Abstract
Background and Aims Pisum sativum L. (pea) seed is a source of carbohydrate and protein for the developing plant. By studying pea seeds inoculated by the cytokinin-producing bacterium, Rhodococcus fascians, we sought to determine the impact of both an epiphytic (avirulent) strain and a pathogenic strain on source–sink activity within the cotyledons during and following germination.
Methods Bacterial spread was monitored microscopically, and real-time reverse transcription–quantitative PCR was used to determine the expression of cytokinin biosynthesis, degradation and response regulator gene family members, along with expression of family members of SWEET, SUT, CWINV and AAP genes – gene families identified initially in pea by transcriptomic analysis. The endogenous cytokinin content was also determined.
Key Results The cotyledons infected by the virulent strain remained intact and turned green, while multiple shoots were formed and root growth was reduced. The epiphytic strain had no such marked impact. Isopentenyl adenine was elevated in the cotyledons infected by the virulent strain. Strong expression of RfIPT, RfLOG and RfCKX was detected in the cotyledons infected by the virulent strain throughout the experiment, with elevated expression also observed for PsSWEET, PsSUT and PsINV gene family members. The epiphytic strain had some impact on the expression of these genes, especially at the later stages of reserve mobilization from the cotyledons.
Conclusions The pathogenic strain retained the cotyledons as a sink tissue for the pathogen rather than the cotyledon converting completely to a source tissue for the germinating plant. We suggest that the interaction of cytokinins, CWINVs and SWEETs may lead to the loss of apical dominance and the appearance of multiple shoots.
Keywords: Apical dominance, amino acid transporter, cell wall invertase, cytokinin, cytokinin oxidase/dehydrogenase, Pisum sativum L., pea, Rhodococcus fascians, seed, sink and source, SWEET, sucrose transporter
INTRODUCTION
Changes in source–sink dynamics are an integral component of the plant life cycle. During its developmental phase, the seed of an annual plant is the major sink for assimilates. In contrast, during germination, the seed changes to function as a source for the developing seedling. However, impacting on the developmental control of source–sink relationships exercised by the plant is the challenge invoked by plant pathogens. Here, cellular, biochemical and physiological changes may occur within the plant, as the pathogen competes with the plant for assimilates.
For many years, the cytokinins have been functionally implicated in source–sink dynamics during plant development (Mothes and Engelbrecht, 1963; Ehness et al., 1997; Balibrea Lara et al., 2004; Werner and Schmülling, 2009; Jameson and Song, 2016), as well as being implicated in plant–pathogen (bacteria, fungal and viral) interactions and particularly in the establishment of a suitable niche for gall-forming pathogens (Jameson, 2000). In plants, cytokinins are biosynthesized directly by isopentenyl transferase (IPT) attaching an isoprenoid side chain to the N6 position of an adenine nucleotide moiety. The cytokinin side chain may become hydroxylated or unsaturated. The cytokinin nucleotides may be activated to the free base forms by LONELY GUY (LOG), or to the riboside forms. The cytokinin molecule may be inactivated by the removal of the side chain by cytokinin oxidase/dehydrogenase (CKX), or metabolized to storage O-glucosides (which can be released to active forms by β-glucosidases) or 7- and 9-glucosides (which appear not to be reactivated). The free bases are considered the active forms perceived by cytokinin receptors (Lomin et al., 2015) and the ribosides the translocated forms. Cytokinins may also be formed by tRNA-IPTs. Cytokinins released from tRNA may supplement the endogenous cytokinin pool. For recent overviews, see Spichal (2012) and Jameson (2017).
Germinating seeds are metabolically highly active, degrading carbohydrate, lipid and proteins stores and mobilizing these to the embryo and from there to the emerging root and shoot (Weitbrecht et al., 2011). Pea seeds, which store starch and protein, exhibit increased respiration and metabolic activity within hours of imbibition (Nawa and Asahi, 1971), followed by reserve mobilization as roots and shoots emerge (Weitbrecht et al., 2011). The embryo axis of germinating lupin seeds is capable of biosynthesizing cytokinin (Nandi et al., 1988; Nandi and Palni, 1989). The cytokinins, which were transported to the cotyledons, were shown to be highly stable and to induce cotyledon expansion and chlorophyll synthesis (Nandi and Palni, 1989).
As our earlier work had noted that the cotyledons of pea (Pisum sativum L.) infected with a virulent strain of Rhodococcus fascians became green and retained their integrity (Eason et al., 1995), we hypothesized that cytokinins emanating from the bacterium induced the chlorophyll synthesis, and that the bacterium also caused the germinating pea to change from being a source of metabolites for the developing root and shoot to becoming a sink of metabolites for the pathogen. Rhodococcus fascians is a Gram-positive, non-host-specific, bacterium that can exist as both epiphytic and endophytic colonies. To determine the impact of R. fascians inoculation of germinating pea seeds on cytokinin biosynthesis and metabolism, we analysed the expression of PsIPT, PsCKX and PsLOG gene family members in P. sativum simultaneously with the expression of the R. fascians fas genes [RfIPT (FasD), RfLOG (FasF) and RfCKX (FasE); Pertry et al., 2010; Stes et al., 2011] using the primers designed specifically to discriminate between the plant and microbial cytokinin genes. The expression of P. sativum response regulator (PsRR) genes was also monitored to assess if the cytokinin signal transduction pathway was affected by R. fascians, and the endogenous cytokinins identified and quantified.
A transcriptome analysis conducted by Depuydt et al. (2009) showed both upregulated and downregulated genes following infection of arabidopsis by virulent strain D188 of R. fascians, including upregulation of genes coding for sugar transporter proteins, but downregulation of amino acid transporter family proteins. Importantly, Depuydt et al. (2009) also showed that the avirulent strain D188-5 had an impact on primary metabolism and, recently, Francis et al. (2016) suggested that D188-5 has a plant growth-promoting effect. To assess the impact of R. fascians on the source–sink dynamics of the germinating pea, expression of sucrose and amino acid transporter gene family members (SUT and AAP), cell wall invertase (CWINV) and SWEET were monitored. Enhanced expression of CWINV has been reported in several plant–pathogen interactions (Berger et al., 2007; Depuydt et al., 2009; Chandran et al., 2010; Siemens et al., 2011). Interaction between cytokinins and CWINV is well documented, in relation to both cell division (Roitsch and González, 2004; Jameson and Song, 2016) and source–sink dynamics (Ehneß and Roitsch, 1997; Albacete et al., 2015). Cytokinins failed to delay senescence when CWINV was inhibited (Balibrea Lara et al., 2004), indicating that nutrient mobilization is essential in cytokinin-induced senescence delay, providing the basis for pathogen-induced ‘Green Island’ formation (Roitsch and González, 2004), and potentially the greening of the infected pea cotyledons.
We also chose to monitor the SWEET (Sugar Will Eventually be Exported Transporter) gene family as it was recently identified as a sugar efflux protein (Chen et al., 2010) and pathogens have been shown to modulate sugar efflux in the host plant to promote their own growth (Chandran, 2015; Eom et al., 2015). The SWEET family members cluster into four clades (Eom et al., 2015). Different SWEET gene family members transport hexoses (Chen et al., 2010) and sucrose (Chen et al., 2012). Clade II members are hexose transporters and Clade III transport sucrose, in both cases across the plasma membrane to the apoplasm, whereas members of Clade IV transport hexoses across the tonoplast into the vacuole (Guo et al., 2014). Clade I members transport hexoses and are localized either to the plasma membrane (Chen et al., 2010) or to the tonoplast (Chen et al., 2015). When SWEET function is blocked, growth and virulence of some pathogens have been shown to be reduced. For example, work with bacterial blight of rice caused by Xanthomonas oryzae pv. oryzae indicates that this pathogen enhances the release of sucrose from the host cells by inducing expression of OsSWEET13, a member of Clade III. Targeted mutation of OsSWEET13 led to reduced pathogenesis (Zhou et al., 2015). Earlier it was shown that two other Clade III SWEETs (OsSWEET11 and OsSWEET14) were also targets of bacterial rice blight (Li et al., 2013; Streubel et al., 2013).
Chandran (2015) suggested that co-option of SWEETs may be a ‘universal strategy adopted by diverse types of pathogens’ and that different pathogens have co-opted different SWEET gene family members to provide carbohydrate. However, in contrast to this, recently Chen et al. (2015) showed that mutants of AtSWEET2 were more susceptible to Pythium infection. They suggested that expression in the roots of the Clade I member, AtSWEET2, restricted the amount of glucose in the apoplast, and by extension the rhizosphere, by sequestering it to the vacuole. Upon infection by Pythium irregulare, expression of AtSWEET2 and AtSWEET10 increased. AtSWEET2 accumulated in the region of the tonoplast of the root cells where the Pythium was concentrated, a mechanism upregulated, the authors suggest, to sequester glucose away from the pathogen and into the vacuole. Additionally, an interaction between sucrose-transporting SWEETs and CWINV has also been invoked for pathogens that take up hexoses preferentially (Chandran, 2015).
In addition to carbon, microbes require a source of nitrogen (Fagard et al., 2014). Pratelli and Pilot (2014) suggested that pathogen infection leads to changes in expression of genes involved in amino acid metabolism and transport. They also tabulated the effect of nematode infection on AtAAP gene family members, showing that these responded differentially (induced or repressed) by infection. As functional studies have shown that AtAAPs can transport a wide range of amino acids (Tegeder, 2012; Tegeder and Ward, 2012), we were interested in whether these transporters were activated following inoculation by R. fascians.
It has been noted that cytokinin-mediated resistance in arabidopsis and tobacco operates differently (Großkinsky et al., 2011), and consequently may also differ in legumes. We chose to work with pea as it has historically been the test organism for R. fascians infection (Lacey, 1936). As R. fascians is a soil-borne microbe, we inoculated pea seeds at the time of imbibition with R. fascians avirulent strain 589 and virulent strain 602. Strain 602 is highly virulent (Eason et al., 1996; Stange et al., 1996), and has a 210 kb linear plasmid containing RfIPT, RfLOG and RfCKX, whereas avirulent strain 589 lacks a linear plasmid and all three genes (Galis et al., 2005; Stange et al., 1996; Supplementary Data Table S1). Both strains extrude multiple cytokinin types into the culture medium, with the production of non-hydroxylated [isopentenyl adenine (iP)-type] cytokinins about 1000 times greater than that of hydroxylated [zeatin (Z)-type] cytokinins (Eason et al., 1996). Along with a microscopic investigation, gene expression and endogenous cytokinins were monitored in the imbibed and germinating seeds inoculated by the R. fascians strains and mock-inoculated controls.
MATERIALS AND METHODS
Plant material and Rhodococcus fascians strains
Pisum sativum variety Bohatyr was used as the host plant. Whole, surface-sterilized seeds were added to mid-log phase cultures of virulent (602) and avirulent (589) strains of R. fascians growing in ‘523’ broth (Kado and Heskett, 1970) and incubated at 26 °C with shaking at 121 rpm for 4 h, a time frame coinciding with phase I water uptake during imbibition (Green and Baisted, 1972). Five to six inoculated seeds, and mock-inoculated controls, were then placed in sterilized 500 mL containers with 0·6 % (w/v) agar and 10 % (w/v) Hoagland’s mineral salts solution (Lawson et al., 1982). The containers were placed in a growth room in a randomized block design, with three treatments (control, avirulent and virulent) and ten sampling times [4 hours post-inoculation (hpi), and 2, 5, 9, 11, 15, 20, 25, 30, 35 and 40 days post-inoculation (dpi)], each with five replications. The plants were grown at 22 °C with a 16 h photoperiod. Within 24 h, the radicle had emerged and by 2 dpi germination was complete: radicles and small plumules had both emerged from the seeds. Samples of whole plants or separated cotyledons, roots and shoots were taken for a variety of purposes, but at 2 dpi the entire germinating seed was extracted. The samples were either flash frozen in liquid nitrogen and stored at −80 °C for gene expression studies, fixed in FAA (10 % formaldehyde:5 % acetic acid:50 % alcohol) for light microscopy or cryopreserved for scanning electron microscopy (SEM). For each sampling, five plants were collected from each treatment (control, avirulent and virulent). The experiment was repeated twice, with the same procedures and sampling.
Light microscopy
A modified procedure of Carletom and Druvy (1957) for fixation, dehydration, embedding and microtoming was followed. Serial sections of the embedded tissue blocks were cut to a thickness of 10–13 μm on a Lecia RM2165 microtome. After de-waxing and hydration, the sections were stained with Mayer’s Haemalum (double strength) for 5 min. Sections were counter-stained with 0·5 % (v/v) Eosin Y (Sigma) in 100 % ethanol. Sections were then treated with 1:1 xylene:absolute ethanol, followed by 100 % xylene at room temperature for 3 min, mounted in Eukitt and dried in a 37 °C oven for a minimum of 2 d (Kitin et al., 2005). The stained sections were viewed using a Zeiss Axio Imager M1 compound microscope with brightfield (BF) and differential interference contrast (DIC) optics/lighting capabilities. Micrographs were captured using a Zeiss AxioCam HRc cooled CCD camera at 3900 × 3090 pixel resolution to facilitate later digital enlargement of specific areas to allow visualization of the fine details of R. fascians.
RNA isolation and cDNA synthesis
RNA was extracted from pea tissues using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. The integrity and quality of isolated RNA were assessed by electrophoresis on a 1 % (w/v) agarose gel. The concentration and purity of RNA were assessed using a Nano Drop™ Spectrophotometer. The RNA was stored in 1× RNA secure™ in a –20 °C freezer. The extracted RNA was converted to cDNA by reverse transcription, using approx. 1 μg of total RNA, 50 U of Expand Reverse Transcriptase (Roche, Mannheim Germany), 50 pmol of oligo(dT) primers, and 100 pmol of random hexamer (pdN6) primers in a 20 μL reaction. The cDNA was diluted 5-fold with TE buffer and stored at –20 °C. The cDNA quality was assessed by real-time reverse transcription–quantitative PCR (RT–qPCR), where two reference genes and a target gene were used to assess the amplification curve and melting point curve of each cDNA sample.
RNA was isolated from cultures of R. fascians strains. Mid-log phase cultures were inoculated into ‘523’ broth and incubated at 26 °C with shaking at 150 rpm for 4 h. The broth cultures were centrifuged at 4 °C, 15 000 rpm for 15 min. The pellets were suspended in TRIzol reagent. The procedure for plant RNA and cDNA synthesis was then followed as described above. Ribosome RNA 18S was used as the reference gene.
Gene isolation and sequence analysis
Sequences of family members of candidate genes of interest were identified from RNA sequencing (RNA-Seq) transcriptomic data. An RNA pool of combined RNA samples, each extracted separately from pea cotyledons, shoots, roots, flowers and pods, and seeds at various developmental stages, was used to construct the cDNA library, which was then sequenced using an Illumina HiSeq2000 genome analyser at the Beijing Genome Institute (BGI) Customer Service.
The RNA-Seq generated a transcriptome of 4·69 Gb clean data and >52 181 000 cleaned pair-end reads, which were assembled into 95 199 contigs. In total, nearly 42 300 unigene sequences were obtained, with a median length of 777 bp. This suggests that the transcriptome data represented adequate sequencing depth and genome coverage to meet the requirement of identifying candidate gene sequences expressed in the tissue samples in the current study.
For the identification of IPT, LOG, CKX, RR, SUT, INV, SWEET and AAP gene sequences in pea, all annotated family members of these multigene families from Arabidopsis thaliana and the available legume orthologues from Glycine max, Medicago truncatula, Lotus japonicus and P. sativum were used as query sequences to BLAST search the transcriptome data. The putative sequences were verified to gene family level by BLAST searching the GenBank database (http://blast.ncbi.nlm.nih.gov) and multiple sequence alignment with representative orthologue sequences.
In the case of R. fascians, the virulent strain (602) was used as the template and primers were designed from the NCBI database to identify IPT, LOG and CKX genes, with other microbial gene orthologues being selected and used for alignment. For phylogenetic analysis, orthologues from arabidopsis, legumes, maize, rice and R. fascians of each gene of interest were used to construct a Maximum Parsimony tree with either 1000 bootstrap replicates (ClustalX; Thompson et al., 1997) or 10 000 bootstrap replicates with MEGA4 (Tamura et al., 2007). Each tree was rooted with an outgroup orthologue.
Polymerase chain reaction
The PCR primers specific to each family member of the genes of interest were designed using Primer Premier 5.0 based on the sequence information obtained from the transcriptome. PCRs were performed in a 20 μL mix containing 1 μL of 25 mm MgCl2, 1 μL of 10 pmol μL–1 of each forward and reverse primer, 2 μL of 10-fold diluted cDNA, 2 μL of 10× Taq buffer and 0·2 μL of 5 U μL–1 Taq polymerase (Roche), using a PCR program consisting of 35 cycles of 94 °C for 40 s, 50–60 °C for 40 s, 72 °C for 40 s followed by one cycle of 72 °C for 5 min, then held at 4 °C. Thermocycling was performed on either a BIOrad DNA Engine Peltier Thermal Cycler or an MJ Research PTC-200 Peltier Thermal Cycler. The PCR products were separated on a 1 % (w/v) agarose gel and all the bands of approximately the expected size were purified with an UltraClean™ 15 DNA Purification Kit and sequenced at MACROGEN Inc., Korea.
Real-time reverse transcription–quantitative PCR
The expression studies were conducted following the RT–qPCR MIQE guidelines (Bustin et al., 2009), with three technical replicates for each of two biological replicates. Primers designed for RT–qPCR were based on the PCR product sequences, and were unique and specific for the gene of interest. Primers were designed using Primer Premier 6.00 to obtain a PCR product length of around 200 bp. Primer sequences are listed in Supplementary Data Table S2. The PCR products were sequenced by MACROGEN Inc. Korea. For each gene of interest, raw sequences of at least two forward and two reverse directions were used to correct sequencing errors (Song et al., 2012). Sequence verification after sequencing was done for each specific target gene with BLAST (http://www.ncbi.nlm.gov/BLAST) searching the GenBank database, and each confirmed target gene was selected and designated to gene family level based on available annotated sequence or putative sequence of the target genes. The nucleotide sequences reported herein are included in Table S2.
The primers for RT–qPCR were optimized by determining the optimal annealing temperature of all the target primers and reference genes. A duplicate, no-template control was included in every run to test for DNA contamination and to assess for primer-dimers. Relative gene expression was obtained using Rotor-Gene Q (Qiagen) using either a home-made SYBR Green master mix or the KAPA SYBR® FAST qPCR Kits (Kapa Biosystems, Boston, MA, USA). A reaction volume of 10 μL was used, consisting of 5 μL of Kapa qPCR buffer (Kapa Biosystems), 2 μL of millipore water, 1 μL of cDNA and 1 μL of each forward and reverse primer. The thermal cycle consisted of an initial hold at 95 °C for 10 min, followed by 40 cycles of 95 °C for 10 s, 58 °C for 15 s and 72 °C for 20 s; then 90 s of pre-melt conditioning at 72 °C followed by melt of 72 °C to 95 °C, raising the temperature by 1 °C every 5 s.
Reference genes were used as internal controls to normalize the data by correcting for differences in quantity of cDNA used as template (Vandesompele et al., 2002; Gutierrez et al., 2008). To achieve accurate normalization, four reference genes, Ps18S, PsEF, PsGAP and PsACT, were used. The reference genes were first compared for their expression stability over different cDNA samples from tissues at different growth stages, with three replicates. The geometric mean of four reference genes was calculated according to the method described by Song et al. (2012), and its correlation to the target gene was compared with every gene of interest.
Relative gene expression was calculated based on the method of Pfaffl and Hageleit (2001) and Song et al. (2012). Gene expression was determined by comparing the expression of the gene of interest/target gene with the average expression of the four reference genes. Due to the fact that all samples could not be run in a single PCR run, an inter-run calibrator was used (Derveaux et al., 2010) which was composed of mixed cDNA from all tissues used in the project. The data analysis was done as described by Song et al. (2012). For each reference gene, a correction factor (CF) for each of the cDNA samples was calculated using the Ct value of this cDNA sample divided by the average Ct value of all cDNA samples of the same experiment. The values of three technical replicates were averaged to form the CF for each biological replicate of each reference gene. The final CF value for each biological replicate was created by averaging the CF value of four reference genes. For each cDNA sample, the Ct value of each target gene was corrected before analysis of its expression level. The expression data are presented as heat maps, with fold differences calculated relative to the mock-inoculated control.
Cytokinin analyses
Cotyledons from four individual plants were ground under liquid nitrogen and freeze-dried. The four biological replicates were extracted and purified using the method published previously by Dobrev and Kamı´nek (2002) with some minor modifications (Antoniadi et al., 2015). Samples of 3–5·5 mg d. wt were extracted in 1 mL of modified Bieleski buffer (60 % MeOH, 10 % HCOOH and 30 % H2O) together with a cocktail of 18 stable isotope-labelled cytokinin internal standards (0·2 pmol of cytokinin bases, ribosides, N-glucosides, 0·5 pmol of O-glucosides and nucleotides) to check recovery during purification and to validate the determination. The samples were purified using a combination of C18 (100 mg mL–1) and MCX cartridges (30 mg mL–1). The eluates were evaporated to dryness and dissolved in 20 μL of the mobile phase used for quantitative analysis. The samples were analysed by the liquid chromatography–tandem mass spectrometry (LC-MS/MS system) consisting of an ACQUITY UPLC® System (Waters, MA, USA) and Xevo® TQ-S (Waters) triple quadrupole mass spectrometer. Quantification was obtained using a multiple reaction monitoring (MRM) mode of selected precursor ions and the appropriate production (Svačinová et al., 2012).
Chlorophyll estimation
The chlorophyll content of cotyledon samples was measured from 4 hpi to 35 dpi using a Nanodrop spectrophotometer (Evans et al., 2012). The total chlorophyll content was calculated using the equation of Wellburn (1994): Chltotal = 7·12A664 + 18·12A647
RESULTS
Sequencing and phylogeny analysis
We used annotated family members of the gene of interest in arabidopsis and leguminous species in GenBank and other publicly available databases as query sequences to BLAST search our pea transcriptome data, and identified three IPT, three LOG, four RR, five CKX, four CWINV, four SUT, 15 SWEET and 13 AAP putative orthologous sequences in pea. Results of sequence verification via BLAST searching the GenBank database showed that most of the identified sequences were confirmed to be the target gene sequences. Phylogenetic analysis and gene family designations are shown in Supplementary Data Figs S1–S9. We have numbered both the PsSWEET gene family members and clades (Fig. S9) as outlined by Chandran (2015) and Eom et al. (2015), and not according to Patil et al. (2015) where gene families and clades are numbered somewhat differently.
Morphological and microscopic differences are evident following inoculation with avirulent and virulent strains of Rhodococcus fascians
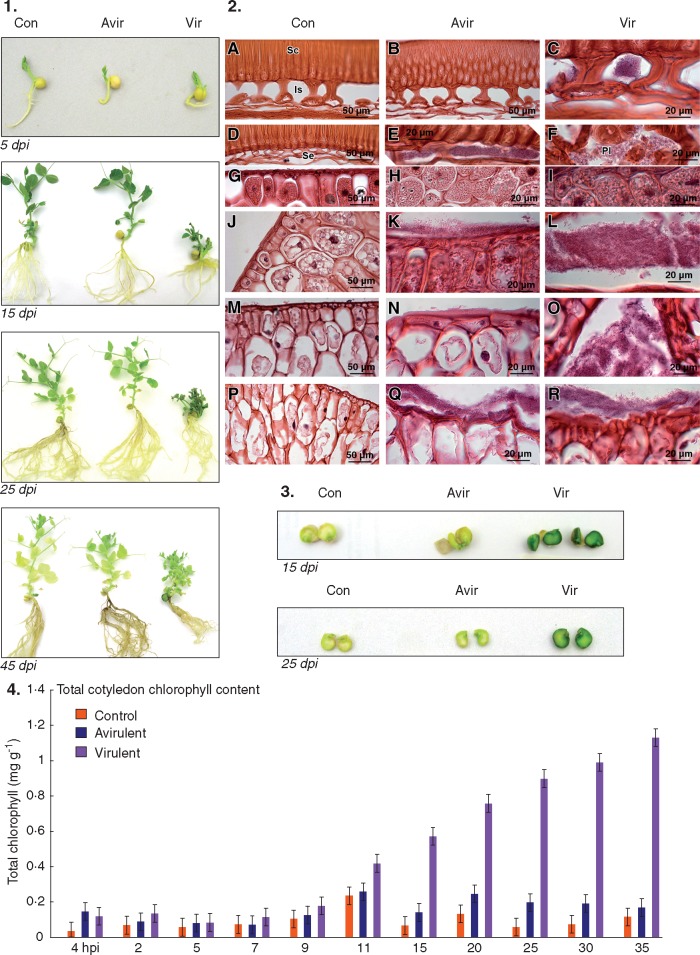
The typical multiple shoot symptom of R. fascians infection was apparent at 5 dpi in plants inoculated with the virulent strain (vir-plants) (Fig. 1-1). By 9 dpi, 80–90 % of the vir-plants had multiple shoots branching at the crown region of the root and shoot, and root growth was less prolific when compared with plants inoculated with the avirulent strain (avir-plants) and the mock-inoculated control plants (con-plants). The reduced leaf expansion, stunted shoot growth with multiple shoots, shortened and thickened primary roots and suppressed lateral root growth were evident from 11 to 45 dpi in vir-plants (Fig. 1-1). By 38 dpi, most of the con-plants started flowering, some avir-plants had flowers but few vir-plants flowered. Leaf senescence was observed in con-plants and avir-plants from 40 dpi but not in the vir-plants. The avir-plants and con-plants had similar morphology throughout (Fig. 1-1).
Fig. 1.
Response of Pisum sativum following seed inoculation with Rhodococcus fascians. (1) Growth stages and morphology of P. sativum inoculated with R. fascians avirulent strain 589 (Avir), virulent strain 602 (Vir) and medium as mock inoculation (Con) grown in sterile agar containers at 5, 15, 25 and 45 days post-inoculation (dpi). (2) Light micrographs of sections of P. sativum seed coat and cotyledon following inoculation with R. fascians at 4 h post-inoculation (hpi) (A–C), 2 dpi (D–L), 5 dpi (J–L), 15 dpi (M–O) and 35 dpi (P–R). Sc, seed coat; Is, intercellular spaces in the seed coat; Se, sub-epidermal layer of the seed coat, and PL, parenchyma layer of the seed coat. Magnification for control (×40; 50 μm scale bar); magnification for R. fascians-inoculated tissues (×100; 20 μm scale bar). (3) Morphological differences between P. sativum cotyledons inoculated with R. fascians avirulent strain 589 (Avir), virulent strain 602 (Vir) and mock inoculated (Con) at 15 and 25 dpi. (4) Total chlorophyll content in cotyledons of P. sativum imbibed for 4 h with R. fascians avirulent strain 589 (Avirulent), virulent strain 602 (Virulent) and medium as mock inoculation (Control) grown in sterile agar containers until 35 dpi. The error bars are ± 1 s.d. of two biological replicates and four technical replicates.
A particularly noticeable response was that cotyledons on the vir-plants became bright green and remained robust throughout, compared with con-plants and avir-plants where the cotyledons were light yellow, becoming shrivelled and reduced in size as the plants grew (Fig. 1-3). The cotyledons inoculated with the virulent strain (vir-cots) showed increasing chlorophyll content from 11 dpi compared with the control (Fig. 1-4).
Light microscopy revealed early differences in infection of the seeds inoculated with either the virulent or the avirulent microbe (Fig. 1-2). Within 4 h of pea seeds being inoculated with the virulent strain 602, purple-stained R. fascians colonies were detectable in the intercellular spaces of the sub-epidermal layer of the seed coat (Fig. 1-2C), whereas bacteria were not detected in the seed coat of seeds inoculated with the avirulent strain 589 (avir-cot) (Fig. 1-2B). Within 2 dpi, virulent bacteria were present in the parenchyma layer of the seed coat (Fig. 1-2F) as well as on the surface of the cotyledon (Fig. 1-2I), whereas the avirulent strain of R. fascians was now detectable in the intercellular spaces of the sub-epidermal layer of the seed coat but not on the surface of the cotyledon (Fig. 1-2E, H). However, by 5 dpi, both strains of bacteria were spread across the surface of the cotyledon (Fig. 1-2K, L). By 15 dpi, the avirulent strain was evident as clumps of cells on the cotyledon surface (Fig. 1-2N). In comparison, the virulent strain appeared as a widespread accumulation of cells (Fig. 1-2O). Microscopically, increased colonization of the cotyledons by the virulent strain was apparent from 15 to 35 dpi (Fig. 1-2O, R), whereas colonization by the avirulent strain became more obvious in later stages of growth, mainly at 25 and 35 dpi (Fig. 1-2Q).
Scanning electron microscopy of the plant surfaces showed R. fascians present on cotyledons, roots, shoots and flower surfaces throughout the life cycle of the plant (Dhandapani, 2014).
Endogenous cytokinins in inoculated pea tissues
Peas imbibed in the presence of virulent and avirulent strains of R. fascians showed differential accumulation of cytokinins, in terms of both quantity and type (Table 1). Of the changes at the free base, riboside and nucleotide level, most noticeable were the strongly elevated levels of iP in the vir-cots relative to con-cots across all experimental time points. iPR levels were strongly increased in vir-cots at 4 hpi, but then fluctuated relative to con-cots. Both Z-type and iP-type cytokinins were significantly elevated in the avir-cots relative to the mock-inoculated controls, consistently at 15 and 25 dpi (Table 1).
Table 1.
Endogenous cytokinin content in Pisum sativum cotyledons following seed inoculation with Rhodococcus fascians
| Cytokinin (pmol g–1 d wt) | Time points | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4hpi | 2 dpi | 5 dpi | 11 dpi | 15 dpi | 25 dpi | |||||||||||||
| Cotyledon | CON | AVIR | VIR | CON | AVIR | VIR | CON | AVIR | VIR | CON | AVIR | VIR | CON | AVIR | VIR | CON | AVIR | VIR |
| tZ | 1·65 | 3·08 | 2·79 | 5·26 | 2·76 | 2·14 | 1·89 | 2·27 | 1·00 | 2·26 | 1·56 | 0·61 | 0·65 | 1·26 | 1·13 | 1·04 | 1·6 | 0·8 |
| tZR | 0·13 | 0·09 | 0·1 | 1·01 | 1·16 | 0·44 | 0·33 | 0·4 | 0·18 | 0·34 | 0·16 | 0·04 | 0·28 | 0·18 | 0·13 | 0·16 | 1·03 | 0·15 |
| tZRMP | <LOD | <LOD | <LOD | 4·46 | 4·02 | 2·09 | 2·71 | 3·09 | 2·06 | 3·05 | 2·31 | 0·42 | 0·81 | 1·13 | 0·44 | 1·85 | 1·41 | 0·72 |
| tZOG | <LOD | <LOD | <LOD | <LOD | <LOD | <LOD | <LOD | 0·33 | 0·2 | 0·36 | 1·10 | 0·23 | 0·66 | 0·69 | 0·67 | 0·4 | 0·78 | 0·21 |
| tZROG | <LOD | <LOD | <LOD | 0·09 | 0·07 | 0·10 | 0·13 | 0·24 | 0·23 | 0·19 | 0·47 | 0·09 | 0·70 | 0·54 | 0·55 | 0·47 | 0·96 | 0·23 |
| iP | 1·66 | 3·75 | 4·15 | 1·76 | 1·74 | 7·59 | 1·61 | 1·93 | 3·44 | 2·28 | 3·29 | 11·95 | 1·39 | 1·86 | 7·93 | 2·22 | 4·54 | 14·92 |
| iPR | 0·22 | 3·41 | 11·66 | 1·52 | 1·03 | 2·33 | 1·32 | 0·92 | 1·01 | 0·96 | 1·15 | 0·51 | 0·89 | 1·97 | 1·92 | 0·82 | 3·27 | 1·73 |
| iPRMP | <LOD | <LOD | <LOD | 21·63 | 16·56 | 11·2 | 23·8 | 21·84 | 13·78 | 18·58 | 8·5 | 1·69 | 5·43 | 19·76 | 14·42 | 2·17 | 7·08 | 2·5 |
LOD, below detection limit; tZ7G, tZ9G, iP7G and iP9G were generally below the detection limits.
The pea seeds were imbibed for 4 h (hpi) with R. fascians strain 589 (AVIR), strain 602 (VIR) and with medium for mock inoculation (CON).
The data are averages of four biological replicates.
Numbers in bold indicate a significant difference from the control, P < 0·05.
Nucleotides were not detected in inoculated cotyledons at 4 hpi, but were generally decreased relative to controls at 2, 5 and 11 dpi in vir-cots, and generally elevated in avir-cots at 15 and 25 dpi (Table 1). The storage O-glucosyl forms were not detected in cotyledons until 5 dpi (Table 1). Both tZOG and tZROG were elevated in all vir-tissues at 5 dpi, but were decreased consistently subsequently relative to controls. In avir-cots the patterns were generally the opposite to those in vir-cots. The 7- and 9-glucoside conjugates of both tZ and iP were generally below the limit of detection. The cis-cytokinins and methyl-thio forms will be presented elsewhere. However, these forms did not correlate with virulence (data not shown).
Expression of RfIPT, RfLOG and RfCKX genes during infection
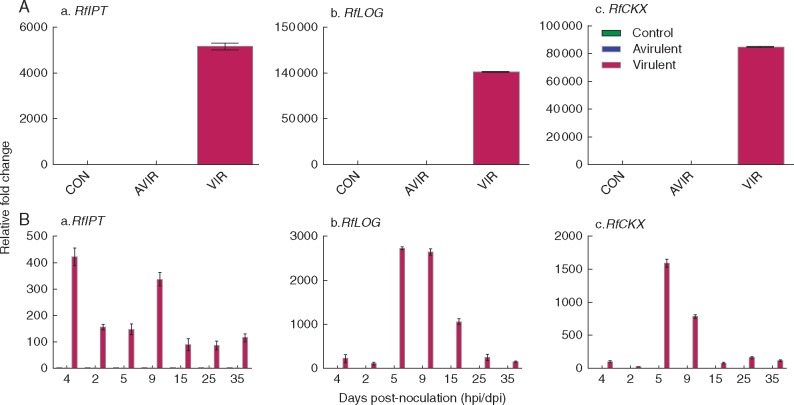
PCR confirmed that not only IPT and CKX, but also LOG is present in 18 virulent strains but not in 18 avirulent strains (Table S1). Primers were designed to discriminate between the pathogen genes and pea genes. Clear expression of the R. fascians genes was shown in cultures of the virulent strain (Fig. 2A). Following inoculation, expression of all three R. fascians genes was detected in the vir-cots but not in avir-cots. RfIPT was expressed strongly across time in the vir-cots. Expression of RfLOG was strongest in vir-cots at 5, 9 and 15 dpi, and that of RfCKX at 5 and 9 dpi (Fig. 2B).
Fig. 2.
Relative expression of RfIPT, RfLOG and RfCKX detected in R. fascians cultures and Pisum sativum cotyledons inoculated with R. fascians. (A) Expression of RfIPT, RfLOG and RfCKX was determined following RNA extraction and cDNA synthesis from R. fascians avirulent strain 589 (AVIR) and virulent strain 602 (VIR). A no-template reaction was used as control (CON). Relative fold change values were calculated using Ps18S as an internal control. The error bars are ± 1 s.d. using four technical replicates for each of two biological replicates in the RT–qPCR. (B) Following seed inoculation with R. fascians avirulent strain 589 (AVIR) and virulent strain 602 (VIR) from 4 h to 35 days post-inoculation. Relative fold change values were calculated using PsEF, Ps18S, PsGAP and PsACT as internal controls. The error bars are ± 1 s.d. using three technical replicates for each of two biological replicates in the RT–qPCR.
Expression of PsIPT, PsLOG and PsCKX genes in cotyledons inoculated with virulent R. fascians
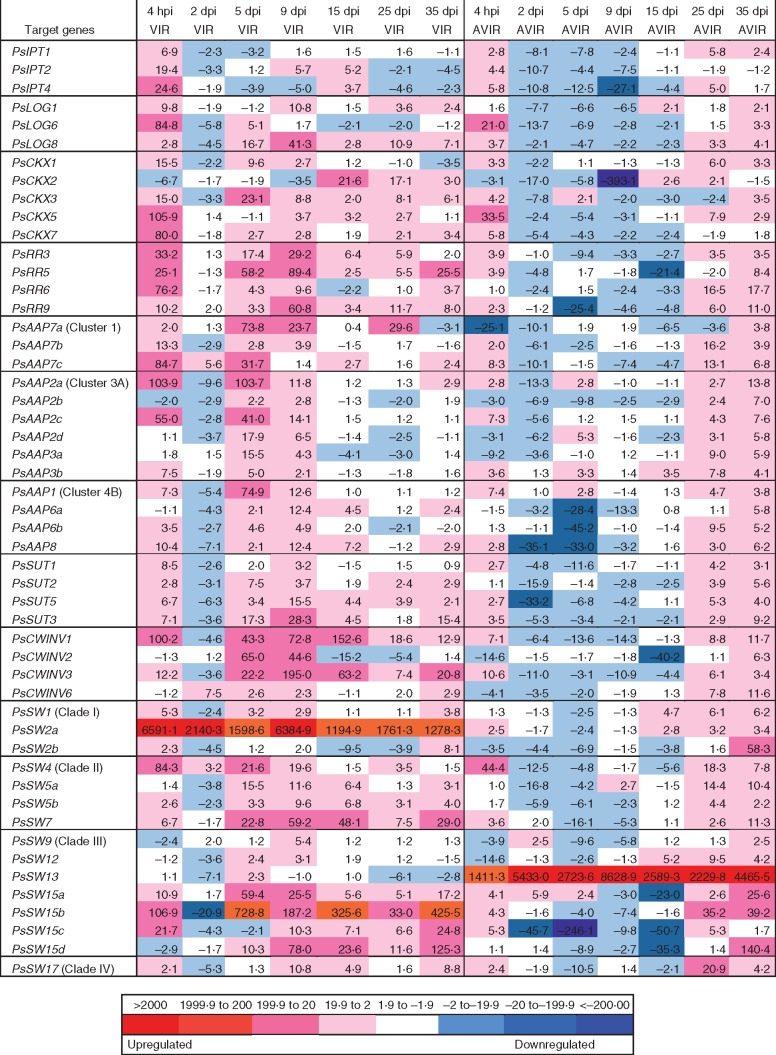
It would appear that virulent R. fascians and, to a lesser extent, the avirulent strain had an early effect on the expression of PsIPT, with all three gene family members showing enhanced expression within 4 hpi (Fig. 3). This was also shown in an independent experiment (Fig. S9). Subsequently, in the avir-cots there was a suppression of PsIPT expression until 25–35 dpi. By 2 dpi, in those plants inoculated with the virulent strain, PsIPT expression was reduced relative to the control, with the exception of PsIPT2 at 9 and 15 dpi in cotyledons. Mirroring the PsIPT expression, PsLOG expression was elevated within 4 hpi in both vir- and avir-cots (Fig. 3). PsLOG expression then reduced in the avir-cots until the latter stages of the experiment. Conversely, PsLOG8 was elevated in vir-cots at most growth stages.
Fig. 3.
Relative expression of cytokinin biosynthesis (PsIPT), degradation (PsCKX) and response regulator (PsRR) gene family members along with PsAAP, PsSUT, PsCWINV and PsSWEET gene family members in Pisum sativum cotyledons following seed inoculation with Rhodococcus fascians strain 589 (AVIR) and virulent strain 602 (VIR) from 4 h to 35 d post-inoculation. Values are fold changes relative to the expression of the mock-inoculated control. Initial fold change values were calculated using PsEF, Ps18S, PsGAP and PsACT as internal controls using three technical replicates for each of two biological replicates in the RT–qPCR. The relative expression level of each gene was then compared with the expression in pea cotyledons inoculated with medium as the mock-inoculated control. The colour scale indicates upregulated expression (red scale), similar (white) and downregulated expression (blue scale) relative to the mock-inoculated control.
With the exception of PsCKX2, the other four PsCKX gene family members showed elevated expression in both vir- and avir-cots within 4 hpi relative to controls, with expression in vir-cots exceeding that in avir-cots (Fig. 3). Throughout the experiment, various PsCKX gene family members showed elevated expression in vir-cots. PsCKX1 and PsCKX5 were mostly elevated in avir-cots at the later stages of the experiment. Increased PsRR activity relative to control was noticeable in vir-cots throughout the experiment, but particularly at 4 hpi and at 5 and 9 dpi (Fig. 3). Expression of several PsRR genes was elevated at 25 and 35 dpi in avir-cots.
Expression of amino acid transporters in inoculated pea tissues
Relative to control, enhanced expression of several of the PsAAP gene family members was apparent in vir-cots at 4 hpi (Fig. 3). Most noticeable was enhanced expression at 5 and 9 dpi and the subsequent lesser expression of most PsAAP gene family members over time in vir-cots. In contrast, elevated expression of PsAAP gene family members relative to controls was consistently shown in avir-cots at 25 and 35 dpi.
Expression of sugar transporters in inoculated pea tissues
With the exception of vir-cots at 2 dpi, expression of Type I PsSUT5 and PsSUT2, and Type II PsSUT3 was enhanced in vir-cots relative to controls throughout the experiment (Fig. 3). Enhanced expression of this gene family relative to controls was apparent in avir-cots at 4 hpi, but decreased expression relative to controls then occurred from 2 to 9 dpi. Enhanced expression was evident in avir-cots at 25 and 35 dpi.
Expression of multiple PsSWEET gene family members in inoculated pea tissues
Constitutively strong expression was shown for PsSW2a, a Clade I SWEET, in vir-cots (Fig. 3). Transcript levels of three of the four Clade II members (PsSW4, 5b and 7) were elevated at 4 hpi, but not relative to controls at 2 dpi, but were subsequently elevated through to 35 dpi. Several of the Clade III SWEETs in vir-cots, particularly PsSW15a, b and d, were elevated relative to the control throughout development. Strong constitutive expression of Clade III PsSW13 was detected in avir-cots from 4 hpi to 35 dpi (Fig. 3). Additionally, several other Clade III SWEET genes that were expressed at 4 hpi were expressed more strongly at 25 and/or 35 dpi in the avir-cots.
Expression of cell wall invertases in inoculated pea tissues
Strong expression of PsCWINV1 and PsCWINV3 was seen in vir-cots at all time points; and in PsCWINV2 at 5 and 9 dpi (Fig. 3). Interestingly, increased expression relative to the control was observed for PsCWINV1 and PsCWINV3 gene family members in avir-cots at 4 hpi and for most family members at 25 and 35 dpi.
DISCUSSION
Infection by virulent R. fascians leads to retention and greening of cotyledons
The marked increase in chlorophyll content of the cotyledons infected by the virulent strain was particularly striking, but not apparent until several days after the appearance of multiple shoots. Cornelis et al. (2001) observed the colonizing behaviour of both D188 and its plasmid-cured strain (D188-5) on both tobacco and arabidopsis seedlings. They suggested that both strains colonized the plant surface extensively and increasingly over time, but with less penetration of cell layers by D188-5 compared with D188 in infected tobacco. Our wild-type virulent strain spread through the seed coat and onto the surface of cotyledons faster than the wild-type avirulent strain, and was clearly visible prior to symptom emergence. There was not strong evidence for penetration of the bacteria beyond the surface layer of cells.
Previously Depudyt et al. (2008, 2009) had shown, in arabidopsis, decreased expression of AtIPT (particularly AtIPT7), increased AtCKX and increased AtRR expression relative to D188-5. They also reported an early increase in iPRMP and iP. We compared expression of both a virulent and an avirulent strain with mock-inoculated controls within 4 h of inoculation, and showed, relative to the mock-inoculated control, that PsIPT gene family members were initially strongly up-regulated in both vir- and avir-cots. This is reflected in the elevated levels of iP, iPR and tZ in the cotyledons. Matching this was strong expression of the cytokinin PsRR genes, and also of PsCKX expression. Interestingly, increased activity of the enzymes of the isoprenoid pathway has been reported during the initial 2–6 h of imbibition (Green and Baisted, 1972), indicating that a supply of precursor was probably available to the PsIPT and RfIPT genes. An early and transient expression of BrIPT genes was also noted by Ando et al. (2005) during clubroot infection in Chinese cabbage. They suggested that this transient induction of BrIPT expression may be the ‘trigger for clubroot development’. Potentially, then, the early upregulation of PsIPT is indicative of recognition by the plant of the presence of the bacteria, irrespective of pathogenic status.
However, subsequently, PsIPT expression was depressed in both vir- and avir-cots. As the iP levels in the vir-cots continued to be greater than in the controls, the source of the iP in the vir-cots was now likely to be from the RfIPT. At the stage when the increased chlorophyll levels were apparent (11 dpi), iPRMP and ZRMP levels were actually less than in the con-cots, and less than in the avir-cots, reflecting earlier results obtained in pea (Eason et al., 1996) and arabidopsis (Depuydt et al., 2008). However, the expression of RfLOG and the elevated expression of PsLOG8 are likely to account for the low level of nucleotides in the vir-cots. The elevated expression of the PsRR genes in the vir-cots is indicative of active signal transduction (Hwang et al., 2012), and the elevated PsCKX expression is indicative of the plant homeostatic mechanisms coming into play – albeit somewhat ineffectually – a situation similar to that occurring in arabidopsis leaves responding to R. fascians D188 (Depudyt et al., 2008; Pertry et al., 2009).
Virulent and avirulent R. fascians strains cause changes in expression of genes regulating source–sink dynamics
As evident in the heat map (Fig. 3), infection by the virulent strain had a marked effect on expression of PsCWINV, PsSUT and SWEET genes – key components in the regulation of source–sink dynamics. Additionally, we showed early changes in gene expression by the plant in response to the avirulent bacterium and consistently after some 25 and 35 dpi, by which time epiphytic colonies were well established.
Depuydt et al. (2009) reported that within 4 dpi, well before symptoms were apparent, AtCWINV was induced by both D188 and D188-5, but enzyme activity was greater and lasted longer following infection by D188. Enhanced expression of PsCWINV genes was particularly noticeable in the vir-cots. Further supporting the establishment of a carbohydrate sink for the bacteria is the upregulation of sucrose transporter genes. Moreover, strong expression of SWEET gene family members was also evident. However, in this case, the SWEET gene family members may have had opposite effects: Clade I AtSWEET2 transports glucose to the vacuole (Chen 2015) and PsSW2a, a Clade I SWEET, was elevated in response to the virulent strain. This aligns with the suggestion by Chen et al. (2015) that expression in the roots of AtSWEET2 restricted the amount of glucose in the apoplast, by sequestering it to the vacuole. In the case of R. fascians, expression of several Clade II and III SWEET genes was also elevated in vir-cots, in which case hexoses and sucrose would have been exported to the apoplasm. These sugars have been detected in tissues of arabidopsis infected with the virulent D188 strain (Depuydt et al., 2009). As sugars have recently been identified as key components of the release of apical dominance (Barbier et al., 2015), a high sugar environment may be the signal for multiple bud formation, as suggested for witches’ broom disease of cacao (Barau et al., 2015).
In contrast, PsSW13, a Clade III SWEET, was strongly upregulated in avir-cots, but not in vir-cots. Additionally, several PsCWINV and PsSUT gene family members were also upregulated later in development, supporting the contention that the epiphytic bacteria are also impacting on metabolism (Depuydt et al., 2009). Along with establishing a carbon supply, a supply of amino acids was secured by the virulent bacterium through upregulation of PsAAP genes in the germinating seeds. However, and particularly later in the experiment, the epiphytic colonies also impacted on amino acid transporters.
Our data add to the earlier arabidopsis microarray analysis by Depuydt et al. (2009) which revealed not only that D188 infection modulated primary metabolism, but also that inoculation by the avirulent D188-5 caused changes relative to the mock-inoculated control. However, our data provide an interesting time component, with both virulent and avirulent strains impacting plant gene expression early during inoculation. The apparent decrease in impact of the bacteria at 2 dpi may reflect the transition in the cotyledons from the initial metabolism occurring during Phase I imbibition and the mobilization of reserves in the germinating seedling by 2 dpi, when the radicle and plumule were emerging (Weitbrecht et al., 2011). Clearly, plant cytokinin homeostasis and sink metabolism were then affected strongly by the virulent strain, with the avirulent strain impacting more modestly later in the experiment as the cotyledon reserves were becoming depleted.
Conclusions
In conclusion, it is clear that both the virulent and avirulent strains affected the expression of genes involved in the transport of carbon and nitrogen, as well as cytokinin biosynthesis and metabolism. Only the virulent strain markedly impacted both morphogenesis and the integrity of the cotyledon. We suggest that the interaction between cytokinins, PsCWINV and PsSWEET genes contributes to the loss of apical dominance and the appearance of multiple shoots emerging from the seeds inoculated with the virulent bacterium, as well as the re-greening and maintenance of the cotyledon.
However, whether the elevated iP alone is sufficient to have such a marked impact on the plant is yet to be determined. Subsequent investigations will focus on expression of RfMT1 and RfMT2 and levels of methylated cytokinins [newly identified from Rhodococcus-infected plant tissues by Radhika et al. (2015)], as well as the methylthio- and cis-forms of the cytokinins. We will then be able to compare, in pea, the impact of the methylated cytokinins (Radhika et al., 2015), with the ‘trick-with-the-mix’ hypothesis developed based on experiments with R. fascians and arabidopsis (Pertry et al., 2009, 2010), with the suggestion by Creason et al. (2014) that only one cytokinin type (the iP-type) is necessary for disease symptoms to be manifested.
SUPPLEMENTARY DATA
Supplementary data are available online at www.aob.oxfordjournals.org and consist of the following. Figure S1: phylogenetic tree of IPT. Figure S2: phylogenetic tree of LOG. Figure S3: phylogenetic tree of CKX. Figure S4: phylogenetic tree of RR. Figure S5: phylogenetic tree of SUT. Figure S6: phylogenetic tree of CWINV/INV. Figure S7: phylogenetic tree of AAP. Figure S8: phylogenetic tree of SWEET. Figure S9: PsIPT, PsLOG, PsCKX and PsRR expression in seeds of P. sativum following seed inoculation with R. fascians at 4 h and 2 days post-inoculation. Table S1: PCR detection of RfIPT, RfLOG and RfCKX genes in R. fascians cultures. Table S2: primer sequences used for RT–qPCR, and GenBank accession numbers.
Supplementary Material
ACKNOWLEDGEMENTS
A UC doctoral scholarship to P.D. is gratefully acknowledged. Thanks to Graeme Bull and Jan McKenzie for histological specimen preparation and light microscopy; Neil Andrews for assisting in preparation and operation of the scanning electron microscope; Hana Martinková and Ivan Petřík for help with cytokinin analyses; Annu Smitha Ninan for isolating some of the transporter gene family members; and Matt Walters for assistance in graphic presentation. The constructive advice from the two anonymous referees is acknowledged with thanks. O.N. was funded by the Ministry of Education, Youth and Sports of the Czech Republic (the National Program for Sustainability I Nr. LO1204) and the “Navrat” program (LK21306).
REFERENCES
- Ando S, Asano T, Tsushima S, Kamachi S, Hagio T, Tabei Y.. 2005. Changes in gene expression of putative isopentenyltransferase during clubroot development in Chinese cabbage (Brassica rapa L.). Physiological and Molecular Plant Pathology 67: 59–67. [Google Scholar]
- Antoniadi I, Plačková L, Simonovik B, et al. 2015. Cell-type specific cytokinin distribution within the Arabidopsis primary root apex. The Plant Cell 27: 1955–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albacete A, Cantero-Navarro E, Großkinsky DK, et al. 2015. Ectopic overexpression of the cell wall invertase gene CIN1 leads to dehydration avoidance in tomato. Journal of Experimental Botany 66: 863–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balibrea Lara ME, Gonzalez Garcia M-C, Fatima T, et al. 2004. Extracellular invertase is an essential component of cytokinin-mediated delay of senescence. The Plant Cell 16: 1276–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barau J, Grandis A, Carvalho VMdA, et al. 2015. Apoplastic and intracellular plant sugars regulate developmental transitions in witches’ broom disease of cacao. Journal of Experimental Botany 66: 1325–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbier FF, Lunn JE, Beveridge CA.. 2015. Ready, steady, go! A sugar hit starts the race to shoot branching. Current Opinion in Plant Biology 25: 39–45. [DOI] [PubMed] [Google Scholar]
- Berger S, Sinha AK, Roitsch T.. 2007. Plant physiology meets phytopathology: plant primary metabolism and plant–pathogen interactions. Journal of Experimental Botany 58: 4019–4026. [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, et al. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry 55: 611–622. [DOI] [PubMed] [Google Scholar]
- Carletom HM, Druvy RAB.. 1957. Histological technique for normal pathological tissues and the identification of parasites, 3rd edn.London: Oxford University Press. [Google Scholar]
- Chandran D. 2015. Co-option of developmentally regulated plant SWEET transporters for pathogen nutrition and abiotic stress tolerance. IUBMB Life 67: 461–71. [DOI] [PubMed] [Google Scholar]
- Chandran D, Inada N, Hather G, Kleindt CK, Wildermuth MC.. 2010. Laser microdissection of Arabidopsis cells at the powdery mildew infection site reveals site-specific processes and regulators. Proceedings of the National Academy of Sciences, USA 107: 460–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H-Y, Huh J-H, Yu Y-C, et al. 2015. The Arabidopsis vacuolar sugar transporter SWEET2 limits carbon sequestration from roots and restricts Pythium infection. The Plant Journal 83: 1046–1058. [DOI] [PubMed] [Google Scholar]
- Chen L-Q, Hou B-H, Lalonde S, et al. 2010. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468: 527–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L-Q, Qu XQ, Hou B-H, et al. 2012. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 335: 207–211. [DOI] [PubMed] [Google Scholar]
- Cornelis K, Ritsema T, Nijsse J, Holsters M, Goethals K, Jaziri M.. 2001. The plant pathogen Rhodococcus fascians colonizes the exterior and interior of the aerial parts of plants. Molecular Plant-Microbe Interactions 14: 599–608. [DOI] [PubMed] [Google Scholar]
- Creason AL, Vandeputte OM, Savory EA, et al. 2014. Analysis of genome sequences from plant pathogenic Rhodococcus reveals genetic novelties in virulence loci. PLoS One 9: e101996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depuydt S, Doležal K, Van Lijsebettens M, Moritz T, Holsters M, Vereecke D.. 2008. Modulation of the hormone setting by Rhodococcus fascians results in ectopic KNOX activation in Arabidopsis. Plant Physiology 146: 1267–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depuydt S, Tenkamp S, Fernie AR, et al. 2009. An integrated genomics approach to define niche establishment by Rhodococcus fascians. Plant Physiology 149: 1366–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derveaux S, Vandesompele J, Hellemans J.. 2010. How to do successful gene expression analysis using real-time PCR. Methods 50: 227–230. [DOI] [PubMed] [Google Scholar]
- Dhandapani P. 2014. Rhodococcus fascians–plant interactions: microbiological and molecular aspects. PhD Thesis, University of Canterbury, New Zealand.
- Dobrev PI, Kamínek M.. 2002. Fast and efficient separation of cytokinins from auxin and abscisic acid and their purification using mixed-mode solid-phase extraction. Journal of Chromatography A 950: 21–29. [DOI] [PubMed] [Google Scholar]
- Eason JR, Jameson PE, Bannister P.. 1995. Virulence assessment of Rhodococcus fascians strains on pea cultivars. Plant Pathology 44: 141–147. [Google Scholar]
- Eason JR, Morris RO, Jameson PE.. 1996. The relationship between virulence and cytokinin production by Rhodococcus fascians (Tilford 1936) Goodfellow 1984. Plant Pathology 45: 323–331. [Google Scholar]
- Ehness R, Ecker M, Godt DE, Roitsch T.. 1997. Glucose and stress independently regulate source and sink metabolism and defense mechanisms via signal transduction pathways involving protein phosphorylation. The Plant Cell 9: 1825–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehneß R, Roitsch T.. 1997. Co-ordinated induction of mRNAs for extracellular invertase and a glucose transporter in Chenopodium rubrum by cytokinins. The Plant Journal 11: 539–548. [DOI] [PubMed] [Google Scholar]
- Eom JS, Chen LQ, Sosso D, et al. 2015. SWEETs, transporters for intracellular and intercellular sugar translocation. Current Opinion in Plant Biology 25: 53–62. [DOI] [PubMed] [Google Scholar]
- Evans T, Song J, Jameson PE.. 2012. Micro-scale chlorophyll analysis and developmental expression of a cytokinin oxidase/dehydrogenase gene during leaf development and senescence. Plant Growth Regulation 66: 95–99. [Google Scholar]
- Fagard M, Launay A, Clément G, et al. 2014. Nitrogen metabolism meets phytopathology. Journal of Experimental Botany 65: 5643–5656. [DOI] [PubMed] [Google Scholar]
- Francis IM, Stes E, Zhang Y, Rangel D, Audenaert K, Vereecke D.. 2016. Mining the genome of Rhodococcus fascians, a plant growth-promoting bacterium gone astray. New Biotechnology (in press). [DOI] [PubMed] [Google Scholar]
- Galis I, Bilyeu K, Wood G, Jameson PE.. 2005. Rhodococcus fascians: shoot proliferation without elevated cytokinins? Plant Growth Regulation 46: 109–115. [Google Scholar]
- Green TR, Baisted DJ.. 1972. Development of the activities of enzymes of the isoprenoid pathway during early stages of pea-seed germination. Biochemical Journal 130: 983–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Großkinsky DK, Naseem M, Abdelmohsen UR, et al. 2011. Cytokinins mediate resistance against Pseudomonas syringae in tobacco through increased antimicrobial phytoalexin synthesis independent of salicylic acid signaling. Plant Physiology 157: 815–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo WJ, Nagy R, Chen HY, et al. 2014. SWEET17, a facilitative transporter, mediates fructose transport across the tonoplast of Arabidopsis roots and leaves. Plant Physiology 164: 777–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez L, Mauriat M, Pelloux J, Bellini C, Van Wuytswinkel O.. 2008. Towards a systematic validation of references in real-time RT–PCR. The Plant Cell 20: 1734–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sheen J, Muller B.. 2012. Cytokinin signaling networks. Annual Review of Plant Biology 63: 353–380. [DOI] [PubMed] [Google Scholar]
- Jameson PE. 2000. Cytokinins and auxins in plant–pathogen interactions – an overview. Plant Growth Regulation 32: 369–380. [Google Scholar]
- Jameson PE. 2017. Cytokinins In: Thomas B, Murray B, Murphy DJ, eds. Encyclopedia of applied plant sciences, Vol 1 Waltham, MA: Academic Press, pp. 391-402. [Google Scholar]
- Jameson PE, Song J.. 2016. Cytokinin: a key driver of seed yield. Journal of Experimental Botany 67: 593–606. [DOI] [PubMed] [Google Scholar]
- Kado CI, Heskett MG.. 1970. Selective media for isolation of Agrobacterium, Corynebacterium, Erwinia, Pseudomonas and Xanthomonas. Phytopatholoy 60: 969–976. [DOI] [PubMed] [Google Scholar]
- Kitin P, Iliev I, Scaltsoyiannes A, Nellas C, Rubos A, Funada R.. 2005. A comparative histological study between normal and fasciated shoots of Prunus avium generated in vitro. Plant Cell, Tissue and Organ Culture 82: 141–150. [Google Scholar]
- Lacey MS. 1936. Studies in bacteriosis. XXII. 1. Isolation of a bacterium associated with ‘fasciaton’ of sweet peas, ‘cauliflower’ strawberry plants and ‘leafy gall’ of various plants. Annals of Applied Biology 23: 302–310. [Google Scholar]
- Lawson E, Gantotti B, Starr M.. 1982. A 78-megadalton plasmid occurs in avirulent strains as well as virulent strains of Corynebacterium fascians. Current Microbiology 7: 327–332. [Google Scholar]
- Li T, Huang S, Zhou J, Yang B.. 2013. Designer TAL effectors induce disease susceptibility and resistance to Xanthomonas oryzae pv. oryzae in rice. Molecular Plant 6: 781–789. [DOI] [PubMed] [Google Scholar]
- Lomin SN, Krivosheev DM, Steklov MY, et al. 2015. Plant membrane assays with cytokinin receptors underpin the unique role of free cytokinin bases as biologically active ligands. Journal of Experimental Botany 66: 1851–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mothes K, Engelbrecht L.. 1963. On the activity of a kinetin-like root factor. Life Sciences 2: 852–857. [Google Scholar]
- Nandi S, Palni L.. 1989. Transport and metabolism of dihydrozeatin riboside in germinating lupin seeds. Journal of Experimental Botany 40: 615–621. [Google Scholar]
- Nandi S, Palni L, Letham D, Knypl J.. 1988. The biosynthesis of cytokinins in germinating lupin seeds. Journal of Experimental Botany 39: 1649–1665. [Google Scholar]
- Nawa Y, Asahi T.. 1971. Rapid development of mitichondria in pea cotyledons during early stage of germination. Plant Physiology 48: 671–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil G, Valliyodan B, Deshmukh R, et al. 2015. Soybean (Glycine max) SWEET gene family: insights through comparative genomics, transcriptome profiling and whole genome re-sequence analysis. BMC Genomics 16: 520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertry I, Václavíková K, Depuydt S, et al. 2009. Identification of Rhodococcus fascians cytokinins and their modus operandi to reshape the plant. Proceedings of the National Academy of Sciences, USA 106: 929–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertry I, Václavíková K, Gemrotová M, et al. 2010. Rhodococcus fascians impacts plant development through the dynamic fas-mediated production of a cytokinin mix. Molecular Plant-Microbe Interactions 23: 1164–1174. [DOI] [PubMed] [Google Scholar]
- Pfaffl M, Hageleit M.. 2001. Validities of mRNA quantification using recombinant RNA and recombinant DNA external calibration curves in real-time RT–PCR. Biotechnology Letters 23: 275–282. [Google Scholar]
- Pratelli R, Pilot G.. 2014. Regulation of amino acid metabolic enzymes and transporters in plants. Journal of Experimental Botany 65: 5535–5556. [DOI] [PubMed] [Google Scholar]
- Radhika V, Ueda N, Tsuboi Y, et al. 2015. Methylated cytokinins from the phytopathogen Rhodococcus fascians mimic plant hormone activity. Plant Physiology 169: 1118–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roitsch T, Gonzalez MC.. 2004. Function and regulation of plant invertases: sweet sensations. Trends in Plant Science 9: 606–613. [DOI] [PubMed] [Google Scholar]
- Siemens J, Gonzalez MC, Wolf S, et al. 2011. Extracellular invertase is involved in the regulation of clubroot disease in Arabidopsis thaliana. Molecular Plant Pathology 12: 247–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Jiang L, Jameson PE.. 2012. Co-ordinate regulation of cytokinin gene family members during flag leaf and reproductive development in wheat. BMC Plant Biology 12: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spichal L. 2012. Cytokinins – recent news and views of evolutionally old molecules. Functional Plant Biology 39: 267–284. [DOI] [PubMed] [Google Scholar]
- Stange RR, Jr, Jeffares D, Young C, Scott DB, Eason JR, Jameson PE.. 1996. PCR amplification of the fas-1 gene for the detection of virulent strains of Rhodococcus fascians. Plant Pathology 45: 407–417. [Google Scholar]
- Stes E, Vandeputte OM, El Jazziri M, Holsters M, Vereecke D.. 2011. A successful bacterial coup d’état: how Rhodococcus fascians redirects plant development. Annual Review of Phytopathology 49: 69–86. [DOI] [PubMed] [Google Scholar]
- Streubel J, Pesce C, Hutin M, Koebnik R, Boch J, Szurek B.. 2013. Five phylogenetically close rice SWEET genes confer TAL effector-mediated susceptibility to Xanthomonas oryzae pv. oryzae. New Phytologist 200: 808–819. [DOI] [PubMed] [Google Scholar]
- Svačinová J, Novák O, Plačková L, et al. 2012. A new approach for cytokinin isolation from Arabidopsis tissues using miniaturized purification: pipette tip solid-phase extraction. Plant Methods 8: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei Ma, Kumar S.. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution 24: 1596–1599. [DOI] [PubMed] [Google Scholar]
- Tegeder M. 2012. Transporters for amino acids in plant cells: some functions and many unknowns. Current Opinion in Plant Biology 15: 315–21. [DOI] [PubMed] [Google Scholar]
- Tegeder M, Ward JM.. 2012. Molecular evolution of plant AAP and LHT amino acid transporters. Frontiers in Plant Science 3: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG.. 1997. The CLUSTALX windows interface: flexible strategies for multiple sequence alignment aided by quality tools. Nucleic Acids Research 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, et al. 2002. Accurate normalization of real-time quantitative RT–PCR data by geometric averaging of multiple internal control genes. Genome Biology 3: research0034.1–research0034.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitbrecht K, Muller K, Leubner-Metzger G.. 2011. First off the mark: early seed germination. Journal of Experimental Botany 62: 3289–3309. [DOI] [PubMed] [Google Scholar]
- Wellburn AR. 1994. The spectral determination of chlorophylls a and b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. Journal of Plant Physiology 144: 307–313. [Google Scholar]
- Werner T, Schmülling T.. 2009. Cytokinin action in plant development. Current Opinion in Plant Biology 12: 527–538. [DOI] [PubMed] [Google Scholar]
- Zhou J, Peng Z, Long J, et al. 2015. Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice. The Plant Journal 82: 632–43. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.