Abstract
Background and Aims Current strategies for increased crop protection of susceptible tomato plants against pathogen infections include treatment with synthetic chemicals, application of natural pathogen-derived compounds or transfer of resistance genes from wild tomato species within breeding programmes. In this study, a series of 45 genes potentially involved in defence mechanisms was retrieved from the genome sequence of inbred reference tomato cultivar Solanum lycopersicum ‘Heinz 1706’. The aim of the study was to analyse expression of these selected genes in wild and cultivated tomato plants contrasting in resistance to the biotrophic pathogen Oidium neolycopersici, the causative agent of powdery mildew. Plants were treated either solely with potential resistance inducers or by inducers together with the pathogen.
Methods The resistance against O. neolycopersici infection as well as RT-PCR-based analysis of gene expression in response to the oomycete elicitor oligandrin and chemical agent β-aminobutyric acid (BABA) were investigated in the highly susceptible domesticated inbred genotype Solanum lycopersicum ‘Amateur’ and resistant wild genotype Solanum habrochaites.
Key Results Differences in basal expression levels of defensins, germins, β-1,3-glucanases, heveins, chitinases, osmotins and PR1 proteins in non-infected and non-elicited plants were observed between the highly resistant and susceptible genotypes. Moreover, these defence genes showed an extensive up-regulation following O. neolycopersici infection in both genotypes. Application of BABA and elicitin induced expression of multiple defence-related transcripts and, through different mechanisms, enhanced resistance against powdery mildew in the susceptible tomato genotype.
Conclusions The results indicate that non-specific resistance in the resistant genotype S. habrochaites resulted from high basal levels of transcripts with proven roles in defence processes. In the susceptible genotype S. lycopersicum ‘Amateur’, oligandrin- and BABA-induced resistance involved different signalling pathways, with BABA-treated leaves displaying direct activation of the ethylene-dependent signalling pathway, in contrast to previously reported jasmonic acid-mediated signalling for elicitins.
Keywords: BABA; defence genes; ethylene, Oidium neolycopersici; oligandrin; powdery mildew; resistance; Solanum lycopersicum; Solanum habrochaites; tomato
INTRODUCTION
Tomato (Solanum lycopersicum) is one of the most important food crops and is widely used as a model plant for scientific purposes. Through domestication, breeding and research activities of the ancestral wild Solanum species, originating from the Andean region, many morphologically different cultivars and forms have been created. During this process, the genomes of cultivated tomatoes have passed through a progressive genetic bottleneck, reducing the genetic diversity in cultivated varieties compared to their wild relatives (Sim et al., 2011). Breeding selection has focused almost exclusively on traits related to desirable agricultural characteristics such as fruit yield and quality, loss of germination inhibition or compact growth habit. However, disease resistance traits have been disregarded, resulting in cultivars highly susceptible to many pathogens (Foolad, 2007). Wild tomato species, in contrast, such as S. chilense, S. peruvianum, S. habrochaites, S. parviflorum, S. pennellii or S. pimpinellifolium, are considered a rich source of disease resistance genes and have been utilized in breeding programmes for the introgression of these genes into cultivated varieties to improve their resistance (Menda et al., 2014).
In addition to inbreeding, the use of resistance inducers is another strategy for the protection of susceptible crop plants. Treatment with natural or synthetic chemicals can establish a primed state of the plant that enables a stronger and faster activation of defence responses and enhanced resistance to challenging stresses (Conrath, 2009). The non-protein amino acid β-aminobutyric acid (BABA) belongs to well-known priming agents. Unlike its isomers α- and γ-aminobutyric acid, BABA induces resistance in many plants against various pathogens (Piękna-Grochala and Kępczyńska, 2013). For instance, BABA induces a strong resistance of tomato towards the tomato late blight Phytophthora infestans (Cohen et al., 1994; Cohen, 2002) and to root-knot nematodes (Oka and Cohen, 2001). However, it has been observed that BABA treatment can undesirably affect plant growth, thus hindering its use for field applications (Wu et al., 2010). Studies on Arabidopsis–Botrytis cinerea interactions have suggested that the protective effects of BABA are mediated through a salicylic acid (SA)-dependent pathway (Zimmerli et al., 2001). Furthermore, a recent study of an Arabidopsis mutant in BABA-induced resistance (BABA-IR) towards Hyaloperonospora arabidopsidis has revealed the Impaired in BABA-induced Immunity 1 (IBI1) gene, which encodes an aspartyl-tRNA synthetase (Luna et al., 2014). Enantiomer-specific binding of the R-enantiomer of BABA to IBI1 disrupts its canonical activity and primes the protein for non-canonical defence activity. This results in tRNAasp accumulation, inducing GCN2-dependent phosphorylation of translation initiation factor eIF2α responsible for the consequent growth inhibition. Moreover, the gcn2-1 Arabidopsis mutant remains unaffected in BABA-IR, demonstrating that BABA-IR and BABA-induced stress are controlled via separate pathways (Luna et al., 2014).
With the development of multiple pesticide-resistant pathogen strains, current research efforts tend to focus on the replacement of chemical usage with alternative agents and substances for sustainable, economically profitable agriculture. A potential biological approach exploits the use of pathogen-derived molecules called elicitors for the activation of defence responses and subsequent systemic acquired resistance (SAR) in host plants. This group of chemically diverse compounds includes elicitins, a class of small proteinaceous molecules secreted by members of Phytophthora and Pythium spp. in the family Pythiaceae. Elicitins are known for their capacity to induce the hypersensitive response and SAR against fungal and bacterial pathogens in some plant species such as tobacco (Kamoun et al., 1993) or radish (Bonnet et al., 1996). Oligandrin is an elicitin-like protein secreted by the oomycete Pythium oligandrum. Apart from its mycoparasitical activity, oligandrin adversely affects pathogen growth and development in the host plant and confers enhanced resistance in many plants against various pathogens, i.e. tomato to Phytophthora parasitica (Picard et al., 2000) and Fusarium oxysporum f. sp. radicis-lycopersici (Benhamou et al., 2001) or tobacco to P. parasitica and to phytoplasma (Lherminier et al., 2003). However, the precise mechanism of the elicitation process remains poorly understood. A study of the effect of INF1 elicitin, secreted by P. infestans, on tomato plants has shown that INF1 establishes resistance to bacterial wilt caused by Ralstonia solanacearum (Kawamura et al., 2009). Moreover, treatment by INF1 induces in tomato leaves the expression of jasmonic acid (JA)-responsive PR-6 encoding proteinase inhibitor II, LeATL6 encoding ubiquitin ligase E3 and LOX-E encoding lipoxygenase. The accumulation of ethylene (ET) and expression of ET-responsive genes are also induced in tomato leaves treated by INF1. Thus, the authors of this study suggest that elicitins induce plant resistance via JA- and ET-mediated signalling pathways (Kawamura et al., 2009).
In the present study, we characterized the defence responses of highly resistant wild tomato genotype S. habrochaites and susceptible domesticated inbred genotype S. lycopersicum ‘Amateur’. We quantified the expression of selected defence genes by real-time quantitative PCR (RT-qPCR) in plants treated with two known inducers of resistance, namely the elicitin oligandrin and the non-protein amino acid BABA, alone or with the economically important biotrophic pathogen Oidium neolycopersici, the causative agent of tomato powdery mildew. We describe here the localization and differences in basal expression levels of selected genes between the resistant and susceptible genotypes. We also show that the oligandrin- and BABA-induced resistance against O. neolycopersici are not based on similar signalling defence pathways. Furthermore, we suggest that BABA-induced resistance in tomato is ET-dependent and not based on SA- or JA-dependent signalling pathways.
MATERIALS AND METHODS
Plant material
Two Solanum spp. genotypes, S. lycopersicum ‘Amateur’ and S. habrochaites f. glabratum (LA 2128), expressing differential phenotypes of resistance to O. neolycopersici (Mieslerová et al., 2000) were selected for the studies. Seeds were sown on moistened perlite (Agroperlite, Nový Jičín, Czech Republic) and seedlings were transferred to a garden soil–peat mixture (2:1, v/v) in plastic pots (7 cm in diameter). Plants were grown in a growth chamber with a 12-h photoperiod (light intensity of 100 μmol m−2) and day/night temperature of 20/18 °C. Plants approx. 10 weeks old and with eight true leaves were employed in all experiments.
Plant inoculation
Oligandrin and BABA were dissolved in Milli-Q deioniaed water. A 10 mm solution of BABA was applied by spraying, whereas a 100 nm solution of oligandrin was directly infiltrated into 4th and 5th tomato leaves. Control plants were treated with an equal volume of Milli-Q water.
Freshly sporulating mycelium (8 d post-inoculation) of O. neolycopersici (isolate C-2) was used as the inoculum source. The 4th and 5th oldest true leaves per plant were inoculated by surface contact (dusting/tapping) with the conidia of O. neolycopersici. The leaves were frozen in liquid nitrogen 48 h after the treatment and stored at –80 °C. Identical plant growth conditions and inoculation procedures were employed in three independent experiments.
Oidium neolycopersici phytoprotection test
Plant resistance was induced by spraying of 5–7-week-old tomato plants with BABA or by direct infiltration of oligandrin into the leaflets as described. Two days after the treatment, leaf discs 12 mm in diameter were cut from treated leaves using a cork borer and inoculated with O. neolycopersici. At 48 h post-inoculation, five leaf discs were transferred to glacial acetic acid for 48 h and then mounted in glycerol prior to observations under a light microscope (Olympus BX50) (Lebeda and Reinink, 1994). Pathogen structures were stained with 1 % Evans Blue. Germination of the conidia was expressed as the percentage of conidia producing germ tubes per 100 conidia counted for each leaf disc. Fungal development was assessed as the number of germ tubes per conidium at 48 h post-inoculation and as the length of germ tubes (1st tube with lobbed appressorium, and 2nd and 3rd tubes with nipple-shaped appressorium). At least 100 conidia were observed per each tomato genotype and experimental setting (Piterková et al., 2011).
Quantification of defence gene expression
Potentially interesting transcript sequences corresponding to known targeted functions involved in defence mechanisms were recovered from the genome sequence of the tomato inbred reference tomato cultivar S. lycopersicum ‘Heinz 1706’. These transcripts are generally considered as defence-responsive genes in other model plants such as Nicotiana tabacum or Arabidopsis thaliana (Sels et al., 2008; Spoel and Dong, 2012) and include defensins (def), germins (germ), β-1,3-glucanases (glu), β-1,4-glucanases (gluco), heveins (hev), chitinases (chit), osmotins (osm), thaumatins (thaum) and systemin (syst). The selection was performed using an assembly generated from the available tomato EST-cDNA at NCBI (nearly 300 000) annotated through BlastX against plant sequences and translated to proteins. Subsequently, the recovered transcripts were analysed by the BlastN tool using the available shotgun genome sequence of the wild tomato S. habrochaites to confirm their presence. The specific primers for individual genes were designed using the program Primer3 (Koressaar and Remm, 2007), with annealing temperatures of 60 °C and expected amplicons of 100–300 bp. Gene expression was analysed by RT-qPCR using the fluorescent intercalating dye SYBR-Green and Light Cycler 480 (Roche Diagnostics, Czech Republic). Total RNA was isolated from 100 mg of leaf tissue using TRI reagent (Ambion, Austin, TX, USA) and purified using the TURBO DNA-free kit (Ambion). Reverse transcriptase reactions were performed with the ImProm-II reverse transcription system (Promega, Madison, WI, USA) using 0·4 μg of total RNA in a volume of 20 μl according to the manufacturer’s instructions. Obtained cDNA was amplified by qPCR using gene-specific primers (Supplementary Data Table S1) and GoTaq qPCR Master Mix (Promega) according to the manufacturer’s instructions. PCR amplification was carried out as follows: 45 cycles of DNA denaturation at 95 °C for 20 s, annealing and extension at 60 °C for 40 s. Three biological and technical replicates were analysed for each sample. To evaluate gene expression relative to an endogenous control, the transcript level of each gene was normalized to that of elongation factor 1α (EF-1α) and TIP41-like family protein (TIP41) genes by the ΔΔCT method. It has been shown previously that expression of the EF-1α and TIP41 genes is not influenced by different stress conditions (Exposito-Rodriguez et al., 2008; Nicot et al., 2005).
Analysis of ethylene production
Analysis was performed using a gas chromatography-flame ionization detector as described previously (Malá et al., 2009). Briefly, 1 mL of air was removed from each test tube containing previously weighed whole tomato leaves and analysed using an Agilent GC 6890 equipped with a flame ionization detector and 50-m capillary column (HP-AL/S stationary phase, 15 μm, i.d. = 0·535 mm). The injection temperature was set to 200 °C, oven temperature to 40 °C and detector temperature to 220 °C. The measurements were conducted four times from four different test tubes of each variant. The final concentrations were calculated from the calibration curve and adjusted to vial volume and 1 g of fresh weight.
RESULTS
Comparison of levels of induced resistance by BABA and oligandrin in tomato genotypes
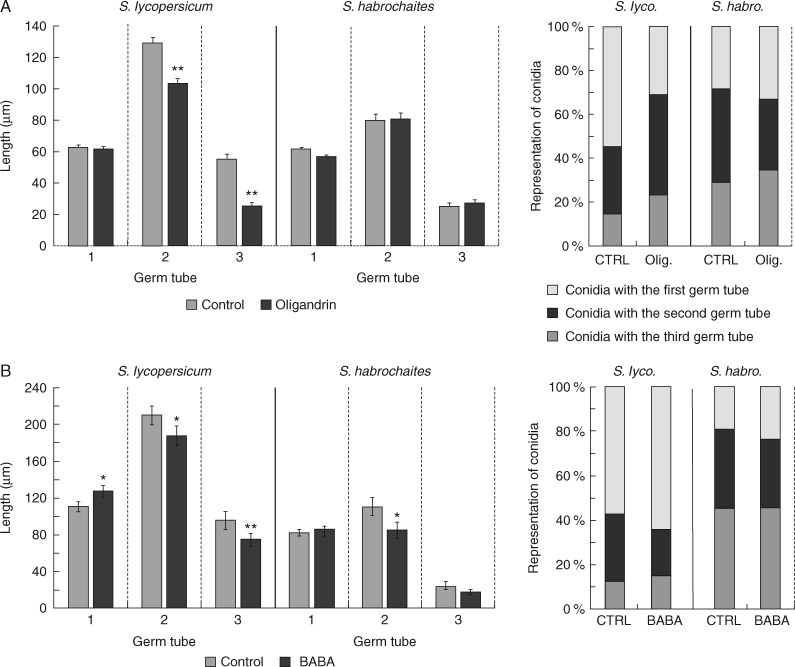
In agreement with previous studies (Mieslerová et al., 2000), we observed a high level of resistance against O. neolycopersici, characterized by decreased lengths and numbers of pathogen 2nd and 3rd germ tubes, in control non-treated tomato plants of wild S. habrochaites compared to susceptible genotype S. lycopersicum ‘Amateur’ (Fig. 1). Treatment of 7-week-old tomato plants with BABA and oligandrin, using optimal concentrations reported in previous publications (Kawamura et al., 2009; Bengtsson et al., 2014a), resulted in significant changes in the plant resistance against the biotrophic pathogen O. neolycopersici (Fig. 1). Oligandrin at a concentration of 100 nm induced a significantly increased resistance of S. lycopersicum ‘Amateur’ against O. neolycopersici characterized by a decreased percentage of 3rd germ tubes and decreased lengths of 2nd and 3rd germ tubes (Fig. 1A). Similarly, the application of 10 mm BABA induced significant resistance against O. neolycopersici in S. lycopersicum ‘Amateur’ in agreement with previous studies (Worrall et al., 2012; Bengtsson et al., 2014a). However, in contrast to oligandrin, BABA application induced significant changes only in the length of the conidia and not their representation (Fig. 1B). On the other hand, both oligandrin and BABA application induced almost no observable changes in resistance in S. habrochaites, which may be a consequence of its considerably higher basal resistance (Fig. 1). The only observed changes related to the resistance response of S. habrochaites plants induced by BABA were slightly decreased lengths of pathogen 2nd germ tubes (Fig. 1B).
Fig. 1.
Oligandrin- and BABA-induced resistance of tomato plants against Oidium neolycopersici. The relative representation of pathogen germ tube lengths and pathogen developmental structures was determined 24 h after the inoculation with O. neolycopersici (isolate C-2) on leaf discs from 7-week-old tomato plants (Solanum lycopersicum ‘Amateur’ and Solanum habrochaites) treated with (A) the elicitin oligandrin (100 nm) and (B) BABA (10 mm). Data are presented as means ± s.e. of three replicates from three independent experiments. Each replicate corresponds to eight inoculated areas on four leaves from one plant. Asterisks denote a significant difference of the mean values compared to the control group determined by Student’s t-test at **P ≤ 0·05 or **P ≤ 0·01.
Selection of genes involved in tomato defence responses
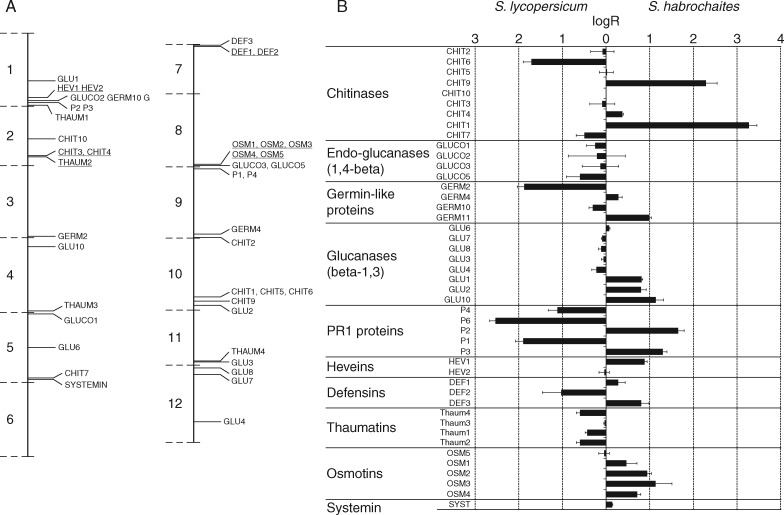
To determine the molecular mechanisms involved in BABA and oligandrin-derived activation of defence in tomato, the expression levels of selected defence gene transcripts were measured by qPCR. Transcript changes were also monitored after plant infection with O. neolycopersici, in both compatible and incompatible interactions. In total, 55 genes generally considered as defence responsive in other model plants such as N. tabacum or A. thaliana were selected. These genes included defensins (def), germins (germ), β-1,3-glucanases (glu), β-1,4-glucanases (gluco), heveins (hev), chitinases (chit), osmotins (osm), systemin (syst) and genes of PR1 proteins. During the optimization of reaction conditions, ten genes were excluded on the basis of very low basal transcript levels, non-specific amplification or primer dimer formation. Thus, only 45 of the 55 genes were finally selected for the ensuing experiments (Table S2). Mapping of these genes on the reference genome of S. lycopersicum ‘Heinz 1706’ showed that considering the number of genes on each chromosome there was a relatively higher concentration of these defence genes on chromosomes 1, 5, 8, 9 and 10. Intriguingly, there was no overlap with previously analysed genomic locations of domestication sweeps, but on chromosomes 1 (2), 2 (3) and 8 (4) there was an overlap with the genomic locations of improvement sweeps analysed and described in another recent study (Lin et al., 2014) (Fig. 2A).
Fig. 2.
Solanum lycopersicum ‘Amateur’ and Solanum habrochaites exhibit differences in the basal levels of defence-related gene transcripts. (A) Location of individual transcripts in the reference genome of Solanum lycopersicum ‘Heinz 1706’. Underlined transcripts overlapped with the genomic locations of improvement sweeps analysed in the study of Lin et al. (2014). (B) Overview of the differential accumulation of candidate defence-related transcripts identified by RT-qPCR in control non-infected leaf tissues of the respective tomato genotypes.
Comparison of basal levels of defence gene transcripts
Initial comparison of basal levels of defence gene transcripts between the highly resistant wild tomato S. habrochaites and the susceptible genotype S. lycopersicum ‘Amateur’ revealed that about half of the analysed genes with expected roles in tomato defence showed different basal expression levels (Fig. 2). However, only 14 transcripts showed an upregulation in S. habrochaites compared to S. lycopersicum ‘Amateur’, while the remaining 11 transcripts were downregulated (Fig. 2). Interestingly, 12 of the 14 transcripts (86 %) upregulated in the highly resistant genotype were found to co-localize on chromosomes 1, 8 and 10 to quantitative trait loci (QTLs) for early blight and P. infestans resistance (Foolad, 2007). These observations provide additional support for the involvement of these genes in resistance mechanisms in the highly resistant genotype, in contrast to upregulated transcripts observed in the susceptible genotype, which showed an almost homogeneous genomic distribution.
Comparison of induction of defence-related transcripts by BABA, oligandrin and pathogen infection
The non-protein amino acid BABA is a well-known priming agent that induces resistance in many plants against various pathogens. The manner by which BABA mediates resistance has recently been unmasked as the disruption of the canonical activity of an aspartyl-tRNA synthetase (Luna et al., 2014). Application of BABA as a foliar spray is one of the most common application methods in laboratory and field conditions. The optimized concentration of 10 mm BABA used in this study was chosen to overcome possible side effects on treated plants manifested as hypersensitive response (HR)-like lesions with subsequent leaf wilting (Cohen et al., 1994; Siegriest et al., 2000; Bengtsson et al., 2014a). During testing of the optimal BABA concentrations on the two tomato genotypes, an interesting phenomenon was observed. Treatment of susceptible S. lycopersicum ‘Amateur’ plants with 50 and 100 mm BABA caused severe side effects, as demonstrated by the formation of HR-like lesions 2 d after treatment, whereas no visible symptoms were observed in S. habrochaites plants even after the application of 100 mm BABA (Fig. 3). In this respect, the behaviour of S. habrochaites plants is in accordance with the study of Cohen et al. (2010) that was conducted on lettuce, with formation of HR-like lesions as the result of plant-specific responses to BABA.
Fig. 3.
Necrotic symptoms triggered by BABA application to tomato leaves. Solanum lycopersicum ‘Amateur’ (A) and Solanum habrochaites (B) plants were sprayed with 10, 50 and 100 mm BABA and the photographs of representative leaves were obtained 48 h later.
Application of 10 mm BABA solution onto leaves of S. lycopersicum ‘Amateur’ resulted in significant (more than three times) upregulation of ten measured transcripts, with the highest inductions being observed in the PR1 class (Fig. 4, Supplementary Data Table S2). On the other hand, no upregulation of defensins, germin-like proteins, β-1,4-glucanases or thaumatin-like proteins was observed. Surprisingly, the application of 10 mm BABA solution onto leaves of S. habrochaites resulted in almost no significant upregulation of any measured transcript, with the exception of P4 and P6 transcripts belonging to the PR1 family (Fig. 4). However, this finding is in agreement with the compromised response to BABA observed on the leaves.
Fig. 4.

Changes in the expression of tomato defence-related genes induced by oligandrin or BABA treatment and after infection by Oidium neolycopersici. Overview of the differential accumulation of candidate defence-related transcripts identified by RT-qPCR 48 h after treatment or inoculation. Yellow and purple colours represent upregulated and downregulated transcripts, respectively. The y-axis shows hierarchical clustering of the corresponding protein sequences by the unweighted pair group method with arithmetic mean (UPGMA) method in MEGA6 software. Water-sprayed leaves (BABA, Oidium neolycopersici) or leaflets infiltrated with water (oligandrin) were used as the control samples.
Infiltration of plant leaves with 100 nm solution of oligandrin triggered the upregulation of multiple transcripts in both genotypes (Fig. 4, Table S2). A higher induction rate (18 transcripts) was observed in the highly resistant genotype S. habrochaites. Moreover, from 13 transcripts upregulated in S. lycopersicum ‘Amateur’, most of the ten transcripts were shared with those upregulated in S. habrochaites, pointing to a similar signalling mechanism involved in plant responses to oligandrin in both studied genotypes. A notable finding was a strong downregulation of several chitinase and osmotin transcripts in both genotypes as well as an upregulation of the systemin transcript, probably due to the mechanical wounding caused by oligandrin infiltration through the leaf abaxial layer (Fig. 4, Table S2).
Infection of tomato plants with the biotrophic pathogen O. neolycopersici clearly demonstrated differences between the resistant and susceptible genotype. While in the infected highly resistant genotype a strong induction of 24 transcripts could be observed, only 14 transcripts were found to be induced in the highly susceptible genotype, the majority of them common for both genotypes. This finding might be related to a generally different extent and timing of transcript induction in the susceptible and resistant tomato genotypes. In our previous studies on these tomato genotypes, we also found differences in the extent and timing of the increased production of defence signalling molecules NO and H2O2 during the time-course of powdery mildew invasion (Piterková et al., 2009; Lebeda et al., 2014). The most remarkable difference between the genotypes studied was in the PR1 and germin-like protein classes (Fig. 4). Moreover, in comparison with a recent study on tomato plants inoculated with Phytophthora capsici (Jupe et al., 2013), there was almost perfect correspondence regarding the downregulation of germ2, gluco5 and thaum2 transcripts caused by O. neolycopersici infection (Fig. 4, Table S2).
Overlap of upregulated genes among BABA-, oligandrin- and pathogen-treated plants
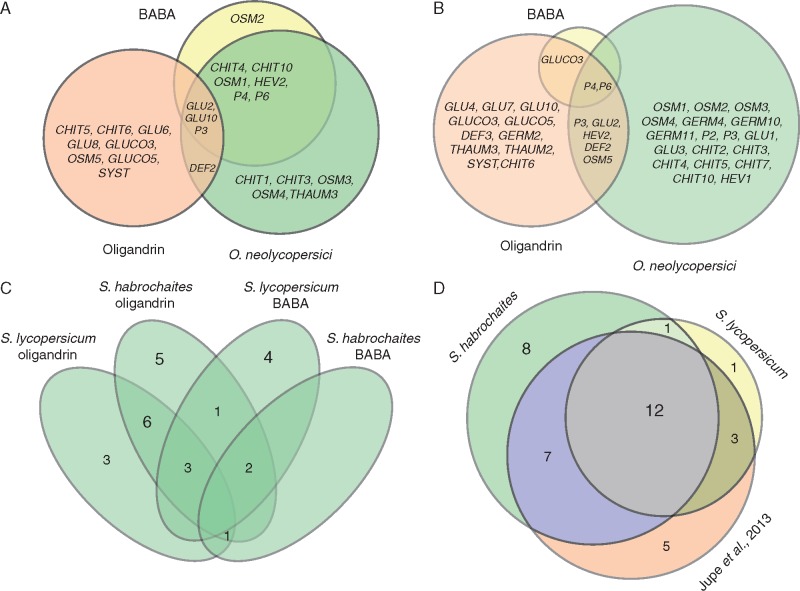
Initial comparison of the transcript profiles revealed a high similarity in the results obtained from BABA-treated and O. neolycopersici-infected S. lycopersicum ‘Amateur’ plants, as well as a very low overlap between oligandrin-treated and O. neolycopersici-infected plants of both studied genotypes (Fig. 5). Moreover, in both genotypes, transcripts of genes encoding GERM2, GLUCO5 or OSM4 demonstrated an opposing regulation between the oligandrin treatment and O. neolycopersici infection. These findings raise a question on the different regulation mechanisms involved in plant responses to oligandrin and BABA or O. neolycopersici. In the case of elicitins, activation of JA- and ET-mediated signalling pathways has been previously demonstrated (Kawamura et al., 2009). For BABA treatment, an SA-dependent pathway has been well described in Arabidopsis plants; however, to the best of our knowledge, no such study showing evidence for SA-dependent signalling in tomato has been published. Therefore, we analysed the promoter regions of BABA-upregulated transcripts spanning about 1000 bp before the start codon using the PlantPAN program (Chang et al., 2008) in the tomato transcription factor library. In the majority of BABA-upregulated transcripts, the sequence for the transcription factor ERELEE4 containing an ET-responsive element was identified (Table S2). Based on this result, the transcript levels of key enzymes involved in the ET synthesis pathway, namely ACC synthase and ACC oxidase, were determined along with quantification of the production of ET. On the basis of previous studies of different ACC synthase isoforms in Arabidopsis (Skottke et al., 2011; Li et al., 2012) and ACC synthase and oxidase in tomato (Lincoln et al., 1993; Kim et al., 1998; Cohn and Martin, 2005; Shi and Zhang, 2014; Yim et al., 2014), three candidate transcripts for each enzyme (ACS1a, ACS2, ACS6 for ACC synthase, and ACO1, ACO 1-like and ACO 4-like for ACC oxidase) with suspected roles in pathogenesis processes were selected.
Fig. 5.
Venn diagrams showing the relationship among treatments. (A, B) Venn diagrams showing the transcript induction overlap of two inducers, namely BABA and oligandrin, and Oidium neolycopersici infection in Solanum lycopersicum ‘Amateur’ (A) and Solanum habrochaites (B) plants measured by RT-qPCR. (C) Overlap of induced transcripts in BABA- and oligandrin-treated plants measured by RT-qPCR. (D) Overlap of Phytophthora capsici-induced transcripts from the study by Jupe et al. (2013) with our results of O. neolycopersici infection of S. lycopersicum ‘Amateur’ and S. habrochaites plants. Transcript levels were measured 48 h after infection by microarray (Jupe et al., 2013) and RT-qPCR and compared to control represented by time zero.
In the BABA-treated plants, a good correlation between the accumulation of defence transcripts after 24 h and the ET production was observed (Fig. 6). In BABA-responsive S. lycopersicum plants, a significant upregulation of ACS2 transcript followed by an increased ET production was observed. In contrast, in BABA low-responsive S. habrochaites plants, there was no upregulation of any analysed ACS transcript nor any increase of ET production (Fig. 6). In both genotypes, an upregulation of ACO 4-like transcript was observed in BABA-treated plants. However, unlike ACC synthase, ACC oxidase is not considered as the key regulatory enzyme in ET synthesis (Iqbal et al., 2013).
Fig. 6.
BABA treatment or Oidium neolycopersici infection activates ET production and enzymes of the ET synthesis pathway in tomato plants. (A, B) Transcript accumulation of genes from the ET biosynthesis pathway was measured 24 h after treatment/inoculation by RT-qPCR. (C) ET accumulation was measured 24 and 48 h after BABA treatment by gas chromatography (n = 4). The control tissue was a water-treated control sample. Each bar represents the mean ± s.e.
Following O. neolycopersici infection, an upregulation of the ACS2 transcript was observed in both genotypes, while the ACO transcripts were upregulated predominantly in S. habrochaites (Fig. 6). This finding is clearly related to the more intense upregulation of defence-related transcripts in S. habrochaites after O. neolycopersici inoculation. Treatment with oligandrin did not trigger upregulation of any analysed ACO transcript. However, it did cause an upregulation of ACS2 transcript in the S. habrochaites genotype, corresponding to a more intense upregulation of defence-related transcripts in this genotype upon oligandrin treatment.
DISCUSSION
In the present study, we attempted to analyse and compare the responses of susceptible and resistant tomato genotypes to resistance inducers oligandrin and BABA and to infection by the powdery mildew fungus O. neolycopersici, which represents a typical biotrophic pathogen of tomato. The proteinaceous elicitor oligandrin from P. oligandrum, representing elicitins as the best known oomycete pathogen-associated molecular pattern (M/PAMPs), induces the HR and SAR in several plants (Picard et al., 2000; Benhamou et al., 2001). BABA represents a non-protein amino acid generally accepted as a potent priming agent conferring increased resistance to biotic and abiotic stresses in many studied plants. As model tomato genotypes, we selected the previously well-characterized highly susceptible S. lycopersicum ‘Amateur’, representing a domesticated inbred cultivar, and S. habrochaites that exhibits a high non-specific resistance (Mieslerová et al., 2000; Tománková et al., 2006; Piterková et al., 2009, 2011; Lebeda et al., 2014). The defence responses were monitored as transcript-level changes of selected genes with proven roles in non-specific resistance in tomato and model plants N. tabacum or A. thaliana.
Interestingly, the localization of the targeted genes to the chromosome map of the inbred reference tomato cultivar S. lycopersicum ‘Heinz 1706’ showed their clustering towards the ends of chromosomes 1, 5, 8, 9 and 10. The subsequent analysis of basal transcript levels in both tested genotypes suggested a role of genes located on chromosomes 1, 8 and 10 in plant disease resistance. Several other studies have confirmed the relevance of genes on these chromosomes to defence processes. Following an infection of tomato with P. infestans, an over-representation of gene sets belonging to chromosome 10 and a similar tendency in their localization towards the ends of chromosomes were recently detected by gene set enrichment analysis (Lopez-Kleine et al., 2013). Moreover, in agreement with our results, QTLs for early blight resistance in tomato using backcross populations of an S. lycopersicum × S. habrochaites cross have been identified in chromosomes 1, 2, 5, 8, 9, 10 and 12 and QTLs for early resistance to P. infestans have also been detected in chromosomes 3, 4, 5, 8, 10 and 11 (reviewed by Foolad, 2007).
Our results suggest an absence of involvement for five genes (gluco1, gluco2, def1, thaum1 and thaum4) in the resistance process with no significant upregulation by any treatment or increased basal levels in the resistant genotype. Even though the numbers of differently expressed genes at basal levels of both genotypes were quite similar, we noticed that the expression of the majority of downregulated genes in the susceptible compared to the resistant genotype was not affected by any treatment.
Pathogenesis experiments in the resistant S. habrochaites plants clearly demonstrated the previously proven high level of basal non-specific resistance to O. neolycopersici, similar to that induced by BABA or oligandrin in the susceptible S. lycopersicum genotype (Mieslerová et al., 2000). This high level of basal non-specific resistance was found to be associated with the upregulation of two chitinases, three β-1,3-glucanases and four osmotins. Transgenic plants overexpressing chitinase alone or in combination with β-1,3-glucanase have been previously shown to display an enhanced resistance when challenged with a powdery mildew infection (Oldach et al., 2001) or bacterial pathogens (Dana et al., 2006). Delayed symptoms of Fusarium head blight have also been reported in these transgenic plants (Anand et al., 2004). Furthermore, overexpression of osmotins acting as antifungal cytotoxic compounds delayed the development of P. infestans and disease symptoms in infected potato plants (Liu et al., 1994; Zhu et al., 1996).
Oligandrin treatment of the susceptible S. lycopersicum plants demonstrated the effectiveness of elicitins in inducing resistance to O. neolycopersici. The observed resistance in BABA-treated S. lycopersicum plants against O. neolycopersici infection has previously been demonstrated for BABA-treated tomato seeds (Worrall et al., 2012). However, our data from BABA-treated leaves show a direct activation of the defence responses, characterized by an enhanced accumulation of pathogenesis-related genes, rather than the priming effect, as observed in the study on tomato seeds. Such activation of defence responses has been reported in A. thaliana, with BABA applied as a foliar spray, in contrast to soil drench, enhancing expression of the PR1 gene (Jakab et al., 2001). Similarly, a global effect on the transcriptome and an increased abundance of secreted proteins related to resistance was observed in tobacco and potato plants only after application of a BABA foliar spray (Siegriest et al., 2000; Bengtsson et al., 2014a, b). It therefore appears that the mode of action of BABA as a priming or resistance-inducing agent may depend upon the application method. A clear explanation of this phenomenon is a matter of debate and deserves further investigation.
Our data obtained from BABA-treated leaves indicate a different resistance mechanism to that observed with oligandrin, with the suppression of germ tube elongation occurring, rather than the inhibition of conidial germination. The very low overlap of BABA- and oligandrin-induced transcripts in S. lycopersicum and opposing trends in their expression support this suggestion of different modes of action. Even though the relationship between different signalling pathways is still a matter of debate (Glazebrook, 2005; Vleeshouwers and Oliver, 2014), the very low overlap between BABA- and oligandrin-induced transcripts may be a result of the previously reported dependence of elicitin INF1-induced resistance on the activation of JA- and ET-mediated signalling but not on SA-mediated signalling (Kawamura et al., 2009). Perhaps the dependence of BABA-induced resistance on both SA- and abscisic acid (ABA)-dependent defence mechanisms might be involved (Ton et al., 2005). However, we found a low representation of ABA-responsive element (ABRE) and a high representation of ET-responsive elements in the promoter regions of BABA-responsive genes (Table S2). This result, together with an increased ET production and a clear upregulation of key enzymes (ACS2 and ACO4-like) of the ET synthesis pathway, supports suggested ET-dependent mechanisms in BABA-induced resistance in tomato. The participation of ET signalling in BABA-primed resistance in tomato is further promoted by the observation that BABA as well as ET increased disease severity caused by the necrotrophs (van Loon et al., 2006; Worrall et al., 2012). Moreover, we found an interesting overlap of transcripts upregulated by BABA and O. neolycopersici with those detected in Phytophthora capsici–tomato interaction and suggesting involvement of ET in the disease development during early and biotrophic infection stages (Jupe et al., 2013). All these findings indicate strongly the involvement of ET in BABA-induced resistance against biotrophic and hemi-biotrophic pathogens in tomato in contrast to Arabidopsis or tobacco plants (Ton et al., 2005).
Interestingly, the virtual absence of upregulation of the analysed genes in S. habrochaites plants upon BABA application is consistent with the lack of observable symptoms after BABA treatment; however, the primary cause of this effect is still unknown. This different response has already been demonstrated in potato cultivars exhibiting different levels of resistance (Bengtsson et al., 2014a). The partial response of resistant S. habrochaites plants to very high concentrations of BABA requires confirmation by screening a larger set of contrasting genotypes. Similarly, whether the tolerance of the resistant genotype to BABA treatment is a consequence of basal resistance requires further investigation.
In conclusion, the genotype S. habrochaites, exhibiting a high level of non-specific resistance in previous studies, showed considerably higher basal levels of transcripts with proven roles in the resistance process against a broad range of pathogens and a more extensive induction of defence-related transcripts following infection. For five genes (gluco1, gluco2, def1, thaum1 and thaum4), the absence of any significant upregulation following any treatment or increased basal expression in the resistant genotype suggests no involvement in the resistance response. In the present study, BABA and oligandrin treatment of the susceptible tomato genotype resulted in induction of defence-related transcripts, accompanied by an enhancement of resistance against the biotrophic pathogen. Nevertheless, different regulation mechanisms for induced resistance were observed for each compound, with ET-dependent signalling, rather than SA- or JA-dependent pathways, appearing to be involved in BABA-induced resistance in tomato. In the context of previous studies on Arabidopsis and tobacco, our findings suggest that diverse signalling mechanisms occur in BABA-induced resistance in plants, dependent on the mode of application and model plant under study.
SUPPLEMENTARY DATA
Supplementary data are available online at www.aob.oxfordjournals.org and consist of the following. Table S1: sequences of the primers used for qPCR. Table S2: results of qPCR analysis of selected genes involved in tomato defence responses.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the Grant Agency of the Czech Republic (P501/12/0590).
LITERATURE CITED
- Anand A, Lei ZT, Sumner LW, et al. 2004. Apoplastic extracts from a transgenic wheat line exhibiting lesion-mimic phenotype have multiple pathogenesis-related proteins that are antifungal. Molecular Plant-Microbe Interactions 17: 1306–1317. [DOI] [PubMed] [Google Scholar]
- Bengtsson T, Holefors A, Witzell J, Andreasson E, Liljeroth E.. 2014a. Activation of defence responses to Phytophthora infestans in potato by BABA. Journal of Experimental Botany 63: 193–202. [Google Scholar]
- Bengtsson T, Weighill D, Proux-Wera E, et al. 2014b. Proteomics and transcriptomics of the BABA-induced resistance response in potato using a novel functional annotation approach. BMC Genomics 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamou N, Belanger RR, Rey P, Tirilly Y.. 2001. Oligandrin, the elicitin-like protein produced by the mycoparasite Pythium oligandrum, induces systemic resistance to Fusarium crown and root rot in tomato plants. Plant Physiology and Biochemistry 39: 681–696. [Google Scholar]
- Bonnet P, Bourdon E, Ponchet M, Blein JP, Ricci P.. 1996. Acquired resistance triggered by elicitins in tobacco and other plants. European Journal of Experimental Botany 102: 181–192. [Google Scholar]
- Chang WC, Lee TY, Huang HD, Huang HY, Pan RL.. 2008. PlantPAN: plant promoter analysis navigator, for identifying combinatorial cis-regulatory elements with distance constraint in plant gene groups. BMC Genomics 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen Y. 2002. β-Aminobutyric acid-induced resistance against plant pathogens. Plant Disease 86: 448–457. [DOI] [PubMed] [Google Scholar]
- Cohen Y, Niderman T, Mosinger E, Fluhr R.. 1994. Beta-aminobutyric acid induces the accumulation of pathogenesis-related proteins in tomato (Lycopersicon esculentum L.) plants and resistance to late blight infection caused by Phytophthora infestans. Plant Physiology 104: 59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen Y, Rubin AE, Kilfin G.. 2010. Mechanisms of induced resistance in lettuce against Bremia lactucae by DL-β-amino-butyric acid (BABA). European Journal of Plant Pathology 126: 553–573. [Google Scholar]
- Cohn JR, Martin GB.. 2005. Pseudomonas syringae pv. tomato type III effectors AvrPto and AvrPtoB promote ethylene-dependent cell death in tomato. The Plant Journal 44: 139–154. [DOI] [PubMed] [Google Scholar]
- Conrath U. 2009. Priming of induced plant defense responses. Plant Innate Immunity 51: 361–395. [Google Scholar]
- Dana MD, Pintor-Toro JA, Cubero B.. 2006. Transgenic tobacco plants overexpressing chitinases of fungal origin show enhanced resistance to biotic and abiotic stress agents. Plant Physiology 142: 722–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exposito-Rodriguez M, Borges AA, Borges-Perez A, Perez JA.. 2008. Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biology 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foolad MR. 2007. Genome mapping and molecular breeding of tomato. International Journal of Plant Genomics 2007: 1–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J. 2005. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annual Review of Phytopathology 43: 205–227. [DOI] [PubMed] [Google Scholar]
- Iqbal N, Trivellini A, Masood A, Ferrante A, Khan NA.. 2013. Current understanding on ethylene signalling in plants: the influence of nutrient availability. Plant Physiology and Biochemistry 73: 128–138. [DOI] [PubMed] [Google Scholar]
- Jakab G, Cottier V, Toquin V, et al. 2001. β-Aminobutyric acid-induced resistance in plants. European Journal of Experimental Botany 107: 29–37. [Google Scholar]
- Jupe J, Stam R, Howden AJM, et al. 2013. Phytophthora capsici-tomato interaction features dramatic shifts in gene expression associated with a hemi-biotrophic lifestyle. Genome Biology 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoun S, Young M, Glascock CB, Tyler BM.. 1993. Extracellular protein elicitors from Phytophthora - host-specificity and induction of resistance to bacterial and fungal phytopathogens. Molecular Plant-Microbe Interactions 6: 15–25. [Google Scholar]
- Kawamura Y, Hase S, Takenaka S, et al. 2009. INF1 elicitin activates jasmonic acid- and ethylene-mediated signalling pathways and induces resistance to bacterial wilt disease in tomato. Journal of Phytopathology 157: 287–297. [Google Scholar]
- Kim YS, Choi D, Lee MM, Lee SH, Kim WT.. 1998. Biotic and abiotic stress-related expression of 1-aminocyclopropane-1-carboxylate oxidase gene family in Nicotiana glutinosa L. Plant and Cell Physiology 39: 565–573. [DOI] [PubMed] [Google Scholar]
- Koressaar T, Remm M.. 2007. Enhancements and modifications of primer design program Primer3. Bioinformatics 23: 1289–1291. [DOI] [PubMed] [Google Scholar]
- Lebeda A, Reinink K.. 1994. Histological characterization of resistance in Lactuca-Saligna to lettuce downy mildew (Bremia-Lactucae). Physiological and Molecular Plant Pathology 44: 125–139. [Google Scholar]
- Lebeda A, Mieslerová B, Petřivalský M, et al. 2014. Resistance mechanisms of wild tomato germplasm to infection of Oidium neolycopersici. European Journal of Experimental Botany 138: 569–596. [Google Scholar]
- Lherminier J, Benhamou N, Larrue J, et al. 2003. Cytological characterization of elicitin-induced protection in tobacco plants infected by Phytophthora parasitica or phytoplasma. Phytopathology 93: 1308–1319. [DOI] [PubMed] [Google Scholar]
- Li GJ, Meng XZ, Wang RG, Mao GH, Han L, Liu YD, Zhang SQ.. 2012. Dual-level regulation of ACC synthase activity by MPK3/MPK6 cascade and its downstream WRKY transcription factor during ethylene induction in Arabidopsis. PLoS Genetics 8: e1002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T, Zhu GT, Zhang JH, et al. 2014. Genomic analyses provide insights into the history of tomato breeding. Nature Genetics 46: 1220–1226. [DOI] [PubMed] [Google Scholar]
- Lincoln JE, Campbell AD, Oetiker J, et al. 1993. Le-Acs4, a fruit ripening and wound-induced 1-aminocyclopropane-1-carboxylate synthase gene of tomato (Lycopersicon esculentum) – Expression in Escherichia coli, structural characterization, expression characteristics, and phylogenetic analysis. Journal of Biological Chemistry 268: 19422–19430. [PubMed] [Google Scholar]
- Liu D, Raghothama KG, Hasegawa PM, Bressan RA.. 1994. Osmotin overexpression in potato delays development of disease symptoms. Proceedings of the National Academy of Sciences of the USA 91: 1888–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Kleine L, Pinzon A, Chaves D, Restrepo S, Riano-Pachon DM.. 2013. Chromosome 10 in the tomato plant carries clusters of genes responsible for field resistance/defence to Phytophthora infestans. Genomics 101: 249–255. [DOI] [PubMed] [Google Scholar]
- Luna E, van Hulten M, Zhang Y, et al. 2014. Plant perception of beta-aminobutyric acid is mediated by an aspartyl-tRNA synthetase. Nature Chemical Biology 10: 450–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malá J, Machová P, Cvrčková H, et al. 2009. Micropropagation of wild service tree (Sorbus torminalis [L.] Crantz): the regulative role of different aromatic cytokinins during organogenesis. Journal of Plant Growth Regulation 28: 341–348. [Google Scholar]
- Menda N, Strickler SR, Edwards JD, et al. 2014. Analysis of wild-species introgressions in tomato inbreds uncovers ancestral origins. BMC Plant Biology 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieslerová B, Lebeda A, Chetelat RT.. 2000. Variation in response of wild Lycopersicon and Solanum spp. against tomato powdery mildew (Oidium lycopersici). Journal of Phytopathology 148: 303–311. [Google Scholar]
- Nicot N, Hausman JF, Hoffmann L, Evers D.. 2005. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. Journal of Experimental Botany 56: 2907–2914. [DOI] [PubMed] [Google Scholar]
- Oka Y, Cohen Y.. 2001. Induced resistance to cyst and root-knot nematodes in cereals by dl-beta-amino-N-butyric acid. European Journal of Plant Pathology 107: 219–227. [Google Scholar]
- Oldach KH, Becker D, Lorz H.. 2001. Heterologous expression of genes mediating enhanced fungal resistance in transgenic wheat. Molecular Plant-Microbe Interactions 14: 832–838. [DOI] [PubMed] [Google Scholar]
- Picard K, Ponchet M, Blein JP, Rey P, Tirilly Y, Benhamou N.. 2000. Oligandrin. A proteinaceous molecule produced by the mycoparasite Pythium oligandrum induces resistance to Phytophthora parasitica infection in tomato plants. Plant Physiology 124: 379–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piękna-Grochala J, Kępczyńska E.. 2013. Induction of resistance against pathogens by beta-aminobutyric acid. Acta Physiologiae Plantarum 35: 1735–1748. [Google Scholar]
- Piterková J, Petřivalský M, Luhová L, Mieslerová B, Sedlářová M, Lebeda A.. 2009. Local and systemic production of nitric oxide in tomato responses to powdery mildew infection. Molecular Plant Pathology 10: 501–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piterková J, Hofman J, Mieslerová B, et al. 2011. Dual role of nitric oxide in Solanum spp.–Oidium neolycopersici interactions. Environmental and Experimental Botany 74: 37–44. [Google Scholar]
- Sels J, Mathys J, De Coninck BM, Cammue BP, De Bolle MF.. 2008. Plant pathogenesis-related (PR) proteins: a focus on PR peptides. Plant Physiology and Biochemistry 46: 941–950. [DOI] [PubMed] [Google Scholar]
- Shi HY, Zhang YX.. 2014. Expression and regulation of pear 1-aminocyclopropane-1-carboxylic acid synthase gene (PpACS1a) during fruit ripening, under salicylic acid and indole-3-acetic acid treatment, and in diseased fruit. Molecular Biology Reports 41: 4147–4154. [DOI] [PubMed] [Google Scholar]
- Siegrist J, Orober M, Buchenauer H.. 2000. β-Aminobutyric acid-mediated enhancement of resistance in tobacco to tobacco mosaic virus depends on the accumulation of salicylic acid. Physiological and Molecular Plant Pathology 56: 95–106. [Google Scholar]
- Sim SC, Robbins MD, Van Deynze A, Michel AP, Francis DM.. 2011. Population structure and genetic differentiation associated with breeding history and selection in tomato (Solanum lycopersicum L.). Heredity 106: 927–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skottke KR, Yoon GM, Kieber JJ, DeLong A.. 2011. Protein phosphatase 2A controls ethylene biosynthesis by differentially regulating the turnover of ACC synthase isoforms. PLoS Genetics 7: e1001370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Dong X.. 2012. How do plants achieve immunity? Defence without specialized immune cells. Nature Reviews Immunology 12: 89–100. [DOI] [PubMed] [Google Scholar]
- Tománková K, Luhová L, Petřivalský M, Peč P, Lebeda A.. 2006. Biochemical aspects of reactive oxygen species formation in the interaction between Lycopersicon spp. and Oidium neolycopersici. Physiological and Molecular Plant Pathology 68: 22–32. [Google Scholar]
- Ton J, Jakab G, Toquin V, et al. 2005. Dissecting the beta-aminobutyric acid-induced priming phenomenon in arabidopsis. The Plant Cell 17: 987–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon LC, Geraats BPJ, Linthorst HJM.. 2006. Ethylene as a modulator of disease resistance in plants. Trends in Plant Science 11: 184–191. [DOI] [PubMed] [Google Scholar]
- Vleeshouwers VGA, Oliver RP.. 2014. Effectors as tools in disease resistance breeding against biotrophic, hemibiotrophic, and necrotrophic plant pathogens. Molecular Plant-Microbe Interactions 27: 196–206. [DOI] [PubMed] [Google Scholar]
- Worrall D, Holroyd GH, Moore JP, et al. 2012. Treating seeds with activators of plant defence generates long-lasting priming of resistance to pests and pathogens. New Phytologist 193: 770–778. [DOI] [PubMed] [Google Scholar]
- Wu CC, Singh P, Chen MC, Zimmerli L.. 2010. l-Glutamine inhibits beta-aminobutyric acid-induced stress resistance and priming in Arabidopsis. Journal of Experimental Botany 61: 995–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim WJ, Kim KY, Lee YW, Sundaram SP, Lee Y, Sa TM.. 2014. Real time expression of ACC oxidase and PR-protein genes mediated by Methylobacterium spp. in tomato plants challenged with Xanthomonas campestris pv. vesicatoria. Journal of Plant Physiology 171: 1064–1075. [DOI] [PubMed] [Google Scholar]
- Zhu BL, Chen THH, Li PH.. 1996. Analysis of late-blight disease resistance and freezing tolerance in transgenic potato plants expressing sense and antisense genes for an osmotin-like protein. Planta 198: 70–77. [DOI] [PubMed] [Google Scholar]
- Zimmerli L, Metraux JP, Mauch-Mani B.. 2001. β-Aminobutyric acid-induced protection of Arabidopsis against the necrotrophic fungus Botrytis cinerea. Plant Physiology 126: 517–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.