Abstract
Background Plants are constantly exposed to evolving pathogens and pests, with crop losses representing a considerable threat to global food security. As pathogen evolution can overcome disease resistance that is conferred by individual plant resistance genes, an enhanced understanding of the plant immune system is necessary for the long-term development of effective disease management strategies. Current research is rapidly advancing our understanding of the plant innate immune system, with this multidisciplinary subject area reflected in the content of the 18 papers in this Special Issue.
Scope Advances in specific areas of plant innate immunity are highlighted in this issue, with focus on molecular interactions occurring between plant hosts and viruses, bacteria, phytoplasmas, oomycetes, fungi, nematodes and insect pests. We provide a focus on research across multiple areas related to pathogen sensing and plant immune response. Topics covered are categorized as follows: binding proteins in plant immunity; cytokinin phytohormones in plant growth and immunity; plant–virus interactions; plant–phytoplasma interactions; plant–fungus interactions; plant–nematode interactions; plant immunity in Citrus; plant peptides and volatiles; and assimilate dynamics in source/sink metabolism.
Conclusions Although knowledge of the plant immune system remains incomplete, the considerable ongoing scientific progress into pathogen sensing and plant immune response mechanisms suggests far reaching implications for the development of durable disease resistance against pathogens and pests.
Keywords: PAMP-triggered immunity, effector-triggered immunity, NLRs, lectins, cytokinins, plant antiviral immunity, phytoplasma, fungi, nematodes, citrus, induced resistance, defensins, terpenoids, source/sink dynamics
INTRODUCTION
Plants are exposed to a vast range of pathogens and pests. In natural ecosystems, the coevolution during millions of years between genetically diverse plant and pathogen populations has resulted in disease being relatively rare and geographically restricted. In contrast, agricultural environments with monoculture cropping systems often provide an environment for the selection of virulent pathogen races, which can result in considerable pre-harvest crop losses, threatening food security (Popp et al., 2011; Boyd et al., 2012; Dangl et al., 2013). This occurs despite the employment of agrochemicals, cultural practices and planting of available commercial disease-resistant crop cultivars. Although traditional crop breeding practices during the last century have resulted in successful introgression of resistance genes (R genes) from wild relatives into commercial crop cultivars, the number of commercial cultivated species containing appropriate R genes remains limited. As pathogen evolution may overcome resistance conferred by individual R genes, a greater understanding of the plant immune system is fundamental for effective disease management. Plant immunity can be defined simply as the capacity of a plant to prevent or withstand biological attack by pathogens. Progress in our understanding of the molecular complexity of this innate immune system in plants has advanced considerably in recent decades. Plant surveillance of pathogen presence is known to involve a large number of receptor proteins, with signal perception occurring at the plasma membrane or within the cytoplasm. In current conceptual models, two key interconnected branches of the immune system are recognized, which are defined principally according to the pathogen molecules recognized by the host plant.
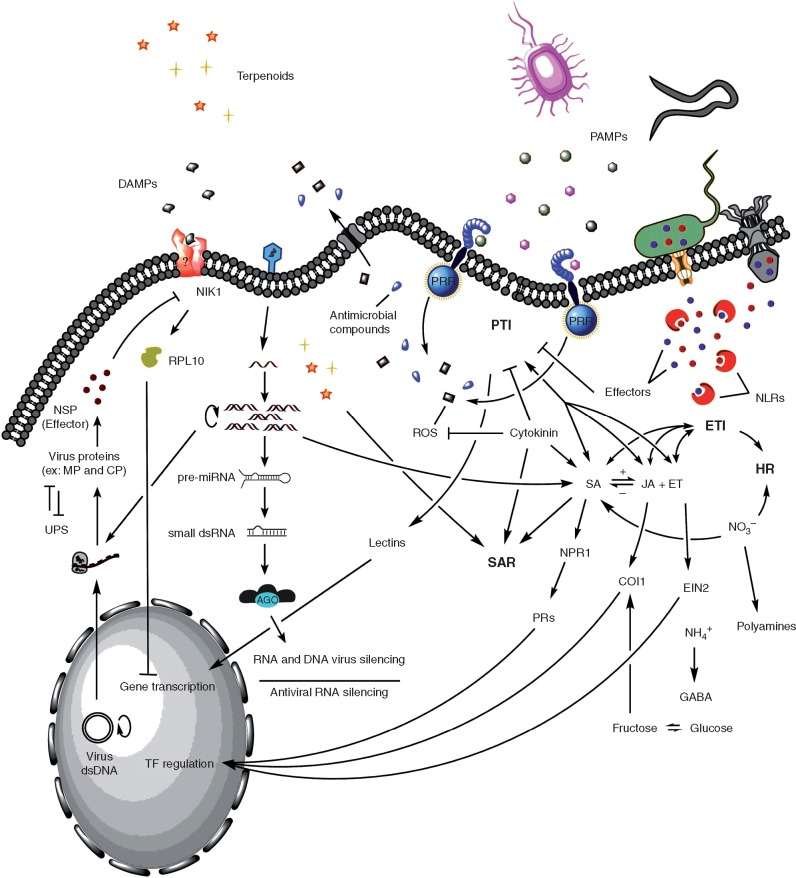
One branch of this system, known as PAMP-triggered immunity (PTI; Fig. 1), is based upon activation of host cell surface transmembrane pattern recognition receptors (PRRs) (Dangl and Jones, 2001; Shiu and Bleecker, 2001; Monaghan and Zipfel, 2012) following recognition of conserved pathogen (or microbial)-associated molecular patterns (PAMPs/MAMPs) (Jones and Dangl, 2006), that are molecules essential for microbial survival. Host molecules that are breakdown products of wounding or infection, known as damage-associated molecular patterns (DAMPs), can also induce PTI following interaction with host PRRs (Yamaguchi et al., 2010). Plants typically show similar responses to recognition of diverse PAMPs, with PRR activation resulting in intracellular signalling and host gene expression modulation that can result in defence responses restricting microbial movement in the host. PTI is considered to be responsible for non-host-specific resistance, occurring at the species level or above.
Fig. 1.
A schematic summary of the innate immunity pathogen sensing and plant immune response mechanisms characterized across pathosystems. PTI, PAMP-triggered immunity; PAMPs, pathogen-associated molecular patterns; PRRs, pattern recognition receptors; DAMPs, damage-associated molecular patterns; ROS, reactive oxygen species; ETI, effector-triggered immunity; NLR, nucleotide-binding oligomerization domain-like receptor; HR, hypersensitive response; JA, jasmonic acid; ET, ethylene; SA, salicylic acid; NPR1, non-expressor of pathogenesis-related genes1; PR, pathogenesis-related protein; COI1, coronatine insensitive protein 1; EIN2, ethylene-insensitive protein2; SAR, systemic acquired resistance; TF, transcription factor; NSP, nuclear shuttle protein; NIK1, NSP-interacting kinase 1; RLP10, receptor-like protein 10; dsDNA, double-stranded DNA; UPS, ubiquitin–proteasome system; MP, movement protein; CP, coat protein; miRNA, microRNA; dsRNA, double-stranded RNA; ARG, Argonaute-containing silencing complex.
In a second branch of the plant immune system, originally characterized genetically as the gene-for-gene model (Flor, 1971), pathogen avirulence (Avr) genes have evolved to code for species-, race- or even strain-specific secreted effector proteins (Avr proteins). Following translocation into the host cell, these can suppress PTI signalling responses and disease results due to this effector-triggered susceptibility (ETS) (Chisholm et al., 2006; Jones and Dangl, 2006; Gohre and Robatzek, 2008). Plants, in response, have coevolved specific cytoplasmic resistance (R) protein receptors to recognize specific pathogen Avr effector proteins and activate effector-triggered immunity (ETI; see Fig. 1) (Ellis et al., 2000; Jones and Dangl, 2006). With greater amplitude than observed in PTI, ETI often leads to defence responses that includes a hypersensitive response and death of infected cells, limiting pathogen advance. In contrast to PTI, ETI operates at the intraspecific level, with resistant host genotypes possessing the necessary polymorphism in R gene resistance determinants (Xiao et al., 2008). A number of distinct plant resistance gene families have now been recognized as involved in ETI, based upon deduced protein domain structure and biochemical function. The largest class encodes polymorphic intracellular receptor proteins within a superfamily containing nucleotide-binding (NB) and leucine-rich repeat (LRR) domains, referred to as NLRs (Dangl and Jones, 2001; Jones and Dangl, 2006). Secreted pathogen effector recognition by such receptors occurs across bacteria, oomycetes, fungi, nematodes and viruses.
Today plant immunity is a multidisciplinary subject area and a rapidly advancing field. Current research activity can be measured by the number of recent publications and special focus editions related to the themes of plant immunity, biotic interactions and plant–microbe cell biology, as seen in recent years in journals such as Current Opinion in Plant Biology (2012), Frontiers in Plant Science (2014, 2015) and New Phytologist (2015).
This issue
The papers collated here highlight advances in specific areas of plant innate immunity, with focus on molecular interactions occurring between plant hosts and viruses, bacteria, phytoplasmas, oomycetes, fungi, nematodes and insect pests. The issue provides snapshots of research at the forefront of plant sciences that embrace pathogen perception, disease development and plant immune response, with topics presented either as timely reviews or as primary research articles. Subjects range from pathogen effector proteins, plant immune receptor signalling in PTI and ETI, plant susceptibility genes, plant signalling networks and phytohormones, downstream defence responses, induced resistance mechanisms, to the role of sugars and nitrogen assimilation in plant defence (Fig. 1).
INNATE IMMUNITY SYSTEMS IN PLANTS
Binding proteins in plant immunity
Structure–function analyses of the domains of plant resistance gene NLRs have so far been limited (Bonardi et al., 2012; Takken and Goverse, 2012). Given that plant NLRs show considerable structural and functional similarities to animal NLRs that are involved in inflammatory and innate immune responses, knowledge of mechanisms involved in activation and regulation of animal NLRs may advance understanding of the action of plant NLRs. In their review, Bentham et al. (2017) explore current understanding of the innate immune pathways in plants and animals, focusing on similarities and differences in structure and biochemistry for both plant and animal NLRs. Important gaps in our knowledge between animal and plant systems are highlighted, with focus on the need for more structural information for plant NLRs. Current knowledge of protein crystal structures is limited to only N-terminal domains and an integrated decoy domain. Similarities and differences between plant and animal NLRs are used to derive a plausible model for plant NLR activation, based on binding by an activating elicitor and ATP. The resultant structural rearrangement of the NLR is followed by signalling through a co-operative process with the formation of a resistosome structure (Nishimura and Dangl, 2010). With current advances in methodologies in structural biology, determination of full-length structures of plant NLRs will increase understanding of regulation and activation, and enable design of NLRs with new specificities.
Plant lectins comprise proteins that specifically bind carbohydrates during essential plant processes. Division into subfamilies can be made according to conserved carbohydrate recognition domains (van Damme et al., 2008). The Nictaba-like family groups proteins with a domain showing homology to the Nicotiana tabacum agglutinin, or Nictaba. Nictaba-like lectins appear to act as signalling molecules that alter gene expression in response to biotic stresses (Chen et al., 2002; Vandenborre et al., 2009; Delporte et al., 2014). The research conducted by Van Holle et al. (2017) focuses on the dynamic evolution of Nictaba-related lectin genes in 15 crop species that represent members of the Fabaceae, Poaceae, Solanaceae, Musaceae, Arecaceae, Malvaceae and Rubiaceae. A bioinformatics approach revealed a total of 360 putative Nictaba-like lectin genes, with distinct domain architecture combinations comprising Nictaba domains with a second Nictaba domain, an F-box domain, a protein kinase domain, a Zeta toxin domain, a TIR domain, a C1 domain, a methyltransferase domain, an NB-ARC domain and/or LRRs. Nictaba-like genes are likely to play diverse roles in plant development and defence.
Cytokinin phytohormones in plant growth and immunity
The cytokinins are N6-substituted adenine derivatives that were first discovered to be involved in cell division regulation in plants. These phytohormones were subsequently recognized to be involved in plant stress tolerance (Argueso et al., 2009), with defence responses to biotrophic pathogens involving hormonal cross-talk between salicylic acid (SA) and cytokinins. While moderate levels of cytokinins have been shown to create favourable physiological conditions for biotrophic pathogen development (Argueso et al., 2012; Hann et al., 2014), higher concentrations of the phytohormone activate plant immunity primarily through SA-dependent processes (Choi et al., 2010; Argueso et al., 2012). The review by Albrecht and Argueso (2017) focuses on current research that highlights fitness costs associated with plant defence activation against pathogens. Cytokinin-regulated physiological and molecular processes are associated with plant growth and response to pathogens, and indicate a role for this class of phytohormone in regulation of growth–defence trade-off in plants. Uncoupling of defence activation from growth reduction offers considerable potential for the development of engineered crops displaying broad-spectrum, durable resistance to biotrophic pathogens, with reduced yield penalties.
Plant–pathogen interactions
Plant–virus interactions.
Papers in this category focus on aspects of the multilayered plant antiviral immune system, with emphasis on recent advances in understanding of antiviral innate immunity, together with viral proteins that modulate antiviral defence mechanisms. In their review, Calil et al. (2017) investigate the mechanisms adopted by plants to overcome viral infections in their continuous coevolution. The authors highlight both the potential of the plant immune system for the development of broad-spectrum tolerance to plant viruses across distinct plant species, as well important gaps that remain today in our understanding of plant–virus interaction dynamics, from virus-derived PAMPs and effectors, to PTI and ETI host receptors. Particular attention is given to recent breakthroughs in mechanisms of receptor signalling in antiviral immunity (NB-LRRs, LRR-RLKs, and NIK1), RNA silencing/interference machinery for antiviral defence, hormone-mediated antiviral pathways and protein degradation pathways in plant defence. Integration of antiviral innate immunity, systemic acquired resistance (SAR) and RNA interference (RNAi) defence layers will enable development of durable defence against plant viruses. Conti et al. (2017) examine the effects of Tobacco mosaic virus-encoded proteins on host plant physiology, focusing on replicase, movement protein (MP) and coat protein (CP). They discuss the effects of each viral component on the modulation of host defence responses, through mechanisms involving hormonal imbalance, innate immunity modulation and antiviral RNA silencing. Individual and combined effects of viral-encoded proteins contribute to viral replication and movement in the host plant.
Plant–phytoplasma interactions.
Phytoplasmas are obligate biotrophic plant pathogenic bacteria that replicate intracellularly in phloem sieve elements of infected plants. Infecting a diversity of monocotyledonous and dicotyledonous wild, ornamental and crop plant species, they cause significant crop yield losses worldwide. Symptoms of phytoplasma-infected plants indicate modulation of fundamental plant developmental processes, which include flower development and fruit production. Whole-genome sequences for phytoplasmas are fundamental for the identification of pathogen genes responsible for developmental changes and subsequent yield losses in affected host plants. In a study by Orlovskis et al. (2017), symptom differences in maize are correlated with gene sequence polymorphisms in isolates of Maize bushy stunt phytoplasma (MBSP), which code for a phase-variable lipoprotein candidate effector and an ATP-dependent lipoprotein ABC export protein. In other pathosystems, these proteins have been shown to activate host defence responses, regulate pathogen attachment to host cells and activate effector secretion systems. Polymorphisms observed in the lipoprotein and exporter genes in MBSP isolates are associated with lateral branching organ proliferation symptoms in maize.
Plant–fungus interactions.
In the study conducted by Powell and colleagues (2017), next-generation sequencing approaches are employed to assess global gene expression responses in wheat genotypes moderately and highly susceptible to Fusarium pseudograminearum, causal agent of Fusarium crown rot. Host responses related to pathogen perception, defence signalling, transport, metabolism, and chemical and enzymatic defence are observed, with potential roles in resistance and/or susceptibility discussed in the context of development of durable disease resistance in wheat. The fungal mycotoxin deoxynivalenol is also shown to play an important role in virulence.
Sugarcane smut, caused by the facultative biotrophic pathogen Sporisorium scitamineum, is a disease in sugarcane, occurring across all global production areas. In their study, Marques et al. (2017) employ a combination of light and electron microscopy methods for advancing basic understanding of this pathosystem, revealing processes involved in infection, host tissue colonization, pathogen sporogenesis, whip-shaped sorus ontogeny and host tissue structural responses to the pathogen during infection. Induced resistance in plants is characterized by a latent defence response which, following induction, is activated only at a later moment, upon attack by a pathogen or insect herbivore (Kue, 1982; Pieterse et al., 2012). This induced state of resistance is expressed systemically at the whole-plant level, not only in plant tissues exposed to the inducer (Durrant and Dong, 2004). Both biological and chemical agents have been employed as inducers, as potential alternatives to agrochemicals. In the study conducted by Satková et al. (2017), the authors investigate the molecular mechanisms underlying host responses in tomato to resistance inducers, identifying modulated genes following treatment with the chemical agent β- aminobutyric acid (BABA) and the pathogen-derived proteinaceous elicitor oligandrin. Differential defence responses against Oidium neolycopersici fungal infection are observed in Solanum genotypes contrasting in resistance, with the involvement of ethylene (ET)-dependent signalling pathways suggested in the BABA-induced resistance.
Plant–nematode interactions.
Papers in this category examine the molecular components involved in complex compatible and incompatible interaction between plants and nematode pathogens. Plant-parasitic nematodes (PPNs) are important pests of many cultivated crop species, with global annual economic losses estimated at over US$120 billion (Chitwood, 2003). Root-knot nematodes (Meloidogyne spp.) attack >3000 plant species and represent a threat to agricultural production worldwide. As information is limited with regard to both host plant resistance responses and root cell development modulation during root knot nematode infection, particularly in the case of monocotyledonous plants, the characterization of both defence responses that may limit parasitism and the identification of potential effector-targeted host genes will be far reaching in the development of genetic improvement-based control measures for such nematodes. Petitot et al. (2017) employ the rice–Meloidogyne graminicola pathosystem as a model for plant–nematode interactions, investigating cytological and molecular mechanisms that underlie resistance to M. graminicola in an African rice cultivar (Oryza glaberrima). Nematode penetration and establishment of galls in root tissues is reduced in this species when compared with observations in a susceptible Oryza sativa cultivar. Catalogued changes in gene expression during host–pathogen interactions reveal differential upregulation of candidate genes for nematode resistance in the resistant line that include ETI NLRs, as well as genes involved in downstream defence responses, phenylpropanoid and phytohormone biosynthetic pathways.
While there has been considerable focus on host mechanisms that are activated during incompatible plant–nematode interactions, our understanding of how root-knot nematodes induce or suppress host genes during infection and giant cell development in compatible interactions remains limited. In this context, Castañeda et al. (2017) examine the transcriptome of banana root tissues in two root-knot nematode-susceptible Musa acuminata genotypes during early stages of infection with Meloidogyne incognita. Early host defence responses that appear to involve reactive oxygen species (ROS) and jasmonic acid (JA)/ET signalling events are later suppressed, with concomitant auxin metabolism and cell wall modification processes activated that are involved in root cell development and giant cell formation (Fig. 2). These specialized feeding cells are the exclusive nutritive source for all stages of the nematode life cycle. Gillet and colleagues (2017) provide an overview of the molecular interplay in plant–nematode interactions, with particular attention given to the early stages of infection and events that occur prior to nematode establishment of successful infection. The authors discuss how PPNs probably orchestrate cytoprotection to resist plant immune responses, focusing on the potential roles played by PPN heterologues of Caenorhabditis elegans dauer abnormal formation (DAF) and SKN transcription factors in overcoming the oxidative stress conditions which are typically produced by the host plant during nematode invasion and migration. As these transcription factor genes may play pivotal roles during parasitism, the authors suggest strategies for control of PPNs through RNAi approaches.
Fig. 2.
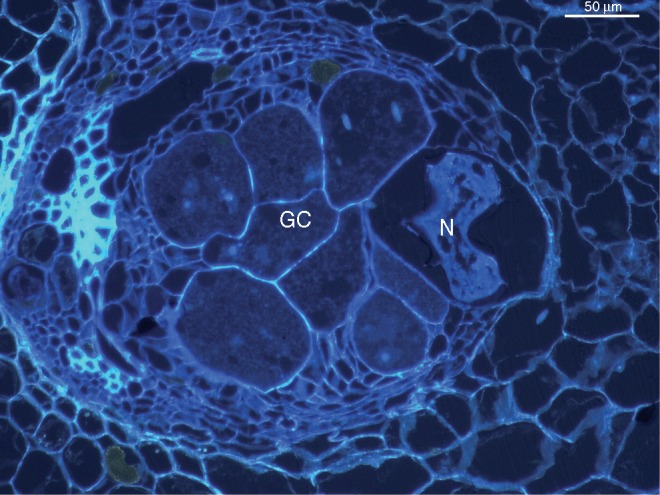
UV fluorescence microscopy-based examination of root sections of Musa acuminata infected with Meloidogyne incognita displaying adult female and giant cells in the central cylinder. GC, giant cell; N, nematode.
Plant immunity in Citrus
As perennial woody plants, Citrus sp. are vulnerable to diseases caused by a diverse range of biotic factors, including viruses, bacteria, fungi, oomycetes and nematodes. In their review, Dalio et al. (2017) provide an overview of current understanding of citrus plant immunity, discussing molecular mechanisms involved in biotic interactions, in terms of recognized and candidate host pathogen recognition receptors and R proteins, together with microbial PAMPs and effectors. Emphasis is given to the economically important diseases tristeza, psorosis, citrus variegated chlorosis, citrus canker, huanglongbing (HLB), brown spot, post-bloom, anthracnose, gummosis and citrus root rot. Candidate host genes are recommended for development of disease-resistant genotypes through conventional breeding, biotechnology-based approaches for development of transgenic or cisgenic citrus, and via genome editing and host-induced gene silencing.
Plant peptides and volatiles
Plant defensins are small basic cysteine-rich peptides that occur throughout the plant kingdom. In their study, Weiller et al. (2017) provide evidence for a link between root border-like cells and defensin peptide secretion in root tips in the Brassicaceae species Heliophila coronopifolia, a South African semi-desert flower. Border-like cells are thought to provide protection to root meristems in a similar manner to classical root border cells. The authors propose that the presence of the Hc-AFP3 defensin peptides in the border-like cells indicates a potentially important role for these cells in root protection. Upon biotic or abiotic stress challenge, plants can emit a blend of volatile organic compounds. Terpenoid volatiles, which are low molecular weight compounds derived from five-carbon building blocks of isopentenyl diphosphate (IPP), play important roles in direct and indirect defences, priming responses within the plant and to neighbouring plants. Plant symbiotic associations involving arbuscular mycorrhiza fungi can increase host secondary metabolism and alter the concentration and composition of terpenoids. In this review, Sharma and colleagues (2017) discuss the roles of terpernoids in plant defences, with focus on defence against insect herbivores. The importance of mycorrhizal plants in alteration of terpenoid production is highlighted, with prospects outlined for sustainable management of pests in agricultural systems through bioengineering of terpenoid-producing plants.
Assimilate dynamics in source/sink metabolism
Plant assimilates, that can be defined as organic carbon and nitrogen, are of fundamental importance for plant development and productivity (Ruan et al., 2010). Assimilate source/sink dynamics, as well as integral components of the life cycle in plants, can also be affected during pathogen infection. Knowing that cytokinins are widely recognized to be implicated in source/sink dynamics during plant development, Dhandapani and colleagues (2017) investigate the impact of both an epiphytic (avirulent) strain and a virulent strain of the cytokinin-producing bacterium, Rhodococcus fascians, on source/sink activity within pea cotyledons during and following germination. Based on their findings, they observe re-greening and maintenance of the cotyledon as a sink tissue during pathogen infection, and consider this to occur through redistribution of sucrose that involves interaction between cytokinins and modulation of a set of PsCWINV and PsSWEETS genes. Plant soluble sugars have been implicated in plant defence against fungal pathogens. In this context, Lecompte et al. (2017) investigate the dynamics of soluble sugars in sink tissues of tomato during the course of infection by the necrotrophic fungus Botrytis cinerea. The results indicate distinct roles for host glucose and fructose in defence response to infection by B. cinerea and strongly suggest that adjustment of the relative fructose content is required for enhanced plant defence. Plants require nitrogen for growth, development and defence against abiotic and biotic stresses. An extensive review is presented by Mur and colleagues (2017) on the agricultural impact of nitrogen nutrition on disease development. Plant disease resistance based upon PTI, ETI or the mobilization of nutrients away from sites of infection is shown to be potentially compromised under different soil nitrogen availability. The authors highlight how nitrogen content and form also play essential roles in defensive primary and secondary metabolism and nitric oxide-mediated defence signalling events.
CONCLUSIONS AND PERSPECTIVES
Meeting the demands for food supply in the context of the current rapid global population increase is a major challenge for sustainable agriculture, with global losses across crop species due to pre-harvest diseases today estimated at approx. 15 %. While plants and their pathogens are continuously coevolving, continuous monoculture planting of cultivars can favour the emergence of virulent pathogen populations that can overcome disease resistance. Agricultural practices can also facilitate pathogen movement to new areas, exacerbated by human migration and movement of contaminated germplasm. The global tendencies for reduction in dependency on pesticide regimes for disease control mean that the development and deployment of pest- and disease-resistant crop cultivars will probably play an ever-increasingly important role in the intensification of sustainable agriculture. As such, continued progress in plant immunity research has far-reaching implications for global sustainable agriculture. This special issue showcases exciting research across multiple areas related to pathogen sensing and plant immune response, with molecules characterized across numerous pathosystems. Furthering our understanding of the plant immune system is fundamental for the development of durable defence against plant pathogens and pests. Increasingly accessible nucleic acid sequencing technologies, together with genome editing and transgenic approaches, offer considerable potential for durable, broad-spectrum disease resistance development. Whole-genome and transcriptome analyses, as widely employed in the presented papers, are advancing the characterization of genes and pathways involved in compatible and incompatible host–pathogen interactions, consolidating knowledge on host receptors and pathogen recognition, signalling and downstream defence molecules. As genome sequencing of pathogen isolates is becoming increasingly accessible, greater understanding of genome adaptation will also probably be achieved, facilitating control of emerging pathogens (Misra and Chaturvedi, 2015). Integration of DNA and RNA ‘omics’ data with mass spectrometry high-throughput technologies for metabolite and proteomic determination is also providing opportunities for systems approaches to advance understanding of plant pathogens and environmental conditions that result in disease. Bioinformatics-based identification of core pathogen effector-coding genes is also a promising route for identifying cognate activated host R gene alleles in the plant genepool (Dangl et al., 2013). Given the recently developed genome editing methods, such as clustered regulatory interspaced short palindromic repeat (CRISPR) (Gaj et al., 2013), both validation of function of candidate R gene alleles as well as editing of susceptibility alleles will also further advance non-conventional crop improvement. Although breeding for disease resistance is an important component of crop improvement programmes, a number of restraints exist. Conventional breeding methods for resistance introgression into commercial cultivars is typically limited to those crop species with available R-genes in germplasm material. Disease resistance may also be short-lived in monoculture cropping systems that carry only single R genes, with evolving pathogen populations often able to overcome host resistance. As progress is made with R gene structure and function determination, however, possibilities emerge for their pyramiding in crops, potentially accelerating durable disease resistance development (Huang et al., 2004; Steuernagel et al., 2016; Bentham et al., 2017). In such strategies, several alleles of an R-gene can be introgressed into a single crop cultivar, such that pathogens must undergo multiple mutations or recombinations in Avr genes to overcome the disease resistance. Spatio-temporal planting of different resistant cultivars can also be appropriate where multiple resistance alleles would negatively affect agronomic characteristics. In addition to biotic stresses, abiotic stresses have a major negative influence on global agricultural production. Drought conditions affect one-third of the world’s agricultural land, with this stress today considered to be one of the greatest obstacles to increasing crop production. Climate change and associated temperature rises have also been shown to increase geographic distribution and reproductive potential of pathogens. Much ongoing research on individual stresses in plants is providing evidence for overlap in receptors, signalling pathways and responses, not only between different biotic stresses, but also between biotic and abiotic stresses. The characterization of hub genes and common components in multiple stress resistance offers considerable potential for the development of multiple stress-resistant crop cultivars, appropriate for sustainable agricultural practices.
ACKNOWLEDGEMENTS
This work was partially funded by the Fundação de Amparo à Pesquisa do Estado de Distrito Federal (FAPDF) (project 193.000.983/2015). G.S.C.A. was supported by a post-doctoral fellowship from CAPES. R.N.G.M. was supported by a fellowship from CNPq. M.-A.V.S. was supported by a fellowship from CNPq, We would like to thank Professor Pat Heslop-Harrison (Chief Editor), Dr Trude Schwarzacher (transitional Managing Editor), Dr Catherine Hyland (Managing Editor) and Dr Stephanie Sacharov (Deputy Managing Editor) at Annals of Botany for all their guidance and support during the development of this Special Issue. Our thanks also go to the authors who contributed their manuscripts for publication in the Special Issue. Lastly, we are very grateful for the important input provided by all the hard-working reviewers who refereed the submitted manuscripts.
LITERATURE CITED
- Albrecht T, Argueso CT.. 2017. Should I fight or should I grow now? The role of cytokinins in plant growth and immunity and in the growth–defence trade-off. Annals of Botany 119: 725–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argueso CT, Ferreira FJ, Epple P, et al. 2012. Two-Component elements mediate interactions between cytokinin and salicylic acid in plant immunity. Plos Genetics 8: e1002448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argueso CT, Ferreira FJ, Kieber JJ.. 2009. Environmental perception avenues: the interaction of cytokinin and environmental response pathways. Plant, Cell & Environment 32: 1147–1160. [DOI] [PubMed] [Google Scholar]
- Bentham A, Burdett H, Anderson PA, Williams SJ, Kobe B.. 2017. Animal NLRs provide structural insights into plant NLR function. Annals of Botany 119: 689–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonardi V, Cherkis K, Nishimura MT, Dangl JL.. 2012. A new eye on NLR proteins: focused on clarity or diffused by complexity? Current Opinion in Immunology 24: 41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd LA, Ridout C, O’Sullivan DM, Leach JE, Leung H.. 2013. Plant–pathogen interactions: disease resistance in modern agriculture. Trends in Genetics 29: 233–240. [DOI] [PubMed] [Google Scholar]
- Calil IP, Fontes EPB.. 2017. Plant immunity against viruses: antiviral immune receptors in focus. Annals of Botany 119: 711–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castañeda NEN, Alves GSC, Mansan-Almeida R, et al. 2017. Gene expression analysis in Musa acuminata during compatible interactions with Meloidogyne incognita. Annals of Botany 119: 915–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Peumans WJ, Hause B, et al. 2002. Jasmonate methyl ester induces the synthesis of a cytoplasmic/nuclear chitooligosaccharide-binding lectin in tobacco leaves. FASEB Journal 16: 905–907. [DOI] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ.. 2006. Host–microbe interactions: shaping the evolution of the plant immune response. Cell 124: 803–814. [DOI] [PubMed] [Google Scholar]
- Chitwood DJ. 2003. Nematicides In: JR Plimmer, ed. Encyclopedia of agrochemicals, Vol. 3 New York: John Wiley & Sons: 1104–1115. [Google Scholar]
- Choi J, Huh SU, Kojima M, Sakakibara H, Paek KH, Hwang I.. 2010. The cytokinin activated transcription factor ARR2 promotes plant immunity via TGA3/NPR1-dependent salicylic acid signaling in Arabidopsis. Developmental Cell 19: 284–295. [DOI] [PubMed] [Google Scholar]
- Conti G, Rodriguez MC, Venturuzzi AL, Asurmendi S.. 2017. Modulation of host plant immunity by Tobamovirus proteins. Annals of Botany 119: 737–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalio RJD, Magalhães DM, Rodrigues CM, et al. 2017. PAMPs, PRRs, effectors, and R-genes associated with citrus–pathogen interactions. Annals of Botany 119: 749–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl JL, Jones JDG.. 2001. Plant pathogens and integrated defence responses to infection. Nature 411: 826–833. [DOI] [PubMed] [Google Scholar]
- Dangl JL, Horvath DM, Staskawicz BJ.. 2013. Pivoting the plant immune system from dissection to deployment. Science 341: 746–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delporte A, De Vos WH, Van Damme EJM.. 2014. In vivo interaction between the tobacco lectin and the core histone proteins. Journal of Plant Physiology 171: 1149–1156. [DOI] [PubMed] [Google Scholar]
- Dhandapani P, Song J, Novak O, Jameson PE.. 2017. Infection by Rhodococcus fascians maintains cotyledons as a sink tissue for the pathogen. Annals of Botany 119: 841–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant WE, Dong X.. 2004. Systemic acquired resistance. Annual Review of Phytopathology 42: 185–209. [DOI] [PubMed] [Google Scholar]
- Ellis J, Dodds P, Pryor T.. 2000. Structure, function and evolution of plant disease resistance genes. Current Opinion in Plant Biology 3: 278–284. [DOI] [PubMed] [Google Scholar]
- Flor H. 1971. Current status of the gene-for-gene concept. Annual Reviews in Phytopathology 9: 275–296. [Google Scholar]
- Gaj T, Gersbach CA, Barbas CF.. 2013. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends in Biotechnology 31: 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillet FX, Bournaud C, de Souza Júnior JDA, Grossi-de-Sa MF.. 2017. Plant-parasitic nematodes: towards understanding molecular players in stress responses. Annals of Botany 119: 775–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohre V, Robatzek S.. 2008. Breaking the barriers: microbial effector molecules subvert plant immunity. Annual Review of Phytopathology 46: 189–215. [DOI] [PubMed] [Google Scholar]
- Hann DR, Dominguez-Ferreras A, Motyka V, et al. 2014. The Pseudomonas type III effector HopQ1 activates cytokinin signaling and interferes with plant innate immunity. New Phytologist 201: 585–598. [DOI] [PubMed] [Google Scholar]
- Huang N, Angeles ER, Domingo J, et al. 2004. Pyramiding of bacterial blight resistance genes in rice: marker-assisted selection using RFLP and PCR. Theoretical and Applied Genetics 95: 313–320. [Google Scholar]
- Jones JDG, Dangl JL.. 2006. The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Kué J. 1982. Induced immunity to plant disease. Bioscience 32: 854–860. [Google Scholar]
- Lecompte F, Nicot PC, Ripoll J, et al. 2017. Reduced susceptibility of tomato stem to the necrotrophic fungus Botrytis cinerea is associated with a specific adjustment of fructose content in the host sugar pool. Annals of Botany 119: 931–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques JPR, Appezzato-da-Glória B, Piepenbring M, Massola NS Jr, Monteiro-Vitorello CB, Carneiro Vieira ML.. 2017. Sugarcane smut: shedding light on the development of the whip shaped sorus. Annals of Botany 119: 815–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra BB, Chaturvedi R.. 2015. When plants brace for the emerging pathogens. Physiological and Molecular Plant Pathology 92: 181–185. [Google Scholar]
- Monaghan J, Zipfel C.. 2012. Plant pattern recognition receptor complexes at the plasma membrane. Current Opinion in Plant Biology 15: 349–357. [DOI] [PubMed] [Google Scholar]
- Mur LAJ, Simpson C, Kumari A, Gupta AK, Gupta KJ.. 2017. Moving nitrogen to the centre of plant defence against pathogens. Annals of Botany 119: 703–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura MT, Dangl JL.. 2010. Arabidopsis and the plant immune system. The Plant Journal 61: 1053–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlovskis Z, Canale MC, Haryono M, Lopes JRS, Kuo CH, Hogenhout S.. 2017. A few sequence polymorphisms among isolates of Maize bushy stunt phytoplasma associate with organ proliferation symptoms in infected maize plants. Annals of Botany 119: 869–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petitot AS, Kyndt T, Haidar R, et al. 2017. Transcriptomic and histological responses of the African rice (Oryza glaberrima) to Meloidogyne graminicola provide new insights into root-knot nematode resistance in monocots. Annals of Botany 119: 885–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CMJ, Zamioudis C, Berendsen RL, et al. 2012. Induced systemic resistance by beneficial microbes. Annual Review of Phytopathology 52: 347–375. [DOI] [PubMed] [Google Scholar]
- Popp J, Hantos K.. 2011. The impact of crop protection on agricultural production. Studies in Agricultural Economics 113: 47–66. [Google Scholar]
- Powell JJ, Carere J, Fitzgerald TL, et al. 2017. The Fusarium crown rot pathogen Fusarium pseudograminearum triggers a suite of transcriptional and metabolic changes in bread wheat (Triticum aestivum L.). Annals of Botany 119: 853–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan YL, Patrick JW, Weber H.. 2010. Assimilate partitioning and plant development. Molecular Plant 6: 941. [DOI] [PubMed] [Google Scholar]
- Satková P, Starý T, Plešková V, et al. 2017. Diverse responses of wild and cultivated tomato to BABA, oligandrin and Oidium neolycopersici infection. Annals of Botany 119: 829–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma E, Anand G, Kapoor R.. 2017. Terpenoids in plant and Arbuscular mycorrhiza-reinforced defence against herbivorous insects. Annals of Botany 119: 791–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB.. 2001. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proceedings of the National Academy of Sciences, USA 98: 10763–10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steuernagel B, Periyannan SK, Hernández-Pinzón, et al. 2016. Rapid cloning of disease-resistance genes in plants using mutagenesis and sequence capture. Nature Biotechnology 34: 652–655. [DOI] [PubMed] [Google Scholar]
- Takken FL, Goverse A.. 2012. How to build a pathogen detector: structural basis of NB-LRR function. Current Opinion in Plant Biology 15: 375–384. [DOI] [PubMed] [Google Scholar]
- Vandenborre G, Miersch O, Hause B, Smagghe G, Wasternack C, Van Damme EJM.. 2009. Spodoptera littoralis-induced lectin expression in tobacco. Plant and Cell Physiology 50: 1142–1155. [DOI] [PubMed] [Google Scholar]
- Van Damme EJM, Lannoo N, Peumans WJ.. 2008. Plant Lectins. Advances in Botanical Research 48: 107–209. [Google Scholar]
- Van Holle S, Rougé P, Van Damme EJM.. 2017. Evolution and structural diversification of Nictaba-like lectin genes in food crops with a focus on soybean (Glycine max). Annals of Botany 119: 901–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiller F, Moore JP, Young P, Driouich A, Vivier MA.. 2017. The Brassicaceae species Heliophila coronopifolia produces root border-like cells that protect the root tip and secrete defensin peptides. Annals of Botany 119: 803–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Wang W, Yang X. 2008. Evolution of resistance genes in plants. innate immunity of plants, animals, and humans. Nucleic Acids and Molecular Biology 21: 1–25. [Google Scholar]
- Yamaguchi Y, Huffaker A, Bryan AC, Tax FE, Ryan CA.. 2010. PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. The Plant Cell 22: 508–522. [DOI] [PMC free article] [PubMed] [Google Scholar]