Abstract
Wolbachia are gram-negative, obligate, intracellular bacteria carried by a majority of insect species worldwide. Here we use a Wolbachia-infected Drosophila cell line and genome-wide RNA interference (RNAi) screening to identify host factors that influence Wolbachia titer. By screening an RNAi library targeting 15,699 transcribed host genes, we identified 36 candidate genes that dramatically reduced Wolbachia titer and 41 that increased Wolbachia titer. Host gene knockdowns that reduced Wolbachia titer spanned a broad array of biological pathways including genes that influenced mitochondrial function and lipid metabolism. In addition, knockdown of seven genes in the host ubiquitin and proteolysis pathways significantly reduced Wolbachia titer. To test the in vivo relevance of these results, we found that drug and mutant inhibition of proteolysis reduced levels of Wolbachia in the Drosophila oocyte. The presence of Wolbachia in either cell lines or oocytes dramatically alters the distribution and abundance of ubiquitinated proteins. Functional studies revealed that maintenance of Wolbachia titer relies on an intact host Endoplasmic Reticulum (ER)-associated protein degradation pathway (ERAD). Accordingly, electron microscopy studies demonstrated that Wolbachia is intimately associated with the host ER and dramatically alters the morphology of this organelle. Given Wolbachia lack essential amino acid biosynthetic pathways, the reliance of Wolbachia on high rates of host proteolysis via ubiquitination and the ERAD pathways may be a key mechanism for provisioning Wolbachia with amino acids. In addition, the reliance of Wolbachia on the ERAD pathway and disruption of ER morphology suggests a previously unsuspected mechanism for Wolbachia’s potent ability to prevent RNA virus replication.
Keywords: Wolbachia, RNAi, Drosophila, oocyte, ubiquitin, virus
WOLBACHIA is a bacterial endosymbiont present in insects and filarial nematodes (Serbus et al. 2008; Werren et al. 2008). Wolbachia resides in both somatic and germline cells of its male and female insect hosts (Pietri et al. 2016). The evolutionary success of Wolbachia depends on efficient vertical transmission through the female germline. This is facilitated by Wolbachia localization to the posterior pole of the oocyte, ensuring its incorporation into the germline of the next generation. To achieve this, Wolbachia rely on host microtubules, motor proteins, and an interaction with the host pole plasm components (Kose and Karr 1995; Ferree et al. 2005; Serbus and Sullivan 2007). The success of Wolbachia also requires regulation of bacterial abundance within host somatic and germline cells. Underreplication of Wolbachia in the oocyte results in inefficient vertical transmission and overreplication of Wolbachia results in disruption of critical host cellular functions (Serbus et al. 2011; Newton et al. 2015). Cytological and PCR-based studies demonstrate that recently caught wild strains of Drosophila exhibit tremendous variability in Wolbachia titer (Unckless et al. 2009). These variations not only occur from one individual to another but also between tissues within an individual (Albertson et al. 2009; Muller et al. 2013).
A combination of host and Wolbachia factors as well as the environment influence Wolbachia abundance. For example, in the Drosophila oocyte, Wolbachia rely on normal host microtubule organization and the Gurken dorsal signaling complex to maintain titer (Ferree et al. 2005; Serbus et al. 2011). Additional evidence for the influence of host factors on Wolbachia titer comes from the finding that the same Wolbachia strain in D. simulans and D. melanogaster exhibits dramatically different titers in the mature oocyte (Poinsot et al. 1998; Serbus and Sullivan 2007). Evidence that factors intrinsic to Wolbachia influence its titer comes from the identification of the Wolbachia variant, wMelPop. The wMelPop strain exhibits extremely high titers in the central nervous system relative to other Wolbachia strains, independent of the host strain or species in which it resides (Min and Benzer 1997). Finally, extrinsic environmental factors such as temperature and diet dramatically influence Wolbachia titer (Mouton et al. 2006; Serbus et al. 2015). These changes are moderated in part through the host insulin signaling pathway (Serbus et al. 2015)
To comprehensively identify host factors that influence Wolbachia titer, we have employed a genome-wide RNA interference (RNAi) screen using a Drosophila cell line infected with Wolbachia. Our approach was motivated by the success of a number of previous cell-based screens using Drosophila cell lines (Mohr et al. 2014). Using Drosophila cells, genome-wide RNAi screens were performed to identify host genes that alter Listeria monocytogenes, Mycobacterium fortuitum, Chlamydia caviae, and Francisella tularensis infection and proliferation (Agaisse et al. 2005; Philips et al. 2005; Derre et al. 2007; Akimana 2010). We specifically assayed for RNAi-mediated gene knockdowns that either up- or down-regulate Wolbachia titer. The cell line was created from primary embryonic cultures of Drosophila melanogaster infected with wMel Wolbachia strain (Serbus et al. 2012). Wolbachia is stably maintained in these cultures and exhibits a close association with microtubules as found in Drosophila somatic and germline tissues (Kose and Karr 1995; Albertson et al. 2009). The cell line expresses a transgene encoding the GFP-tagged gene Jupiter, which encodes a microtubule-associated protein that labels microtubules and facilitates high-throughput, cell-based screening approaches (Karpova et al. 2006). The Wolbachia-infected cells were originally employed for high-throughput, cell-based screens to identify small molecule inhibitors of Wolbachia (Serbus et al. 2012; Van Voorhis et al. 2016). By combining genome-wide RNAi approaches with automated microscopy, we were able to screen the majority of the Drosophila genome for those genes that influence Wolbachia titer. As described below, this analysis yielded a number of host genes critical for regulating intracellular Wolbachia titer and revealed that the host ubiquitin and proteolysis pathways play an especially critical role in maintaining Wolbachia titer.
Materials and Methods
Generation of cultured cells
The JW18 cell line bearing the Jupiter-GFP transgene was generated according to previously described methods (Karpova et al. 2006; Serbus et al. 2012; Debec et al. 2016). Two- to 15-hour-old embryos derived from Wolbachia-infected flies carrying a Jupiter-GFP transgene were homogenized and plated in flasks. During the next 6 months of maintenance, 5 of the initial 20 seed flasks converted into immortal tissue culture lines. The JW18 cell line was selected for further pursuit due to its planar growth pattern and stably abundant Wolbachia infection. Cells were maintained at 25–26° in Sang and Shields media containing 10% fetal bovine serum, split weekly at a 1:2 dilution. A cured version of the JW18 line, referred to as JW18DOX, was generated by treating the cells with doxycycline. Electron microscopy was primarily performed on a second Wolbachia-infected cell line that expressess Jupiter-GFP and Histone-RFP (LDW1) generated as described above.
Screening approach
Two different Drosophila RNAi libraries were used in this screen: UCSF DmRNAi library version 1 and UCSF DmRNAi library version 2. These libraries have been used previously in Echard et al. (2004) and Goshima et al. (2007). Both libraries are available in 96-well format from Open Biosystems, now a part of GE Healthcare. Three out of four wells from each 384-well screening plate (Griener Bio-one) were set aside for experimental use. The fourth quadrant of wells was used for controls to define baselines for (1) untreated wells with standard levels of infected cells and (2) wells carrying significant reductions in infection, as induced by pararosaniline pamoate, administered to 100 μM final concentration (Serbus et al. 2012). JW18 cells without serum were added at a concentration of 6500 cells per well to a plate precoated with 0.5 mg/ml Concanavalin A. After the cells adhered to the plates for 50 min, double-stranded RNA (dsRNA) was transferred into the plate of JW18 cells using a Janus MDT pin tool and media containing 40% fetal bovine serum (FBS) was added for a final concentration of 10% FBS. The average concentration of dsRNA was 0.223 μg/well. All treatments were distributed into two plate replicates. After a 5-day incubation with the dsRNAs at 25°, the cells were prepared for imaging. Cells were fixed for 20 min in 4% formaldehyde and rinsed with PBS using an automated BioTek liquid handler. All staining solutions were administered using a Multidrop robot, with extensive rinsing between treatments. Mouse anti-histone (MAB052, Millipore, Bedford, MA) and goat anti-mouse Alexa 594 (Invitrogen, Carlsbad, CA) was diluted to 1:1250 in PBS/0.1% Triton. A stock solution saturated with DAPI was used at a final concentration of 1:40. After staining, PBS+sodium azide was added to all wells of the plates.
Screen data analysis
Stained plates were imaged using an ImageXpress Micro system (Molecular Devices, Sunnyvale, CA). Ten images were acquired per well at ×40 magnification. These images were analyzed using customized analysis software provided by Molecular Devices. The software routine masks any areas where clumps of cells are detected, then detects boundaries of the cells and nuclei based upon Jupiter-GFP and anti-histone stains. A mask was applied to the nuclei to obscure fluorescent signal from those areas. Thresholds for DAPI fluorescence detection were set to maximize Wolbachia detection while minimizing the background DAPI signal in pararosaniline pamoate-treated control cells, then applied to analysis of the entire plate. Cytoplasmic DAPI attributable to Wolbachia nucleoids was scored in each cell to classify it as Wolbachia-infected or uninfected. A spreadsheet from the data analysis software indicated the quantity of Wolbachia-infected cells vs. total cells measured in each well. Average and standard deviation values were calculated for the frequency of Wolbachia infection in JW18 and pararosaniline pamoate-treated JW18 control cells, using the descriptive statistics function in Statistical Package for the Social Sciences (SPSS by IBM). These values were used to calculate a Z′ factor for each plate. The Z′ factor represents one – (three multiplied by the absolute value of the sum of the SD for each control divided by the absolute value of the difference between mean values for each control) (Zhang et al. 1999). Z′ factors regarded as acceptable in this field range from 0 to 1. Our Z′ factors ranged from 0.25 to 0.85 per plate, confirming that the controls were clearly distinguishable by the assay. To identify preliminary “hit” compounds that substantially reduced intracellular Wolbachia titer, an initial hit range was calculated to lie between the JW18 average infection frequency − 3 SD, and the average pararosaniline pamoate-treated JW18 infection frequency + 3 SD. To enable comparison of hits identified on different treatment plates, the scaling of this initial hit range was next reset to span from 0 to 1, thus applying a uniform, normalized hit range to all plates. To further increase the stringency for identifying hits, we also calculated an average infection frequency for all JW18 cells on the plate (treated or not), as most treatment wells are expected to be indistinguishable from untreated controls. Wells that lay within 3 SD of the mean were excluded from the hit list. Hit wells identified in only one of two replicates were also removed.
Ubiquitin foci staining and quantification
For analysis of ubiquitin foci in tissue culture cells, chamber slides were coated with 0.5 mg/ml Concanavalin A, followed by addition of JW18 cells. After a 24-hr incubation at 25°, cells were fixed for 20 min in 4% formaldehyde, blocked using 1% BSA, and stained with anti-ubiquitin [Enzo Mono- and Polyubiquitinylated Conjugates (FK2) BML-PW8805] for 1 hr at 1:100 dilution. Goat anti-mouse Alexa 594 (Invitrogen) was used at a 1:1250 dilution in PBS/0.1% Triton. Cells were imaged on a Leica DMI6000B wide-field inverted microscope equipped with a Hamamatsu EM CCD camera (ORCA C9100-02) at ×100 magnification. Areas to image were selected randomly without bias. Foci were quantified by eye from cells with all boundaries visible in the image. Chi-squared analysis in SPSS was used to determine significance, and post hoc tests performed as previously described (Beasley and Schumacker 1995).
For analysis of ubiquitin foci in D. melanogaster oogenesis, flies of the genotype w; Sp/Cyo; Sb/TM6B were reared on fly food consisting of 0.5% agar, 7% molasses, 6% cornmeal, and 0.8% killed yeast. Newly eclosed flies were collected and reared for 5 days. Ovaries were dissected in EBR (Ringer’s solution) and fixed for 5 min in 600 μl heptane and 100 μl devitellinizing solution (100 μl Buffer B, 112.5 μl 32% paraformaldehyde, and 387.5 μl of diH2O) (Verheyen and Cooley 1994). Ovaries were then rinsed, treated with RNAse overnight, blocked with 1% BSA, and incubated with anti-ubiquitin [Enzo Mono- and Polyubiquitinylated Conjugates (FK2) BML-PW8805] overnight at a 1:50 dilution. Last, the ovaries were incubated with goat anti-mouse:Alexa 488 at a 1:500 dilution and resuspended overnight in mounting media containing propidium iodide (PI). All samples were then imaged on a Leica SP2 confocal microscope at ×63 magnification with ×1.5 zoom. Experimental samples verified to exhibit the same degree of compression as the control sample were pursued further, while any experimental samples deviating from that were discarded. Z-series images were acquired from oocytes at 1.5-μm intervals. Uniform intensity settings were applied to all egg chambers imaged within each replicate. A minimum of 7–10 oocytes were ultimately imaged from each condition per replicate with all experimental oocytes matched for morphological consistency against control oocytes of the same replicate. Significance was determined by ANOVA.
Drug treatments in Drosophila
Newly eclosed flies of the genotype w; Sp/Cyo; Sb/TM6B were collected, reared for 2 days, and then exposed to food containing compounds of interest for 3 days. Drugs were diluted to a final concentration of 100 μM in food. Equivalent amounts of carrier DMSO diluted into these nutrient sources were used as a control.
Generation of Wolbachia-infected RNAi lines in Drosophila
wMel Wolbachia were crossed from w; Sp/Cyo; Sb/TM6B into a germline triple driver stock (Mazzalupo and Cooley 2006) to ultimately generate infected flies of the genotype w; P{GAL4-Nos.NGT}40; P{GAL4::VP16-Nos.UTR}MVD1 (Serbus et al. 2015). Females from this infected driver stock were then crossed to males from responder stocks that carried the following upstream activation sequence (UAS) dsRNA transgenes: the Ubc6 Valium20 TRiP line: y,v; P{TRiP.HMS02466}attP40; the psGEF Valium20 TRiP line: y, sc,v; P{TRiP.HMS00320}attP2; the mtt Valium20 TRiP line: y,sc,v; P{TRiP.HMS00367}attP2; the ND-B17.2 Valium20 line: y,sc,v; P{TRiP.HMS01584}attP2; and the CG18324 Valium20 line: y,sc,v; P{TRiP.HMS01199}attP2/TM3, Sb. Wolbachia-infected driver females were also outcrossed to OreR uninfected males in parallel for analysis as a control.
Quantification of Wolbachia nucleoids in oogenesis
PI staining of Drosophila ovarian tissue was done using established methods (Serbus et al. 2012). Data collection was conducted as previously. All tissues were imaged using a Leica SP2 confocal microscope. Wolbachia were quantified in single focal planes of stage 10 oocytes using established methods (Serbus et al. 2015). Average Wolbachia titer values associated with drug treatments were normalized against their respective DMSO controls as previously to ensure comparability between experiments in displays of the data (Serbus et al. 2015). Statistical analysis was conducted on raw data only using ANOVA, as described previously (Serbus et al. 2012). A minimum of 2–3 experimental replicates were performed for the germline staining experiments described in this study.
Transmission electron microscopy
LDW1 cells and the cured equivalent LDW1DOX were fixed with 2% glutaraldehyde and 0.5% paraformaldehyde in Cacodylate buffer 0.075M and postfixed with 2% osmium tetroxide. Samples were dehydrated through a graded series of ethanol and embedded in epoxy resin. Ultrathin (70 nm) sections (Ultracut UC6, Leica) were collected on formvar/carbon-coated copper grids. Sections were then poststained by aqueous 4% uranyl acetate and lead citrate. All samples were observed in a Tecnai12 (FEI, The Netherlands) transmission electron microscope at 80 kV equipped with a 1K×1K Keen View camera. Chi-square analysis was used to determine significance, with post hoc tests performed as previously (Beasley and Schumacker 1995).
Data availability
All primary data can be found in Supplemental Material, File S1. Additional information about in vivo validation of genes that alter Wolbachia quantity can be found in Figure S1.
Results
A genome-wide RNAi cell-based screen yields host genes that either enhance or suppress Wolbachia titer
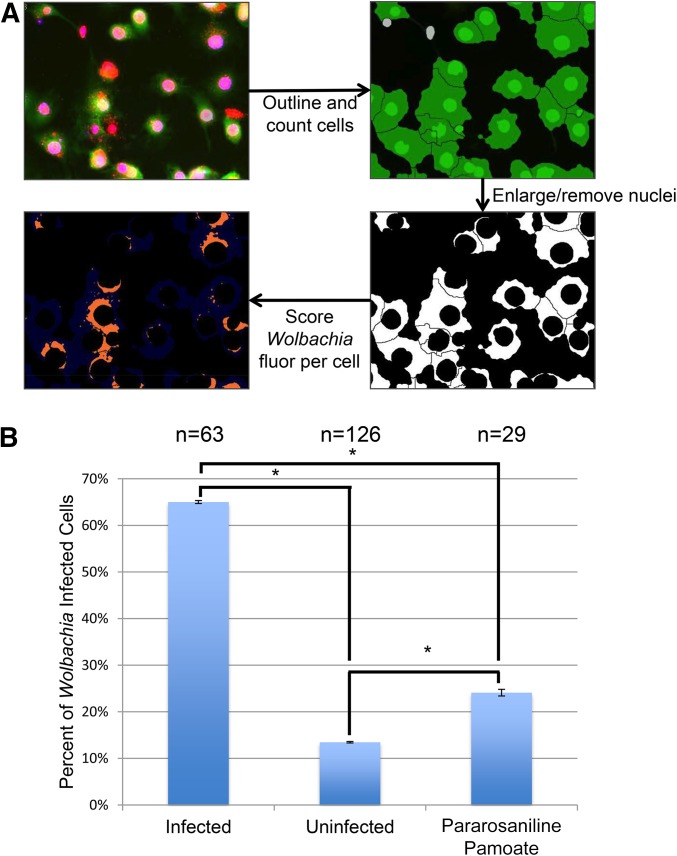
To identify host components that influence Wolbachia infection of insect cells, we took advantage of the ability to perform genome-wide RNAi screens in Drosophila tissue culture cells. Because Drosophila cell lines are particularly amenable to RNAi-based screening, this approach has been successfully used for studying a number of cellular processes including the cellular basis of host–pathogen interactions (Moser et al. 2010). Here we used a well-characterized Wolbachia-infected cell line known as JW18 (Serbus et al. 2012). Our analysis revealed that 90% of the JW18 host cells are infected and Wolbachia have no obvious effects on cell viability or division (Serbus et al. 2012). Quantification of Wolbachia infection was based on fluorescent images of Wolbachia-infected and cured JW18 cell lines using automated microscopy and journaling software (Figure 1).
Figure 1.
Automated microscope-based quantification of Wolbachia titer. (A) Flow chart showing screening methodology. Top left panel: Wolbachia-infected JW18 cells showing fluorescence from the markers DAPI (blue) and anti-histone (red), and the GFP-Jupiter (green). Top right panel: The journaling algorithm outlines cell borders based upon Jupiter-GFP. Bottom right panel: The algorithm digitally removes the nucleus from each cell based upon anti-histone staining. Bottom left panel: The total cytoplasmic DAPI signal in each cell is scored. (B) Graph depicts the percentage of cells scored by this algorithm as “Wolbachia-infected” in control JW18 cells, cured JW18 cells, and pararosaniline pamoate-treated JW18 cells. Though the algorithm can mis-identify fluorescent debris in the well as “infected cells,” there are consistent significant differences by ANOVA between all of the control conditions tested (P < 0.0001). Error bars represent the SEM.
By using the JW18 cell line in the 384-well plate format, we screened an RNAi library that targets 15,699 Drosophila protein-encoded genes (Goshima et al. 2007). Wolbachia-infected JW18 cells were plated into individual wells, each containing a unique dsRNA, and allowed to incubate for 5 days. Robotics and automated microscopy were used to fix, stain, and image the cells (see Materials and Methods). The JW18 cell line expresses a GFP-Jupiter fusion protein that labels the cytoskeleton (Karpova et al. 2006), thus highlighting the entire cytoplasm and indicating cell boundaries. Anti-histone staining labeled the host nuclei, and DAPI was used to stain both host nuclei and cytoplasmic Wolbachia. Through customized journaling software, the overlap of anti-histone and DAPI nuclear staining enabled digital removal of the nucleus. This facilitated quantification of cytoplasmic DAPI, representing Wolbachia density per cell (Figure 1) (Serbus et al. 2012). Each cell was scored as Wolbachia positive or negative based on a predetermined cytoplasmic DAPI intensity cut-off value. Ten images per well were recorded and analyzed, and two independent screens of the RNAi library were performed. RNAi knockdowns that significantly reduced or increased the proportion of Wolbachia-infected cells per well without dramatically altering cell viability in both independent replicates were scored as hits (Figure 2). Because we are starting with a population in which 90% or more of the cells are infected, RNAi-induced alterations in the ability of Wolbachia to infect cells are expected to exert only a minor impact on titer.
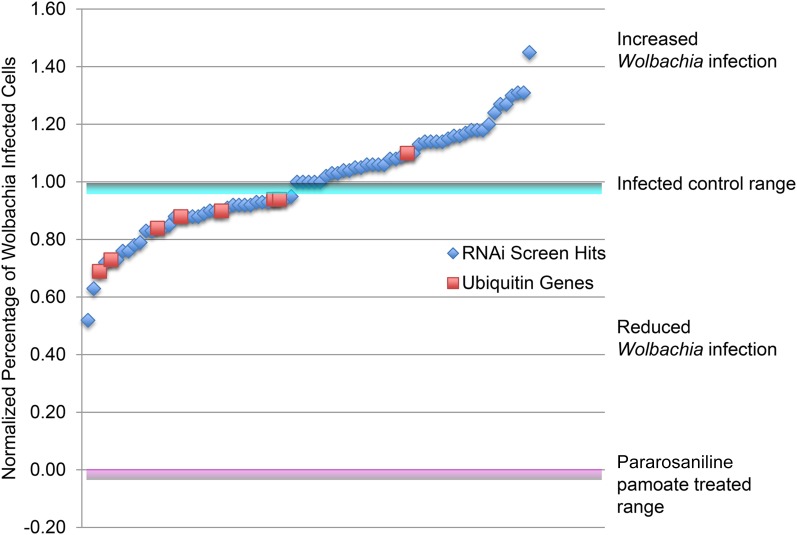
Figure 2.
RNAi knockdowns that decreased or increased Wolbachia infection rate. Of 15,699 transcribed host genes, 36 candidate genes were identified that dramatically reduced Wolbachia titer and 41 that increased Wolbachia titer when knocked down via RNAi. Blue diamonds below or above the infected control range indicates dsRNA treatments targeting a single host gene that reduced or increased the proportion of Wolbachia-infected tissue culture cells. Red boxes indicate ubiquitin-related dsRNA treatment outcomes.
The screen yielded 36 host genes that when knocked down through RNAi resulted in a significant drop in Wolbachia titer. These included hits in two genes involved in host lipid metabolism: CG9243, a phospholipase D; and CG1718, a gene involved in sterol uptake and esterification. In addition, hits in three mitochondrial metabolism components were recovered: CG3214, an NADH dehydrogenase; CG14757, a succinate dehydrogenase; and CG18324, a mitochondrial transporter. Knockdown of host genes encoding ATPases, GTPases, and ribosomal proteins also reduced titer (Table 1 and Table 2).
Table 1. List of host genes that reduce Wolbachia infection rates when knocked down.
| CG# | Effect on titer | Gene name | Normalized value | Functional category | Additional functional category |
|---|---|---|---|---|---|
| CG14835 | Reduced | CG14835 | 0.52 | ||
| HDC10201 | Reduced | HDC10201 | 0.63 | ||
| CG12725 | Reduced | CG12725 | 0.69 | Ubiquitin | |
| CG9777 | Reduced | CG9777 | 0.72 | ||
| CG12218 | Reduced | mei-P26 | 0.73 | Ubiquitin transferase | Ligase |
| HDC05374 | Reduced | HDC05374 | 0.73 | ||
| CG1527 | Reduced | RpS14b | 0.76 | Ribosomal | |
| CG14579 | Reduced | CG14579 | 0.76 | ||
| CG10970 | Reduced | CG10970 | 0.78 | Acylphosphatase | |
| CG13738 | Reduced | CG13738 | 0.79 | ||
| CG2916 | Reduced | Sep5 | 0.83 | Septin | GTP-binding protein |
| CG14047 | Reduced | PsGEF | 0.83 | RhoGEF | GTPase |
| CG2960 | Reduced | RpL40 | 0.84 | Ubiquitin | Ribosomal |
| CG32100 | Reduced | CG32100 | 0.84 | ||
| CG30372 | Reduced | Asap | 0.85 | ArfGap | GTPase activator |
| CG2958 | Reduced | lectin-24Dd | 0.88 | C-type lectin | Carbohydrate binding |
| CG3450 | Reduced | ubl | 0.88 | Ubiquitin | |
| CG4688 | Reduced | GstE14 | 0.88 | Glutathione S-transferase | |
| CG32399 | Reduced | CG32399 | 0.88 | ||
| CG7320 | Reduced | CG7320 | 0.88 | Hemocyanin | |
| CG12172 | Reduced | Spn43Aa | 0.89 | Serpin | Endopeptidase inhibitor |
| CG1344 | Reduced | CG1344 | 0.90 | Kinase | |
| CG3214 | Reduced | ND-B17.2 | 0.90 | Mitochondrial | NADH dehydrogenase |
| CG2013 | Reduced | Ubc6 | 0.90 | Ubiquitin | Ubiquitin-conjugating enzyme |
| HDC15882 | Reduced | HDC15882 | 0.91 | ||
| CG33196 | Reduced | dpy | 0.92 | EGF domain | Structural constituent |
| CG18324 | Reduced | CG18324 | 0.92 | Mitochondrial carrier | Transport |
| CG14757 | Reduced | CG14757 | 0.92 | Mitochondrial | Succinate dehydrogenase |
| CG12807 | Reduced | Spn85F | 0.92 | Serpin | Endopeptidase inhibitor |
| CG1070 | Reduced | Alh | 0.93 | Transcription | |
| CG1332 | Reduced | CG1332 | 0.93 | Endocytosis | |
| CG1718 | Reduced | CG1718 | 0.93 | ABC transporter | ATPase |
| CG8715 | Reduced | lig | 0.94 | Ubiquitin | |
| CG32744 | Reduced | Ubiquitin-5E | 0.94 | Ubiquitin | |
| CG1394 | Reduced | CG1394 | 0.94 | ||
| CG43345 | Reduced | CG43345 | 0.95 | Phospholipase D | Catalytic activity |
Hits are ordered from strongest at the top to weakest at the bottom.
Table 2. List of host genes that significantly alter Wolbachia infection rates ordered by function.
| CG# | Effect on titer | Gene name | Normalized value | Functional category | Additional Functional Category | |
|---|---|---|---|---|---|---|
| ATP/GTP | CG1718 | Reduced | CG1718 | 0.93 | ABC transporter | ATPase |
| CG30372 | Reduced | Asap | 0.85 | ArfGap | GTPase activator | |
| CG14047 | Reduced | PsGEF | 0.83 | RhoGEF | GTPase | |
| CG2916 | Reduced | Sep5 | 0.83 | Septin | GTP-binding protein | |
| CG1657 | Increased | CG1657 | 1.20 | GTPase activating | Activates Rab GTPases | |
| Kinase | CG1344 | Reduced | CG1344 | 0.90 | Kinase | |
| CG5813 | Increased | chif | 1.14 | Kinase | DBF zinc finger | |
| Mitochondrial | CG3214 | Reduced | ND-B17.2 | 0.90 | Mitochondrial | NADH dehydrogenase |
| CG14757 | Reduced | CG14757 | 0.92 | Mitochondrial | Succinate dehydrogenase | |
| CG18324 | Reduced | CG18324 | 0.92 | Mitochondrial carrier | Transport | |
| Ribosomal | CG2960 | Reduced | RpL40 | 0.84 | Ubiquitin | Ribosomal |
| CG1527 | Reduced | RpS14b | 0.76 | Ribosomal | ||
| Serpin | CG12172 | Reduced | Spn43Aa | 0.89 | Serpin | Endopeptidase inhibitor |
| CG12807 | Reduced | Spn85F | 0.92 | Serpin | Endopeptidase inhibitor | |
| Ubiquitin | CG2960 | Reduced | RpL40 | 0.84 | Ubiquitin | Ribosomal |
| CG8715 | Reduced | lig | 0.94 | Ubiquitin | ||
| CG32744 | Reduced | Ubiquitin-5E | 0.94 | Ubiquitin | ||
| CG3450 | Reduced | ubl | 0.88 | Ubiquitin | ||
| CG2013 | Reduced | Ubc6 | 0.90 | Ubiquitin | Ubiquitin-conjugating enzyme | |
| CG12725 | Reduced | CG12725 | 0.69 | Ubiquitin | ||
| CG12218 | Reduced | mei-P26 | 0.73 | Ubiquitin transferase | Ligase | |
| CG31807 | Increased | CG31807 | 1.10 | Ubiquitin transferase | RING finger domain | |
| Ungrouped | CG10970 | Reduced | CG10970 | 0.78 | Acylphosphatase | |
| CG1629 | Increased | mal | 1.04 | Aminotransferase | MOSC domain | |
| CG2958 | Reduced | lectin-24Dd | 0.88 | C-type lectin | Carbohydrate binding | |
| CG33196 | Reduced | dpy | 0.92 | EGF domain | Structural constituent | |
| CG1332 | Reduced | CG1332 | 0.93 | Endocytosis | ||
| CG30361 | Increased | mtt | 1.31 | G-protein-coupled receptor | ||
| CG4688 | Reduced | GstE14 | 0.88 | Glutathione S-transferase | ||
| CG7968 | Increased | CG7968 | 1.03 | Hemolymph juvenile hormone-binding protein | ||
| CG7320 | Reduced | CG7320 | 0.88 | Hemocyanin | ||
| CG32005 | Increased | CG32005 | 1.14 | High-Mobility group box | DNA binding | |
| CG43345 | Reduced | CG43345 | 0.95 | Phospholipase D | Catalytic activity | |
| CG7988 | Increased | CG7988 | 1.30 | Regulation of circadian clock | ||
| CG4484 | Increased | Slc45-1 | 1.45 | Sucrose transporter | ||
| CG1070 | Reduced | Alh | 0.93 | Transcription |
Particularly striking, the screen yielded eight hits in the ubiquitin-related pathways, seven of which reduced the proportion of Wolbachia-infected tissue culture cells. DsRNA targeting the Ubiquitin-60S ribosomal protein L40 (CG2960), lingerer (CG8715), Ubiquitin-5E (CG32744), the ubiquitin transferase mei-P26 (CG12218), Ubiquitin-like protein 5 (CG3450), Ubiquitin-conjugating enzyme Ubc6 (CG2013), and Ubiquitin-like protein (CG12725) reduced infection, and targeting of the ubiquitin transferase CG31807 increased infection (Table 1 and Table 3). Ubiquitin is a small protein with a variety of functions including the marking of proteins for degradation and is targeted by a number of intracellular bacteria (Zhou and Zhu 2015). Previous inspection of the Wolbachia genome revealed that Wolbachia lack the ability to synthesize key amino acids and thus rely on the host as a source (Wu et al. 2004; Foster et al. 2005). Thus, it is possible that Wolbachia require high levels of host proteolysis to supply sufficient amino acids for growth and reproduction.
Table 3. List of host genes that increase Wolbachia infection rates when knocked down.
| CG# | Effect on titer | Gene name | Normalized value | Functional category | Additional functional category |
|---|---|---|---|---|---|
| CG10589 | Increased | CG10589 | 1.00 | Domain of unknown function | |
| CG11231 | Increased | CG11231 | 1.00 | ||
| CG14442 | Increased | CG14442 | 1.00 | ||
| CG32736 | Increased | CG32736 | 1.00 | Uncharacterized protein family | |
| CG32718 | Increased | CG32718 | 1.00 | ||
| CG15480 | Increased | CG15480 | 1.02 | ||
| CG7968 | Increased | CG7968 | 1.03 | Hemolymph juvenile hormone-binding protein | |
| HDC17458 | Increased | HDC17458 | 1.03 | ||
| CG1629 | Increased | mal | 1.04 | Aminotransferase | MOSC domain |
| CG34434 | Increased | CG34434 | 1.04 | ||
| CG1971 | Increased | CG1971 | 1.05 | ||
| HDC16920 | Increased | HDC16920 | 1.05 | ||
| CG18656 | Increased | CG18656 | 1.06 | ||
| CG4455 | Increased | CG4455 | 1.06 | ||
| CG4631 | Increased | CG4631 | 1.06 | Domain of unknown function | |
| CG11260 | Increased | CG11260 | 1.06 | ||
| CG32790 | Increased | CG32790 | 1.08 | ||
| CG3568 | Increased | CG3568 | 1.08 | Domain of unknown function | |
| CG18404 | Increased | CG18404 | 1.09 | ||
| CG31807 | Increased | CG31807 | 1.10 | Ubiquitin transferase | RING finger domain |
| CG43117 | Increased | CG43117 | 1.10 | ||
| CG14673 | Increased | CG14673 | 1.13 | ||
| CG5813 | Increased | chif | 1.14 | Kinase | DBF zinc finger |
| CG32005 | Increased | CG32005 | 1.14 | High-mobility group box | DNA binding |
| HDC12508 | Increased | HDC12508 | 1.14 | ||
| HDC12757 | Increased | HDC12757 | 1.14 | ||
| HDC19233 | Increased | HDC19233 | 1.15 | ||
| CG9263 | Increased | CG9263 | 1.16 | ||
| CG31819 | Increased | CG31819 | 1.16 | ||
| CG31817 | Increased | CG31817 | 1.17 | ||
| CG32988 | Increased | CG32988 | 1.18 | ||
| CG31813 | Increased | CG31813 | 1.18 | ||
| CG42749 | Increased | CG42749 | 1.18 | ||
| CG1657 | Increased | CG1657 | 1.20 | GTPase activating | Activates Rab GTPases |
| CG16852 | Increased | CG16852 | 1.24 | ||
| CG4666 | Increased | CG4666 | 1.27 | ||
| CG13365 | Increased | CG13365 | 1.27 | ||
| CG7988 | Increased | CG7988 | 1.30 | Regulation of circadian clock | |
| CG30361 | Increased | mtt | 1.31 | G-protein-coupled receptor | |
| CG16853 | Increased | CG16853 | 1.31 | ||
| CG4484 | Increased | Slc45-1 | 1.45 | Sucrose transporter |
Strongest to weakest hits are ordered from top to bottom
The RNAi screen also yielded 41 host genes that when knocked down significantly increased the proportion of Wolbachia-infected cells. These included hits in: CG4484, a sucrose transporter; CG30361, a G-protein-coupled receptor; CG1657, a GTPase-activating protein (GAP); CG7968, a hemolymph juvenile hormone-binding protein. Of the 41 hits that increased Wolbachia titer, 32 were of unknown function. Unlike the hits that reduced titer, we did not recover multiple hits in known specific biological processes or pathways (Table 3).
The presence of Wolbachia increases the quantity and distribution of ubiquitinated proteins in infected host cells and oocytes
Given RNAi knockdowns of host components involved in the ubiquitin/proteolysis pathway resulted in a reduction in Wolbachia titer, we examined whether the presence of Wolbachia might influence the ubiquitination state of host proteins. This was accomplished through immunofluorescence analysis with an antibody that specifically recognizes ubiquitin-conjugated host proteins. The antibodies were developed to recognize the multi-ubiquitinated chains of polyubiquitinated proteins but not free ubiquitin (see Materials and Methods) (Fujimuro et al. 1994; Everett 2000). This analysis revealed a significant increase in the number of ubiquitin foci in the Wolbachia-infected cells compared to the uninfected controls (Figure 3). Figure 3B presents DAPI-stained nuclei (blue), microtubule-labeled cytoplasm (green), and ubiquitin foci (red) in JW18 infected and uninfected cells. The images reveal that the number, size, and intensity of the ubiquitin foci are much greater in the former. For example, 85% (n = 142) of the infected cells had one or more ubiquitin foci compared to 45% (n = 138) for the uninfected cells. Additionally, 51% of infected cells had five or more ubiquitin foci (n = 72), which was significantly different (P = 0.0001) compared to 7% of uninfected cells with that outcome (n = 10) (Figure 3A). Significance was determined using chi-square analysis.
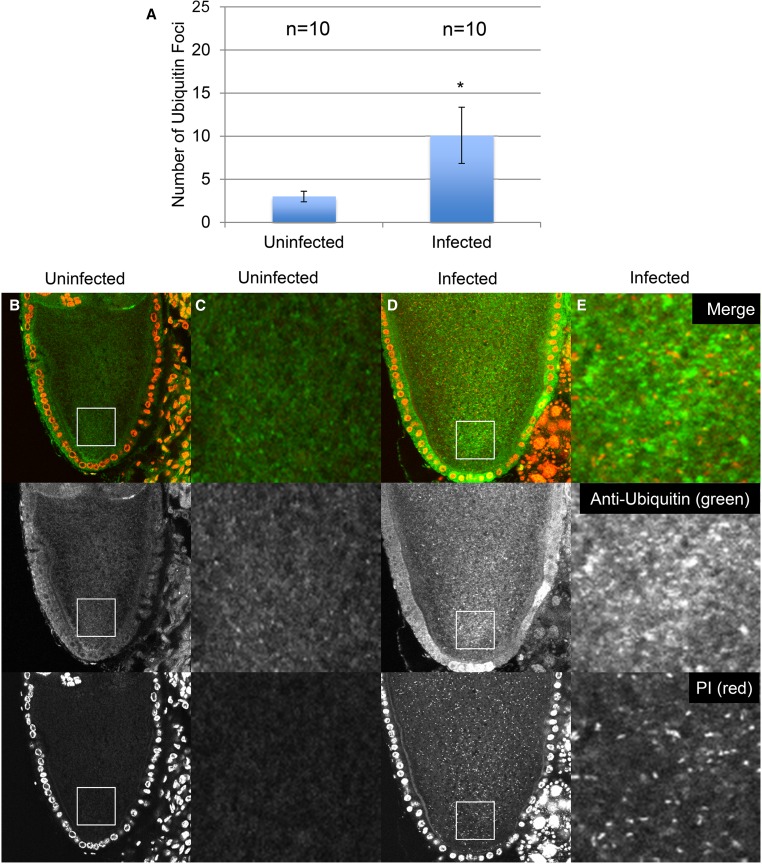
Figure 3.
Increase in ubiquitin foci in Wolbachia-infected tissue culture cells. (A) Quantification of the number of ubiquitin foci in infected and uninfected cells. All P values were below 0.0001, and adjusted α value was 0083333. (B and C) Low and high magnification images of stained uninfected cells. (D and E) Low and high magnification images of stained Wolbachia-infected cells. Staining indicates DNA (blue), microtubules (GFP-Jupiter) (green), and ubiquitin foci (red). Significance was determined using chi-square analysis.
We also performed this analysis on infected and uninfected Drosophila oocytes. The images shown in Figure 4 depict oocytes from infected and uninfected adult D. melanogaster females. PI (red) stains host nuclei in uninfected cells and both the host nuclei and Wolbachia in infected cells. The anti-ubiquitin antibody (green in top panels and black-and-white in bottom panels) stains the ubiquitin foci. Quantification was performed as previously described (Serbus et al. 2015) (see also Materials and Methods). Ubiquitin foci counts were accomplished by selecting a defined area at the posterior pole from an image of a medial section of each oocyte. This revealed an average of 10 ± 3 (n = 10) posterior ubiquitin foci in infected oocytes, in contrast to 3 ± 1 (n = 10) posterior ubiquitin foci detected in uninfected oocytes.
Figure 4.
Increase in ubiquitin foci in Wolbachia-infected Drosophila oocytes. Immmunofluorescence analysis using an anti-ubiquitin antibody indicated more ubiquitin foci in Wolbachia-infected oocytes. (A) Quantification of ubiquitin foci per area in medial planes of infected and uninfected oocytes. Average values were significantly different according to ANOVA (P = 0.046). Error bars indicate SEM. (B and C) Low and high magnification images of uninfected oocytes. (D and E) Low and high magnification images of Wolbachia-infected oocytes. DNA (red) and ubiquitin foci (green).
Drug-induced inhibition of the proteasome reduces Wolbachia titer in the Drosophila oocyte
To determine if the maintenance of Wolbachia titer relies on high levels of host proteasome activity, Wolbachia-infected Drosophila females were treated with Lactacystin, a proteasome inhibitor that targets the 20S subunit of the proteasome (Yang et al. 2008). After being kept on a regular diet for 2 days, the flies were exposed to food containing 100 μM lactacystin for 3 days. Control females were exposed to food with an equivalent amount of carrier DMSO. Ovarian tissues were dissected and stained with PI to label Wolbachia DNA. As previously described, Wolbachia abundance was determined by confocal imaging of the medial plane of PI-stained stage 10 oocytes (Serbus et al. 2015) (see also Materials and Methods). In these images, the PI stains the nuclei of the host follicle cells that border the oocyte and the Wolbachia nucleoids within the oocyte (Figure 5). This analysis indicated significant reduction in Wolbachia titer in lactacystin-treated oocytes compared to untreated control oocytes (Figure 5, A and B). Control oocytes averaged 700 ± 54 Wolbachia (n = 29), while the treated oocytes averaged 534 ± 35.0 (n = 29) (P = 0.012). Significance was determined using ANOVA.
Figure 5.
Disruption of host proteasome reduces Wolbachia titer in the Drosophila oocyte. Adult females were fed food containing either DMSO (A) or lactacystin (B). (C) Quantification of the Wolbachia in medial plane of stage 10 oocytes indicated a significant difference by ANOVA (P = 0.012). Control (D) and Ubc6 (E) knocked down using GAL4-UAS dsRNA expression. Quantification of oocyte titers (F) indicated a significant difference by ANOVA (P < 0.001). Error bars indicate SEM.
RNAi knockdown of proteasome component Ubc6 results in a dramatic decrease in Wolbachia titer in the oocyte
We next genetically tested the impact of the host ubiquitin/proteasome pathway on oocyte Wolbachia titer. We used the GAL4:UAS system to knockdown specific components of the ubiquitin/proteasome pathway in the female germline (see Materials and Methods) (Hudson and Cooley 2014). Because Wolbachia is intimately associated with the ER in our cell lines (see next section), we focused on the host factor Ubc6 recovered in our screen (Table 1). Ubc6 is an ER integral membrane protein that functions as an E2 conjugating enzyme in the ERAD pathway: endoplasmic reticulum (ER)-associated degradation of mis-folded proteins (Lemus and Goder 2014). Ubc6 is specifically required for the degradation of cytosolic domains of membrane proteins (ERAD-C) (Metzger et al. 2008). Previous studies have successfully knocked down Ubc6 using UAS Ubc6 RNAi fly lines (Ashton-Beaucage et al. 2016). By crossing in the maternal nanos-GAL4 driver into the UAS-Ubc6 RNAi line, we specifically knocked down the level of Ubc6 in Drosophila female germline cells. This RNAi knockdown of Ubc6 in the Drosophila germline resulted in an approximately fourfold reduction in oocyte Wolbachia titer (Figure 5). The wild-type oocytes averaged 368 ±133 (n = 19) Wolbachia while the Ubc6 knockdowns averaged 93 ± 43 (n = 19) (P < 0.01) (Figure 5). These results suggest that a fully functional host ERAD pathway is required to maintain Wolbachia titer in the Drosophila oocytes and cell lines.
Wolbachia is closely associated with and alters the morphology of the ER
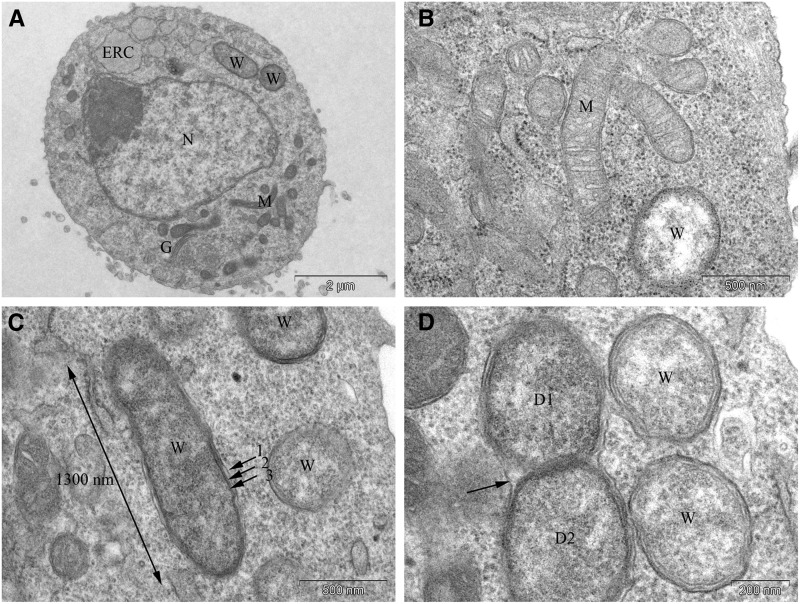
Given the dependence of Wolbachia titer on ERAD, we performed ultrastructural analysis on the Wolbachia-infected cell lines to determine its subcellular localization. Ultrastructural images of the Wolbachia-infected cell line are shown in Figure 6 and Figure 7. The Drosophila cells are round with a mean diameter of 6–8 μm. Wolbachia (W), golgi (G), the endoplasmic reticulum cisternae (ERC), mitochondria (M), and nuclei (N) are readily visualized in these cells (Figure 6, A and B). Wolbachia tend to exhibit an approximate diameter of 0.5 μm and occasionally observe lengths >1 μm (Figure 6C). As previously described, Wolbachia are often encompassed by a double membrane, enclosing the bacteria within a small vacuole (Voronin et al. 2004; Chagas-Moutinho et al. 2015). In some instances, multiple Wolbachia occupy the same vacuole. Dividing Wolbachia are depicted in Figure 6D. In this case, both daughter bacteria lie in the same vacuole although it is expected that each daughter bacterium will reside within its own host vacuole after completion of division (Figure 6D).
Figure 6.
Ultrastructural analysis of Wolbachia in Drosophila cultured cells. (A) Cross-section of a Wolbachia-infected Drosophila cell in which the nucleus (N), endoplasmic reticulum cisternae (ERC), golgi (G), mitochondria (M), and Wolbachia (W) are readily visualized. Cells are spherical with a mean diameter of 6–8 μm. (B) Mitochondria and Wolbachia are readily distinguished as the former exhibit distinct internal tubular structures and are narrower in width. (C) The maximum size observed for Wolbachia is a 1300 nm length and 460 nm width. Each Wolbachia bacterium is encompassed by three membranes, the outermost derived from the host (arrows 1, 2, 3). (D) Image of dividing Wolbachia. The two daughter cells (D1 and D2) lie in the same vacuole (arrow), which ultimately will abscise.
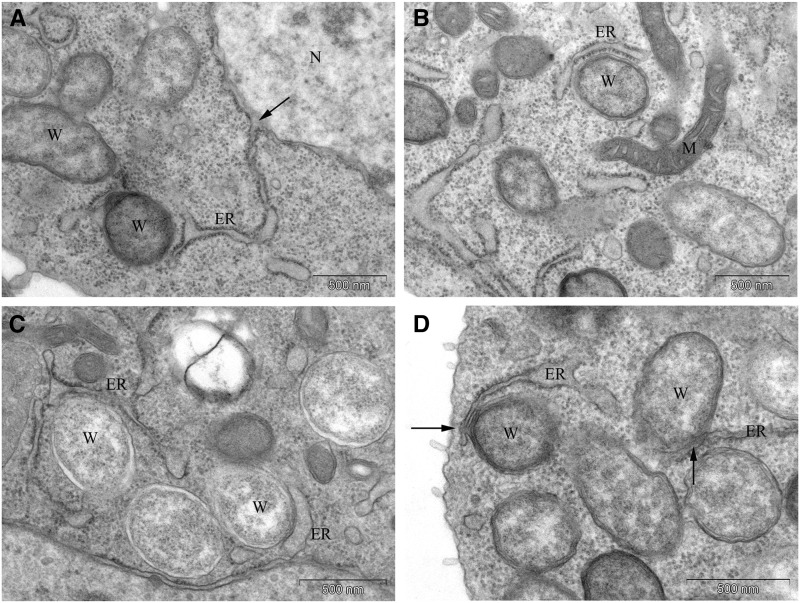
Figure 7.
Wolbachia closely associate and alter the morphology of the ER. (A) The presence of Wolbachia results in an ER extension from the nuclear envelope (arrow) to Wolbachia. (B and C) Examples of ER extensions wrapping around and in close association with Wolbachia. (D) In some instances, Wolbachia appear to communicate with the ER lumen (arrows). W, Wolbachia; ER, endoplasmic reticulum; N, nucleus; M, mitochondria.
Our ultrastructural analysis reveals an intimate association between Wolbachia and the host ER. Figure 7A depicts an elaboration of the ER extending from the nuclear envelope to a Wolbachia located in the cytoplasm. Figure 7, B and C depict similar ER extensions that encompass individual Wolbachia. In some instances, there is a direct contact and continuity between the host ER and Wolbachia membrane vacuole. This is especially evident in Figure 7D. These images suggest that Wolbachia are connected to the luminal space of the ER.
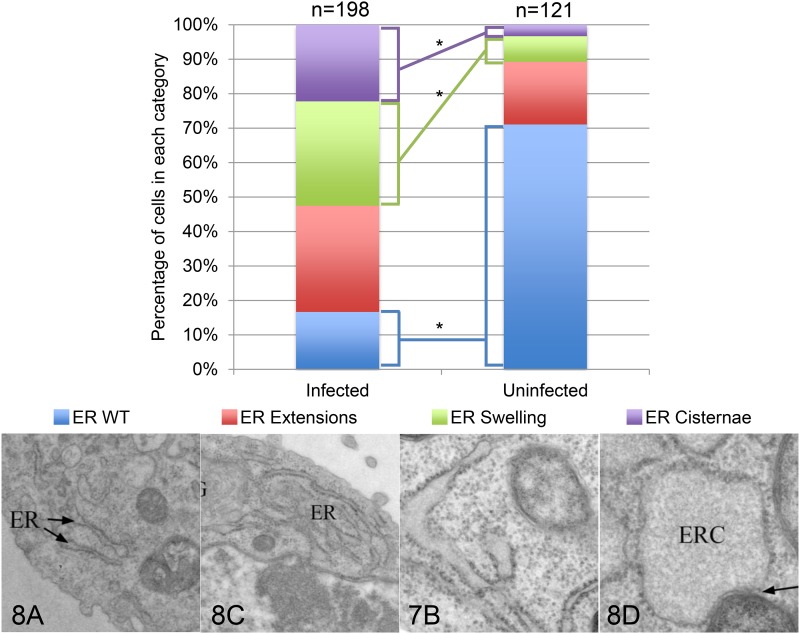
In addition to the intimate association between Wolbachia and the ER, we find that the presence of Wolbachia induces dramatic alterations in ER morphology. In the uninfected cell line, the ER exhibits classic tubule morphology with ribosomes decorating its entire length (Figure 8, A and B). By contrast, in the Wolbachia-infected cells, the ER is highly elaborated forming a complex network of extensions (Figure 8, C and D). In addition, the ER tubules have expanded to form large cisternae. As seen in the images, Wolbachia are often associated with these cisternae. To quantify the effect of Wolbachia on ER morphology, we analyzed randomly selected EM images from Wolbachia-infected and cured cell lines (n = 128 and 121, respectively) (Figure 9). Quantification revealed a dramatic increase in all classes of abnormal ER morphology, including the ER network extensions, ER swelling, and formation of large ER cisternae. Particularly striking is the 10-fold increase in the formation of ER cisternae in Wolbachia-infected cells (Figure 9).
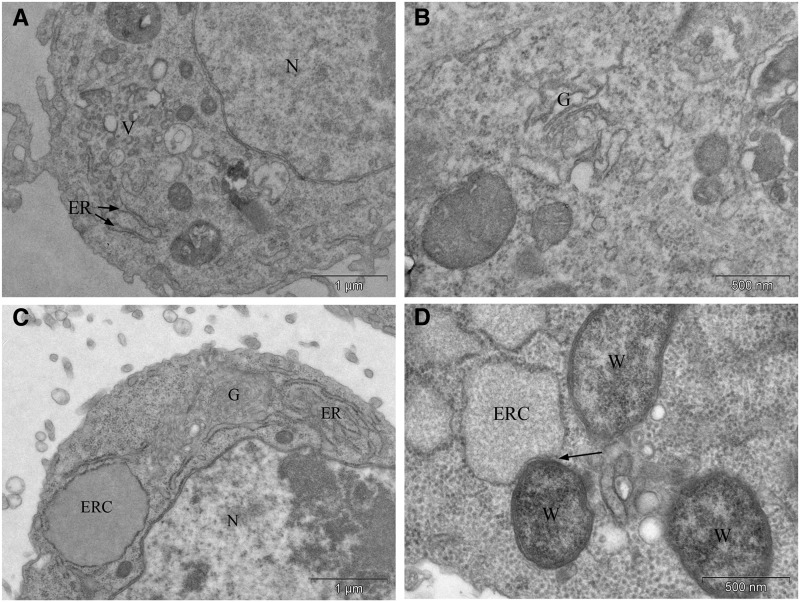
Figure 8.
Wolbachia alter the morphology of the ER. (A and B) Images from uninfected (doxycycline cured) Drosophila cell lines. Under these conditions, the ER compartments exhibited a normal tubular and discrete shape. Additionally, only few Golgi bodies were found. (C and D) Images from Wolbachia-infected Drosophila cell lines. (C) The presence of Wolbachia resulted in the swelling of the ER to form either ER cisternae or a highly elaborate complex of ER extensions and golgi. (D) Wolbachia are often observed in close contact with enlarged ER cisternae (ERC, arrow). W, Wolbachia; ER, endoplasmic reticulum; N, nucleus; V, vesicles; G, golgi.
Figure 9.
Quantification of ER morphology in Wolbachia-infected and cured cells. ER morphology in randomly selected EM sections from 198 Wolbachia-infected cells and 121 cured cells was classified into four distinct categories of ER morphology: wild type (WT), extensions, swelling, and cisternae. Quantification revealed the presence of Wolbachia significantly decreased the percentage of cells exhibiting wild-type ER and significantly increased the percentage of cells exhibiting ER swelling and ER cisternae (P < 0.0001 for all, with adjusted α = 0.00625). Significance was determined using chi-square analysis. Images below illustrate each category. The number and letter in the bottom left of the images indicate which previous figure the image originated from. The ER and ER cisternae (ERC) are labeled where appropriate.
Discussion
Performing the genome-wide, cell-based RNAi screen enabled us to comprehensively identify host factors that influenced Wolbachia titer. Of the 15,699 targets tested by RNAi disruption (Goshima et al. 2007), the screen yielded 36 RNAi treatments that decreased Wolbachia titer and 41 RNAi treatments that increased titer, implicating 77 host genes. The fact that only a small fraction of the RNAi knockdowns influenced titer suggests that Wolbachia possesses robust mechanisms of maintaining specific intracellular titers. Not surprisingly, hits that reduced Wolbachia titer spanned a broad array of cellular functions including host transcriptional and translational machinery, mitochondrial proteins, cell signaling, and metabolism (Table 1).
Of the 77 gene knockdowns that significantly altered Wolbachia titer, eight genes were involved in the ubiquitin/proteolysis pathway. These included RpL40, lig, Ubiquitin-5E, ubl, Ubc6, mei-P26, CG12725, and CG31807. These studies indicate that maintenance of Wolbachia titer requires fully functional host ubiquitin and proteolysis pathways. A likely explanation for this dependence comes from genomic studies demonstrating that Wolbachia lacks many of the pathways for amino acid production (Wu et al. 2004; Brownlie and O’Neill 2005). A robust ubiquitin/proteasome pathway would ensure an adequate pool of amino acids in order for Wolbachia to thrive. Similarly, RNAi screens in Drosophila infected with L. monocytogenes, M. fortuitum, and F. tularensis revealed that bacteria titer uptake and proliferation was highly dependent on the ubiquitin and proteasome pathways (Agaisse et al. 2005; Philips et al. 2005; Akimana 2010).
We used immunofluorescence to determine if the presence of Wolbachia influenced the ubiquitination state of proteins in these cells by taking advantage of an antibody that broadly recognizes mono and poly-ubiquitinated proteins (Everett 2000). Our analysis of ubiquitin staining in tissue culture cells and Drosophila oogenesis revealed significantly more foci in the Wolbachia-infected conditions than uninfected conditions. Previous studies using an antibody that broadly recognizes poly-ubiquitinated proteins revealed that the herpes simplex virus immediate-early protein ICPO induces proteasome-dependent degradation of host proteins and results in a similar increase in the number and extent of ubiquitin foci, which are interpreted as sites of concentrated poly-ubiquitinated proteins (Everett 2000).
Function disruption tests also support a role for the ubiquitin-proteasome system in regulating Wolbachia titer in Drosophila oogenesis. Disruption of proteasome activity with the small molecule inhibitor lactacystin also significantly reduced oocyte Wolbachia titer, indicating that maintenance of Wolbachia titer in oogenesis is dependent on host proteasome activity. We also knocked down Ubc6, an E2 conjugating enzyme involved in the ERAD pathway (Metzger et al. 2008). Ubc6 is specifically associated with the ERAD-C complex that monitors the folding state of the cytosolic domains of membrane proteins. These results suggest that Wolbachia preferentially rely on the ERAD-C protein degradation pathway as an amino acid source. In support of this conclusion, our EM analysis of Wolbachia-infected tissue culture cells reveals that Wolbachia exhibits a particularly close association with the ER (Figure 6, Figure 7, and Figure 8).
Our results are in accord with proteomic and genomic studies indicating that Wolbachia in Aedes and B. malayi must rely on host-derived amino acids (Foster et al. 2005; Baldridge et al. 2014). In addition, studies demonstrate that amino acids may be limiting because Wolbachia competes with its host for amino acids (Caragata et al., 2014). Our findings are also in line with work demonstrating that mosquito cell lines newly infected with Wolbachia exhibit up-regulation of the host 26S proteasome and a general increase in ubiquitinated proteins (Fallon and Witthuhn 2009). However, a key difference between these studies in mosquito cells and our work in Drosophila is the timeline of the effects. Differences in the proteasome and ubiquitination levels were not observed between long-term Wolbachia-infected and cured mosquito cell lines, nor was it observed in cured cell lines reinfected with Wolbachia. This suggests that the up-regulation was a transient host response to new infection. In contrast, we found that up-regulation of the proteasome occurs in Drosophila cell lines and oocytes with long-term Wolbachia infections. Whether this is the result of a difference in the fundamental biology of mosquito and Drosophila cells is unclear.
Our ultrastructural studies of the infected cell lines demonstrate a close association between Wolbachia and the ER. As shown in Figure 6 and Figure 7, Wolbachia shares membrane with the ER and is often found embedded in the ER. A similar close association between Wolbachia and the ER has been documented in early Drosophila embryos (Voronin et al. 2004), nurse cells (Serbus et al. 2011), and in the central nervous system (Strunov and Kiseleva 2014). These observations are intriguing in light of our finding that the ERAD pathway is required for maintaining Wolbachia titer both in cells lines and Drosophila oocytes. By being embedded in the ER, Wolbachia is in prime position to utilize the products derived from ERAD-mediated protein degradation.
An alternative interpretation of these results is based on studies of apicoplasts, organelles present in apicomlexan parasites including Toxoplasma gondii and Plasmodium falciparum. This organelle originated from an alga that underwent secondary endocytosis (Vaishnava and Striepen 2006). Apicoplasts have many properties in common with Wolbachia including maternal inheritance, multiple membranes, and a close association with the ER (Vaishnava and Striepen 2006). In apicoplasts, the ERAD pathway has been usurped and modified to facilitate import of key required proteins from the host cytoplasm (Agrawal et al. 2013). Strikingly, it also relies on the ubiquitin pathway marking proteins for import to the apicoplast. These findings raise the intriguing possibility that Wolbachia may also rely on the ERAD pathway for import of host cytoplasmic proteins.
Two other functional categories have been validated in vivo in addition to Ubiquitin: “ATP/GTP” and “Mitochondrial.” Five hits were identified in the ATP/GTP functional category. We found that knockdown of protostome-specific GEF (CG14047) in the Drosophila germline significantly decreased oocyte titer (Supplemental Material, Figure S1A). Three hits were also recovered in mitochondrial metabolism: a NADH dehydrogenase (CG3214), a succinate dehydrogenase (CG14757), and a mitochondrial carrier protein (CG18324). We found that knockdown of CG3214 and CG18324 in the Drosophila oocyte resulted in significant increases and decreases in Wolbachia titer, respectively (Figure S1, C and D). We suspect that the reduction of cellular ATP levels expected from knockdown of these genes limits the ability of Wolbachia to replicate within the cells. Alternatively, because specific Wolbachia and mitochondrial strains have coevolved, Wolbachia may be highly sensitive to functional changes in its companion mitochondria (Perlman et al. 2015). It should be pointed out that some of these RNAi hits might be false positives because secondary screens have not confirmed them.
Two hits that reduced Wolbachia titer were in genes regulating lipid metabolism. CG9243 encodes phospholipase D that catalyzes breakdown of phosphatidylcholine to phosphatidic acid and choline. CG1718 is an ABC transporter that functions in sterol uptake and esterification. This reliance on host lipid metabolism to maintain titer may be due to the fact that Wolbachia is encompassed by an outer host-derived membrane and lacks key fatty acid and cholesterol metabolic pathways (Voronin et al. 2004; Wu et al. 2004). Thus, replication of Wolbachia is likely to place exceptional demands on host lipid metabolism. Our results are also in accord with a recent study demonstrating that the presence of Wolbachia significantly alters lipid metabolism of Aedes albopictus mosquito cells (Molloy et al. 2016). Significantly, the titer of Spiroplasma, another maternally inherited insect endosymbiont, is also limited by the levels of host lipids (Herren et al. 2014). In addition, replication of mammalian pathogens M. tuberculosis and C. trachomatis requires host-derived lipids (Ehrt and Schnappinger 2007; Robertson et al. 2009).
It is now well documented that the presence of Wolbachia confers resistance against a number of positive-strand RNA viruses including dengue and Zika (Johnson 2015). There is evidence that multiple host mechanisms may be involved in inhibiting viral replication including synthesis of reactive oxygen and cholesterol and induction of host autophagy (Le Clec’h et al. 2012; Pan et al. 2012; Caragata et al. 2013; Wong et al. 2015). Our finding that Wolbachia dramatically alters host ER morphology and relies on the ERAD pathway to maintain titer suggests the involvement of a previously unsuspected mechanism in preventing the replication of positive-strand RNA viruses. For a number of positive-strand RNA viruses, the ER is the preferred site of replication and assembly (Romero-Brey and Bartenschlager 2016). Viral targeting of host ER induces dramatic changes in the morphology of the ER including invaginations of the ER membrane, swelling of the ER into large cisternae, and formation of virus-containing vesicles in the lumen (Romero-Brey and Bartenschlager 2016). These alterations are believed to create membrane-based sites of viral replication and assembly. Particularly striking are the recent findings that positive-strand RNA viruses specifically target and subvert the ERAD pathway to facilitate replication and assembly (Noack et al. 2014). For example, the ERAD pathway was identified in a recent genome-wide CRISPR (clustered regularly interspaced short palindromic repeats) screen to identify host factors essential to the dengue virus (Marceau et al. 2016). Our finding that Wolbachia also relies on the ERAD pathway for replication and produces dramatic alterations in ER morphology suggests that Wolbachia and positive-strand RNA viruses may be competing for the same intracellular niche. The intimate association between Wolbachia and the ER may physically prevent viruses from associating with ERAD sites. Alternatively, the extensive disruption in ER morphology may nonspecifically prevent the interactions necessary for viral replication. A satisfying aspect of this explanation of viral suppression is that it readily explains the ability of Wolbachia to suppress RNA viruses, and not DNA viruses, which preferentially replicate in the nucleus (den Boon et al. 2010). A number of other intracellular bacteria also exhibit a close association with the ER (Roy et al. 2006). It will be of great interest to determine whether these also suppress positive-strand RNA viral replication.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.198903/-/DC1.
Acknowledgments
We thank the Sullivan laboratory members present and past for helpful discussion and suggestions as well as the Bloomington Drosophila Stock Center, the Drosophila community for reagents. We also thank Nadine Gassner, Frederic Landmann, Catharina Lindley, William C. Rice, and Gilles Hickson for assistance and advice. We thank Rémi Le Borgne from the ImagoSeine core facility of the Institut Jacques Monod, also associated with Infrastructure en Biologie Santé et Agronomie and France BioImaging infrastructures. Funding from National Institutes of Health grant GM-104486, University of California, Santa Cruz Faculty Research Award, and Florida International University supported this work. A.D. and A.G. were supported by the Centre National de la Recherche Scientifique, the Association de la Recherche sur le Cancer grant PJA 20141201756, and the “Ligue Contre le Cancer” grant RA11/75-34.
Author contributions: P.M.W., L.R.S., A.D., A.G., W.B., R.S.L., and W.S. designed the experiments. P.M.W., L.R.S., A.D., and A.C. conducted the experiments. P.M.W., L.R.S., A.D., and W.S. wrote the manuscript.
Footnotes
Communicating editor: D. A. Barbash
Literature Cited
- Agaisse H., Burrack L. S., Philips J. A., Rubin E. J., Perrimon N., et al. , 2005. Genome-wide RNAi screen for host factors required for intracellular bacterial infection. Science 309: 1248–1251. [DOI] [PubMed] [Google Scholar]
- Agrawal S., Chung D. W., Ponts N., van Dooren G. G., Prudhomme J., et al. , 2013. An apicoplast localized ubiquitylation system is required for the import of nuclear-encoded plastid proteins. PLoS Pathog. 9: e1003426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akimana C, Al-Khodor S., Abu Kwaik Y., 2010. Host factors required for modulation of phagosome biogenesis and proliferation of Francisella tularensis within the cytosol. PLoS One 5: e11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson R., Casper-Lindley C., Cao J., Tram U., Sullivan W., 2009. Symmetric and asymmetric mitotic segregation patterns influence Wolbachia distribution in host somatic tissue. J. Cell Sci. 122: 4570–4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashton-Beaucage D., Lemieux C., Udell C. M., Sahmi M., Rochette S., et al. , 2016. The deubiquitinase USP47 stabilizes MAPK by counteracting the function of the N-end rule ligase POE/UBR4 in Drosophila. PLoS Biol. 14: e1002539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldridge G. D., Baldridge A. S., Witthuhn B. A., Higgins L., Markowski T. W., et al. , 2014. Proteomic profiling of a robust Wolbachia infection in an Aedes albopictus mosquito cell line. Mol. Microbiol. 94: 537–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beasley T. M., Schumacker R. E., 1995. Multiple regression approach to analyzing contingency tables: post hoc and planned comparison procedures. J. Exp. Educ. 64(1): 79–93. [Google Scholar]
- Brownlie J. C., O’Neill S. L., 2005. Wolbachia genomes: insights into an intracellular lifestyle. Curr. Biol. 15: R507–R509. [DOI] [PubMed] [Google Scholar]
- Caragata E. P., Rances E., Hedges L. M., Gofton A. W., Johnson K. N., et al. , 2013. Dietary cholesterol modulates pathogen blocking by Wolbachia. PLoS Pathog. 9: e1003459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caragata E. P., Rances E., O’Neill S. L., McGraw E. A., 2014. Competition for amino acids between Wolbachia and the mosquito host, Aedes aegypti. Microb. Ecol. 67: 205–218. [DOI] [PubMed] [Google Scholar]
- Chagas-Moutinho V. A., Silva R., de Souza W., Motta M. C., 2015. Identification and ultrastructural characterization of the Wolbachia symbiont in Litomosoides chagasfilhoi. Parasit. Vectors 8: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debec A., Megraw T. L., Guichet A., 2016. Methods to establish Drosophila cell lines. Methods Mol. Biol. 1478: 333–351. [DOI] [PubMed] [Google Scholar]
- den Boon J. A., Diaz A., Ahlquist P., 2010. Cytoplasmic viral replication complexes. Cell Host Microbe 8: 77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derre I., Pypaert M., Dautry-Varsat A., Agaisse H., 2007. RNAi screen in Drosophila cells reveals the involvement of the Tom complex in Chlamydia infection. PLoS Pathog. 3: 1446–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echard A., Hickson G. R., Foley E., O’Farrell P. H., 2004. Terminal cytokinesis events uncovered after an RNAi screen. Curr. Biol. 14(18): 1685–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrt S., Schnappinger D., 2007. Mycobacterium tuberculosis virulence: lipids inside and out. Nat. Med. 13: 284–285. [DOI] [PubMed] [Google Scholar]
- Everett R. D., 2000. ICP0 induces the accumulation of colocalizing conjugated ubiquitin. J. Virol. 74: 9994–10005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallon A. M., Witthuhn B. A., 2009. Proteasome activity in a naive mosquito cell line infected with Wolbachia pipientis wAlbB. In Vitro Cell. Dev. Biol. Anim. 45: 460–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferree P. M., Frydman H. M., Li J. M., Cao J., Wieschaus E., et al. , 2005. Wolbachia utilizes host microtubules and dynein for anterior localization in the Drosophila oocyte. PLoS Pathog. 1: e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster J., Ganatra M., Kamal I., Ware J., Makarova K., et al. , 2005. The Wolbachia genome of Brugia malayi: endosymbiont evolution within a human pathogenic nematode. PLoS Biol. 3: e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimuro M., Sawada H., Yokosawa H., 1994. Production and characterization of monoclonal antibodies specific to multi-ubiquitin chains of polyubiquitinated proteins. FEBS Lett. 349: 173–180. [DOI] [PubMed] [Google Scholar]
- Goshima G., Wollman R., Goodwin S. S., Zhang N., Scholey J. M., et al. , 2007. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science 316: 417–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herren J. K., Paredes J. C., Schupfer F., Arafah K., Bulet P., et al. , 2014. Insect endosymbiont proliferation is limited by lipid availability. eLife 3: e02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson A. M., Cooley L., 2014. Methods for studying oogenesis. Methods 68: 207–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K. N., 2015. The impact of Wolbachia on virus infection in mosquitoes. Viruses 7: 5705–5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpova N., Bobinnec Y., Fouix S., Huitorel P., Debec A., 2006. Jupiter, a new Drosophila protein associated with microtubules. Cell Motil. Cytoskeleton 63: 301–312. [DOI] [PubMed] [Google Scholar]
- Kose H., Karr T. L., 1995. Organization of Wolbachia pipientis in the Drosophila fertilized egg and embryo revealed by an anti-Wolbachia monoclonal antibody. Mech. Dev. 51: 275–288. [DOI] [PubMed] [Google Scholar]
- Le Clec’h W., Braquart-Varnier C., Raimond M., Ferdy J. B., Bouchon D., et al. , 2012. High virulence of Wolbachia after host switching: when autophagy hurts. PLoS Pathog. 8: e1002844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemus L., Goder V., 2014. Regulation of endoplasmic reticulum-associated protein degradation (ERAD) by ubiquitin. Cells 3: 824–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marceau C. D., Puschnik A. S., Majzoub K., Ooi Y. S., Brewer S. M., et al. , 2016. Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens. Nature 535: 159–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzalupo S., Cooley L., 2006. Illuminating the role of caspases during Drosophila oogenesis. Cell Death Differ. 13(11): 1950–1959. [DOI] [PubMed] [Google Scholar]
- Metzger M. B., Maurer M. J., Dancy B. M., Michaelis S., 2008. Degradation of a cytosolic protein requires endoplasmic reticulum-associated degradation machinery. J. Biol. Chem. 283: 32302–32316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min K. T., Benzer S., 1997. Wolbachia, normally a symbiont of Drosophila, can be virulent, causing degeneration and early death. Proc. Natl. Acad. Sci. USA 94: 10792–10796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr S. E., Smith J. A., Shamu C. E., Neumuller R. A., Perrimon N., 2014. RNAi screening comes of age: improved techniques and complementary approaches. Nat. Rev. Mol. Cell Biol. 15: 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molloy J. C., Sommer U., Viant M. R., Sinkins S. P., 2016. Wolbachia modulates lipid metabolism in Aedes albopictus mosquito cells. Appl. Environ. Microbiol. 82: 3109–3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser T. S., Sabin L. R., Cherry S., 2010. RNAi screening for host factors involved in vaccinia virus infection using Drosophila cells. JoVE 42: e2137–e2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouton L., Henri H., Bouletreau M., Vavre F., 2006. Effect of temperature on Wolbachia density and impact on cytoplasmic incompatibility. Parasitology 132: 49–56. [DOI] [PubMed] [Google Scholar]
- Muller M. J., Dorr N. C., Depra M., Schmitz H. J., Valiati V. H., et al. , 2013. Reevaluating the infection status by the Wolbachia endosymbiont in Drosophila Neotropical species from the willistoni subgroup. Infect. Genet. Evol. 19: 232–239. [DOI] [PubMed] [Google Scholar]
- Newton I. L., Savytskyy O., Sheehan K. B., 2015. Wolbachia utilize host actin for efficient maternal transmission in Drosophila melanogaster. PLoS Pathog. 11: e1004798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noack J., Bernasconi R., Molinari M., 2014. How viruses hijack the ERAD tuning machinery. J. Virol. 88: 10272–10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Zhou G., Wu J., Bian G., Lu P., et al. , 2012. Wolbachia induces reactive oxygen species (ROS)-dependent activation of the Toll pathway to control dengue virus in the mosquito Aedes aegypti. Proc. Natl. Acad. Sci. USA 109: E23–E31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S. J., Hodson C. N., Hamilton P. T., Opit G. P., Gowen B. E., 2015. Maternal transmission, sex ratio distortion, and mitochondria. Proc. Natl. Acad. Sci. USA 112: 10162–10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips J. A., Rubin E. J., Perrimon N., 2005. Drosophila RNAi screen reveals CD36 family member required for mycobacterial infection. Science 309: 1251–1253. [DOI] [PubMed] [Google Scholar]
- Pietri J. E., DeBruhl H., Sullivan W., 2016. The rich somatic life of Wolbachia. MicrobiologyOpen 5: 923–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poinsot D., Bourtzis K., Markakis G., Savakis C., Merçot H., 1998. Wolbachia transfer from Drosophila melanogaster into D. simulans: host effect and cytoplasmic incompatibility relationships. Genetics 150(1): 227–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson D. K., Gu L., Rowe R. K., Beatty W. L., 2009. Inclusion biogenesis and reactivation of persistent Chlamydia trachomatis requires host cell sphingolipid biosynthesis. PLoS Pathog. 5: e1000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Brey I., Bartenschlager R., 2016. Endoplasmic reticulum: the favorite intracellular niche for viral replication and assembly. Viruses 8: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy C. R., Salcedo S. P., Gorvel J. P., 2006. Pathogen-endoplasmic-reticulum interactions: in through the out door. Nat. Rev. Immunol. 6: 136–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serbus L. R., Sullivan W., 2007. A cellular basis for Wolbachia recruitment to the host germline. PLoS Pathog. 3: e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serbus L. R., Casper-Lindley C., Landmann F., Sullivan W., 2008. The genetics and cell biology of Wolbachia-host interactions. Annu. Rev. Genet. 42: 683–707. [DOI] [PubMed] [Google Scholar]
- Serbus L. R., Ferreccio A., Zhukova M., McMorris C. L., Kiseleva E., et al. , 2011. A feedback loop between Wolbachia and the Drosophila gurken mRNP complex influences Wolbachia titer. J. Cell Sci. 124: 4299–4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serbus L. R., Landmann F., Bray W. M., White P. M., Ruybal J., et al. , 2012. A cell-based screen reveals that the albendazole metabolite, albendazole sulfone, targets Wolbachia. PLoS Pathog. 8: e1002922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serbus L. R., White P. M., Silva J. P., Rabe A., Teixeira L., et al. , 2015. The impact of host diet on Wolbachia titer in Drosophila. PLoS Pathog. 11: e1004777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunov A., Kiseleva E., 2014. Drosophila melanogaster brain invasion: pathogenic Wolbachia in central nervous system of the fly. Insect Sci. 23: 253–264. [DOI] [PubMed] [Google Scholar]
- Unckless R. L., Boelio L. M., Herren J. K., Jaenike J., 2009. Wolbachia as populations within individual insects: causes and consequences of density variation in natural populations. Proc. Biol. Sci. 276: 2805–2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnava S., Striepen B., 2006. The cell biology of secondary endosymbiosis – how parasites build, divide and segregate the apicoplast. Mol. Microbiol. 61: 1380–1387. [DOI] [PubMed] [Google Scholar]
- Van Voorhis W. C., Adams J. H., Adelfio R., Ahyong V., Akabas M. H., et al. , 2016. Open source drug discovery with the malaria box compound collection for neglected diseases and beyond. PLoS Pathog. 12: e1005763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheyen E., Cooley L., 1994. Looking at oogenesis. Methods Cell Biol. 44: 545–561. [PubMed] [Google Scholar]
- Voronin D. A., Dudkina N. V., Kiseleva E. V., 2004. A new form of symbiotic bacteria Wolbachia found in the endoplasmic reticulum of early embryos of Drosophila melanogaster. Dokl. Biol. Sci. 396: 227–229. [DOI] [PubMed] [Google Scholar]
- Werren J. H., Baldo L., Clark M. E., 2008. Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol. 6: 741–751. [DOI] [PubMed] [Google Scholar]
- Wong Z. S., Brownlie J. C., Johnson K. N., 2015. Oxidative stress correlates with Wolbachia-mediated antiviral protection in Wolbachia-Drosophila associations. Appl. Environ. Microbiol. 81: 3001–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Sun L. V., Vamathevan J., Riegler M., Deboy R., et al. , 2004. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2: E69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Landis-Piwowar K. R., Chen D., Milacic V., Dou Q. P., 2008. Natural compounds with proteasome inhibitory activity for cancer prevention and treatment. Curr. Protein Pept. Sci. 9: 227–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. H., Chung T. D., Oldenburg K. R., 1999. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4(2): 67–73. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Zhu Y., 2015. Diversity of bacterial manipulation of the host ubiquitin pathways. Cell. Microbiol. 17: 26–34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All primary data can be found in Supplemental Material, File S1. Additional information about in vivo validation of genes that alter Wolbachia quantity can be found in Figure S1.