Abstract
Preimplantation embryogenesis encompasses several critical events including genome reprogramming, zygotic genome activation (ZGA), and cell-fate commitment. The molecular basis of these processes remains obscure in primates in which there is a high rate of embryo wastage. Thus, understanding the factors involved in genome reprogramming and ZGA might help reproductive success during this susceptible period of early development and generate induced pluripotent stem cells with greater efficiency. Moreover, explaining the molecular basis responsible for embryo wastage in primates will greatly expand our knowledge of species evolution. By using RNA-seq in single and pooled oocytes and embryos, we defined the transcriptome throughout preimplantation development in rhesus monkey. In comparison to archival human and mouse data, we found that the transcriptome dynamics of monkey oocytes and embryos were very similar to those of human but very different from those of mouse. We identified several classes of maternal and zygotic genes, whose expression peaks were highly correlated with the time frames of genome reprogramming, ZGA, and cell-fate commitment, respectively. Importantly, comparison of the ZGA-related network modules among the three species revealed less robust surveillance of genomic instability in primate oocytes and embryos than in rodents, particularly in the pathways of DNA damage signaling and homology-directed DNA double-strand break repair. This study highlights the utility of monkey models to better understand the molecular basis for genome reprogramming, ZGA, and genomic stability surveillance in human early embryogenesis and may provide insights for improved homologous recombination-mediated gene editing in monkey.
Although preimplantation embryo development extends over only a few days in mammals, it encompasses successive critical events, including paternal and maternal genome reprogramming, zygotic genome activation (ZGA), and the two earliest cell-fate determination events. In addition, embryos possess unique cell cycles, particular metabolism, and a critical dependence on genomic stability during preimplantation development (Jaroudi and SenGupta 2007; Niakan et al. 2012; Lee et al. 2014). Prior to ZGA, embryonic activities rely entirely on maternal factors that also are essential for ZGA to facilitate the smooth transition from maternal-to-zygotic control of embryogenesis (Lee et al. 2014). At the 16- to 32-cell stage, blastomeres acquire apical-basal polarity, and inside cells eventually become the inner cell mass while outside cells become extraembryonic trophectoderm. Following the first cell-fate decision, the inner cell mass continues to segregate into the extra-embryonic primitive endoderm and the pluripotent epiblast that gives rise to the embryo proper (Stephenson et al. 2012). This complex preimplantation embryo development is accompanied by drastic changes in transcriptome profiles and epigenetic modifications (Niakan et al. 2012).
In mouse, considerable information has been obtained regarding maternal factors involved in genome reprogramming (Nakamura et al. 2007, 2012; Shen et al. 2014) and the maternal-to-zygotic transition (MZT) (Bultman et al. 2006; Li et al. 2008b; Messerschmidt et al. 2012; Lee et al. 2014; Yu et al. 2016a). Several key transcription factors (TFs) and signaling pathways have been implicated in initiating and/or maintaining the first and second cell-fate segregations (Home et al. 2012; Cockburn et al. 2013; Do et al. 2013; Frum et al. 2013). However, due to ethical constraints, much less is known about regulation of preimplantation embryonic development in human. This is beginning to change with several recent studies reporting on the dynamic transcriptome, DNA methylome, and lineage determination of human preimplantation embryos (Vassena et al. 2011; Xue et al. 2013; Yan et al. 2013; Guo et al. 2014; Smith et al. 2014; Petropoulos et al. 2016), but there remains a dire need for more in depth information.
The rhesus monkey (Macaca mulatta) and human share a high degree of similarity in genome sequence (92.5%–95.0%) and physiology (Yan et al. 2011). Rhesus monkeys have long been considered a reliable model to study human physiology and diseases (Hewitson and Schatten 2002; Roberts et al. 2012), as well as to evaluate preclinical safety of medical treatments (Duncan 2001; Han et al. 2009). Recent success of gene editing with CRISPR/Cas9 and TALENs in monkeys (Liu et al. 2014; Niu et al. 2014) suggests potential development of human disease models and a platform for investigating preimplantation development in nonhuman primates. However, current protocols for genome editing by homologous recombination (HR)–mediated DNA replacement in monkey one-cell embryos are extremely inefficient (Doudna and Charpentier 2014). Thus, a comprehensive map of dynamic transcriptome changes during preimplantation development is urgently needed for studying the molecular mechanisms of monkey embryogenesis, for evaluating the suitability of rhesus monkey as a model for early human embryo development, and for better understanding the deficiencies in DNA recombination. Here, we used RNA-seq to analyze the transcriptome profile of individual and pooled oocytes and preimplantation embryos in rhesus monkey.
Results
The dynamic transcriptome changes during the preimplantation development of rhesus monkey
We collected fully grown germinal vesicle (GV)–stage oocytes, metaphase II (MII)–stage oocytes, one-cell embryos at the pronucleus (PN) stage, two-cell embryos, four-cell embryos, eight-cell embryos, morulae, and blastocysts (Fig. 1A; Supplemental Table S1). RNA sequencing was conducted in each single oocyte/embryo, with two or more biological replicates at each stage. We also pooled five to 23 oocytes or embryos collected from one to 10 female monkeys at each developmental stage for RNA-seq analysis (Supplemental Table S1). These two types of RNA-seq data were analyzed separately to avoid potential batch effects, and the results were compared against each other.
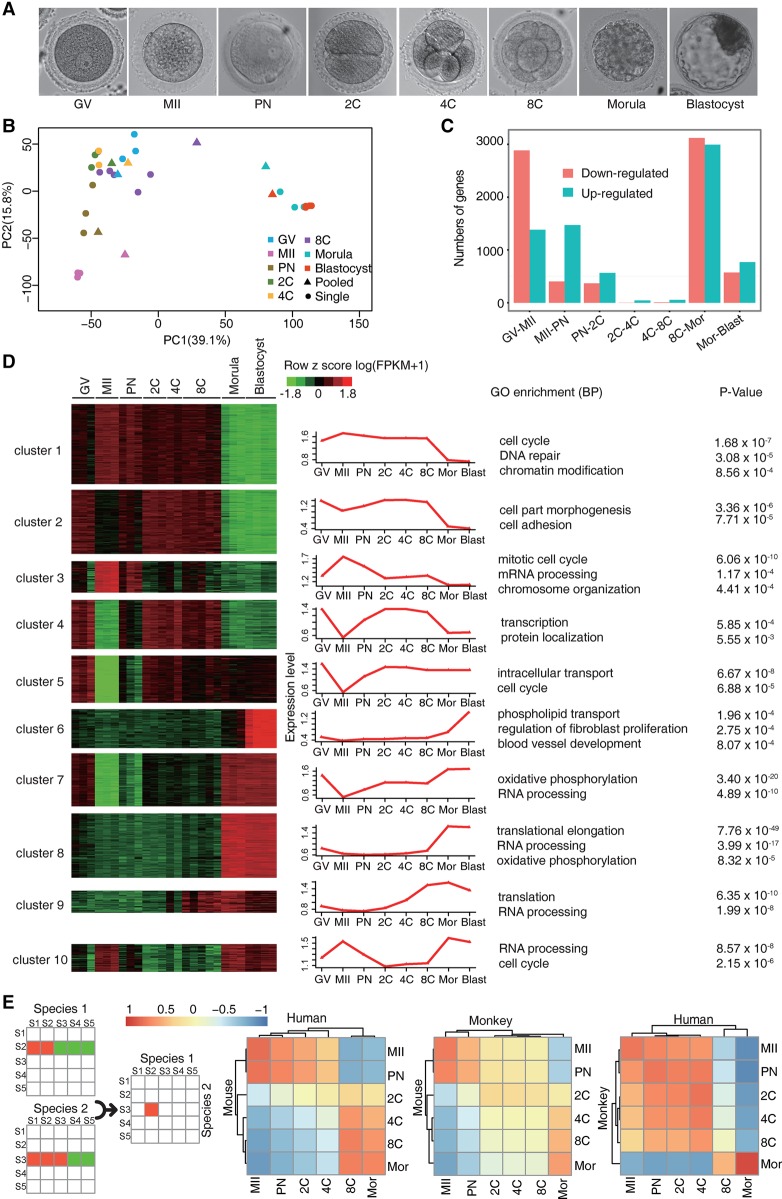
Figure 1.
Gene expression profiling of the rhesus monkey oocytes and preimplantation embryos. (A) Brightfield images of the GV oocyte (GV), mature oocyte (MII), one-cell embryo at the pronucleus stage (PN), two-cell embryo (2C), four-cell embryo (4C), eight-cell embryo (8C), morula, and blastocyst. (B) Principal component analysis (PCA) of the transcriptomes of individual- and pooled-oocyte/embryo samples. Two major clusters were identified, suggesting two discrete developmental stages. (C) Histogram showing up- and down-regulated genes between each of the two adjacent stages identified by DESeq2 (FDR < 10−5). (D) DESeq2 identified 6963 differentially expressed genes (DEGs). These DEGs were then clustered into 10 groups by BICSKmeans. Their average log transformed expression values, representative Gene Ontology (GO) terms, and corresponding enrichment P-values are listed. (E) Comparative transcriptome analysis and Pearson correlation coefficient (PCC) between species of pairwise PCCs within each species. (C–E) Based on monkey individual-oocyte/embryo sequencing data. Similar results for the pooled-embryo data are shown in Supplemental Figure S2.
By using the Illumina sequencing platform, we generated 282.3 million paired-end reads of high quality, with an average of 10.1 million reads per sample for individual-oocyte/embryo sequencing, and 28.5 million reads per sample for pooled-sample sequencing. The read quality was examined by fastQC (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc). Filtered reads were aligned to the rhesus monkey genome database (Zimin et al. 2014). In individual-oocyte/embryo samples, the numbers of detectable genes (defined as fragment per kilobase per million [FPKM] > 1) in each stage ranged from 7588–9940 within the 16,049 RefSeq genes (Supplemental Fig. S1A; Supplemental Tables S1, S2). In pooled-oocyte/embryo samples, we detected 4700–10,000 genes (Supplemental Fig. S1A; Supplemental Table S1). Examination of the FPKM distribution by violin plot (Supplemental Fig. S1B) or density plot (Supplemental Fig. S1C) revealed that the pooled-oocyte/embryo sequencing detected more weakly expressed genes than individual-oocyte/embryo sequencing, as previously reported (Kharchenko et al. 2014). We then conducted principal component analysis (PCA) on all the individual and pooled samples (Fig. 1B). The individual and pooled samples at the same developmental stage were generally clustered together. These samples fell into two clusters. One cluster contained GV, MII, PN, two-cell, four-cell, and eight-cell stage samples; the other, morula and blastocyst stage samples.
The segregation into two clusters could reflect the maternal RNA degradation as well as the major wave of ZGA at MZT, which takes place at about the eight-cell stage in monkey (Schramm and Bavister 1999). To test this hypothesis, we examined the up- and down-regulated genes between each two adjacent stages by DESeq2 (FDR < 10−5) in individual-embryo samples or by GFOLD (generalized fold change; P-values <0.01 and GFOLD > 4) (Feng et al. 2012) in pooled-embryo samples. We found about 3000 genes down-regulated during meiotic maturation (GV to MII) in both the individual- (Fig. 1C; Supplemental Table S3) and pooled-embryo data sets (Supplemental Fig. S2A). On the other hand, over 1000 genes were up-regulated at the eight-cell–morula transition in both the individual- and pooled-embryo data sets (Fig. 1C; Supplemental Fig. S2A), corresponding to the major ZGA. Thus, like in the mouse (Tadros and Lipshitz 2009), MZT in the monkey is characterized by drastic maternal mRNA decay as well as ZGA, both of which contribute to the segregation of the two clusters.
We next performed a pairwise comparison across all time points, that is, each time point is compared to every other time point, or all versus all n*(n−1)/2 comparisons, where n is the number of time points. For individual-embryo samples, we identified a total of 6963 differentially expressed genes (DEGs) between any two stages by DESeq2 (FDR < 10−5). These DEGs are clustered into 10 distinct groups by BICSKmeans (Zhang et al. 2013). The representative Gene Ontology (GO) terms for each cluster were listed (Fig. 1D; Supplemental Fig. S1D; Supplemental Table S4). Clusters 1–4 are enriched in maternal genes (specifically expressed prior to the major wave of ZGA), whereas clusters 6, 8, and 9 are enriched in zygotic genes (only expressed at/after the eight-cell stage). Genes in clusters 5, 7, and 10 are expressed both maternally and zygotically. The same analysis for the pooled-sample sequencing data identified 5560 DEGs between any two stages. These genes were grouped into 12 clusters (Supplemental Fig. S2B; Supplemental Table S5). Despite the technical differences with detection limits and batch effects, the maternal and zygotic genes identified from the single and pooled embryos show significant overlap (Fisher's exact test, P < 2.2 × 10−16) (Supplemental Fig. S2C).
To compare the similarities and differences in the overall gene expression profiles of early embryonic development among monkey, human, and mouse, we used public RNA-seq data sets (http://www.ncbi.nlm.nih.gov/geo/; accession number: GSE44183) (Xue et al. 2013) and performed comparative analysis using the reported method in Tirosh et al. (2007). We also conducted hierarchical clustering to quantitatively assess the similarities and differences. Samples were separated into two large clusters by their similarity in gene expression, corresponding to the maternal and zygotic expression clusters. By using either individual- or pooled-embryo data sets, we consistently observed the similarity in the timing of MZT between human and monkey (Fig. 1E; Supplemental Fig. S2D). These data support that rhesus monkey is more suitable than mouse in studying very early human embryonic development. In summary, the individual- and pooled-embryo data showed high consistency in their dynamic gene expression changes and DEG sets. In the following analyses, we show the individual-embryo results in the main text as they contain two or more biological replicates for each time point and, whenever relevant, show the pooled-embryo results in the Supplemental Information as further confirmation.
Maternal gene classification and stage-specific functional prediction
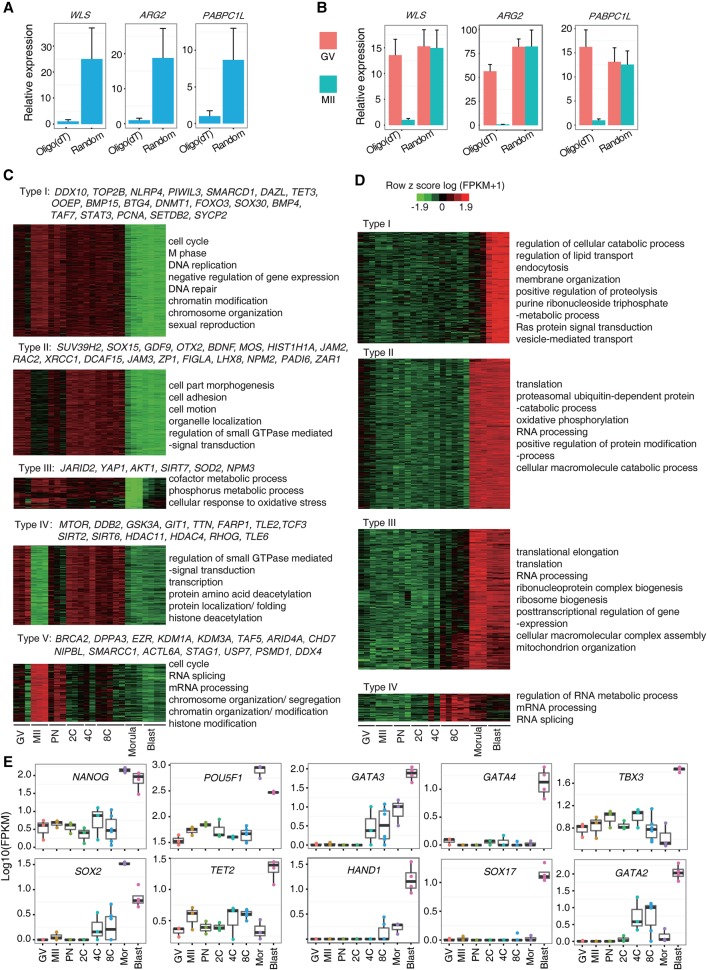
Maternal mRNAs are subject to poly(A) tail adenylation and deadenylation for stage-specific translation and functions (Stitzel and Seydoux 2007; Subtelny et al. 2014; Yu et al. 2016b). Due to the scarcity of the material, we could not directly measure the poly(A) tail length of maternal mRNAs through the PAT assay (Murray and Schoenberg 2008). But, notably, adenylation increases the efficiency of reverse transcription using oligo(dT) as primers. We therefore selected eight genes that showed lower FPKM values in MII eggs than in GV oocytes (WLS, ARG2, PABPC1L, ALKBH5, NPM2, AKR1B1, HIP1R, and LAD1 in clusters 2 and 4 in Fig. 1D). Real-time PCR using cDNA samples of MII oocytes showed that all gene expression levels were significantly higher in samples reverse-transcribed with random primers than in those prepared with oligo(dT) primers (Fig. 2A; Supplemental Fig. S3A). Consistently, gene expression levels were comparable in the GV and MII oocytes when cDNAs were prepared with random primers, whereas the expression levels were higher in GV oocytes than in MII oocytes with oligo(dT) primers (Fig. 2B; Supplemental Fig. S3B). We also tested two genes (AURKC and TMEM70 in clusters 1 and 3 in Fig. 1D) that displayed higher expression in MII oocytes than in GV oocytes in RNA-seq data. AURKC expression was slightly higher in the MII stage than in the GV stage when cDNAs were prepared with oligo(dT) primers, but was comparable at both stages when cDNAs were prepared with random primers (Supplemental Fig. S3C). Taken together, the results suggested that the poly(A) tail length was regulated in a proportion of maternal mRNAs. When using oligo(dT) reverse-transcription–based sequencing, a dramatic increase in the expression of maternal genes at stages prior to ZGA indicates active regulation of RNA polyadenylation and translation of these genes.
Figure 2.
Classification of maternal and zygotic genes. (A) Real-time PCR examination in cDNA samples of MII oocytes. Gene expression levels were significantly higher in samples reverse-transcribed with random primers than in those prepared with oligo(dT) primers. (B) Real-time PCR examination in cDNA samples of GV and MII oocytes. Gene expression levels were comparable between GV and MII oocytes when cDNAs were prepared with random primers, but showed significant difference when cDNAs were prepared with oligo(dT) primers. (C) Five expression patterns of maternal genes were identified. Typical genes and representative GO terms are listed. (D) Four expression patterns of zygotic genes were identified. Their representative GO terms are shown. (E) The expression patterns of several zygotic genes implicated in the cell-fate determination. (C–E) Based on monkey individual-oocyte/embryo sequencing data. Similar results for the pooled-embryo data are shown in Supplemental Figures S4 and S5.
By using the individual-embryo data set, we then classified the maternal genes into five different types based on their expression change patterns from the GV through to the eight-cell stages (Fig. 2C; Supplemental Tables S6, S7). Specifically, type I maternal genes might play fundamental roles in meiosis resumption and early embryo development prior to ZGA. For examples, Btg4 has been shown to directly regulate the maternal RNA decay in mouse (Yu et al. 2016b), and Tet3 is a critical enzyme involved in demethylation of the paternal and maternal genome (Shen et al. 2014). Type III genes displayed a similar pattern as type I genes, and some of them (e.g., YAP1) were shown to be important for ZGA in the mouse (Yu et al. 2016a). We also conducted maternal gene classification on the pooled-embryo data set (Supplemental Fig. S4A), and several clusters overlapped with the ones classified in the single-embryo sequencing data.
Genes with similar expression patterns in different species may have conserved functions. By comparing the monkey maternal genes identified in the individual-embryo data set to those genes reported in human and mouse (Xue et al. 2013), we found more maternal genes were shared between human and monkey (1203) than between primate and mouse (546 and 646 for human vs. mouse and monkey vs. mouse, respectively). A total of 326 common maternal genes was shared among the three species (Supplemental Fig. S4B; Supplemental Tables S8, S9), including some functionally annotated genes in the mouse, such as TP63, ZP1, ZP2, ZP3, WEE2, OOEP, BRCA1, NLRP4, TET3, GDF9, FOXO1, TCF3, and DNMT1 (Rankin et al. 2001; Su et al. 2004; Bultman et al. 2006; Suh et al. 2006; Cole et al. 2008; Li et al. 2008a; Hanna et al. 2010; Cui et al. 2012; Tsai et al. 2012; Shen et al. 2014). Similar results were obtained when using the pooled-embryo data for maternal gene conservation analysis among the three species (Supplemental Fig. S4C; Supplemental Table S10).
Differential expression patterns of zygotic genes
To study the functional time frames of zygotic genes, based on the individual-embryo data set, we classified the zygotic genes into four types and summarized their enriched GO terms or signaling pathways (Fig. 2D; Supplemental Tables S11, S12). Type IV genes showed expression activation at the eight-cell stage and might be involved in regulating ZGA. Notably, several histone variants, which play critical roles in modulating chromosome accessibility to TFs, fall into this class (Supplemental Table S11). Type I–III zygotic genes were activated after the eight-cell stage, and genes for protein synthesis, energy production, and the post-transcriptional regulation were enriched. Specifically, type I genes were highly expressed in blastocyst and might play roles in the cell-fate determination and acquisition of pluripotency. Several key TFs regulating the specification of the trophectoderm (e.g., HAND1, GATA2, and GATA3) (Bai et al. 2011) and primitive endoderm (e.g., SOX17 and GATA4) (Artus et al. 2011) belonged to type I. The pluripotency regulators TET2 and TBX3 (Han et al. 2010; Costa et al. 2013) were also highly expressed at the blastocyst stage (Fig. 2E). These data suggested that the two cell-fate determination events (trophectoderm segregation from the inner cell mass and primitive endoderm segregation from the epiblast) may take place at the blastocyst stage. Similarly, studies on human preimplantation development proposed the simultaneous segregation of the three cell lineages (trophectoderm, primitive endoderm, and epiblast) at the blastocyst stage (Petropoulos et al. 2016). However, TFs associated with pluripotency (e.g., NANOG, POU5F1, and SOX2) started their high expression at the morula stage, suggesting their roles in initiating and maintaining pluripotency (Fig. 2E).
We next classified the zygotic genes and examined the expression patterns of the above key TFs using the pooled-embryo data set. The clusters (Supplemental Fig. S4D) and the expression patterns (Supplemental Fig. S5) similar to those in individual-embryo data were obtained. We also examined the conservation of zygotic genes in the three species. The results from the individual-embryo (Supplemental Fig. S4E; Supplemental Table S13) and pooled-embryo (Supplemental Fig. S4F; Supplemental Table S14) data sets showed a similar pattern of conserved zygotic genes.
Potential TFs regulating preimplantation development
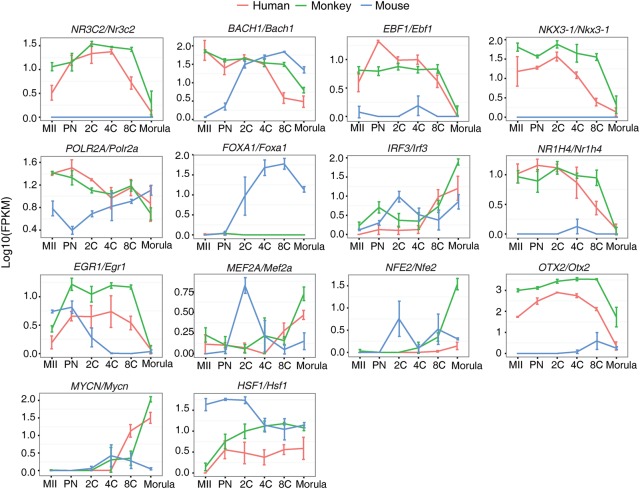
TFs play critical roles in regulating embryo development. We employed the software Hypergeometric Optimization of Motif Enrichment (HOMER) on the above-identified 6963 DEGs to find the potential TFs implicated in early development regulation. We only focused on the highly expressed TFs (FPKM > 5 in at least one developmental stage among the three species). We identified 60 TFs in our monkey individual-embryo data and 50 and 40 TFs in human and mouse data (Supplemental Table S15; Xue et al. 2013). To understand the differences and similarities of the developmental regulation by TFs among the three species, we then compared the expression patterns of these 142 TFs. Distribution of pairwise Pearson correlation coefficient (PCC) between any two species showed that human and monkey were more similar (Kolmogorov-Smirnov test, P-value <1 × 10−5) (Supplemental Fig. S6A). We further identified 19 TFs with expression highly correlated between human and monkey (P-value <0.05 by permutation), but not between primate and mouse (P-value >0.05) (Fig. 3; Supplemental Fig. S6B; Supplemental Table S16). Additional analysis with rank correlation coefficient (RCC), which is insensitive to outliers, revealed that 11 out of 19 TFs were identified by both PCC and RCC (IRF3, NR3C2, NR2C2, TFDP1, E2F1, MYCN, NFE2, EGR1, POLR2A, BACH1 and NKX3-1). Among the 19 TFs, many are involved in the regulation of apoptosis, stress response and epigenetic modification, such as HSF1 (Dayalan Naidu et al. 2015), IRF3 (Freaney et al. 2013), BACH1 (Cantor et al. 2001; Litman et al. 2005), EGR1 (Virolle et al. 2001), NKX3-1, MYCN (Slack et al. 2005; Lei et al. 2006), and EBF1 (Gao et al. 2009).
Figure 3.
Transcription factors (TFs) with differential expression patterns between primate and rodent. The pairwise PCC was calculated. TFs, which have P-value <0.05 between human and monkey and P-value >0.05 between human and mouse or between monkey and mouse, were considered to be differentially expressed between primate and rodent. This figure is based on monkey individual-oocyte/embryo sequencing data. Similar results for the pooled-embryo data are shown in Supplemental Figure S7.
We performed the same analyses using the pooled-sample data set. Consistent with the single-embryo results, human and monkey are more similar in the expression patterns of motif-enriched TFs (Kolmogorov-Smirnov test, P-value <0.001) (Supplemental Fig. S7A). Among the 13 TFs correlatively expressed between human and monkey (P-value <0.05 by permutation), but not between primate and mouse (P-value >0.05) (Supplemental Fig. S7B), almost half of them are also involved in the regulation of apoptosis, stress response, and epigenetic modification (HSF1, ATF3, IRF3, BCLAF1, BMI1 and SUZ12) (Sarras et al. 2010; Lee et al. 2012; Benetatos et al. 2014). Taken together, the differential expression patterns of these TFs between primate and rodent suggest that primate and rodent might possess distinct adaptive and protective abilities to cope with cytotoxic and genotoxic stress during the early stages of embryogenesis.
Interaction networks between the maternal and zygotic control of embryonic development
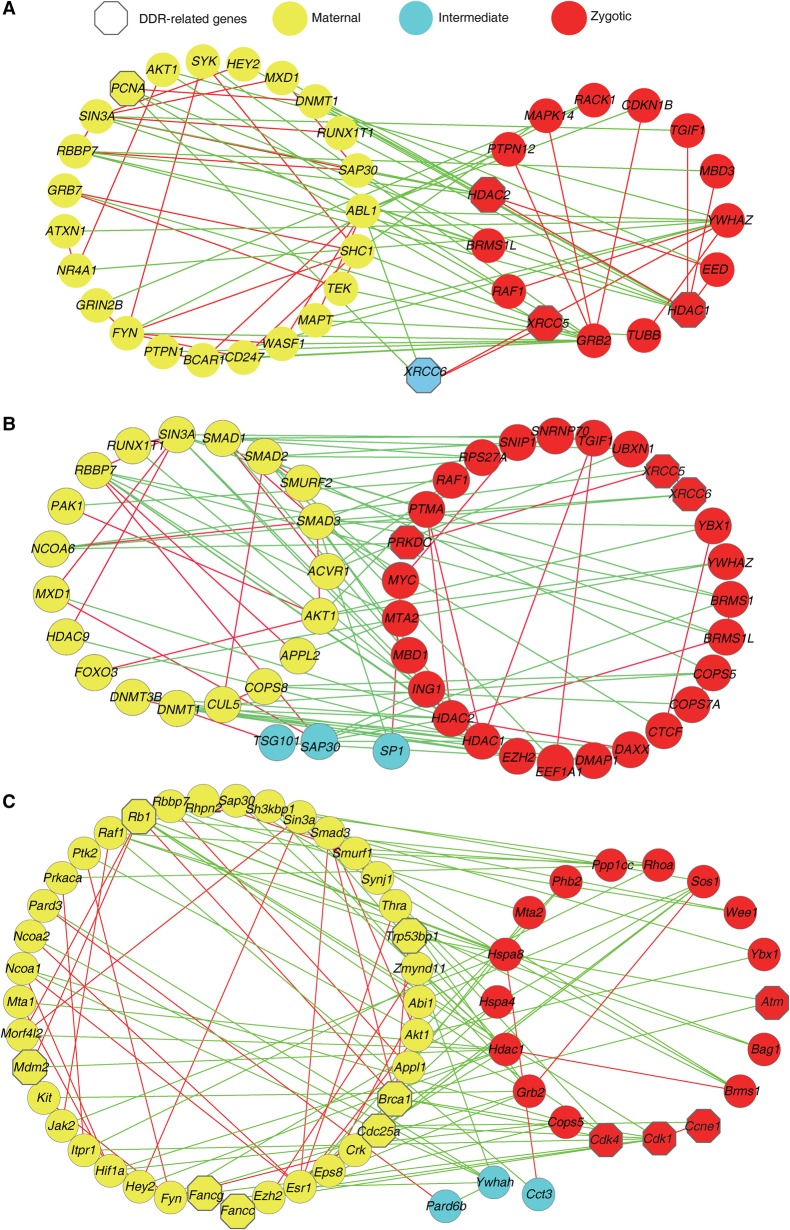
To investigate the regulatory circuitry controlling MZT, we employed a negative-positive (NP) network approach (Xia et al. 2006; Xue et al. 2007; Huang et al. 2015), which is particularly useful for extracting network modules that may regulate the switch between two alternative temporal phases, in our case, the transition from maternal-to-zygotic control. The NP network is essentially a subnetwork within the template protein–protein interaction (PPI) network, where only PPIs between genes that are positively or negatively transcriptionally correlated during a temporal switch (in our case, during the time course of MZT) are included. Based on the PPIs from Human Protein Reference Database (HPRD), we used the individual-sample data set and constructed a monkey NP network comprising 763 correlated interactions (PCC > 0.8, P-value <0.05 by permutation) and 433 anti-correlated interactions (PCC < −0.8, P-value <0.05) among 1064 genes. This interaction network is composed of three modules: a maternal module containing maternal genes, a zygotic module containing zygotic genes, and an intermediate module containing genes expressed in both the maternal and zygotic stages (Supplemental Fig. S8A; Supplemental Table S17). Notably, more positive edges were observed within the modules, while negative edges were dominant between the modules, especially between the maternal and zygotic modules. This NP network reflected the mutually exclusive relationship between the maternal and zygotic genes. The overrepresented GO terms of the genes in maternal and zygotic modules are listed in Supplemental Figure S8A. Comparison of these functional enrichments suggested that embryos prior to and post ZGA were different in their DNA damage responses (DDRs).
Genes that are anti-correlated and interact between alternatively expressed modules are defined as interface genes in the NP network and are found to be enriched in regulatory functions (Xue et al. 2007). Indeed, the interface genes between the maternal and zygotic modules are primarily involved in cell cycle, RNA splicing, negative regulation of protein metabolic process, intracellular transport, protein localization, and cellular response to stress (Supplemental Table S18). As regulatory interactions frequently form feedback loops, we explored all the three-node loops traversing the maternal-zygotic interface by requiring at least one negative edge within the loop. A total of 72 genes formed 158 feedback loops, which in turn formed four subnetworks (Fig. 4A; Supplemental Fig. S8B; Supplemental Table S19). In these subnetworks, 22 genes (HDAC1, HDAC2, SMAD4, EP300, RPS27A, SMAD2, DNMT1, ABL1, LEF1, XRCC6, YWHAZ, FYN, SIN3A, PCNA, TGFBR1, SMAD7, SHC1, SMURF2, BCAR1, SAP30, RBBP7, and GRB2) have connections with at least four other genes, implying that they might be hubs and play critical roles in early development. Analysis of the largest subnetwork (Fig. 4A) showed that the zygotic hub gene GRB2 was negatively connected with 10 maternal genes and positively correlated with three zygotic or intermediate genes. GRB2 is highly expressed in the morula and blastocyst. It represses NANOG and is essential for the establishment of primitive endoderm lineage in the mouse blastocyst (Cheng et al. 1998; Chazaud et al. 2006; Hamazaki et al. 2006). The negative correlation of GRB2 with maternal genes suggests that a subset of maternal genes cooperate to suppress the inappropriate initiation of cell-fate commitment genes. Interestingly, GRB2 is positively correlated with MAPK14. Suppression of MAPK14 facilitates the derivation of human naive pluripotent stem cells (Gafni et al. 2013). Maternal hub PCNA is negatively connected with zygotic and intermediate genes. PCNA is a cofactor of DNA polymerase and plays important roles in DNA replication and cell proliferation (Kelman 1997; Strzalka and Ziemienowicz 2011). Of interest, most of PCNA’s negatively correlated genes (e.g., XRCC5, XRCC6, and HDAC1) are involved in DDR and DNA damage repair (Gu et al. 1997; Schulte-Uentrop et al. 2008; Miller et al. 2010), implying that monkey oocytes and early embryos prior to ZGA might undergo progression and proliferation at the expense of efficient DNA damage repair. Moreover, only one core component gene of DDR and repair was expressed prior to ZGA in this network (XRCC6), whereas more core DDR genes were detected post ZGA (XRCC5, XRCC6, HDAC1, and HDAC2).
Figure 4.
Feedback loops extracted from NP networks of monkey, human, and mouse. (A) The largest feedback loop in the monkey. The three-node loops were explored by requiring at least one negative edge within the loop. Red or green edges represent that the two nodes are transcriptionally correlated or anti-correlated, respectively. Node color indicates the module to which the gene belongs. Node shape indicates whether a gene is related (hexagon) or unrelated (circle) to the DNA damage response (DDR). (B) The largest feedback loop in humans. (C) The largest feedback loop in the mouse. This figure is based on monkey individual-oocyte/embryo sequencing data. Similar results for the pooled-embryo data are shown in Supplemental Figure S9.
To confirm these findings, we performed the same analyses on the pooled-sample data set. The functional enrichment of genes in each module is highly consistent between the individual- and pooled-sample data sets (Supplemental Fig. S9A). Similar hub genes including GRB2 and PCNA were identified in subnetworks, and more DDR hub genes were expressed post ZGA (Supplemental Fig. S9B). Thus, these data collectively suggested that monkey embryo development prior to ZGA might have less robust control over genomic stability.
Distinct capacity for HR-mediated DNA double-strand break repair between primate and rodent
To examine if similar regulatory circuitry exists in human and mouse, we performed NP network analysis and identified the three-node loops traversing the maternal-zygotic interfaces in human (Fig. 4B; Supplemental Fig. S10) and in mouse (Fig. 4C; Supplemental Fig. S11). Like in monkey (Fig. 4A; Supplemental Figs. S8B, S9), few maternally expressed node genes in the feedback loops of human are involved in the DDR (Fig. 4B; Supplemental Fig. S10). In contrast, many maternally expressed hub genes in mouse NP network are key components of the DDR and repair (e.g., Fancc, Fancg, Brca1, Trp53bp1, Mdm2, and Cdc25a) (Fig. 4C; Supplemental Fig. S11). These results suggested that a rodent might have a superior competence to that of a primate in maintaining genomic stability in oocytes and early embryos prior to ZGA.
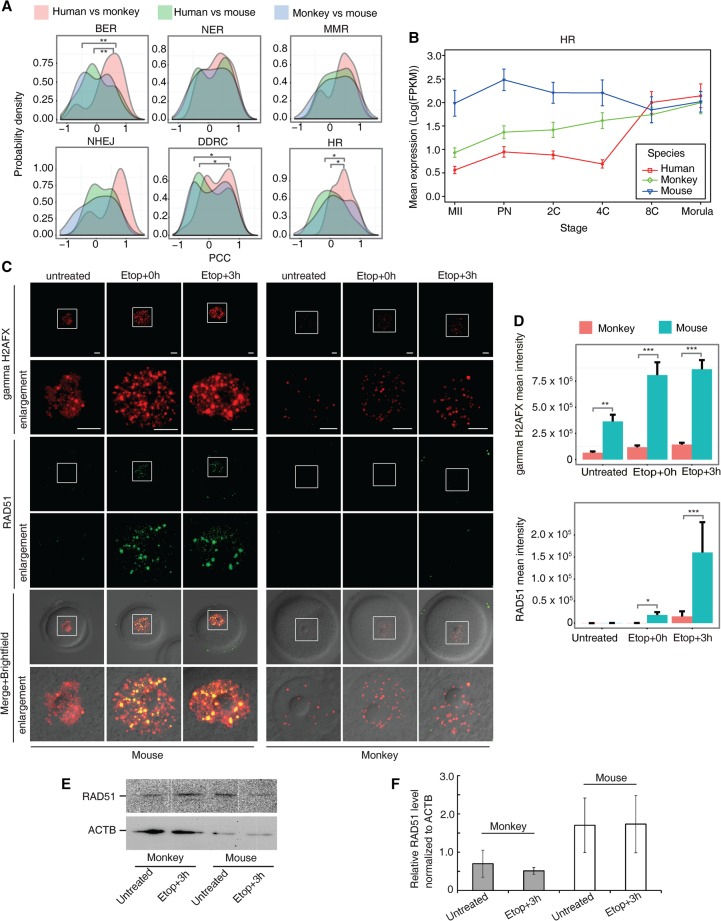
To test this hypothesis, we examined the expression patterns of 190 “DNA damage response”–related genes in the three species. Genes showing high expression level (FPKM ≥ 5) in any two developmental stages within each of the three species were included in the analysis. Based on their functions and involved pathways in DDR as previously described (Beerman et al. 2014), these genes were classified into six groups, including DNA damage response and checkpoints (DDRC), HR-mediated DNA repair, nonhomologous end-joining DNA repair (NHEJ), nucleotide excision repair (NER), mismatch repair (MMR), and base excision repair (BER). By utilizing the monkey individual-embryo sequencing data set, 152 out of 190 genes were analyzed (Supplemental Fig. S12). Comparisons among the three species revealed that the overall expression patterns of genes in the BER, DDRC, and HR groups were statistically different between primates and mouse (P < 0.05, Kolmogorov-Smirnov test) but were similar between monkey and human (Fig. 5A; Supplemental Table S20). Similarly, when using the monkey pooled-sample sequencing data set, 174 out of the 190 DDR genes were analyzed (Supplemental Fig. S13). Again, genes involved in DDRC and HR groups were significantly different between primates and mouse (P < 0.05, Kolmogorov-Smirnov test) but were similar between monkey and human (Supplemental Fig. S14A). These results not only support the hypothesis that primate and rodent embryos possess distinct competence in DNA damage repair but also pinpoint the pathways in which they differ. Of interest, the key HR genes (EME1, RAD51, RAD54L, RECQL, SHFM1, UBA2, and XRCC2), which play central roles in HR-mediated DNA double-strand break (DSB) repair, showed distinct expression patterns between primates and mouse (Supplemental Table S20). These genes were highly expressed post ZGA in monkey and human but equally expressed prior to and post ZGA in mouse (Fig. 5B; Supplemental Fig. S14B). This suggested that oocytes and early cleavage embryos prior to ZGA in primates have poorer HR-mediated DNA DSB repair capacity than in the mouse.
Figure 5.
Comparison of six groups of DDR and HR-mediated repair genes among three species. (A) Distribution of pair-wise PCC. Genes in the BER, DDRC, and HR groups were statistically different between the primates and mouse (P-value <0.05, Kolmogorov-Smirnov test). (B) Mean log-transformed expression pattern of HR-related genes (EME1/Eme1, RAD51/Rad51, RAD54L/Rad54l, RECQL/Recql, SHFM1/Shfm1, UBA2/Uba2, and XRCC2/Xrcc2) in human, monkey, and mouse. (C) Examination of gamma H2AFX and RAD51 foci in untreated and etoposide-treated GV oocytes recovered for 0 h (Etop+0h) and 3 hr (Etop+3h). More gamma H2AFX foci were observed in mouse GV oocytes than in monkey GV oocytes after etoposide treatment. Consistently, more mouse oocytes accumulated RAD51 on damage sites, whereas fewer monkey oocytes had RAD51 foci formation. Images in squares are enlarged in the enlargement panels. (D) Quantification of fluorescence foci intensity of gamma H2AFX (top) and RAD51 (bottom). Intensity was normalized by the number of oocytes examined. (E) Immunoblotting analysis of RAD51 and ACTB protein level in mouse and monkey GV oocytes untreated or treated with etoposide followed by 3-h recovery. (F) Quantification of RAD51 protein level by normalization to ACTB. Data are represented as mean ± SEM. Scale bar, 10 μm. (*) P-value <0.05, (**) P-value <0.001, (***) P-value <0.0001, two-tailed t-test. (A,B) Based on monkey individual-oocyte/embryo sequencing data. Similar results for the pooled-embryo data are shown in Supplemental Figure S14.
To further validate this finding, we compared the response of GV oocytes to DNA DSB between monkey and mouse by immunostaining with antibodies against gamma H2AFX (phosphorylated histone H2AFX at serine 139) and RAD51. Upon DSB, gamma H2AFX is induced and required for eliciting downstream damage responses (Rai et al. 2007; Jackson and Bartek 2009). RAD51 is a key recombinase in HR-mediated DSB repair and recruited to DSB sites (Shrivastav et al. 2008). GV oocytes from the monkey and mouse were subject to the same etoposide treatment to induce a similar level of DSB (Nagy and Soutoglou 2009). As shown in Figure 5C, in the untreated condition, a few gamma H2AFX foci were detected in both the monkey and mouse GV oocytes, and no RAD51 foci were observed (more than 60 oocytes were examined in each species). Interestingly, etoposide treatment elicited different damage responses in mouse and monkey oocytes. More gamma H2AFX foci were induced in mouse oocytes than in monkey oocytes at both 0 and 3 h post treatment. Of note, immediately after treatment, 71.4% (n = 28) mouse oocytes but only 28.1% (n = 32) monkey oocytes contained more than five RAD51 foci colocalized with gamma H2AFX. At 3 h post treatment, 83.6% mouse oocytes (n = 61) versus 38.7% (n = 31) monkey oocytes displayed RAD51 accumulation on damage sites. Foci intensity quantification further confirmed more focal gamma H2AFX and RAD51 in mouse oocytes than in monkey oocytes (Fig. 5D). Moreover, immunoblotting analysis showed that RAD51 protein expression was higher in mouse oocytes than in monkey oocytes in either untreated or etoposide-treated conditions (Fig. 5E,F). These results indicate that in response to similar level of DSB treatment, monkey oocytes have a lower ability to evoke efficient DNA damage signaling and HR-mediated DSB repair than mouse oocytes.
Discussion
Several recent studies have reported on the gene expression dynamics of human preimplantation embryos by microarray or single blastomere RNA sequencing (Vassena et al. 2011; Xue et al. 2013; Yan et al. 2013; Petropoulos et al. 2016). But no effort has been made to dissect the genome-wide RNA expression profiling and molecular characteristics of preimplantation embryos of rhesus monkey, the most suitable nonhuman primate animal model. In the present study, all representative stages of oocytes and preimplantation embryos, including GV oocyte through to blastocyst, were investigated by individual- and pooled-oocyte/embryo RNA sequencing. For the first time, we report the genome-wide gene expression dynamics in rhesus monkey oocytes and preimplantation embryos and evaluate the feasibility of utilizing the rhesus monkey in studying early human embryonic development. We also explore the gene expression features of MZT. Moreover, we provide a list of maternal as well as zygotic genes, and the classification of these genes helps to narrow down the stages where they might function.
The comparison of human, rhesus monkey, and mouse embryos reveals similarities and differences between primate and rodent. Fundamental biological processes of embryo development prior to and post ZGA are preserved among the species. For instance, regulation of protein localization and transport, cell cycle, mitosis, and cytoskeleton organization is prominent in developmental stages prior to ZGA, whereas regulation of RNA processing/splicing, translation, mitochondria, and energy metabolism is predominant in the stages post ZGA. We looked carefully at the differences between primate and rodent and emphasize that oocytes and early embryos of primate and rodent possess different DNA damage response and repair abilities. DDR is an evolutionary conserved mechanism to preserve genome stability and ensure the success of organism development. Maintaining the genome integrity is of particular significance to oocytes and early embryos. DNA strand break, especially DSB, is lethal to cells and must be repaired immediately and properly. DSB can be repaired by the NHEJ or HR pathway. The NHEJ pathway is error-prone and cell cycle independent, whereas the HR pathway has high fidelity but is restricted to the S and G2 phases (Rai et al. 2007; Jackson and Bartek 2009). In this study, our results show that HR repair of DSB in oocytes and early embryos is more efficient in mouse than in primates. First, NP network analysis identifies more maternal hub genes involved in DNA damage response and repair in the mouse than in primates. Second, genes participating in DNA damage signaling, cell cycle checkpoint, and HR-mediated DNA DSB repair display a differential expression pattern between primates and mouse. Specifically, core components of the HR repair pathway exhibit a zygotic expression pattern in the monkey and human but are equally expressed in all stages of oocytes and preimplantation embryos in mouse. Third, a similar level of DSB damage elicits lower levels of gamma H2AFX production in monkey GV oocytes than in mouse GV oocytes. Consistently, a lower levels of RAD51 is expressed and recruited to DSB sites in monkey oocytes than in mouse oocytes. Overall the evidence suggests that monkey GV oocytes have a reduced ability to conduct DDR and HR-directed repair. The HR pathway is the primary mode of DNA DSB repair in oocytes (Sung and Klein 2006). Accumulating evidence supports a causal relationship between the decrease in the HR-mediated DNA DSB repair and the decline of oocyte quantity and quality in mouse and human (Perry et al. 2013; Titus et al. 2013; Day et al. 2015). Therefore, HR-based DNA DSB repair in oocytes and early embryos plays a key role in regulating female reproductive performance. DNA DSB are often associated with chromosome instability (Roukos and Misteli 2014). Our study provides an explanation for the higher rates of embryo chromosome instability and wastage in primates than in any other species ever studied (Dupont et al. 2009; Vanneste et al. 2009). In addition, this finding implies that the efficiency of precise gene editing by HR-based gene replacement in monkey is intrinsically lower than that in mouse. Thus, increasing HR efficiency (e.g., by forced expression of recombinase RAD51) in one-cell embryos may help to improve the efficiency of precise gene editing in monkey.
Methods
Animals
Young adult rhesus monkeys (7 to 14 yr old) with a successful reproductive history were used in this study. All experimental procedures and animal care were performed according to the protocols approved by the ethics committee of the Kunming Institute of Zoology, Chinese Academy of Sciences.
Identification of DEGs
In the individual-embryo samples, DEGs were identified by the package DESeq2 (Love et al. 2014). For the pooled-embryo sequencing, DEGs were identified by GFOLD (Feng et al. 2012). Details of parameters and cutoffs used in the present study are included in the Supplemental Methods.
Enrichment of TF motifs
We used HOMER (Heinz et al. 2010) to identify TFs whose motifs are enriched in each cluster using the following criteria: P-values <0.01 based on hypergeometric distribution, and offset from transcription start site is −1000 to 500. Motif length is set to six, eight, 10, or 12.
NP network analysis
The NP networks were constructed according to the method described previously (Xia et al. 2006; Xue et al. 2007; Huang et al. 2015). To extract the NP network from the HPRD database, we first calculated the PCC of each pair of PPIs based on gene expression level. PCC > 0.8 or PCC < −0.8 is used as positive or negative correlation cutoff, based on permutation (P-value <0.05). To extract feedback loops within this NP network, we explored all the three-node loops traversing the maternal-zygotic interface by requiring at least one negative edge within the loop.
Comparison of gene expression among three species
Comparison among the three species was conducted according to the methods previously described (Tirosh et al. 2007) with the procedures illustrated in Figure 1E. Details are included in Supplemental Methods.
Other methods
Details of the methods on sample collection, low input cDNA amplification, RNA sequencing, read mapping and gene quantification, BICSKmeans clustering, quantitative RT-PCR, etoposide treatment of GV oocytes, immunofluorescence staining, and immunoblotting are included in Supplemental Methods.
Data access
RNA-seq data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE86938. Source codes, including normalization, DEG analysis, cross-species comparison, and NP-network analysis, are available at Supplemental_Source codes file.
Supplementary Material
Acknowledgments
We thank Prof. Jurrien Dean at the National Institutes of Health for critical comments on the manuscript; and Prof. Ji-rong Long at Vanderbilt University and Dr. Ian Logan for their assistance with editing the paper. This work was supported by the “Strategic Priority Research Program” of the CAS, grant numbers XDA01010203 and XDB13000000 to P.Z., and grants from CAS XDA01010303, China Ministry of Science and Technology 2015CB964803, and National Natural Science Foundation of China 91329302, 31210103916, and 91519330 to J.-D.J.H.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.198044.115.
References
- Artus J, Piliszek A, Hadjantonakis AK. 2011. The primitive endoderm lineage of the mouse blastocyst: sequential transcription factor activation and regulation of differentiation by Sox17. Dev Biol 350: 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai H, Sakurai T, Someya Y, Konno T, Ideta A, Aoyagi Y, Imakawa K. 2011. Regulation of trophoblast-specific factors by GATA2 and GATA3 in bovine trophoblast CT-1 cells. J Reprod Dev 57: 518–525. [DOI] [PubMed] [Google Scholar]
- Beerman I, Seita J, Inlay MA, Weissman IL, Rossi DJ. 2014. Quiescent hematopoietic stem cells accumulate DNA damage during aging that is repaired upon entry into cell cycle. Cell Stem Cell 15: 37–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benetatos L, Vartholomatos G, Hatzimichael E. 2014. Polycomb group proteins and MYC: the cancer connection. Cell Mol Life Sci 71: 257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultman SJ, Gebuhr TC, Pan H, Svoboda P, Schultz RM, Magnuson T. 2006. Maternal BRG1 regulates zygotic genome activation in the mouse. Genes Dev 20: 1744–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor SB, Bell DW, Ganesan S, Kass EM, Drapkin R, Grossman S, Wahrer DCR, Sgroi DC, Lane WS, Haber DA, et al. 2001. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell 105: 149–160. [DOI] [PubMed] [Google Scholar]
- Chazaud C, Yamanaka Y, Pawson T, Rossant J. 2006. Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Dev Cell 10: 615–624. [DOI] [PubMed] [Google Scholar]
- Cheng AM, Saxton TM, Sakai R, Kulkarni S, Mbamalu G, Vogel W, Tortorice CG, Cardiff RD, Cross JC, Muller WJ, et al. 1998. Mammalian Grb2 regulates multiple steps in embryonic development and malignant transformation. Cell 95: 793–803. [DOI] [PubMed] [Google Scholar]
- Cockburn K, Biechele S, Garner J, Rossant J. 2013. The Hippo pathway member Nf2 is required for inner cell mass specification. Curr Biol 23: 1195–1201. [DOI] [PubMed] [Google Scholar]
- Cole MF, Johnstone SE, Newman JJ, Kagey MH, Young RA. 2008. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev 22: 746–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa Y, Ding J, Theunissen TW, Faiola F, Hore TA, Shliaha PV, Fidalgo M, Saunders A, Lawrence M, Dietmann S, et al. 2013. NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature 495: 370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Li Y, Zhu L, Liu D, Songyang Z, Wang HY, Wang RF. 2012. NLRP4 negatively regulates type I interferon signaling by targeting the kinase TBK1 for degradation via the ubiquitin ligase DTX4. Nat Immunol 13: 387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day FR, Ruth KS, Thompson DJ, Lunetta KL, Pervjakova N, Chasman DI, Stolk L, Finucane HK, Sulem P, Bulik-Sullivan B, et al. 2015. Large-scale genomic analyses link reproductive aging to hypothalamic signaling, breast cancer susceptibility and BRCA1-mediated DNA repair. Nat Genet 47: 1294–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayalan Naidu S, Kostov RV, Dinkova-Kostova AT. 2015. Transcription factors Hsf1 and Nrf2 engage in crosstalk for cytoprotection. Trends Pharmacol Sci 36: 6–14. [DOI] [PubMed] [Google Scholar]
- Do DV, Ueda J, Messerschmidt DM, Lorthongpanich C, Zhou Y, Feng B, Guo G, Lin PJ, Hossain MZ, Zhang W, et al. 2013. A genetic and developmental pathway from STAT3 to the OCT4–NANOG circuit is essential for maintenance of ICM lineages in vivo. Genes Dev 27: 1378–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. 2014. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346: 1258096. [DOI] [PubMed] [Google Scholar]
- Duncan J. 2001. An adaptive coding model of neural function in prefrontal cortex. Nat Rev Neurosci 2: 820–829. [DOI] [PubMed] [Google Scholar]
- Dupont C, Froenicke L, Lyons LA, Bavister BD, Brenner CA. 2009. Chromosomal instability in rhesus macaque preimplantation embryos. Fertil Steril 91: 1230–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Meyer CA, Wang Q, Liu JS, Shirley Liu X, Zhang Y. 2012. GFOLD: a generalized fold change for ranking differentially expressed genes from RNA-seq data. Bioinformatics 28: 2782–2788. [DOI] [PubMed] [Google Scholar]
- Freaney JE, Kim R, Mandhana R, Horvath CM. 2013. Extensive cooperation of immune master regulators IRF3 and NFκB in RNA Pol II recruitment and pause release in human innate antiviral transcription. Cell Rep 4: 959–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frum T, Halbisen MA, Wang C, Amiri H, Robson P, Ralston A. 2013. Oct4 cell-autonomously promotes primitive endoderm development in the mouse blastocyst. Dev Cell 25: 610–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafni O, Weinberger L, Mansour AA, Manor YS, Chomsky E, Ben-Yosef D, Kalma Y, Viukov S, Maza I, Zviran A, et al. 2013. Derivation of novel human ground state naive pluripotent stem cells. Nature 504: 282–286. [DOI] [PubMed] [Google Scholar]
- Gao H, Lukin K, Ramirez J, Fields S, Lopez D, Hagman J. 2009. Opposing effects of SWI/SNF and Mi-2/NuRD chromatin remodeling complexes on epigenetic reprogramming by EBF and Pax5. Proc Natl Acad Sci 106: 11258–11263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Seidl KJ, Rathbun GA, Zhu C, Manis JP, van der Stoep N, Davidson L, Cheng HL, Sekiguchi JM, Frank K, et al. 1997. Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity 7: 653–665. [DOI] [PubMed] [Google Scholar]
- Guo H, Zhu P, Yan L, Li R, Hu B, Lian Y, Yan J, Ren X, Lin S, Li J, et al. 2014. The DNA methylation landscape of human early embryos. Nature 511: 606–610. [DOI] [PubMed] [Google Scholar]
- Hamazaki T, Kehoe SM, Nakano T, Terada N. 2006. The Grb2/Mek pathway represses Nanog in murine embryonic stem cells. Mol Cell Biol 26: 7539–7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Qian X, Bernstein JG, Zhou HH, Franzesi GT, Stern P, Bronson RT, Graybiel AM, Desimone R, Boyden ES. 2009. Millisecond-timescale optical control of neural dynamics in the nonhuman primate brain. Neuron 62: 191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Yuan P, Yang H, Zhang J, Soh BS, Li P, Lim SL, Cao S, Tay J, Orlov YL, et al. 2010. Tbx3 improves the germ-line competency of induced pluripotent stem cells. Nature 463: 1096–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna CB, Yao S, Patta MC, Jensen JT, Wu X. 2010. WEE2 is an oocyte-specific meiosis inhibitor in rhesus macaque monkeys. Biol Reprod 82: 1190–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitson L, Schatten G. 2002. The use of primates as models for assisted reproduction. Reprod Biomed Online 5: 50–55. [DOI] [PubMed] [Google Scholar]
- Home P, Saha B, Ray S, Dutta D, Gunewardena S, Yoo B, Pal A, Vivian JL, Larson M, Petroff M, et al. 2012. Altered subcellular localization of transcription factor TEAD4 regulates first mammalian cell lineage commitment. Proc Natl Acad Sci 109: 7362–7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Yu X, Sun N, Qiao N, Cao Y, Boyd-Kirkup JD, Shen Q, Han JD. 2015. Single-cell-level spatial gene expression in the embryonic neural differentiation niche. Genome Res 25: 570–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SP, Bartek J. 2009. The DNA-damage response in human biology and disease. Nature 461: 1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaroudi S, SenGupta S. 2007. DNA repair in mammalian embryos. Mutat Res 635: 53–77. [DOI] [PubMed] [Google Scholar]
- Kelman Z. 1997. PCNA: structure, functions and interactions. Oncogene 14: 629–640. [DOI] [PubMed] [Google Scholar]
- Kharchenko PV, Silberstein L, Scadden DT. 2014. Bayesian approach to single-cell differential expression analysis. Nat Methods 11: 740–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YY, Yu YB, Gunawardena HP, Xie L, Chen X. 2012. BCLAF1 is a radiation-induced H2AX-interacting partner involved in γH2AX-mediated regulation of apoptosis and DNA repair. Cell Death Dis 3: e359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MT, Bonneau AR, Giraldez AJ. 2014. Zygotic genome activation during the maternal-to-zygotic transition. Annu Rev Cell Dev Biol 30: 581–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Q, Jiao J, Xin L, Chang C-J, Wang S, Gao J, Gleave ME, Witte ON, Liu X, Wu H. 2006. NKX3.1 stabilizes p53, inhibits AKT activation, and blocks prostate cancer initiation caused by PTEN loss. Cancer Cell 9: 367–378. [DOI] [PubMed] [Google Scholar]
- Li L, Baibakov B, Dean J. 2008a. A subcortical maternal complex essential for preimplantation mouse embryogenesis. Dev Cell 15: 416–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Ito M, Zhou F, Youngson N, Zuo X, Leder P, Ferguson-Smith AC. 2008b. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev Cell 15: 547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litman R, Peng M, Jin Z, Zhang F, Zhang J, Powell S, Andreassen PR, Cantor SB. 2005. BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell 8: 255–265. [DOI] [PubMed] [Google Scholar]
- Liu H, Chen Y, Niu Y, Zhang K, Kang Y, Ge W, Liu X, Zhao E, Wang C, Lin S, et al. 2014. TALEN-mediated gene mutagenesis in rhesus and cynomolgus monkeys. Cell Stem Cell 14: 323–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messerschmidt DM, de Vries W, Ito M, Solter D, Ferguson-Smith A, Knowles BB. 2012. Trim28 is required for epigenetic stability during mouse oocyte to embryo transition. Science 335: 1499–1502. [DOI] [PubMed] [Google Scholar]
- Miller KM, Tjeertes JV, Coates J, Legube G, Polo SE, Britton S, Jackson SP. 2010. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol 17: 1144–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray EL, Schoenberg DR. 2008. Assays for determining poly(A) tail length and the polarity of mRNA decay in mammalian cells. Methods Enzymol 448: 483–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy Z, Soutoglou E. 2009. DNA repair: easy to visualize, difficult to elucidate. Trends Cell Biol 19: 617–629. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Arai Y, Umehara H, Masuhara M, Kimura T, Taniguchi H, Sekimoto T, Ikawa M, Yoneda Y, Okabe M, et al. 2007. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat Cell Biol 9: 64–71. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Liu YJ, Nakashima H, Umehara H, Inoue K, Matoba S, Tachibana M, Ogura A, Shinkai Y, Nakano T. 2012. PGC7 binds histone H3K9me2 to protect against conversion of 5mC to 5hmC in early embryos. Nature 486: 415–419. [DOI] [PubMed] [Google Scholar]
- Niakan KK, Han J, Pedersen RA, Simon C, Pera RA. 2012. Human pre-implantation embryo development. Development 139: 829–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang L, Kang Y, Zhao X, Si W, Li W, et al. 2014. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 156: 836–843. [DOI] [PubMed] [Google Scholar]
- Perry JR, Corre T, Esko T, Chasman DI, Fischer K, Franceschini N, He C, Kutalik Z, Mangino M, Rose LM, et al. 2013. A genome-wide association study of early menopause and the combined impact of identified variants. Hum Mol Genet 22: 1465–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petropoulos S, Edsgard D, Reinius B, Deng Q, Panula SP, Codeluppi S, Plaza Reyes A, Linnarsson S, Sandberg R, Lanner F. 2016. Single-cell RNA-seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell 165: 1012–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai R, Peng G, Li K, Lin SY. 2007. DNA damage response: the players, the network and the role in tumor suppression. Cancer Genomics Proteomics 4: 99–106. [PubMed] [Google Scholar]
- Rankin TL, O'Brien M, Lee E, Wigglesworth K, Eppig J, Dean J. 2001. Defective zonae pellucidae in Zp2-null mice disrupt folliculogenesis, fertility and development. Development 128: 1119–1126. [DOI] [PubMed] [Google Scholar]
- Roberts VH, Rasanen JP, Novy MJ, Frias A, Louey S, Morgan TK, Thornburg KL, Spindel ER, Grigsby PL. 2012. Restriction of placental vasculature in a non-human primate: a unique model to study placental plasticity. Placenta 33: 73–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roukos V, Misteli T. 2014. The biogenesis of chromosome translocations. Nat Cell Biol 16: 293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarras H, Alizadeh Azami S, McPherson JP. 2010. In search of a function for BCLAF1. ScientificWorldJournal 10: 1450–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm RD, Bavister BD. 1999. Onset of nucleolar and extranucleolar transcription and expression of fibrillarin in macaque embryos developing in vitro. Biol Reprod 60: 721–728. [DOI] [PubMed] [Google Scholar]
- Schulte-Uentrop L, El-Awady RA, Schliecker L, Willers H, Dahm-Daphi J. 2008. Distinct roles of XRCC4 and Ku80 in non-homologous end-joining of endonuclease- and ionizing radiation-induced DNA double-strand breaks. Nucleic Acids Res 36: 2561–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Inoue A, He J, Liu Y, Lu F, Zhang Y. 2014. Tet3 and DNA replication mediate demethylation of both the maternal and paternal genomes in mouse zygotes. Cell Stem Cell 15: 459–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastav M, De Haro LP, Nickoloff JA. 2008. Regulation of DNA double-strand break repair pathway choice. Cell Res 18: 134–147. [DOI] [PubMed] [Google Scholar]
- Slack A, Chen Z, Tonelli R, Pule M, Hunt L, Pession A, Shohet JM. 2005. The p53 regulatory gene MDM2 is a direct transcriptional target of MYCN in neuroblastoma. Proc Natl Acad Sci 102: 731–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Chan MM, Humm KC, Karnik R, Mekhoubad S, Regev A, Eggan K, Meissner A. 2014. DNA methylation dynamics of the human preimplantation embryo. Nature 511: 611–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson RO, Rossant J, Tam PP. 2012. Intercellular interactions, position, and polarity in establishing blastocyst cell lineages and embryonic axes. Cold Spring Harb Perspect Biol 4: a008235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitzel ML, Seydoux G. 2007. Regulation of the oocyte-to-zygote transition. Science 316: 407–408. [DOI] [PubMed] [Google Scholar]
- Strzalka W, Ziemienowicz A. 2011. Proliferating cell nuclear antigen (PCNA): a key factor in DNA replication and cell cycle regulation. Ann Bot 107: 1127–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y-Q, Wu X, O'Brien MJ, Pendola FL, Denegre JN, Matzuk MM, Eppig JJ. 2004. Synergistic roles of BMP15 and GDF9 in the development and function of the oocyte–cumulus cell complex in mice: genetic evidence for an oocyte–granulosa cell regulatory loop. Dev Biol 276: 64–73. [DOI] [PubMed] [Google Scholar]
- Subtelny AO, Eichhorn SW, Chen GR, Sive H, Bartel DP. 2014. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature 508: 66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh EK, Yang A, Kettenbach A, Bamberger C, Michaelis AH, Zhu Z, Elvin JA, Bronson RT, Crum CP, McKeon F. 2006. p63 protects the female germ line during meiotic arrest. Nature 444: 624–628. [DOI] [PubMed] [Google Scholar]
- Sung P, Klein H. 2006. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat Rev Mol Cell Biol 7: 739–750. [DOI] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD. 2009. The maternal-to-zygotic transition: a play in two acts. Development 136: 3033–3042. [DOI] [PubMed] [Google Scholar]
- Tirosh I, Bilu Y, Barkai N. 2007. Comparative biology: beyond sequence analysis. Curr Opin Biotechnol 18: 371–377. [DOI] [PubMed] [Google Scholar]
- Titus S, Li F, Stobezki R, Akula K, Unsal E, Jeong K, Dickler M, Robson M, Moy F, Goswami S, et al. 2013. Impairment of BRCA1-related DNA double-strand break repair leads to ovarian aging in mice and humans. Sci Transl Med 5: 172ra121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C-C, Su P-F, Huang Y-F, Yew T-L, Hung S-C. 2012. Oct4 and Nanog directly regulate Dnmt1 to maintain self-renewal and undifferentiated state in mesenchymal stem cells. Mol Cell 47: 169–182. [DOI] [PubMed] [Google Scholar]
- Vanneste E, Voet T, Le Caignec C, Ampe M, Konings P, Melotte C, Debrock S, Amyere M, Vikkula M, Schuit F, et al. 2009. Chromosome instability is common in human cleavage-stage embryos. Nat Med 15: 577–583. [DOI] [PubMed] [Google Scholar]
- Vassena R, Boue S, Gonzalez-Roca E, Aran B, Auer H, Veiga A, Izpisua Belmonte JC. 2011. Waves of early transcriptional activation and pluripotency program initiation during human preimplantation development. Development 138: 3699–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virolle T, Adamson ED, Baron V, Birle D, Mercola D, Mustelin T, de Belle I. 2001. The Egr-1 transcription factor directly activates PTEN during irradiation-induced signalling. Nat Cell Biol 3: 1124–1128. [DOI] [PubMed] [Google Scholar]
- Xia K, Xue H, Dong D, Zhu S, Wang J, Zhang Q, Hou L, Chen H, Tao R, Huang Z, et al. 2006. Identification of the proliferation/differentiation switch in the cellular network of multicellular organisms. PLoS Comput Biol 2: e145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue H, Xian B, Dong D, Xia K, Zhu S, Zhang Z, Hou L, Zhang Q, Zhang Y, Han JD. 2007. A modular network model of aging. Mol Syst Biol 3: 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Z, Huang K, Cai C, Cai L, Jiang CY, Feng Y, Liu Z, Zeng Q, Cheng L, Sun YE, et al. 2013. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 500: 593–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan G, Zhang G, Fang X, Zhang Y, Li C, Ling F, Cooper DN, Li Q, Li Y, van Gool AJ, et al. 2011. Genome sequencing and comparison of two nonhuman primate animal models, the cynomolgus and Chinese rhesus macaques. Nat Biotechnol 29: 1019–1023. [DOI] [PubMed] [Google Scholar]
- Yan L, Yang M, Guo H, Yang L, Wu J, Li R, Liu P, Lian Y, Zheng X, Yan J, et al. 2013. Single-cell RNA-seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol 20: 1131–1139. [DOI] [PubMed] [Google Scholar]
- Yu C, Ji SY, Dang YJ, Sha QQ, Yuan YF, Zhou JJ, Yan LY, Qiao J, Tang F, Fan HY. 2016a. Oocyte-expressed yes-associated protein is a key activator of the early zygotic genome in mouse. Cell Res 26: 275–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Ji SY, Sha QQ, Dang Y, Zhou JJ, Zhang YL, Liu Y, Wang ZW, Hu B, Sun QY, et al. 2016b. BTG4 is a meiotic cell cycle–coupled maternal-zygotic-transition licensing factor in oocytes. Nat Struct Mol Biol 23: 387–394. [DOI] [PubMed] [Google Scholar]
- Zhang W, Liu Y, Sun N, Wang D, Boyd-Kirkup J, Dou X, Han JD. 2013. Integrating genomic, epigenomic, and transcriptomic features reveals modular signatures underlying poor prognosis in ovarian cancer. Cell Rep 4: 542–553. [DOI] [PubMed] [Google Scholar]
- Zimin AV, Cornish AS, Maudhoo MD, Gibbs RM, Zhang X, Pandey S, Meehan DT, Wipfler K, Bosinger SE, Johnson ZP, et al. 2014. A new rhesus macaque assembly and annotation for next-generation sequencing analyses. Biol Direct 9: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.