Abstract
Animals can adapt to unfavorable environments through changes in physiology or behavior. In the nematode, Caenorhabditis elegans, environmental conditions perceived early in development determine whether the animal enters either the reproductive cycle, or enters into an alternative diapause stage named dauer. Here, we show that endogenous RNAi pathways play a role in dauer formation in crowding (high pheromone), starvation, and high temperature conditions. Disruption of the Mutator proteins or the nuclear Argonaute CSR-1 result in differential dauer-deficient phenotypes that are dependent upon the experienced environmental stress. We provide evidence that the RNAi pathways function in chemosensory neurons for dauer formation, upstream of the TGF-β and insulin signaling pathways. In addition, we show that Mutator MUT-16 expression in a subset of individual pheromone-sensing neurons is sufficient for dauer formation in high pheromone conditions, but not in starvation or high temperature conditions. Furthermore, we also show that MUT-16 and CSR-1 are required for expression of a subset of G proteins with functions in the detection of pheromone components. Together, our data suggest a model where Mutator-amplified siRNAs that associate with the CSR-1 pathway promote expression of genes required for the detection and signaling of environmental conditions to regulate development and behavior in C. elegans. This study highlights a mechanism whereby RNAi pathways mediate the link between environmental stress and adaptive phenotypic plasticity in animals.
Keywords: dauer, RNAi, Mutators, CSR-1, Caenorhabditis elegans
ANIMALS are capable of undergoing physiological and behavioral changes to adapt to adverse environments, a process known as allostasis (Sterling and Eyer 1988). One such adaptation is the expression of polyphenism, or alternative developmental morphs, in a population of genetically identical organisms (Michener 1961; Mayr 1963). Sex determination in alligators (Woodward and Murray 1993), caste determination in insects (Nijhout 1998, 1999), seasonal differences in phenotypes of adult butterflies Precis Almana (Nijhout 1999), and growth of a “helmet-like” structure in Daphnia pulex (water flea) in the presence of predators (Brewer et al. 1999) are examples of polyphenisms in response to unfavorable environmental conditions.
Given that polyphenisms can occur in isogenic populations of animals, epigenetic mechanisms, such as RNA interference (RNAi) and DNA methylation, are hypothesized to regulate the expression of alternative phenotypic morphs in response to environmental conditions (West-Eberhard 2003; Wang et al. 2006; Kronforst et al. 2008; Kucharski et al. 2008; Moczek and Snell-Rood 2008; Hunt et al. 2010; Bonasio 2012; Humann et al. 2013). For example, in pea aphids, unwinged and winged morphs develop in response to favorable or unfavorable environments, respectively (Müller et al. 2001; Brisson 2010). Although genetically identical, winged and unwinged female populations exhibit differential DNA methylation patterns and transcriptional profiles of genes implicated in wing polyphenism (Brisson et al. 2007, 2010; Walsh et al. 2010). In addition, polyphenic transitions of locusts from solitary phase to gregarious (swarm formation) depends upon the differential accumulation of small RNAs in the two phases (Wei et al. 2009). Despite these examples, the molecular mechanisms that govern gene targeting and regulation by epigenetic pathways in response to environmental conditions are not well understood.
C. elegans is an excellent model system in which to study the molecular mechanisms regulating polyphenism as their developmental trajectory is determined by the environmental conditions experienced after hatching. Under favorable growth conditions, worms proceed through four larval stages (L1, L2, L3, and L4) to become reproductive adults. Under unfavorable conditions, such as low food availability, high temperatures, or high pheromone concentrations, L1 larvae will enter an alternative developmental stage called dauer (Cassada and Russell 1975; Golden and Riddle 1982; 1984a,b,c). Dauer larvae are stress-resistant and nonaging, and thought to facilitate dispersal in environmental conditions unfavorable for reproduction (Klass and Hirsh 1976; Larsen 1993; Frézal and Félix 2015). Once conditions improve, animals will exit dauer and resume continuous development as L4 larvae. Although adults that passed through, or bypassed, the dauer stage appear morphologically similar, we previously showed that C. elegans retain a cellular memory of their developmental history that is reflected in changes in gene expression, genome-wide chromatin states, and life history traits (Hall et al. 2010). Furthermore, we have shown that RNAi pathways are a major contributor to developmental history-dependent phenotypes in adults (Hall et al. 2013). However, the role of RNAi pathways in regulating environmentally induced phenotypic plasticity during early larval stages is unknown.
Environmental cues sensed by G protein coupled receptors (GPCRs) residing in the ciliary endings of sensory neurons differentially regulate the TGF-β and insulin signaling dauer formation pathways (Fielenbach and Antebi 2008). Mutations in genes operating in these signaling pathways can result in dauer constitutive (daf-c) or dauer deficient (daf-d) phenotypes (Riddle et al. 1981; Vowels and Thomas 1992). Animals that form significantly fewer dauer larvae than wild-type in response to environmental stress are considered daf-d, while daf-c animals can form dauers even in the absence of environmental stress. The primary cue for dauer entry in C. elegans is high local concentrations of dauer pheromone, which is a blend of hydrophilic ascaroside (ascr) molecules containing three to nine carbon side chains (Golden and Riddle 1982, 1984a,b,c; Jeong et al. 2005; Butcher et al. 2007, 2008). Exposure to unfavorable environmental conditions has been shown to inhibit DAF-7 TGF-β production in ASI sensory neurons during the L1 larval stage, resulting in dauer formation (Ren et al. 1996; Schackwitz et al. 1996). In addition, unfavorable conditions can result in diminished insulin signaling, causing reduced expression of daf-28/insulin in ASI and ASJ neurons during the L1 and dauer stages and ins-6/insulin in ASI neurons during the L2d stage (Li et al. 2003; Cornils et al. 2011; Neal et al. 2015). These two pathways converge onto the DAF-12 steroid hormone receptor, which acts a master regulator of dauer formation programs (Riddle et al. 1981; Vowels and Thomas 1992; Thomas et al. 1993; Antebi et al. 2000).
In this study, we sought to characterize the role of endogenous RNAi pathways in the regulation of dauer formation in C. elegans. In worms, small interfering RNAs (siRNAs) are characterized by their biogenesis and associated Argonautes (AGOs). Primary siRNAs are low in abundance, have Dicer-dependent biogenesis, and are 26 nucleotides long with a 5′ guanine (26G-siRNAs) (Bernstein et al. 2001; Grishok et al. 2001; Ketting et al. 2001; Knight and Bass 2001; Han et al. 2009; Pavelec et al. 2009; Vasale et al. 2010). Through an unknown mechanism, 26G-siRNAs stimulate the production of highly abundant siRNAs that are 22 nucleotides long with a 5′ guanine (22G-siRNAs), and are synthesized through the action of RNA-dependent RNA polymerases (RdRPs) (Smardon et al. 2000; Ketting et al. 2001; Knight and Bass 2001; Simmer et al. 2002; Ambros et al. 2003; Maine et al. 2005; Vought et al. 2005; Aoki et al. 2007; Pak and Fire 2007; She et al. 2009; Gent et al. 2010; Vasale et al. 2010; Pak et al. 2012). In addition, a group of proteins called the Mutators were shown to play a role in siRNA amplification of both 26G- and 22G-siRNAs classes (Zhang et al. 2011; Phillips et al. 2012). Specific small RNA classes bind to one or more of the 26 AGO proteins in C. elegans, which are characterized by their expression patterns, and whether they function in the cytoplasm or nucleus (Yigit et al. 2006). Although much progress has been made in characterizing the biogenesis of endogenous siRNAs, little is known about how RNAi pathways target and regulate endogenous genes.
Here, we show that endogenous RNAi pathways are required for dauer formation in adverse environmental conditions. Mutations in the Mutator proteins and nuclear AGO CSR-1 pathway result in differential daf-d phenotypes that are dependent upon the experienced environmental stress. Mutator protein, MUT-16, acts in sensory neurons, likely upstream of the TGF-β and insulin signaling pathways, for dauer formation in high pheromone, starvation, or high temperature conditions. Our results suggest that MUT-16 and CSR-1 function to positively regulate expression levels of genes with sensory signaling functions, mediating the link between RNAi and formation of polyphenism in stressful environments in C. elegans.
Materials and Methods
C. elegans strains and maintenance
All worm strains were cultivated on NGM plates with Escherichia coli OP50 as the food source at either 15 or 20° according to standard methods (Brenner 1974; Stiernagle 2006). Worm strains used in this study are listed in Supplemental Material, File S1. Due to the propensity of high spontaneous mutation through transposon mobilization in a subset of the RNAi strains used (i.e., Mutators) (Vastenhouw et al. 2003), we limited the number of propagated generations after thawing, and genetically backcrossed strains to wild-type when necessary.
Dauer formation assays
Different dauer formation assays were used to test daf phenotypes under high pheromone, starvation, or high temperature conditions. To test dauer formation in the presence of high pheromone concentrations, we conducted assays using crude pheromone as previously described with a few modifications (Neal et al. 2013; Zhang et al. 2013). For each independent batch of crude pheromone, one activity unit was defined as the amount of pheromone that resulted in 33% dauers in wild-type animals (Zhang et al. 2013). Each dauer formation plate contained four activity units of pheromone that were mixed into the medium during plate preparation. Water was used instead of pheromone for the control plates. Each assay plate was seeded with 20 µl of 8 mg/ml (0.16 mg) E. coli OP50 that was heat-killed by incubating at 95° for 30 min.
Next, five well-fed 2-day-old adults were placed on assay plates for 3–6 hr at room temperature until ∼60–80 eggs were laid on the plates, after which the adults were removed. For mutant strains that exhibited severe sterility phenotypes [mut-2(ne298), mut-7(pk720), csr-1 hypomorph, csr-1(tm892)/nT1(qIs51), mut-16(pk710); csr-1(tm892)/nT1(qIs51), and drh-3(ne2453)], eight adults were placed on the assay plates, and the amount of heat-killed E. coli OP50 was doubled to prevent starvation during the extended egg-laying period. No significant change in dauer formation was observed for wild-type animals using these modified conditions (Student’s t-test, P = 0.17). The assay plates were incubated at 25° for 3 days, and then scored for dauers. For strains carrying a chromosome balancer, the daf phenotype of only non-GFP progeny was scored.
To perform dauer formation assays using high temperatures, a similar protocol was used as described above, with a few modifications. First, assay plates were prepared without pheromone. Second, mutant embryos were allowed to hatch at room temperature for 10–12 hr before transfer to 25 or 27° to prevent embryonic lethality or L1 diapause. Assay plates were scored for the presence of dauers after 4 days.
Starvation assays were conducted using assay plates prepared without pheromone that were seeded with 0.04 mg of heat-killed E. coli OP50. Embryos were allowed to hatch at room temperature before transfer to 25°. Dauer formation was scored after 5 days.
For all assays, dauers were distinguished from nondauers by the presence of pharynx pumping and dauer alae as previously described (Cassada and Russell 1975; Popham and Webster 1979; Albert and Riddle 1988; Riddle and Albert 1997; Hu 2007). For mutants that exhibited an increased number of L2d larvae or partial dauers, assay plates were re-examined after 24 hr (File S2). The progeny of all the animals assayed were scored at the same time. All dauer formation assays were performed in duplicate, in at least three biological replicates (File S1). Statistical significance of data were determined using one-way ANOVA with LSD or Tukey’s HSD post hoc tests using SPSS (v. 23).
MUT-16 rescues
Genomic DNA template was used to amplify the full-length mut-16 gene. The following tissue-specific promoters were used: rab-3 (pan-neuronal), ges-1 (intestine), trx-1 (ASJ), unc-130 (ASG), gpa-4 (ASI), sre-1 (ADL), srh-142 (ADF), and odr-10 (AWA) (Egan et al. 1995; Troemel et al. 1995; Sengupta et al. 1996; Nonet et al. 1997; Jansen et al. 1999; Sagasti et al. 1999; Sarafi-Reinach and Sengupta 2000; Lanjuin et al. 2003; Miranda-Vizuete et al. 2006).
The genomic mut-16 gene was fused to tissue-specific promoters and the gfp gene using fusion PCR (Hobert 2002). ADL- and ASI-specific mut-16 rescue constructs were described previously (Sims et al. 2016). Rab-3p::mut-16::gfp, trx-1p::mut-16::gfp, unc-130p::mut-16::gfp, gpa-4p::mut-16::gfp, and sre-1p::mut-16::gfp were cloned into the TOPO-XL (Life Technologies) and injected into mut-16(pk710) at a concentration of 8 ng/μl. For srh-142p::mut-16::gfp, odr-10p::mut-16::gfp, and ges-1p::mut-16::gfp, purified PCR products were directly injected into mut-16(pk710) at concentrations of 8, 4, and 1 ng/μl, respectively. Unc-122p::dsRed (30 ng/μl) was used as the coinjection marker. Two independent transgenic lines were used in all dauer formation assays. See File S3 for primer sequences.
RNA preparation and reverse transcription-real time PCR
Embryos were obtained by bleaching well-fed adult hermaphrodites as described by Stiernagle (2006). Three biologically independent populations of L1 larvae were collected from wild-type, csr-1 hypomorph, and mut-16(pk710) strains. Total RNA was extracted using TRIzol reagent (Life Technologies) according to the manufacturer’s protocol, and reverse transcribed using SuperScript IV Reverse Transcriptase (Life Technologies). Quantitative real-time PCR was performed in triplicate using iTaq Universal SYBR Green Supermix (Bio-Rad) using a Bio-Rad CFX Connect Real-Time System. Normalization of gene expression for flp-21, gpa-1, gpa-3, and gpc-1 was done using the mRNA levels of y45f10d.4, a somatically expressed gene that is not a documented CSR-1 target (Claycomb et al. 2009), and does not experience gene expression changes due to dauer-inducing conditions (M. C. Ow, K. Borziak, S. Dorus, S. E. Hall unpublished results). Primer sequences are listed in File S3.
DiI staining
Well-fed wild-type and mut-16(pk710) young adults were washed with M9 buffer (Stiernagle 2006), and transferred into 1.5 ml microcentrifuge tubes. Worms were washed in M9 buffer, and resuspended in M9 buffer containing 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (DiI) (Molecular Probes) at 1:200 dilution prepared from a 2 mg/ml solution dissolved in N, N-dimethylformamide (Sigma) as previously described (Shaham 2006). The animals were rotated in the dark at room temperature for ∼1 hr, and imaged using a Leica DM5500 B microscope and ORCA-R2 Digital C10600 camera (Hamamatsu).
Data availability
The worm strains used in this study are listed in File S1 and are available on request. The primer sequences used for cloning and qPCR experiments are listed in File S3.
Results
Endogenous RNAi pathways are required for dauer formation
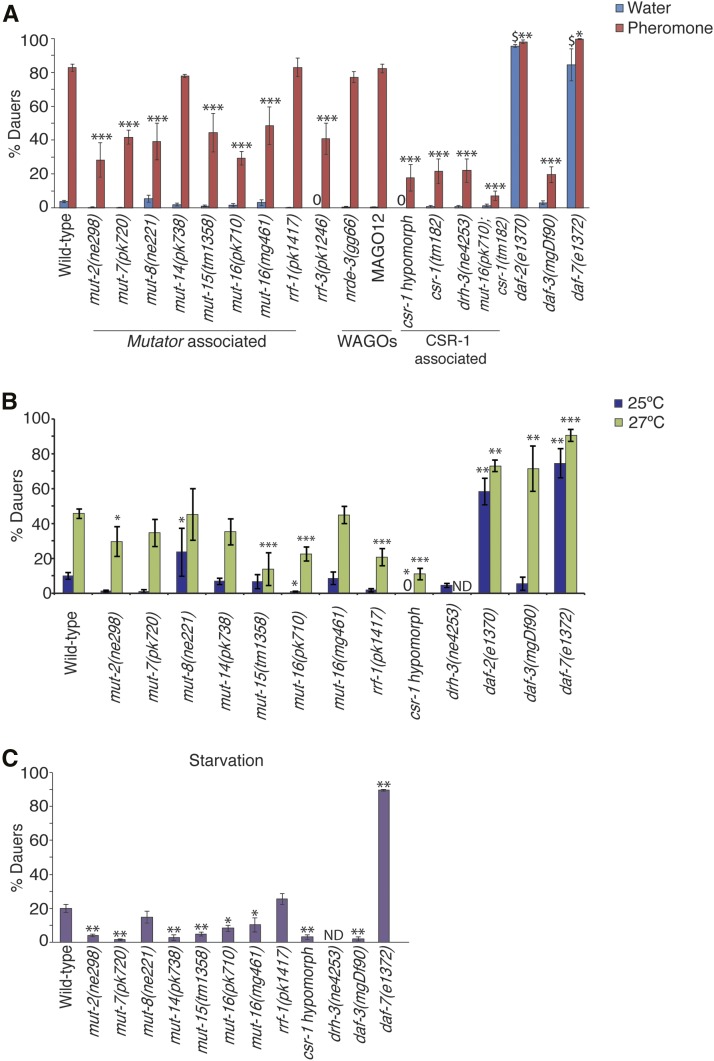
In order to characterize how RNAi pathways regulate polyphenism in response to environmental stress, we conducted dauer formation assays using strains carrying mutations in a subset of genes with functions in endogenous RNAi pathways. First, we subjected newly hatched larvae to high concentrations of crude dauer pheromone as previously described, and scored the percentage of animals that formed dauers (Neal et al. 2013). As expected, wild-type larvae populations formed significantly more dauers in high pheromone conditions (82.2 ± 2.4%) compared to control water plates (4.1 ± 0.8%) (Figure 1A). As controls, we also verified that strains with mutations in TGF-β and insulin-signaling pathways exhibited previously characterized daf-d (daf-3(mgDf90)) and daf-c (daf-7(e1372), daf-2(e1370)) phenotypes (Riddle et al. 1981; Vowels and Thomas 1992; Gottlieb and Ruvkun 1994). Second, we examined dauer formation phenotypes of strains carrying mutations in genes encoding proteins associated with the Mutator foci. In the germline, Mutator proteins form foci that are dependent on MUT-16 and localize adjacent to P granules (Phillips et al. 2012). However, the localization and interaction of Mutator proteins in the soma are not well characterized. We observed that mutations in a majority of the Mutator genes, mut-2(ne298), mut-7(pk720), mut-8/rde-2(ne221), mut-15(tm1358), and two alleles of mut-16(pk710 and mg461), resulted in significantly fewer dauers compared to wild-type (Figure 1A). The pk710 allele is a null mutation located within the coding sequence of mut-16, while the mg461 allele has a small deletion in the upstream regulatory sequences that disrupts somatic RNAi (Zhang et al. 2011). The mut-16(pk710) strain exhibited a daf-d phenotype that is comparable to the negative control daf-3(mgDf90), whereas the other strains, including mut-16(mg461), exhibited an intermediate daf-d phenotype (Figure 1A). However, strains with mutations in mut-14(pk738) and the Mutator-associated RdRP, rrf-1(pk1417), formed dauers comparable to wild-type levels, indicating that these proteins are not required for dauer formation in high pheromone conditions (Figure 1A). Interestingly, we found that mutation in another somatic RdRP gene, rrf-3 (Simmer et al. 2002), results in a daf-d phenotype similar to the Mutators, suggesting that RRF-3 may also interact with Mutator proteins in the soma (Figure 1A). These results indicate that a majority of the Mutator proteins, which are required for siRNA amplification, are necessary for dauer formation in high pheromone conditions.
Figure 1.
Mutator proteins and the CSR-1 AGO pathway are required for dauer formation. (A) Proportion of animals forming dauers in response to high pheromone conditions or water plates is shown. * P < 0.05, ** P < 0.005, *** P < 0.0005 compared to wild-type on pheromone plates, $ P < 0.0005 compared to wild-type on water plates, one-way ANOVA with LSD post hoc test. N ≥ 3 trials; n ≥ 217 animals. (B) Proportion of animals forming dauers when cultivated at 25° and 27°. * p < 0.05, ** p < 0.005, *** p < 0.0005 compared to wild-type, one-way ANOVA with LSD post hoc test. N ≥ 3 trials; n ≥ 164 animals. (C) Proportion of animals forming dauers when subjected to starvation condition. * p < 0.005, ** p < 0.0005 compared to wild-type, one-way ANOVA with LSD post hoc test. N ≥ 3 trials; n ≥ 300 animals. “ND” indicates not determined; “0” indicates no dauers were formed. All error bars represent the standard error of the mean, S.E.M.
Next, we asked which endogenous RNAi pathway contributes to dauer formation by examining the daf phenotypes of strains with mutations in various AGO genes. Since dauer formation occurs during early larval stages, we tested AGOs that are expressed in somatic tissue throughout C. elegans development. Previous work has shown that mutations in Mutator proteins drastically reduce the abundance of siRNAs that associate with worm-specific AGO proteins (WAGOs), including the nuclear AGO NRDE-3 (Zhang et al. 2011). Thus, we tested whether nrde-3(gg66) and MAGO12 (carrying mutations in all 12 wago genes) mutant strains exhibited daf phenotypes in high pheromone conditions. We found that both strains formed dauers similar to wild-type, indicating that Mutator-amplified siRNAs critical for dauer formation are not associated with the WAGO RNAi pathways (Figure 1A). Next, we tested whether the nuclear AGO CSR-1 pathway was playing a role in dauer formation using a null mutant, csr-1(tm182), and a hypomorph strain that expresses CSR-1 only in the germline (Claycomb et al. 2009). CSR-1 associated 22G-siRNAs are only slightly reduced for a subset of target genes in a mut-16(pk710) strain (Zhang et al. 2011). Surprisingly, we observed that csr-1(tm182) and hypomorph strains exhibited significantly decreased dauer formation levels, similar to the mut-16(pk710) and daf-3(mgDf90) strains (Figure 1A). In the germline, the CSR-1 pathway requires RdRP EGO-1, Dicer-related helicase DRH-3, and Tudor-domain protein EKL-1; however, CSR-1, DRH-3, and EKL-1 are also expressed in somatic tissue during all larval stages (Claycomb et al. 2009). We found that the drh-3(ne4253) mutant strain also exhibited a significant daf-d phenotype consistent with that seen in the csr-1(tm182) and hypomorph strains in high pheromone conditions (Figure 1A). We were unable to test an ekl-1 mutant strain due to its sterility phenotype. To examine whether MUT-16 and CSR-1 are functioning in the same pathway, we measured dauer formation of a csr-1(tm182); mut-16(pk710) double mutant. Interestingly, the double mutant did not show a significant difference in dauer formation compared to csr-1(tm182) (Figure 1A) (P = 0.09, Student’s t-test), suggesting that MUT-16 and CSR-1 are functioning in the same pathway. Our results suggest that Mutator-amplified siRNAs associating with the CSR-1 pathway are required for dauer formation in high pheromone conditions.
Mutators exhibit stress-specific daf-d phenotypes
Since starvation and high temperature can also trigger dauer formation, we questioned if the Mutator and CSR-1 pathway proteins also exhibited a daf-d phenotype in different environmental stresses. We subjected newly hatched L1 worms to elevated temperatures (25 and 27°), and measured the proportion of dauer formation (Figure 1B and File S2). At 27°, mut-2(ne298), mut-15(tm1358), mut-16(pk710), rrf-1(pk1417), and csr-1 hypomorph strains formed significantly fewer dauers compared to wild-type (Figure 1B). However, except for mut-16(pk710) and csr-1 hypomorph, these mutants formed dauers similar to wild-type levels when exposed to a moderately high temperature (25°) (Figure 1B). The mut-8(ne221) strain, on the other hand, formed significantly more dauers at 25° compared to wild-type. We also noted that the daf-3(mgDf90) strain exhibited a daf-d phenotype at 25°, but a daf-c phenotype at 27°, as reported previously (Ailion and Thomas 2000). These results suggest that distinct subsets of Mutators are required for dauer formation at different temperatures.
Next, we measured dauer formation in starvation conditions by subjecting newly hatched worms to depleted amounts of E. coli (see Materials and Methods). We observed that mut-2(ne298), mut-7(pk720), mut-14(pk738), mut-15(tm1358), mut-16(pk710), mut-16(mg461), and csr-1 hypomorph strains exhibited a daf-d phenotype compared to wild-type when exposed to starvation conditions early in development (Figure 1C). However, mut-8(ne221) and rrf-1(pk1417) formed dauers not significantly different from wild-type. From these results, we can conclude that a subset of Mutators are also required in starvation conditions for dauer formation.
Together, our results indicate that different subsets of RNAi proteins are required for dauer formation in distinct environmental conditions. We have shown that MUT-16 and CSR-1 AGO are required for dauer formation regardless of the environmental stress. However, MUT-2, MUT-7, MUT-8/RDE-2, MUT-14, MUT-15, RRF-1, and RRF-3 are necessary for dauer formation only for specific conditions. These findings indicate that dauer formation phenotypes can be distinct for different environmental stresses, and suggest the possibilities that endogenous RNAi pathways are regulating different subsets of genes, and/or are acting in different cells and tissue types required for dauer formation in high pheromone, starvation, or high temperature conditions.
MUT-16 functions upstream or parallel to DAF-7 TGF-β and DAF-2 insulin receptor for dauer formation
To address the question of how endogenous RNAi pathways are required for dauer formation, we performed epistasis experiments using RNAi, TGF-β, and insulin signaling mutants. Prior studies have ordered genes in the TGF-β and insulin signaling dauer formation pathways through epistasis analysis using daf-d and daf-c phenotypes (Riddle et al. 1981; Vowels and Thomas 1992; Thomas et al. 1993; Gottlieb and Ruvkun 1994; Malone and Thomas 1994; Larsen et al. 1995); thus, we used a similar approach to identify the possible points of interaction between dauer formation and endogenous RNAi pathways. We should note here that MUT-16-dependent siRNAs antisense to the coding regions of genes in the insulin signaling and TGF-β dauer formation pathways (including daf-2, daf-7, daf-3, daf-16, and daf-12) have been detected (Zhang et al. 2011), suggesting the possibility that RNAi pathways are regulating dauer formation pathways directly. Only daf-2 has also been identified as a CSR-1 target, although its mRNA levels do not significantly change in csr-1 mutants compared to wild-type (Claycomb et al. 2009; Cecere et al. 2014). Thus, the potential points of interaction between RNAi pathways and dauer formation pathway genes downstream of the sensory detection of adverse conditions, and how that could impact the dauer formation decision, remains unclear.
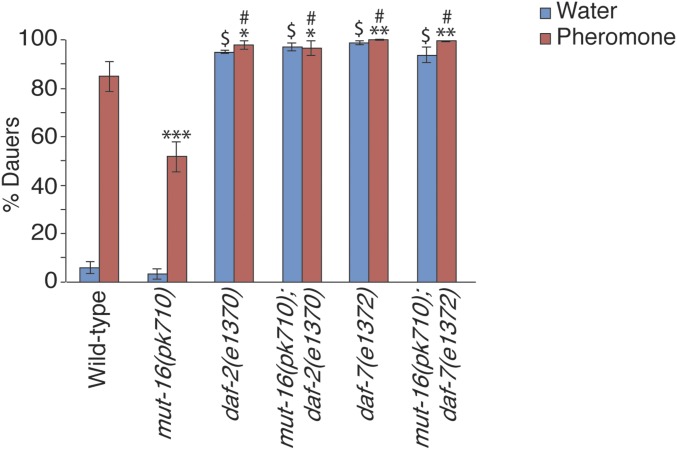
To perform epistasis analysis, we genetically crossed the mut-16 (daf-d) mutant with daf-7 (daf-c) and daf-2 (daf-c) mutants in the TGF-β and insulin signaling pathways, respectively, and tested the resulting mut-16(pk710); daf-2(e1370) and mut-16(pk710); daf-7(e1372) double mutants for dauer formation phenotypes in high pheromone conditions. We observed that both double mutant strains exhibited percentages of dauer formation (96.6 ± 3.0% and 99.3 ± 0.4%) significantly greater than mut-16(pk710) alone (51.7 ± 6.2%), and similar to the individual daf-2(e1370) (97.6 ± 1.7%) and daf-7(e1372) (100.0 ± 0%) mutant strains (Figure 2). Since MUT-16-amplified siRNAs target daf-2 and daf-7, and the e1370 allele of daf-2 is a hypomorph (Gems et al. 1998), we cannot conclusively interpret these results without additional experiments that are beyond the scope of this work. However, these results suggest that MUT-16 is not functioning downstream of DAF-2 and DAF-7 for dauer formation. Since DAF-2 insulin receptor and DAF-7 TGF-β function early in the dauer formation decision (Fielenbach and Antebi 2008), this observation is consistent with the possibility that Mutators and CSR-1 AGO pathway are required in neurons for dauer formation in adverse environmental conditions.
Figure 2.
MUT-16 does not function downstream of DAF-7 TGF-β and DAF-2 insulin receptor. Epistasis analysis was performed using dauer formation assays in high pheromone conditions with mut-16(pk710), daf-2(e1370), daf-7(e1372), mut-16(pk710); daf-2(e1370), and mut-16(pk710); daf-7(e1372) strains. * P < 0.05, ** P < 0.005, *** P < 0.0005 compared to wild-type on pheromone plates. # P < 0.0005 compared to mut-16(pk710) on pheromone plates. $ P < 0.0005 compared to wild-type on water plates, one-way ANOVA with LSD post hoc test. N ≥ 3 trials; n ≥ 274 animals. Error bars represent SEM.
Endogenous RNAi pathways are required in neurons for dauer formation
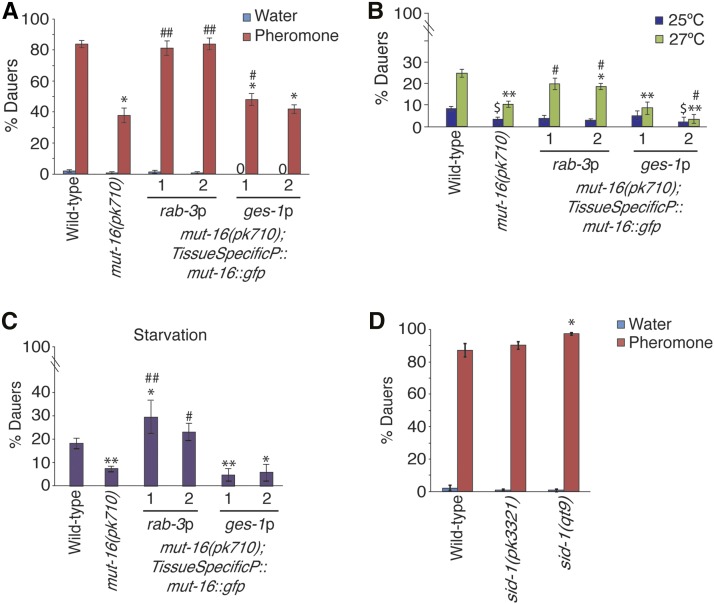
Next, we sought to determine the site of action of RNAi pathways for dauer formation. Based on our results thus far, we hypothesize that Mutators and the CSR-1 pathway are functioning in sensory neurons for dauer formation, since the detection of environmental stresses and the resulting differential regulation of TGF-β and insulin signaling pathways occurs in neurons (Golden and Riddle 1982, 1984a; Ren et al. 1996; Schackwitz et al. 1996; Li et al. 2003). To test our hypothesis, we constructed strains expressing a mut-16::gfp translational fusion driven by a pan-neuronal promoter (rab-3) in the mut-16(pk710) background. We also constructed a mut-16::gfp rescue transgene that is driven by an intestinal promoter (ges-1), since a previous study found that DAF-2 functions in the intestine to regulate dauer formation (Hung et al. 2014). Since mut-16(pk710) exhibited the most significant daf-d phenotype in high pheromone conditions, we first subjected the mut-16 tissue-specific rescue strains to dauer formation assays using high pheromone conditions. As predicted, two independent transgenic lines carrying the rab-3p::mut-16::gfp transgene exhibited levels of dauer formation significantly greater than mut-16(pk710) and similar to wild-type, completely rescuing the daf-d phenotype of mut-16(pk710) (Figure 3A). In contrast, only one transgenic line expressing ges-1p::mut-16::gfp showed a slight but significant increase in dauer formation compared to mut-16(pk710), but remained significantly lower than wild-type (Figure 3A). These results indicate that MUT-16 functions primarily in neurons for dauer formation in high pheromone conditions.
Figure 3.
MUT-16 is required in neurons for dauer formation. (A) Proportion of animals forming dauers in response to high pheromone and water plates is shown for pan-neuronal (rab-3p) and intestinal (ges-1p) mut-16 rescue strains. * P < 0.0005 compared to wild-type on pheromone plates; # P < 0.05 and ## P < 0.0005 compared to mut-16(pk710), respectively, on pheromone plates; one-way ANOVA with LSD post hoc test. N ≥ 3 trials; n ≥ 213 animals. (B) Proportion of animals forming dauers in response to 25 and 27° is shown for pan-neuronal and intestinal mut-16 rescue strains. * P < 0.05, ** P < 0.0005 compared to wild-type at 27°; # P < 0.005 compared to mut-16(pk710) at 27°; $ P < 0.05 compared to wild-type at 25°; one-way ANOVA with LSD post hoc test. N = 3 trials; n ≥ 119 animals. (C) Proportion of animals forming dauers in response to starvation conditions for pan-neuronal and intestinal mut-16 rescue strains. * P < 0.05 and ** P < 0.005 compared to wild-type; # P < 0.005 and ## P < 0.0005 compared to mut-16(pk710); one-way ANOVA with LSD post hoc test. N = 3 trials; n ≥ 196 animals. (D) Proportion of sid-1 animals forming dauers in response to pheromone. * P < 0.05, one-way ANOVA with LSD post hoc test. N = 3 trials; n ≥ 480 animals. All error bars represent SEM.
Next, we subjected the mut-16::gfp tissue-specific rescues to high temperature and starvation dauer formation assays. The pan-neuronal mut-16::gfp strains exhibited complete, or almost complete, rescue of the mut-16(pk710) daf-d phenotype to wild-type levels of dauer formation in high temperature (27°) conditions (Figure 3B). Similarly, the pan-neuronal mut-16 rescue transgenes completely restored dauer formation in starvation conditions (Figure 3C). In contrast, the intestinal mut-16 rescue transgenes failed to increase dauer formation levels compared to mut-16(pk710), for both high temperature and starvation conditions (Figure 3, B and C). These results indicate that MUT-16 functions in neurons for dauer formation regardless of experienced environmental stress.
We next asked whether small RNAs generated in neurons are acting cell autonomously, or spreading to other tissue types to affect dauer formation. C. elegans animals experience systemic RNAi, a process by which dsRNA can spread throughout cells and tissue types to regulate gene expression (Fire et al. 1998; Tabara et al. 1998; Timmons and Fire 1998; Timmons et al. 2001; Winston et al. 2002). Although neurons are resistant to systemic RNAi, they can experience autonomous RNAi, and can export dsRNA to other tissue types in the worm (Tavernarakis et al. 2000; Timmons et al. 2001; Kamath et al. 2003; Calixto et al. 2010; Devanapally et al. 2015). Systemic RNAi is dependent upon the dsRNA uptake channel, SID-1, which is expressed throughout the worm in non-neuronal cells (Winston et al. 2002; Feinberg and Hunter 2003; Jose et al. 2009). Thus, to examine if siRNAs are acting cell autonomously in neurons to regulate dauer formation, we performed dauer formation assays using high pheromone concentrations with the sid-1(pk3321) and sid-1(qt9) mutant strains. The pk3321 allele of sid-1 resulted in wild-type levels of dauer formation, while the qt9 allele exhibited a slight but significant increase in dauer formation compared to wild-type (Figure 3D), indicating that systemic RNAi is not required for dauer formation. Together, these results indicate that endogenous RNAi pathways are required in neurons for dauer formation in response to high pheromone and starvation conditions, and, at least in part, for response to high temperature conditions.
RNAi is required in distinct subsets of neurons for dauer formation in different environmental stresses
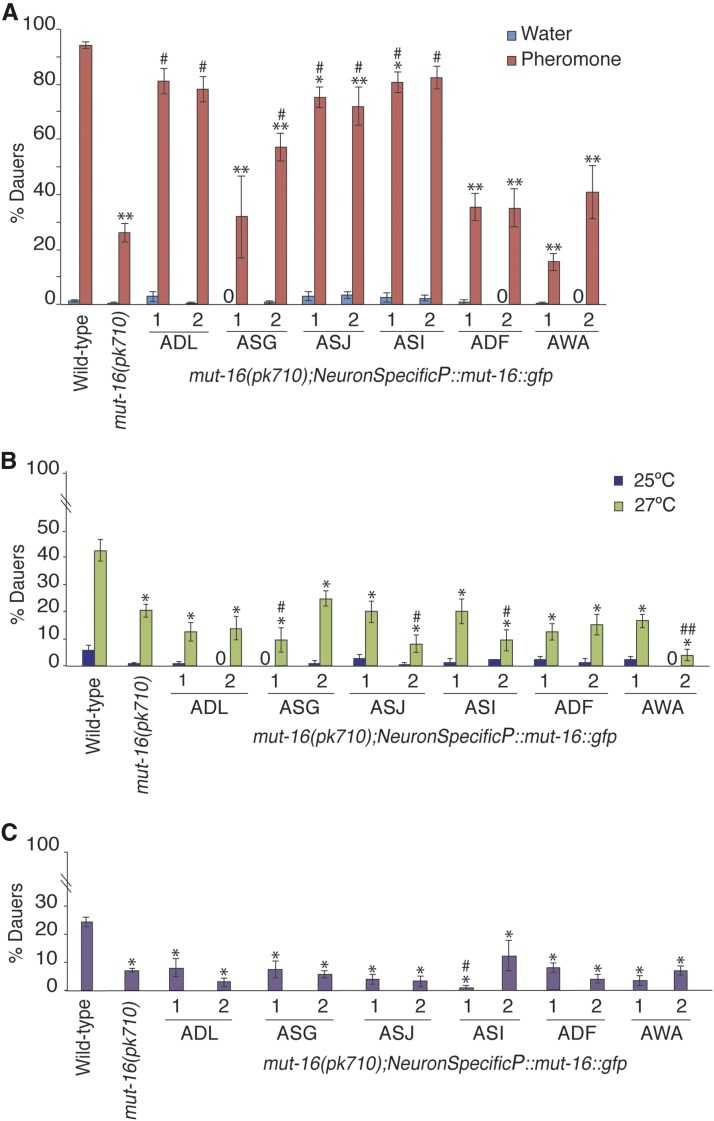
To further characterize the site of action for the RNAi pathways in the regulation of dauer formation, we examined whether MUT-16 function is required in specific sensory neurons for high pheromone, starvation, and high temperature conditions. Previous work using laser ablation has shown that a subset of amphid sensory neurons (ASI, ADF, and ASG) prevent dauer formation, while others (including ASJ, ASK, and ADL) promote dauer entry (Bargmann and Horvitz 1991; Schackwitz et al. 1996). To determine if MUT-16 functions in all, or subsets, of amphid neurons for dauer formation, we generated additional worm strains that express the mut-16::gfp transgene driven by ASJ, ASI, ADF, or ASG specific promoters, and subjected them to dauer formation assays using high pheromone conditions (Figure 4A). We found that expression of mut-16::gfp in ASI and ASJ neurons in the mut-16(pk710) strain resulted in significantly greater number of dauers compared to mut-16(pk710) without the transgene. We also observed a partial rescue of the mut-16 daf-d phenotype for one transgenic line expressing mut-16::gfp in ASG neurons, and no rescue for expression of mut-16::gfp in ADF neurons (Figure 4A). Although ASJ, ASI, ADF, and ASG neurons have previously been implicated in dauer formation (Bargman and Horvitz 1991), these results indicate that expression of MUT-16 in either ASI or ASJ neurons is sufficient to rescue the daf-d phenotype of mut-16(pk710) in high pheromone conditions. Work by others has shown that ASI and ASJ neurons respond to specific pheromone components upstream of DAF-7 TGF-β signaling (Schackwitz et al. 1996; McGrath et al. 2011; Park et al. 2012; Neal et al. 2015). Since our epistasis results are consistent with the possibility that RNAi pathway components function upstream of dauer formation pathways, we reasoned that rescue of the daf-d phenotypes in high pheromone conditions by expression of MUT-16 in ASI and ASJ neurons may be due to restoration of their ability to sense and/or signal the presence of dauer-inducing pheromone levels. To test this hypothesis, we first verified that these neurons exhibited normal development in mut-16(pk710) strains compared to wild-type based on their ability to uptake a lypophilic, florescent dye (see Materials and Methods and Figure S1). Next, we constructed strains that expressed MUT-16 in the ADL neurons, as they have been shown to detect the pheromone component ascr#3 in adults, and the AWA neuron as a negative control, since it has no known role in regulating dauer formation or sensing pheromone components in hermaphrodites (Srinivasan et al. 2008; Jang et al. 2012). As predicted, we observed that expression of mut-16::gfp in ADL neurons also resulted in a significant rescue of the mut-16 daf-d phenotype, while expression in AWA neurons resulted in no rescue (Figure 4A). Based on these results, we would also predict that ASK-specific expression of mut-16 would rescue the mut-16 daf-d phenotype, since detection of high concentrations of ascr#3 by ASK also promotes dauer formation (Kim et al. 2009). ASI, ASJ, and ADL neurons respond to ascr#2 and ascr#3 pheromone components (McGrath et al. 2011; Jang et al. 2012; Park et al. 2012; Neal et al. 2015), which are the two most abundant and potent components of crude dauer pheromone (Butcher et al. 2007, 2008). Our results are consistent with a model that functional RNAi in just one of these pheromone-sensing neurons is sufficient for detection and/or signaling of high pheromone conditions to result in dauer formation.
Figure 4.
MUT-16 is required in different subsets of neurons for dauer formation in response to different environmental stresses. (A) Proportion of animals forming dauers in response to high pheromone and water control is shown for wild-type, mut-16(pk710), and neuron-specific rescue strains. N ≥ 3 trials; n ≥ 173 animals. * P < 0.05, ** P < 0.0005 compared to wild-type; # P < 0.0005 compared to mut-16(pk710); one-way ANOVA with Tukey’s HSD post hoc test. (B) Proportion of animals forming dauers in response to 25 and 27° is shown for neuron-specific mut-16 rescue strains. * P < 0.0005 compared to wild-type at 27°; # P < 0.005 and ## P < 0.0005 compared to mut-16(pk710) at 27°; one-way ANOVA with LSD post hoc test. N ≥ 3; n ≥ 120 at 27° and N ≥ 3; n ≥ 146 at 25°. (C) Proportion of animals forming dauers in response to starvation conditions for neuron-specific mut-16 rescue strains. * P < 0.0005 compared to wild-type; # P < 0.05 compared to mut-16(pk710) using one-way ANOVA with LSD post hoc test. N ≥ 3; n ≥ 160. No dauer formation is indicated by “0.” All error bars represent SEM. Two independent transgenic lines for each neuron-specific rescue were tested. The dye-filling experiment to verify normal structure of the amphid neurons in mut-16(pk710) is shown in Figure S1.
Since our current model predicts that endogenous RNAi pathways are regulating genes functioning at the sensory detection and/or signaling level of dauer-inducing environmental conditions, we hypothesized that MUT-16 would be required in different neurons for dauer formation in starvation and high temperature compared to high pheromone conditions. The neuronal and molecular mechanisms regulating dauer formation in response to these environmental stresses are not as well characterized as for high pheromone conditions (Hu 2007). Recent work has shown that feeding state is encoded by the calcium/calmodulin-dependent protein kinase I (CMKI) in the AWC and ASI sensory neurons to modulate dauer formation (Neal et al. 2015). In addition, AFD neurons have been shown to play a major role in thermotactic behaviors, which are modulated by the AWC chemosensory neurons (Garrity et al. 2010); however, their role in the regulation of dauer formation in high temperatures, if any, is unknown. To address whether MUT-16 is required in ASI, ASJ, or ADL neurons for dauer formation in general or specifically in high pheromone conditions, we tested whether the neuron-specific MUT-16 rescue strains exhibited increased dauer formation in high temperature or starvation conditions. At 27°, all of the neuron specific rescues tested formed percentage dauers that were not significantly greater than mut-16(pk710) levels (Figure 4B). Surprisingly, we observed that some of the neuron-specific MUT-16 rescues exhibited a further decrease in the percentage of dauer formation at 27°. Similarly, we observed no rescue of mut-16(pk710) daf-d phenotype in starvation conditions for the neuron-specific MUT-16 expression strains (Figure 4C), and a significant decrease in dauer formation in one line of ASI-specific MUT-16 expression. These results indicate that MUT-16 is not sufficient in ASJ, ASI, ADF, ASG, ADL, or AWA alone to regulate dauer formation in high temperature or starvation conditions, but may be required in single neurons that remain untested or in subsets of these neurons (Figure 4, B and C). These findings are consistent with a model that the Mutator and CSR-1 pathways are required in specific subsets of neurons for dauer formation in different environmental stresses.
CSR-1 promotes expression of G proteins required for dauer formation
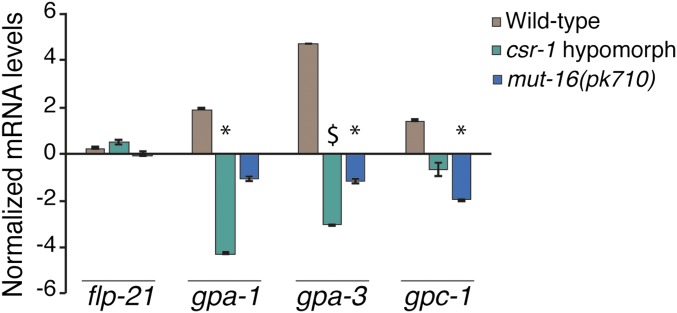
Our results thus far predict that Mutator-amplified siRNAs regulate the expression of genes that play a role in the detection and signaling of high pheromone concentrations via the CSR-1 pathway. In C. elegans neurons, G protein coupled receptors (GPCRs) span the plasma membrane of cilia, and function to detect specific environmental stimuli, including individual pheromone components. When bound to a ligand, the cytoplasmic portion of GPCRs activate their associated G proteins, which function to transduce the sensory signal intracellularly (Koelle 2016). In order to understand the potential molecular mechanism by which endogenous RNAi is required for pheromone sensation, we sought to identify candidate genes involved in sensory signaling that are expressed in ASI, ASJ, and ADL neurons, and exhibit a daf-d phenotype when mutated. The CSR-1 pathway has been shown to positively regulate the transcription of endogenous genes (Avgousti et al. 2012; Conine et al. 2013) through interactions with RNA polymerase II (Cecere et al. 2014), and maintenance of a euchromatic chromatin state at target gene loci (Claycomb et al. 2009; Seth et al. 2013; Wedeles et al. 2013). Thus, we hypothesize that gene expression of CSR-1 targets required for dauer formation would be downregulated in the mut-16 and csr-1 hypomorph strains, resulting in their observed daf-d phenotype. We identified three genes encoding G protein subunits, gpa-1, gpa-3, and gpc-1, that are expressed in ASI, ASJ, and ADL and exhibit a daf-d phenotype in their respective loss-of-function mutant strains in high pheromone concentrations (Zwaal et al. 1997; Lans and Jansen 2007; Kim et al. 2009; Hobert et al. 2016). To examine if Mutator proteins and CSR-1 are playing a role in the regulation of these genes, we measured their mRNA levels in larval L1 stage animals in wild-type, csr-1 hypomorph, and mut-16(pk710) strains cultivated in favorable conditions. We found that expression levels of each of these genes were significantly reduced in either the csr-1 hypomorph or mut-16(pk710) strains, or both, compared to wild-type, indicating that endogenous RNAi promotes expression of these G proteins during the L1 larval stage (Figure 5). In contrast, the expression of the neuropeptide flp-21 gene, which is also expressed in ASI, ASJ, and ADL neurons, and exhibits no known daf-d phenotype when mutated (Li and Kim 2008; Hobert et al. 2016), did not exhibit significant changes in mRNA levels in either mut-16(pk710) or csr-1 hypomorph strains (Figure 5). These results are consistent with our hypothesis that the daf-d phenotype exhibited by the csr-1 hypomorph and mut-16(pk710) strains in high pheromone conditions are a result of decreased G protein signaling in neurons that detect high concentrations of ascaroside molecules. Together, our results indicate that endogenous RNAi pathways are required for the basal level of expression of gpa-1, gpa-3, and gpc-1 genes essential for sensory signaling early in C. elegans development. The reduced expression of these genes in mut-16(pk710) and csr-1 hypomorph animals would result in decreased ability to detect and respond appropriately to environmental cues, and likely contributes to their inability to form dauers in adverse conditions.
Figure 5.
Mutator MUT-16 and CSR-1 AGO promote expression of chemosensation genes required for dauer formation. Log2 mRNA levels for flp-21, gpa-1, gpa-3, and gpc-1 genes in wild-type, csr-1 hypomorph, and mut-16(pk710) strains, normalized to y45f10d.4, are shown. * P < 0.05 and $ P < 0.005 compared to wild-type, one-way ANOVA with LSD post hoc test. N ≥ 3 biological replicates. Error bars represent propagated SD.
Discussion
In this study, we implicate endogenous RNAi as playing a role in the dauer formation behavior in C. elegans. We showed that strains carrying mutations in Mutator proteins and the nuclear CSR-1 AGO exhibit a daf-d phenotype in adverse environmental conditions. In addition, we provide evidence that MUT-16 is required in distinct subsets of neurons for dauer formation in high pheromone concentrations, high temperature, or starvation conditions. In high pheromone conditions, our results indicate that ASI, ASJ, and ADL neurons act as a distributed circuit for promoting dauer formation in high pheromone conditions upstream of the TGF-β and insulin signaling pathways. Furthermore, we provide evidence that suggests that CSR-1 positively regulates expression of a subset of genes that are essential for sensory signaling in neurons that have been shown to detect individual ascarosides. Together, our results suggest a model where Mutator-amplified siRNAs acting in the CSR-1 pathway promote expression of genes that are required for the detection and signaling of different environmental stresses in individual sensory neurons, which then signal to promote dauer formation.
RNAi is required in neurons for behavioral adaptation to environment
Since the publication in 1998 reporting that dsRNA has the ability to silence endogenous genes, much progress has been made defining the genetic pathways and gene targets of RNAi in C. elegans (Fire et al. 1998; Youngman and Claycomb 2014). Although at least half of the protein-coding genes in the C. elegans genome are targets of an RNAi pathway (Gu et al. 2009), we know little about how these pathways regulate endogenous genes, particularly in neurons. Previous work from our laboratory and others has shown that Mutator proteins and the nuclear AGO NRDE-3 play a role in regulating neuronal genes in response to environmental and developmental history. For example, stable downregulation of the guanylyl cyclase gene odr-1 in AWC neurons is required for experience-dependent olfactory adaptation to the odorant butanone, and is dependent upon MUT-7 and NRDE-3-mediated heterochromatin formation at the odr-1 locus (Juang et al. 2013). Similarly, our recent work has shown that downregulation of the osm-9 TRPV channel gene in ADL neurons of animals that passed through the dauer stage is dependent upon a majority of the Mutator proteins and NRDE-3 AGO, resulting in altered responses to ascr#3 in adults (Sims et al. 2016).
While the previous examples illustrate how RNAi can silence neuronal genes in response to environmental history, our results presented here indicate that MUT-16 and CSR-1 AGO are required in L1 larvae, regardless of their history, to promote transcription of genes with functions in sensory signaling, including gpa-1, gpa-3, and gpc-1. These genes are expressed in ASI, ASJ, and ADL sensory neurons, among others, and loss-of-function mutations in these genes result in defects in dauer formation in response to pheromone (Zwaal et al. 1997; Jansen et al. 1999; Lans and Jansen 2007; Kim et al. 2009). Since we have shown that expression of MUT-16 in individual ASI, ASJ, or ADL neurons can rescue the daf-d phenotype of the mut-16(pk710) mutants in high pheromone conditions, we propose a model where Mutator-amplified siRNAs associated with CSR-1 promote expression of sensory signaling genes in each of these neurons, which detect high pheromone stress, such as high levels of ascr#5 by ASI neurons (McGrath et al. 2011). Detection of high levels of a single ascaroside or ascaroside blends by a single neuron would be sufficient to induce dauer formation, as observed when individual ascarosides added to media can induce dauer formation in wild-type animals (Butcher et al. 2007). However, since we have not shown that the ASI, ASJ, and ADL neurons have pheromone-specific defects in mut-16 and csr-1 hypomorph strains, the possibility exists that these neurons have an overall reduced function in response to other stimuli as well.
Furthermore, we have shown that pan-neuronal expression of MUT-16 also rescues the daf-d phenotype of mut-16(pk710) in starvation and high temperature conditions. We have not yet identified the specific neurons where MUT-16 is required for dauer formation during exposure to these stresses; however, our model predicts that it would be in the neurons that detect low food availability or high temperature. Although the chemical identity of the “food signal” is yet to be identified, the presence of food (E. coli) is integrated by CAMKII in ASI and non-cell autonomously in AWC (Neal et al. 2015). However, unlike the pheromone conditions, we have shown that MUT-16 expression in ASI neurons is not sufficient to rescue the daf-d phenotype of mut-16(pk710) in starvation conditions. Furthermore, the AFD and AWC neurons have been shown to respond to temperature (Garrity et al. 2010), which we did not test in our mut-16 rescue assays. Further experiments are necessary to determine how MUT-16 function is required in low food conditions and high temperatures for dauer formation.
A distributed chemosensory circuit promotes dauer formation in high pheromone concentrations
Our finding that expression of MUT-16 in a subset of individual sensory neurons rescued the daf-d phenotype of mut-16(pk710) suggests that a distributed chemosensory circuit functions to detect and promote dauer formation in high pheromone concentrations. Distributed neural circuits have been described previously in C. elegans as a mechanism for aggregation in response to low oxygen and pheromones exhibited by npr-1(lf) strains (Chang et al. 2006; Macosko et al. 2009). In addition, detection of chemical attractants at different concentrations, such as benzaldehyde and isoamyl alcohol, is distributed among a similar subset of chemosensory neurons (Yoshida et al. 2012; Leinwand et al. 2015). The observation that dauer formation in high pheromone conditions is distributed among multiple neurons would be expected, given that GPCRs that detect individual ascaroside components are expressed in different neurons: SRBC-64 and SRBC-66 in ASK, and SRG-36, SRG-37, and DAF-37 in ASI (Kim et al. 2009; McGrath et al. 2011; Park et al. 2012). However, what was unexpected was that we rescued mut-16(pk710) dauer formation defects by expression of MUT-16 in ADL alone, which has not previously been implicated in pheromone detection during early larval stages. ADL has been shown to contribute to dauer formation in response to noxious chemicals, and exhibited a minor contribution to dauer formation when ablated with ASJ, suggesting that it might have a modulatory function for promoting dauer formation (Schackwitz et al. 1996; Neal et al. 2016). Alternatively, ADL may detect unidentified molecules in crude pheromone that are sufficient to promote dauer formation. This result is consistent with the observation that expression of SRBC-64 and SRBC-66 in ASK alone can rescue the dauer formation defects of their respective null mutants, and contribute to the detection of ascr#1–3 (Kim et al. 2009). Based on our results, we speculate that detection of high pheromone concentrations by ADL, ASI, and ASJ results in downstream modulation of TGF-β and insulin signaling pathways to promote dauer formation. Further experimentation is required to test if expression of MUT-16 in ASK also rescues the mut-16(pk710) daf-d phenotypes as predicted by our model.
Furthermore, we propose that a distributed chemosensory circuit to detect the presence of dauer-inducing pheromone concentrations provides an adaptive advantage to C. elegans animals. The concentrations of the major ascaroside molecules isolated from C. elegans populations cultivated at 20° differs from populations cultivated at 25°, indicating that pheromone composition of crude dauer pheromone is dependent upon environmental conditions (Jeong et al. 2005; Butcher et al. 2007, 2008). Thus, the ability of C. elegans larva to detect a high concentration of any one ascaroside component in early development allows for phenotypic plasticity in a crowded area, regardless of other environmental conditions such as temperature.
In this study, we provide evidence that RNAi pathways function early in the dauer formation decision in high pheromone concentrations, by promoting expression of G proteins in neurons that are essential for the detection of individual ascaroside components. Although dauer-inducing environmental conditions are detected by a complex network of chemosensory neurons found in the head of worms, little is known about the concurrent integration of these signals, and whether the dauer formation triggers may have distinct effects at a molecular level. We have shown previously that postdauer adults that experienced high pheromone conditions are phenotypically distinct from adults that bypassed the dauer stage, and that these effects are partially dependent upon RNAi pathways (Hall et al. 2010, 2013). Our data presented here suggest the intriguing possibility that RNAi pathways may mediate distinct effects on gene expression in postdauer animals that experienced different environmental stresses. Since other animals, such as pea aphids, form polyphenisms due to crowding or starvation as well, we predict that this work may have broader implications for the RNAi-mediated regulation of phenotypic plasticity.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.195438/-/DC1.
Acknowledgments
We thank Maria Ow for use of the ADL- and ASI-specific mut-16 transgenes, construction of the double mutant strain, and for proofreading the manuscript. We also thank Scott Neal, Michael O’Donnell, and Kyuhyung Kim for helpful advice and critical reading of the manuscript. Some strains were provided by the Caenorhabditis Genetics Center (CGC), which is funded by National Institutes of Health (NIH) Office of Research Infrastructure Programs (P40 OD010440).
Footnotes
Communicating editor: O. Hobert
Literature Cited
- Ailion M., Thomas J. H., 2000. Dauer formation induced by high temperatures in Caenorhabditis elegans. Genetics 156(3): 1047–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert P. S., Riddle D. L., 1988. Mutants of Caenorhabditis elegans that form dauer-like larvae. Dev. Biol. 126(2): 270–293. [DOI] [PubMed] [Google Scholar]
- Ambros V., Lee R. C., Lavanway A., Williams P. T., Jewell D., 2003. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr. Biol. 13(10): 807–818. [DOI] [PubMed] [Google Scholar]
- Antebi A., Yeh W. H., Tait D., Hedgecock E. M., Riddle D. L., 2000. daf-12 encodes a nuclear receptor that regulates the dauer diapause and developmental age in C. elegans. Genes Dev. 14(12): 1512–1527. [PMC free article] [PubMed] [Google Scholar]
- Aoki K., Moriguchi H., Yoshioka T., Okawa K., Tabara H., 2007. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J. 26(24): 5007–5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avgousti D. C., Palani S., Sherman Y., Grishok A., 2012. CSR‐1 RNAi pathway positively regulates histone expression in C. elegans. EMBO J. 31(19): 3821–3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann C. I., Horvitz H. R., 1991. Control of larval development by chemosensory neurons in Caenorhabditis elegans. Science 251(4998): 1243–1246. [DOI] [PubMed] [Google Scholar]
- Bernstein E., Caudy A. A., Hammond S. M., Hannon G. J., 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409(6818): 363–366. [DOI] [PubMed] [Google Scholar]
- Bonasio R., 2012. Emerging topics in epigenetics: ants, brains, and noncoding RNAs. Ann. N. Y. Acad. Sci. 1260(1): 14–23. [DOI] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77(1): 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer M. C., Dawidowicz P., Dodson S. I., 1999. Interactive effects of fish kairomone and light on Daphnia escape behavior. J. Plankton Res. 21(7): 1317–1335. [Google Scholar]
- Brisson J. A., 2010. Aphid wing dimorphisms: linking environmental and genetic control of trait variation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365(1540): 605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisson J. A., Davis G. K., Stern D. L., 2007. Common genome‐wide patterns of transcript accumulation underlying the wing polyphenism and polymorphism in the pea aphid (Acyrthosiphon pisum). Evol. Dev. 9(4): 338–346. [DOI] [PubMed] [Google Scholar]
- Brisson J. A., Ishikawa A., Miura T., 2010. Wing development genes of the pea aphid and differential gene expression between winged and unwinged morphs. Insect Mol. Biol. 19(s2): 63–73. [DOI] [PubMed] [Google Scholar]
- Butcher R. A., Fujita M., Schroeder F. C., Clardy J., 2007. Small-molecule pheromones that control dauer development in Caenorhabditis elegans. Nat. Chem. Biol. 3(7): 420–422. [DOI] [PubMed] [Google Scholar]
- Butcher R. A., Ragains J. R., Kim E., Clardy J., 2008. A potent dauer pheromone component in Caenorhabditis elegans that acts synergistically with other components. Proc. Natl. Acad. Sci. USA 105(38): 14288–14292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calixto A., Chelur D., Topalidou I., Chen X., Chalfie M., 2010. Enhanced neuronal RNAi in C. elegans using SID-1. Nat. Methods 7(7): 554–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassada R. C., Russell R. L., 1975. Dauerlarva, a post-embryonic developmental variant of nematode Caenorhabditis elegans. Dev. Biol. 46(2): 326–342. [DOI] [PubMed] [Google Scholar]
- Cecere G., Hoersch S., O’Keeffe S., Sachidanandam R., Grishok A., 2014. Global effects of the CSR-1 RNA interference pathway on the transcriptional landscape. Nat. Struct. Mol. Biol. 21(4): 358–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A. J., Chronis N., Karow D. S., Marletta M. A., Bargmann C. I., 2006. A distributed chemosensory circuit for oxygen preference in C. elegans. PLoS Biol. 4(9): e274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claycomb J. M., Batista P. J., Pang K. M., Gu W., Vasale J. J., et al. , 2009. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell 139(1): 123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conine C. C., Moresco J. J., Gu W., Shirayama M., Conte D., et al. , 2013. Argonautes promote male fertility and provide a paternal memory of germline gene expression in C. elegans. Cell 155(7): 1532–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornils A., Gloeck M., Chen Z., Zhang Y., Alcedo J., 2011. Specific insulin-like peptides encode sensory information to regulate distinct developmental processes. Development 138(6): 1183–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devanapally S., Ravikumar S., Jose A. M., 2015. Double-stranded RNA made in C. elegans neurons can enter the germline and cause transgenerational gene silencing. Proc. Natl. Acad. Sci. USA 112(7): 2133–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan C. R., Chung M. A., Allen F. L., Heschl M. F. P., Van Buskirk C. L., et al. , 1995. A gut-to-pharynx/tail switch in embryonic expression of the Caenorhabditis elegans ges-1 gene centers on two GATA sequences. Dev. Biol. 170(2): 397–419. [DOI] [PubMed] [Google Scholar]
- Feinberg E. H., Hunter C. P., 2003. Transport of dsRNA into cells by the transmembrane protein SID-1. Science 301(5639): 1545–1547. [DOI] [PubMed] [Google Scholar]
- Fielenbach N., Antebi A., 2008. C. elegans dauer formation and the molecular basis of plasticity. Genes Dev. 22(16): 2149–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A., Xu S., Montgomery M. K., Kostas S. A., Driver S. E., et al. , 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391(6669): 806–811. [DOI] [PubMed] [Google Scholar]
- Frézal L., Félix M.-A., 2015. The natural history of model organisms: C. elegans outside the Petri dish. eLife 4: e05849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrity P. A., Goodman M. B., Samuel A. D., Sengupta P., 2010. Running hot and cold: behavioral strategies, neural circuits, and the molecular machinery for thermotaxis in C. elegans and Drosophila. Genes Dev. 24(21): 2365–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gems D., Sutton A. J., Sundermeyer M. L., Albert P. S., King K. V., et al. , 1998. Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans. Genetics 150(1): 129–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent J. I., Lamm A. T., Pavelec D. M., Maniar J. M., Parameswaran P., et al. , 2010. Distinct phases of siRNA synthesis in an endogenous RNAi pathway in C. elegans soma. Mol. Cell 37(5): 679–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden J. W., Riddle D. L., 1982. A pheromone influences larval development in the nematode Caenorhabditis elegans. Science 218: 578–580. [DOI] [PubMed] [Google Scholar]
- Golden J. W., Riddle D. L., 1984a A pheromone-induced developmental switch in Caenorhabditis elegans: temperature-sensitive mutants reveal a wild-type temperature-dependent process. Proc. Natl. Acad. Sci. USA 81(3): 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden J. W., Riddle D. L., 1984b A Caenorhabditis elegans dauer-inducing pheromone and an antagonistic component of the food supply. J. Chem. Ecol. 10(8): 1265–1280. [DOI] [PubMed] [Google Scholar]
- Golden J. W., Riddle D. L., 1984c The Caenorhabditis elegans dauer larva: developmental effects of pheromone, food, and temperature. Dev. Biol. 102(2): 368–378. [DOI] [PubMed] [Google Scholar]
- Gottlieb S., Ruvkun G., 1994. daf-2, daf-16 and daf-23: genetically interacting genes controlling dauer formation in Caenorhabditis elegans. Genetics 137(1): 107–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A., Pasquinelli A. E., Conte D., Li N., Parrish S., et al. , 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106(1): 23–34. [DOI] [PubMed] [Google Scholar]
- Gu W., Shirayama M., Conte D., Vasale J., Batista P. J., et al. , 2009. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol. Cell 36(2): 231–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall S. E., Beverly M., Russ C., Nusbaum C., Sengupta P., 2010. A cellular memory of developmental history generates phenotypic diversity in C. elegans. Curr. Biol. 20(2): 149–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall S. E., Chirn G.-W., Lau N. C., Sengupta P., 2013. RNAi pathways contribute to developmental history-dependent phenotypic plasticity in C. elegans. RNA 19(3): 306–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T., Manoharan A. P., Harkins T. T., Bouffard P., Fitzpatrick C., et al. , 2009. 26G endo-siRNAs regulate spermatogenic and zygotic gene expression in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 106(44): 18674–18679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O., 2002. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques 32: 728–730. [DOI] [PubMed] [Google Scholar]
- Hobert O., Glenwinkel L., While J., 2016. Revisiting neuronal cell type classification in Caenorhabditis elegans. Curr. Biol. 26(22): R1197–R1203. [DOI] [PubMed] [Google Scholar]
- Hu, P. J., 2007 Dauer, WormBook, ed. The C. elegans Research Community WormBook, /10.1895/wormbook.1.144.1, http://www.wormbook.org.
- Humann F. C., Tiberio G. J., Hartfelder K., 2013. Sequence and expression characteristics of long noncoding RNAs in honey bee caste development–potential novel regulators for transgressive ovary size. PLoS One 8(10): e78915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung W. L., Wang Y., Zhen M., 2014. A Caenorhabditis elegans developmental decision requires insulin signaling-mediated neuron-intestine communication. Development 141(8): 1767–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt B. G., Brisson J. A., Soojin V. Y., Goodisman M. A., 2010. Functional conservation of DNA methylation in the pea aphid and the honeybee. Genome Biol. Evol. 2: 719–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H., Kim K., Neal S. J., Macosko E., Kim D., et al. , 2012. Neuromodulatory state and sex specify alternative behaviors through antagonistic synaptic pathways in C. elegans. Neuron 75(4): 585–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen G., Thijssen K. L., Werner P., Hazendonk E., Plasterk R. H., 1999. The complete family of genes encoding G proteins of Caenorhabditis elegans. Nat. Genet. 21(4): 414–419. [DOI] [PubMed] [Google Scholar]
- Jeong P. Y., Jung M., Yim Y. H., Kim H., Park M., et al. , 2005. Chemical structure and biological activity of the Caenorhabditis elegans dauer-inducing pheromone. Nature 433(7025): 541–545. [DOI] [PubMed] [Google Scholar]
- Jose A. M., Smith J. J., Hunter C. P., 2009. Export of RNA silencing from C. elegans tissues does not require the RNA channel SID-1. Proc. Natl. Acad. Sci. USA 106(7): 2283–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juang B. T., Gu C., Starnes L., Palladino F., Goga A., et al. , 2013. Endogenous nuclear RNAi mediates behavioral adaptation to odor. Cell 154(5): 1010–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R. S., Fraser A. G., Dong Y., Poulin G., Durbin R., et al. , 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421(6920): 231–237. [DOI] [PubMed] [Google Scholar]
- Ketting R. F., Fischer S. E., Bernstein E., Sijen T., Hannon G. J., et al. , 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 15(20): 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Sato K., Shibuya M., Zeiger D. M., Butcher R. A., et al. , 2009. Two chemoreceptors mediate developmental effects of dauer pheromone in C. elegans. Science 326(5955): 994–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klass M., Hirsh D., 1976. Non-ageing developmental variant of Caenorhabditis elegans. Nature 260: 523–525. [DOI] [PubMed] [Google Scholar]
- Knight S. W., Bass B. L., 2001. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 293(5538): 2269–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koelle, M. R., 2016 Neurotransmitter signaling through heterotrimeric G proteins: insights from studies in C. elegans, WormBook, ed. The C. elegans Research Community WormBook, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Kronforst M. R., Gilley D. C., Strassmann J. E., Queller D. C., 2008. DNA methylation is widespread across social Hymenoptera. Curr. Biol. 18(7): R287–R288. [DOI] [PubMed] [Google Scholar]
- Kucharski R., Maleszka J., Foret S., Maleszka R., 2008. Nutritional control of reproductive status in honeybees via DNA methylation. Science 319(5871): 1827–1830. [DOI] [PubMed] [Google Scholar]
- Lanjuin A., VanHoven M. K., Bargmann C. I., Thompson J. K., Sengupta P., 2003. Otx/otd homeobox genes specify distinct sensory neuron identities in C. elegans. Dev. Cell 5(4): 621–633. [DOI] [PubMed] [Google Scholar]
- Lans H., Jansen G., 2007. Multiple sensory G proteins in the olfactory, gustatory and nociceptive neurons modulate longevity in Caenorhabditis elegans. Dev. Biol. 303(2): 474–482. [DOI] [PubMed] [Google Scholar]
- Larsen P. L., 1993. Aging and resistance to oxidative damage in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 90(19): 8905–8909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen P. L., Albert P. S., Riddle D. L., 1995. Genes that regulate both development and longevity in Caenorhabditis elegans. Genetics 139(4): 1567–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinwand S. G., Yang C. J., Bazopoulou D., Chronis N., Srinivasan J., et al. , 2015. Circuit mechanisms encoding odors and driving aging-associated behavioral declines in Caenorhabditis elegans. eLife 4: e10181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C., and K. Kim, 2008 Neuropeptides, WormBook, ed. The C. elegans Research Community WormBook, /10.1895/wormbook.1.142.1, http://www.wormbook.org.
- Li W., Kennedy S. G., Ruvkun G., 2003. daf-28 encodes a C. elegans insulin superfamily member that is regulated by environmental cues and acts in the DAF-2 signaling pathway. Genes Dev. 17(7): 844–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko E. Z., Pokala N., Feinberg E. H., Chalasani S. H., Butcher R. A., et al. , 2009. A hub-and-spoke circuit drives pheromone attraction and social behaviour in C. elegans. Nature 458(7242): 1171–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maine E. M., Hauth J., Ratliff T., Vought V. E., She X., et al. , 2005. EGO-1, a putative RNA-dependent RNA polymerase, is required for heterochromatin assembly on unpaired DNA during C. elegans meiosis. Curr. Biol. 15(21): 1972–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone E. A., Thomas J. H., 1994. A screen for nonconditional dauer-constitutive mutations in Caenorhabditis elegans. Genetics 136(3): 879–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr E., 1963. Animal Species and Evolution, Vol. 797 Belknap Press of Harvard University Press, Cambridge, MA. [Google Scholar]
- McGrath P. T., Xu Y., Ailion M., Garrison J. L., Butcher R. A., et al. , 2011. Parallel evolution of domesticated Caenorhabditis species targets pheromone receptor genes. Nature 477(7364): 321–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michener C. D., 1961. Social polymorphism in Hymenoptera. Insect Polymorphism 1: 115. [Google Scholar]
- Miranda-Vizuete A., González J. C. F., Gahmon G., Burghoorn J., Navas P., et al. , 2006. Lifespan decrease in a Caenorhabditis elegans mutant lacking TRX‐1, a thioredoxin expressed in ASJ sensory neurons. FEBS Lett. 580(2): 484–490. [DOI] [PubMed] [Google Scholar]
- Moczek A. P., Snell-Rood E. C., 2008. The basis of bee-ing different: the role of gene silencing in plasticity. Evol. Dev. 10(5): 511–513. [DOI] [PubMed] [Google Scholar]
- Müller C. B., Williams I. S., Hardie J., 2001. The role of nutrition, crowding and interspecific interactions in the development of winged aphids. Ecol. Entomol. 12: 330–340. [Google Scholar]
- Neal S. J., Kim K., Sengupta P., 2013. Quantitative assessment of pheromone-induced dauer formation in Caenorhabditis elegans. Methods Mol. Biol. 1068: 273–283. [DOI] [PubMed] [Google Scholar]
- Neal S. J., Takeishi A., O’Donnell M. P., Park J., Hong M., et al. , 2015. Feeding state-dependent regulation of developmental plasticity via CaMKI and neuroendocrine signaling. eLife 4: e10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal S. J., Park J., DiTirro D., Yoon J., Shibuya M., et al. , 2016. A forward genetic screen for molecules involved in pheromone-induced dauer formation in Caenorhabditis elegans. G3 (Bethesda) 6(5): 1475–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijhout H. F., 1998. Insect Hormones, Princeton University Press, Princeton, NJ. [Google Scholar]
- Nijhout H. F., 1999. Control mechanisms of polyphenic development in insects: in polyphenic development, environmental factors alter some aspects of development in an orderly and predictable way. Bioscience 49(3): 181–192. [Google Scholar]
- Nonet M. L., Staunton J. E., Kilgard M. P., Fergestad T., Hartweig E., et al. , 1997. Caenorhabditis elegans rab-3 mutant synapses exhibit impaired function and are partially depleted of vesicles. J. Neurosci. 17(21): 8061–8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak J., Fire A., 2007. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 315(5809): 241–244. [DOI] [PubMed] [Google Scholar]
- Pak J., Maniar J. M., Mello C. C., Fire A., 2012. Protection from feed-forward amplification in an amplified RNAi mechanism. Cell 151(4): 885–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D., O’Doherty I., Somvanshi R. K., Bethke A., Schroeder F. C., et al. , 2012. Interaction of structure-specific and promiscuous G-protein–coupled receptors mediates small-molecule signaling in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 109(25): 9917–9922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelec D. M., Lachowiec J., Duchaine T. F., Smith H. E., Kennedy S., 2009. Requirement for the ERI/DICER complex in endogenous RNA interference and sperm development in Caenorhabditis elegans. Genetics 183(4): 1283–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips C. M., Montgomery T. A., Breen P. C., Ruvkun G., 2012. MUT-16 promotes formation of perinuclear Mutator foci required for RNA silencing in the C. elegans germline. Genes Dev. 26(13): 1433–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popham J. D., Webster C. M., 1979. Aspects of the fine structure of the dauer larva of the nematode Caenorhabditis elegans. Can. J. Zool. 57(I): 794–800. [Google Scholar]
- Ren P., Lim C.-S., Johnsen R., Albert P. S., Pilgrim D., et al. , 1996. Control of C. elegans larval development by neuronal expression of a TGF-β homolog. Science 274(5291): 1389–1391. [DOI] [PubMed] [Google Scholar]
- Riddle, D. L., and P. S. Albert, 1997 Genetic and environmental regulation of dauer larva development, pp. 739–768 in C. Elegans II, Chapter 26, edited by D. L. Riddle, T. Blumenthal, B. J. Meyer, and J. R. Priess. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- Riddle D. L., Swanson M. M., Albert P. S., 1981. Interacting genes in nematode dauer larva formation. Nature 290: 668–671. [DOI] [PubMed] [Google Scholar]
- Sagasti A., Hobert O., Troemel E. R., Ruvkun G., Bargmann C. I., 1999. Alternative olfactory neuron fates are specified by the LIM homeobox gene lim-4. Genes Dev. 13(14): 1794–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarafi-Reinach T. R., Sengupta P., 2000. The forkhead domain gene unc-130 generates chemosensory neuron diversity in C. elegans. Genes Dev. 14(19): 2472–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schackwitz W. S., Inoue T., Thomas J. H., 1996. Chemosensory neurons function in parallel to mediate a pheromone response in C. elegans. Neuron 17(4): 719–728. [DOI] [PubMed] [Google Scholar]
- Sengupta P., Chou J. H., Bargmann C. I., 1996. odr-10 encodes a seven transmembrane domain olfactory receptor required for responses to the odorant diacetyl. Cell 84(6): 899–909. [DOI] [PubMed] [Google Scholar]
- Seth M., Shirayama M., Gu W., Ishidate T., Conte D., et al. , 2013. The C. elegans CSR-1 argonaute pathway counteracts epigenetic silencing to promote germline gene expression. Dev. Cell 27(6): 656–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaham, S., 2006 WormBook: methods in cell biology, WormBook, ed. The C. elegans Research Community WormBook, /10.1895/wormbook.1.49.1, http://www.wormbook.org.
- She X., Xu X., Fedotov A., Kelly W. G., Maine E. M., 2009. Regulation of heterochromatin assembly on unpaired chromosomes during Caenorhabditis elegans meiosis by components of a small RNA-mediated pathway. PLoS Genet. 5(8): e1000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer F., Tijsterman M., Parrish S., Koushika S. P., Nonet M. L., et al. , 2002. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr. Biol. 12(15): 1317–1319. [DOI] [PubMed] [Google Scholar]
- Sims J. R., Ow M. C., Nishiguchi M. A., Kim K., Sengupta P., et al. , 2016. Developmental programming modulates olfactory behavior in C. elegans via endogenous RNAi pathways. eLife 5: e11642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smardon A., Spoerke J. M., Stacey S. C., Klein M. E., Mackin N., et al. , 2000. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr. Biol. 10(4): 169–178. [DOI] [PubMed] [Google Scholar]
- Srinivasan J., Kaplan F., Ajredini R., Zachariah C., Alborn H. T., et al. , 2008. A blend of small molecules regulates both mating and development in Caenorhabditis elegans. Nature 454(7208): 1115–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle, T., 2006 Maintenance of C. elegans, WormBook, ed. The C. elegans Research Community WormBook, /10.1895/wormbook. 1.101. 1, http://www.wormbook.org.
- Sterling P., Eyer J., 1988. Allostasis: a new paradigm to explain arousal pathology, pp. 629–639 in Handbook of Life Stress, Cognition and Health, Wiley, New York. [Google Scholar]
- Tabara H., Grishok A., Mello C. C., 1998. RNAi in C. elegans: soaking in the genome sequence. Science 282(5388): 430–431. [DOI] [PubMed] [Google Scholar]
- Tavernarakis N., Wang S. L., Dorovkov M., Ryazanov A., Driscoll M., 2000. Heritable and inducible genetic interference by double-stranded RNA encoded by transgenes. Nat. Genet. 24(2): 180–183. [DOI] [PubMed] [Google Scholar]
- Thomas J. H., Birnby D. A., Vowels J. J., 1993. Evidence for parallel processing of sensory information controlling dauer formation in Caenorhabditis elegans. Genetics 134(4): 1105–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L., Fire A., 1998. Specific interference by ingested dsRNA. Nature 395(6705): 854. [DOI] [PubMed] [Google Scholar]
- Timmons L., Court D. L., Fire A., 2001. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263(1–2): 103–112. [DOI] [PubMed] [Google Scholar]
- Troemel E. R., Chou J. H., Dwyer N. D., Colbert H. A., Bargmann C. I., 1995. Divergent seven transmembrane receptors are candidate chemosensory receptors in C. elegans. Cell 83(2): 207–218. [DOI] [PubMed] [Google Scholar]
- Vasale J. J., Gu W., Thivierge C., Batista P. J., Claycomb J. M., et al. , 2010. Sequential rounds of RNA-dependent RNA transcription drive endogenous small-RNA biogenesis in the ERGO-1/Argonaute pathway. Proc. Natl. Acad. Sci. USA 107(8): 3582–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw N. L., Fischer S. E., Robert V. J., Thijssen K. L., Fraser A. G., et al. , 2003. A genome-wide screen identifies 27 genes involved in transposon silencing in C. elegans. Curr. Biol. 13: 1311–1316. [DOI] [PubMed] [Google Scholar]
- Vought V. E., Ohmachi M., Lee M. H., Maine E. M., 2005. EGO-1, a putative RNA-directed RNA polymerase, promotes germline proliferation in parallel with GLP-1/notch signaling and regulates the spatial organization of nuclear pore complexes and germline P granules in Caenorhabditis elegans. Genetics 170(3): 1121–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vowels J. J., Thomas J. H., 1992. Genetic analysis of chemosensory control of dauer formation in Caenorhabditis elegans. Genetics 130(1): 105–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh T. K., Brisson J. A., Robertson H. M., Gordon K., Jaubert-Possamai S., et al. , 2010. A functional DNA methylation system in the pea aphid, Acyrthosiphon pisum. Insect Mol. Biol. 19(S2): 215–228. [DOI] [PubMed] [Google Scholar]
- Wang Y., Jorda M., Jones P. L., Maleszka R., Ling X., et al. , 2006. Functional CpG methylation system in a social insect. Science 314(5799): 645–647. [DOI] [PubMed] [Google Scholar]
- Wedeles C. J., Wu M. Z., Claycomb J. M., 2013. Protection of germline gene expression by the C. elegans Argonaute CSR-1. Dev. Cell 27(6): 664–671. [DOI] [PubMed] [Google Scholar]
- Wei Y., Chen S., Yang P., Ma Z., Kang L., 2009. Characterization and comparative profiling of the small RNA transcriptomes in two phases of locust. Genome Biol. 10(1): 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West-Eberhard M. J., 2003. Developmental Plasticity and Evolution. Oxford University Press, New York. [Google Scholar]
- Winston W. M., Molodowitch C., Hunter C. P., 2002. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science 295(5564): 2456–2459. [DOI] [PubMed] [Google Scholar]
- Woodward D. E., Murray J. D., 1993. On the effect of temperature-dependent sex determination on sex ratio and survivorship in Crocodilians. Proc. Biol. Sci. 252: 149–155. [Google Scholar]
- Yigit E., Batista P. J., Bei Y., Pang K. M., Chen C. C. G., et al. , 2006. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 127(4): 747–757. [DOI] [PubMed] [Google Scholar]
- Youngman E. M., Claycomb J. M., 2014. From early lessons to new frontiers: the worm as a treasure trove of small RNA biology. Front. Genet. 5: 416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K., Hirotsu T., Tagawa T., Oda S., Wakabayashi T., et al. , 2012. Odour concentration-dependent olfactory preference change in C. elegans. Nat. Commun. 3: 739. [DOI] [PubMed] [Google Scholar]
- Zhang C., Montgomery T. A., Gabel H. W., Fischer S. E., Phillips C. M., et al. , 2011. mut-16 and other mutator class genes modulate 22G and 26G siRNA pathways in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 108(4): 1201–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Noguez J. H., Zhou Y., Butcher R. A., 2013. Analysis of ascarosides from Caenorhabditis elegans using mass spectrometry and NMR spectroscopy. Methods Mol. Biol. 1068: 71–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaal R. R., Mendel J. E., Sternberg P. W., Plasterk R. H., 1997. Two neuronal G proteins are involved in chemosensation of the Caenorhabditis elegans Dauer-inducing pheromone. Genetics 145(3): 715–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The worm strains used in this study are listed in File S1 and are available on request. The primer sequences used for cloning and qPCR experiments are listed in File S3.