Butterfly wing patterns are a model system for studying the evolution and development of adaptive traits. Zhang et al. combine RNA-seq and CRISPR/Cas9...
Keywords: butterfly wing, pigmentation, melanin, CRISPR/Cas9 genome editing, RNA-seq, evo–devo
Abstract
Despite the variety, prominence, and adaptive significance of butterfly wing patterns, surprisingly little is known about the genetic basis of wing color diversity. Even though there is intense interest in wing pattern evolution and development, the technical challenge of genetically manipulating butterflies has slowed efforts to functionally characterize color pattern development genes. To identify candidate wing pigmentation genes, we used RNA sequencing to characterize transcription across multiple stages of butterfly wing development, and between different color pattern elements, in the painted lady butterfly Vanessa cardui. This allowed us to pinpoint genes specifically associated with red and black pigment patterns. To test the functions of a subset of genes associated with presumptive melanin pigmentation, we used clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 genome editing in four different butterfly genera. pale, Ddc, and yellow knockouts displayed reduction of melanin pigmentation, consistent with previous findings in other insects. Interestingly, however, yellow-d, ebony, and black knockouts revealed that these genes have localized effects on tuning the color of red, brown, and ochre pattern elements. These results point to previously undescribed mechanisms for modulating the color of specific wing pattern elements in butterflies, and provide an expanded portrait of the insect melanin pathway.
BUTTERFLIES are a canvas upon which evolution paints with many varied hues. Indeed, for these highly visual creatures, pigmentation is the primary medium used to communicate with other animals: whether to attract mates, to deter predators, or to escape detection. If we are to understand butterfly diversity, we must work to better understand the origin and diversification of pigments themselves. Here, we expand work on the genetic basis of butterfly pigmentation for several reasons. First, there is a large diversity of pigment types in butterflies, including both characterized and uncharacterized pigments, which provide an excellent model of how genetic biosynthetic mechanisms evolve in concert with physiology and morphology (Nijhout 1991). Second, several butterfly species are highly accessible laboratory models; they can be reared in large numbers, their wing tissues are large enough for functional genomics work, wings produce enough pigments for chemical characterization, and recently developed clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 genome editing methods now allow experimental validation of gene function (Li et al. 2015; Perry et al. 2016; Zhang and Reed 2016). Third, in many species, pigments play important adaptive functions and show clear phylogenetic patterns of gain and loss, thus providing a link between developmental genetics and evolutionary biology. Finally, ongoing work by multiple research groups continues to define genes and regulatory networks involved in wing pattern evolution (Beldade and Brakefield 2002; Kronforst and Papa 2015; Monteiro 2015; Wallbank et al. 2016), thus providing a foundation for work aimed at understanding the upstream processes that regulate pigmentation genes. In this study, we directly test the function of a suite of pigmentation genes in the painted lady butterfly, Vanessa cardui, the buckeye Junonia coenia, the squinting bush brown Bicyclus anynana, and the Asian swallowtail Papilio xuthus. All of these are popular model species that display a wide range of many pigment and pattern types and are valuable systems for studying the evolution and development of wing patterns.
This study has three specific goals: (1) to characterize candidate genes implicated in ommochrome and melanin pigmentation using comparative transcriptomics, (2) to test if melanin genes characterized in other contexts in other insects (Figure 1) also play a role in butterfly wing pigmentation, and (3) to functionally characterize previously unknown genes potentially involved in pigmentation. To achieve these goals, we employed a two-step process of transcript characterization followed by genome editing of candidate loci. Specifically, using RNA sequencing (RNA-seq) in V. cardui, we were able to identify both known and novel candidate wing pigmentation genes expressed during wing development. Using these sequences, we then employed CRISPR/Cas9 genome editing to induce targeted knockouts in eight selected candidate genes of the melanin synthesis pathway (MSP), which is associated with yellow, brown, and black pigment synthesis. Six of the eight targeted genes yielded wing pigmentation phenotypes across a total of four species. In addition to significantly improving our mechanistic understanding of butterfly wing pattern development, this work also expands our current knowledge of the insect melanin pathway. From a technical perspective, our work demonstrates how combining high-throughput transcript characterization with Cas9-mediated genome editing can be a powerful tool for discovering novel genes underlying morphological trait development in nontraditional model systems.
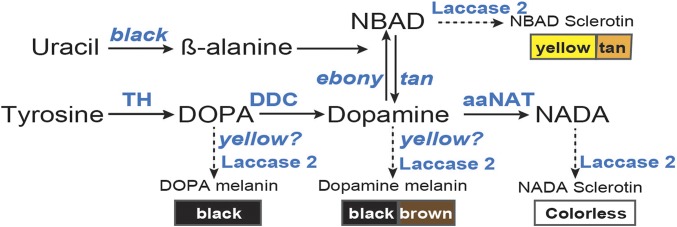
Figure 1.
Genetic model of the insect melanin biosynthesis pathway (Wright 1987). Tyrosine is the initial precursor for all insect melanins. Tyrosine hydroxylase (TH, encoded by pale) and DDC (encoded by Ddc) convert tyrosine to dihydroxyphenylalanine (DOPA) and dopamine (dihydroxyphenylethylamine), which are the precursors for black melanin synthesis, respectively. yellow is required for synthesizing DOPA and dopamine melanins, which are eumelanins. Laccase 2 is a phenoloxidase gene required for cuticular pigmentation. Yellowish–tan hues are produced by N-β-alanyl dopamine (NBAD) sclerotin, which requires the function of ebony, black, and tan. N-acetyl dopamine (NADA), which is catalyzed by aaNAT, is a major constituent of colorless or transparent cuticle.
Materials and Methods
Animals
V. cardui and J. coenia were maintained in a 16:8 hr light/dark cycle at 28°, P. xuthus were reared in a 14:10 light/dark cycle at 28°, while B. anynana were reared at 27° and 60% humidity in a 12:12 light/dark cycle. V. cardui butterflies originated from Carolina Biological Supplies, were fed on a multi-species artificial diet (Southland), and were induced to oviposit on leaves of Malva parviflora. J. coenia butterflies were derived from a laboratory colony maintained at Duke University (kind gift from F. Nijhout), were fed on a multi-species artificial diet (Southland) supplemented with dried leaves of the host plant Plantago lanceolata, and were induced to oviposit on leaves of P. lanceolata. P. xuthus were a line derived from wild-caught females from Kanagawa, Japan. Larvae were reared in the laboratory on fresh citrus leaves. B. anynana, originally collected in Malawi, have been reared in the lab since 1988. Larvae were fed on young corn plans and adults on mashed banana.
RNA isolation and library construction
Hindwings from five different stages of V. cardui were sampled and used in this study, including last instar larvae, 72 hr after pupation, 5 days after pupation (prepigment pupae), ommochrome development (∼5.5 days after pupation when red–orange ommochorome pigments started to be expressed), and melanin development (∼6 days after pupation when black melanin pigments began to show up). Wings were rapidly dissected in PBS and stored in RNAlater at −80° (Life Technologies). Red and black regions from the melanin stage forewings were further separated as two samples (Figure 2A). RNA was extracted from each sample using an Ambion Purelink RNA Mini Kit (Life Technologies). Due to the small size of the source tissue, each last instar larval wing disc sample consisted of hindwings taken from three individuals that were pooled together in equimolar amounts of RNA. For pupal wings, each sample represented RNA taken from a single individual. Two biological replicates were sampled for RNA-seq library construction from each developmental stage, resulting in 12 RNA-seq samples.
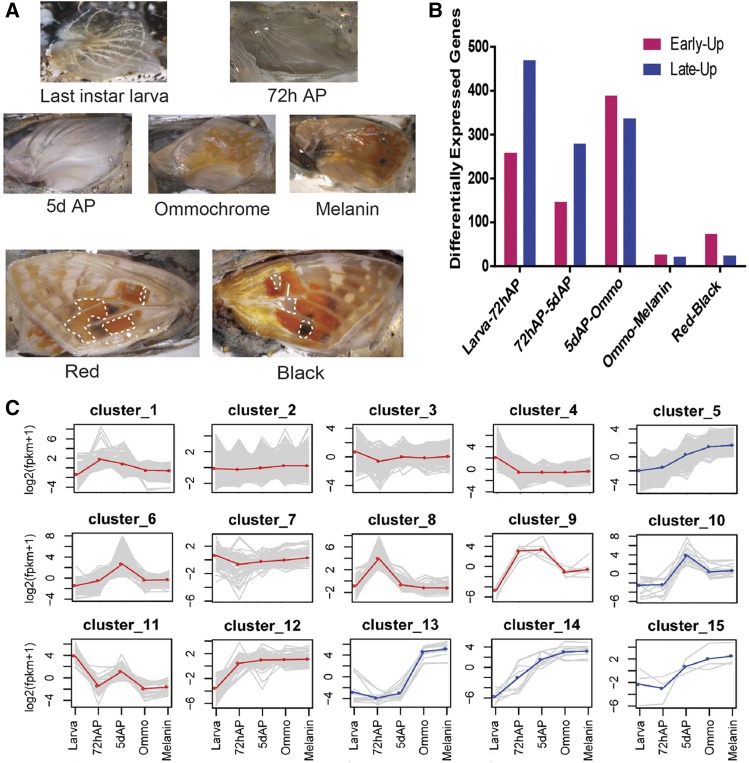
Figure 2.
RNA-seq analysis of the developing V. cardui butterfly wing. (A) The five developmental stages and two color regions sampled in this study (see Table S1 in File S1 for details). “Black” and “Red” tissues were dissected at the melanin stage in forewings from areas outlined by white dashed lines. (B) Number of differentially expressed unigenes in pairwise comparisons of consecutive stages (pink: upregulation in first, early stage; blue: upregulation in second, later stage). (C) Grouping of 13,875 unigenes into 15 clusters with similar expression profiles. Each gene is plotted (gray) in addition to the mean expression profile for that cluster (blue or red). Clusters with upregulated expression levels in later pigmentation stage pupae (5 days AP, Ommo, and Melanin) are marked in blue. AP, after pupation; RNA-seq, RNA sequencing.
Poly(A) RNA was isolated with Oligo d(T)25 beads (New England Biolabs, Beverly, MA) from 1 μg total RNA of each sample and fragmented to size ∼350–450 bp. First- and second-strand cDNA (complementary DNA) synthesis, end repair, 3′dA tailing, and adaptor ligation were performed using a NEBNext Ultra RNA library Prep kit for Illumina (New England Biolabs). All samples were tagged with a unique Illumina adapter index. The adapter-ligated sample was size-selected to ∼500-bp fragments by AMPure XP beads (Beckman, Fullerton, CA). After 15 PCR cycles, libraries were quantified, multiplexed, and sequenced on two Illumina HiSeq 2500 lanes with rapid run mode as 150 bp paired-end reads. Sequencing was performed at Cornell University’s Genomics Facility.
Analysis of transcript expression data
After removing PCR primers, adapters, and low-quality reads, data sets from 12 samples were merged and an in silico normalization was performed to remove high coverage reads. Trinity (Grabherr et al. 2011), which can take advantage of paired-end information, was used for assembly. The assembled sequences with putative isoforms identified by Trinity were filtered and only the longest isoform was kept. Cluster database at high identity with tolerance (CD-HIT) was used for further clustering and redundancy removal with minimum similarity cut-off of 95%. TransDecoder, a package included in the Trinity software, was used for the prediction of coding regions. Assembled contigs were searched in the NR, Swissprot, and FlyBase protein databases using BLASTX with a cutoff E value of 1e−5. Predicted proteins were further searched against InterProScan and Gene Ontology (GO) databases for domain and GO annotation. TBLASTN and BLASTP packages were used to search for homologies with pigmentation genes in Drosophila melanogaster as query sequences. Candidate genes were also aligned back to NR (protein) and NT (nucleotide) databases for additional annotation.
Filtered reads from each sample were mapped to the assembly using Bowtie (Langmead and Salzberg 2012). Gene expression levels for transcripts were measured by FPKM (fragments per kilobase of transcript per million mapped reads) and normalized as suggested (Chen et al. 2010). Differential expression analyses were performed for any pairwise comparison among five different developmental stages, last instar larva (Larva), 72 hr after pupation (72h AP), 5 days after pupation (5d AP), ommochrome (Ommo), and melanin developmental stages (Melanin) using edge R with biological replicates and a cutoff FDR (false discovery rate) of 0.001 (Robinson et al. 2010). Functional enrichment was further performed by identifying GO terms enriched in differentially expressed gene data sets (Chen et al. 2010). We next conducted K-means clustering analyses to identify coexpressed genes during wing development. Clusters represented by genes highly upregulated in 5d AP, Ommo, and Melanin stages were extracted for further manual annotation. Pigmentation candidate genes were first identified by screening unregulated genes during pupal pigment maturation and then further categorized into two groups based on sequence homology and expression patterns. For example, ommochrome-associated genes are upregulated genes during pupal pigmentation stages that are either orthologs of known ommochrome pathway genes or other ommochrome-rich color region-specific expressed genes.
Cas9-mediated genome editing
Our approach for producing G0 mosaic knockouts in V. cardui and J. coenia butterfly wings followed the protocol of Zhang and Reed (2016). In general, we designed sgRNAs (single-guide RNAs) following the GGN18NGG or N20NGG rule on the sense or antisense strand of the DNA. Two sgRNAs, aiming to induce long deletions, were used in V. cardui and P. xuthus for target genes including Ddc, black, yellow, ebony, yellow-d, yellow-h2, and yellow-h3, depending on the species, while single sgRNAs, aiming to induce small indels, were used for pale, yellow, and ebony in J. coenia and B. anynana (Supplemental Material, Table S5 in File S1). A sgRNA template was produced by PCR amplification with a forward primer encoding a T7 polymerase-binding site and a sgRNA target site, and a reverse primer encoding the remainder of the sgRNA sequence (Bassett et al. 2013; Bassett and Liu 2014). sgRNAs were in vitro-transcribed using the MegaScript or MegaShortScript T7 Kits (Ambion) and purified by phenol/chloroform extraction followed by isopropanol precipitation or using MEGAClear columns (Ambion). Recombinant Cas9 protein was purchased from PNABio (catalog number CP01). We mixed 1 μg of Cas9 protein and 375 ng of each sgRNA in a 5 μl volume prior to injection. V. cardui eggs were collected from Malva leaves for 3 hr, lined up on double-sided adhesive tape onto a microscope slide, and put into desiccant for 15 min. Slight variations in the method used across species can be found in Table S6 in File S1. Microinjection of butterfly embryos was conducted using a pulled borosilicate glass needle (Sutter Instrument, I.D.:0.5 mm) at an injection pressure of 20 psi. Treated embryos were then incubated in a chamber at 28° and 70% humidity for the remainder of development. To confirm that our sgRNAs produced mutations at the desired target sites, genomic DNA was extracted from single caterpillars or adult butterfly legs using proteinase K in digestion buffer (Bassett et al. 2013). Fragments flanking the Cas9 target regions were amplified by PCR using primers flanking the deletion (Table S6 in File S1). For the double sgRNA strategy, mutant PCR fragments were detected by agarose gel electrophoresis assay. For single sgRNA experiments, PCR fragments were detected with a T7 endonuclease I (T7E1) assay as previously described (Guschin et al. 2010).
In B. anynana, sgRNA target sequences were selected based on their GC content (∼60%) and number of mismatch sequences relative to other sequences in the genome (>3 sites). In addition, we picked target sequences that started with a guanidine for subsequent in vitro transcription by T7 RNA polymerase. sgRNA templates were produced by PCR amplification (Bassett et al. 2013; Bassett et al. 2014). In vitro transcription of sgRNAs was performed using T7 RNA polymerase (New England Biolabs) and purified by ethanol precipitation. We injected Cas9 protein (PNABio) or Cas9 messenger RNA (mRNA) as a source of CRISPR RNA-guided nuclease. We used the pT3TS-nCas9n vector (Addgene) for Cas9 mRNA in vitro synthesis. The template for Cas9 mRNA was generated by cutting the pT3TS-nCas9n vector with XbaI. Capped Cas9 mRNA was in vitro-transcribed using a mMESSAGE mMACHINE T3 Kit (Thermo Fisher), and a poly(A) tail was added using a Poly(A) Tailing Kit (Thermo Fisher). We co-injected 0.5 µg/µl final concentration of gRNA and 0.5 µg/µl final concentration of either Cas9 mRNA or protein into embryos within 1 hr after egg laying. Food dye was added to the injection solution for visualization. Injected embryos were incubated at 27° and 60% humidity. After hatching, larvae were moved to corn leaves, and reared at 27° with a 12:12 hr light/dark cycle and 60% relative humidity. To confirm that our sgRNAs produced mutations at the desired target sites, genomic DNA was extracted from a pool of ∼5 injected embryos that did not hatch via the SDS and Proteinase K method, and was used for T7 endonuclease I (New England Biolabs) assay and sequence analyses. Primers for these analyses are listed on Table S6 in File S1.
Microspectrophotometry
Scale reflectance of wild-type and CRISPR mutants was measured at two specific regions (arrowheads in Figure 7, A and C) of ventral forewings using an Ocean Optics USB 2000 spectrophotometer (Ocean Optics) attached to a compound light microscope.
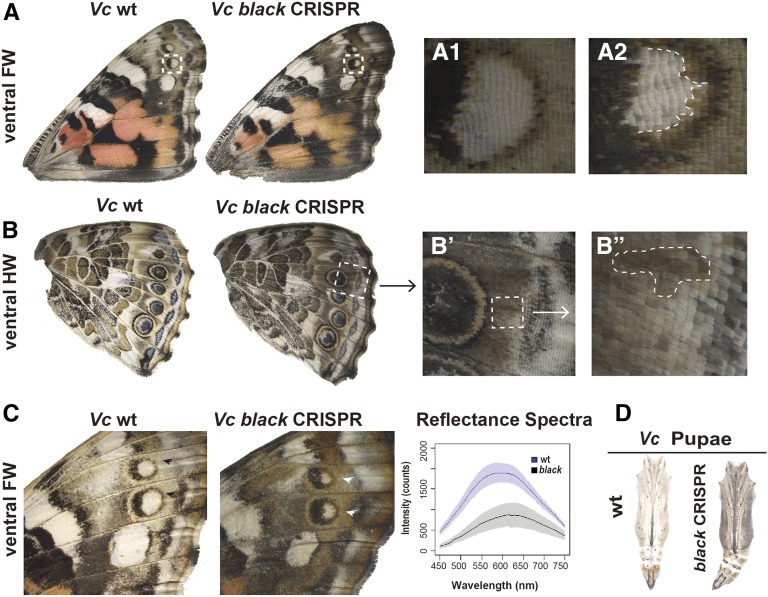
Figure 7.
Wing pattern-specific effects of black loss-of-function in Vc. (A) Ventral view of wt and black mutation wings in Vc shows enhanced melanization phenotypes. Darker yellow ring of eyespot (A2) can be observed compared to wt (A1). (B–B”) black knockout causes hypermelanization of a ventral HW in Vc. (C) black knockout in Vc results in local changes in pigment hue. Graphs of reflectance spectra of ventral FW (white arrowheads) show significant shift in coloration between wt and CRISPR mutants. (D) black knockout results in hyperpigmented pupae in Vc. CRISPR, clustered regularly interspaced short palindromic repeats; FW, forewing; HW, hindwing; Vc, V. cardui; wt, wild-type.
Imaging
Wing phenotypes were photographed using a Nikon DSLR camera equipped with a Micro-Nikkor 105 mm – F2/8 Macro Lens, a Keyence VHX-5000 digital microscope, or a Zeiss Axiocam 506 color camera with a Plan Apo S 1.0 × FWD 60 mm Macro Lens on a SteREO Discovery.V20 microscope.
Data availability
V. cardui wing RNA-seq raw reads are available as NCBI SRP062599. Full transcript assembly and expression profiles are available at the NCBI Gene Expression Omnibus (GEO) database with accession number GSE78119. The transcript assembly and unigenes are also available for searching at butterflygenome.org.
Results
Transcriptome analyses
We used RNA-seq to generate a comprehensive profile of transcript abundance during wing developmental stages (last instar larva, 72 hr early stage pupa, 5 days prepigment pupa, ommochrome, and melanin development papa) and from different color patterns as a first step to investigate the molecular basis of wing pattern development in V. cardui (Figure 2A and Table S1 in File S1). We first performed a de novo assembly based on a total of 174 million 150 bp paired-end Illumina HiSeq 2500 reads (GSE78119). The Trinity assembler generated 74,995 transcripts with longest ORF and a N50 length of 2062 bp. Consequently, 13,875 unigenes (i.e. predicted transcripts output from the assembly software), with a minimum length of 100 bp and a N50 length of 1626, were predicted. A search against Swissprot and FlyBase databases provided annotations for 13,064 and 13,636 unigenes, respectively, based on an E-value of 1e−5. InterProScan and GO analyses assigned 10,455 and 7938 unigenes, respectively, into functional domains or categories.
We next aimed to identify general transcription patterns during wing development. Overall, 2114 unigenes were found to be significantly differentially expressed (FDR < 0.001) by pairwise comparison among the five stages (Figure 2B). Of these, 1505 unigenes were upregulated by comparing consecutive stages, while 1106 unigenes were downregulated. A total of 80 and 29 unigenes were identified to be specifically upregulated in red and black color regions, respectively. Final instar larvae (Larvae)-vs.-72h AP comparison showed the highest number of upregulated genes (n = 673), perhaps reflecting major tissue state changes after metamorphosis. GO enrichment analyses of differential expression genes indicated dominant biological process during each representative developmental stage including larvae, early pupal, and pigmentation-stage pupal wings (Table S2 in File S1). Interestingly, the day 5 stage, characterized by early development of wing scales, was enriched for the GO terms “structural constituent of cuticle.” We also note that the GO category “transporter activity” was significantly enriched during the pigmentation phase. No functional term was enriched in comparison of ommochrome-stage and melanin-stage pupal wings (P > 0.01).
K-means clustering of coexpressed genes revealed the dynamic nature of expression patterns throughout wing development (Figure 2C and Table S3 in File S1). We divided expression series data into 15 clusters to identify genes with similar expression dynamics. Many sets of genes showed clear stage-specific expression patterns: clusters 3, 4, 7, and 11 showed the highest gene expression in last instar imaginal wing discs. Clusters 1, 8, and 9 showed highest expression at 72 hr early pupae, and clusters 5, 10, 12, 13, 14, and 15 showed highest expression in late pupal development, including at stages that show the visible deposition of pigments. These latter clusters include MSP genes such as pale, Ddc, tan, black, and ebony (Table S3 in File S1), consistent with previous results (Koch et al. 1998; Ferguson et al. 2011b; Hines et al. 2012; Daniels et al. 2014; Connahs et al. 2016). In clusters 3 and 13, many known ommochrome pigmentation genes including cinnabar and kynurenine formamidase showed upregulated expression patterns in both last instar larvae and pigmentation stages, consistent with previous results based on quantitative PCR in V. cardui (Reed and Nagy 2005). Overall, these clustering results show that sets of genes involved in similar types of pigmentation tend to be regulated in similar temporal patterns.
Identification of pigmentation candidate genes by RNA-seq
To identify genes potentially involved in wing pigment development, we screened transcript expression based on two criteria. First, we selected transcripts belonging to gene families previously implicated in pigmentation if they showed upregulation during pupal pigment maturation. Second, if a transcript was not related to a previously known pigmentation gene family, we selected it if it showed differential expression between red and black color regions (Figure 2A) and also during pupal pigment maturation.
Ommochrome-associated genes:
We identified 26 genes associated with presumptive ommochrome pigment patterns, following the criteria described above (Table S4 in File S1). All of the 26 genes were upregulated in late-stage red color patterns, and three were previously suspected to play a role in pigmentation. One of these, optix is a transcription factor that promotes red wing patterns in Heliconius butterflies (Reed et al. 2011; Martin et al. 2014). We also found high red-associated expression levels of the known ommochrome genes cinnabar and kynurenine formamidase. Many transcripts coding for poorly characterized transporters also showed red-associated expression patterns (Table S4 in File S1). Importantly, seven major facilitator superfamily (MFS) transporter transcripts (Lemieux 2007) showed significant upregulation during pigment development. Of these, four showed strong and specific associated expression patterns with red regions. One transcript coding for an ATP-binding cassette transporter C family member transcript also showed a strong red-associated expression pattern (Table S4 in File S1). Surprisingly, we also found two transcripts belonging to juvenile hormone-binding protein family genes (Hiruma et al. 1984) expressed in extremely strong association with ommochrome pigmentation. Another notable surprise was that the expression levels of the known D. melanogaster ommochrome transporters scarlet and white did not show differential expression patterns associated with pigmentation (Table S4 in File S1).
Melanin-associated genes:
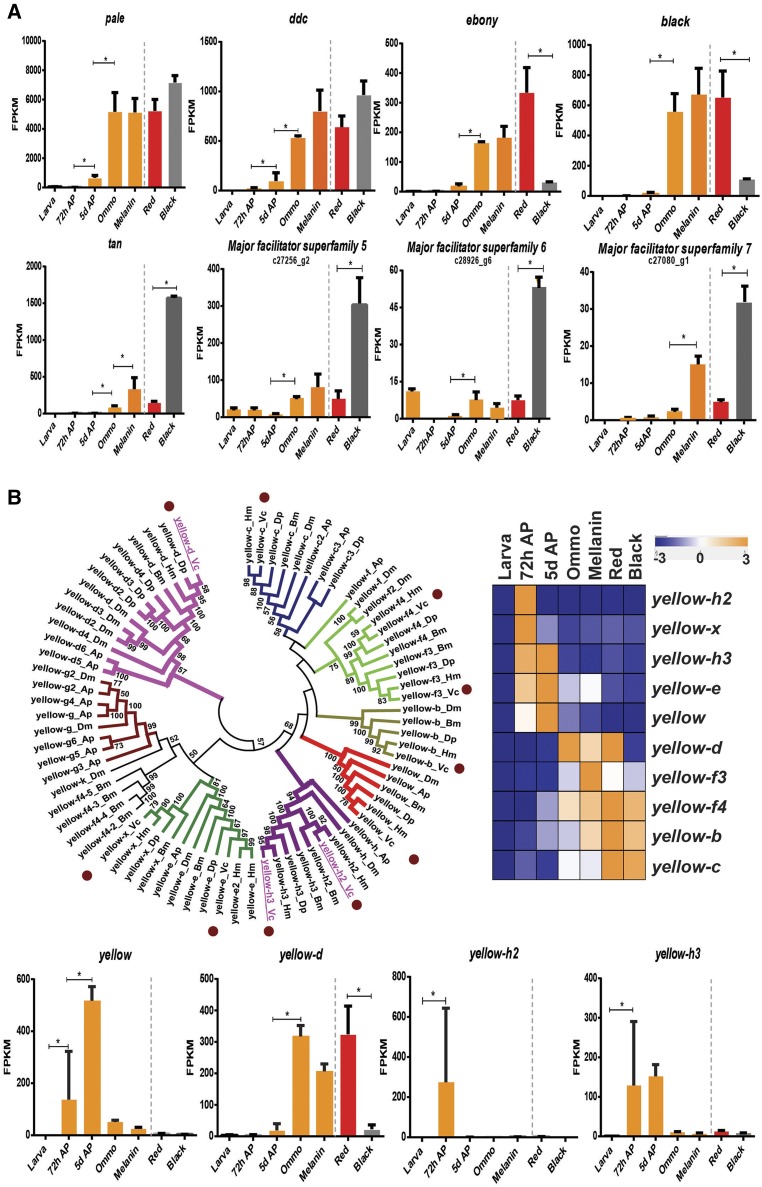
We identified 27 genes associated with presumptive melanin pigment patterns (Table S4 in File S1), including six previously recognized melanin synthesis genes, all of the members of the yellow gene family that have been previously characterized in other insects (Ferguson et al. 2011a), and six transporters. Of these, 15 showed color pattern-specific expression where transcripts were upregulated in black vs. red patterns late in pupal development. We observed particularly strong upregulation of the melanin-associated genes pale, Ddc, tan, ebony, and black during both ommochrome- and melanin-stage pupae, with expression peaking at the onset of melanin pigment deposition (Figure 3A and Table S4 in File S1). These results show that both ommochrome- and melanin-associated genes are activated at the ommochrome stage; however, melanin gene expression persists longer. Similar to our results from the ommochrome candidates, we also observed three uncharacterized MFS transporter transcripts that showed significant upregulation in black patterns.
Figure 3.
Expression patterns of major genes of the melanin biosynthetic pathway in V. cardui (Vc). (A) Gene expression levels of known, major genes of the melanin biosynthetic pathway. The selected genes are those significantly differentially expressed during pupal pigment maturation and between black and red color patterns. Expression levels at developmental stages are indicated in orange bars, red regions in red bars, and black regions in gray bars. Genes detected with significant upregulation expression by pairwise comparisons of consecutive stages are represented by an asterisk. (B) Phylogenetic analysis of insect yellow gene family members and their expression patterns in Vc. Neighbor joining tree of predicted yellow proteins from D. melanogaster (Dm), A. pisum (Ap), B. mori (Bm), H. melpomene (Hm), D. plexippus (Dp), and Vc. Putative orthologs in different species are denoted with the same color. Sequences from Vc are marked with brown circles. CRISPR deletion targets are underlined. The expression profiles of Vc yellow gene family members during different developmental stages as well as between red and black regions are indicated. AP, after pupation.
The yellow family of genes showed great diversity in expression profiles so we examined genes in greater depth. We identified 10 V. cardui yellow family gene members via phylogenetic comparisons of protein sequences with those in Heliconius melpomene, Danaus plexippus, B. mori, Acyrthosiphon pisum, and D. melanogaster (Figure 3B). All 10 genes showed upregulation during mid- or late-pupal development. Specifically, yellow, yellow-e, yellow-h2, yellow-h3, and yellow-x showed high expression levels in prepigmentation early pupae, while yellow-b, yellow-c, yellow-d, yellow-f4, and yellow-f3 showed higher expression during later pigment synthesis stages. Interestingly, yellow-d was observed with a significantly higher expression level in red regions (P < 0.001). These diverse expression patterns indicate that yellow gene family members might be involved in multiple, previously uncharacterized aspects of butterfly pigment development.
Functional validation of melanin pathway genes
We focused functional analyses on melanin biosynthetic pathway genes, in part because the functions of many of these genes are understood thanks to previous work in other insects (Figure 1) and thus provide clear predictions for potential phenotypes. We selected eight candidate melanin genes for functional assessment by CRISPR/Cas9 somatic mutagenesis: pale, Ddc, ebony, black, yellow, yellow-d, yellow-h2, and yellow-h3 (Table S5 in File S1). This represents a range of genes that would be predicted to have a general negative effect on melanization (pale, Ddc, and yellow), that should alter melanin level and/or type (ebony and black), or are of unknown function (yellow-d, yellow-h2, and yellow-h3). As described below, six out of eight gene loss-of-function experiments yielded clear G0 generation mosaic phenotypes, and PCR-amplified fragment length genotyping validated the efficacy of our approach for producing mutations at the targeted sites (Figure S1 and Tables S5 and S6 in File S1).
pale:
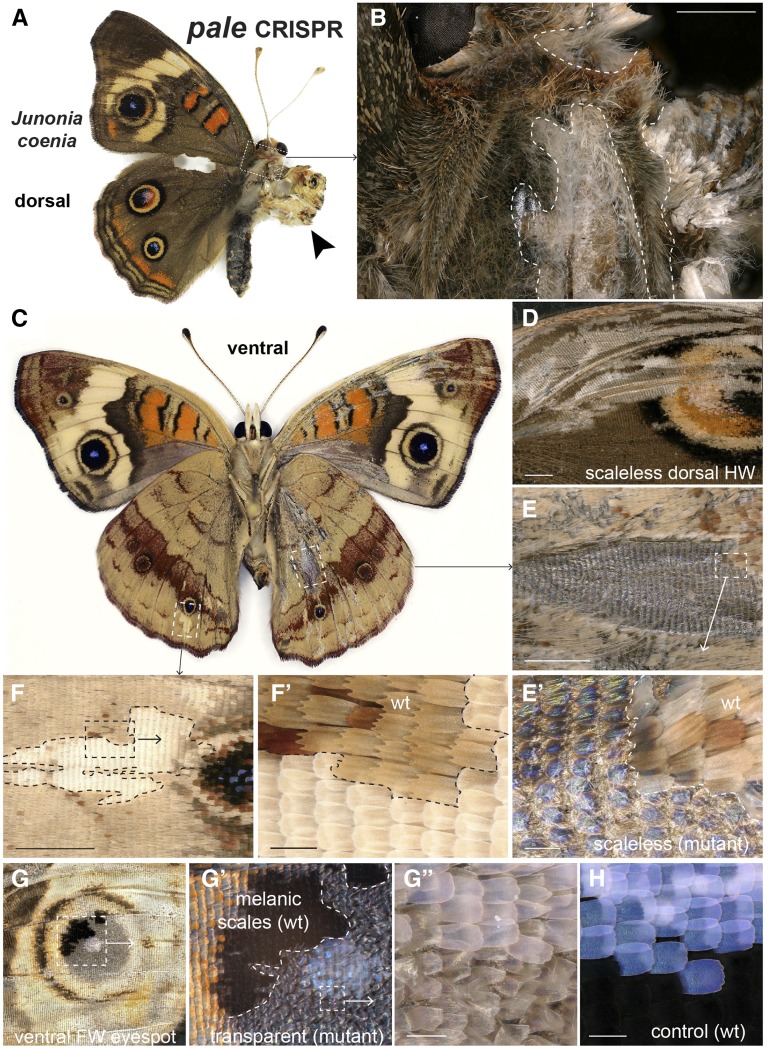
pale encodes the tyrosine hydroxylase enzyme, which catalyzes the formation of dihydroxyphenylalanine (DOPA) from tyrosine at the earliest metabolic step of the melanin synthesis cascade (Figure 1). We produced pale mutations in J. coenia using a single sgRNA. pale mutations resulted in severe cuticular defects, including improperly expanded wings and patches showing failed scale development on the thorax and wings (Figure 4, A–E). Tyrosine hydroxylase deficiency also triggered complete amelanism in scales that emerged properly, with effects varying depending on their ground state: in the thorax, mutant scales shifted from dark brown to white (Figure 4B); most wing mutant scales showed a lighter coloration and an altered morphology such as a rounded aspect, instead of a marked scalloping (Figure 4, F and F’); and finally, melanic scales of the eyespot inner rings became completely transparent, also taking a disheveled aspect due to an apparent structural defect or thickness reduction (Figure 4, F and F’). Interestingly, mutant eyespots also lost the characteristic blue color of their foci (central dots, Figure 4, G and H), and inspection at high magnification reveals that this is associated with a transformation of black ground scales into transparent scales. In other words, the loss of the blue hue is not due to a structural defect in blue scales; rather, the cover scales responsible for the blue coloration lack a black background in the mutant context that is necessary for blue reflectance by superimposition, an effect described in blue-iridescent Morpho butterflies (Giraldo and Stavenga 2016).
Figure 4.
Effects of pale somatic mutagenesis in J. coenia. (A and B) Mosaic pale mutant showing severe one-sided defects in sclerotization and pigmentation (dorsal view), including improper wing expansion (arrowhead) and missing or amelanic scales (dotted lines). (C–F’) Somatic pale mutant showing varied effects on scale morphology [(C) ventral view; (D) dorsal HW], including scaleless patches (D–E’) and depigmented round scales lacking scalloped ridges (F and F’). (G–G”) Mutant ventral FW eyespot showing the transformation to thin transparent scales. Residual black color in the mutant patch is due to the opposite wing surface. The white color of the mutant eyespot focus compared to the wt blue focus (H) is due to the transformation of black ground scales into transparent scales. Bars, 1000 μm (B and D–F); 100 μm (F’, E’, G”, and H). FW, forewing; HW, hindwing; wt, wild-type.
yellow:
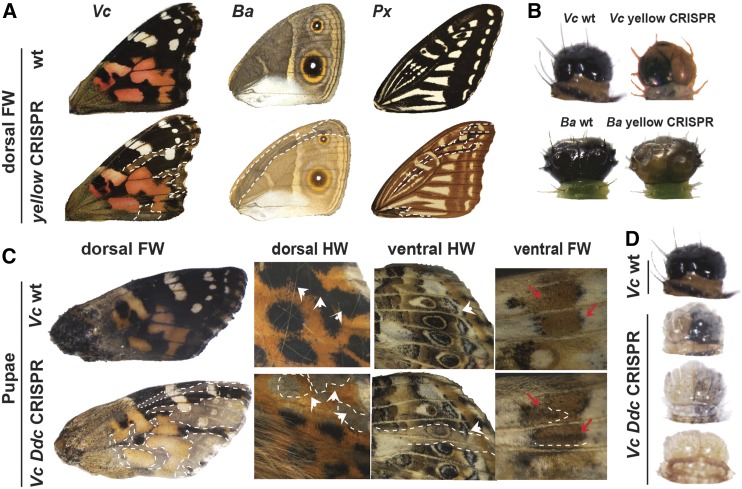
yellow encodes a secreted extracellular protein required for production of black melanin pigments in Drosophila (Biessmann 1985) (Figure 1). We produced yellow mutations in V. cardui, B. anynana, and P. xuthus using single or double sgRNAs. yellow knockout mosaics revealed strong reduction of presumptive melanins in both V. cardui and B. anynana wings, and in P. xuthus wings as previously reported (Perry et al. 2016) (Figure 5A). In B. anynana, overall brown regions became yellow and the black scales became brown. We also observed marked larval phenotypes in V. cardui and B. anynana, including larvae with depigmented head capsules (Figure 5B). Unlike for pale, yellow mutations appeared to have no obvious effect on red wing scales, thus suggesting a primary role for this gene in the synthesis of black melanin pigments, with little or no effect on scale cuticularization or nonmelanin pigment deposition.
Figure 5.
yellow and Ddc loss-of-function induces wing depigmentation mutants. (A) Dorsal view of FWs in wt and yellow mutation butterflies (left-right: Vc, Ba, and Px). Dotted lines: mosaic depigmentation phenotypes. (B) yellow knockout result in depigmentation of larvae head in Vc and Ba. (C) Examples of Ddc knockout Vc phenotypes in black region of dorsal FW, black eyespots, and parafocal elements of dorsal HW (white arrowhead), colored rings of ventral HW eyespot (white arrowhead), and gray strip of ventral FW (red arrow). (D) Different levels of depigmentation in mosaic larval heads of V. cardui. Ba, B. anynana; CRISPR, clustered regularly interspaced short palindromic repeats; FW, forewing; HW, hindwing; Px, P. xuthus; Vc, V. cardui; wt, wild-type.
Ddc:
Ddc encodes the melanin synthesis enzyme DOPA-decarboxylase (Figure 1). We produced Ddc mutations in V. cardui using double sgRNAs. As previously reported (Zhang and Reed 2016), Ddc mutations produced a phenotype similar to yellow with strong depigmentation of black scales and no effect on red scales (Figure 5, C and D). However, unlike for yellow, Ddc mutations also changed brown and tan pigments on the ventral surface (Figure 5C). Ddc knockout larvae were typically unable to break through their eggshells, perhaps due to an incomplete sclerotization of their mouthparts as seen in D. melanogaster mutants (Wright et al. 1976b). For this particular locus, it is important to note the caveat that knockouts resulted in such high mortality rates that we generated only two adults with wing phenotypes. However, we did observe a large number of larvae with reduced melanin phenotypes (Figure 5D), providing clear evidence of Ddc’s role in melanin synthesis.
ebony:
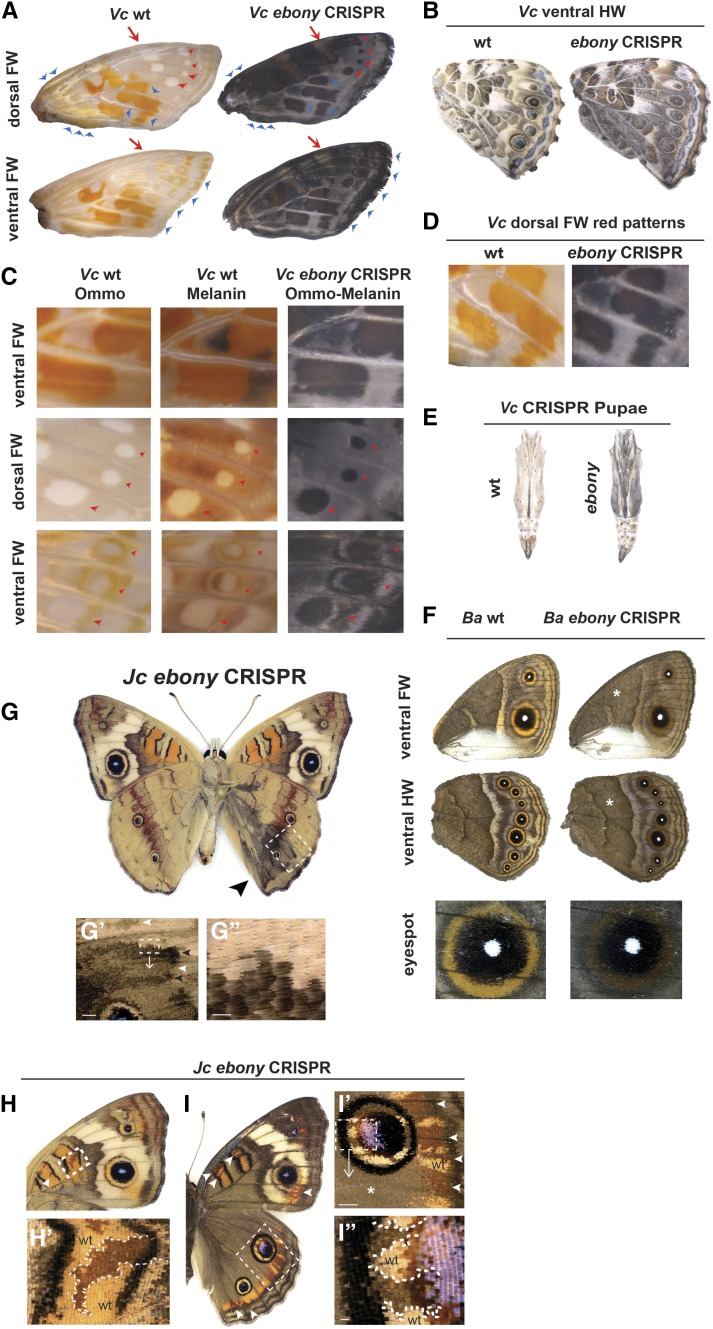
ebony encodes the N-β-alanyldopamine (NBAD) synthase enzyme, which promotes the formation of light-color NBAD pigment derivatives (Figure 1). We produced ebony mutations in V. cardui using double sgRNAs, and J. coenia and B. anynana with a single sgRNA. Generally, ebony knockout leads to strong darkening of wing phenotypes, with some interesting stage-dependent nuances (Figure 6, A–D). Late-stage pupal wings from V. cardui with ebony mutations showed marked, dramatic phenotypes where red (e.g., central band), white (e.g., eyespots), and brown (e.g., marginal band) color patterns turned black, producing an almost inverse coloration of the wild-type wing (Figure 6, A–D). We did not recover this phenotype in fully-emerged adult V. cardui, leading us to speculate these very strong phenotypes resulted in pupal mortality before normal melanin pigment development. In fact, ebony mutations resulted in many pupae being hyperpigmented (Figure 6E). This excessive melanization of internal tissues reflects ebony deficiency across large somatic clones, which would be predicted to have multiple deleterious effects on nervous system function, based on Drosophila knockout phenotypes (Pérez et al. 2010). However, we did recover adults with darker mutant clones in both B. anynana and V. cardui (Figure 6, B and F). In B. anynana, the brown background color (asterisk) was unaffected, but the white band regions and the yellow rings of the eyespots became brown (Figure 6F). We also recovered mosaic J. coenia adults that showed presumptive ebony mutant clones consisting of dark scale patches (Figure 6, G–G”). The strength was variable across a wing surface, probably reflecting different allelic dosages between clones (Figure 6G’). In all J. coenia (Figure 6, H and I), V. cardui (Figure 6D), and B. anynana (Figure 6F), these hypermelanic states affected red–orange (e.g., Basalis, Discalis I and II elements in J. coenia and central band in V. cardui) and buff/yellow scales (e.g., dorsal hindwing eyespot ring in J. coenia and ventral band and eyespot rings in B. anynana) that are traditionally considered to reflect an ommochrome composition (Nijhout and Koch 1991). Interestingly, we found no effect of ebony knockouts on the J. coenia dorsal brown background color field (asterisk, Figure 6I’) and B. anynana ventral brown background color field scales (asterisks, Figure 6F), in spite of predictable mutant clone extensions into these wing territories (Figure 6, F, I, and I’). Thus, it is possible that this gene is transcriptionally inactive in the corresponding scale cell precursors. In contrast, because mutant scales of the yellow eyespot rings of the dorsal surface in J. coenia show a similar color to the brown background (Figure 6I”), we speculate that ebony expression and NBAD synthesis are important for making this inner ring lighter than the dark color on the rest of this wing surface.
Figure 6.
ebony loss-of-function results in hyperpigmented wing phenotypes. (A) Dorsal and ventral view of FWs in wt and ebony knockout pupal wings in Vc. Obvious patches of mutant tissues can be observed (arrow and arrowhead). (B) Ventral view of HWs in wt and ebony knockout adults in Vc. (C) Detailed morphology of FWs reveals wing scales turn black at the initiation of ommochrome stage in ebony knockout Vc mutants. Arrowhead: obvious mutant tissues. (D) Red colors are disturbed in Vc ebony mutants. (E) Vc wt and ebony knockout pupae. (F) Ventral view of wt and ebony mutation adults in Ba. Darker scales were observed in the yellow ring of Ba eyespots in ebony knockout mutants. (G–G”) Mosaic hypermelanization of a ventral HW following ebony somatic knockout in Jc. Variable darkness of the mutant clones may reflect different allelic states [black and white arrowheads in (G’)]. Bars, 500 μm (G’) and 100 μm (G”). (H and H’) Ventral FW mutant clones showing darkening of orange patterns in Jc (arrowheads and dotted lines). (I–I”) Effects of ebony mutation on dorsal patterns in Jc, including the darkening of orange patterns (arrowheads) and of the eyespot inner ring (dotted lines). Mutant scales of the yellow ring resemble the brown background scales (asterisk), which are not affected by ebony mutation. Bars, 100 μm (I”) and 1000 μm (H’ and I’). Ba, B. anynana; CRISPR, clustered regularly interspaced short palindromic repeats; FW, forewing; HW, hindwing; Jc, J. coenia; Px, P. xuthus; Vc, V. cardui; wt, wild-type.
black:
black encodes a protein for synthesizing β-alanine, which is enzymatically conjugated to dopamine to form NBAD (Figure 1). We produced black mutations using double sgRNAs in V. cardui. We found that black knockout caused darkening of a subset of color fields on the ventral wings (Figure 7, A and B), especially the distal ventral forewing (Figure 7C). The hypermelanic state affects most of the brown and buff-colored pattern elements including a stripe between MI and EIII, marginal elements, forewing wing eyespot yellow rings (Figure 7, A1 and A2), hindwing basal, and hindwing central regions. To investigate the quantitative nature of the pigmentation difference, we measured reflectance spectra from 10 individual scales from the same regions of both wild-type and black knockout butterflies (Figure 7C). We found significant changes of reflectance spectra, with peak reflectance at 575–625 nm in wild-type and 600–650 nm in black mutants. black mutants also showed a dramatic decrease in light reflectance, consistent with an overall darkening of color. We also recovered hyperpigmented pupae (Figure 7D).
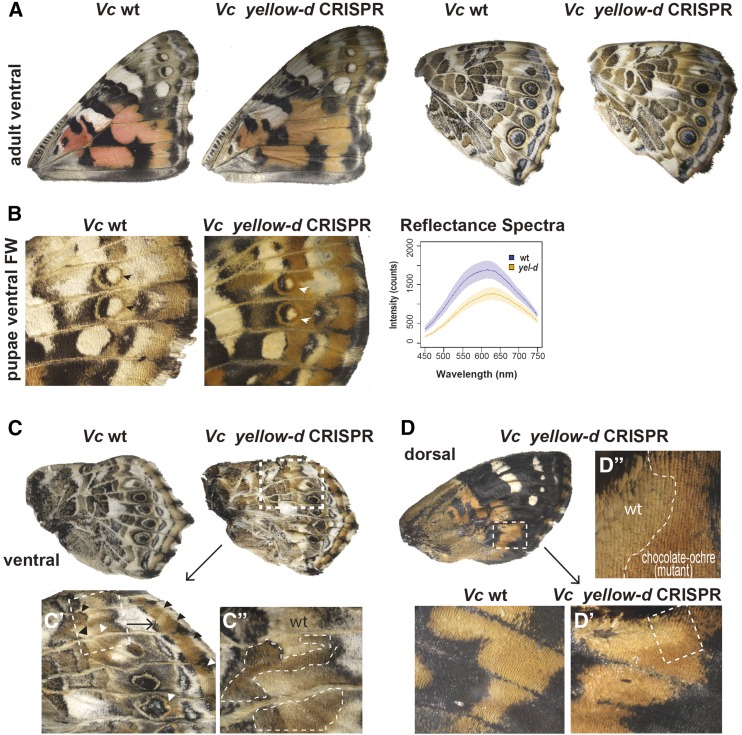
yellow-d:
yellow-d encodes a protein of unknown function. We produced yellow-d mutations using double sgRNAs in V. cardui. We observed that yellow-d mutation promoted orange–brown pigmentation in specific ventral forewing patterns, especially the submarginal and marginal elements between EI and EII, and the stripe between MI and EIII including the rings around border ocelli (Oc) (Figure 7A). Optical reflectance spectra of affected pattern elements on the ventral forewing show significant differences in absorbance between wild-type butterflies and yellow-d knockouts, as well as subtle differences in the shapes of spectra (Figure 7B). Mosaic orange pigmentation was also found in ventral hindwings (Figure 7, C–C”), e.g., marginal elements between EI and EII, eyespot rings, and Discalis I element, and red patterns in dorsal forewings (Figure 7, D–D”).
yellow-h2 and yellow-h3:
yellow-h2 and yellow-h3 encode proteins whose transcripts have shown interesting color-related expression patterns in other species, but that have poorly understood function (Ferguson et al. 2011a; Futahashi et al. 2012). We produced yellow-h2 and yellow-h3 mutations using double sgRNAs in V. cardui. All injected animals that survived to pupation (15 yellow-h2 and 26 yellow-h3 pupae) died during late pupal pigment development. PCR genotyping validated the existence of long deletions in all dead pupae (Figure S1); however, detection of possible pigment phenotypes was confounded by tissue necrosis.
Discussion
In this study, we significantly expand our knowledge of butterfly wing pattern development by functionally characterizing six genes underlying wing pigmentation. Our strategy was to use comparative RNA-seq to identify candidate pigment gene transcripts, and then to use Cas9-mediated targeted knockout to test the function of a subset of these candidates. RNA-seq is widely employed in nonmodel organisms to characterize candidate genes involved in the development of interesting traits; however, such studies are often frustrated by long lists of candidate genes that are difficult to interpret due to a lack of functional validation. Our work shows how combining comparative transcriptomics and genome editing can provide a powerfully synergistic approach for exploring the evolution and development of traits in emerging model systems.
Our comparative transcriptomic work identified over 2000 transcripts showing significant expression differences between wing development stages and color patterns. Using stringent screening criteria, we identified 26 candidate ommochrome-associated genes and 27 candidate melanin-associated genes, complementing previous wing transcriptome studies (Futahashi et al. 2012; Hines et al. 2012; Daniels et al. 2014; Connahs et al. 2016) by providing a more detailed portrait of pigment-specific expression patterns in V. cardui, a species that is highly amenable to genome editing. Although we will focus most of our discussion below on functional results, there are two surprising expression results that we would like to highlight. First, finding that optix is upregulated in red patterns in V. cardui contradicts conclusions from previous comparative work at earlier stages of pupal development, which suggested that the red color pattern-related expression of this gene was limited to heliconiine butterflies (Reed et al. 2011; Martin et al. 2014). Our new results imply that optix may indeed play a color patterning role at later stages of development in nonheliconiine butterflies, although confirmation of this will require further work. Second, we identified a number of MFS transporters that showed highly specific associations with either red or black color patterns, suggesting that these genes might play roles in both ommochrome and melanin pigmentation. Indeed, MFS genes have been previously associated with insect ommochrome biosynthesis in Bombyx eggs, eyes, and larvae (Osanai-Futahashi et al. 2012; Zhao et al. 2012), and melanin pigmentation in Bombyx larval cuticles (Ito et al. 2012). Interestingly, however, these previously characterized MFS genes are not orthologous to the ones identified in this study. Therefore, we speculate that many members of the diverse MFS gene family may play roles in the development of different pigment types across insects. We are currently working to better characterize these and related genes.
Taking advantage of our transcriptome data, we were able to use Cas9-mediated genome editing to test the wing pigmentation roles of eight candidate melanin synthesis genes. Of these, pale, yellow, and Ddc are previously known as melanin-promoting factors. CRISPR-induced knockouts in these three genes produced melanin repression phenotypes in accordance with expectations from Drosophila (Morgan 1916; Wright et al. 1976a; True et al. 1999; Wittkopp et al. 2002b) and other insects including P. xuthus (Futahashi and Fujiwara 2005), Manduca sexta (Gorman et al. 2007), Bombyx mori (Futahashi et al. 2008; Liu et al. 2010), Tribolium castaneum (Gorman and Arakane 2010), and Oncopeltus fasciatus (Liu et al. 2016). Our findings support the idea that pale, yellow, and Ddc play deeply conserved roles in insect melanin pigmentation. The structural defects we observed in pale knockout butterflies are also consistent with the known role of DOPA in sclerotization (Andersen 2010). It is worth noting that yellow mutants in V. cardui, P. xuthus, and B. anynana all resulted in widespread defect clones in both forewing and hindwing, suggesting potential similar involvement of yellow between forewing and hindwing. This is consistent with findings in Drosophila but in contrast to those in Tribolium and Oncopeltus. In Oncopeltus, RNAi (RNA interference) knockdown of yellow caused much greater defects in hindwing than forewing (Liu et al. 2016), and only hindwing defects (not elytra) were observed in Tribolium RNAi knockdowns (Arakane et al. 2010). Such differences indicate that region-specific deployment of yellow might have changed through insect evolution. In summary, our findings support a growing body of literature to suggest that there is a highly conserved set of genes involved in insect melanin pigmentation, and that these genes have been deployed in multiple tissue types throughout evolutionary history.
Our results with yellow-d, ebony, and black were of particular interest to us because they showed that these genes can affect the hue of color patterns that do not appear to be typical black DOPA-derived eumelanins. One of the most novel findings from our study relates to the function of yellow-d, which is a protein of previously uncharacterized function. Previous work has shown an association between yellow-d expression and dark pupal cuticles in B. mori (Xia et al. 2006), and red wing patterns in Heliconius spp. (Ferguson et al. 2011a; Hines et al. 2012), but thus far there has been no functional evidence for a role in pigmentation and these previous associations were potentially contradictory. Thus, we were interested to find that V. cardui knockout of yellow-d resulted in specific buff-colored color pattern elements converting to a chocolate–ochre hue (Figure 7C and Figure 8B). Although determining the chemical basis of this color variation is beyond the scope of this study, it is notable that this hue is not one that is normally associated with eumelanin pigments. We tentatively speculate that this color may be generated by a pigment or pigments in the NBAD sclerotin or pheomelanin pathways, and/or even perhaps ommochromes, although additional work will be required to confirm this. Unlike the other yellow gene family member yellow, loss of yellow-d function not only affects melanin patterns but also presumptive ommochrome patterns. This finding is consistent with the different expression profiles of these genes, where yellow-d shows red-specific expression while no differential expression was detected for yellow. We further speculate that coexpression of yellow-d and the other two melanin-suppressing genes ebony and black (Table S4 in File S1) may indicate that these genes can be recruited together to color tune-specific pattern elements. In any case, our yellow-d knockout results clearly demonstrate that the activity of this single gene is sufficient to tune the color of specific pattern elements by switching pigments.
Figure 8.
Wing pattern-specific effects of yellow-d loss-of-function in Vc. (A) Ventral view of wt and yellow-d knockout wings in Vc shows promoted orange–brown pigmentation. (B) yellow-d mutation in Vc results in yellowing of pigments. Graphs of reflectance spectra of ventral FW (white arrowhead) show significant shift in coloration between wt and CRISPR mutants. (C–C”) Ventral hindwing view of yellow-d knockout mosaic clones showing mutant ochre pigmentation in normally brown and gray color patterns. (D–D”) yellow-d loss-of-function mosaic clones showing increased ochre pigmentation in dorsal FW red patterns. CRISPR, clustered regularly interspaced short palindromic repeats; FW, forewing; Vc, V. cardui; wt, wild-type.
Our ebony results were also of significant interest, again due to the dramatic effect of ebony gene knockout on noneumelanin wing patterns. Work in Drosophila has shown that ebony suppresses melanin development in some tissue types, presumably because the enzyme encoded by ebony, NBAD-synthase, depletes dopamine that would otherwise be used for eumelanin synthesis. Therefore, Drosophila ebony mutants show an overall increase in melanization, while ectopic expression of ebony inhibits melanization (Wittkopp et al. 2002a). Supporting this, ebony mutagenesis in P. xuthus showed enhanced melanic pigmentation in larvae (Li et al. 2015) and dark wing patches in B. anynana (Beldade and Peralta 2017). Based on these previous results, we were unsurprised to see darkened wing clones in adults of V. cardui, B. anynana, and J. coenia (Figure 6). Interestingly, however, this darkening occurred across a range of color pattern types in all species, including buffs, tans, and light and dark browns. The most marked knockout phenotype was seen in a subset of V. cardui pupae when dissected, where developing pupal wings showed an almost complete conversion of all nonmelanic color patterns to black, including red ommochrome and colorless (white) pattern. These results strongly suggest that ebony is a strong repressor of melanin synthesis in all wing scales, regardless of specific pigment type. ebony knockout in ommochrome stage wings caused darkening phenotypes in the scales that would become black in the melanin stage (Figure 6A), and we know that ebony is significantly upregulated at the ommochrome stage. Together, these findings suggest that ebony suppresses melanin synthesis in black patterns before black pigmentation is produced. Furthermore, the phenotype of increased melanin in red scales correlates with the transcription profile where ebony showed red-specific expression. This is consistent with previous speculation based on correlations between ebony expression and red ommochrome color patterns in Heliconius spp. (Ferguson et al. 2011b; Hines et al. 2012). These results are also consistent with Oncopeltus ebony RNAi knockdown phenotypes in forewings, where ebony appears to suppress melanization in the orange pattern. The aberrant red patterns in ebony CRISPR knockout butterflies are known to show strong incorporation of ommochrome-precursor tryptophan (Nijhout and Koch 1991). However, we notice that radiolabeling experiments in J. coenia also show weak levels of tyrosine incorporation in these patterns. Our current hypothesis is thus that orange scales synthesize light-color melanin derivatives such as NBAD sclerotin, and that ebony loss-of-function triggers the accumulation of dopamine, which is in turn converted into dark melanin (Figure 1). It is also plausible that an unidentified type of tyrosine–tryptophan hybrid pigment, such as a papiliochrome, may occur in J. coenia, although further work would be required to test this.
A third gene that had notable effects on wing coloration was black. Disruption of this gene is known to result in β-alanine deficiencies, therefore limiting NBAD sclerotin production to result in an excess of dopamine. This excess dopamine is thought to underlie the mutant phenotypes observed in various insects, an overall increase in cuticular melanization (Hori et al. 1984; Roseland et al. 1987; Wright 1987). In butterflies, we also observed similar enhanced melanization phenotypes in wing scales and pupal cuticles. While the apparently conserved function of black may not be remarkable by itself, the wing pattern specificity of the effect is indeed notable. Specifically, black knockout seemed to have a highly specific effect on the same color pattern elements that yellow-d affected, and resulted in a similar darkening of hue, though without the marked yellow overtones (Figure 8). These results tentatively suggest that local activity of black may be implicated in color-tuning specific color pattern elements.
The RNA-seq and genome editing results in this study indicate previously undescribed genetic mechanisms underlying pigment development in butterflies. However, it is important to point out that, even though our approach is quick and powerful, use of G0 mosaics does have some limitations. One limitation that is general to all somatic mosaic studies, especially in smaller organisms such as insects, is that it is challenging to characterize specific causative alleles in specific clones. This is especially difficult in adult butterfly wings where wing scales are dead cuticle with degraded genomic DNA. Because of this, our genotyping is limited to nonwing tissue, and thus primarily serves to confirm the accuracy and efficiency of our sgRNAs. Further, since we cannot perform molecular genotyping on specific wing clones, we cannot confirm whether a clone is monoallelic or biallelic for induced lesions, and thus we can only speculate about potential dosage effects. We also cannot exclude off-target effects, in the absence of genetic crosses and further allele-specific validation. However, despite these limitations, the easy-to-visualize nature of butterfly wing pigment phenotypes allowed us to unambiguously validate the roles of multiple pigmentation genes in wing pattern development. Some gene functions were found to be deeply conserved in insects, while others were found to play novel or expanded roles in butterfly wing coloration. Understanding the genetic basis of adaptive features is one of the key goals in evolutionary and developmental biology, and our results provide a foundation for future work on the development and evolution of butterflies and other organisms as well.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.196451/-/DC1.
Acknowledgments
We thank Joseph Fetcho and Joe DiPietro at Cornell University for assistance with microinjection, Kyle DeMarr and Benjamin John Brack for help raising butterflies, and Ellis Loew for help with microspectrophotometry. Finally, we are indebted to members of the Reed and Monteiro Labs, as well as to Nipam Patel and his team for experimental assistance and discussions. This work was supported by National Science Foundation grants IOS-1354318 and IOS-1557443 to R.D.R. and Ministry of Singapore grant MOE2015-T2-2-159 to A. Monteiro.
Author contributions: R.D.R., L.Z., A. Martin, M.W.P., and Y.M. conceived the experiments. L.Z. performed RNA-seq work and data analysis. Genome editing and associated genotyping were performed by L.Z. (V. cardui), A. Martin, and K.R.L.v.d.B. (J. coenia), Y.M. (B. anynana), and M.W.P. (V. cardui, P. xuthus). R.D.R. and A. Monteiro supervised experiments. L.Z., A. Martin, and R.D.R. wrote the manuscript with input from all coauthors.
Footnotes
Communicating editor: D. M. Parichy
Literature Cited
- Andersen S. O., 2010. Insect cuticular sclerotization: a review. Insect Biochem. Mol. Biol. 40: 166–178. [DOI] [PubMed] [Google Scholar]
- Arakane Y., Dittmer N. T., Tomoyasu Y., Kramer K. J., Muthukrishnan S., et al. , 2010. Identification, mRNA expression and functional analysis of several yellow family genes in Tribolium castaneum. Insect Biochem. Mol. Biol. 40: 259–266. [DOI] [PubMed] [Google Scholar]
- Bassett A., Liu J.-L., 2014. CRISPR/Cas9 mediated genome engineering in Drosophila. Methods 69: 128–136. [DOI] [PubMed] [Google Scholar]
- Bassett A. R., Tibbit C., Ponting C. P., Liu J.-L., 2013. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 4: 220–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beldade P., Brakefield P. M., 2002. The genetics and evo–devo of butterfly wing patterns. Nat. Rev. Genet. 3: 442–452. [DOI] [PubMed] [Google Scholar]
- Beldade P., Peralta C. M., 2017. Developmental and evolutionary mechanisms shaping butterfly eyespots. Curr. Opin. Insect Sci. 19: 22–29. [DOI] [PubMed] [Google Scholar]
- Biessmann H., 1985. Molecular analysis of the yellow gene (y) region of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 82: 7369–7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Yang P., Jiang F., Wei Y., Ma Z., et al. , 2010. De novo analysis of transcriptome dynamics in the migratory locust during the development of phase traits. PLoS One 5: e15633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connahs H., Rhen T., Simmons R. B., 2016. Transcriptome analysis of the painted lady butterfly, Vanessa cardui during wing color pattern development. BMC Genomics 17: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels E. V., Murad R., Mortazavi A., Reed R. D., 2014. Extensive transcriptional response associated with seasonal plasticity of butterfly wing patterns. Mol. Ecol. 23: 6123–6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson L. C., Green J., Surridge A., Jiggins C. D., 2011a Evolution of the insect yellow gene family. Mol. Biol. Evol. 28: 257–272. [DOI] [PubMed] [Google Scholar]
- Ferguson L. C., Maroja L., Jiggins C. D., 2011b Convergent, modular expression of ebony and tan in the mimetic wing patterns of Heliconius butterflies. Dev. Genes Evol. 221: 297–308. [DOI] [PubMed] [Google Scholar]
- Futahashi R., Fujiwara H., 2005. Melanin-synthesis enzymes coregulate stage-specific larval cuticular markings in the swallowtail butterfly, Papilio xuthus. Dev. Genes Evol. 215: 519–529. [DOI] [PubMed] [Google Scholar]
- Futahashi R., Sato J., Meng Y., Okamoto S., Daimon T., et al. , 2008. yellow and ebony are the responsible genes for the larval color mutants of the silkworm Bombyx mori. Genetics 180: 1995–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futahashi R., Shirataki H., Narita T., Mita K., Fujiwara H., 2012. Comprehensive microarray-based analysis for stage-specific larval camouflage pattern-associated genes in the swallowtail butterfly, Papilio xuthus. BMC Biol. 10: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo M., Stavenga D., 2016. Brilliant iridescence of Morpho butterfly wing scales is due to both a thin film lower lamina and a multilayered upper lamina. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 202: 381–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman M. J., Arakane Y., 2010. Tyrosine hydroxylase is required for cuticle sclerotization and pigmentation in Tribolium castaneum. Insect Biochem. Mol. Biol. 40: 267–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman M. J., An C., Kanost M. R., 2007. Characterization of tyrosine hydroxylase from Manduca sexta. Insect Biochem. Mol. Biol. 37: 1327–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., et al. , 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guschin D. Y., Waite A. J., Katibah G. E., Miller J. C., Holmes M. C., et al. , 2010. A rapid and general assay for monitoring endogenous gene modification. Methods Mol. Biol. 649: 247–256. [DOI] [PubMed] [Google Scholar]
- Hines H. M., Papa R., Ruiz M., Papanicolaou A., Wang C., et al. , 2012. Transcriptome analysis reveals novel patterning and pigmentation genes underlying Heliconius butterfly wing pattern variation. BMC Genomics 13: 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiruma K., Matsumoto S., Isogai A., Suzuki A., 1984. Control of ommochrome synthesis by both juvenile hormone and melanization hormone in the cabbage armyworm, Mamestra brassicae. J. Comp. Physiol. B 154: 13–21. [Google Scholar]
- Hori M., Hiruma K., Riddiford L. M., 1984. Cuticular melanization in the tobacco hornworm larva. Insect Biochem. 14: 267–274. [Google Scholar]
- Ito K., Kidokoro K., Katsuma S., Shimada T., Yamamoto K., et al. , 2012. Positional cloning of a gene responsible for the cts mutation of the silkworm, Bombyx mori. Genome 55: 493–504. [DOI] [PubMed] [Google Scholar]
- Koch P. B., Keys D. N., Rocheleau T., Aronstein K., Blackburn M., et al. , 1998. Regulation of dopa decarboxylase expression during colour pattern formation in wild-type and melanic tiger swallowtail butterflies. Development 125: 2303–2313. [DOI] [PubMed] [Google Scholar]
- Kronforst M. R., Papa R., 2015. The functional basis of wing patterning in Heliconius butterflies: the molecules behind mimicry. Genetics 200: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux M. J., 2007. Eukaryotic major facilitator superfamily transporter modeling based on the prokaryotic GlpT crystal structure. Mol. Membr. Biol. 24: 333–341. [DOI] [PubMed] [Google Scholar]
- Li X., Fan D., Zhang W., Liu G., Zhang L., et al. , 2015. Outbred genome sequencing and CRISPR/Cas9 gene editing in butterflies. Nat. Commun. 6: 8212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Yamamoto K., Cheng T.-C., Kadono-Okuda K., Narukawa J., et al. , 2010. Repression of tyrosine hydroxylase is responsible for the sex-linked chocolate mutation of the silkworm, Bombyx mori. Proc. Natl. Acad. Sci. USA 107: 12980–12985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Lemonds T. R., Marden J. H., Popadić A., 2016. A pathway analysis of melanin patterning in a Hemimetabolous insect. Genetics 203: 403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A., McCulloch K. J., Patel N. H., Briscoe A. D., Gilbert L. E., et al. , 2014. Multiple recent co-options of Optix associated with novel traits in adaptive butterfly wing radiations. Evodevo 5: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro A., 2015. Origin, development, and evolution of butterfly eyespots. Annu. Rev. Entomol. 60: 253–271. [DOI] [PubMed] [Google Scholar]
- Morgan T. H., 1916. Sex-Linked Inheritance in Drosophila. Carnegie Institution of Washington, Washington. [Google Scholar]
- Nijhout H. F., 1991. The Development and Evolution of Butterfly Wing Patterns (Smithsonian Series in Comparative Evolutionary Biology). Smithsonian Institution Scholarly Press, Washington, DC. [Google Scholar]
- Nijhout H. F., Koch P., 1991. The distribution of radiolabeled pigment precursors in the wing patterns of nymphalid butterflies. J. Res. Lepid. 30: 1–13. [Google Scholar]
- Osanai-Futahashi M., Tatematsu K.-I., Yamamoto K., Narukawa J., Uchino K., et al. , 2012. Identification of the Bombyx red egg gene reveals involvement of a novel transporter family gene in late steps of the insect ommochrome biosynthesis pathway. J. Biol. Chem. 287: 17706–17714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez M. M., Schachter J., Berni J., Quesada-Allué L. A., 2010. The enzyme NBAD-synthase plays diverse roles during the life cycle of Drosophila melanogaster. J. Insect Physiol. 56: 8–13. [DOI] [PubMed] [Google Scholar]
- Perry M., Kinoshita M., Saldi G., Huo L., Arikawa K., et al. , 2016. Molecular logic behind the three-way stochastic choices that expand butterfly colour vision. Nature 535: 280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R. D., Nagy L. M., 2005. Evolutionary redeployment of a biosynthetic module: expression of eye pigment genes vermilion, cinnabar, and white in butterfly wing development. Evol. Dev. 7: 301–311. [DOI] [PubMed] [Google Scholar]
- Reed R. D., Papa R., Martin A., Hines H. M., Counterman B. A., et al. , 2011. optix drives the repeated convergent evolution of butterfly wing pattern mimicry. Science 333: 1137–1141. [DOI] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J., Smyth G. K., 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseland C. R., Kramer K. J., Hopkins T. L., 1987. Cuticular strength and pigmentation of rust-red and black strains of Tribolium castaneum: correlation with catecholamine and β-alanine content. Insect Biochem. 17: 21–28. [Google Scholar]
- True J. R., Edwards K. A., Yamamoto D., Carroll S. B., 1999. Drosophila wing melanin patterns form by vein-dependent elaboration of enzymatic prepatterns. Curr. Biol. 9: 1382–1391. [DOI] [PubMed] [Google Scholar]
- Wallbank R. W., Baxter S. W., Pardo-Diaz C., Hanly J. J., Martin S. H., et al. , 2016. Evolutionary novelty in a butterfly wing pattern through enhancer shuffling. PLoS Biol. 14: e1002353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp P. J., True J. R., Carroll S. B., 2002a Reciprocal functions of the Drosophila Yellow and Ebony proteins in the development and evolution of pigment patterns. Development 129: 1849–1858. [DOI] [PubMed] [Google Scholar]
- Wittkopp P. J., Vaccaro K., Carroll S. B., 2002b Evolution of yellow gene regulation and pigmentation in Drosophila. Curr. Biol. 12: 1547–1556. [DOI] [PubMed] [Google Scholar]
- Wright T. R., 1987. The genetics of biogenic amine metabolism, sclerotization, and melanization in Drosophila melanogaster. Adv. Genet. 24: 127. [PubMed] [Google Scholar]
- Wright T. R., Bewley G. C., Sherald A. F., 1976a The genetics of dopa decarboxylase in Drosophila melanogaster. II. Isolation and characterization of dopa-decarboxylase-deficient mutants and their relationship to the α-methyl-dopa-hypersensitive mutants. Genetics 84: 287–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright T. R., Hodgetts R. B., Sherald A. F., 1976b The genetics of dopa decarboxylase in Drosophila melanogaster I. Isolation and characterization of deficiencies that delete the dopa-decarboxylase-dosage-sensitive region and the α-methyl-dopa-hypersensitive locus. Genetics 84: 267–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia A.-H., Zhou Q.-X., Yu L.-L., Li W.-G., Yi Y.-Z., et al. , 2006. Identification and analysis of YELLOW protein family genes in the silkworm, Bombyx mori. BMC Genomics 7: 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Reed R. D., 2016. Genome editing in butterflies reveals that spalt promotes and Distal-less represses eyespot colour patterns. Nat. Commun. 7: 11769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Zhang H., Li Z., Duan J., Jiang J., et al. , 2012. A major facilitator superfamily protein participates in the reddish brown pigmentation in Bombyx mori. J. Insect Physiol. 58: 1397–1405. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
V. cardui wing RNA-seq raw reads are available as NCBI SRP062599. Full transcript assembly and expression profiles are available at the NCBI Gene Expression Omnibus (GEO) database with accession number GSE78119. The transcript assembly and unigenes are also available for searching at butterflygenome.org.