Abstract
Reversing DNA methylation abnormalities and associated gene silencing, through inhibiting DNA methyltransferases (DNMTs) is an important potential cancer therapy paradigm. Maximizing this potential requires defining precisely how these enzymes maintain genome-wide, cancer-specific DNA methylation. To date, there is incomplete understanding of precisely how the three DNMTs, 1, 3A, and 3B, interact for maintaining DNA methylation abnormalities in cancer. By combining genetic and shRNA depletion strategies, we define not only a dominant role for DNA methyltransferase 1 (DNMT1) but also distinct roles of 3A and 3B in genome-wide DNA methylation maintenance. Lowering DNMT1 below a threshold level is required for maximal loss of DNA methylation at all genomic regions, including gene body and enhancer regions, and for maximally reversing abnormal promoter DNA hypermethylation and associated gene silencing to reexpress key genes. It is difficult to reach this threshold with patient-tolerable doses of current DNMT inhibitors (DNMTIs). We show that new approaches, like decreasing the DNMT targeting protein, UHRF1, can augment the DNA demethylation capacities of existing DNA methylation inhibitors for fully realizing their therapeutic potential.
There is a growing focus on the use of “epigenetic therapy” approaches in cancer (Baylin and Jones 2011; Azad et al. 2013) including targeting the three enzymes that catalyze DNA methylation, DNA methyltransferases (DNMTs) 1, 3A, and 3B to reverse the DNA methylation abnormalities inherent to virtually all cancer types (Baylin and Jones 2011; Shen and Laird 2013). Optimally pursuing this requires a basic understanding of how these proteins function to establish and especially maintain DNA methylation in cancer cells. To address the maintenance roles for all three DNMTs in human cancer cells, one can take advantage of expanding databases, gathered by ever deeper genome-wide DNA methylation analyses of normal and tumor cells (Lister et al. 2009; Varley et al. 2013; Roadmap Epigenomics Consortium et al. 2015). Cancer of all types have widespread losses of DNA methylation, some in large regional blocks (Baylin and Jones 2011; Berman et al. 2012; Hon et al. 2012), within which there are simultaneous focal gains of methylation in normally nonmethylated CpG islands in gene promoters (Berman et al. 2012; Varley et al. 2013). These latter can associate with abnormal gene silencing of key tumor suppressor genes (Baylin and Jones 2011), and both the gains and losses can involve other gene regulatory regions, including gene bodies and enhancers (Blattler et al. 2014; Yang et al. 2014) and chromatin insulator sequences (Flavahan et al. 2016). By blocking DNMTs, the focal gains in cancer are potentially reversed to allow reexpression of abnormally silenced genes to reprogram cancer cells (Tsai and Baylin 2011; Easwaran et al. 2014). For this purpose, two current inhibitors (DNMTIs), 5-azacytidine (5-AZA) and 2′-deoxy-5-azacytidine (DAC), have been approved by FDA for use in a preleukemic syndrome, myelodysplasia myelodysplastic syndrome (MDS) (Baylin and Jones 2011; Issa et al. 2015), and there are emerging signs for efficacy in solid tumors also (Azad et al. 2013; Ahuja et al. 2014).
The maintenance roles of the three biologically active DNMTs are somewhat different between normal embryonic stem cells (ESC) from mice and humans (for summary, see Supplemental Table S1; Chen et al. 2003; Li et al. 2015; Liao et al. 2015). Genetic depletion of Dnmt1 in mouse ESC largely abolishes DNA methylation, but cells survive as long as maintained in an undifferentiated state (Chen et al. 2003). Simultaneous genetic disruption of de novo DNMTs Dnmt3a and 3b in mouse ESC also leads to profound losses of DNA methylation, with cell passaging suggesting a strong maintenance role for both enzymes (Chen et al. 2003; Li et al. 2015). Yet, for human ESC, disrupting DNMT1 is lethal and disrupting DNMT3A and 3B has a lesser impact for maintaining DNA methylation (Liao et al. 2015). Results of disrupting DNMTs in human colorectal cancer cells (CRC) has been both informative and confusing (Supplemental Table S1; Rhee et al. 2000, 2002; Chen et al. 2007) and initially challenged the classic view that maintenance of DNA methylation in human cells is solely through DNMT1. Thus, most DNA methylation remains intact with genetic disruption of DNMT1 (1KO cells) in HCT116 (CRC) cells (Rhee et al. 2000). Simultaneously genetically disrupting DNMT3B in this setting (HCT116 DKO cells), resulted in virtual depletion of genomic DNA methylation, suggesting a strong maintenance role for this enzyme (Rhee et al. 2002). However, subsequently, the 1KO cells actually were found to be severely hypomorphic for DNMT1, containing ∼5%–15% levels of a truncated DNMT1 isoform that can apparently maintain the bulk of DNA methylation. This raises questions of why the complete knockout of DNMT3B in 1KO cells (HCT116 DKO) removes virtually all DNA methylation (Rhee et al. 2002). Moreover, our attempts and those of others to deplete DNMT1 in HCT116 and other cancer cells suggested that key tumor suppressor genes cannot be demethylated and/or reexpressed (Ting et al. 2004; Chen et al. 2007), whereas others found this is not the case (Robert et al. 2003). Finally, the role of DNMT3A is important to dissect because mutations in this protein are frequent in human leukemias (Ley et al. 2010; Yan et al. 2011), but the consequences for DNA methylation maintenance are not fully clear.
To address the preceding questions, we now match genetic and shRNA disruption of DNMTs in cancer cells and map the consequences for DNA methylation and gene expression patterns. Finally, we match these results with what clinically relevant doses of DNMTIs can achieve and, as a potential combinatorial cancer treatment strategy, add to this the targeting of UHFR1, a major protein for recruiting DNMTs to DNA (Bostick et al. 2007; Sharif et al. 2007; Rothbart et al. 2013).
Results
Role of DNMT1 threshold in maintenance of DNA methylation in cancer cells
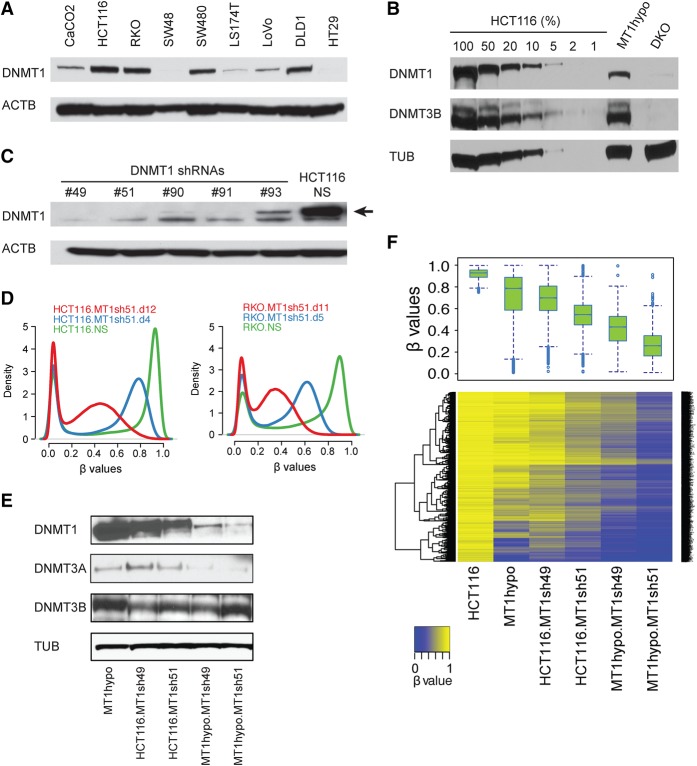
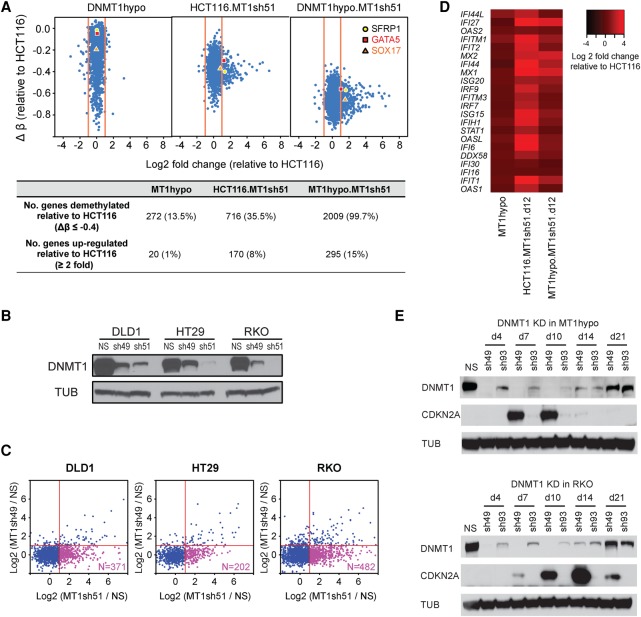
In previous genetic knockout studies of DNMT1 in human HCT116 CRC cells, 80% of DNA methylation is retained (Rhee et al. 2000) through function of a remaining truncated form of DNMT1 with 2%–6% residual DNMT activity (Rhee et al. 2000; Egger et al. 2006; Spada et al. 2007), including the cancer-specific hypermethylation in promoters of key tumor suppressor genes like CDKN2A (Ting et al. 2004). Others reported that DNMT1 depletion by anti-sense RNA in HCT116 induced complete DNA demethylation and reexpression of CDKN2A (Robert et al. 2003). To further explore DNMT1 maintenance of DNA methylation, we first screened several human CRC lines finding no correlation of the widely different numbers of cancer-specific hypermethylated genes (Schuebel et al. 2007) with a wide range of basal DNMT1 levels (Fig. 1A; Supplemental Fig. S1A). The remaining truncated DNMT1 protein in HCT116 DNMT1 hypomorph cells is about 15% or less of the amount of wild-type DNMT1 in HCT116 parent cells (Fig. 1B). Reducing levels of DNMT1, using effective shRNAs for 4–12 d, (Fig. 1C, numbers 49 and 51) in HCT116 and other CRC, breast, and lung cancer cells (Fig. 1D; Supplemental Fig. S1B), plus using the same approach to further reduce levels of the truncated DNMT1 protein in HCT116 1KO cells, resulted in a progressive, incremental decline in overall DNA methylation relative to WT HCT116 cells (Fig. 1E,F). With the deepest reduction of the truncated DNMT1 in 1KO cells, extensive loss of DNA methylation occurred in promoter, hypermethylated CpG islands, gene bodies, and intergenic regions (Supplemental Fig. S1C), indicating the major role for DNMT1 in maintaining genomic DNA methylation. Importantly, this very deep threshold for DNMT1 reduction is required to achieve, in HCT116 cells, DNA demethylation and reexpression of key tumor suppressor genes including CDKN2A. Thus, for promoter CpG island sites with very high starting beta values of 0.75 or greater for Infinium 450 probes, the frequency of genes with increased expression jumps from 1% to 15% with beta value decreases of 0.4 or greater (Fig. 2A) as exemplified for hypermethylated genes in other human CRC cells (Fig. 2B,C). Examples include the intestinal epithelium differentiating inducing transcription factor GATA5, and the anti-WNT genes, SFRP1, and SOX17 (Fig. 2A; Akiyama et al. 2003; Suzuki et al. 2004; Zhang et al. 2008) and others (Supplemental Fig. S2A). Recently, we and others found that DNMTIs produced demethylation and increased transcription of endogenous retroviruses associated with up-regulated expression of viral defense genes (Chiappinelli et al. 2015; Roulois et al. 2015). These are involved in a pathway sensing cytosolic double-stranded RNA, and this drug effect has been termed “viral mimicry” (Chiappinelli et al. 2015; Roulois et al. 2015). Many of these viral defense genes show low basal expression in DNMT1 hypomorph cells, but are sharply up-regulated by reaching a deeper, intermediate DNMT1 threshold in HCT116.MT1sh51.d12 cells and also, but slightly less, with the deepest threshold reached in MT1hypo.MT1sh51.d12 cells (Fig. 2D).
Figure 1.
Relationships of DNMT1 levels to the maintenance of DNA methylation in colon cancer cells. (A) Western blot analysis of DNMT1 in a panel of colorectal cancer cell lines: (ACTB) beta actin. (B) Western blot analysis of DNMT1 and DNMT3B from HCT116 DNMT1 hypomorph cells (MT1hypo) and compared to dilutions of extracts of wild-type HCT116 cells: (TUB) alpha tubulin; (DKO) HCT116 double knockout cells (carrying a DNMT1 hypomorph and DNMT3B knockout). (C) Western blot analysis of DNMT1 in HCT116 cells subject to DNMT1 shRNA knockdowns: (NS) nonsilencing shRNA control. An arrow denotes the DNMT1-specific band. (D) Genome-wide methylation profiles measured by Illumina Infinium HumanMethylation450K array in HCT116 and RKO subjected to DNMT1 shRNA knockdown (MT1sh51) for different lengths of time. The density plots include all probes on individual arrays for each sample. The x-axis indicates β values: scores in the range between 0 and 1 indicating the level of DNA methylation (lowest to highest = increasing DNA methylation). The y-axis indicates the probability densities which describe the distribution of β values for all probes. (E) Western blot analysis of DNMTs in MT1hypo and HCT116 cells subject to DNMT1 knockdowns with different shRNAs. (F) DNA methylation analyses by Illumina Infinium HumanMethylation 450K array in HCT116 cells with decreasing DNMT1 levels shown in E. A whisker box plot of the resultant β values is shown (upper) with a heatmap of the β values (lower). Note the step-wise reduction of DNMT1 that correlates with progressive demethylation. The analysis includes methylated promoter CpGi (CpG island) probes with β values greater than or equal to 0.75 in wild-type HCT116 cells.
Figure 2.
Effects of DNMT1 depletion on gene expression in colon cancer cells. (A) Relationships of gene expression to DNA methylation in HCT116 cells with decreasing DNMT1 protein levels. The y-axis denotes changes in average β values for individual genes with respect to those in the wild-type HCT116 cells. Only genes with hypermethylated promoter CpG islands (β values ≥0.75) in HCT116 cells are shown. The x-axis denotes log2 fold changes in gene expression relative to the wild-type HCT116 cells. Numbers of demethylated and up-regulated genes are summarized in the lower table. Three known tumor suppressor genes—SFRP1, GATA5 and SOX17—are marked in the plot. (B) Western blot analysis of DNMT1 in colorectal cancer cell lines, DLD1, HT29, and RKO, following DNMT1 knockdown using shRNAs of higher (sh51) and lower (sh49) efficiencies: (NS) nonsilencing shRNA control; (TUB) alpha tubulin. (C) Genome-wide gene expression analyses of genes with promoter CpG island methylation in cell lines shown in B, subject to DNMT1 knockdowns. Genes with hypermethylated promoter CpG islands (β values ≥0.75) in the parental lines are included in the analysis for each cell line. The x-axis denotes relative gene expression change (log2 fold change of sh51 treatment/control) of the hypermethylated genes. The y-axis denotes log2 fold change of sh49 treatment/control of the hypermethylated genes. (D) Heatmap of log2 fold expression changes for viral defense genes (y-axis; see text for details of genes) in wild-type HCT116 or MT1hypo cells treated with DNMT1 shRNA51 for 12 d. (E) Western blot analysis of CDKN2A and DNMT1 in HCT116 MT1hypo cells (upper) and RKO cells (lower) at different time points following DNMT1 knockdown using shRNAs of higher (sh49) and lower (sh93) efficiencies: (NS) nonsilencing shRNA control.
The threshold effect is especially dramatic for CDKN2A (also known as p16), one of the most important tumor suppressor genes frequently hypermethylated and silenced in cancer (Myöhänen et al. 1998; Esteller et al. 2001). In HCT116 cells, this gene has one unmethylated, expressed mutant allele producing no functional protein and a fully methylated and silenced wild-type allele (Myöhänen et al. 1998), whereas RKO cells have two hypermethylated, silenced alleles (Herman et al. 1995). In both cell types, CDKN2A protein appears by day 7 when extremely low levels of DNMT1 are achieved with shRNA knockdown (Fig. 2E), then reach a plateau from day 10 to day 14 before decreasing when DNMT1 protein rebounds on day 21 despite continued shRNA treatment (Fig. 2E). These dynamics correlate well with bisulfite sequencing of the CDKN2A promoter CpG island in RKO and HCT116 DNMT1 hypomorph cells showing decreases in DNA methylation level by day 7, a nadir at day 14, and increases by day 21 (Supplemental Fig. S2B,C).
Role of DNMTs 3A and 3B in maintenance of DNA methylation in CRC cells
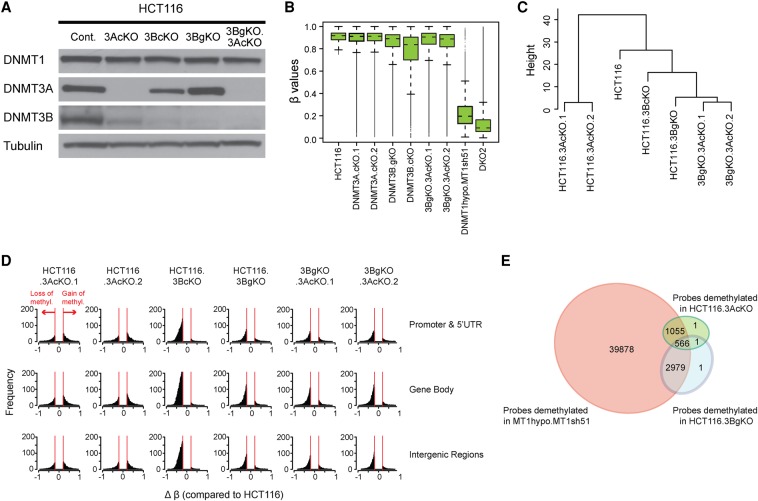
Having firmly established a dominant role and threshold effects for DNMT1 to maintain DNA methylation patterns in cancer cells, what roles do DNMTs 3A and 3B play? We find their maintenance role is minor but is operative at some specific sites. In previously generated genetic knockouts of DNMT3B (3BgKO) (Rhee et al. 2002) and acutely generated CRISPR disruption of DNMT3A and DNMT3B in HCT116 cells (Fig. 3A), overall loss of DNA methylation is dramatically less than in DKO or DNMT1 hypomorph cells with DNMT1 shRNA knockdown (Fig. 3B). Only individual CpG sites are affected in individual genes (Supplemental Fig. S3A). Interestingly, hypomethylated sites can differ overall between 3A and 3B CRISPR KO cells (Fig. 3C); surprisingly, for the two different 3A CRISPR knockout clones, there are as many, or more, gains than losses in proximal promoter, gene body and intergenic regions (Fig. 3D; Supplemental Figs. S3A,B, S4). In contrast, loss of 3B induces mostly DNA methylation losses at all of these regions and losses fully dominate in the 3A/3B double knockout cells (Fig. 3D), suggesting that DNMT3A may block sites that DNMT3B could otherwise methylate. Although DNMT3A and DNMT3B targets are overlapping since 35% of the probes demethylated in 3A KO cells are also demethylated in 3B KO cells, there are distinct CpG sites for each (Fig. 3E). Finally >99% of approximately 4600 probes demethylated in either 3A KO cells or 3B KO cells also get demethylated in MT1hypo.MT1sh51 cells (Fig. 3E). Thus, the maintenance of a subset of CpG sites requires both DNMT1 and DNMT3s, since loss of any one of them will result in DNA hypomethylation. Similarly, in another human CRC cell line SW480, again, DNMT3 deletions produce minor overall gains and losses of methylation (Supplemental Fig. S5A–D). However, for unknown reasons, unlike in HCT116 cells, both gains and losses occur in single 3B KO and 3A/3B double KO cells (Fig. 3D; Supplemental Fig. S5D). The ramifications for gene expression of disrupting 3A or 3B are also minimal, and both reductions and increases occur with little changes in DNA methylation (Supplemental Fig. S3C). These latter changes may reflect that, as for DNMT1, DNMTs 3A and 3B can affect gene expression through scaffolding functions independent of their catalytic sites (Bachman et al. 2001), and there may also be effects resulting from secondary and tertiary changes induced in the DNMT3 knockouts.
Figure 3.
Effects of depleting DNMTs 3A and 3B on DNA methylation maintenance. (A) Western blot analysis of DNMTs in HCT116 cells subject to depletion of DNMT3s using CRISPR (3AcKO, 3BcKO) or genetic knockout approach (3BgKO). DNMT3 double knockout cells are created by CRISPR knockout of DNMT3A in 3BgKO cells (3BgKO.3AcKO). (B) Whisker box plots of DNA methylation levels assayed by Infinium 450K array in HCT116 and the derivative cell lines depleted of individual DNMTs (x-axis). All probes with β values greater than 0.75 were included in the analyses. (C) Hierarchical cluster analysis of DNA methylation patterns in HCT116 WT and DNMT mutant cells. Euclidean distance matrixes were used for the clustering based on complete linkage agglomerative algorithm. The top 10,000 most variable probes were included in the analysis. (D) Histograms of frequency distribution (y-axis) of differentially methylated (Δβ ≥ 0.2) Infinium 450K CpGi probes (x-axis) in DNMT3 knockout cells compared to the HCT116 parental cells. (E) Venn diagram of Infinium 450K probes demethylated by DNMT1 shRNA knockdown (MT1hypo.MT1sh51), DNMT3A CRISPR knockout (common between HCT116.3AcKO.1 and HCT116.3AcKO.2), and DNMT3B genetic knockout (HCT116.3BgKO). The analysis includes promoter probes with β values greater than or equal to 0.75 in parental HCT116 cells and showing a decrease by more than 0.2 upon depletion of individual DNMTs.
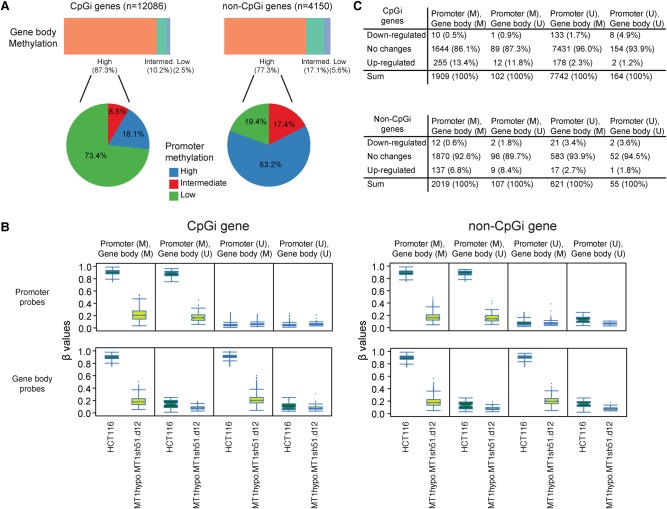
One singularly defined function for DNMT3B concerns reports that DNA methylation in gene body regions facilitates gene expression for a small group of genes, and this activity is well seen when DNMT3B is required for gene body remethylation after DAC treatment (Yang et al. 2014; Baubec et al. 2015). However, again, DNMT1 is the dominant protein for maintaining gene body methylation, since only a few such sites are demethylated by disruption of 3B alone (Fig. 4A,B), whereas shRNA knockdown of DNMT1 in HCT116 DNMT1 hypomorph cells causes severe gene body demethylation for genes with or without methylated gene promoters (Fig. 4A,B; Supplemental Fig. S6A). Importantly, in genes with high basal DNA methylation in both promoter and gene body regions, promoter methylation plays a more dominant role for transcriptional regulation, since genes with hypermethylated promoters have up- and not down-regulation. Also, only a few genes with unmethylated promoters (Fig. 4C), including those most highly expressed (Supplemental Fig. S6B), show decreased expression when gene body methylation is severely reduced by DNMT1 depletion.
Figure 4.
DNMT1 depletion disrupts gene body DNA methylation maintenance. (A) The top bars represent the percentages of gene body probes with different basal DNA methylation values (β ≥ 0.75) of intermediate (0.25 < β < 0.75) and low (β ≤ 0.25) in genes with and without promoter CpG islands (CpGi) in HCT116 cells. The pie charts represent promoter methylation profiles—high (average β ≥ 0.75), intermediate (0.25 < β < 0.75), low (β ≤ 0.25) for genes with high gene body methylation. (B) Box and whisker plots showing methylation level decreases in HCT116 cells with the lowest DNMT1 threshold (β values; y-axis) relative to wild-type HCT116 cells (x-axis designations). Changes in β values of promoter (upper) and gene body (lower) probes before and after DNMT1 depletion are graphed for genes with (left) and without (right) promoter CpG islands, respectively. The analyses include four subgroups of genes in HCT116: (M) methylated = average β ≥ 0.75; (U) unmethylated = average β ≤ 0.25. (C) Numbers (percentages) of genes with expression changes of twofold in HCT116 cells with the lowest DNMT1 threshold comparing to wild-type HCT116 cells. Genes are categorized based on their promoter and gene body methylation status. (M) average β ≥ 0.75; (U) average β ≤ 0.25.
Dissecting the molecular characteristics of HCT116 DKO cells
The deep reduction of DNA methylation in HCT116 DNMT1 mutant cells is a passive loss not associated with any changes in 5-hydroxymethylcytosine (5-hmc) (Supplemental Fig. S7), a product formed with active DNA demethylation. Since disruption of DNMTs 3A, 3B, or both causes minimal maintenance methylation changes, what then explains the severe loss of DNA methylation when DNMT3B is deleted in the HCT116 DNMT1 hypomorph to produce DKO cells? First, analysis of four of the eight original DKO clones (Rhee et al. 2002) reveals that at the protein level, DKO clones 2, 3, and 4, are virtually triple DNMT knockouts or “TKO” cells. Thus, these have much lower DNMT1 protein levels than even the DNMT1 hypomorph (1KO) cells (Supplemental Fig. S8A) and also have virtual absence of DNMT3A (Supplemental Fig. S8A). It is hypothesized this loss of DNMT3A results from the fact that presence of DNA methylation is required for stability of this protein (Sharma et al. 2011). The fourth clone, DKO8, as shown previously (Rhee et al. 2002) has much higher DNA methylation than all the rest (Rhee et al. 2002) and, as found by others (Egger et al. 2006; Sharma et al. 2011), a much higher level of the truncated DNMT1 protein (Supplemental Fig. S8A,B). Importance of truncated DNMT1 in these “TKO” cells is again illustrated for the CDKN2A protein. As previously reported, CDKN2A is reexpressed in early passages of DKO2 when its growth is starkly slower than wild-type HCT116 cells, but is silenced again by passages 80 and beyond when these cells begin to grow much more rapidly (Bachman et al. 2003). We now find these dynamics are fully explained by levels of truncated DNMT1 as its resilencing is accompanied by a marked increase in later passages of truncated DNMT1 (Supplemental Fig. S8C). Moreover, if the DNMT1 levels of late passage DKO cells are again reduced by DNMT1 shRNA knockdown, levels of CDKN2A protein reappear proportional to the degree of DNMT1 reduction (Supplemental Fig. S8D). Selected other important genes are also down-regulated in late passage DKO cells, although many are not (Fig. 2A).
When DNMT3A is completely disrupted with CRISPR technology in DNMT1 hypomorph cells, truncated DNMT1 levels remain the same (Supplemental Fig. S8F). In contrast, DNMT1 hypomorph cells with absent DNMT3B could not be obtained. We hypothesize that the full DKO phenotype for clones 2, 3, and 4 results from rare capture during selection for DNMT1 deletion in the DNMT3B knockout cells, or the “TKO” cells defined above. These have the very lowest levels of truncated DNMT1 and absence of DNMT3A. Only in these cells does simultaneous disruption of 3B appear to play some role for DNA methylation maintenance.
Maintenance of DNA methylation in enhancer sequences
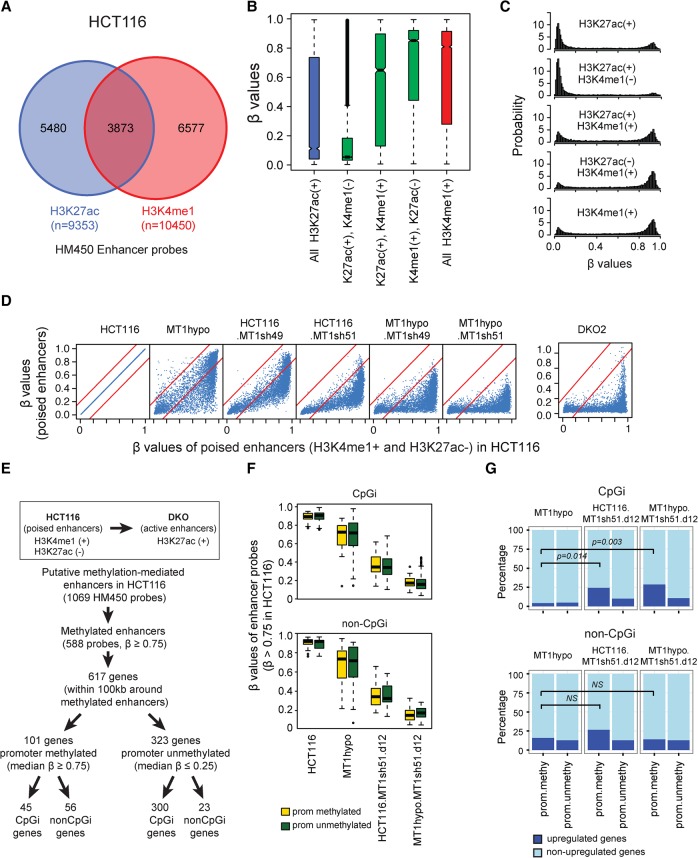
In addition to the genomic regions queried above, DNA methylation may play regulatory roles for enhancer function (Ziller et al. 2013). Although these may be more complex than for promoters, DNA methylation is generally higher in inactive or poised enhancers and lower in active ones (Roadmap Epigenomics Consortium et al. 2015). Enhancers in cancer cells can retain normal patterns or acquire abnormal gains or losses of DNA methylation (Varley et al. 2013) but the functional significance of these changes are less well studied than those for proximal promoter sequences. We matched DNA methylation at putative enhancer sequences with expression of associated genes in HCT116 wild-type and DKO cells (Blattler et al. 2014) with the histone mark, H3K4me1. This latter mark is characteristically found alone as a mark for inactive and/or poised enhancers and H3K27ac—alone is more characteristic of active enhancers (Creyghton et al. 2010; Roadmap Epigenomics Consortium et al. 2015). For approximately 6600 of these matches (Fig. 5A), most enhancer sequences, characterized by either H3K4me1 alone or also having both above marks, have high DNA methylation levels (Fig. 5B,C). In contrast, a group of about 5500 enhancers (Fig. 5A) marked only by H3K27ac (Creyghton et al. 2010) have dramatically lower DNA methylation levels (Fig. 5B,C). These have been noted to generally be active enhancers (Creyghton et al. 2010) but have not been previously associated with low DNA methylation. Moreover, these nonmethylated regions are distant from transcription start sites (TSS), which often are H3K27ac-only marked regions (Supplemental Fig. S9A,B), indicating they are putative distal enhancers. Importantly, for each aforementioned punitive enhancer, as in other genomic regions (Fig. 5D; Supplemental Fig. S9C), incremental decreases in DNA methylation accompany incremental decreases in DNMT1, with most methylation being severely disrupted in HCT116 DNMT1 hypomorph cells treated with DNMT1 shRNAs and in the DKO cells (Fig. 5D). Only minor changes occur with disruption of the other two DNMTs (Supplemental Fig. S9C).
Figure 5.
DNA methylation profiles in gene enhancers. (A) Infinium Methylation 450K (HM450) probes that overlap with putative enhancers marked by either H3K27ac or H3K4me1. (B,C) DNA methylation levels of putative enhancers in HCT116 depicted in whisker box plots (B) and histograms (C). In B, β values (y-axis) are matched to enhancer histone modification status as determined by ENCODE (encodeproject.org) (The ENCODE Project Consortium 2012) (x-axis). (D) DNA methylation levels of poised enhancers (H3K4me1+ and H3K27ac−) in HCT116-derived clones with various DNMT1 protein levels. The x-axis shows β values for wild-type HCT116 cells (range; first panel); the y-axis shows β values for DNMT1 knockdowns or knockouts and DKO cells. (E) Flow chart for selection of putative enhancers stratified by methylation analyses and that have been denoted by ENCODE to be poised in HCT116 wild-type cells and that became active in HCT116-DKO cells (Blattler et al. 2014). The putatively controlled genes for these enhancers (50 kb upstream or downstream) have been determined by informatics analyses outlined in the text. HM450 probes that are located within the poised enhancers and heavily methylated (β values ≥0.75) are included in the analyses. (F) Whisker box plots for methylation levels (y-axis) of poised enhancer probes (β values ≥0.75) in HCT116 cells with different DNMT1 levels. Genes are grouped according to their promoter CpG islands and methylation status. The yellow box represents genes with methylated promoters (average β values ≥0.75), and the green box represents genes with unmethylated promoters (average β values ≤0.25). (G) Percentage of genes (y-axis) with up-regulated (defined by being 1.4-fold or greater than basal values in wild-type HCT116 cells; dark blue portion of bar) versus unchanged expression (light blue portion of bar) following the DNMT1 depletions outlined in F (shown at the tops of the panels). The analysis included 617 genes defined in E. P-values, indicated by the bars in the panels, are calculated for Fisher's exact test.
How do cancer-specific gains in enhancers balance with those in promoter CpG islands for gene expression consequences? We queried, for 617 genes in HCT116 cells, the DNA methylation status of enhancer regions lying ∼100 kb outside of proximal promoter regions that gain active H3K27ac marks in DKO cells (Blattler et al. 2014) and split the associated genes into those with and without hypermethylation of their promoters (Fig. 5E). Although all of these DNA methylated enhancer sequences equally lose DNA methylation with decreasing DNMT1 levels and are predominantly unmethylated in DNMT1 hypomorph cells treated with DNMT1 shRNAs (Fig. 5F), associated gene expression differs with the basal status of methylation in CpG island and CpG poor promoters (Fig. 5G). Importantly, for CpG island promoters, the number of up-regulated genes significantly increases with both intermediate and full methylation losses in the DNMT1 shRNA-treated cells (Fig. 5G). Also, the percentage of up-regulated genes within this subgroup are significantly higher than for genes without methylated promoters (Fig. 5G). In contrast, the percentage of up-regulated genes does not increase significantly for non-CpG island genes with methylated enhancers and promoters for intermediate or full losses of DNA methylation (Fig. 5G). We interpret these findings as suggesting that, in tumor cells, cancer-specific de novo gene promoter CpG island methylation may be dominant over enhancer methylation alone for maintaining abnormal transcriptional repression of target genes.
Comparing levels of DNA methylation reduction by targeting UHRF1 as a means to improve clinically relevant use of DNMTIs
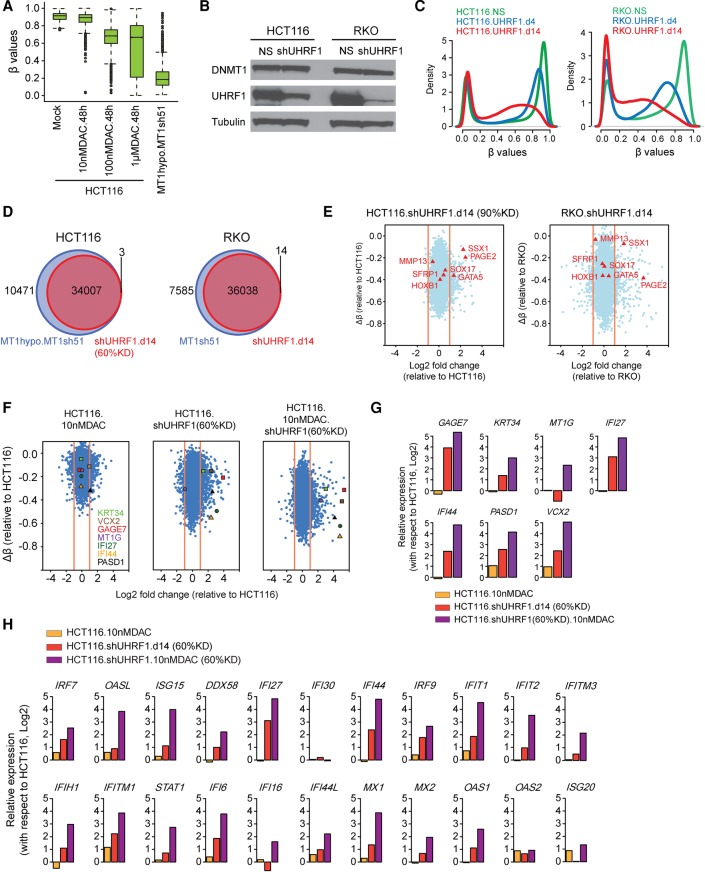
A key translational question is how the previously mentioned degrees of DNA demethylation achieved with DNMT1 depletion compares to that induced by DNMTIs currently used for cancer management. All three DNMTs are blocked at their catalytic sites by these drugs and also transiently degraded (Weisenberger et al. 2004). In the laboratory, high DNMTI doses can easily reverse DNA methylation and reexpress many genes with abnormal promoter CpG island DNA methylation (Schuebel et al. 2007). However, such doses produce severe toxicities in the clinic (Issa 2005). Low, more clinically realistic doses can induce reprogramming-like, anti-tumor effects in hematopoietic and solid tumor cells, but the accompanying degrees of DNA demethylation are low and likely very heterogeneously affect the total population of hypermethylated alleles and their reexpression (Tsai et al. 2012; Easwaran et al. 2014). This challenge for reversing cancer-specific DNA hypermethylation with DNMTIs is seen for HCT116 cells in a dose response of DAC for 48 h. DNA demethylation for hypermethylated promoter CpG island probes plateaus above 100 nM (Fig. 6A; Supplemental Fig. S10A). Even a 1 µM dose, not achievable in the clinic without severe toxicity, induces only ∼40% decrease compared to >80% in HCT116 DNMT1 hypomorph cells treated with DNMT1 shRNA (cf. Fig. 6A and Fig. 1F). Low doses of AZA over much longer time periods, in several CRC lines, cannot induce demethylation of hypermethylated promoter CpG islands to the degree achieved with shRNA depletion of DNMT1 (Supplemental Fig. S10B–D).
Figure 6.
UHRF1 inhibition augments the DNA demethylation effect of low-dose DAC. (A) Whisker box plots for methylation levels (y-axis) of promoter probes in HCT116 cells treated with different concentrations of decitabine (DAC) for 48 h and in HCT116 MT1hypo cells treated with DNMT1 shRNA (sh51) for 12 d. The analysis includes methylated promoter CpGi probes (β values greater ≥0.75) in HCT116 cells. (B) Western blot analysis of UHRF1 and DNMT1 in HCT116 and RKO, with alpha tubulin as loading controls: (NS) nonsilencing shRNA control. (C) Genome-wide DNA methylation profiles at days 4 and 14 following UHRF1 knockdown (β values between 0 and 1; x-axis) versus probability densities that describe the distribution of β values for all probes (y-axis). (D) Venn diagram of Infinium 450K probes demethylated by DNMT1 and UHRF1 knockdowns. The analysis includes promoter probes with β values greater than or equal to 0.75 in HCT116 and RKO cells and that show a decrease of more than 0.2 upon knockdown of DNMT1 or UHRF1. (E) Expression-methylation plots of HCT116 and RKO cells subject to UHRF1 shRNA knockdown (∼90% reduction of UHRF1). The y-axis denotes changes in average β values for individual genes with respect to those in the parental HCT116 or RKO cells. Only genes with hypermethylated promoter (β values ≥0.75) are examined. The x-axis denotes log2 fold changes in gene expression relative to the parental HCT116 or RKO cells. (F) Expression-methylation plots of HCT116 following DAC treatment, 60% UHRF1 knockdown or a combination of both. (G) Quantitated gene expression changes of the seven representative genes marked in F in HCT116 cells following DAC treatment (yellow bars), UHRF1 knockdown (red bars), or a combination of both (purple bars). The y-axis denotes log2 fold changes of gene expression relative to that in the untreated HCT116 cells. (H) Gene expression changes of immune-related, viral defense genes following DAC treatment (yellow bars), UHRF1 knockdown (red bars), or a combination of both (purple bars).
The preceding data suggest that combinatorial approaches are needed to achieve full reexpression of DNA hypermethylated genes in cancer cells. To date, these have mainly utilized low doses of DNMTIs with inhibitors of repressive chromatin modifications, mostly studied for histone deacetylase inhibitors (HDACIs) (Suzuki et al. 2004; Schuebel et al. 2007; Kelly et al. 2010). However, these approaches relieve transcriptional repression without causing DNA demethylation (Cameron et al. 1999). One alternative means of reversing maintenance of cancer-specific DNA hypermethylation might be to block the proteins that recruit DNMTs to DNA, such as UHRF1 (Bostick et al. 2007; Sharif et al. 2007; Rothbart et al. 2013). Indeed, an initial timed, 60% UHRF1 shRNA knockdown in HCT116 cells gives an intermediate decrease of methylation, and 90% UHRF1 knockdown in RKO cells yields a further decrease (Fig. 6B,C) in CpG sites encompassing those hypomethylated by DNMT1 knockdown (Fig. 6D). Neither UHRF1 knockdown level reproduced the low DNA methylation threshold achieved in DNMT1 hypomorph cells treated with DNMT1 shRNA or in DKO cells (cf. Fig. 6E and Figs. 2A, 3B). Similar knockdown (90%) of UHRF1 in HCT116 to that in RKO cells (Supplemental Fig. S11A) induces reexpression of multiple promoter hypermethylated genes in both cell lines (Fig. 6E). When reduction of UHRF1 levels is paired with simultaneous low-dose DAC treatment, there is a complex effect on promoter, hypermethylated genes that arises because UHRF1 is likely required to covalently trap DNMT1 at sites where DAC has incorporated next to guanosines in the genome. Thus, with 60% versus 90% UHRF1 depletion in the HCT116 cells, there is augmentation of low-dose DAC effects on overall DNA methylation reduction only for the former (Fig. 6F,G; Supplemental Figs. S11, S12), and reexpression of hypermethylated genes generally is only augmented for DAC treatment by this 60%, not 90% UHRF1 knockdown (Fig. 6F,G; Supplemental Fig. S11B–F).
Interestingly, low-dose DAC plus 60% UHRF1 knockdown alters promoter methylation preferentially for genes involved in immune attraction and those with CpG-poor promoters. Thus, five of seven selected genes, marked in Supplemental Figure S11C,D, which had little or no increased expression with either DAC or UHRF1 knockdown alone have densely methylated, CpG island promoters. Four are known tumor suppressors (SFRP1, SOX17, GATA5, HOXB1), one (MMP13) has a CpG-poor promoter, and the other two are both cancer testis antigen genes, one with a promoter CpG island (PAGE2) and one (SSX1) without. These genes all had increased reexpression only with combined DAC plus UHRF1 knockdown. In contrast, for seven promoter methylated genes for which reexpression did not occur with DAC alone, each had reexpression to a high degree with the 60% UHRF1 knockdown alone, and this was further augmented by the combination (cf. Fig. 6F,G with Supplemental Fig. S12A,B for lack of augmentation with 90% UHRF1 knockdown). Six of these have CpG poor promoters and, again four are linked to immune responses, with IFI27 and IFI44 being among the viral defense genes studied previously in Figure 2D; two, PASD1 and GAGE7, are X-linked cancer testis genes, the former having a CpG-poor promoter and the latter being the only one of the seven genes having a CpG island promoter (Kao et al. 2014; Michael et al. 2015). The others are a cytoskeletal differentiation gene KRT34, a tumor suppressor gene MT1G (Fu et al. 2013), and a novel cancer testis antigen gene family member VCX2 (Taguchi et al. 2014). Thus, targeting UHRF1 for an intermediate level of inhibition in combination with low dose DNMTIs might be best considered for the clinically important purpose of up-regulating expression of viral defense genes and cancer testis antigens to enhance tumor-associated immune cell attraction (Chiappinelli et al. 2015; Roulois et al. 2015). In support of this, when we examine the entire panel of viral defense genes initially studied in Figure 2D, virtually all are minimally up-regulated by low dose DAC alone, further by both the 60% (Fig. 6H) and 90% (Supplemental Fig. S12C) UHRF1 knockdowns alone, but to the greatest degree when DAC treatment was combined with the 60% UHRF1 reduction (Fig. 6H).
Discussion
Our data further support the dominant role of DNMT1 for maintaining DNA methylation in human cancer cells. Thus, even very small levels of a truncated hypomorphic DNMT1 protein in HCT116 can sustain most DNA methylation (Egger et al. 2006; Spada et al. 2007). Only breaching an extremely low DNMT1 threshold level yields robust demethylation of many genes, with hypermethylated promoter CpG islands to give maximum gene reexpression. As discussed, existing DNMTIs in the clinic cannot achieve such severe DNMT1 reductions in patients without incurring severe toxicity. Chronic low dosing may achieve this in subsets of tumor cells and individual gene alleles. This may be a major part of therapy efficacies in MDS/AML and also account for detectable gene reexpression of hypermethylated genes by PCR analyses (Daskalakis et al. 2002; Cechova et al. 2012). However, this scenario is probably far from fully optimal in terms of maximizing epigenetic therapy with DNMTIs, and decreasing DNA methylation further will likely require combinatorial approaches to existing DNMTIs to block DNMT1 methylation activity, such as we now show for targeting UHRF1. Other options might be to block DNMT1 interactions with other proteins recruiting it to DNA such as PCNA. Most other approaches target DNMT1 interactions that interact with the potential scaffolding functions of DNMT1 to recruit transcriptional corepressors, such as HDACs, EZH2, EHMT2, and KDM1A (Rountree et al. 2000; Bachman et al. 2001; Qin et al. 2011) but these will not alter DNA methylation levels per se.
The role of the other two DNMTs, 3A and 3B, in maintenance of DNA methylation in CRC cells is more complex than for DNMT1; what determines their targets is not clear, although they overlap with DNMT1 for methylating CpG sites (Fig. 3E). Also, our data and those of others (Sharma et al. 2011) show a positive correlation between DNA methylation levels and DNMT3A stability. This complex interaction between DNMTs 3A and 3B has translational relevance. It appears that 3A may block the activity of 3B at some CpG sites and vice versa. Thus in SW480 and HCT116 DNMT3 single KO cells, we see both losses and gains of DNA methylation in all genomic sites queried. These dynamics may be very relevant to mouse DNMT3 knockout studies in which similar gains and losses have been observed (Challen et al. 2014). This antagonism between DNMT3s also varies across cell lines, because there is much more gain of DNA methylation in SW480 versus HCT116 3B KO cells; the reasons for these small but distinct differences between these cell lines are not fully known. However, these interactive dynamics may well help explain why it has been difficult to chronicle the exact alterations in DNA methylation, and consequences for therapy, which accompany the frequent DNMT3A R882H mutation in AML (Russler-Germain et al. 2014). This mutation is thought to be a dominant-negative one; thus, both the gains and losses of DNA methylation we see in our results might be induced. Therefore, examining any one genomic region alone might lead to false overall conclusions. Ironically, the important gains of DNA methylation in cancer, such as those in promoter CpG islands, might not only be maintained in AML with 3A mutations, but even show increases. Such gains indeed are seen in mouse Dnmt3a knockout studies in hematopoietic lineages (Challen et al. 2014).
Our data now also provide further understanding of why DNMT3B deletions may help DNMT1 disruption in chronically inducing severe losses of DNA methylation in HCT116 DKO cells. These cells were originally created by targeting DNMT1 in the DNMT3B knockout cells by using the same DNMT1 targeting vector previously used to generate DNMT1 hypomorph cells (Rhee et al. 2002). At the end of the day, this creates multiple DKO clones with much lower levels of DNMT1 protein than the DNMT1 hypomorph cells (Supplemental Fig. S8A). In the present studies, our attempts to recapitulate the DKO phenotype with acute DNMT3B CRISPR knockout in the DNMT1 hypomorph cells were unsuccessful because cells probably cannot survive this. We surmise that the original chronic selection of DKO cells with the very low levels of truncated DNMT1, below a threshold level for maintenance of DNA methylation in DNMT1 hypomorph cells, has accentuated the minor maintenance roles of DNMT3B. Also, our data emphasize how HCT116 DKO clones 2–4 with the lowest levels of DNMT1 and DNA methylation are functionally TKO cells with virtual absence of DNMT3A protein (Supplemental Fig. S8A,B), probably due to dependency for stability of this protein on its binding to sites of existing DNA methylation (Sharma et al. 2011). We hypothesize that DKO cells were then obtained as rare cells that, during selection, adapted to progressive decreases in DNA methylation but with very slow growth. Indeed, in the initial recombinant approaches to creating HCT116 DKO cells, extensive selection was required (Rhee et al. 2002) to obtains clones like DKO 2, 3, and 4.
A final important set of observations emerging in our present study concerns the maintenance of DNA methylation in enhancers and the relative importance of this in juxtaposition to other DNA methylation abnormalities such as promoter CpG island hypermethylation. Clearly in normal cells and cancer cells, enhancer sequences are key for control of gene expression patterns and overall control of cell phenotypes (Grimmer and Farnham 2014; Smith and Shilatifard 2014; Clermont et al. 2016). Abnormal losses or gains of enhancer DNA methylation associate with cancer risk, including for CRC (Akhtar-Zaidi et al. 2012), and features of tumor progression and aggressiveness (Kron et al. 2014). Our current results indicate, however, that there may be a complex balance in individual cancer types of abnormalities of DNA methylation in enhancers linked to specific genes that have methylated promoter CpG islands. Thus, in discrete regions such as we have studied, when abnormal gene silencing is present and DNA methylation involves both enhancers and their putative target gene promoters, the maximal reexpression of these genes requires the demethylation of both genomic elements. Thus, in cancer cells, DNA hypermethylation in proximal promoter CpG islands and distal enhancers may function independently or synergistically for transcriptional repression; this suggests the importance of detailed mapping of both promoter and enhancer DNA methylation patterns to fully decipher this balance.
Methods
Cell lines and drug treatment
HCT116 WT and isogenic DNMT mutant cell lines were maintained in McCoy's 5A medium. All other cancer cell lines were obtained from ATCC and maintained under recommended conditions. Drug treatments were performed as described (Schuebel et al. 2007), and detailed protocols are described in the Supplemental Methods.
Western blot and antibodies
Western blots were performed as described (Tsai et al. 2012). Briefly, cells were lysed with 4% SDS, and the lysates were homogenized with QIAshredder columns (Qiagen) for 2 min at 14,000 rpm before being used in immunoblotting. Antibodies are described in the Supplemental Methods.
shRNA knockdown and CRISPR knockout
All lentiviral shRNA clones were ordered from Sigma. For CRISPR genome-editing, gRNAs were cloned into the lentiCRIPSR v2 vector (Addgene plasmid #52961) based on the protocol recommended by Addgene. The generation of CRISPR knockout constructs and the production of lentivirus were performed according to the protocol recommended by Addgene. shRNA and gRNA sequences were shown in Supplemental Table S2.
Nucleic acid extraction and bisulfite sequencing
Nucleic acid extraction and bisulfite sequencing were performed as described elsewhere (Tsai et al. 2012). Genomic DNA was extracted with Wizard Genomic DNA purification kit (Promega). One microgram of genomic DNA was treated with sodium bisulfite with the EZ DNA Methylation Kit (Zymo Research). A detailed protocol is described in the Supplemental Methods.
Dot blot assay
Dot blots of 5-methylcytosine and 5-hydroxymethylcytosine were performed according to the protocol recommended by Diagenode. A detailed protocol is described in the Supplemental Methods.
Genome-wide methylation and gene expression analyses
Genome-wide DNA methylation is measured by Infinium HumanMethylation450 (HM450) BeadChip array (Illumina) according to the manufacturer's instructions. Global gene expression profiles were analyzed with Agilent Human 4×44K expression arrays (Agilent Technologies). Analyses were performed as detailed in the Supplemental Methods.
Data access
All genome-wide DNA methylation and gene expression data generated in this study have been submitted to NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE93136.
Supplementary Material
Acknowledgments
Research reported in this publication was supported by National Institutes of Environmental Health Sciences under award number R01ES011858, MDxHealth, Hodson Trust, and Samuel Waxman Cancer Research Foundation to S.B.B. and the Ministry of Science and Technology of Taiwan grant MOST 105-2628-B-002-040-MY3 to H.-C.T. We wish to thank Kathy Bender for manuscript preparation.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.208108.116.
References
- Ahuja N, Easwaran H, Baylin SB. 2014. Harnessing the potential of epigenetic therapy to target solid tumors. J Clin Invest 124: 56–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar-Zaidi B, Cowper-Sal-lari R, Corradin O, Saiakhova A, Bartels CF, Balasubramanian D, Myeroff L, Lutterbaugh J, Jarrar A, Kalady MF, et al. 2012. Epigenomic enhancer profiling defines a signature of colon cancer. Science 336: 736–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama Y, Watkins N, Suzuki H, Jair KW, van Engeland M, Esteller M, Sakai H, Ren CY, Yuasa Y, Herman JG, et al. 2003. GATA-4 and GATA-5 transcription factor genes and potential downstream antitumor target genes are epigenetically silenced in colorectal and gastric cancer. Mol Cell Biol 23: 8429–8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad N, Zahnow CA, Rudin CM, Baylin SB. 2013. The future of epigenetic therapy in solid tumours—lessons from the past. Nat Rev Clin Oncol 10: 256–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman KE, Rountree MR, Baylin SB. 2001. Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J Biol Chem 276: 32282–32287. [DOI] [PubMed] [Google Scholar]
- Bachman KE, Park BH, Rhee I, Rajagopalan H, Herman JG, Baylin SB, Kinzler KW, Vogelstein B. 2003. Histone modifications and silencing prior to DNA methylation of a tumor suppressor gene. Cancer Cell 3: 89–95. [DOI] [PubMed] [Google Scholar]
- Baubec T, Colombo DF, Wirbelauer C, Schmidt J, Burger L, Krebs AR, Akalin A, Schübeler D. 2015. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature 520: 243–247. [DOI] [PubMed] [Google Scholar]
- Baylin SB, Jones PA. 2011. A decade of exploring the cancer epigenome—biological and translational implications. Nat Rev Cancer 11: 726–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman BP, Weisenberger DJ, Aman JF, Hinoue T, Ramjan Z, Liu Y, Noushmehr H, Lange CP, van Dijk CM, Tollenaar RA, et al. 2012. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina–associated domains. Nat Genet 44: 40–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattler A, Yao L, Witt H, Guo Y, Nicolet CM, Berman BP, Farnham PJ. 2014. Global loss of DNA methylation uncovers intronic enhancers in genes showing expression changes. Genome Biol 15: 469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostick M, Kim JK, Estève PO, Clark A, Pradhan S, Jacobsen SE. 2007. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317: 1760–1764. [DOI] [PubMed] [Google Scholar]
- Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. 1999. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet 21: 103–107. [DOI] [PubMed] [Google Scholar]
- Cechova H, Lassuthova P, Novakova L, Belickova M, Stemberkova R, Jencik J, Stankova M, Hrabakova P, Pegova K, Zizkova H, et al. 2012. Monitoring of methylation changes in 9p21 region in patients with myelodysplastic syndromes and acute myeloid leukemia. Neoplasma 59: 168–174. [DOI] [PubMed] [Google Scholar]
- Challen GA, Sun D, Mayle A, Jeong M, Luo M, Rodriguez B, Mallaney C, Celik H, Yang L, Xia Z, et al. 2014. Dnmt3a and Dnmt3b have overlapping and distinct functions in hematopoietic stem cells. Cell Stem Cell 15: 350–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Ueda Y, Dodge JE, Wang Z, Li E. 2003. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol Cell Biol 23: 5594–5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Hevi S, Gay F, Tsujimoto N, He T, Zhang B, Ueda Y, Li E. 2007. Complete inactivation of DNMT1 leads to mitotic catastrophe in human cancer cells. Nat Genet 39: 391–396. [DOI] [PubMed] [Google Scholar]
- Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, Hein A, Rote NS, Cope LM, Snyder A, et al. 2015. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 162: 974–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clermont PL, Parolia A, Liu HH, Helgason CD. 2016. DNA methylation at enhancer regions: novel avenues for epigenetic biomarker development. Front Biosci (Landmark Ed) 21: 430–446. [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci 107: 21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daskalakis M, Nguyen TT, Nguyen C, Guldberg P, Kohler G, Wijermans P, Jones PA, Lübbert M. 2002. Demethylation of a hypermethylated P15/INK4B gene in patients with myelodysplastic syndrome by 5-Aza-2′-deoxycytidine (decitabine) treatment. Blood 100: 2957–2964. [DOI] [PubMed] [Google Scholar]
- Easwaran H, Tsai HC, Baylin SB. 2014. Cancer epigenetics: tumor heterogeneity, plasticity of stem-like states, and drug resistance. Mol Cell 54: 716–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger G, Jeong S, Escobar SG, Cortez CC, Li TW, Saito Y, Yoo CB, Jones PA, Liang G. 2006. Identification of DNMT1 (DNA methyltransferase 1) hypomorphs in somatic knockouts suggests an essential role for DNMT1 in cell survival. Proc Natl Acad Sci 103: 14080–14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M, Corn PG, Baylin SB, Herman JG. 2001. A gene hypermethylation profile of human cancer. Cancer Res 61: 3225–3229. [PubMed] [Google Scholar]
- Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, Suvà ML, Bernstein BE. 2016. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529: 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Lv H, Guan H, Ma X, Ji M, He N, Shi B, Hou P. 2013. Metallothionein 1G functions as a tumor suppressor in thyroid cancer through modulating the PI3K/Akt signaling pathway. BMC Cancer 13: 462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimmer MR, Farnham PJ. 2014. Can genome engineering be used to target cancer-associated enhancers? Epigenomics 6: 493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JG, Merlo A, Mao L, Lapidus RG, Issa JP, Davidson NE, Sidransky D, Baylin SB. 1995. Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res 55: 4525–4530. [PubMed] [Google Scholar]
- Hon GC, Hawkins RD, Caballero OL, Lo C, Lister R, Pelizzola M, Valsesia A, Ye Z, Kuan S, Edsall LE, et al. 2012. Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res 22: 246–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa JP. 2005. Optimizing therapy with methylation inhibitors in myelodysplastic syndromes: dose, duration, and patient selection. Nat Clin Pract Oncol 2Suppl 1: S24–S29. [DOI] [PubMed] [Google Scholar]
- Issa JP, Roboz G, Rizzieri D, Jabbour E, Stock W, O'Connell C, Yee K, Tibes R, Griffiths EA, Walsh K, et al. 2015. Safety and tolerability of guadecitabine (SGI-110) in patients with myelodysplastic syndrome and acute myeloid leukaemia: a multicentre, randomised, dose-escalation phase 1 study. Lancet Oncol 16: 1099–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao CS, Badve SS, Ulbright TM. 2014. The utility of immunostaining for NUT, GAGE7 and NY-ESO-1 in the diagnosis of spermatocytic seminoma. Histopathology 65: 35–44. [DOI] [PubMed] [Google Scholar]
- Kelly TK, De Carvalho DD, Jones PA. 2010. Epigenetic modifications as therapeutic targets. Nat Biotechnol 28: 1069–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kron KJ, Bailey SD, Lupien M. 2014. Enhancer alterations in cancer: a source for a cell identity crisis. Genome Med 6: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J, et al. 2010. DNMT3A mutations in acute myeloid leukemia. N Engl J Med 363: 2424–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Dai H, Martos SN, Xu B, Gao Y, Li T, Zhu G, Schones DE, Wang Z. 2015. Distinct roles of DNMT1-dependent and DNMT1-independent methylation patterns in the genome of mouse embryonic stem cells. Genome Biol 16: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J, Karnik R, Gu H, Ziller MJ, Clement K, Tsankov AM, Akopian V, Gifford CA, Donaghey J, Galonska C, et al. 2015. Targeted disruption of DNMT1, DNMT3A and DNMT3B in human embryonic stem cells. Nat Genet 47: 469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael AK, Harvey SL, Sammons PJ, Anderson AP, Kopalle HM, Banham AH, Partch CL. 2015. Cancer/testis antigen PASD1 silences the circadian clock. Mol Cell 58: 743–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myöhänen SK, Baylin SB, Herman JG. 1998. Hypermethylation can selectively silence individual p16ink4A alleles in neoplasia. Cancer Res 58: 591–593. [PubMed] [Google Scholar]
- Qin W, Leonhardt H, Pichler G. 2011. Regulation of DNA methyltransferase 1 by interactions and modifications. Nucleus 2: 392–402. [DOI] [PubMed] [Google Scholar]
- Rhee I, Jair KW, Yen RW, Lengauer C, Herman JG, Kinzler KW, Vogelstein B, Baylin SB, Schuebel KE. 2000. CpG methylation is maintained in human cancer cells lacking DNMT1. Nature 404: 1003–1007. [DOI] [PubMed] [Google Scholar]
- Rhee I, Bachman KE, Park BH, Jair KW, Yen RW, Schuebel KE, Cui H, Feinberg AP, Lengauer C, Kinzler KW, et al. 2002. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature 416: 552–556. [DOI] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, et al. 2015. Integrative analysis of 111 reference human epigenomes. Nature 518: 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert MF, Morin S, Beaulieu N, Gauthier F, Chute IC, Barsalou A, MacLeod AR. 2003. DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. Nat Genet 33: 61–65. [DOI] [PubMed] [Google Scholar]
- Rothbart SB, Dickson BM, Ong MS, Krajewski K, Houliston S, Kireev DB, Arrowsmith CH, Strahl BD. 2013. Multivalent histone engagement by the linked tandem Tudor and PHD domains of UHRF1 is required for the epigenetic inheritance of DNA methylation. Genes Dev 27: 1288–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roulois D, Loo Yau H, Singhania R, Wang Y, Danesh A, Shen SY, Han H, Liang G, Jones PA, Pugh TJ, et al. 2015. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell 162: 961–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rountree MR, Bachman KE, Baylin SB. 2000. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat Genet 25: 269–277. [DOI] [PubMed] [Google Scholar]
- Russler-Germain DA, Spencer DH, Young MA, Lamprecht TL, Miller CA, Fulton R, Meyer MR, Erdmann-Gilmore P, Townsend RR, Wilson RK, et al. 2014. The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer Cell 25: 442–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuebel KE, Chen W, Cope L, Glockner SC, Suzuki H, Yi JM, Chan TA, Van Neste L, Van Criekinge W, van den Bosch S, et al. 2007. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet 3: 1709–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, et al. 2007. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450: 908–912. [DOI] [PubMed] [Google Scholar]
- Sharma S, De Carvalho DD, Jeong S, Jones PA, Liang G. 2011. Nucleosomes containing methylated DNA stabilize DNA methyltransferases 3A/3B and ensure faithful epigenetic inheritance. PLoS Genet 7: e1001286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Laird PW. 2013. Interplay between the cancer genome and epigenome. Cell 153: 38–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E, Shilatifard A. 2014. Enhancer biology and enhanceropathies. Nat Struct Mol Biol 21: 210–219. [DOI] [PubMed] [Google Scholar]
- Spada F, Haemmer A, Kuch D, Rothbauer U, Schermelleh L, Kremmer E, Carell T, Längst G, Leonhardt H. 2007. DNMT1 but not its interaction with the replication machinery is required for maintenance of DNA methylation in human cells. J Cell Biol 176: 565–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Watkins DN, Jair KW, Schuebel KE, Markowitz SD, Chen WD, Pretlow TP, Yang B, Akiyama Y, Van Engeland M, et al. 2004. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet 36: 417–422. [DOI] [PubMed] [Google Scholar]
- Taguchi A, Taylor AD, Rodriguez J, Celiktas M, Liu H, Ma X, Zhang Q, Wong CH, Chin A, Girard L, et al. 2014. A search for novel cancer/testis antigens in lung cancer identifies VCX/Y genes, expanding the repertoire of potential immunotherapeutic targets. Cancer Res 74: 4694–4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting AH, Jair KW, Suzuki H, Yen RW, Baylin SB, Schuebel KE. 2004. CpG island hypermethylation is maintained in human colorectal cancer cells after RNAi-mediated depletion of DNMT1. Nat Genet 36: 582–584. [DOI] [PubMed] [Google Scholar]
- Tsai HC, Baylin SB. 2011. Cancer epigenetics: linking basic biology to clinical medicine. Cell Res 21: 502–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai HC, Li H, Van Neste L, Cai Y, Robert C, Rassool FV, Shin JJ, Harbom KM, Beaty R, Pappou E, et al. 2012. Transient low doses of DNA-demethylating agents exert durable antitumor effects on hematological and epithelial tumor cells. Cancer Cell 21: 430–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varley KE, Gertz J, Bowling KM, Parker SL, Reddy TE, Pauli-Behn F, Cross MK, Williams BA, Stamatoyannopoulos JA, Crawford GE, et al. 2013. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res 23: 555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenberger DJ, Velicescu M, Cheng JC, Gonzales FA, Liang G, Jones PA. 2004. Role of the DNA methyltransferase variant DNMT3b3 in DNA methylation. Mol Cancer Res 2: 62–72. [PubMed] [Google Scholar]
- Yan XJ, Xu J, Gu ZH, Pan CM, Lu G, Shen Y, Shi JY, Zhu YM, Tang L, Zhang XW, et al. 2011. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet 43: 309–315. [DOI] [PubMed] [Google Scholar]
- Yang X, Han H, De Carvalho DD, Lay FD, Jones PA, Liang G. 2014. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell 26: 577–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Glöckner SC, Guo M, Machida EO, Wang DH, Easwaran H, Van Neste L, Herman JG, Schuebel KE, Watkins DN, et al. 2008. Epigenetic inactivation of the canonical Wnt antagonist SRY-box containing gene 17 in colorectal cancer. Cancer Res 68: 2764–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller MJ, Gu H, Müller F, Donaghey J, Tsai LT, Kohlbacher O, De Jager PL, Rosen ED, Bennett DA, Bernstein BE, et al. 2013. Charting a dynamic DNA methylation landscape of the human genome. Nature 500: 477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.