Abstract
Background
A wide range of response rates have been reported in HER2-positive gastric cancer (GC) patients treated with trastuzumab. Other HER2-targeted therapies for GC have yet to show efficacy in clinical trials. These findings raise question about the ability of standard HER2 diagnostics to accurately distinguish between GC patients who would and would not benefit from anti-HER2 therapies.
Patients and methods
GC patients (n = 237), including a subset from the Trastuzumab in GC (ToGA) trial were divided into three groups based on HER2 status and history of treatment with standard chemotherapy or chemotherapy plus trastuzumab. We applied mass spectrometry-based proteomic analysis to quantify HER2 protein expression in formalin-fixed tumor samples. Using HER2 expression as a continuous variable, we defined a predictive protein level cutoff to identify which patients would benefit from trastuzumab. We compared quantitated protein level with clinical outcome and HER2 status as determined by conventional HER2 diagnostics.
Results
Quantitative proteomics detected a 115-fold range of HER2 protein expression among patients diagnosed as HER2 positive by standard methods. A protein level of 1825 amol/µg was predicted to determine benefit from the addition of trastuzumab to chemotherapy. Trastuzumab treated patients with HER2 protein levels above this cutoff had twice the median overall survival (OS) of their counterparts below the cutoff (35.0 versus 17.5 months, P = 0.011). Conversely, trastuzumab-treated patients with HER2 levels below the cutoff had outcomes similar to HER2-positive patients treated with chemotherapy. (Progression-free survival = 7.0 versus 6.5 months: P = 0.504; OS = 17.5 versus 12.6 months: P = 0.520). HER2 levels were not prognostic for response to chemotherapy.
Conclusions
Proteomic analysis of HER2 expression demonstrated a quantitative cutoff that improves selection of GC patients for trastuzumab as compared with current diagnostic methods.
Keywords: HER2, gastric cancer, proteomics, trastuzumab, molecular characterization
Introduction
Gastric cancer (GC) accounts for 10% of cancer deaths worldwide [1]; median survival is 10–12 months in advanced cases [2]. Chemotherapy is the standard treatment of the majority of GC patients, however a subset of patients who harbor HER2 amplification are candidates for HER2-targeted therapy. In the ToGA trial, the addition of trastuzumab to chemotherapy improved overall survival (OS) among patients whose tumors tested HER2 positive (HER2+) by immunohistochemistry (IHC) and/or fluorescence in situ hybridization (FISH). The benefit was greatest among FISH+ patients with highest levels of HER2 protein expression as determined by IHC [2]. Based on these results, trastuzumab was approved for HER2+ GC, defined in the United States and Asia as a HER2 IHC score of 3+ or a positive HER2 FISH test. In Europe, HER2 positivity is defined as a HER2 IHC 3+ or IHC 2+ with a positive FISH test.
Subsequent trials of trastuzumab in HER2+ GC have yielded response rates ranging between 32% and 68% [2–4]. Broad variability of anti-HER2 therapy response rates has been attributed to lack of standardized testing methods and scoring criteria, gastric tumor molecular heterogeneity, and tissue sampling errors [5]. This is underscored by variable incidence of HER2 positivity in patient cohorts, as well as discordance between IHC and FISH. In the ToGA trial, nearly one-quarter of patients (131/584) were FISH+ but had low IHC scores (0 or 1+) and received moderate benefit from trastuzumab [2]. In the TyTAN trial, ∼35% of FISH+ patients had low IHC scores [6].
Trials of additional HER2-targeted therapies in GC have yielded disappointing results [6–8]. Lapatinib failed to demonstrate efficacy in the TyTAN and LOGiC trials [6, 7]. T-DM1 fared worse than taxane in the GATSBY trial of HER2+ gastroesophageal adenocarcinoma patients including those who had failed trastuzumab (OS: 7.9 versus 8.6 months) [8]. These findings have cast doubt on standard HER2 diagnostic methods and fueled debate regarding how best to select GC patients for anti-HER2 therapy.
The emergence of clinical mass spectrometry has advanced molecular diagnostics by enabling multiplexed, quantitative analysis of a large number of proteins in a small, formalin-fixed, paraffin embedded (FFPE) tissue samples. Targeted proteomics using selected reaction monitoring (SRM) mass spectrometry is currently being used in CAP/CLIA laboratories to quantify protein targets in FFPE tumor tissues [9–12]. We have developed a HER2 SRM assay that measures a single peptide unique to HER2 and provides accurate quantitation of HER2 protein in a solubilized lysate prepared from laser microdissected tumor tissue [10]. Another laboratory has confirmed the ability of targeted proteomics to accurately quantitate HER2 and other oncogenic proteins in FFPE tissue [13].
We previously established HER2 protein cutoff levels which are highly correlated with HER2 gene amplification: 750 amol/µg in GC [11] and 740 amol/µg in breast cancer [12]. We have also demonstrated that trastuzumab-treated breast cancer patients with high HER2 expression (>2200 amol/µg) survived longer than patients with lower HER2 expression [12].
In the current study, we applied quantitative proteomic analysis to FFPE tumor samples of GC patients (including a subset from the ToGA trial) whose HER2 status was determined by IHC/FISH testing. As these patients were treated using standard chemotherapy with or without trastuzumab, we hypothesized that mass spectrometric measurement of HER2 protein could define a quantitative proteomic cutoff to identify patients who would benefit from the addition of trastuzumab to chemotherapy.
Patients and methods
Study population
FFPE tissue samples were retrospectively selected from those of patients with histologically confirmed recurrent or metastatic GC at Seoul National University Hospital (SNUH) between September 2005 and May 2014. The inclusion criteria were documented HER2 status by IHC and/or FISH, and documented treatment with standard chemotherapy (with or without trastuzumab) during or after the ToGA trial. HER2+ patients were defined as those with either IHC 3+ or HER2/CEP17 ratio >2. FISH score had been determined using a standard DNA probe kit and by counting HER2 and CEP 17 signals from 20 nuclei of each sample. The study was approved by the institutional review board at SNUH.
Protein quantitation using targeted proteomics
HER2 protein level was quantified by SRM mass spectrometry as previously described [9–11]. Briefly, one section was cut for hematoxylin and eosin (H&E) staining, and multiple sections were cut on to Director® microdissection slides and stained with eosin. The H&E was used to guide tumor area selection on the slide sections, which were then microdissected (Molecular Machines & Industries, Eching, Germany). Collected tumor tissue was solubilized using Liquid Tissue® according to manufacturer’s instructions. Total protein concentration from each sample was measured by a micro bicinchoninic acid assay (Thermo Fisher Scientific Inc, Waltham, MA).
A mixture of stable isotope-labeled synthetic peptides was added to the liquefied tumor samples as internal standards. All samples were analyzed in triplicate using a triple quadrupole mass spectrometer (TSQ Quantiva™, Thermo Scientific, Waltham, MA) interfacing with a nanoACQUITY liquid chromatography system (Waters Corporation, Milford, MA). A 100 µm inner diameter chromatographic column packed with C18 resin (ProntoSIL 200-5-C18AQ; Bischoff Chromatography, Germany) was used for peptide separation before mass spectrometry analysis. For protein quantitation, peak areas from each endogenous and internal standard peptide were calculated and ratios were determined using PinPoint 1.3 (Thermo Scientific, Waltham, MA).
Statistical analysis
Progression-free survival (PFS) and OS were estimated by the Kaplan–Meier method. Cox regression models that included an interaction term between HER2 protein levels and treatment groups (trastuzumab with chemotherapy or chemotherapy alone) were fit to evaluate the effect of HER2 on the comparison (hazard ratio) between treatment groups. Association between clinicopathologic parameters and HER2 protein data was determined with a chi-square test. Statistical analyses were carried out using R version 3.1.2, STATA version 12, and Prism.
Results
Patient characteristics
Of 957 GC patients who had been tested for HER2, 247 (25.8%) were HER2+ and the remainder HER2−. From these patients, a total of 237 tumor samples were included for analysis (supplementary Table S1 and Figure S2, available at Annals of Oncology online). These included all available tumor samples of HER2+ patients who received trastuzumab plus chemotherapy (Cohort 1, n = 95) or chemotherapy alone before trastuzumab became the standard of care (Cohort 2, n = 58), and a sample of HER2− patients who received chemotherapy (selected by propensity score-based matching) (Cohort 3, n = 84). The proportions of tumor samples from the ToGA trial were 14/95 and 11/58 in Cohorts 1 and 2, respectively.
Comparison of HER2 protein level with IHC and FISH
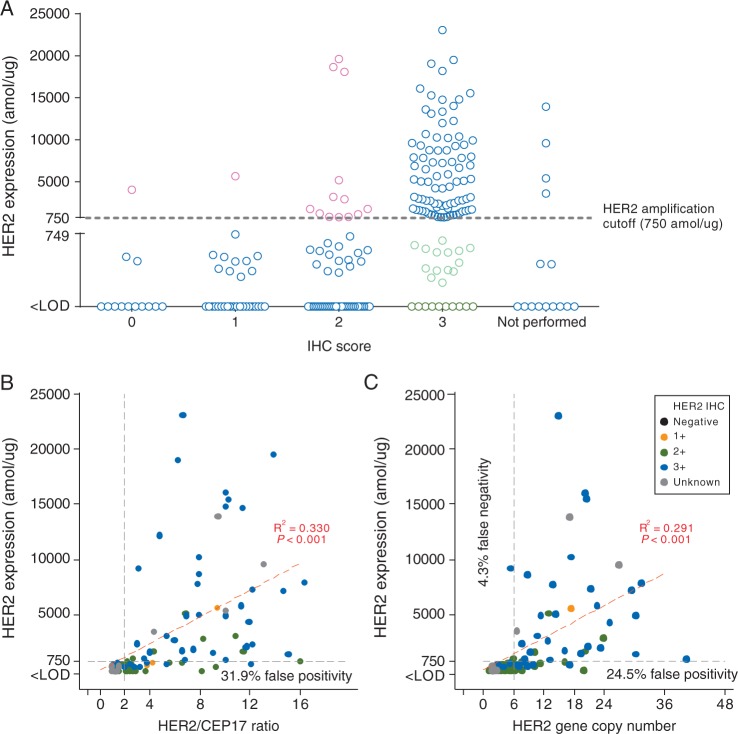
Targeted proteomics demonstrated a 115-fold range of HER2 protein expression (range: ≤200 [limit of detection]-23 055 amol/µg) among patients diagnosed as HER2+ by standard methods (n = 153). Among samples with HER2 IHC scores (n = 222), HER2 protein expression generally increased with IHC score, with the highest protein levels found in samples scored as IHC 3+ (Figure 1A). When patients were stratified by the HER2 protein level cutoff of 750 amol/µg (previously determined as highly correlated with HER2 amplification in GC [11]), 24 of 110 tumors (21.8%) in the IHC 3+ group expressed protein levels below the cutoff (Figure 1A, green circles), suggesting that these samples may be falsely positive by IHC. In contrast, 15 of 112 samples (13.4%) with IHC scores of 0, 1+ and 2+ had HER2 expression above the cutoff and may be falsely negative (Figure 1A, pink circles).
Figure 1.
Quantitated HER2 protein levels according to HER2 assessment by immunohistochemistry (IHC) and fluorescence in situ hybridization (FISH). (A) In each HER2 IHC category, a wide range of HER2 protein levels were quantitated. Among gastric cancer tumor samples diagnosed as IHC3+, 24/110 (21.8%) expressed low HER2 protein levels (<750 amol/µg) and thus may be falsely positive by IHC (green circles). Among samples with IHC scores of 0, 1, and 2+, 15/112 (3.4%) expressed high HER2 levels (>750 amol/µg) and may be falsely negative by IHC (pink circles). (B and C) Linear regression suggests that quantitative HER2 protein levels are loosely correlated with FISH. HER2 protein levels below the 750 amol/µg threshold were measured in tumors that had tested FISH-positive by HER2/CEP17 ratio (23/72; 31.9%) and by gene copy number (GCN) (12/49; 24.5%). These tumors may be false positive by FISH. Of tumors considered FISH-negative by GCN, 3/70 (4.3%) may be false negatives.
Among samples with available HER2 FISH scores [n = 135 for HER2/CEP17 ratio and n = 119 for HER2 gene copy number (GCN)], HER2 protein levels generally correlated with the degree of HER2 amplification (adjusted R2 0.330 and 0.291, respectively) (Figure 1B and C), consistent with previous reports [11]. Of 72 FISH-positive (HER2/CEP17 ≥2) patients, 23 expressed HER2 protein levels below the 750 amol/µg cutoff, suggesting a false positive rate of 31.9% (Figure 1B). In contrast, none of the patients with HER2/CEP17 <2 expressed HER2 protein levels above the cutoff. Of 49 patients with HER2 GCN ≥6, 12 patients expressed <750 amol/µg of protein (false positive rate: 24.5%) and of 70 patients who were HER2− by GCN, 3 patients expressed >750 amol/µg of HER2 (false negative rate: 4.3%) (Figure 1C). The sensitivity and specificity of the 750 amol/µg cutoff for predicting HER2 positivity were 68.6% and 98.8%, respectively, when positivity was defined as IHC 3+ or FISH+, and 70.7% and 93.8%, respectively, when positivity was defined as IHC 3+ or IHC 2+ with confirmation by FISH.
Of 84 samples that were HER2− by conventional testing, proteomic analysis identified a single patient expressing >750 amol/µg of HER2 protein (false negative rate: 1.2%); 49 of 153 conventionally HER2+ samples expressed HER2 below the cutoff (false positive rate of 32.0%) (Table 1).
Table 1.
Number of patients above and below the proteomic HER2 amplification cutoff, by status of conventional HER2 test
| HER2 conventional test (IHC/FISH) | HER2 <750 amol/µg | HER2 ≥750 amol/µg | Total | |
|---|---|---|---|---|
| (SRM) | (SRM) | |||
| HER2+ | IHC3+ and FISH− | 5 | 0 | 5 |
| IHC3+ and FISH+ | 5 | 36 | 41 | |
| IHC3+ and no FISH | 14 | 50 | 64 | |
| FISH+ and IHC1+ | 6 | 1 | 7 | |
| FISH+ and IHC2+ | 17 | 13 | 30 | |
| FISH+ and no IHC | 2 | 4 | 6 | |
| Total | 49 | 104 | 153 | |
| HER2− | FISH− | 58 | 0 | 58 |
| No FISH and IHC0 | 9 | 1 | 10 | |
| No FISH and IHC1+ | 14 | 0 | 14 | |
| No FISH and IHC 2+ | 2 | 0 | 2 | |
| Total | 83 | 1 | 84 | |
SRM, selected reaction monitoring; IHC, immunohistochemistry; FISH, fluorescence in situ hybridization.
Patient selection for trastuzumab using HER2 SRM quantity
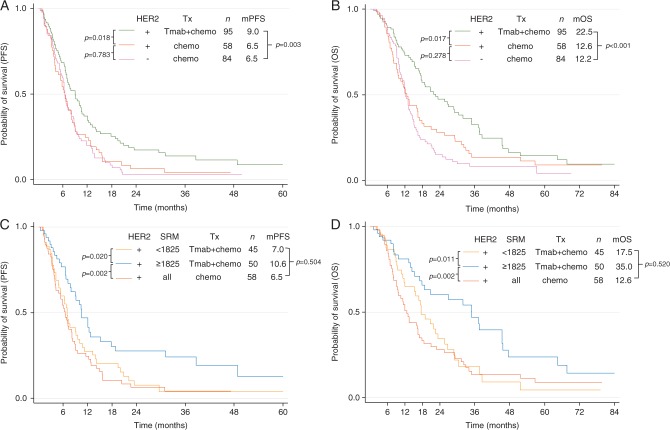
As expected, HER2+ patients benefited from the addition of trastuzumab to chemotherapy [median PFS 9.0 versus 6.5 months, P = 0.018 (Figure 2A); median OS 22.5 versus 12.6 months, P = 0.017 (Figure 2B)]. The median PFS and OS of HER2− patients treated with chemotherapy were 6.5 and 12.2 months respectively. Trastuzumab-treated patients with a HER2/CEP17 ratio >4.7 had greater OS than patients with lower HER2/CEP17 ratios, although the difference was not statistically significant (25.7 versus 15.2 months, P = 0.160).
Figure 2.
HER2 selected reaction monitoring (SRM) identifies gastric cancer patients who would benefit from trastuzumab. With conventional HER2 testing methods, HER2-positive patients selected for treatment with trastuzumab plus chemotherapy had better (A) progression-free survival and (B) overall survival than HER2-positive patients who received chemotherapy alone and HER2-negative patients who received chemotherapy. (C and D) The survival benefits are greater with HER2 assessment by quantitative proteomics. Trastuzumab-treated patients with HER2 protein expression above the cutoff (1825 amol/µg) have twice the overall survival of patients whose HER2 levels are below the cutoff. Trastuzumab-treated patients with HER2 levels below the cutoff had survival outcomes similar to patients treated with chemotherapy alone. P-value is for the log rank test. Tx, treatment; Tmab, trastuzumab; chemo, chemotherapy.
Upon Cox regression analysis of PFS and OS in the HER2 IHC/FISH-positive groups, a statistically significant interaction was detected between treatment group and HER2 protein level (P = 0.007) (supplementary Figure S3, available at Annals of Oncology online). Patients expressing <1825 amol/µg had statistically similar PFS and OS regardless of therapy group, while patients with HER2 protein expression >1825 amol/µg showed significant benefit when treated with trastuzumab in addition to chemotherapy.
We applied 1825 amol/µg as a HER2 protein level cutoff to determine the magnitude of differential benefit observed in the trastuzumab-treated population. As compared with patients expressing HER2 levels below the cutoff, patients with levels above the cutoff had a 3.6 month improvement in PFS (10.6 versus 7.0 months, P = 0.020) (Figure 2C) and double the OS (35.0 versus 17.5 months, P = 0.011) (Figure 2D). Notably, the survival benefit in trastuzumab-treated patients with HER2 expression below the cutoff was not statistically different from that of HER2+ patients treated with chemotherapy alone (mPFS: 7.0 versus 6.5 months, P = 0.504; mOS: 17.5 versus 12.6 months, P = 0.520) (Figure 2C and D). The HER2 cutoff was not prognostic within the chemotherapy-treated cohort (mPFS: 6.1 versus 6.6 months, P = 0.870 for PFS; mOS: 11.9 versus 13.0 months, P = 0.803) (supplementary Figure S4, available at Annals of Oncology online).
Univariate and multivariate survival analysis by clinicopathologic characteristics known to be prognostic factors in GC was conducted to determine whether the 1825 amol/µg cutoff was relevant to subgroups of patients within cohort 1 (supplementary Table S5, available at Annals of Oncology online). In nearly all subgroups, patients with HER2 protein levels above the cutoff fared better on trastuzumab treatment than patients below the cutoff (supplementary Figure S6, available at Annals of Oncology online).
Discussion
The ToGA trial was the first demonstration outside of breast cancer that HER2+ tumors can respond to HER2-targeted therapy. This led to optimism that additional HER2-targeted therapies would show efficacy in GC, but unfortunately this has not happened. It seems likely that the HER2 diagnostics which demonstrated the benefit of trastuzumab in the ToGA trial have failed to identify responsive patients in randomized trials of the HER2-targeted agents lapatinib and T-DM1. Even for trastuzumab, response rates across trials ranged from 32% to 68% [2–4] suggesting that in a ‘responsive’ population, many patients are not benefiting from therapy. Clearly, the development and validation of novel HER2 diagnostics is critical to improve patient selection, both for existing agents and for those in development. Since all of these agents directly bind to HER2 protein, we used SRM mass spectrometry to perform direct, quantitative analysis of HER2 expression in the tumors of GC patients. This data was then used to define quantitative cutoffs of HER2 expression, identifying patients likely to benefit from the addition of trastuzumab to chemotherapy, as well as those patients in whom the addition of trastuzumab offered no benefit over chemotherapy alone. Previous studies have similarly demonstrated that outcomes following trastuzumab treatment clearly differ according to HER2 gene amplification level [14, 15].
Targeted proteomics demonstrated a very wide range of HER2 protein expression within a supposedly homogenous group of patients identified as HER2+ by IHC and FISH. These patients expressed quantities of HER2 protein ranging from non-detectable to ‘super-expression’ (>20 000 amol/µg tumor protein). In current clinical practice, and based upon existing diagnostics, all HER2+ patients would be treated with trastuzumab. Based upon the response rate of trastuzumab in GC, we hypothesize that the bulk of non-responders have low or absent HER2 protein expression, although there are other possible reasons for non-response, including molecular alterations in the downstream pathways of HER2 as reported in breast cancer (e.g. activation of cyclin E and D, loss of PTEN, mutations in PI3KCA, p95HER2) [16–18]. Unlike IHC, SRM mass spectrometry is quantitative over a broad expression range of at least 5 orders of magnitude. This allows mass spectrometry to see differences in expression even within ‘correctly’ diagnosed IHC3+, and affords the opportunity to identify quantitative cutoffs within an otherwise homogeneous IHC3+ population.
In this study we see some discordance between FISH and quantitative proteomics; roughly 32% of 72 patients with apparent gene amplification failed to express HER2 protein, consistent with a previous comparison of HER2 amplification and targeted proteomics in breast cancer [12]. In that study, discordances between gene amplification and protein levels were attributed to a gene amplification pattern known as ‘double minutes’ consisting of extrachromosomal fragments of DNA harboring HER2 amplicons. Such tumors are properly diagnosed as HER2+ based on GCN and HER2/CEP17 ratio, but fail to express HER2 protein, and are theorized to be non-responsive to HER2-targeted therapy. The relationship between HER2 protein expression and amplification pattern (e.g. homogeneously staining region versus double minutes) in GC should be investigated.
Both FISH and IHC have flaws that can result in overidentification of HER2+ patients and the consequent administration of trastuzumab to patients who are unlikely to respond due to low or negative HER2 expression. Using HER2 expression as a continuous variable, we determined a protein cutoff level to identify likely responders to trastuzumab. Application of this cutoff retrospectively to HER2+ patients doubled the median OS as compared with traditional HER2 diagnostics (17.5 versus 35.0 months). As expected, the cutoff had no predictive value in the HER2+ or HER2− patients treated with chemotherapy alone, suggesting that it truly identified a subpopulation of HER2+ patients who stand to benefit from trastuzumab treatment.
Of the 95 patients in the trastuzumab-treated cohort, 22 had HER2 IHC scores of 1+ or 2+. Four of these cases had elevated levels of HER2 protein (>1825 amol/µg) and notable responses to trastuzumab (mOS: 46.6 months; range: 37.3–101.2 months). One patient, originally scored as IHC 1+ had very high levels of HER2 protein (5662 amol/µg) and an OS of 45.6 months on trastuzumab; this patient would not have received trastuzumab based on the IHC score using current pathology algorithms.
The results of this study are similar to those of a retrospective study in breast cancer which showed that proteomic HER2 levels correlated with durable response to trastuzumab [12]. In metastatic and adjuvant treatment settings, breast cancer patients with HER2 protein levels above 2200 amol/µg had improved response [12]. The breast cancer study was limited as it lacked a HER2+ chemotherapy-treated arm. That said, we wonder if the improved prognosis seen for breast cancer patients with HER2 expression >2200 amol/µg and a predictive benefit >1825 amol/µg for GC are a class effect, and may be predictive of trastuzumab response in additional indications where HER2 is overexpressed. Different HER2-targeted therapies use different mechanisms to block HER2 oncogene activity. We speculate that each agent will have different optimal HER2 expression based upon targeted proteomic analysis. For example, antibody drug conjugates such as T-DM1 should have a threshold for activity based on the minimum expression of HER2 which delivers sufficient DM1 to kill cells. Preliminary data from breast cancer suggests that lapatinib shows a biphasic efficacy response; patients with expression below the threshold are non-responsive because HER2 expression is insufficient to drive tumor growth, and expression above a separate threshold overwhelms the inhibitory ability of lapatinib [19]. This retrospective study demonstrated the ability of HER2 protein quantitation by mass spectrometry to better predict trastuzumab treatment outcomes in GC patients, compared with current diagnostic methods. Unlike previous studies, this study compared HER2 expression and outcome against a control arm, strengthening the connection between expression and benefit of trastuzumab. Most importantly, we also showed that the benefit of trastuzumab in low HER2 expressors was not statistically different than in the HER2− group treated with chemotherapy alone, suggesting an opportunity to spare such patients from an ineffective and cardiotoxic course of treatment. Indeed, these findings require further validation in independent studies. The current study does support the feasibility of targeted proteomics to better inform and guide HER2 therapy for HER2+ GC patients. Targeted proteomics may provide a better method of identifying patients for treatment with trastuzumab and other HER2-targeted agents.
Supplementary Material
Acknowledgements
The authors thank Sang Mee Lee and Theodore Karrison for assistance with the statistical analysis in supplementary Figure S3, available at Annals of Oncology online, David Krizman and Shankar Sellappan for manuscript editing, and Paul Bhar for independent statistical analysis using SAS.
Funding
This research was supported in part by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (Grant No. 2013R1A1A2008705, to Dr. Do-Youn Oh) and part by NIH K23 award (CA178203-01A1), UCCCC (University of Chicago Comprehensive Cancer Center) Award in Precision Oncology- CCSG (Cancer Center Support Grant) (P30 CA014599), Oncoplex Dx Collaborative Research Agreement, LLK (Live Like Katie) Foundation Award, and the Sal Ferrara II Fund for PANGEA (to Dr. Daniel V.T. Catenacci).
Disclosure
Parts of the study were presented at the Annual Meeting of 2015 American Society of Clinical Oncology, Chicago, IL, USA.
EA, WLL, FC, AB, and ST: Employment and stock ownership of NantOmics.
TH: Leadership position and stock ownership of NantOmics.
DC: Advisory role, research funding, expert testimony, and honoraria from NantOmics.
References
- 1. Torre LA, Bray F, Siegel RL. et al. Global cancer statistics, 2012. CA Cancer J Clin 2015; 65: 87–108. [DOI] [PubMed] [Google Scholar]
- 2. Bang YJ, Van Cutsem E, Feyereislova A. et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet 2010; 376: 687–697. [DOI] [PubMed] [Google Scholar]
- 3. Gravalos C, Gomez-Martin C, Rivera F. et al. Phase II study of trastuzumab and cisplatin as first-line therapy in patients with HER2-positive advanced gastric or gastroesophageal junction cancer. Clin Transl Oncol 2011; 13: 179–184. [DOI] [PubMed] [Google Scholar]
- 4. Kurokawa Y, Sugimoto N, Miwa H. et al. Phase II study of trastuzumab in combination with S-1 plus cisplatin in HER2-positive gastric cancer (HERBIS-1). Br J Cancer 2014; 110: 1163–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Warneke VS, Behrens HM, Boger C. et al. Her2/neu testing in gastric cancer: evaluating the risk of sampling errors. Ann Oncol 2013; 24: 725–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Satoh T, Xu RH, Chung HC. et al. Lapatinib plus paclitaxel versus paclitaxel alone in the second-line treatment of HER2-amplified advanced gastric cancer in Asian populations: TyTAN—a randomized, phase III study. J Clin Oncol 2014; 32: 2039–2049. [DOI] [PubMed] [Google Scholar]
- 7. Hecht JR, Bang YJ, Qin SK. et al. Lapatinib in combination with capecitabine plus oxaliplatin in human epidermal growth factor receptor 2-positive advanced or metastatic gastric, esophageal, or gastroesophageal adenocarcinoma: TRIO-013/LOGiC—a randomized phase III trial. J Clin Oncol 2016; 34: 443–451. [DOI] [PubMed] [Google Scholar]
- 8. Kang Y, Shaw M, Ohtsu A. A randomized, open-label, multicenter, adaptive phase 2/3 study of trastuzumab emtansine (T-DM1) versus a taxane (TAX) in patients (pts) with previously treated HER2-positive locally advanced or metastatic gastric/gastroesophageal junction adenocarcinoma (LA/MGC/GEJC). J Clin Oncol 34(suppl 4S); abstr 5. http://meetinglibrary.asco.org/content/159174-173 (3 November 2016, date last accessed). [Google Scholar]
- 9. Hembrough T, Thyparambil S, Liao WL. et al. Selected reaction monitoring (SRM) analysis of epidermal growth factor receptor (EGFR) in formalin fixed tumor tissue. Clin Proteomics 2012; 9: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hembrough T, Thyparambil S, Liao WL. et al. Application of selected reaction monitoring for multiplex quantification of clinically validated biomarkers in formalin-fixed, paraffin-embedded tumor tissue. J Mol Diagn 2013; 15: 454–465. [DOI] [PubMed] [Google Scholar]
- 11. Catenacci DV, Liao WL, Zhao L. et al. Mass-spectrometry-based quantitation of Her2 in gastroesophageal tumor tissue: comparison to IHC and FISH. Gastric Cancer 2015; 19: 1066-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nuciforo P, Thyparambil S, Aura C. et al. High HER2 protein levels correlate with increased survival in breast cancer patients treated with anti-HER2 therapy. Mol Oncol 2016; 10: 138–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Steiner C, Tille JC, Lamerz J. et al. Quantification of HER2 by targeted mass spectrometry in formalin-fixed paraffin-embedded breast cancer tissues. Mol Cell Proteomics 2015; 14: 2786–2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gomez-Martin C, Plaza JC, Pazo-Cid R. et al. Level of HER2 gene amplification predicts response and overall survival in HER2-positive advanced gastric cancer treated with trastuzumab. J Clin Oncol 2013; 31: 4445–4452. [DOI] [PubMed] [Google Scholar]
- 15. Ock CY, Lee KW, Kim JW. et al. Optimal patient selection for trastuzumab treatment in HER2-positive advanced gastric cancer. Clin Cancer Res 2015; 21: 2520–2529. [DOI] [PubMed] [Google Scholar]
- 16. Goel S, Wang Q, Watt AC. et al. Overcoming therapeutic resistance in HER2-positive breast cancers with CDK4/6 inhibitors. Cancer Cell 2016; 29: 255–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Berns K, Horlings HM, Hennessy BT. et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell 2007; 12: 395–402. [DOI] [PubMed] [Google Scholar]
- 18. Sergina NV, Rausch M, Wang D. et al. Escape from HER-family tyrosine kinase inhibitor therapy by the kinase-inactive HER3. Nature 2007; 445: 437–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nuciforo P, Thyparambil S, Galvan P. Quantitative HER family proteins assessment as prognostic and predictive biomarkers in the EGF30008 clinical trial. Cancer Research 2016; 76(Suppl): abstract P3-07-08. http://cancerres.aacrjournals.org/content/76/4_Supplement/P3-07-08 (3 November 2016, date last accessed). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.