Structural Rearrangements can have unexpected effects on quantitative phenotypes. Surprisingly, these rearrangements can also be considered as...
Keywords: structural variation, Arabidopsis, quantitative trait locus, heritability, low-coverage sequencing
Abstract
To understand the population genetics of structural variants and their effects on phenotypes, we developed an approach to mapping structural variants that segregate in a population sequenced at low coverage. We avoid calling structural variants directly. Instead, the evidence for a potential structural variant at a locus is indicated by variation in the counts of short-reads that map anomalously to that locus. These structural variant traits are treated as quantitative traits and mapped genetically, analogously to a gene expression study. Association between a structural variant trait at one locus, and genotypes at a distant locus indicate the origin and target of a transposition. Using ultra-low-coverage (0.3×) population sequence data from 488 recombinant inbred Arabidopsis thaliana genomes, we identified 6502 segregating structural variants. Remarkably, 25% of these were transpositions. While many structural variants cannot be delineated precisely, we validated 83% of 44 predicted transposition breakpoints by polymerase chain reaction. We show that specific structural variants may be causative for quantitative trait loci for germination and resistance to infection by the fungus Albugo laibachii, isolate Nc14. Further we show that the phenotypic heritability attributable to read-mapping anomalies differs from, and, in the case of time to germination and bolting, exceeds that due to standard genetic variation. Genes within structural variants are also more likely to be silenced or dysregulated. This approach complements the prevalent strategy of structural variant discovery in fewer individuals sequenced at high coverage. It is generally applicable to large populations sequenced at low-coverage, and is particularly suited to mapping transpositions.
WHILE genome resequencing can readily determine variations such as Single Nucleotide Polymorphisms (SNPs) and small indels, it remains challenging to identify structural variants (SVs) and rearrangements, despite improvement in algorithms for calling SVs. The current gold standard for determining SVs between individuals is by de novo assembly (Simpson and Pop 2015). This requires high-coverage paired-end sequence over a range of insert sizes, together with long-range information such as from long-read technologies (Chaisson and Tesler 2012; Jain et al. 2015) for scaffolding. The high cost and low throughput of de novo assembly limit its use, and leaves open two important questions. First, whether an SV is identified in an individual frequently enough to contribute to phenotypic heritability in a population. Second, whether an SV represents a local rearrangement, such as a deletion, inversion or tandem copy-number variant (CNV), or is long-range, such as a transposition (Cao et al. 2011; Mills et al. 2011).
SVs are frequently revealed by the anomalous alignment of short-reads to the reference genome. Specific anomaly signatures characterize different types of SVs (Table 1). Thus, same-strand pairs indicate inversion, high read coverage duplications, abnormal insert sizes, and unpaired reads indels (Figure 1). These anomalies arise, often in combination, because the reads have been aligned to the wrong genome—the anomalies disappear if instead the reads are aligned to the true genome. This idea is used by algorithms such as GATK (McKenna et al. 2010) and Platypus (Rimmer et al. 2014) that identify small indels by local realignment, and in whole-genome reassembly by iterative realignment (Gan et al. 2011).
Table 1. MAGIC SV-QTL classified by read pair anomaly type and QTL type, after removing duplicates.
| SV-QTL | Unique | Cis | Trans | |
|---|---|---|---|---|
| Trait type | ||||
| IP | 1,997 | 833 | 1617 | 380 |
| ER | 184 | 165 | 112 | 72 |
| LIS | 2,051 | 585 | 1677 | 374 |
| SS | 1,950 | 1887 | 1358 | 592 |
| U | 2,060 | 1998 | 1530 | 530 |
| U+LIS | 2,033 | 431 | 1661 | 372 |
| Total | 10,275 | 5899 | 7955 | 2320 |
| SV type | ||||
| Duplication | 175 | 109 | 66 | |
| Indel | 3,035 | 3035 | 0 | |
| Inversion | 1,976 | 1373 | 603 | |
| Other | 1,316 | 381 | 935 | |
| Total | 6,502 | 4898 | 1604 |
SV-QTL: total number of QTL detected using each anomaly type. If the same QTL is detected by multiple anomalies then it is counted multiple times in this column). Unique: number of QTL detected after counting duplicates only once. cis: number of cis SV-QTL (source and sink within 2 Mb from each other), trans: number of trans SV-QTLS. Note that the total number of SV-QTL is 10,275, of which 6502 are distinct after removing overlapping events, and 5899 unique to a single anomaly type. IP, improperly paired; ER, excess reads; LIS, large insert size; SS, same strand; U, unmapped; U_LIS, unmapped or large insert size.
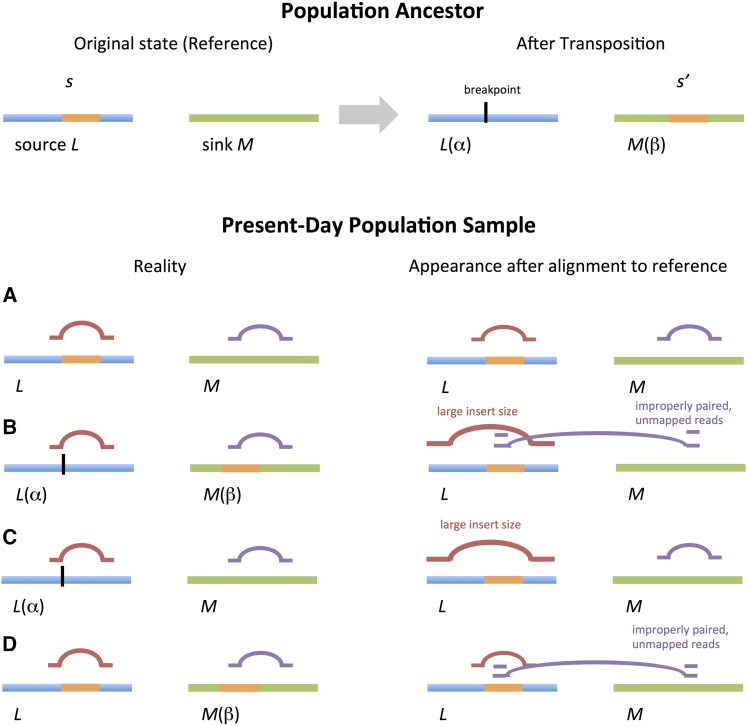
Figure 1.
Effects of a transposition on short-read mapping. Chromosomes are horizontal bars and read pairs are pairs of horizontal lines linked by curves. Upper panel: a population ancestor corresponding to the reference genome (left) undergoing a transposition (right), in which a segment at source locus with haplotype context is copied to at recipient sink locus with haplotype context Lower panel: all four possible combinations (A–D) of source and sink haplotype in descendants. On left are shown the alignment of reads to the true haplotypes, where there are no read-mapping anomalies. On right are shown the read-mapping anomalies that arise, depending on the true haplotype backgrounds at source and sink, upon alignment to the reference genome.
Many SV-calling algorithms utilize read-anomaly signatures to identify SVs segregating in individuals sequenced at high coverage (Chen et al. 2009; Manske and Kwiatkowski 2009; Ye et al. 2009; Simpson et al. 2010; Rausch et al. 2012; Sindi et al. 2012; Layer et al. 2014; Kronenberg et al. 2015). They focus on short-range SVs because of the difficulties in distinguishing long-range rearrangements from read mapping errors. They also work best when calling SVs in individuals sequenced at intermediate to high coverage; for example, LUMPY (Layer et al. 2014) and WHAM (Kronenberg et al. 2015) are most sensitive at coverage >10×. In other applications, e.g., cancer resequencing, typical coverage is even higher, at 30× or above.
Further challenges arise when calling SVs in large samples of population sequence data, for the purpose of testing genetic association. Population sequencing provides an alternative to genotyping by SNP arrays, simultaneously providing both haplotype reference panels for imputation (Durbin et al. 2010), and cohorts for disease mapping (Cai et al. 2015; Nicod et al. 2016). As the sample size increases, the coverage of each individual may be reduced without affecting imputation accuracy (Davies et al. 2016). Although the information present in each sample is then sparse, and therefore it would be difficult to call SVs (and even SNPs) on an individual basis, by pooling information across samples it might be possible to determine common SVs analogously to the way SNPs are imputed.
In addition to simple indels, inversions, and transpositions, where a segment with well-defined breakpoints is affected, many SVs are composites of multiple events (Yalcin et al. 2011), often driven by transposons and other mobile elements. These complex SVs resist simple classification, and the precise sequence of mutations that occurred may be unrecoverable. While current algorithms for calling SVs in simulated high-coverage human data can identify simple SVs with sensitivities of ∼90% depending on the type of SV (Kronenberg et al. 2015), they are less accurate when applied to real data, and their performance on complex SVs is unreported.
Despite this, there may still be strong evidence from read-mapping anomalies that an SV of some sort segregates at a locus. Furthermore, if the intensity of its anomaly signature can be used as a proxy for the purposes of testing genetic association, then one need not call the SV precisely. It then follows that information encoded by these anomalies across the genome could be used to compute relationships between individuals based on their structural profiles alone, and hence to estimate the heritability attributable to structural variation directly.
Here, we show how low-coverage population sequencing provides new ways for mapping SVs and estimating heritability, complementing the sequencing of fewer individuals at high coverage. As an illustration, we investigate the architecture and phenotypic impact of structural variation in Arabidopsis thaliana. Among natural accessions of Arabidopsis, structural variation is plentiful (Cao et al. 2011). The extent of rDNA repeats (Hu et al. 2011) and mobile transposable elements (Quadrana et al. 2016) vary between accessions, and variation in the overall amounts of both classes of repetitive sequence elements are complex traits, partially under genetic control. In this study we investigate all types of structural variation in Arabidopsis, including those not mediated by mobile elements. We show that long-range transpositions are common, and that structural variation has a significant impact on particular quantitative trait loci (QTL) and on trait heritability, distinct from that explained by other types of sequence variation.
Materials and Methods
DNA extraction and sequencing
Multiparent Advanced Generation Inter-Cross (MAGIC) lines were grown at Bath (laboratory of P.K.) or Oxford (laboratory of N.P.H.) in greenhouses or growth chambers, respectively. Leaves were harvested for DNA extraction. DNA isolation was performed at the John Innes Centre, in 96-well plates using the DNeasy 96 Plant Kit and DNeasy 96 Protocol (http://www.quiagen.com). Sequencing was performed by the Oxford Genomics Centre.
Genomic DNA library construction and multiplexing
Samples were quantified using the Quant-iT PicoGreen dsDNA Kits (Invitrogen, Carslbad, CA) and a Genios plate scanner (Tecan, Männedorf, Switzerland) according to manufacturer specifications. Sample integrity was assessed using 1% agarose gel. DNA (∼300 ng) was fragmented using a Covaris S2 system with the following settings: Intensity: 5, Duty Cycle: 20, Cycles per Burst: 200, Time: 60 sec. Distribution of fragments after shearing was determined using a Tapestation D1200 system (Agilent/Lab901, Santa Clara, CA). DNA Libraries were constructed using the NEBNext DNA Sample Prep Master Mix Set 1 Kit (New England Biolabs, Beverly, MA), with minor modifications, and a custom automated protocol on a Biomek FX (Beckman, Fullerton, CA). Ligation of adapters was performed using Illumina Adapters (Multiplexing Sample Preparation Oligonucleotide Kit). Ligated libraries were size selected using Ampure magnetic beads (Agencourt, Beckman, Fullerton, CA). Each library was PCR enriched with 25 µM each of the following custom primers:
Multiplex PCR primer 1.0
5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT-3′
Index primer
5′-CAAGCAGAAGACGGCATACGAGAT[INDEX]CAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT-3′
Indexes used were 8 bp long. Enrichment and adapter extension of each preparation was obtained using 5 µl of size-selected library in a 50 µl PCR reaction. After 10 cycles of amplification (cycling conditions as per Illumina recommendations), the reactions were purified with Ampure beads (Agencourt/Beckman). The final size distribution was determined using a Tapestation 1DK system (Agilent/Lab901). The concentrations used to generate the multiplex pool were determined by Picogreen. The library resulting from the pooling was quantified using the Agilent qPCR Library Quantification Kit and a MX3005P instrument (Agilent) before sequencing on an Illumina GAIIx as 50 or 100 bp paired-end reads. All steps for library construction, including the setup of the PCR reaction were performed on a Biomek FX (Beckman). Post PCR cleanup was carried out on a Biomek NXp (Beckman) whereas a Biomek 3000 (Beckman) was used to generate the pools of 96 indexed libraries.
Processing sequence reads and SNP calling
The Illumina reads were mapped to the A. thaliana reference (TAIR10) using Stampy v1.0.20 (Li and Durbin 2010; Lunter and Goodson 2011). Alignments were stored in a separate BAM file for each MAGIC line. Previous sequencing for the 18 MAGIC line progenitors had produced a catalog of 3,316,270 segregating SNPs (Gan et al. 2011). We ran GATK v2.6 (McKenna et al. 2010) on the segregating SNPs to call variants for the 19 founders, setting the following read filters: Allele Balance, BaseQualityRankSumTest, Clipping RankSumTest, Coverage, DepthPerAlleleBySample, FisherStrand, GCContent, HaplotypeScore, LowMQ, MappingQualityRankSumTest, MappingQualityZero, MappingQualityZeroBySample, RMSMappingQuality, and ReadPosRankSumTest. We filtered out SNPs that were triallelic, within annotated transposons, or heterozygous for any founders.
Definition of structural variant traits
We divided the TAIR10 reference genome into 11,915 abutting 10 kb segments. Within each segment, we computed six measures of anomalously mapped reads that are signatures of SVs. Let be the set of all reads mapped to a genome of length is the number of reads in and the number of reads mapped to a segment of length 10 kb. The read anomaly measures computed in each segment are:
High read coverage: where is the expected read coverage of the segment
Unpaired reads: number of reads mapping to the segment whose pair is not mapped
Pairs on the same strand: number of reads with pair on the same strand
Reads with large insert size: number of read pairs with insert size outside the range , or mapped to different chromosomes, being the median and interquartile range of insert sizes of all the reads in the sample.
Unpaired reads or with large insert size:
Improperly paired reads:
The last two traits are combination of others—certain SV types can cause multiple anomaly signatures, so merging them may increase power. Each type of read pair anomaly was measured in each of the 11,915 10 kb segments, determining 71,490 traits in total.
Genome scans
We treated the SV traits like a gene expression eQTL study, performing a genome scan for each one. Association was tested by fitting SV trait vectors to the imputed ancestral haplotype at each locus in the 488 genome mosaics. In combination, the mosaics partitioned the genome into 16,700 haplotype blocks, with the ancestral haplotype of all lines unchanged in each one. Let be the number of anomalous reads of a certain type at source segment in line At every haplotype block we fitted the linear model:
is the average trait value at is a binary indicator of whether line carries haplotype at is the effect of founder haplotype and the error. Founder effects were estimated by ANOVA to test the null hypothesis of no SV-QTL: returning the P-value for each block Let be the genome wide maximum logP for the scan.
To determine genome-wide statistical significance, controlling for the number of tests and for associations driven by outliers or any non-normality in the SV-traits, we performed 100 permutations of each trait vector , and repeated the genome scan for each one. The distribution of the 100 genome-wide maxima of each of the permutations was used to determine the significance of the observed logPs of the original SV-trait. We fitted a parametric generalized extreme value distribution to the permuted maxima (Dudbridge and Koeleman 2004), using the EVD R package to estimate a genome wide corrected logP:
where are the MLEs of respectively. This procedure was performed separately for each SV-trait. Study-wide SV-QTL were selected at corresponding to
Prediction of SV allele frequency
We predicted which founder haplotypes carried a given SV, reasoning they have more anomalous reads compared to those without the SV. For each SV-QTL the founders’ contributions were arranged as a table, , containing the read anomaly count (of a certain type) at the source for all lines carrying haplotype at the sink and haplotype at the source. A founder was classified as carrying the SV if its corresponding row was generally higher than the rest of the table (for cis SV-QTL, the matrix is almost diagonal).
The contribution of founder is defined as We reordered the founders such that We reasoned that, if the SV is biallelic with founders carrying the SV, then would be much larger than expected compared to the null hypothesis of no SV, when The z-score for is,
where and are estimated by 1000 permutations of denoted as We choose to maximize We declare to be significant—and that there is a partitioning of the founders into two groups at the SV—if <1% of permutations generate a value of exceeding that observed.
Validation of SVs by paired-end data
We used high and low coverage paired-end reads from the 19 founders (Gan et al. 2011), and from the MAGIC lines, respectively, to search for enrichment of read pairs linking the source and sink. First, for the high-coverage analysis of the founders, we restricted attention to the 2391 SV-QTL in which we had predicted which founders carried the SV. Using this partitioning, we used Fisher’s exact test to compare the numbers of anomalous read pairs (with one read mapped to the source and the other within a variably sized window of kb from the sink) in the founders predicted to carry the SV to the other founders. Second, in the low coverage MAGIC data, we performed the same test comparing the 100 lines with the highest read anomaly trait value to the rest of the population.
Validation by de novo contigs
We used BLAT (Kent 2002) to align 5,524,143 short de novo assembly contigs (lengths 50–1000 bp) of the 18 nonreference founders to TAIR10 to identify contigs split between the source and sink (where disjoint pieces aligned to each), or shared (where a common piece aligned to both). We excluded genomic regions with annotated repeats or transposons, and alignments that mapped to over five genomic loci. We randomized the SV-QTL by circular genome permutation (Cabrera et al. 2012) to determine whether such split and shared contig alignments are overrepresented near SV-QTL. For SV-QTL if are the original position of the source and sink, respectively, then a permuted SV-QTL is defined as where We required to be on the same chromosome for cis SV-QTL. We then computed one-sided permutation P-values
Validation by PCR
We designed PCR primers for 77 breakpoints from 44 SV-QTL predicted from de novo contigs. We conducted 96 type I experiments (one for each of the 77 breakpoints) that used primers corresponding to remote or inverted sink loci, so PCR should produce a product in only in SV genomes and not in the reference. We also performed 19 type II control experiments that should produce a PCR product in the reference, but not in SV genomes.
We designed 20–30 bp primer oligos based on the reference (TAIR10), using Primer3 (Rozen and Skaletsky 2000), after masking out repeats, transposons, and known polymorphisms. SVs tend to be near such sequence features, so we relaxed the default Primer3 criteria to detect oligos, and required: (i) Maximum allowed product 1.5 kb, (ii) Annealing temperature 10–90°, (iii) GC-content 10–90%, and (iv) Self-complementarity 8 bp. Primer specificity was tested by BLAT (Kent 2002).
Association with physiological phenotypes
For each of the six read anomaly categories, we computed Pearson correlations and corresponding P-values between nine physiological phenotypes, and the 11,915 SV-traits. We selected significant correlations with logP > 4 (so we expect about one false positive result per scan). After filtering correlations driven by outliers (i.e., in which removal of the three most extreme samples reduced the correlation below the significance threshold), we found 549 SV-traits associated with 40 phenotypes. Each physiological phenotype had, on average, 1.56 associated SV traits of the same anomaly type.
The effect of SVs on each phenotype was measured by a heritability-like measure, estimated by linear models. Let be the vector of phenotypic values for a physiological phenotype with correlated SV traits (of the same type): represented by the matrix The phenotype is modeled as:
The parameters were estimated using the R function glm(). We also computed the individual effect sizes of SV-traits, by fitting simple linear regression models. We mapped QTL for the phenotype residuals after regressing all/each associated SV traits of the same type, and compared them to the phenotype QTL.
Published phenotypes
We used flowering time and rosette diameter data from a field experiment (Springate and Kover 2014), as well as phenotypes described previously in Kover et al. (2009).
Phenotyping resistance
Three replicates of each MAGIC line were grown at the University of Bath in 2.5-inch plastic pots. Plants were monitored daily, and germination and bolting day recorded. After plants senesced, the inflorescence height and the total number of branches were measured. In a separate experiment, MAGIC lines were grown in growth chambers in P24 plastic trays ,and sprayed with A. laibachii race Nc14 (Thines et al. 2009) when plants were 21 days old. Nc14 zoospores were suspended in water at a concentration of 105 spores/ml and incubated on ice for 30 min. The spore suspension was then sprayed on plants using a spray gun, and plants were incubated in a coldroom in the dark overnight. Infected plants were kept in a growth chamber under 10-hr light and 14-hr dark cycles with a 20° day and 16° night temperature (Kemen et al. 2011). Resistance was defined as absence of pustules on the leaves at 7 days after inoculation. To minimize errors in scoring, resistant plants were monitored up to 14 days after inoculation. The experiment was reproduced twice.
Collection of RNA
We obtained seeds of MAGIC lines from the Nottingham Arabidopsis Stock Centre (NASC) and grew 209 lines at 20° in Percival environmental chambers (Perry, IA), in the laboratory of R.M.C., Salt Lake City. We prepared total RNA from pools of 20 aerial rosettes from seedlings at the fourth true leaf stage (Gan et al. 2011). RNAseq library construction and sequencing was performed at the Oxford Genomics Centre (Oxford, UK) to produce 2 × 100 bp reads using the Illumina nonstrand specific method. Per Illumina HiSeq lane, samples were barcoded and run in 13-plexes to give ∼14 million reads per sample.
Alignment of RNAseq reads and expression quantification
All libraries were aligned to the TAIR10 reference gene set augmented by any novel genes reported in Gan et al. (2011) using PALMapper v0.6 (Jean et al. 2010), following a variation-aware alignment approach. Genome variants collected from the 19 founder strains, as well as variants reported for a diverse natural population (Long et al. 2013) were integrated and provided to the aligner as known variants to prevent reference biases in RNAseq read mapping (Gan et al. 2011). To facilitate accurate alignments, we provided splice junctions in the founder strains (Gan et al. 2011) and from the TAIR10 genome annotation. The full alignment parameter set for PALMapper was: -M 3 -G 0 -E 3 -l 12 -L 14 -K 12 -C 14 -I 5000 -NI 1 -SA 5 -UA 50 -CT 50 -JA 15 -JI 1 -z 10 -S -seed-hit-truncate-threshold 100 -report-map-read -report-spliced-read -report-map-region -report-splice-sites 0.9 -filter-max-mismatches 0 -filter-max-gaps 0 -filter-splice-region 5 -min-spliced-segment-len 1 -qpalma-use-map-max-len 10 -f bam -qpalma-prb-offset-fix -junction-remapping <junction_file> -score-annotated-splice-sites <junction_file> -max-dp-deletions 2 -use-variants-editop-filter -use-variants <variant_file> -filter-variants-minuse 1 -merge-variant-source-ids -use-iupac-snp-variants -filter-variants-map-window 20 -iupac-genome -filter-variants-maxlen 100 -index-precache
Gene expression quantification
We used a custom python script that counted the number of reads overlapping with at least one exonic position of an annotated gene feature. For each read, only the best alignment was considered for counting, and we excluded alignments if the alignment (i) overlapped an annotated intron, (ii) was entirely in a region where two or more annotated genes overlap, or (iii) did not start at a position inside an exon in all annotated isoforms. For each gene feature, the number of reads passing these filters was used as the expression count.
Effects of SVs on gene expression
We considered only SVs with accurate breakpoints (see section Validation by de novo contigs). A total of 119 TAIR10 genes spanned SV breakpoints (i.e., were disrupted by SVs), and 6909 were inside them (transposed, inverted, or duplicated). Genes were divided into three categories: disrupted by breakpoints, within SV-regions, and outside SVs, and compared with respect to mean and variance using t-tests. We also computed the correlation of these genes with their local read anomaly values (for the six read anomaly types), i.e., with the 10 kb source region that contains the gene, and compared the mean and variance (by a t-test and an F-test, respectively) of the Pearson correlation coefficients across categories.
Heritability
We computed genetic relationship matrices between MAGIC lines three ways:
Identity by descent (haplotype-based)
Each MAGIC chromosome is a mosaic of the 19 founders, which we used to determine identity by descent (IBD). Across MAGIC lines, we identified the union of all mosaic breakpoints, and then segmented the genome of each MAGIC line according to these breakpoints. Thus, by construction, the founder haplotype for each line is constant within each segment. The founder haplotype in segment in line is represented by an indicator matrix which is if the founder is and otherwise. Then indicates whether lines are IBD at and if is the fraction of the genome covered by then the fraction of the genome IBD for lines is
This matrix is then standardized to take the form of a genetic relationship matrix. Let be the probability that, given the observed population-wide founder haplotype fractions at two randomly sampled lines are IBD, i.e.,
Define and hence standardize the IBD matrix as
which has main diagonal 1, and off diagonal elements in the range Note that, in a small fraction of cases, and the corresponding values of all take the same minimum (the horizontal line of points in Figure 8C).
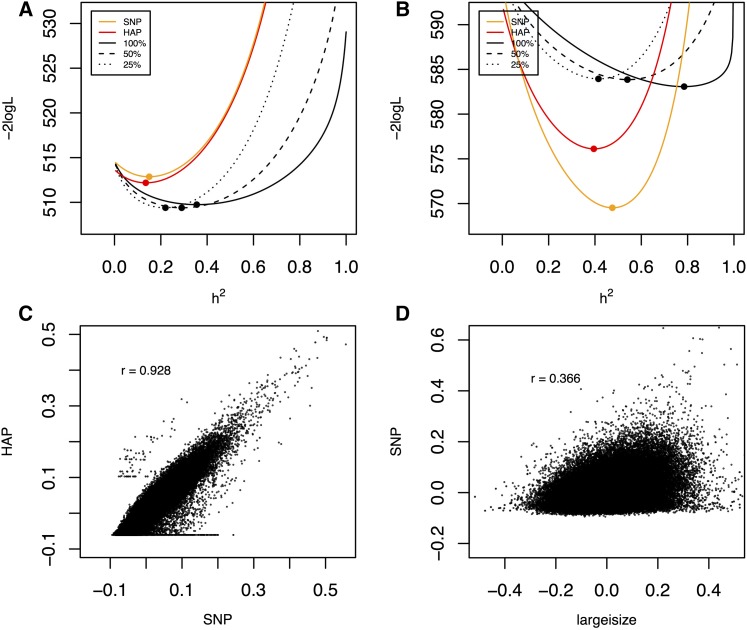
Figure 8.
(A, B) log-likelihood curves for two phenotypes Days.to.germ and Bolting.Bath (both with large insert size anomalies), illustrating contrasting behavior of heritability estimates based on structural variants, SNPs, and haplotypes. Log-likelihood curves as functions of heritability are plotted for the GRMs estimated from SNPs, haplotypes, and various fractions of anomalies. The maximum likelihood estimates of each heritability measure correspond to the minima of the corresponding curves, and are marked with dots. (C, D) Scatter plots comparing the off-diagonal elements of genetic relationship matrices. (C) vs. (D) vs.
Identity by state (SNP-based)
SNPs were imputed in the MAGIC lines by using the haplotype mosaics and the filtered set of SNP variants in the 19 founders. If encodes the homozygous SNP genotype in individual and SNP and, if is the allele frequency at then the normalized genotype is
since the MAGIC lines are almost fully inbred the normalization differs slightly from an outbred population under Hardy-Weinberg equilibrium. The SNP-based GRM is the positive semi-definite matrix with elements or .
Read Anomalies :
We constructed read-anomaly GRMs by analogy to SNP-based GRMs. Let be the read anomaly trait for individual at locus Let and be the sample means and variances. Define the standardized trait matrix with elements
where is the number of loci. The relationship between individuals is i.e., . We computed a matrix for each of the six measures of read anomaly. The choice of contributing loci was varied as described in the Results. In addition, we ignored loci with no anomaly variation, or where only a small fraction (<3%) of individuals varied.
For a phenotype measured in the MAGIC lines, and a given matrix, the variance matrix is represented by the mixed model where is the identity matrix, and are the genetic and environmental components of variance and the phenotypic heritability is We estimated heritability by minimizing the negative log-likelihood with respect to using purpose-written R code based on the eigen-decomposition in (Kang et al. 2008a).
Data availability
Source codes and supporting data are available from http://mtweb.cs.ucl.ac.uk/mus/www/19genomes/magic.html. MAGIC Genomic short read sequence data are available from ENA under study accession PRJEB19252. MAGIC RNAseq data are available from GEO under series number GSE94107.
Results
Structural variants as quantitative traits
We combined ideas from signature-based SV identification and quantitative genetics to analyze structural variation in a population. The following scenario motivates our reasoning: suppose an SV arose in a certain population ancestor, α, transposing a genomic segment, s, originating at a “source” locus, L, and targeting to a “sink” locus, M. Source and sink can be coincident or unlinked, but, for the moment, suppose they are unlinked. If the event is transposon-mediated, then the segment s is duplicated to s′ at M, and possibly altered, leaving the original s at L. Among the descendent population, random chromosomal assortment and recombination ensures there will be a mix of individuals carrying the segment at neither, one, or both loci.
Among the descendants, one individual is sequenced, and chosen as the reference genome. Depending on the choice of reference individual, and the mechanism of transposition, the reference might carry zero or one copies of s at the source, and of s′ at the sink. Assume the reference has one copy of s, and zero copies of s′. In a population sample, only individuals that inherited the haplotype descended from α at the sink carry the transposed segment, regardless of their haplotype at the source. The sample is sequenced with short-reads, and the reads are mapped to the reference genome. Individuals carrying the transposition s′ at the sink will have reads spanning the breakpoint that split between source and sink. Hence, read mapping anomalies apparently originating at the source will be enriched in those individuals carrying the sink haplotype α; genotypes that tag α at the sink will be associated with anomalies at the source.
If, on the other hand, the reference contains both s at the source and s′ at the sink, then those individuals that did not inherit the haplotype α at the sink will appear to carry a deletion there. Reads with anomalously large insert sizes will map to the sink, and will be associated with genotypes tagging the haplotype α at the sink—the generative role played by the source will be invisible.
Similarly, by considering other situations—for example, tandem duplications—where the source and sink are coincident in a population, we would expect to encounter a mix of short-range cis and long-range trans associations between various classes of read-mapping anomalies and genotypes, depending on the history of each structural variant.
To apply these ideas in practice, we count the numbers of anomalous reads mapping to each source L in a population sample, treat it as a quantitative trait, and identify genetic loci whose genotypes correlate with variation in the SV-trait. This defines an SV-QTL associating the number of anomalous reads mapping to locus in individual , and the haplotype at sink locus in individual (Figure 1 and Materials and Methods). cis SV-QTL are where the source and sink overlap and indicate local structural variants such as CNVs, deletions and inversions; trans SV-QTL indicate transpositions (insertional translocations) or larger scale rearrangements. In this way, we may determine whether an SV is in trans, its originating haplotype, which individuals now carry it (Supplemental Material, Figure S1), and its frequency (Figure S2).
Structural variation in Arabidopsis
We used our strategy to map SVs in the 120 Mb genome of the plant A. thaliana. We sequenced 488 of the Arabidopsis MAGIC recombinant inbred lines (Kover et al. 2009) at ∼0.3× coverage using 51 bp paired-end Illumina reads. The MAGIC lines descend from 19 ancestral founder accessions that have been previously sequenced at high coverage (Gan et al. 2011) (Table S1), such that each MAGIC chromosome is a mosaic of the 19 founder haplotypes. Consequently, we expect most SVs segregating in MAGIC to also segregate in the founders, thereby providing a means of verifying any SVs we detect. The choice of MAGIC lines rather than natural accessions means that the confounding effects of population structure, and of selection, are largely absent from the population. Very rare alleles with frequency below 1/19 = 4.5% are uncommon, increasing the power to detect QTL. However, MAGIC QTL mapping resolution is also poorer, at ∼200 kb, compared to ∼10 kb in natural accessions.
We mapped the reads to the TAIR10 reference using Stampy (Li and Durbin 2010; Lunter and Goodson 2011), and inferred the mosaic of each line using a hidden Markov model (HMM) implemented in the software “reconstruction” available from http://mtweb.cs.ucl.ac.uk/mus/www/19genomes/magic.html. The algorithm uses as input SNP calls for each MAGIC genome, and 1.2 M biallelic variants segregating in the 19 founders (excluding loci tagged as within transposons, and those sites called as heterozygous or multi-allelic in the founders) (Gan et al. 2011), and finds the most likely sequence of haplotype assignments for each chromosome. Because the lines were called at low coverage, most SNP sites were not covered by reads in any given line; consequently we called on average 301k SNPs per line (using GATK; McKenna et al. 2010) (i.e., a randomly sampled of ∼25% of the 1.2 M sites). However, these data are sufficient for the HMM to determine the founder mosaic accurately; we estimated by simulation that mosaic breakpoints (which correspond to recombination events) were mapped to within ∼2 kb (data not shown).
Using this procedure, we reconstructed each MAGIC genome into ∼34 haplotype blocks, on average, with mean size 3.48 Mb, representing contributions from ∼11 founder haplotypes (Table S2). We imputed the full variant catalog into each line. Comparison of imputed SNPs with 782 GoldenGate SNP genotypes measured in 370 of the MAGIC lines (Kover et al. 2009) showed 98% concordance.
To map SVs, we divided the reference genome into 11,915 abutting source loci, each 10 kb wide, and computed six measures of anomalous read mapping in each locus (6 × 11,915 = 71,490 SV trait vectors) (Materials and Methods, Table 1). Four of these measures address different types of anomalous read mapping, diagnostic of specific anomalies, namely high read coverage for duplications, strandedness of reads for inversions, anomalously large insert size for translocations, and unpaired reads for deletions. The remaining two measures are linear combinations of other measures.
Genetic association between each of the SV-trait vectors and the local haplotype space was determined using a one-way ANOVA. We chose to determine association at the level of haplotypes rather than SNPs for two reasons. First, the founder haplotype space in the MAGIC lines is well-defined, and measuring association with haplotypes can capture relationships invisible at the level of SNPs. Second, the set of haplotype tests—defined by the union of all the breakpoints across the lines, comprising 16,700 haplotype blocks, such that the ancestral haplotype of all lines is unchanged within each block—means ∼75× fewer tests are performed, thereby speeding up the procedure (Materials and Methods). To determine genome-wide significance thresholds for SV-QTL, we performed 100 phenotype permutations for each trait, and then fitted extreme value distributions (evd) to the genome-wide maxima of the permutations (Dudbridge and Koeleman 2004) (Materials and Methods).
At this 1% FDR (evd P < 0.001), we mapped 10,275 SV-QTL in total. Table S3 shows mapped QTL per read anomaly category; 3773 SV-QTL had coincident sources and sinks, probably corresponding to the same SV, and were merged, leaving 6502 SV-QTL, tabulated in Table S4. Of these, 1604 (25%) were trans, defined as mapping >2 Mb from the source. Overall, 4073/11,915 (34.2%) source loci harbored structural variants. While we have greater power to detect larger SVs, 2379 overlapped annotated indels <2 kb (Gan et al. 2011).
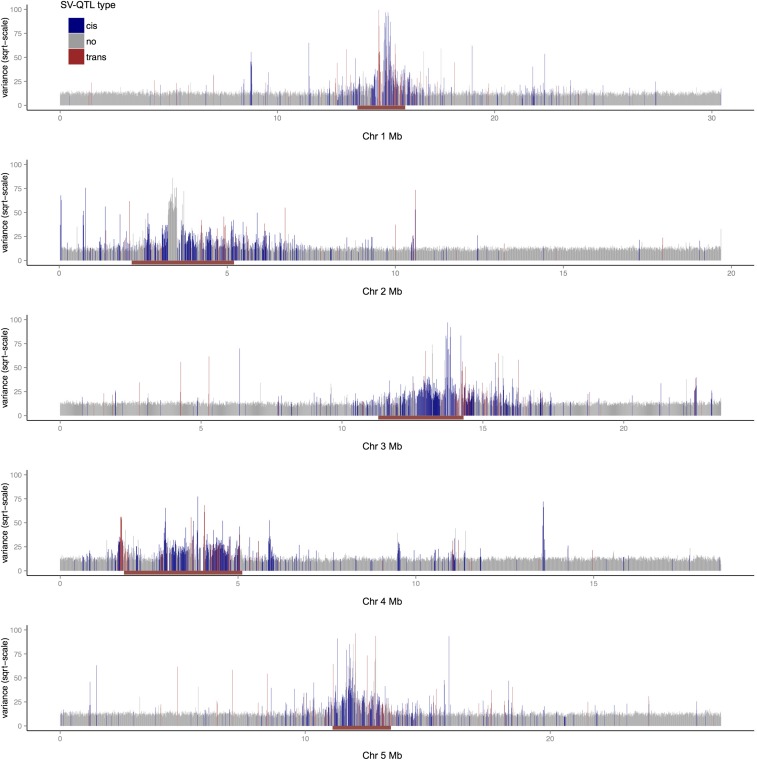
The likelihood that a structural trait vector has an SV-QTL increases with its variance (Figure 2). SV-QTL are enriched around centromeres, as expected. Away from the centromeres, Figure 2 also shows that bins with variable SV traits are isolated, rather than in clusters. Figure 3A shows the genome-wide distribution of SV-QTL segregating in one MAGIC founder, Ler-0. Figure 3 and Table S3 show that trans SV-QTL link all five chromosomes.
Figure 2.
Genome-wide distribution of the variance for the trait “improperly paired reads,” computed in 10-kb windows. The x-axis shows genomic position and the y-axis the variance of each trait vector scaled by its mean. Each vertical line corresponds to a window; those with SV-QTL are blue (cis) and red (trans). The centromeres are marked by brown horizontal bars.
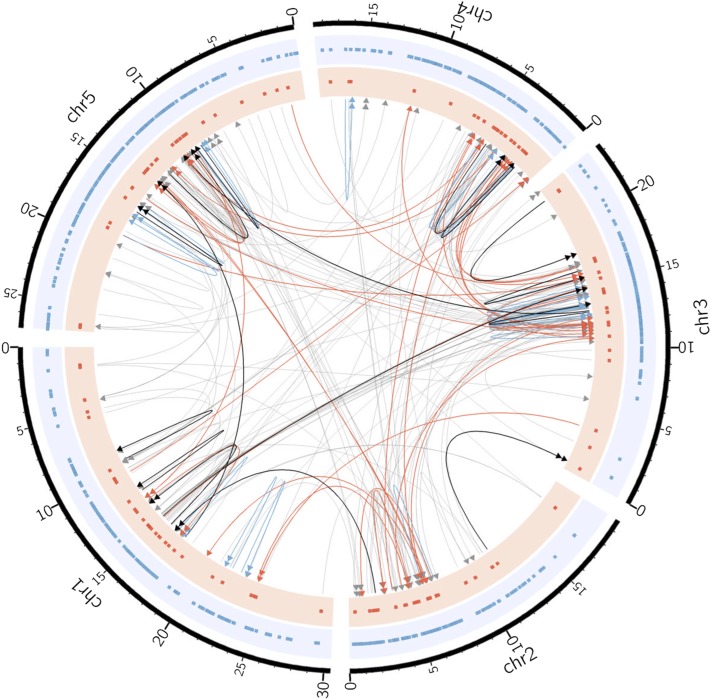
Figure 3.
Structural variants segregating in the accession Ler-0. The gray directed lines show SV-QTL, with the arrows pointing toward the sink locus. Red and blue links indicate 37 trans and 30 cis SV-QTL, confirmed by de novo contigs. The black links show 16 SVs confirmed by PCR (seven cis, nine trans). Double arrows in links indicate inversions. The dots in the red and blue tracks mark the sources (trans and cis, respectively) of all SVs associated with the Ler-0 haplotype.
In 319 SVs, we were able to pinpoint both breakpoints exactly (see Validation) Mean SV size was 53 kb in these SVs, and the largest was 189 kb. Thus, many of the SVs we discovered are too large to be due to insertions of small transposable elements. This probably reflects our lack of power to detect very small events, but also emphasizes that not all SVs are driven by mobile elements.
Validation
Genome-wide confirmation of SVs using short-read sequence is challenging because SV breakpoints often associate with transposons and repeats that hinder read-mapping and reassembly. However, among our SV-QTL are several known rearrangements. These include trans SV-QTL linking a cluster of rDNA repeats at ∼14.2 Mb on chromosome 3, to clusters at the ends of chromosome 2. Polymorphisms in these clusters are implicated in massive genome size variations among Arabidopsis accessions (Long et al. 2013; Rabanal et al. 2016). We also identified the known knob inversion on chromosome 4 as reciprocal transpositions linking 1.61 and 2.65 Mb (Fransz et al. 2000), and a 93 kb inverted transposition identified previously in a cross between Ler-0 and Col-0 (Wijnker et al. 2013), and found it was present in 12 MAGIC founders.
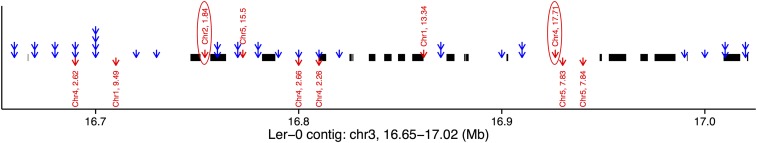
To validate further SVs, we compared our SV calls for the founder accession Ler-0 against two Ler-0 contigs (chr3:16.65–17.02 Mb, chr5:25.06–25.23 Mb) that were independently resequenced and manually reassembled (Lai et al. 2011), thereby constituting a gold standard for comparison. The chromosome 3 contig (Figure 4) is enriched in SVs (83 indels, 31 > 100 bp), consistent with our analysis: 42 SV-QTL sources (36 cis and six trans) are in this region, and four trans SV-QTL map into it. As would be expected, the sources of these SV-QTL are within gaps in the contig. Furthermore, alignment revealed two long-range SVs within the contig (a transposition and a duplication that align to chromosomes 4 and 2, respectively), which coincide with the source and sink of two trans SV-QTL mapped within the contig. Similarly, in the chromosome 5 contig, six cis SV-QTL correspond to deletions (Figure S3).
Figure 4.
Alignment of a manually assembled contig from Ler-0, chr3:16.65–17.02 Mb to the reference annotated with SV-QTL. Thick black lines show alignments to reference genome. Blue arrows show the sources of cis SV-QTL; stacked arrows mean multiple read anomaly traits had SV-QTL. Red arrows display trans QTL with arrows starting from the source, and pointing toward the sink. Gaps in the contig alignment indicate loci where Ler-0 did not align to the reference, with the exception of two transposed segments that mapped to chromosomes 2 and 4 at positions concordant with the sources of two trans SV-QTL (circled).
We also analyzed an independent de novo assembly of Ler-0 built from long PAC-BIO reads, GenBank accession GCA_000835945.1 (Berlin et al. 2015) to validate our trans SV predictions. This assembly was constructed algorithmically without manual revisions, and so is not guaranteed to be correct. Further, the Ler-0 individual sequenced in the PAC-BIO assembly was different from the individual that founded the MAGIC population, and therefore might carry private structural variations. Nonetheless, we expect it to be more accurate and contiguous than a Ler-0 assembly built from short Illumina reads alone. We took those 3080 Illumina paired-end reads for Ler-0 from Gan et al. (2011) that carried large insert size mapping anomalies when mapped to TAIR10, and that mapped to the sources of our predicted Ler-0 trans SV-QTL, and then mapped them to the PAC-BIO assembly using BWA (Li and Durbin 2010). These Illumina reads are from an individual grown from the same batch of seeds used to found the MAGIC population in ∼2007, and should therefore share the same structural variants. Read anomalies with correct SV predictions should map contiguously to the PAC-BIO assembly, if the latter assembly accurately portrays the Ler-0 genome. We found 2460 (80%) of these formerly split Illumina read pairs now mapped contiguously, defined as both members of a read-pair mapping to the PAC-BIO assembly with an insert size below 600 bp.
With the exception of these manually assembled Ler-0 contigs, and the provisional Ler-0 PAC-BIO assembly, the MAGIC founders are not contiguously reassembled into a genome-wide gold standard reference panel. Nevertheless, they provide information to test our predictions: at each SV-QTL, we predicted which founder haplotypes carried SVs at the origination of the population, under the assumption the SV was biallelic. Using the low coverage data for the 488 MAGIC lines, at each SV-QTL, we then predicted which founders carried the SV allele, based on correspondence between their SV-trait value and predicted founder allele, using the fact that SV haplotypes have elevated anomalous reads. We did this confidently at 2391 SVs where the founders partitioned into two groups, the remainder having complex multi-allelic SV predictions (Materials and Methods). Examples of founder partitions for cis and trans SV-QTL are shown in Figure S1.
We then examined the independently collected high-coverage reads in each of the 19 MAGIC founders (Gan et al. 2011) for read-mapping signatures that supported the predicted grouping of founders at each SV. We counted the read pairs linking source and sink at each of the 2391 SVs in the 19 high coverage founders. At 1585/2391 (66.3%, FDR 7.5%) SVs, we observed significant differences in anomalies between the predicted groupings of founders (Figure S4, which also shows that the majority of SVs were mapped within 50 kb). In the founders, the mean SV allele frequency was 6/19 = 31%. Only 387 (12%) were private to a single founder (Figure S2), in contrast to the fraction of SNPs (45%) that are private to a single founder (Gan et al. 2011). This may reflect our lack of power to detect private SVs.
Independent analysis of low-coverage reads from the 488 MAGIC genomes (Materials and Methods) supported 1228/2391 (51.3%, most also supported by the founder genomes) and 1631/4111 (39.7%) of those remaining SVs without founder predictions. In total, 2965/6502 (45.6%) SVs were supported by either method.
Breakpoint prediction and confirmation
To estimate SV sizes and identify SV breakpoints to test by PCR, we next de novo assembled the high-coverage sequence data for the MAGIC founders into high-quality short contigs, each up to a few kilobases long. We aligned these contigs to the reference to find alignments split between sources and sinks. We found 2619 contigs with alignments split into disjoint pieces across 420 QTL sources and sinks, suggesting a cut-and-paste mechanism (Materials and Methods, and Table S4). Of these, at 319 SV-QTL both breakpoints were identified. We found 460,656 (8.3%) shared contigs whose alignments overlapped between source and sink regions.
We designed PCR primers at 77 breakpoints from 45 predicted SVs (both breakpoints in seven SVs, and one in each of the remaining 38). In 30 (66.6%) SVs, 46 type 1 experiments (designed to amplify in the presence of the predicted SV but not the reference, Materials and Methods), at least one breakpoint was confirmed, i.e., there was at least one type 1 experiment that amplified in founders predicted to carry the SV, while not producing a product in the reference, as expected. In a further seven SV-QTL (15.6%) (15 type 1 experiments) the founders carrying an SV-QTL were amplified, but, unexpectedly, the reference genome was also amplified. This suggests the presence of duplicated sequence nearby, causing unexpected binding of one of the primers. It might also indicate errors in the reference assembly. In 10/15 cases, we observed duplications (multiple PCR bands) in >2 founders. However, in all 15 cases at least three founder genomes amplified differently from the reference, indicating that the locus is structurally variant, but not exactly as predicted. The remaining 16 type 1 experiments failed to amplify in any founder. Of the 19 type 2 experiments (designed to amplify in the presence of the reference but not the SV), 16 amplified as expected, two were ambiguous, and one failed.
Overall, we confirmed at least 30 (66.6%) SVs at either or both breakpoints, and, at a further 7 (15.6%), we found evidence of structural variation. Among the total of 37 SVs supported by PCR, we confirmed 61 (79%) breakpoints. There were 14 cis (six inversions, seven transpositions, and one indel) and 23 trans (13 with inversions) SV-QTL (Table S5). Consistent with our difficulties in predicting biallelic founder alleles, in 11 SVs, the breakpoints were polymorphic among the founders carrying the SV, and, in five transpositions, the orientation of the SV differed between founders.
Effects of SVs on phenotypic QTL and gene expression
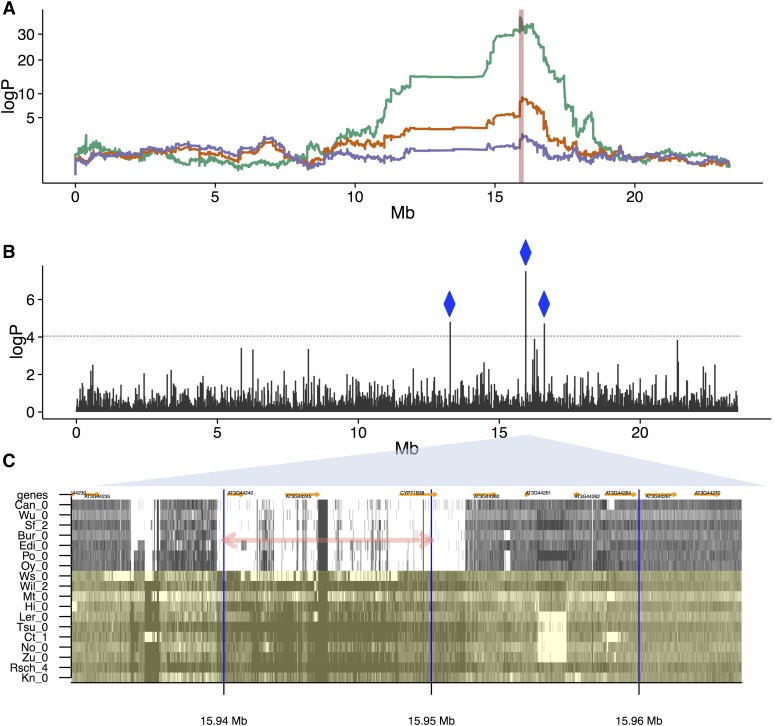
We next investigated associations between SVs and nine physiological phenotypes, either previously published (Kover et al. 2009; Springate and Kover 2014), or first reported in this study (Table S6). We found 16 distinct SV-QTL (eight in trans, Table S7) that overlap physiological QTL. In some cases, regressing the SV-trait from the physiological trait ablated the physiological QTL, consistent with, albeit not proving, that the SV is causal. This is illustrated by a QTL for germination time (Kover et al. 2009) on chromosome 3, which is ablated by a cis SV-QTL for unpaired reads at ∼15,936,650–15,951,640 bp (Figure 5, A and B). Our analysis predicts seven founders carry a deletion at this locus, which was confirmed by the independent founder sequences (Figure 5C), revealing a 15 kb deletion containing three genes, AT3G44240 (Polynucleotidyl transferase, ribonuclease H-like superfamily protein), AT3G44245 (pseudogene of cytochrome P450, family 71, subfamily B, polypeptide 21), and CYP71B38 (AT3G44250, cytochrome P450, family 71, subfamily B, polypeptide 38). Other SVs segregate nearby, but with allelic patterns inconsistent with the trait, and therefore unlikely to be causal. It is probable that the deletion contains the causal variant(s). The deleted genes are not known to affect germination, although a mutant of another polynucleotidyl transferase, AHG2 (AT1G55870) does (Nishimura et al. 2009).
Figure 5.
Effects of SVs on germination time. (A) Genome scans over chromosome 3 (x-axis: genomic position, y-axis: logP of association). Orange: association of local haplotype with germination time (days), peaking at 15.93 Mb. Green: association of local haplotype with the SV trait unpaired reads at the source locus 15.94–15.95 Mb (indicated by the vertical red line), explaining 8.13% of the variance in germination time, with an SV-QTL mapped at the same position as the germination QTL. Purple: residuals of germination time after regressing out the SV trait, ablating the QTL. (B) Chromosome-wide Pearson correlations between germination time and the numbers of unpaired reads measured at each 10 kb source locus (x-axis: genomic position, y-axis: −log10 P-value of test that the correlation is zero). Three source loci correlate strongly with germination (logP > 4), all with cis SV-QTL (blue diamonds). (C) Structural variation in the MAGIC founders. Shown is the read coverage in 18 accessions (labeled on y-axis),covering ∼30 kb surrounding ∼15.94 Mb (x-axis). Dark shades indicate high coverage, light shades low coverage. The 10 kb intervals used to define source loci are delineated by vertical blue lines. The source locus giving rise to the SV-QTL in (A), (B) is marked with a pink double-arrow. Those founder accessions predicted to carry the reference allele (No-0, Ct-1, Mt-0, Wil-2, Ler-0, Tsu-0, Rsch-4, Kn-0, Zu-0, Hi-0, and Ws-0) are in green, those predicted to carry the SV are in gray. Genes are annotated in orange.
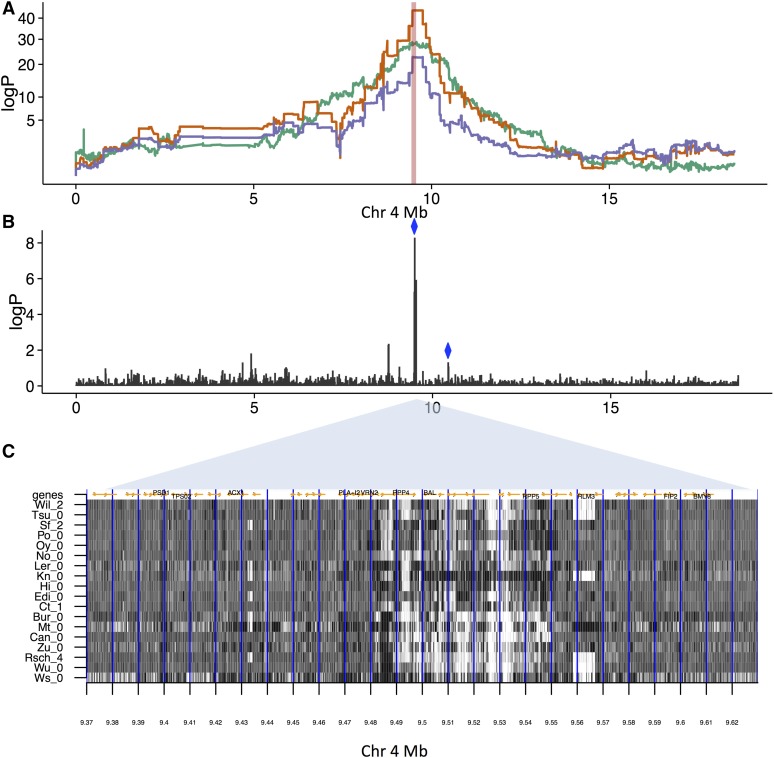
We found similar effects on the chromosome 4 QTL for resistance to the fungal pathogen A. laibachii, isolate Nc14 (Thines et al. 2009) (Figure 6 and Table S7). Variation in the number of unpaired reads at 9.50–9.51 Mb explains 18.3% of the variance in resistance, and is adjacent to a cluster of leucine-rich repeat genes, and the genes RPP4 (Van Der Biezen et al. 2002), BAL (Yi and Richards 2009), and RPP5. This locus is rearranged in some Arabidopsis accessions, and is known to affect disease resistance (Yi and Richards 2009); Figure 6 confirms the founder genomes have complex, polymorphic, SVs in this region. Since the resistance QTL is not completely ablated by the SV traits associated with it, additional nonstructural variants likely contribute to it.
Figure 6.
Effects of SVs on resistance to Albugo laibachii infection, (A) Genome scans on chromosome 4. Orange: Association with resistance. The peak of association for is at 9.50 Mb. Green: Association with SV-trait improperly paired reads at source 9.50–9.51. Purple: Resistance after two SV traits have been regressed out measuring improperly paired reads [sources chr4, 9.50–9.51 Mb (green line) and chr4, 10.44–10.45 Mb (not shown), both marked with blue diamonds in (B)] that together explain 24.7% of the phenotypic variance. (B) logP of association between SV traits for improperly paired reads and the resistance trait. There is a cluster of associated traits near 9.50 Mb, in addition to the more weakly associated trait at 10.44–10.45 Mb. (C) Structural variation in high-coverage sequence in the MAGIC founders ∼9.50 Mb. Shown is the number of improperly paired reads (dark: high values, light: low values) in 18 accessions (labeled on y-axis), between 9.37 and 9.63 Mb (x-axis). The 10 kb intervals used to define source loci are delineated by vertical blue lines. There is a region of complex structural variation spanning ∼9.48–9.55 Mb, with considerable variation between the founder accessions. Genes are marked by orange arrows, and selected genes, some implicated in disease resistance at this locus, are labeled.
Importantly, Figure 5B and Figure 6B show that correlations between SV traits and phenotypes are tightly localized, generally within the width of a single SV trait window, in contrast with wider linkage disequilibrium decay seen in QTL genetic mapping (Figure 5A). Therefore, correlations between SV traits and physiological traits pinpoint causal variants within physiological QTL that are otherwise too broad to localize [mapping resolution in MAGIC is ∼200 kb (Kover et al. 2009)].
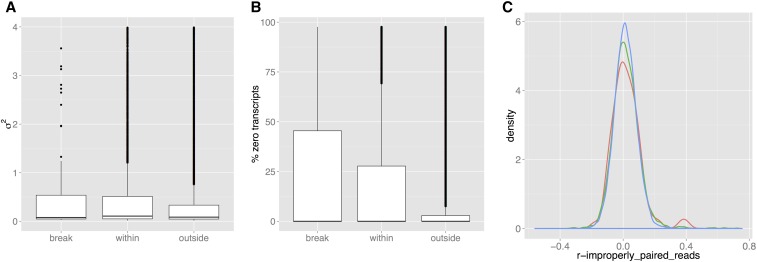
We also corroborated studies (Yalcin et al. 2011; Quadrana et al. 2016) showing SVs associate with gene dysregulation, even when the gene sequence is undisturbed. Within those SVs with mapped breakpoints, 119 genes spanned the breakpoints, 6909 lay inside the SVs (Table S8), and 21,747 outside. Using RNA-seq from 200 MAGIC aerial seedlings, scaled expression variance increased among genes spanning breakpoints (t-test: ) and within SVs () (Figure 7A). Similarly, more lines exhibited silenced transcripts for genes spanning breakpoints (t-test ), or within SVs () (Figure 7B). Expression within SVs was more correlated with local SV traits than outside SVs (F-test ) (Figure 7C).
Figure 7.
Variation of expression in 200 MAGIC leaf transcriptomes, in genes spanning SV breakpoints, within SVs or outside SVs. (A) Boxplots of transcript variance (scaled by the mean). (B) Boxplots of the fractions of silenced genes (C) Distributions of the Pearson correlations between gene expression and number of improperly-paired reads in the locus containing the gene (red: spanning breakpoints, green: within SVs, blue: outside SVs).
Effects of SV-traits on heritability
Finally, we treated the SV traits as if they were quantitative, noisy genotypes, to compute pairwise correlations between MAGIC lines, as weighted correlations of their SV traits (Materials and Methods). We constructed SV genetic relationship matrices (GRMs) which we used to compute the SV-heritability of each of the physiological traits mapped above by analogy with the mixed models used for estimating SNP-based heritability (Kang et al. 2008a). This idea resembles the use of gene expression data to model intersample relationships (Kang et al. 2008b). We also compared these SV-heritabilities with those obtained from “classical” haplotype or SNP-based GRMs (Table 2). was computed from the identity between haplotype mosaics (i.e., IBD), while and were computed from the correlations of 1.2 M imputed SNPs or 12k SV-traits, respectively (Materials and Methods). We also computed SV heritability when only the most variable 50 or 25% of SV-traits were included, to test if heritability was concentrated at the most structurally variable loci.
Table 2. Estimates of heritability.
| Phenotype | |||||||
|---|---|---|---|---|---|---|---|
| IP | LIS | SS | U | U + LIS | |||
| Resistance (resistance to A. laibachii) | 0.000 (0.139) | 0.258 (0.085) | 0.490 (0.335) | 0.511 (0.307) | 0.000 (NA) | 0.673 (0.504) | 0.503 (0.314) |
| RosetteLeafNumber.LongDay (number of leaves in a rosette for plants grown under long daylight | 0.228 (0.081) | 0.322 (0.076) | 0.463 (0.148) | 0.456 (0.146) | 1.000 (NA) | 1.000 (0.377) | 0.447 (0.146) |
| RosetteLeafNumber.ShortDay (number of leaves in a rosette for plants grown under short daylight) | 0.038 (0.060) | 0.047 (0.062) | 0.000 (NA) | 0.000 (NA) | 0.000 (NA) | 0.000 (NA) | 0.000 (NA) |
| Bolting.Bath (bolting time in a greenhouse) | 0.426 (0.064) | 0.476 (0.048) | 0.783 (0.093) | 0.783 (0.093) | 0.952 (0.047) | 0.989 (0.025) | 0.785 (0.092) |
| Days.to.germ. (germination time) | 0.220 (0.068) | 0.149 (0.063) | 0.385 (0.116) | 0.357 (0.113) | 0.598 (0.165) | 0.835 (0.146) | 0.365 (0.114) |
| FieldFT.pl (flowering time in the field) | 0.000 (0.068) | 0.095 (0.076) | 0.000 (0.179) | 0.000 (0.130) | 0.000 (0.913) | 0.000 (NA) | 0.000 (0.145) |
| FieldRD.pl (rosette diameter plasticity) | 0.000 (NA) | 0.000 (0.063) | 0.000 (0.085) | 0.000 (0.084) | 0.000 (0.239) | 0.166 (0.220) | 0.000 (0.085) |
| Leaves.day.28.given.days.to.germ (residuals for number of leaves at day 28 regressed on germination) | 0.193 (0.081) | 0.299 (0.066) | 0.391 (0.146) | 0.362 (0.140) | 0.836 (0.189) | 0.675 (0.272) | 0.366 (0.142) |
| ttl_branch.BATH (total number of branches of plants) | 0.106 (0.048) | 0.196 (0.054) | 0.276 (0.104) | 0.275 (0.100) | 0.419 (0.193) | 0.616 (0.214) | 0.279 (0.102) |
is haplotype-based heritability. is SNP-based heritability. is the heritability estimated from structural variant anomaly traits. Numbers in brackets are the standard errors (SEs) of the heritability estimates above. Heritability for excess reads are not reported because the fraction of bins in any individual containing nonzero entries was too small. IP, Improperly-paired; LIS, Large Insert Size; SS, Same Strand; U, Unpaired; U + LIS, Unpaired or Large Insert Size.
As expected, SNP-based heritability resembles haplotype-based heritability for all phenotypes tested. However, the heritability captured by the six measures of SV anomaly is more variable, sometimes being close to zero, but sometimes exceeding classical heritability considerably (Table 2). The SE of was typically about twice that of or (∼0.1 compared to 0.05), presumably reflecting greater uncertainty in SV-traits. Therefore the larger estimates should be treated with caution. Nonetheless, for phenotypes such as time to germination or bolting, the SEs of all estimates are ∼0.05, and it is possible to compare them. Figure 8, A and B illustrates likelihood curves for the times to germination (A) and bolting (B), for SNP, haplotype and large insert-size anomalies. Visualizing the entire curves gives a better sense of the uncertainty of the maximum likelihood estimates at the curves’ minima (the SEs in Table 2 are asymptotic estimates based on the curvature at these minima). Figure 8B shows that, for bolting time, the heritability attributable to all largeisize SV-traits, is close to 80%, compared to 40–50% for haplotype or SNP-based estimates. As the fraction of SV traits is reduced by progressively removing those traits with lower variance, reduces to that of SNPs or haplotypes. This suggests that there is genome-wide structural variation that is not tagged by standard genetic variation, and which has important effects on specific phenotypes. These effects are not universal, as Figure 8A shows for germination time, where heritability is similar for all GRMs.
The independence of the heritability estimates is borne out by low correlations between the corresponding elements of SNP and SV-based GRMs, which range ∼0.3 depending on the anomaly type (Figure 8D shows the relationship between GRMs computed from SNPs vs. large insert size anomalies), compared to the correlation of 0.93 between SNP and haplotype based GRMs (Figure 8C).
Discussion
We have combined analysis of the read-mapping signatures commonly used to detect SVs in individuals sequenced at high coverage, with association mapping in populations (Durkin et al. 2012). Related ideas based on linkage disequilibrium have been used for mapping unlocalized contigs into reference assemblies (Genovese et al. 2013). In doing so, we have generated a partial catalog of SVs in Arabidopsis, although our purpose is not to call SVs systematically, a task that remains challenging with short reads. Rather, we have shown how the impact of SVs can be assayed without necessarily calling them, or mapping their breakpoints.
In this way, we can distinguish transpositions from local SVs, and determine the approximate locations of transpositions. The privileged role of the reference genome in the analysis means that some transpositions appear as deletions, so we probably have underestimated their true frequency. Nevertheless, a quarter of the SVs we detected are transpositions. Given the large numbers of transposable elements in Arabidopsis [>11,000 from >300 families are annotated in the reference (Quadrana et al. 2016)], this is unsurprising. However, many of the SVs we mapped are too large, covering tens of kilobases, to be single transposon-mediated events.
In the minority of cases where we could delineate breakpoints exactly, we often found SVs are complex combinations of different SV types. More often, breakpoints are not simple cut-and-paste transformations of the reference genome, as illustrated in Figure 6C. Indeed, it is frequently impossible to determine precisely the changes that led to many observed structural variants.
Because we used ultra-low-coverage 0.3× sequence data, we divided the Arabidopsis genome into 10 kb bins when counting read-mapping anomalies. With higher coverage and a larger sample size, it would be possible to use a larger number of narrower bins, thereby improving resolution. The release of >3000 rice genomes sequenced at ∼14× (Li et al. 2014), and >1000 Arabidopsis accessions sequenced at over ∼20× (Alonso-Blanco et al. 2016) means that there are now large collections of inbred plant genomes available for analysis. Both of these sets are worldwide surveys of germplasm, in which we expect SVs to contribute significantly to, and be confounded with, their extensive population structure, in contrast to the MAGIC population used here. Disentangling these effects will be a challenging but important task.
Similarly, extending this methodology to outbred individuals, either descended from inbred strains [such as the mouse diversity outcross, DO (Svenson et al. 2012)], or natural populations, such as humans, requires modification. Heterozygous loci may carry different SVs with different SV-QTL. It is likely that a population with a limited haplotype space, like the DO, will be less challenging to map than will natural populations containing many low-frequency SVs associated with mobile elements. Large populations of sequenced humans are now available (Cai et al. 2015), making such investigations possible.
Mapping SVs as traits in a population brings new insights to the problem of QTL analysis. First, an SV trait inside a QTL may entirely explain the genetic effect at the QTL, and hence provide support for being the causal variant (e.g., Figure 5). Second, SV traits are much more tightly localized than are QTL: there is little or no correlation between neighboring SV traits, so there are no effects of linkage disequilibrium. Our analysis also shows that expression of genes is often dysregulated or even silenced within large SVs, suggesting that an SV causes multiple regulatory and phenotypic effects.
Finally, even in a population like Arabidopsis MAGIC where the local haplotype space is known, structural variation has an impact on heritability that cannot be explained by standard genetic variation. This is unexpected given the breeding history and genetic architecture of the MAGIC lines, for, if an SV segregated among the founders of the MAGIC lines, then it should be tagged by the local haplotype context, and therefore contribute to both and
One possible explanation is that structural variation at loci rich in mobile elements accumulates independently within each lineage, leading to SVs that are private to each MAGIC line but tend to occur at the same loci, thereby creating similar phenotypic effects. Supporting this, in our analysis, the SV-relationship matrix is calculated empirically, without regard to the ancestry of the MAGIC lines, being solely a function of the correlations of read-mapping anomalies. Therefore, recalling that the history of each MAGIC line includes a private lineage of at least five generations of selfing, were SVs to accumulate recurrently, but independently, in different lineages, then these could generate phenotypic associations invisible to SNP or haplotype variation. In Arabidopsis, some mobile elements are methylated, often in response to environmental cues, and this methylation plays a role in the epigenetic control of certain phenotypes (Ito and Kakutani 2014). This effect might contribute to the heritability associated with structural variation observed here. Testing this hypothesis in Arabidopsis MAGIC lines would require complete and precise reassembly of each genome using long reads, annotation of mobile elements, and determination of their methylation status.
The role that recurrent, but independent, genomic rearrangements might play in Arabidopsis, and in other species remains to be seen, but there is no a priori reason why it should not drive phenotypic variation. The unstable inheritance of 45S rDNA genes in Arabidopsis lends support to this view (Rabanal et al. 2017). The approach used here may therefore have wider application to other populations to characterize the impact of cryptic structural variation on phenotypes.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.192823/-/DC1.
Acknowledgments
We thank Fernando Rabanal for comments on the manuscript. M.I. and R.M. were supported by the Wellcome Trust Core Award grant 090532/Z/09/Z. M.I. was supported by a grant from the UK Engineering and Physical Sciences Research Council. R.M.C. was supported by National Science Foundation grant 0929262. E.J.O. and R.G. were supported by National Institutes of Health Genetics training grant T32 GM-007464.
Footnotes
Communicating editor: T. F. C. Mackay
Literature Cited
- Alonso-Blanco C., Andrade J., Becker C., Bemm F., Bergelson J., et al. , 2016. 1,135 Genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 166: 481–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin K., Koren S., Chin C.-S., Drake J. P., Landolin J. M., et al. , 2015. Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat. Biotechnol. 33: 623–630 (erratum: Nat Biotechnol. 33: 1109). [DOI] [PubMed] [Google Scholar]
- Cabrera C. P., Navarro P., Huffman J. E., Wright A. F., Hayward C., et al. , 2012. Uncovering networks from genome-wide association studies via circular genomic permutation. G3 2: 1067–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai N., Bigdeli T. B., Kretzschmar W., Li Y., Liang J., et al. , 2015. Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature 523: 588–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Schneeberger K., Ossowski S., Gunther T., Bender S., et al. , 2011. Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat. Genet. 43: 956–963. [DOI] [PubMed] [Google Scholar]
- Chaisson M. J., Tesler G., 2012. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics 13: 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Wallis J. W., McLellan M. D., Larson D. E., Kalicki J. M., et al. , 2009. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat. Methods 6: 677–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies R. W., Flint J., Myers S., Mott R., 2016. Rapid genotype imputation from sequence without reference panels. Nat. Genet. 48: 965–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudbridge F., Koeleman B. P. C., 2004. Efficient computation of significance levels for multiple associations in large studies of correlated data, including genomewide association studies. Am. J. Hum. Genet. 75: 424–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin R. M., Abecasis G. R., Altshuler D. L., Auton A., Brooks L. D., et al. , 2010. A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durkin K., Coppieters W., Drögemüller C., Ahariz N., Cambisano N., et al. , 2012. Serial translocation by means of circular intermediates underlies colour sidedness in cattle. Nature 482: 81–84. [DOI] [PubMed] [Google Scholar]
- Fransz P. F., Armstrong S., de Jong J. H., Parnell L. D., van Drunen C., et al. , 2000. Integrated cytogenetic map of chromosome arm 4S of A. thaliana: structural organization of heterochromatic knob and centromere region. Cell 100: 367–376. [DOI] [PubMed] [Google Scholar]
- Gan X., Stegle O., Behr J., Steffen J. G., Drewe P., et al. , 2011. Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature 477: 419–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese G., R. E. Handsaker, H. Li, N. Altemose, A. M. Lindgren et al., 2013 Using population admixture to help complete maps of the human genome. Nat. Genet. 45: 406–414, 414e1–414e2. [DOI] [PMC free article] [PubMed]
- Hu T. T., Pattyn P., Bakker E. G., Cao J., Cheng J.-F., et al. , 2011. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat. Genet. 43: 476–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Kakutani T., 2014. Control of transposable elements in Arabidopsis thaliana. Chromosome Res. 22: 217–223. [DOI] [PubMed] [Google Scholar]
- Jain M., Fiddes I. T., Miga K. H., Olsen H. E., Paten B., et al. , 2015. Improved data analysis for the MinION nanopore sequencer. Nat. Methods 12: 351–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean, G., A. Kahles, V. T. Sreedharan, F. De Bona, and G. Rätsch, 2010 RNA-Seq read alignments with PALMapper. Curr. Protoc. Bioinformatics. 11: Unit 11.6. [DOI] [PubMed] [Google Scholar]
- Kang H. M., Zaitlen N. A., Wade C. M., Kirby A., Heckerman D., et al. , 2008a Efficient control of population structure in model organism association mapping. Genetics 178: 1709–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H. M., Ye C., Eskin E., 2008b Accurate discovery of expression quantitative trait loci under confounding from spurious and genuine regulatory hotspots. Genetics 180: 1909–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemen E., Gardiner A., Schultz-Larsen T., Kemen A. C., Balmuth A. L., et al. , 2011. Gene gain and loss during evolution of obligate parasitism in the white rust pathogen of Arabidopsis thaliana. PLoS Biol. 9: e1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W. J., 2002. BLAT–the BLAST-like alignment tool. Genome Res. 12: 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kover P. X., Valdar W., Trakalo J., Scarcelli N., Ehrenreich I. M., et al. , 2009. A multiparent advanced generation inter-cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet. 5: e1000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronenberg Z. N., Osborne E. J., Cone K. R., Kennedy B. J., Domyan E. T., et al. , 2015. Wham: identifying structural variants of biological consequence. PLoS Comput. Biol. 11: e1004572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai A. G., Denton-Giles M., Mueller-Roeber B., Schippers J. H., Dijkwel P. P., 2011. Positional information resolves structural variations and uncovers an evolutionarily divergent genetic locus in accessions of Arabidopsis thaliana. Genome Biol. Evol. 3: 627–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layer R. M., Chiang C., Quinlan A. R., Hall I. M., 2014. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol. 15: R84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R., 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26: 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.-Y., Wang J., Zeigler R. S., 2014. The 3,000 rice genomes project: new opportunities and challenges for future rice research. Gigascience 3: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q., Rabanal F. A., Meng D., Huber C. D., Farlow A., et al. , 2013. Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nat. Genet. 45: 884–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunter G., Goodson M., 2011. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 21: 936–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manske H. M., Kwiatkowski D. P., 2009. LookSeq: a browser-based viewer for deep sequencing data. Genome Res. 19: 2125–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., et al. , 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills R. E., Walter K., Stewart C., Handsaker R. E., Chen K., et al. , 2011. Mapping copy number variation by population-scale genome sequencing. Nature 470: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicod J., Davies R. W., Cai N., Hassett C., Goodstadt L., et al. , 2016. Genome-wide association of multiple complex traits in outbred mice by ultra-low-coverage sequencing. Nat. Genet. 48: 912–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N., Okamoto M., Narusaka M., Yasuda M., Nakashita H., et al. , 2009. ABA hypersensitive germination2–1 causes the activation of both abscisic acid and salicylic acid responses in Arabidopsis. Plant Cell Physiol. 50: 2112–2122. [DOI] [PubMed] [Google Scholar]
- Quadrana L., Bortolini Silveira A., Mayhew G. F., LeBlanc C., Martienssen R. A., et al. , 2016. The Arabidopsis thaliana mobilome and its impact at the species level. Elife 5: e15716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabanal F. A., Viktoria N., Terezie M., Novikova P. Y., Lysak M., Mott R., Nordborg M., 2017. Unstable Inheritance of 45S rRNA genes in Arabidopsis thaliana. G3: Genes, Genomes, Genetics DOI: 10.1534/g3.117.040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch T., Zichner T., Schlattl A., Stütz A. M., Benes V., et al. , 2012. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 28: i333–i339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimmer A., Phan H., Mathieson I., Iqbal Z., Twigg S. R. F., et al. , 2014. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat. Genet. 46: 912–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H., 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132: 365–386. [DOI] [PubMed] [Google Scholar]
- Simpson J. T., Pop M., 2015. The theory and practice of genome sequence assembly. Annu. Rev. Genomics Hum. Genet. 16:153–172. [DOI] [PubMed] [Google Scholar]
- Simpson J. T., McIntyre R. E., Adams D. J., Durbin R., 2010. Copy number variant detection in inbred strains from short read sequence data. Bioinformatics 26: 565–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sindi S. S., Onal S., Peng L. C., Wu H.-T., Raphael B. J., 2012. An integrative probabilistic model for identification of structural variation in sequencing data. Genome Biol. 13: R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springate D. A., Kover P. X., 2014. Plant responses to elevated temperatures: a field study on phenological sensitivity and fitness responses to simulated climate warming. Glob. Chang. Biol. 20: 456–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenson K. L., Gatti D. M., Valdar W., Welsh C. E., Cheng R., et al. , 2012. High-resolution genetic mapping using the Mouse Diversity outbred population. Genetics 190: 437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines M., Choi Y. J., Kemen E., Ploch S., Holub E. B., et al. , 2009. A new species of Albugo parasitic to Arabidopsis thaliana reveals new evolutionary patterns in white blister rusts (Albuginaceae). Persoonia 22: 123–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Biezen E. A., Freddie C. T., Kahn K., Parker J. E., Jones J. D. G., 2002. Arabidopsis RPP4 is a member of the RPP5 multigene family of TIR-NB-LRR genes and confers downy mildew resistance through multiple signalling components. Plant J. 29: 439–451. [DOI] [PubMed] [Google Scholar]
- Wijnker E., Velikkakam James G., Ding J., Becker F., Klasen J. R., et al. , 2013. The genomic landscape of meiotic crossovers and gene conversions in Arabidopsis thaliana. Elife 2: e01426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin B., Wong K., Agam A., Goodson M., Keane T. M., et al. , 2011. Sequence-based characterization of structural variation in the mouse genome. Nature 477: 326–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K., Schulz M. H., Long Q., Apweiler R., Ning Z., 2009. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 25: 2865–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H., Richards E. J., 2009. Gene duplication and hypermutation of the pathogen Resistance gene SNC1 in the Arabidopsis bal variant. Genetics 183: 1227–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source codes and supporting data are available from http://mtweb.cs.ucl.ac.uk/mus/www/19genomes/magic.html. MAGIC Genomic short read sequence data are available from ENA under study accession PRJEB19252. MAGIC RNAseq data are available from GEO under series number GSE94107.