Abstract
This paper presents a history of the changing meanings of the term “gene,” over more than a century, and a discussion of why this word, so crucial to genetics, needs redefinition today. In this account, the first two phases of 20th century genetics are designated the “classical” and the “neoclassical” periods, and the current molecular-genetic era the “modern period.” While the first two stages generated increasing clarity about the nature of the gene, the present period features complexity and confusion. Initially, the term “gene” was coined to denote an abstract “unit of inheritance,” to which no specific material attributes were assigned. As the classical and neoclassical periods unfolded, the term became more concrete, first as a dimensionless point on a chromosome, then as a linear segment within a chromosome, and finally as a linear segment in the DNA molecule that encodes a polypeptide chain. This last definition, from the early 1960s, remains the one employed today, but developments since the 1970s have undermined its generality. Indeed, they raise questions about both the utility of the concept of a basic “unit of inheritance” and the long implicit belief that genes are autonomous agents. Here, we review findings that have made the classic molecular definition obsolete and propose a new one based on contemporary knowledge.
Keywords: gene, structure, function, gene networks, regulation, theory
IN 1866, Gregor Mendel, a Moravian scientist and Augustinian friar, working in what is today the Czech Republic, laid the foundations of modern genetics with his landmark studies of heredity in the garden pea (Pisum sativum) (Mendel 1866). Though he did not speak of “genes”—a term that first appeared decades later—but rather of elements, and even “cell elements” (original German Zellelemente p. 42), it is clear that Mendel was hypothesizing the hereditary behavior of miniscule hidden factors or determinants underlying the stably inherited visible characteristics of an organism, which today we would call genes. This is apparent throughout his publication in his use of abstract letter symbols for hereditary determinants to denote the physical factors underlying the inheritance of characteristics. There is no doubt that he considered the mediators of heredity to be material entities, though he made no conjectures about their nature.
The word “gene” was not coined until early in the 20th century, by the Danish botanist Johannsen (1909), but it rapidly became fundamental to the then new science of genetics, and eventually to all of biology. Its meaning, however, has been evolving since its birth. In the beginning, the concept was used as a mere abstraction. Indeed, Johannsen thought of the gene as some form of calculating element (a point to which we will return), but deliberately refrained from speculating about its physical attributes (Johannsen 1909). By the second decade of the 20th century, however, a number of genes had been localized to specific positions on specific chromosomes, and could, at least, be treated, if not thought of precisely, as dimensionless points on chromosomes. Furthermore, groups of genes that showed some degree of coinheritance could be placed in “linkage groups,” which were the epistemic equivalent of the cytological chromosome. We term this phase the “classical period” of genetics. By the early 1940s, certain genes had been shown to have internal structure, and to be dissectable by genetic recombination; thus, the gene, at this point, had conceptually acquired a single dimension, length. Twenty years later, by the early 1960s, the gene had achieved what seemed like a definitive physical identity as a discrete sequence on a DNA molecule that encodes a polypeptide chain. At this point, the gene had a visualizable three-dimensional structure as a particular kind of molecule. We will call this period—from roughly the end of the 1930s to the early 1960s—the “neoclassical period.”
The 1960s definition of the gene is the one most geneticists employ today, but it is clearly out-of-date for deoxyribonucleic acid (DNA)-based organisms. (We will deal only with the latter; RNA viruses and their genes will not be discussed.) Here, we review the older history of the terminology, and then the findings from the 1970s onwards that have undermined the generality of the 1960s definition. We will then propose a contemporary definition of the “gene” that accounts for the complexities revealed in recent decades. This publication is a follow-on paper to an earlier paper by one of us (Portin 2015).
The Classical Period of Genetics
The development of modern genetics began in 1900, when three botanists—the German Carl Correns, the Dutchman Hugo de Vries, and the Austrian Erich von Tschermak—independently cited and discussed the experiments of Mendel as basic to understanding the nature of heredity. They presented results similar to Mendel’s though using different plants as experimental material (Correns 1900; de Vries 1900; Tschermak 1900). Their conceptual contributions as “rediscoverers” of Mendel, however, were probably not equivalent. De Vries and Correns claimed that they had discovered the essential facts and developed their interpretation before they found Mendel’s article, and they demonstrated that they fully understood the essential aspects of Mendel’s theory (Stern 1970). In contrast, Tschermak’s analysis of his own data was inadequate, and his paper lacked an interpretation. Thus, while he sensed the significance of Mendel’s work, Tschermak should not be given credit equal to that due to de Vries and Correns.
In 1900, chromosomes were already known, and they were soon seen to provide a concrete basis for Mendel’s abstract hereditary factors. This postulated connection between genes and chromosomes, which later came to be known as the chromosome theory of inheritance, was initially provided by the German biologist T. H. Boveri and the American geneticist and physician W. S. Sutton during the years 1902–1903. Boveri first demonstrated the individuality of chromosomes with microscopic observations on the sea urchin Paracentrotus lividus (Boveri 1902). He went on to demonstrate the continuity of chromosomes through cell divisions with studies of Ascaris megalocephala, a parasitic nematode worm (Boveri 1903). These two characteristics—individuality and continuity—are necessary, although not sufficient, characteristics of the genetic material. Sutton’s contribution (Sutton 1903), on the basis of his studies on the spermatogenesis of Brachystola magna, a large grasshopper, was to demonstrate a clear equivalence between the behavior of chromosomes at the meiotic divisions and Mendel’s postulated separation and independent inheritance of character differences at gamete formation. Thus, this early version of the chromosome theory of inheritance suggests an explanation for Mendel’s laws of inheritance: the law of segregation and the law of independent assortment. It was not until 1916, however, that it could be considered to be proven. In that year, C. B. Bridges, an American geneticist, showed in Drosophila melanogaster that nondisjunction, a rare exceptional behavior of genetic makers (lack of segregation) during gamete formation, was always associated with an analogous exceptional behavior of a given chromosome pair during meiosis (Bridges 1916).
Shortly after the birth of the chromosome theory, however, a new phenomenon had been discovered that appeared to contradict Mendel’s law of independent assortment. This was the phenomenon of linkage, initially found in the sweet pea (Lathyrus odoratus), in which some genes were found to exhibit “coupling,” violating independent assortment (Bateson et al. 1905a,b). This exception to the rule, however, became the basis of an essential extension of the chromosome theory when it was realized that genes showing linkage are located on the same chromosome, and genes showing independent assortment are located on different chromosomes.
According to the canonical history of genetics, it was the American geneticist T. H. Morgan who was the first to propose in 1910 this extension of the chromosome theory (Morgan 1910, 1917). Recent studies on the history of genetics (Edwards 2013), however, show that, most likely, Morgan was influenced by the first textbook of genetics in English written by R. H. Lock, a British botanist associated with Bateson and Punnet, published in 1906, where the possibility that linkage might result from genes lying on the same chromosome was first suggested (Lock 1906). Thus, it is Lock to whom the credit of explaining linkage must be given.
It was soon understood that genes sufficiently far apart on the chromosome can also show independent assortment, due to extensive genetic recombination during meiosis, while genes that are closer to each other show a degree of coinheritance, the frequency of their separation by recombination being directly related to the distance between them. Owing to the work of Morgan and his group on the fruit fly (D. melanogaster), the phenomenon of linkage and its breakdown via crossing over became the essential basis for the mapping of genes (Morgan 1919, 1926; Morgan et al. 1915). The first map, of the Drosophila X-chromosome, was constructed by Alfred Sturtevant, one of Morgan’s students (Sturtevant 1913). The linear sequence of genes he diagrammed was the abstract genetic epistemic equivalent of the chromosome itself.
The genetic maps of the linkage groups were subsequently followed by cytological maps of the chromosomes. These were first constructed by showing that X-ray-induced changes of the order of the genes in the linkage groups, such as translocations and deletions, were associated with corresponding changes in the structure of chromosomes (Dobzhansky 1929; Muller and Painter 1929; Painter and Muller 1929). This was followed by detailed cytological mapping of genes, made possible by the existence of the “giant” chromosomes of the salivary glands of the fruit fly, in which genes identified by their inheritance patterns could be localized to specific (visible) locations on chromosomes (Painter 1934; Bridges 1935, 1938).
Morgan conceived the cytological explanation for the genetical phenomenon of crossing over by adopting the chiasmatype theory of Frans Alfons Jannsens, a Belgian cytologist, that was based on his observations of meiosis at spermatogenesis in the salamander Batrachoseps attenuatis (Janssens 1909; see also Koszul et al. 2012). Janssens observed cross figures at synapsis in meiotic chromosome preparations of this amphibian that resembled the Greek letter chi (χ). Accordingly, he called such a junction “chiasma” (pl. chiasmata). Janssens interpreted each of these as due to fusion at one point between two of the four strands of the tetrad of chromatids at the pachytene stage of the meiotic prophase. According to the chiasmatype theory, chiasmata were due to breakage and reunion of one maternal and one paternal chromatid of the tetrad. Consequently, the formation of each chiasma leads to an exchange of equal and corresponding regions of two of the four chromatids. This mechanism of exchange provided the needed physical explanation for the partial genetic linkage of genes that Morgan had observed. In other words, chiasmata are cytological counterparts of the genetical crossover points.
An alternative explanation for the origin of chiasmata was the so called classical hypothesis, which did not require breakage and rejoining of chromosomes, but assumed that chiasmata were simply a result of the paternal and maternal chromatids going across each other, forming a cross-like configuration at the pachytene stage of meiosis (McClung 1927; Sax 1932a,b). This hypothesis did not explain the phenomenon of genetic recombination, but was preferred by most cytologists of that time because it did not threaten the permanence and individuality of the chromosomes, which the chiasmatype theory initially seemed to do. During subsequent years, many cytological facts, as reviewed, for example, in Whitehouse (1973), supported the chiasmatype theory, but not the classical theory.
Thus, by the early 1930s, the concept of the gene had become more concrete. Genes were regarded as indivisible units of inheritance, each located at a specific point on a specific chromosome. Furthermore, they could be defined in terms of their behavior as fundamental units on the basis of four criteria: (1) hereditary transmission, (2) genetic recombination, (3) mutation, and (4) gene function. Moreover, it was believed, albeit without any empirical evidence, that these four ways of defining the gene fully agreed with one another (reviewed in Portin 1993; Keller 2000). As A. Sturtevant and G. W. Beadle wrote in (1939), near the end of what we are calling the classical period of genetics, it was also clear that genes determine the nature of developmental reactions and thus, ultimately, the visible traits they generate. But how genes do these things was unknown; indeed, that was considered one of the major unsolved problems in biology, and it remained so for two decades (Sturtevant and Beadle 1962, p. 335). Further, it was believed that the integration of genetics with such fields as biochemistry, developmental physiology, and experimental embryology would lead to a deep understanding of the nature and role of genes, and that this integration would add to our understanding of those processes that make up development (Sturtevant and Beadle 1962, p. 357; see also Sturtevant 1965).
The significance of this perspective was initially elaborated by H. J. Muller, an American geneticist and a student of Morgan’s, who had done important work on several key aspects of the subject: the mapping of genes (Muller 1920), the relation between genes and characteristics of organisms (Muller 1922), and the nature of gene mutation (Muller 1927; also see Carlson 1966). In his classic paper dealing with the effect of changes in individual genes on the variation of the organism, Muller (1922) published arguments that can be viewed as a theoretical summary of the essence of the classical period of genetics. On the basis of a considerable body of earlier work, he put forward an influential theory that genes are molecules with three essential capacities: autocatalysis (self-reproduction), heterocatalysis (production of nongenetic material or effects), and ability to mutate (while retaining the first two properties). In this view, genes were undoubted physical entities, three-dimensional ultramicroscopic ones, possessing individuated heritable structures, with some capacity for change that itself could be passed on.
In another visionary paper, Muller (1926) connected the concept of the gene to the theory of evolution, while he described the gene as the basis of evolution and the origin of life itself, indeed as the basis of life itself. These profound views of Muller strongly influenced the direction of much future research, not only in genetics, but in biology as a whole (Carlson 1966 p. 82).
The Neoclassical Period of Genetics
Whatever the speculations of Muller and a few others, the classical period of genetics was one in which the gene could be treated effectively as a dimensionless point on a chromosome. It was followed, however, by what we are calling the neoclassical period, in which the gene first acquired an unambiguous spatial dimension, namely length, and later a likewise linear chemical identity, in the form of the DNA molecule. This period of genetics involved two different, but complementary, research programs: on the one hand, it was demonstrated, using the classic genetic tool of recombinational mapping, that genes have an internal structure; on the other hand, the basic molecular nature of the gene and its function began to be revealed. These two streams fused in the late 1950s.
The neoclassical period began in the early 1940s, with work in formal genetics showing that genes could be dissected into contiguous segments by genetic recombination. Hence, they were not dimensionless points but entities with length. These observations were made first in D. melanogaster (Oliver 1940; Lewis 1941; 1945; Green and Green 1949), and then in microbial fungi (Bonner 1950; Giles 1952; Pontecorvo 1952; Pritchard 1955).
If genes had length, however, they must be long molecules of some sort, and the question was whether those molecules were proteins or DNA, the two major molecular constituents of the chromosomes. Critically important work in the early 1940s, in the laboratory of Oswald Avery at Rockefeller University, answered the question. Avery and his colleagues showed that DNA is the hereditary material by demonstrating that the causative agent in bacterial transformation, which entailed a heritable change in the morphology of the bacterial cells (Griffith 1928), was DNA (Avery et al. 1944). Though this work was published in 1944, it would take nearly a decade for this to become universally accepted. The experimental proof that convinced the scientific community was the experiment of Hershey and Chase (1952), in which these authors showed that the DNA component of bacteriophages was the one responsible for their multiplication.
The most critical and final breakthrough for the DNA theory of inheritance, however, was the revelation of the double-helical structure of DNA (Watson and Crick 1953a, 1954), and the realization of the genetic implications of that (Watson and Crick 1953b). This was followed by demonstrations in the early 1960s that genes are first transcribed into messenger RNA (mRNA), which transmitted the genetic information from the nucleus to the protein synthesis machinery in the cytoplasm (reviewed in Portin 1993; Judson 1996). Earlier work in the 1940s had established the connection between genes and proteins, in the “one gene-one enzyme” hypothesis of Beadle and Tatum (1941) (see also Srb and Horowitz 1944; reviewed in Strauss 2016). By the late 1950s, there was thus a satisfying molecular theory of both the nature of the gene, and the connections between genes and proteins.
Crucial further work involved the genetic fine structure mapping of genes—a research program that reached its culmination with work by S. Benzer and C. Yanofsky. Benzer, using the operational cis-trans test, originated by E. B. Lewis in Drosophila, defined the unit of genetic complementation, i.e., the basic unit of gene function, which he called the cistron (Box 1). He also defined the smallest units of genetic recombination and gene mutation: the recon and muton, respectively (Benzer 1955, 1959, 1961). The postulate of the classical period that the gene was a fundamental unit not only of function, but also of recombination and mutation, was definitively disproved by Benzer’s work showing that the “gene” had many mutons and recons. Yanofsky and his coworkers validated the material counterparts of these formal concepts of Benzer. The equivalent of the cistron is a sequence of nucleotide pairs in DNA that contain information for the synthesis of a polypeptide, and determines its amino acid sequences, an idea known as the colinearity hypothesis. Furthermore, the physical DNA equivalent of the recon and the muton was shown to be one nucleotide pair (Crawford and Yanofsky 1958; Yanofsky and Crawford 1959; Yanofsky et al. 1964, 1967). The period of neoclassical genetics culminated in the cracking of the universal genetic code by several teams, revealing that nucleotide sequences specify the sequence of polypeptide chains (reviewed in Ycas 1969; Judson 1996).
The neoclassical concept of the gene, outlined above, can be summarized in the formulation “one gene—one mRNA—one polypeptide,” which combines the idea of mRNA, as developed by Jacob and Monod (1961a); Gros et al. (1961); Brenner et al. (1961), and the earlier “one gene—one enzyme” hypothesis of Beadle and Tatum (1941) (and see Srb and Horowitz 1944). Another version of this hypothesis is that of “one cistron—one polypeptide” (Crick 1963), which emerged as a slogan in the 1960s–1980s. Altogether, the conceptual journey from Johannsen’s totally abstract entities termed “genes” to a defined, molecular idea of what a gene is, and how it works, had taken a little over half a century.
The Breakdown of the Neoclassical Concept of the Gene and the Beginning of the Modern Period of Genetics
Deviations from the one gene—one mRNA—one polypeptide hypothesis
The hypothesis of “one gene—one mRNA—one polypeptide” as a general description of the gene and how it works started to expire, however, when it was realized that a single gene could produce more than one mRNA, and that one gene can be a part of several transcription units. This one-to-several relationship of genes to mRNAs occurs by means of complex promoters and/or alternative splicing of the primary transcript.
Multiple transcription initiation sites, i.e., alternative promoters, have been found in all kingdoms of organisms, and they have been classified into six classes (Schibler and Sierra 1987). All of them can produce transcripts that do not obey the rule of one-to-one correspondence between the gene and the transcription unit, since transcription can be initiated at different promoters. The result is that a single gene can produce more than one kind of transcript (Schibler and Sierra 1987).
The discovery of alternative splicing as a way of producing different transcripts from one gene had a more complex history. In the late 1970s it was discovered, first in animal viruses and then in eukaryotes, that genes have a split structure. That is, genes are interrupted by introns (see review by Portin 1993). Split genes produce one pre-mRNA molecule, from which the introns are removed during the maturation of the mRNA by pre-mRNA splicing. Depending on the gene, the splicing pattern can be invariant (“constitutive”) or variable (“alternative”). In constitutive splicing, all the exons present in a transcript are incorporated into one mature mRNA through invariant ligation of consecutive exons, yielding a single kind of mRNA from the gene. In alternative splicing, nonconsecutive exons are joined by the processing of some, but not all, transcripts from a gene. In other words, individual exons can be excluded from the mature mRNA in some transcripts, but they can be included in others (Leff et al. 1986; Black 2003). Alternative splicing is a regulated process, being tissue-specific and developmental-stage-specific. Nevertheless, the colinearity of the gene and the mRNA is preserved, since the order of the exons in the gene is not changed.
In addition to alternative splicing, two other phenomena are now known that contradict a basic tenet of the neoclassical gene concept, namely that amino-acid sequences of proteins, and consequently their functions, are always derivable from the DNA of the corresponding gene. These are the phenomena of RNA editing (reviewed by Brennickle et al. 1999; Witzany 2011) and of gene sharing originally found by J. Piatigorsky (reviewed in Piatigorsky 2007). The term RNA editing describes post-transcriptional molecular processes in which the structure of an RNA molecule is altered. Though a rare event, it has been observed to occur in eukaryotes, their viruses, archaea and prokaryotes, and involves several kinds of base modifications in RNA molecules. RNA editing in mRNAs effectively alters the amino acid sequence of the encoded protein so that it differs from that predicted by the genomic DNA sequence (Brennickle et.al. 1999). The concept of gene sharing describes the fact that different cells contain identically sequenced polypeptides, derived from the same gene, but so differently configured in different cellular contexts that they perform wildly different functions. This phenomenon, facetiously called “protein moonlighting,” means that a gene may acquire and maintain a second function without gene duplication, and without loss of the primary function. Such genes are under two or more entirely different selective constraints (Piatigorsky and Wistow 1989).
Despite these observations, showing the potential one-to-many relationships of genes to mRNAs and their encoded proteins, the concept of the gene remained intact; the gene itself could still be seen as a defined and localized nucleotide sequence of DNA even though it could contain information for more than one kind of polypeptide chain. Matters changed, however, when the sequencing projects revealed still more bizarre phenomena.
Severe cracks in the concept of the gene
These new findings have shown that there are multiple possible relationships between DNA sequences and the molecular products they specify. The net result has been the realization that the basic concept of the gene as some form of generic, universal “unit of heredity” is too simple, and correspondingly, that, a new definition or concept of “the gene” is needed (Keller 2000; Falk 2009; Portin 2009). Several observations have been crucial to this re-evaluation, and one of us has reviewed these relatively recently (Portin 2009). They are worth summarizing here:
In eukaryotic organisms, there are few if any absolute boundaries to transcription, making it impossible to establish simple general relationships between primary transcripts and the ultimate products of those transcripts.
Hence, the structural boundaries of the gene as the unit of transcription are often far from clear, as documented particularly well in mammals (reviewed by Carninci 2006). In reality, whole chromosomes, if not the whole genome, seem to be continuums of transcription (Gingeras 2007). Furthermore, the genome is full of overlapping transcripts, thus making it impossible to draw 1:1:1 relationship between specific DNA sequences, transcripts and functions (Pearson 2006). Indeed, convincing evidence indicates that the human genome is comprehensively transcribed from both DNA strands, so that the majority of its bases can be found in primary transcripts that compendiously overlap one another (The FANTOM Consortium and RIKEN Genome Exploration Group 2005; The ENCODE Project Consortium 2007; 2012). Both protein coding and noncoding transcripts may be derived from either or both DNA strands, and they may be overlapping and interlaced. Furthermore, different transcripts often include the same coding sequences (Mattick 2005). The functional significance of these overlaps is still largely unclear, but there is an increasing number of examples in which both transcripts are known to have protein-coding exons from one position in the genome combined with exons from another part of the genome hundreds of thousands of nucleotides away (Kapranov et al. 2007). This was wholly unanticipated when the 1960s definition of the gene was formulated.
2. Exons of different genes can be members of more than one transcript.
Gene fusion, at the level of transcripts, is a reality, and is completely at odds with the “one gene—one mRNA—one protein” hypothesis. And this is not a rare phenomenon. It has been estimated that at least 4–5% of the tandem gene pairs in the human genome can be transcribed into a single RNA sequence, called chimeric transcripts, encoding a putative chimeric protein (Parra et al. 2006).
3. Comparably, in the organelles of microbial eukaryotes, many examples of “encrypted” genes are known: genes are often in pieces that can be found as separate segments around the genome.
Hence, in addition to the fusion of two adjacent genes at the level of transcription, different building blocks of a given mRNA molecule can often be located, as modules, on different chromosomes (reviewed in Landweber 2007). Some evidence indicates that, even in multicellular eukaryotes, protein-coding transcripts are derived from different nonhomologous chromosomes (reviewed in Claverie 2005).
4. In contradiction to the neoclassical definition of a gene, which posits that the hereditary information resides solely in DNA sequences, there is increasing evidence that the functional status of some genes can be inherited from one generation of individuals to the next, a phenomenon known as transgenerational epigenetic inheritance (Holliday 1987; Gerhart and Kirschner 2007; Jablonka and Raz 2009).
One example is mouse epigenetic changes mediated by RNA that are inherited between generations in a non-Mendelian fashion (Rassoulzadegan et al. 2006). On the other hand, many of the epigenetic changes, or so called epimutations, are inherited otherwise in a Mendelian fashion, except that, in contrast to conventional mutations, they are not always inherited with the same stability, but can be swept away during the course of some generations (e.g., Jablonka and Raz 2009).
5. “Genetic restoration” a mechanism of non-Mendelian inheritance of extragenomic information, first found in Arabidopsis thaliana, may also take place (Lolle et al. 2005).
It was observed that several independent mutant strains yielded apparently normal progeny at a high frequency of a few percent, which is higher than could be expected if it were a question of random mutations. It seems neither to be a question of epigenetic changes, but rather healing of fixed mutations. Lolle et al. (2005) suggested that this is due to precise reversion of the original DNA with a mechanism that involves template-directed restoration of ancestral DNA passed on in an RNA cache. This phenomenon, called the “RNA cache” hypothesis, means that organisms can sometimes rewrite their DNA on the basis of RNA messages inherited from generations past (Lolle et al. 2005). The RNA cache hypothesis has, however, been disputed by several authors (Comai and Cartwright 2005; Mercier et al. 2008; Miyagawa et al. 2013).
6. Finally, in addition to protein coding genes, there are many RNA-encoding genes that produce diverse RNA molecules that are not translated to proteins.
That there are special genes that specify only RNA products was recognized in the early 1960s; these are the ribosomal RNA and tRNA genes, vital for protein synthesis. Yet, it is now apparent that there are many transcripts that do not encode proteins, and that are not the classic structural RNAs of protein synthesis (tRNAs and rRNAs). Those sequences that specify long noncoding RNAs (lncRNAs), and which serve some biological function, surely deserve to be called genes. In contrast, sequences specifying lncRNAs or transcripts from defunct mobile elements, which are made constitutively in all or most cell types probably do not have biological function and should not be designated as genes. The surprisingly large multitude of different noncoding RNA genes and their function has been reviewed by several authors (e.g., Eddy 2001; Carninci and Hayashizaki 2007; Carninci et al. 2008).
Current Status and Future Perspectives Regarding the Concept of the Gene
The observations summarized above, together with many others, have created the interesting situation that the central term of genetics— “the gene”—can no longer be defined in simple terms. The neoclassical molecular definition of the gene does not capture the bewildering variety of hereditary elements, all based in DNA, that collectively specify the organism, and which therefore deserve the appellation of “genes.” Even the classical notion of the gene simply as a fundamental “unit of heredity” is itself problematic. After all, if it is difficult or impossible to generalize about the nature of such “units,” it is probably not very helpful to speak about them. Unsurprisingly, this realization has called forth various attempts to redefine the gene, in terms of both DNA sequence properties, and those of the products specified by those sequences. A number of proposed definitions are listed in Table 1. A detailed discussion of these ideas will not be given here, but they have been summarized, classified, and characterized (see Waters 2013). These definitions, however, all tend to neglect one central, albeit implicit, aspect of the earlier notions of the “gene”: its presumed autonomy of action. We return to this matter below.
Table 1. Abridged list of different propositions for a definition of the gene in the current era given by different authors.
| Essential Content or Character of the Proposition | Classification | Author(s) |
|---|---|---|
| These three first operational definitions give criteria, formal, experimental and computational, for identifying genes in the DNA sequences of genomes, annotation of genomes, and for specifying the function of genes | Operational | Snyder and Gerstein (2003) |
| Operational | Pesole (2008) | |
| Operational | Stadler et al. (2009) | |
| In these three following definitions, classified as molecular, the structural and the functional gene are conceptually distinguished and separated | Molecular | Scherrer and Jost (2007) |
| Molecular | Keller and Harel (2007) | |
| Molecular | Burian (2004) | |
| In this definition two gene concepts, “gene-P (preformationist)” and “gene-D (developmental)”, are distinguished | Complex | Moss (2003) |
| This definition presents three different concepts of the gene: instrumental, nominal and postgenomic | Complex | Griffiths and Stotz (2006) |
| This definition aims at to define the gene on the basis of its products and separates it from DNA | A new kind of redefinition | Waters (1994) |
How should geneticists deal with this situation? Should we simply invoke a plurality of different kinds of genes and leave it at that? In effect, we could settle for using the collective term “the genes” as a synonym for the genome, and not fuss over the seeming impossibility of defining the singular form, the “gene.” This, however, would seem to be more of an evasion of the problem than its solution. Alternatively, would it be preferable to accept the inadequacy of the notion of a simple general “unit of heredity,” and foreswear the use of the term “gene” altogether?
The problem with that last suggestion, junking the term “gene,” is not just that the word is used ubiquitously by geneticists and laymen alike, but that it seems indispensable to the discipline’s discourse. This is apparent in the foundations of several subdisciplines of genetics, such as many fields of applied genetics, like medical genetics and plant and animal breeding, that frequently deal in genes identified solely by their nonmolecular mutant phenotypes. It also applies to quantitative genetics and population genetics, which operate using mathematical modeling, and in which the gene is often regarded merely as an abstract unit of calculation (not dissimilarly to the view of Johannsen described below), but one that is vital to conceptualizing the genetic compositions of populations and their changes. In those fields, the molecular intricacies and complications of the genetic material can be largely ignored, at least initially, but the term “gene” itself seems irreplaceable. It is hard to imagine those disciplines abandoning it, whatever the range of molecular complexities that the word both hides and embraces.
In other subdisciplines, such as developmental genetics and molecular genetics, however, there is an urgent need to redefine the gene because the molecular details are often crucial to understanding the phenomena being investigated. The definitions that have been attempted so far (Table 1), however, seem inadequate; for the most part, they focus on either structural or functional aspects, yet it is ultimately meaningless to separate structure and function, even though both can initially be studied in isolation from one another. One attempt to unite the structural and functional aspects of the gene in a single definition has been made by P. E. Griffiths and E. M. Neumann-Held, who introduced the “molecular process” gene concept. In this idea, the word “gene” denotes not some structural “unit of heredity” but the recurring process that leads to the temporally and spatially regulated expression of a particular polypeptide product (Griffiths and Neumann-Held 1999; Neumann-Held 1999, 2001). One difficulty with this redefinition is that it neglects all the nonconventional genes that specify only RNA products. More fundamentally, it has nothing to say about hereditary transmission, which was the original and fundamental impetus for coining the term “gene.”
Perhaps the way forward is to take a step backward in history, and focus on the initial concerns of Johannsen. He not only coined the term “gene,” but was also responsible for the words “genotype” and “phenotype,” and the crucial distinction between them in heredity. Though he could say nothing about how genes (genotype) specified or determined traits (phenotype), he clearly saw this as a crucial question. Indeed, that issue has been at the heart of genetics since the 1930s, in contrast to the questions about how genes are transmitted in heredity, which dominated the first decades of 20th-century genetics. It is apparent, however, that Johannsen thought that the genotype is primary, and that genes are minute computational devices whose precise material nature could be left for solution to a later time. He wrote: “Our formulas, as used here for not directly observable genotypic factors—genes as we used to say—are and remain computational-formulas, placement-devices that should facilitate our overview. It is precisely therefore that the little word “gene” is in place; no imagination of the nature of this “construction” is prejudiced by it, rather the different possibilities remain open from case to case.” (Johannsen 1926 p. 434, English translation in Falk 2009 p. 70).
The initial expectations were that the connections between genes and phenes would be fairly direct, an expectation bolstered initially by findings about pigmentation genetics, and later by mutations affecting nutritional requirements in microbial cells. In both situations, the connection between the mutant effects and the known biochemistry were often direct and easy to understand. Furthermore, the early success of Mendelian genetics had been based, in large part, on the fact that many of the genetic variants initially studied had constant, unambiguous effects; this was vital to the work of Mendel and to the early 20th-century Mendelians. As the field matured, however, it became apparent that the phenotypic effects of many alleles could be influenced by other genes, influencing both the degree of severity of a mutation’s expression (its “expressivity”), and the proportion of individuals possessing the mutation that expressed it at all (its “penetrance”).
To illustrate the differences in the manifestation of a given gene’s function caused by genetic background effects, take the various degrees of expression of the gene regulating the size and shape of incisors in man. Copies of one dominant gene, identical by descent, caused missing, or peg-shaped, or strongly mesio-distally reduced upper lateral incisors in subsequent generations (Alvesalo and Portin 1969). Though the precise nature of the gene involved is not known, the example shows that the same gene can have different manifestations in different individuals, i.e., in different genetic backgrounds. There is an enormous number of documented examples of such genetic background effects in all organisms that have been investigated genetically.
The phenomenon of genetic background effects was already well recognized by geneticists in the second decade of the 20th century, as illustrated, for example, in the multi-part series of papers, dealing with coat color inheritance in mammals by S. Wright, published in Journal of Genetics (Wright 1917a,b). (Wright would later achieve eminence as one of the key founders of population genetics, but he started his career in what was then known as “physiological genetics.”) The whole matter, however, was raised to a new conceptual level in the 1930s, by C. H. Waddington, a British developmental biologist and geneticist, who called the totality of interactions among genes and between genes and the environment “the epigenotype.”
The epigenotype consists of the total developmental system lying between the genotype and the phenotype through which the adult form of an organism is realized (Waddington 1939). Although a clear concept of “gene regulation” did not exist in the 1940s and 1950s, Waddington, with this concept, was clearly edging toward it. When the Jacob-Monod model of gene regulation came forth in the early 1960s, Waddington promptly saw its relevance for development (Waddington 1962; 1966) as, of course, did Jacob and Monod themselves (Jacob and Monod 1961a,b; Monod and Jacob 1961). The crucial point, with respect to the definition of the gene, is that genes are not autonomous, independent agents—as was implicit in much of the early treatment of genes, and which indeed remains potent in much contemporary thinking, as exemplified in R. Dawkins’ still influential book, “The Selfish Gene” (Dawkins 1976). Rather, they exert their effects within, or as the output of, complex systems of gene interactions. Today, we term such systems “genetic networks” or “genetic regulatory networks” (GRNs). Sewall Wright, along with Waddington, was an early exponent of such network thinking (Wright 1968), but the modern concept of GRNs reached its fruition only in the late 1990s (reviewed in Davidson 2001; Wilkins 2002; Davidson and Erwin 2006; Wilkins 2007).
The conceptual consequences of viewing individual genes not as autonomous actors but as interactive elements or outputs of networks are profound. For one thing, it becomes relatively easy to think about the nature of genetic background effects in terms of the structure of GRNs (Box 2). While much of the thinking of the 20th century about genes was based on the premise that the route from gene to phenotype was fairly direct, and often deducible from the nature of the gene product, the network perspective envisages far more complexity and indirectness of effects. In general, the path from particular genes to specific phenes is long, and the role of many gene products seems to be the activation or repression of the activities of other genes. As a result, for most of these interactive effects, the normal (wild-type) function of the gene can only rarely be deduced directly from the mutant phenotype, which often involves complicated secondary effects resulting from the disrupted operation of the GRN within which the gene acts. Hence, the widely held popular belief that particular genes govern or “determine” particular traits, including complex psychological ones (e.g., risk-taking, gender identity, autism), as inferred from studies of genetic variants, is a gross oversimplification, hence distortion, of a complex reality.
In effect, genes do not have independent “agency”; for the most part they are simply cogs in the complex machinery of GRNs, and interpreting their mutant phenotypes is often difficult. In contrast, the genes for which there is an obvious connection between the mutant form and an altered phenotype are usually ultimate outputs of GRNs, such as pigmentation genes, hemoglobins, and enzymes of intermediary metabolism. These genes, however, also lack true autonomy, being activated in response to the operation of GRNs. Therefore, to fully understand how a gene functions, one must comprehend the larger systems in which they operate. Genetics, in this sense, is becoming systems biology, a point that has also been made by others (see, for example, Keller 2005). In effect, since genes can only be defined with respect to their products, and those products are governed by GRNs, the particular cellular and regulatory (GRN) contexts involved may be considered additional “dimensions” vital to specifying a gene’s function and identity. The examples of “gene sharing,” in which the function of the gene is wholly a function of its cellular context, illustrate this in a particularly vivid way. The “gene”—however it comes to be defined—can therefore be seen not as a three-dimensional entity but as a multi-dimensional one.
Putting it all Together: Toward a New Definition of the “Gene”
Where do all these considerations leave us? It took approximately half a century to go from Johannsen’s wholly abstract formulation of the term “gene” as a “unit of heredity,” to reach the early 1960s concept of the gene as a continuous segment of DNA sequence specifying a polypeptide chain. A further half century’s worth of experimental investigation has brought us to the realization that the 1960s definition is no longer adequate as a general one. Yet the term “gene” persists as a vaguely understood generic description. It is, to say the least, an anomalous situation that the central term of genetics should now be shrouded in confusion and ambiguity. That is not only intellectually unsatisfactory for the discipline, but has detrimental effects on the popular understanding of genetics. Such misunderstanding is seen most starkly in the situation noted earlier, the commonly held view that there are individual genes responsible “for” certain complex conditions, e.g., schizophrenia, alcoholism, etc. A clearer definition of the term would thus help both the field of genetics, and, ultimately, public understanding.
Here, therefore, we will propose a definition that we believe comes closer to doing justice to the idea of the “gene,” in light of current knowledge. It makes no reference to “the unit of heredity”—the long-standing sense of the term—because we feel that it is now clear that no such generic universal unit exists. By referring to DNA sequences, however, our definition embodies the hereditary dimension of genes (in a way that pure “process”-centered definitions focused on gene expression do not). Furthermore, in its emphasis on the ultimate molecular products and reference to GRNs as both evokers and mediators of the actions of those products, it recognizes the long causal chains that often operate between genes and their effects. Our provisional definition is this:
A gene is a DNA sequence (whose component segments do not necessarily need to be physically contiguous) that specifies one or more sequence-related RNAs/proteins that are both evoked by GRNs and participate as elements in GRNs, often with indirect effects, or as outputs of GRNs, the latter yielding more direct phenotypic effects.
This is an explicitly “molecular” definition, but we think that is what is needed now. In contrast, “genes” that are identified purely by their phenotypic effects, as for example in genome-wide association study (GWAS) experiments, would, in our view, not deserve such a characterization until found to specify one or more RNAs/proteins. The genetic effects picked up in such work often identify purely regulatory elements, and these should not qualify as genes, only as part of genes. Our definition, like the classic 1960s’ formulation, makes identifying the product(s) crucial to delimiting, hence identifying, the genes themselves. It, however, also emphasizes the molecular and cellular context in which those products form and function. Those larger contexts, in effect, become necessary to define the function of the specifying gene(s).
The new definition, however, is slightly cumbersome. We therefore offer it only as a tentative solution, hence as a challenge to the field to find a better formulation but one that does justice to the complex realities of the genetic material uncovered in the past half-century.
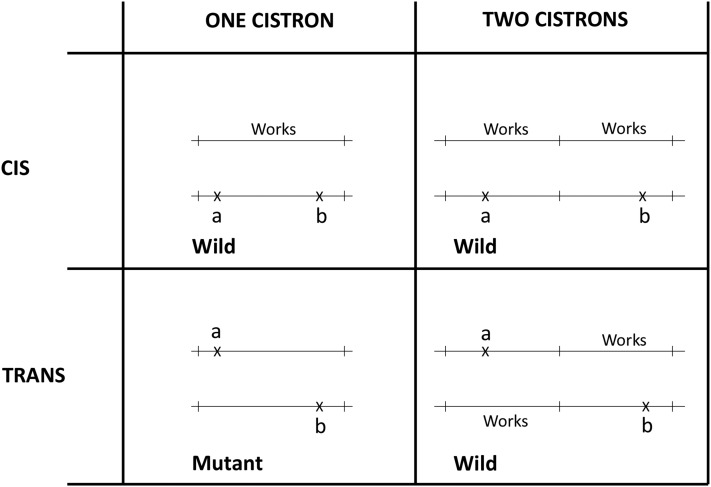
Box 1. The cis-trans test.
Of fundamental importance in the operational definition of the gene is the cis-trans test (Lewis 1951; Benzer 1957). To test whether mutations a and b belong to the same gene or cistron (Benzer 1957), or different cistrons, the cis-heterozygote a b/+ + and the trans-heterozygote a +/+ b are compared. If the cis-heterozygotes, and the trans-heterozygotes are phenotypically similar (usually wild type), they are said to “complement” one another, and the mutations are inferred to fall into different cistrons. If, however, the cis-heterozygotes and the trans-heterozygotes are phenotypically different, the trans-heterozygote being (usually) mutant, and the cis-heterozygote (usually) of wild type, the mutations do not complement, and are inferred to belong to the same cistron. The attached figure clarifies the idea.
The principle of the cis-trans test. If mutations a and b belong to the same cistron, the phenotypes of the cis- and trans-heterozygotes are different. If, however, the cis- and trans-heterozygotes are phenotypically similar, the mutations a and b belong to different cistrons. The notation “works” on the Figure means that the cistron is able to produce a functional polypeptide. Mutations a and b are recessive mutations that both affect the same phenotypic trait, such as the eye color of D. melanogaster, for example.
Box 2. Interpreting “genetic background” effects in terms of GRNs.
Genetic background effects typically exhibit either of two forms, when a pre-existing mutation, with an associated phenotypic manifestation, is crossed into a different strain: the reduction (“suppression”) of the mutant phenotype or its increase (“enhancement”). The effects involve either changes in the degree (“expressivity”) of the mutant effect, or the number of individuals) affected (its “penetrance”), or both. When analyzed genetically, these effects could often be traced to specific “suppressor” or “enhancer” loci, which could be either tightly linked or distant in the genome from the original mutant locus. Typically regarded as an unnecessary complication in analysis of the original mutation, they were usually not pursued further. Yet, in terms of current understanding of GRNs, they are not, in principle, mysterious. Each gene that is part of a GRN can be thought of as either transmitting a signal for the activation or repression of one or more other “downstream” genes in that network, but, given the hierarchical nature of GRNs, it follows that a mutational alteration in a specific gene in the network can be either strengthened or reduced by other mutational changes in the network, either upstream or downstream of the original mutation. The particular effect achieved will depend on the characteristics of each of the two mutations involved—whether they are loss-of- or gain-of-function mutations—and the precise nature of their connectivity. Such effects are most readily illustrated with linear sequences of gene actions, genetic pathways (Wilkins 2007), but can be understood in networks, when the network structure and the placement of the two genes within them is known. Some genetic background effects, in principal, however, might involve partially redundant networks, in which the effects of the two pathways are additive. In those cases, a mutant effect in one pathway may be either compensated, hence suppressed, or exacerbated, by a second mutation in the other pathway, the precise effects again depending upon the specific characteristics of the mutations and the degree of redundancy between the two GRNs.
Acknowledgments
We thank Mark Johnston and Richard Burian for many helpful suggestions, both editorial and substantive, on previous drafts. A.W. would also like to acknowledge earlier conversations with Jean Deutsch on the subject of this article; we disagreed on much but the process was stimulating and helpful. P.P. wants to thank his friends Marja Vieno, M.Sc. for linguistic aid at the very first stages of this project, and Harri Savilahti, Ph.D. for a fruitful discussion, and Docent Mikko Frilander, Ph.D. for consultation. The authors declare no conflict of interest.
Footnotes
Communicating editor: M. Johnston
Literature Cited
- Alvesalo L., Portin P., 1969. The inheritance pattern of missing, peg-shaped, and strongly mesio-distally reduced upper lateral incisors. Acta Odontol. Scand. 27: 563–575. [DOI] [PubMed] [Google Scholar]
- Avery O. T., MacLeod C. M., MacCarty M., 1944. Studies on the chemical nature of the substance inducing transformation of Pneumococcal types. Induction of transformation by a deoxyribonucleic acid fraction isolated from Pneumococcus type III. J. Exp. Med. 79: 137–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateson W., Saunders E. R., Punnett R. C., 1905a Experimental studies in the physiology of heredity. Reports to the Evolution Committee of the Royal Society, Report II. pp. 4–99 1–55.
- Bateson W., Saunders E. R., Punnett R. C., 1905b Experimental studies in the physiology of heredity. Reports to the Evolution Committee of the Royal Society, Report II. pp. 80–99.
- Beadle G. W., Tatum E. L., 1941. Genetic control of biochemical reactions in Neurospora. Proc. Natl. Acad. Sci. USA 27: 499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benzer S., 1955. Fine structure of a genetic region in bacteriophage. Proc. Natl. Acad. Sci. USA 41: 344–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benzer S., 1957. The elementary units of heredity, pp. 70–93 in The Chemical Basis of Heredity, edited by McElroy W. D., Glass B. Johns Hopkins Press, Baltimore. [Google Scholar]
- Benzer S., 1959. On the topology of the genetic fine structure. Proc. Natl. Acad. Sci. USA 45: 1607–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benzer S., 1961. On the topography of the genetic fine structure. Proc. Natl. Acad. Sci. USA 47: 403–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black D. L., 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72: 291–336. [DOI] [PubMed] [Google Scholar]
- Bonner D. M., 1950. The Q locus of Neurospora. Genetics 35: 655–656. [Google Scholar]
- Boveri T., 1902. Über mehrpolige Mitosen als Mittel zur Analyse des Zellkerns. Verh. phys-med. Ges. Würzb. 35: 60–90. [Google Scholar]
- Boveri T., 1903. Über die Konstitution der chromatischen Kernsubstanz. Verh. deutsch. zool. Ges. Würzb. 13: 10–33. [Google Scholar]
- Brenner S., Jacob F., Meselson M., 1961. An unstable intermediate carrying information from genes to ribosomes for protein synthesis. Nature 190: 576–581. [DOI] [PubMed] [Google Scholar]
- Brennickle A., Marchfelder A., Binder S., 1999. RNA editing. FEMS Microbiol. Rev. 23: 297–316. [DOI] [PubMed] [Google Scholar]
- Bridges C. B., 1916. Non-disjunction as proof of the chromosome theory of heredity. Genetics 1: 1–52, 107–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges C. B., 1935. Salivary chromosome maps with a key to the banding of the chromosomes of Drosophila melanogaster. J. Hered. 26: 60–64. [Google Scholar]
- Bridges C. B., 1938. A revised map of the salivary gland X-chromosome of Drosophila melanogaster. J. Hered. 29: 11–13. [Google Scholar]
- Burian R. M., 2004. Molecular epigenesis, molecular pleiotropy, and molecular gene definitions. Hist. Philos. Life Sci. 26: 59–80. [DOI] [PubMed] [Google Scholar]
- Carlson E. A., 1966. The Gene: A Critical History. W. B. Saunders Company, Philadelphia. [Google Scholar]
- Carninci P., 2006. Tagging the mammalian transcription complexity. Trends Genet. 22: 501–510. [DOI] [PubMed] [Google Scholar]
- Carninci P., Hayashizaki Y., 2007. Noncoding RNA transcription beyond annotated genes. Curr. Opin. Genet. Dev. 17: 139–144. [DOI] [PubMed] [Google Scholar]
- Carninci P., Yasuda J., Hayashizaki Y., 2008. Multifaceted mammalian transcriptome. Curr. Opin. Cell Biol. 20: 274–280. [DOI] [PubMed] [Google Scholar]
- Claverie J.-M., 2005. Fewer genes, more noncoding RNA. Science 309: 1529–1530. [DOI] [PubMed] [Google Scholar]
- Comai L., Cartwright R. A., 2005. A toxic mutator and selection alternative to the non-Mendelian RNA cache hypothesis for hothead reversions. Plant Cell 17: 2856–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correns C. G., 1900. Mendels Regel über das Verhalten der Nachkommenschaft der Rassenbastarde. Ber. Deut. Bot. Ges. 18: 158–168. [Google Scholar]
- Crawford I. P., Yanofsky C., 1958. On the separation of the tryptophan synthetase of Escherichia coli into two protein components. Proc. Natl. Acad. Sci. USA 44: 1161–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F. H. C., 1963. On the genetic code. Science 139: 461–464. [DOI] [PubMed] [Google Scholar]
- Davidson E. H., 2001. Genomic Regulatory Systems: Development and Evolution. Academic Press, San Diego. [Google Scholar]
- Davidson E. H., Erwin D. H., 2006. Gene regulatory networks and the evolution of animal body plans. Science 311: 796–800. [DOI] [PubMed] [Google Scholar]
- Dawkins R., 1976. The Selfish Gene. Oxford University Press, Oxford. [Google Scholar]
- de Vries H., 1900. Sur la loi de disjonction des hybrides. CR. Acad. Sci. Paris. 130: 845–847. [Google Scholar]
- Dobzhansky Th., 1929. Genetical and cytological proof of translocations involving the third and fourth chromosome in Drosophila melanogaster. Biol. Zentralbl. 49: 408–419. [Google Scholar]
- Eddy S. R., 2001. Non-coding RNA genes and the modern RNA world. Nat. Rev. Genet. 2: 919–929. [DOI] [PubMed] [Google Scholar]
- Edwards A. W. F., 2013. Robert Heath Lock and his textbook of genetics, 1906. Genetics 194: 529–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk R., 2009. Genetic Analysis. A History of Genetic Thinking. Cambridge University Press, Cambridge. [Google Scholar]
- Gerhart J., Kirschner M., 2007. The theory of facilitated variation. Proc. Natl. Acad. Sci. USA 104(Suppl. 1): 8582–8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles N. H., 1952. Studies on the mechanism of reversion in biochemical mutants of Neurospora crassa. Cold Spring Harb. Symp. Quant. Biol. 16: 283–313. [DOI] [PubMed] [Google Scholar]
- Gingeras T. R., 2007. Origin of phenotypes: genes and transcripts. Genome Res. 17: 682–690. [DOI] [PubMed] [Google Scholar]
- Green M. M., Green K. C., 1949. Crossing over between alleles of the lozenge locus in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 35: 586–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith F., 1928. The significance of pneumococcal types. J. Hyg. (Lond.) 27: 113–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths P. E., Neumann-Held E. M., 1999. The many faces of the gene. Bioscience 49: 656–662. [Google Scholar]
- Griffiths P. E., Stotz K., 2006. Genes in the postgenomic era. Theor. Med. Bioeth. 27: 499–521. [DOI] [PubMed] [Google Scholar]
- Gros F., Gilbert W., Hiatt H. H., Attardi G., Spahr D. F., et al. , 1961. Molecular and biological characterization of messenger RNA. Cold Spring Harb. Symp. Quant. Biol. 26: 111–132. [DOI] [PubMed] [Google Scholar]
- Hershey A. D., Chase M., 1952. Independent functions of viral protein and nucleic acid in growth of bacteriophage. J. Gen. Physiol. 36: 39–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R., 1987. The inheritance of epigenetic defects. Science 238: 163–170. [DOI] [PubMed] [Google Scholar]
- Jablonka E., Raz G., 2009. Transgenerational epigenetic inheritance: prevalence, mechanisms, and implications for the study of heredity and evolution. Q. Rev. Biol. 84: 131–176. [DOI] [PubMed] [Google Scholar]
- Jacob F., Monod J., 1961a Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 3: 318–356. [DOI] [PubMed] [Google Scholar]
- Jacob F., Monod J., 1961b On the regulation of gene activity. Cold Spring Harb. Symp. Quant. Biol. 26: 193–211. [DOI] [PubMed] [Google Scholar]
- Janssens F. A., 1909. La théorie de la chiasmatypie, nouvelle interpretation des cinéses de maturation. Cellule 25: 387–406. [Google Scholar]
- Johannsen W., 1909. Elemente der exakten Erblichkeitslehre. Gustav Fischer, Jena. [Google Scholar]
- Johannsen W., 1926. Elemente der exakten Erblichkeitslehre, Ed. 3rd Gustav Fischer, Jena. [Google Scholar]
- Judson H. F., 1996. The Eighth Day of Creation: Markers of the Revolution in Biology. Expanded Edition. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. [Google Scholar]
- Kapranov P., Willingham A. T., Gingeras T. R., 2007. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 8: 413–423. [DOI] [PubMed] [Google Scholar]
- Keller E. F., 2000. The Century of the Gene. Harvard University Press, Cambridge, MA. [Google Scholar]
- Keller E. F., 2005. The century of the gene. J. Biosci. 30: 3–10. [DOI] [PubMed] [Google Scholar]
- Keller E. F., Harel D., 2007. Beyond the gene. PLoS One 2(11): e1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszul R., Meselson M., Van Donick K., Vandenhaute J., Zickler D., 2012. The centenary of Janssens’s chiasmatype theory. Genetics 191: 309–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landweber L. F., 2007. Why genomes in pieces? Science 318: 406–407. [DOI] [PubMed] [Google Scholar]
- Leff S. E., Rosenfeld M. G., Evans R. M., 1986. Complex transcriptional units: diversity in gene expression by alternative RNA processing. Annu. Rev. Biochem. 55: 1091–1117. [DOI] [PubMed] [Google Scholar]
- Lewis E. B., 1941. Another case of unequal crossing over in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 27: 31–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis E. B., 1945. The relation of repeats to position effect in Drosophila melanogaster. Genetics 30: 137–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis E. B., 1951. Pseudoallelism and gene evolution. Cold Spring Harb. Symp. Quant. Biol. 16: 159–174. [DOI] [PubMed] [Google Scholar]
- Lock R. H., 1906. Recent Progress in the Study of Variation, Heredity and Evolution. Murray, London. [Google Scholar]
- Lolle S. J., Victor J. L., Young J. M., Pruitt R. E., 2005. Genome-wide non-Mendelian inheritance of extra-genomic information in Arabidopsis. Nature 434: 505–509. [DOI] [PubMed] [Google Scholar]
- Mattick J. S., 2005. The functional genomics of noncoding RNA. Science 309: 1527–1528. [DOI] [PubMed] [Google Scholar]
- McClung C. E., 1927. The chiasmatype theory of Janssens. Q. Rev. Biol. 2: 344–366. [Google Scholar]
- Mendel G., 1866. Versuche über Pflanzen-Hybriden. Verh. naturf. Ver. Brünn 4: 3–47. [Google Scholar]
- Mercier R., Jolivet S., Vignard J., Durand S., Drouaud J., et al. , 2008. Outcrossing as an explanation of the apparent unconventional genetic behavior of Arabidopsis thaliana hth mutants. Genetics 180: 2295–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagawa Y., Ogawa J., Iwata Y., Koizumi N., Mishiba K.-I., 2013. An attempt to detect siRNA-mediated genomic DNA modification by artificially induced mismatch siRNA in Arabidopsis. PLoS One 8(11): e81326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monod J., Jacob F., 1961. General conclusions: teleonomic mechanisms in cellular metabolism, growth and differentiation. Cold Spring Harb. Symp. Quant. Biol. 26: 389–401. [DOI] [PubMed] [Google Scholar]
- Morgan T. H., 1910. Chromosomes and heredity. Am. Nat. 44: 449–496. [Google Scholar]
- Morgan T. H., 1917. The theory of the gene. Am. Nat. 51: 513–544. [Google Scholar]
- Morgan T. H., 1919. The Physical Basis of Heredity. Yale University Press, New Haven. [Google Scholar]
- Morgan T. H., 1926. The Theory of the Gene. Yale University Press, New Haven. [Google Scholar]
- Morgan T. H., Sturtevant A. H., Muller H. J., Bridges C. B., 1915. The Mechanism of Mendelian Heredity. Henry Holt, New York. [Google Scholar]
- Moss L., 2003. What Genes Can’t Do. MIT Press, Cambridge, MA. [Google Scholar]
- Muller H. J., 1920. Are the factors of heredity arranged in a line? Am. Nat. 54: 97–121. [Google Scholar]
- Muller H. J., 1922. Variation due to change in the individual gene. Am. Nat. 56: 32–50. [Google Scholar]
- Muller H. J., 1926. The gene as the basis of life. Proc. Internat. Cong. Plant Sci. 1: 897–921. [Google Scholar]
- Muller H. J., 1927. Artificial transmutation of the gene. Science 66: 84–87. [DOI] [PubMed] [Google Scholar]
- Muller H. J., Painter T. S., 1929. The cytological expression of changes in gene alignment produced by X-rays in Drosophila. Am. Nat. 63: 193–200. [Google Scholar]
- Neumann-Held E. M., 1999. The gene is dead – Long live the gene: Conceptualizing genes the constructionist way, pp. 105–137 in Sociobiology and Bioeconomics. The Theory of Evolution in Biological and Economic Theory, edited by Koslowski P. Springer-Verlag, Berlin. [Google Scholar]
- Neumann-Held E. M., 2001. Let’s talk about genes: The process molecular gene concept and Its context, pp. 69–73 in Cycles of Contingency, edited by Oyama S., Griffiths P. E., Gray R. D. Bradford, MIT Press, Cambridge, MA. [Google Scholar]
- Oliver P., 1940. A reversion to wild type associated with crossing over in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 26: 452–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter T. S., 1934. A new method for the study of chromosome aberrations and the blotting of chromosome maps in Drosophila melanogaster. Genetics 19: 175–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter T. S., Muller H. J., 1929. Parallel cytology and genetics of induced translocations and deletions in Drosophila. J. Hered. 20: 287–298. [Google Scholar]
- Parra G., Reymond A., Dabbousch N., Dermitzakis E. T., Castelo R., et al. , 2006. Tandem chimerism as a means to increase protein complexity in the human genome. Genome Res. 16: 37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson H., 2006. What is a gene? Nature 441: 399–401. [DOI] [PubMed] [Google Scholar]
- Pesole G., 2008. What is a gene? An updated operational definition. Gene 417: 1–4. [DOI] [PubMed] [Google Scholar]
- Piatigorsky J., 2007. Gene Sharing and Evolution: The Diversity of Protein Functions. Harvard University Press, Cambridge, MA. [Google Scholar]
- Piatigorsky J., Wistow G. J., 1989. Enzyme/crystallins: gene sharing as an evolutionary strategy. Cell 57: 197–199. [DOI] [PubMed] [Google Scholar]
- Pontecorvo G., 1952. The genetic formulation of gene structure and action. Adv. Enzym. 13: 121–149. [DOI] [PubMed] [Google Scholar]
- Portin P., 1993. The concept of the gene: short history and present status. Q. Rev. Biol. 68: 173–223. [DOI] [PubMed] [Google Scholar]
- Portin P., 2009. The elusive concept of the gene. Hereditas 146: 112–117. [DOI] [PubMed] [Google Scholar]
- Portin P., 2015. The development of genetics in the light of Thomas Kuhn’s theory of scientific revolutions. Recent Adv. DNA Gene Seq. 9: 14–25. [DOI] [PubMed] [Google Scholar]
- Pritchard R. H., 1955. The linear arrangement of a series of alleles of Aspergillus nidulans. Heredity 9: 343–371. [Google Scholar]
- Rassoulzadegan M., Grandjean V., Gounon P., Vincent S., Gillot I., 2006. RNA-mediated non-Mendelian inheritance of an epigenetic change in the mouse. Nature 441: 469–474. [DOI] [PubMed] [Google Scholar]
- Sax K., 1932a The cytological mechanism of crossing over. J. Arnold Arbor. 13: 180–212. [Google Scholar]
- Sax K., 1932b Meiosis and chiasma formation in Paeonia suffruticosa. J. Arnold Arbor. 13: 375–384. [Google Scholar]
- Scherrer K., Jost J., 2007. Gene and genon concept: coding vs. regulation. A conceptual and information-theoretic analysis of genetic storage and expression in the light of modern molecular biology. Theory Biosci. 126: 65–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibler U., Sierra F., 1987. Alternative promoters in developmental gene expression. Annu. Rev. Genet. 21: 237–257. [DOI] [PubMed] [Google Scholar]
- Snyder M., Gerstein M., 2003. Defining genes in the genomics era. Science 300: 258–260. [DOI] [PubMed] [Google Scholar]
- Srb A. M., Horowitz N. H., 1944. The ornithine cycle in Neurospora and its genetic control. J. Biol. Chem. 154: 129–139. [Google Scholar]
- Stadler P. F., Prohaska S. J., Frost C. V., Krakauer D. C., 2009. Defining genes: a computational framework. Theory Biosci. 128: 165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern C., 1970. The continuity of genetics. Daedalus 99: 882–908. [PubMed] [Google Scholar]
- Strauss B. S., 2016. Biochemical genetics and molecular biology: the contributions of George Beadle and Edward Tatum. Genetics 203: 13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant A. H., 1913. The linear arrangement of the six sex-linked factors in Drosophila, as shown by their mode of association. J. Exp. Zool. 14: 43–59. [Google Scholar]
- Sturtevant A. H., 1965. A History of Genetics. Harper & Row, New York. [Google Scholar]
- Sturtevant A. H., Beadle G. W., 1939. An Introduction to Genetics. W. B. Saunders, Philadelphia. [Google Scholar]
- Sturtevant A. H., Beadle G. W., 1962. An Introduction to Genetics. The Dover edition of the work first published by W. B. Saunders Company in 1939. Dover Publications, New York. [Google Scholar]
- Sutton W. S., 1903. The chromosomes in heredity. Biol. Bull. 4: 231–251. [Google Scholar]
- The ENCODE Project Consortium , 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium , 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The FANTOM Consortium and RIKEN Genome Exploration Research Group (Genome Network Project Core Group) , 2005. The transcriptional landscape of the mammalian genome. Science 309: 1559–1563. [DOI] [PubMed] [Google Scholar]
- Tschermak E., 1900. Über künstliche Kreuzung bei Pisum sativum. Ber. Deut. Bot. Ges. 18: 232–239. [Google Scholar]
- Waddington C. H., 1939. An Introduction to Modern Genetics. Allen & Unwin, London. [Google Scholar]
- Waddington C. H., 1962. New Patterns in Genetics and Development. Columbia University Press, New York. [Google Scholar]
- Waddington C. H., 1966. Principles of Development and Differentiation. Macmillan Company, New York. [Google Scholar]
- Waters C. K., 1994. Genes made molecular. Philos. Sci. 61: 163–185. [Google Scholar]
- Waters, K., 2013 Molecular genetics. in The Stanford Encyclopedia of Philosophy (Fall 2013 Edition), edited by E. N. Zalta. Stanford University Press, Redwood City, CA. Available at: <http://plato.stanford.edu/archives/fall2013/entries/molecular-genetics/>. Accessed: October 27, 2015.
- Watson J. D., Crick F. H. C., 1953a Molecular structure of nucleic acids. A structure for deoxyribose nucleic acid. Nature 171: 737–738. [DOI] [PubMed] [Google Scholar]
- Watson J. D., Crick F. H. C., 1953b Genetical implications of the structure of deoxyribonucleic acid. Nature 171: 964–967. [DOI] [PubMed] [Google Scholar]
- Watson J. D., Crick F. H. C., 1954. The structure of DNA. Cold Spring Harb. Symp. Quant. Biol. 18: 123–131. [DOI] [PubMed] [Google Scholar]
- Whitehouse H. L. K., 1973. Towards an Understanding of the Mechanism of Heredity, Ed. 3rd Edward Arnold, London. [Google Scholar]
- Wilkins A. S., 2002. The Evolution of Developmental Pathways, Sinauer Associates, Sunderland, MA. [Google Scholar]
- Wilkins A. S., 2007. Between “design” and “bricolage”: genetic networks, levels of selection, and adaptive evolution. Proc. Natl. Acad. Sci. USA 104: 8590–8596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witzany G., 2011. The agents of natural genome editing. J. Mol. Cell Biol. 3: 181–189. [DOI] [PubMed] [Google Scholar]
- Wright S., 1917a Color inheritance in mammals. III: the rat—few variations of factors known until recently—castle’s selection experiment—any interpretation of it demonstrates the efficacy of Darwinian selection. J. Hered. 8: 426–430. [Google Scholar]
- Wright S., 1917b Color inheritance in mammals. V. The guinea-pig—great diversity in coat-pattern, due to interaction of many factors in development—some factors hereditary, others of the nature of accidents in development. J. Hered. 8: 476–480. [Google Scholar]
- Wright S., 1968. Evolution and the Genetics of Populations. Vol. 1. Genetic and Biometric Foundations. University of Chicago Press, Chicago. [Google Scholar]
- Yanofsky C., Crawford I. P., 1959. The effects of deletions, point mutations on the two components of the tryptophan synthetase of Escherichia coli. Proc. Natl. Acad. Sci. USA 45: 1016–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky C., Carlton B. C., Guest J. R., Helinski D. R., Henning U., 1964. On the colinearity of gene structure and protein structure. Proc. Natl. Acad. Sci. USA 51: 266–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky C., Drapeau G. R., Guest J. R., Carlton B. C., 1967. The complete amino acid sequence of the tryptophan synthetase A protein (a subunit) and its colinear relationship with the genetic map of the A gene. Proc. Natl. Acad. Sci. USA 57: 296–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ycas M., 1969. The Biological Code. North-Holland Publishing Company, Amsterdam. [Google Scholar]