Abstract
Carbonic anhydrase IX (CA9) is a membrane‐associated carbonic anhydrase that regulates cellular pH, is upregulated in various solid tumors, and is considered to be a therapeutic target. Here, we describe the essential role of CA9 in the tumorigenicity of cells derived from human adult T‐cell leukemia/lymphoma (ATL). We previously established the highly tumorigenic ST1‐N6 subline from the ATL‐derived ST1 cell line by serial xenotransplantation in NOG mice. In the present study, we first show that CA9 expression is strongly enhanced in ST1‐N6 cells. We then sorted ST1 cells by high or low CA9 expression and established ST1‐CA9high and ST1‐CA9low sublines. ST1‐CA9high cells, like ST1‐N6 cells, were more strongly tumorigenic than ST1‐CA9low or parental ST1 cells when injected into NOG mice. Knockdown of CA9 with shRNAs suppressed the ability of ST1‐CA9high cells to initiate tumors, and the tumorigenicity of ST1 cells was significantly enhanced by introducing wild‐type CA9 or a CA9 mutant with deletion of an intracytoplasmic domain. However, a CA9 with point mutations in the catalytic site did not increase the tumorigenicity of ST1 cells. Furthermore, we detected a small population of CA9+ CD25+ cells in lymph nodes of ATL patients. These findings suggest that CA9, and particularly its carbonic anhydrase activity, promotes the tumorigenicity of ATL‐derived cells and may be involved in malignant development of lymphoma‐type ATL.
Keywords: Adult T‐cell leukemia/lymphoma, carbonic anhydrase IX, human T‐cell leukemia virus type 1, tumorigenicity, xenotransplantation
Adult T‐cell leukemia/lymphoma (ATL) is an aggressive disorder characterized by clonal propagating mature CD4‐positive T cells. ATL develops decades after infection by human T‐cell leukemia virus type 1 (HTLV‐1).1 Most HTLV‐1 carriers are asymptomatic, and of these carriers approximately 2.1% of women and 6.6% of men will develop ATL.2 The clinical manifestations of ATL fall into four subtypes: acute, lymphomatous, chronic, and smoldering; acute and lymphoma‐type ATL are aggressive and have a poor prognosis.3, 4
Although HTLV‐1 infection is of primary importance in the pathogenesis of ATL, the molecular basis of ATL development after HTLV‐1 infection is uncertain. There is growing evidence that ATL‐derived cells and HTLV‐1‐infected T cells carry genetic and epigenetic alterations in the host genome; these include deletions, mutations, and upregulated expression of genes such as the CDKN2A family (p15 and p16), p53, Rb, Fas, BCL11B, CARMA1, c‐Met, BMP6, and miR‐31.5 Some of these alterations may deregulate signaling pathways that control cell cycling, proliferation, and apoptosis.5 Integrated molecular analyses in 426 ATL cases identified molecular alterations associated with ATL,6 but it is not clear which of these alterations are critical for the malignancy of ATL cells, and particularly that of ATL cancer stem cells (CSCs).
The crucial role of CSCs in tumor initiation and growth was first identified in acute myeloid leukemia,7 and subsequently in several other human cancers, in experiments in non‐obese diabetic/severe combined immunodeficient (NOD/SCID) mice.8 We previously established the immunodeficient mouse strain NOD/Shi‐scid‐IL‐2Rγnull (NOG) by crossing NOD‐SCID and γc‐deficient mice.9 Both NOD/SCID and NOG mice have high engraftment rates for human hematopoietic stem cells (HSCs). We used the NOG mice to establish humanized mice with a rebuilt human immune system,10 and a two‐hit mouse model of human leukemia.11 NOG mice can be used to select CSC populations from some types of human cancer, such as colon,12 hypopharyngeal,13 bile‐duct,14 and pancreatic15 cancers. ATL‐derived and HTLV‐1‐infected cells also develop tumors in NOG mice, although their tumorigenic cell populations are not well characterized.16 HTLV‐1 Tax‐transgenic mice form lymphomas and leukemia resembling human ATL and carry a CSC‐like population.17, 18 The side‐population cells detected in ATL cell lines are resistant to interferon (IFN)‐α treatment and chemotherapy,19 and CD44 expression in primary ATL cells is closely related to CSC‐like phenotypes.20, 21 These observations suggest the presence of ATL‐associated CSCs that define the malignancy of ATL.
We previously established the highly tumorigenic ST1‐N6 subline by performing serial xenografts of the ATL ST1‐cell line in NOG mice, and demonstrated that elevated AKT activation contributes to the tumorigenic potential of ST1‐N6 cells.22 In the present study, we show that ST1‐N6 cells express elevated levels of carbonic anhydrase IX (CA9), a membrane‐associated metalloenzyme that catalyzes hydration of carbon dioxide to bicarbonate and protons and thereby regulates cellular pH.23 CA9 overexpression is closely associated with aggressive behaviors and a poor prognosis in human solid tumors.24 In particular, CA9 critically mediates expansion of breast‐cancer stem cells in hypoxic niches by sustaining the mesenchymal and stemness phenotypes of these cells.25 Here, we show that CA9 promotes the tumorigenicity of ATL‐derived cells, and that ATL patients carry small populations of CA9‐positive ATL cells in their lymphoid tissues.
Materials and Methods
Clinical samples and ethics statements
Peripheral blood mononuclear cells (PBMCs) from ATL patients and healthy individuals, and lymph node specimens from ATL patients, were collected at Miyagi Cancer Center, Tohoku University Hospital, and the University Hospital of the Ryukyus after obtaining informed written consent from each donor. This study conforms to the Declaration of Helsinki and was approved by the ethics committee of Miyagi Cancer Center.
Cell lines and culture
The ATL‐derived cell line ST1 established by Dr. Yamada (Nagasaki University)26 was used in the study. We previously established the ST1‐N6 subline by performing serial xenografts of ST1 cells in NOG mice.22 In this study, we established two other ST1 sublines, ST1‐CA9high and ST1‐CA9low. These cell lines were maintained in RPMI‐1640 medium containing 10% fetal bovine serum (Gibco, Grand Island, NY, USA) and penicillin/streptomycin (Nacalai, Tokyo, Japan) under 5% CO2 at 37°C in a humidified condition. Cells were also cultured in a hypoxic condition under 1% O2, 5% CO2, and 94% N2.
Mice and in vivo tumorigenesis assay
NOD/Shi‐scid/IL‐2Rγnull (NOG) mice purchased from the Central Institute for Experimental Animals (Kawasaki, Japan) were used for in vivo tumorigenesis assays, in which each cell sample (1 × 103 or 1 × 104 cells) was suspended in RPMI‐1640 medium, mixed with an equal volume of Matrigel (BD Biosciences, San Jose, CA, USA), and injected subcutaneously into two sites on the back of three NOG mice (8–10 weeks old). Tumor formation was monitored weekly by palpation and tumor volumes were calculated as length × width2/2. The tumors became palpable in our hands when their volumes reached at 50 mm3 (4–5 mm in diameter). Our animal care and experimental protocols followed the procedures and guidelines established by the Miyagi Cancer Center Animal Care and Use Committee.
Microarray analysis
Total RNA was extracted from cells with a mirVana miRNA Isolation Kit (Life Technologies, Carlsbad, CA, USA) and labeled with a Low Input Quick Amp Labeling Kit (Agilent Technologies, Santa Clara, CA, USA). The labeled probes were hybridized with Sure Print G3 Human GE 8×60K microarray slides (Agilent Technologies) following the manufacturer's instructions, and the hybridized probes were scanned with an Agilent microarray scanner. Data were processed by Feature Extraction Software (Agilent Technologies) followed by the Rank Products method in the R package.27 Gene expression was scaled by Z‐score and is presented as a heatmap.
Quantitative real‐time PCR
Total RNA was prepared with a RNeasy Mini Kit (Qiagen, Valencia, CA, USA), and cDNA was synthesized using a PrimeScript II cDNA Synthesis Kit (Takara Bio, Kusatsu, Japan). PCR was performed with Brilliant III Ultra‐Fast SYBR Green QPCR Master Mix (Agilent Technologies). Primer sequences are shown in Table S1. PCR reactions were carried out with an LC480 II (Roche Diagnostics, Mannheim, Germany) using beta‐actin expression as an internal standard.
Flow cytometry analysis and cell sorting
The following antibodies were used: allophycocyanin (APC)‐conjugated mouse monoclonal anti‐human CA9 (clone: FAB2188A; R&D Systems, Minneapolis, MN, USA), FITC‐conjugated mouse monoclonal anti‐human CD25 (clone: M‐A251; BD Pharmingen, San Diego, CA, USA), PE‐conjugated mouse monoclonal anti‐human CD4 (clone: RPA‐T4; BD Pharmingen), and APC‐conjugated mouse monoclonal anti‐human CD45RA (clone: 1F6; Nichirei Biosciences, Tokyo, Japan). APC‐conjugated mouse IgG (clone: MOPC‐173; BioLegend, San Diego, CA, USA) and FITC‐conjugated mouse IgG (clone: 555748; BioLegend) were used as isotype controls, and 7‐AAD was used to exclude dead cells. Stained cells were analyzed with a FACSCanto II (BD Biosciences) or sorted with a FACSAria II (BD Biosciences) following the manufacturer's protocol.
Enzyme‐linked immunosorbent assay
Soluble CA9 concentrations in cell‐culture supernatants were measured using a Quantikine enzyme‐linked immunosorbent assay (ELISA) kit for Human Carbonic Anhydrase IX (R&D Systems). Cells (1 × 105/mL) were cultured for 48 h and the culture supernatants were clarified by brief centrifugation (12 000 g, 5 min at 4°C) and assayed for CA9 concentration.
Expression vectors
Methods for preparing expression vectors for wild‐type CA9, CA9 mutants, and CA9 shRNAs are described in Data S1.
Western blotting
Western blots were performed as described previously.28 In brief, cells were lysed in lysis buffer (50 mM Hepes pH 7.5, 150 mM NaCl, 1% Triton X‐100, 2 mM EDTA, 10 mM NaF, 2 mM sodium orthovanadate, 2 mM phenylmethanesulfonyl fluoride, 7.5 μg/mL aprotinin, and 10 μg/mL leupeptin) and the protein concentration of the lysates was measured with a Bio‐Rad protein assay kit (Bio‐Rad, Hercules, CA, USA). Lysate protein (10 μg) was loaded into each lane of a 4–20% SDS‐polyacrylamide gel (Wako, Osaka, Japan), separated by electrophoresis, and transferred onto polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA). After blocking with 5% nonfat milk in phosphate‐buffered saline containing 0.1% Tween 20 (PBST), the membranes were probed with a rabbit monoclonal anti‐human CA9 antibody (clone: D10C10; Cell Signaling Technology, Danvers, MA, USA) at 1:1000. A horseradish peroxidase‐conjugated anti‐rabbit antibody (#7074P2; Cell Signaling Technology) was applied at 1:1000 and detected with SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific, Rockford, IL, USA). Identical filters were probed with a mouse anti‐α‐tubulin monoclonal antibody (Sigma‐Aldrich, St. Louis, MO, USA) followed by 1:1000 horseradish peroxidase‐conjugated anti‐mouse antibody (#7076S; Cell Signaling Technology) to verify equal protein loading and transfer. Signals were visualized with a LAS‐4000mini CCD Imager (Fujifilm, Tokyo, Japan).
Immunohistochemistry
Paraffin‐embedded lymph node tissue samples were sectioned at 3 μm, placed on adhesive slide glass (Matsunami, Kishiwada, Japan), deparaffinized in xylene, and rehydrated through ethanol to distilled water. Heat‐induced epitope retrieval was performed in a pH 9.0 target‐retrieval solution (Dako, Carpinteria, CA, USA) at 96°C for 45 min. Endogenous peroxidase was removed with 0.3% H2O2. Specimens were blocked with 5% normal goat serum in TBST and incubated with a rabbit monoclonal anti‐human CA9 antibody (clone: D10C10; Cell Signaling Technology) and a mouse monoclonal anti‐human CD25 antibody (clone: 4C9; Thermo Fisher Scientific) at 4°C overnight. Bound antibodies were probed with HRP‐conjugated anti‐mouse IgG and a Tyramide Signal Amplification Kit (Molecular Probes, Eugene, OR, USA) and with Alexa 594 anti‐rabbit IgG (Molecular Probes). Specimens were stained with DAPI (Invitrogen, Carlsbad, CA, USA), mounted with Antifade Gold (Molecular Probes), and observed by confocal microscopy (LSM 510 META; Carl Zeiss, Jena, Germany).
Statistics
Most experiments were repeated at least three times. Data were evaluated by Student's t‐test.
Results
CA9 is strongly expressed in ST1‐N6 cells
The ST1‐N6 subline was established by six serial xenografts of the ATL‐derived ST1 cell line into NOG mice and is more tumorigenic than the parental ST1 cell line.22 In this study, we identified genes related to this tumorigenic potential in a gene expression microarray using ST1‐N6 and parental ST1 cells. ST1‐N6 cells were analyzed with ST1‐N7, ST1‐N8, and ST1‐N9 cells, which had been serially transplanted into NOG mice seven, eight and nine times, respectively. Compared to ST1 cells, ST1‐N6 and the other serially xenografted ST1 sublines expressed significantly higher levels of at least 20 genes, with CA9 being one of the most notable (Fig. 1a). The difference in CA9 expression between ST1‐N6 and ST1 cells was confirmed by flow cytometry: only 13.9% of ST1 cells were CA9‐positive, while 99.2% of ST1‐N6 cells expressed CA9 on their surface (Fig. 1b). These results suggest that CA9‐positive cells selectively proliferate from ST1 cells to form tumors in NOG mice.
Figure 1.
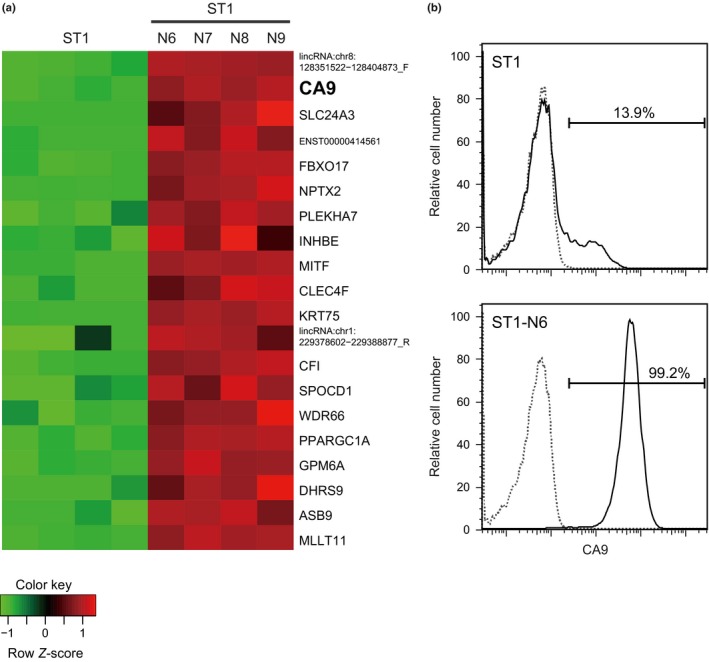
Enhanced CA9 expression in xenografted ST1 cells. (a) Gene expression microarray analysis of cells from xenografted ST1 sublines (ST1‐N6, ‐N7, ‐N8, and ‐N9) and the parental ST1 line (four independent samples). The heatmap shows Z‐scores of gene expression levels, with high expression indicated in red and low expression in green. Data are shown for the 20 most strongly upregulated genes in sublines compared with parental ST1 cells. (b) Flow cytometry results showing the percentage of ST1 and ST1‐N6 cells positive for cell‐surface CA9. Cells were stained with anti‐CA9 (solid lines) or isotype‐matched control (dotted lines) antibodies.
Establishment of CA9high and CA9low ST1 sublines
To investigate the relationship between CA9 expression and the tumorigenic capacity of ST1 cells, sublines with high and low CA9 expression were established by immunostaining ST1 cells, FACS‐sorting the CA9‐positive and CA9‐negative cell populations, and culturing each cell population in vitro. This cell‐sorting procedure was repeated at least four times to obtain ST1‐CA9high and ST1‐CA9low sublines, and CA9 expression in these sublines was assessed by immunostaining, RT‐PCR, and ELISA. Flow cytometry showed that 97.1% of ST1‐CA9high cells were CA9‐positive, while only 3.6% of ST1‐CA9high cells and 13.6% of parental ST1 cells were CA9‐positive (Fig. 2a). RT‐PCR showed higher CA9‐mRNA levels in ST1‐CA9high and ST1‐N6 cells than in ST1‐CA9low or ST1 cells (Fig. 2b). ELISA showed that the culture supernatant from ST1‐CA9high and ST1‐N6 cells contained much higher amounts of soluble CA9 than that of ST1‐CA9low or parental ST1 cells (Fig. 2c). Thus, CA9 expression is increased at both the gene and protein levels in ST1‐CA9high and ST1‐N6 cells, and these cells produce extracellular soluble CA9 molecules.
Figure 2.
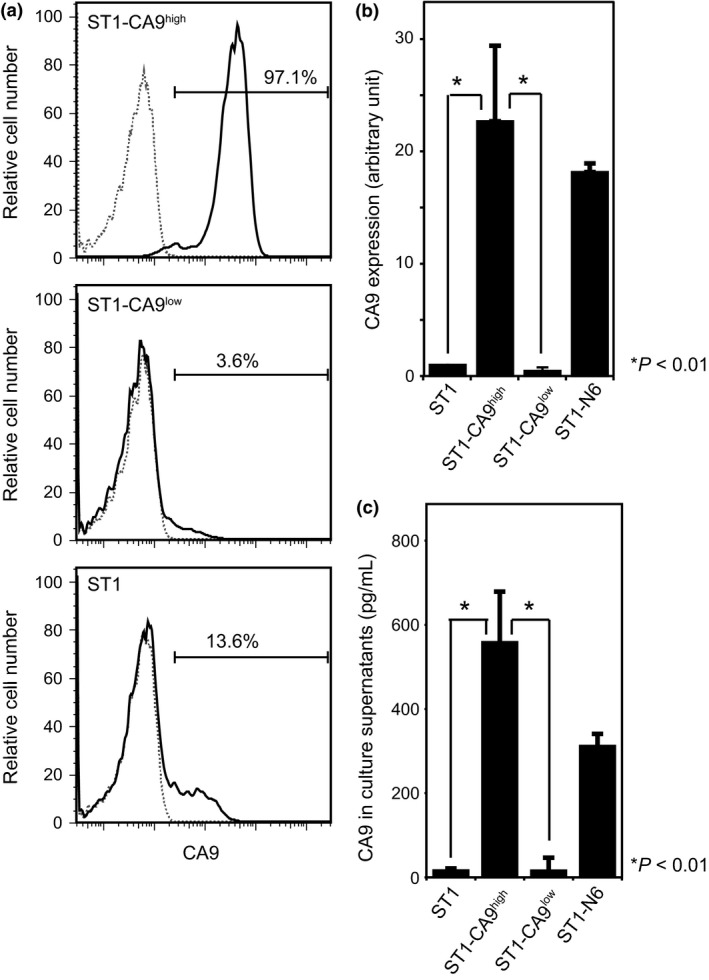
CA9 expression in ST1 and subline cells. (a) Flow cytometry analysis showing the percentage of CA9‐positive cells in ST1 cells and ST1‐CA9high and ST1‐CA9low sublines. Cells were stained with anti‐CA9 (solid lines) or isotype‐matched control (dotted lines) antibodies. (b) RT‐PCR analysis of CA9 expression in ST1 cells and sublines. (c) Concentration of soluble CA9 in culture supernatants of ST1 cells and sublines, measured by ELISA.
Tumorigenicity of ST1 cells and sublines
We next assessed the tumorigenic capacity of the ST1‐CA9high, ST1‐CA9low, and parental ST1 cells in NOG mice. The cells were injected subcutaneously into NOG mice and the resultant tumor formation was assessed weekly. Palpable tumors developed at all six ST1‐CA9high injection sites within 5 weeks, but at only one of six ST1‐CA9low or ST1 injection sites within 7 weeks (Fig. 3a). Tumor growth from ST1‐CA9high cells appeared to be more rapid than that from the other cell lines (Fig. 3b). Flow cytometry showed that 92.5% of cells from ST1‐CA9high tumors expressed CA9; this percentage was similar to that in ST1‐CA9high cells before injection (Fig. 3c). However, 45.3% and 48.2% of cells from ST1‐CA9low and ST1 tumors, respectively, were CA9‐positive; these percentages were significantly higher than those found in the cells before injection (Fig. 3c). These results suggest that the CA9 expression level is well correlated with the tumorigenic capacity of ST1 cells or sublines, and that CA9‐positive ST1 cells may preferentially proliferate in NOG mice. We next asked whether CA9 expression was related to in vitro cell proliferation. There was no significant difference in proliferation rates among ST1‐CA9high, ST1‐CA9low, and ST1 cells in vitro (Fig. S1), indicating that CA9 expression did not affect in vitro cell growth.
Figure 3.
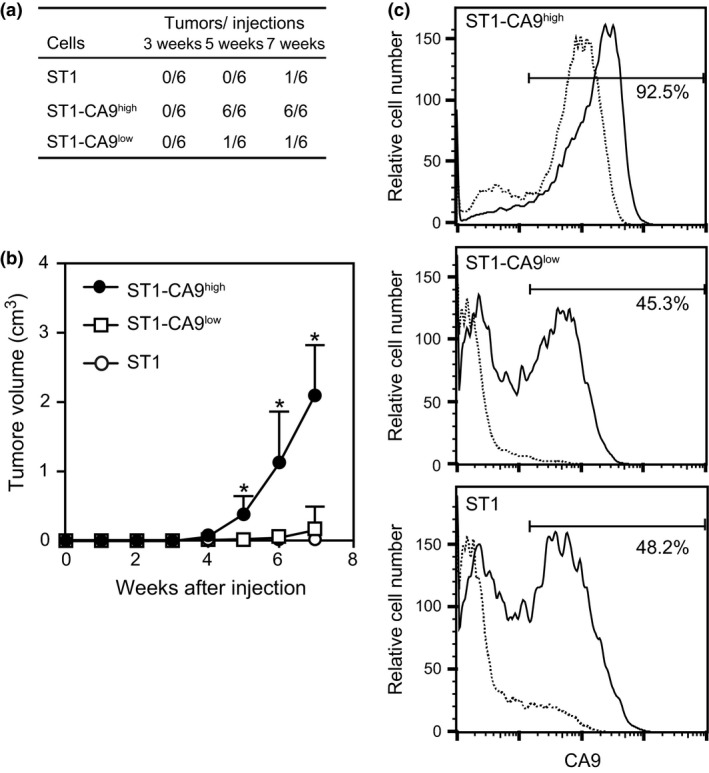
Tumor development from ST1‐CA9high, ST1‐CA9low, and ST1 cells. (a) Cells were injected subcutaneously into NOG mice at two dorsal sites (1 × 103 cells/site) in three mice for each cell line. Tumor sizes were monitored weekly. (b) Tumor growth over time. Data are the mean with the SD of the tumor sizes at the six injection sites. *P < 0.01. (c) Flow cytometry results showing the percentage of CA9‐positive cells among single cells from dissected tumors (solid lines) and cells prior to injection (dotted lines).
CA9 confers tumor‐forming ability on ST1 cells
We then addressed the role of CA9 expression in tumor formation by ST1 cells by introducing a full‐length, wild‐type CA9 expression vector (CA9wt) or an empty vector (EV) into these cells. CA9 was first confirmed to be expressed more abundantly on the surface of CA9wt‐transfected ST1 (ST1/CA9wt) cells than on EV‐transfected ST1 (ST1/EV) cells (Fig. 4a). ST1/CA9wt and ST1/EV cells were then injected subcutaneously into NOG mice and subsequent tumor development was observed. ST1/CA9wt cells produced tumors at all six injection sites within 7 weeks after injection, whereas ST1/EV cells produced tumors at only three of the six injection sites (Fig. 4b). The ST1/CA9wt tumors, like the ST1‐CA9high tumors, grew more quickly than the ST1/EV tumors (Fig. 4c). These results suggested that CA9 contributes to ST1‐cell tumor development. Enhanced tumorigenicity by CA9 overexpression was also observed in another ATL‐cell line, TL‐Om1 (Fig. S2), although the effect was not statistically significant. To confirm this effect of CA9, retroviral expression vectors containing CA9 shRNA (shCA9‐1 or shCA9‐2) were introduced into ST1‐CA9high cells. Transfection of shCA9‐1 or shCA9‐2 significantly decreased CA9 in ST1‐CA9high cells (Fig. 4d) and suppressed tumor development of ST1‐CA9high cells more strongly than EV transfection, although the suppressive effect of shCA9‐2 was weaker than that of shCA9‐1 (Fig. 4e,f). These results indicate that CA9 has a critical role in eliciting the tumorigenic potential of ST1‐CA9high cells.
Figure 4.
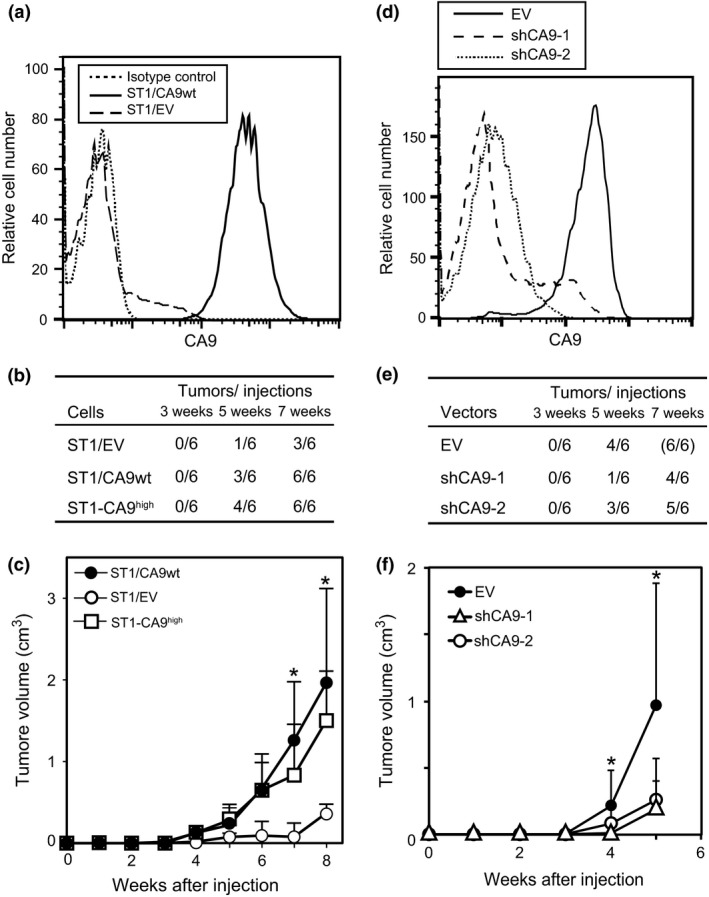
Induction of ST1‐cell tumorigenicity by CA9. (a) ST1 cells were transfected with a wild‐type CA9 expression vector (CA9wt) or an empty vector (EV). CA9 expression on two transfected cell lines, ST1/CA9wt and ST1/EV, was verified by FACS. (b) ST1/CA9wt, ST1/EV, and ST1‐CA9high cells were injected into six dorsal sites (1 × 104 cells/site) in NOG mice and the number of palpable tumors that developed was monitored. (c) Tumor growth was observed weekly. Data are the mean with the SD of the tumor sizes at the six injection sites. *P < 0.05. (d) Vectors expressing CA9 shRNAs (shCA9‐1 or shCA9‐2) or an empty control vector (EV) were introduced into ST1‐CA9high cells and CA9 expression was examined by FACS. (e) After introduction of shRNA, the cells were injected into six dorsal sites (1 × 104 cells/site) of NOG mice and the number of palpable tumors that developed was monitored. (f) Tumor growth was assessed weekly. Data are the mean with the SD of the tumor sizes at the six injection sites. *P < 0.05.
CA9 carbonic anhydrase activity is required for ST1 cells to form tumors
To investigate the functional role of CA9 in the tumor‐forming ability of ST1 cells, ST1 cells were stably transfected with three CA9 mutants: CA9mt, which has two point mutations (E238A/T333A) in an active site that is conserved in all carbonic anhydrase family members (sees Data S1), resulted in decreased extracellular acidification activity (Fig. S3); CA9∆PG, which has a deletion in the proteoglycan‐like domain; and CA9∆IC, which has a deletion in the intracytoplasmic domain (Fig. 5a). These mutants were transfected into ST1 cells to establish three transfectant cell lines: ST1/CA9mt, ST1/CA9∆PG, and ST1/CA9∆IC. Western blots showed that the cell lines all expressed CA9 at comparable levels (Fig. 5b). The deduced molecular weight of CA9 is 45 823 in a mature form (signal peptide‐processed form), but many bands of different sizes were detected, presumably because of glycosylation on the proteoglycan‐like domain and proteolytic processing of the protein. None of these bands were detected after shRNA‐mediated knockdown, indicating the specificity of the antibody used (Fig. S4). Cells expressing wild type or mutant CA9 were then injected into NOG mice to assess their tumorigenic activity. ST1/CA9mt and ST1/EV cells produced palpable tumors at one and two out of six injection sites, respectively, within 5 weeks; these rates were lower than those of ST1/CA9∆PG, ST1/CA9∆IC, and ST1/CA9wt cells (Fig. 5c). Furthermore, 7 weeks after injection, the average sizes of the tumors produced by ST1/CA9mt and ST1/EV cells were extremely small compared to those produced by ST1/CA9wt and ST1/CA9∆IC cells, and ST1/CA9∆PG tumors were of intermediate size (Fig. 5d). These results indicate that CA9 carbonic anhydrase activity plays a critical role in the tumorigenicity of ST1 cells, and that while the CA9 proteoglycan‐like domain is involved to some degree, the intracytoplasmic domain is not involved.
Figure 5.
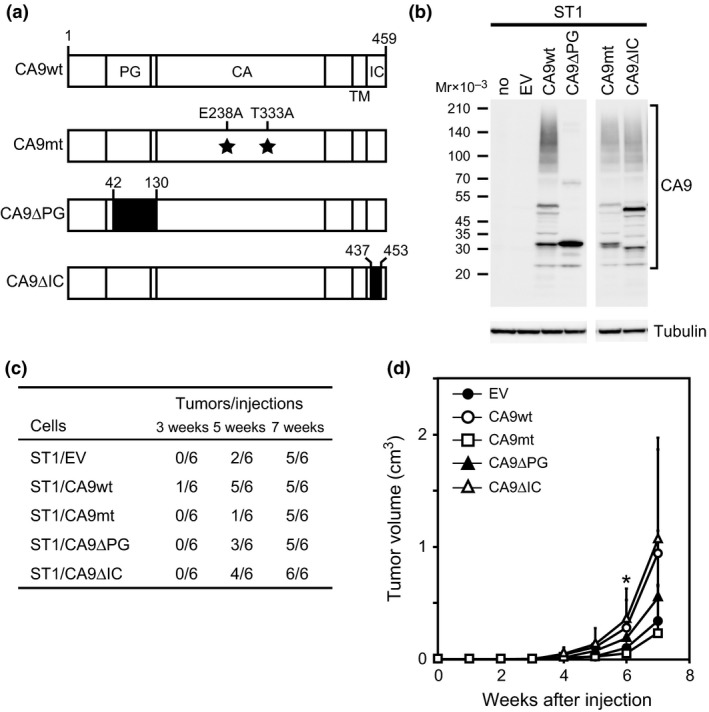
Effects of CA9 mutations on ST1‐cell tumorigenicity. (a) Schematic representation of CA9 and its mutants. Amino acid numbers are shown above each construct. Deleted regions are shown by filled boxes. PG, proteoglycan‐like domain; CA, catalytic domain; TM, transmembrane domain; IC, intracytoplasmic domain. (b) ST1 cells were transfected with vectors expressing CA9wt, CA9mt, CA9∆PG, and CA9∆IC, or an empty control vector (EV). CA9wt, CA9∆CA, CA9∆PG, and CA9∆IC expression levels in transfected cells were verified by Western blot. (c) Transfected cells were injected into six dorsal sites (1 × 104 cells/site) in NOG mice and the number of palpable tumors that developed was monitored. (d) Tumor growth was observed weekly. Data are the mean with the SD of the tumor sizes at the six injection sites. *P < 0.05 for CA9wt versus CA9mt.
CA9‐positive cells in lymph node tissues of ATL patients
To investigate CA9 expression in primary ATL cells, we prepared PBMCs from four patients with acute‐type ATL and from four healthy individuals, and immunostained the cells for CA9 and CD25 or CD4. Flow cytometry showed that PBMCs from the ATL patients and healthy donors contained few CA9+CD25+ or CA9+CD4+ cells (data not shown). Next, we examined lymph node tissue specimens obtained from four patients with lymphoma‐type ATL by immunostaining for CA9 and CD25. The lymph node tissues contained large populations of CD25+ cells (Fig. 6). The CA9+ population was smaller, but almost all CA9+ cells were also positive for CD25 (Fig. 6). These results suggest that primary ATL cells in lymph node tissues contain small but significant CA9+ cell populations.
Figure 6.
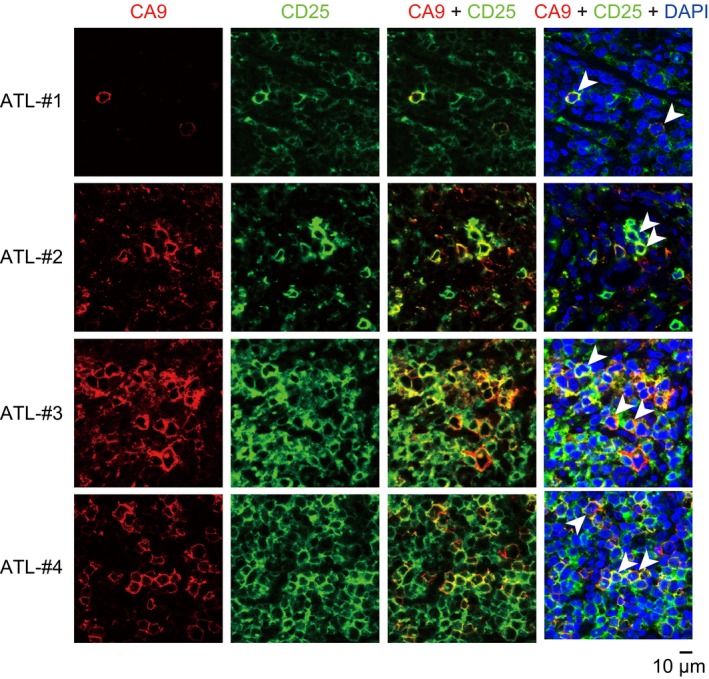
CA9+ CD25+ cells in lymph node tissues of ATL patients. Paraffin‐embedded lymph node samples from four ATL patients (ATL‐1, ATL‐2, ATL‐3 and ATL‐4) were immunohistochemically stained for CA9, CD25, and DAPI. Typical CA9+ CD25+ double‐positive cells are indicated by arrowheads.
Discussion
Serial xenotransplantation of human cancer cells into immunodeficient NOG mice has enabled selection of CSC‐like cell populations from a variety of human solid tumors.12, 13, 14, 15 We previously used this xenotransplantation method to select tumorigenic ATL cells and to establish the highly tumorigenic ST1‐N6 subline from ATL‐derived ST1 cells.22 Here, we compared the whole gene expression in ST1‐N6 and parental ST1 cells, and found that ST1‐N6 expressed CA9 far more strongly than the parental cell line. Based on this observation, ST1‐CA9high and ST1‐CA9low lines were established from ST1 cells by repeated cell sorting. Using these sublines, we showed that ST1‐CA9high cells, like ST1‐N6 cells, are more strongly tumorigenic than ST1‐CA9low or parental ST1 cells when transplanted into NOG mice. This finding indicates that CA9 marks highly tumorigenic ST1 cells. We next investigated the functional contribution of CA9 to the tumorigenicity of ST1‐CA9high cells, and showed that suppression of CA9 expression by shRNA significantly decreased the tumorigenic potential of these cells. In contrast, transfecting ST1 cells with a wild‐type CA9 expression vector significantly enhanced their tumorigenicity. These observations indicate that CA9 plays a critical role in ST1‐cell tumor formation in NOG mice.
CA9 is associated with malignancy in human solid tumors, and CA9 carbonic anhydrase activity is thought to be important for malignancy.24 We used four CA9 mutants to address the possible involvement of CA9 in ST1‐cell tumorigenicity (Fig. 5). ST1 cells transfected with a CA9 mutant with defective catalytic activity (CA9mt) formed significantly fewer tumors than ST1 cells transfected with wild‐type CA9 (CA9wt). Like CA9wt, a CA9 mutant with an intracytoplasmic‐domain deletion (CA9∆IC) conferred high tumorigenicity on ST1 cells. Since CA9mt has no carbonic anhydrase activity (Fig. S3), these results indicate that CA9 carbonic anhydrase activity is critical for eliciting the tumorigenicity of ST1 cells in NOG mice. This confirms that CA9 carbonic anhydrase activity endows ST1 cells with high tumor‐forming activity in NOG mice. Similarly, B‐cell lymphomas express CA9, which is correlated with extracellular acidosis in xenograft tumors.29 Murine T‐cell acute lymphoblastic leukemia/lymphoma cells overexpress the CA9 isoform CA12, and inhibiting CA12 decreases cell proliferation and induces apoptosis in these cells.30 Based on these observations, increased CA9 expression may enhance the ST1‐cell tumor‐initiating capacity in NOG mice by controlling extracellular and intracellular pH dynamics.
Since CA9 expression is mediated by the hypoxia‐inducible factor HIF‐1α in human solid tumors,31 we examined HIF‐1α expression in ST1 cells and sublines. We found no correlation between HIF‐1α and CA9 expression, suggesting that the elevated CA9 expression in ST1‐CA9high and ST1‐N6 cells is not entirely regulated by HIF‐1α (data not shown). We also showed that CA9 expression was not correlated with expression of the HTLV‐1 oncogenes Tax and HBZ in these cell lines (data not shown). Hence, it appears that CA9 is upregulated by an as‐yet‐unknown mechanism in ST1‐CA9high and ST1‐N6 cells, and this mechanism may be important for sustaining the highly tumorigenic capacity of these cells in NOG mice.
We previously showed that AKT signaling, which is activated in ST1‐N6 cells, is required for the tumorigenicity of ST1‐N6 cells.22 AKT phosphorylation at T308 and S473 is a marker of AKT activation in T cells.32 T308 phosphorylation was similar in ST1‐CA9high and ST1‐CA9low cells cultured in vitro, and S473 phosphorylation appeared only to be slightly elevated in ST1‐CA9high cells (Fig. S5). However, AKT phosphorylation was significantly elevated at both T308 and S473 in ST1‐CA9high, ST1‐CA9low, and parental ST1 cells freshly prepared from tumors in NOG mice (Fig. S5). Thus, AKT may be activated during tumor development in these cell lines. We found that overexpression of CA9 in ST1 cells did not affect AKT phosphorylation (data not shown), indicating that there is no direct relationship between CA9 expression and AKT activation, and that AKT activation is not a prerequisite for CA9‐mediated tumor formation by ST1 cells.
CD44 expression in primary ATL cells is reported to be correlated with severity of disease and cell malignancy.20, 21 Hence, we compared CD44 expression among ST1 cells and sublines, but we found no correlation between CD44 and CA9 expression; moreover, there was no difference in the tumorigenicity of CD44+ versus CD44− ST1 cells (data not shown). Primary ATL cells contain a rare CD4+CD45RA+ T memory stem (TSCM) cell population that can induce identical ATL clones in NOG mice.33 We detected similar numbers of CD4+CD45RA+ cells in ST1 cells and sublines (data not shown), suggesting that neither the CD44+ nor CD45RA+ populations contribute to CA9‐mediated tumorigenicity of ST1 cells.
The presence of CA9 in primary ATL cells is a reason for concern. Since most ATL cells express CD4 and CD25,1 we looked for CA9+CD25+ cells or CA9+CD4+ cells in PBMCs and lymph node tissues from ATL patients. We found few CA9+CD25+ or CA9+CD4+ cells in PBMCs from ATL patients or healthy donors, but there was a small but significant CA9+CD25+ population, possibly ATL cells, in lymph node tissues from all four ATL patients. These observations suggest that primary ATL cells in lymphoid tissues of ATL patients contain a small CA9+ ATL‐cell population. Since CA9 is functionally involved in tumor formation of ATL‐derived ST1 cells, CA9+ ATL cells may possess malignant phenotypes for tumor growth and invasiveness into lymphoid organs, reminiscent of putative CSCs in ATL.
Disclosure statement
The authors have no conflict of interest.
Abbreviations
- ATL
adult T‐cell leukemia/lymphoma
- CA9
carbonic anhydrase IX
- CIC
cancer‐initiating cell
- CSC
cancer stem cell
- EV
empty vector
- HSC
hematopoietic stem cell
- HTLV‐1
human T‐cell leukemia virus type 1
- NOD/SCID
non‐obese diabetic/severe combined immunodeficient
- NOG
NOD/Shi‐scid‐IL‐2Rγnull
- qPCR
quantitative PCR
- TSCM
T memory stem cell
Supporting information
Fig. S1. The in vitro growth of ST1 and its subline cells.
Fig. S2. Tumorigenicity of TL‐Om1 cells overexpressing CA9.
Fig. S3. Evaluation of CA activity of ectopically expressed CA9 or its mutant.
Fig. S4. Multiple forms of CA9 in ST1 cells.
Fig. S5. AKT phosphorylation in ST1‐CA9high, ST1‐CA9low, and ST1 cells.
Table S1. Oligonucleotide sequences for quantitative RT‐PCR.
Data S1. Supplementary methods.
Acknowledgments
We thank Dr. M. Ito (Central Institute for Experimental Animals) for the NOG mice, Dr. T. Kitamura (Tokyo Univ.) for the Plat A cells, and Dr. S. Kamihira (Nagasaki Univ.) for the ST1 cells. This study was supported in part by JSPS KAKENHI grants to K. Y. (#24570142), S. I. (#26830087), and K. S. (#25290047).
Cancer Sci 108 (2017) 435–443
Funding Information
JSPS KAKENHI (#24570142, #26830087, #25290047).
References
- 1. Matsuoka M, Jeang K‐T. Human T‐cell leukemia virus type 1 (HTLV‐1) infectivity and cellular transformation. Nat Rev Cancer 2007; 7: 270–80. [DOI] [PubMed] [Google Scholar]
- 2. Arisawa K, Soda M, Endo S et al Evaluation of adult T‐cell leukemia/lymphoma incidence and its impact on non‐Hodgkin lymphoma incidence in southwestern Japan. Int J Cancer 2000; 85: 319–24. [DOI] [PubMed] [Google Scholar]
- 3. Shimoyama M. Diagnostic criteria and classification of clinical subtypes of adult T‐cell leukaemia‐lymphoma. A report from the Lymphoma Study Group (1984–87). Br J Haematol 1991; 79: 428–37. [DOI] [PubMed] [Google Scholar]
- 4. Katsuya H, Ishitsuka K, Utsunomiya A et al Treatment and survival among 1594 patients with ATL diagnosed in the 2000s: a report from the ATL‐PI project performed in Japan. Blood 2015; 126: 2570–7. [DOI] [PubMed] [Google Scholar]
- 5. Yamagishi M, Watanabe T. Molecular hallmarks of adult T cell leukemia. Front Microbiol 2012; 3: doi: 10.3389/fmicb.2012.000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kataoka K, Nagata Y, Kitanaka A et al Integrated molecular analysis of adult T cell leukemia/lymphoma. Nat Genet 2015; 47: 1304–15. [DOI] [PubMed] [Google Scholar]
- 7. Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med 1997; 3: 730–7. [DOI] [PubMed] [Google Scholar]
- 8. Sugihara E, Saya H. Complexity of cancer stem cells. Int J Cancer 2013; 132: 1249–59. [DOI] [PubMed] [Google Scholar]
- 9. Ito M, Hiramatsu H, Kobayashi K et al NOD/SCID/gamma(c)(null) mouse: an excellent recipient mouse model for engraftment of human cells. Blood 2002; 100: 3175–82. [DOI] [PubMed] [Google Scholar]
- 10. Suzuki M, Takahashi T, Katano I et al Induction of human humoral immune responses in a novel HLA‐DR‐expressing transgenic NOD/Shi‐scid/gammacnull mouse. Int Immunol 2012; 24: 243–52. [DOI] [PubMed] [Google Scholar]
- 11. Moriya K, Suzuki M, Watanabe Y et al Development of a multi‐step leukemogenesis model of MLL‐rearranged leukemia using humanized mice. PLoS ONE 2012; 7: e37892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kobayashi S, Yamada‐Okabe H, Suzuki M et al LGR5‐positive colon cancer stem cells interconvert with drug‐resistant LGR5‐negative cells and are capable of tumor reconstitution. Stem Cells 2012; 30: 2631–44. [DOI] [PubMed] [Google Scholar]
- 13. Imai T, Tamai K, Oizumi S et al CD271 defines a stem cell‐like population in hypopharyngeal cancer. PLoS ONE 2013; 8: e62002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tamai K, Nakamura M, Mizuma M et al Suppressive expression of CD274 increases tumorigenesis and cancer stem cell phenotypes in cholangiocarcinoma. Cancer Sci 2014; 105: 667–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Matsuda Y, Yoshimura H, Ueda J et al Nestin delineates pancreatic cancer stem cells in metastatic foci of NOD/Shi‐scid IL2Rgamma(null) (NOG) mice. Am J Pathol 2014; 184: 674–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dewan MZ, Terashima K, Taruishi M et al Rapid tumor formation of human T‐cell leukemia virus type 1‐infected cell lines in novel NOD‐SCID/gammac(null) mice: suppression by an inhibitor against NF‐kappaB. J Virol 2003; 77: 5286–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hasegawa H, Sawa H, Lewis MJ et al Thymus‐derived leukemia‐lymphoma in mice transgenic for the Tax gene of human T‐lymphotropic virus type I. Nat Med 2006; 12: 466–72. [DOI] [PubMed] [Google Scholar]
- 18. Yamazaki J, Mizukami T, Takizawa K et al Identification of cancer stem cells in a Tax‐transgenic (Tax‐Tg) mouse model of adult T‐cell leukemia/lymphoma. Blood 2009; 114: 2709–20. [DOI] [PubMed] [Google Scholar]
- 19. Kayo H, Yamazaki H, Nishida H et al Stem cell properties and the side population cells as a target for interferon‐alpha in adult T‐cell leukemia/lymphoma. Biochem Biophys Res Commun 2007; 364: 808–14. [DOI] [PubMed] [Google Scholar]
- 20. Miyatake Y, Sheehy N, Ikeshita S et al Anchorage‐dependent multicellular aggregate formation induces CD44 high cancer stem cell‐like ATL cells in an NF‐kappaB‐ and vimentin‐dependent manner. Cancer Lett 2015; 357: 355–63. [DOI] [PubMed] [Google Scholar]
- 21. Chagan‐Yasutan H, Tsukasaki K, Takahashi Y et al Involvement of osteopontin and its signaling molecule CD44 in clinicopathological features of adult T cell leukemia. Leuk Res 2011; 35: 1484–90. [DOI] [PubMed] [Google Scholar]
- 22. Yamaguchi K, Takanashi T, Nasu K et al Xenotransplantation elicits salient tumorigenicity of adult T‐cell leukemia‐derived cells via aberrant AKT activation. Cancer Sci 2016; 107: 638–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Swietach P, Hulikova A, Vaughan‐Jones RD. New insights into the physiological role of carbonic anhydrase IX in tumour pH regulation. Oncogene 2010; 29: 6509–21. [DOI] [PubMed] [Google Scholar]
- 24. McDonald PC, Winum JY, Supuran CT. Recent developments in targeting carbonic anhydrase IX for cancer therapeutics. Oncotarget 2012; 3: 84–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lock FE, McDonald PC, Lou Y et al Targeting carbonic anhydrase IX depletes breast cancer stem cells within the hypoxic niche. Oncogene 2013; 32: 5210–9. [DOI] [PubMed] [Google Scholar]
- 26. Yamada Y, Sugahara K, Tsuruda K et al Fas‐resistance in ATL cell lines not associated with HTLV‐I or FAP‐1 production. Cancer Lett 1999; 147: 215–9. [DOI] [PubMed] [Google Scholar]
- 27. Breitling R, Armengaud P, Amtmann A et al Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett 2004; 573: 83–92. [DOI] [PubMed] [Google Scholar]
- 28. Kimura O, Takahashi T, Ishii N et al Characterization of the epithelial cell adhesion molecule (EpCAM)+ cell population in hepatocellular carcinoma cell lines. Cancer Sci 2010; 101: 2145–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen LQ, Howison CM, Spier C et al Assessment of carbonic anhydrase IX expression and extracellular pH in B‐cell lymphoma cell line models. Leuk Lymphoma 2015; 56: 1432–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lounnas N, Rosilio C, Nebout M et al Pharmacological inhibition of carbonic anhydrase XII interferes with cell proliferation and induces cell apoptosis in T‐cell lymphomas. Cancer Lett 2013; 333: 76–88. [DOI] [PubMed] [Google Scholar]
- 31. Wykoff CC, Beasley NJ, Watson PH et al Hypoxia‐inducible expression of tumor‐associated carbonic anhydrases. Cancer Res 2000; 60: 7075–83. [PubMed] [Google Scholar]
- 32. Cantrell D. Protein kinase B (Akt) regulation and function in T lymphocytes. Semin Immunol 2002; 14: 19–26. [DOI] [PubMed] [Google Scholar]
- 33. Nagai Y, Kawahara M, Hishizawa M et al T memory stem cells are the hierarchical apex of adult T‐cell leukemia. Blood 2015; 125: 3527–35. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. The in vitro growth of ST1 and its subline cells.
Fig. S2. Tumorigenicity of TL‐Om1 cells overexpressing CA9.
Fig. S3. Evaluation of CA activity of ectopically expressed CA9 or its mutant.
Fig. S4. Multiple forms of CA9 in ST1 cells.
Fig. S5. AKT phosphorylation in ST1‐CA9high, ST1‐CA9low, and ST1 cells.
Table S1. Oligonucleotide sequences for quantitative RT‐PCR.
Data S1. Supplementary methods.


