knapp AB). Oxford, UK: Blackwell. [Google Scholar]
- 63.Price TD. 2000. Europe's first farmers. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 64.Chandler H, Sykes B, Zilhão J. 2005. Using ancient DNA to examine genetic continuity at the Mesolithic-Neolithic transition in Portugal. In Congreso del Neolítico en la Península Ibérica (eds Arias P, Ontañón R, García-Moncó C), pp. 781–786. Santander, UK: Monografías del Instituto Internacional de Investigaciones Prehistóricas de Cantabria. [Google Scholar]
- 65.Sampietro ML, Lao O, Caramelli D, Lari M, Pou R, Marti M, Bertranpetit J, Lalueza-Fox C. 2007. Palaeogenetic evidence supports a dual model of Neolithic spreading into Europe. Proc. R. Soc. B 274, 2161–2167. ( 10.1098/rspb.2007.0465) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gamba C, et al. 2012. Ancient DNA from an Early Neolithic Iberian population supports a pioneer colonization by first farmers. Mol. Ecol. 21, 45–56. ( 10.1111/j.1365-294X.2011.05361.x) [DOI] [PubMed] [Google Scholar]
- 67.Gunther T, et al. 2015. Ancient genomes link early farmers from Atapuerca in Spain to modern-day Basques. Proc. Natl Acad. Sci. USA 112, 11 917–11 922. ( 10.1073/pnas.1509851112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zilhão J. 2001. Radiocarbon evidence for maritime pioneer colonization at the origins of farming in west Mediterranean Europe. Proc. Natl Acad. Sci. USA 98, 14 180–14 185. ( 10.1073/pnas.241522898) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gamble C. 2009. Human display and dispersal: a case study from biotidal Britain in the Middle and Upper Pleistocene. Evol. Anthropol. 18, 144–156. ( 10.1002/evan.20209) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting this article have been uploaded as part of the supplementary material, supplementary data S1, supplementary data S2, and supplementary data S3. The sequences data are deposited in GenBank under accessions numbers KX440187-KX440389.