Abstract
Background
Nontuberculous mycobacterial (NTM) species are assuming public health importance in pulmonary diseases; they are increasingly being isolated, and importantly, most NTMs do not respond to routine tuberculosis (TB) drugs. This study aimed to identify NTMs isolated from pulmonary TB cases and also determine their susceptibility to streptomycin (STR), isoniazid (INH), and rifampicin (RIF).
Methods
A total of 1755 mycobacterial isolates, obtained between August 2012 and July 2014, from 2036 smear-positive pulmonary cases were identified using polymerase chain reaction amplification of IS6110, and hsp65 gene sequencing analysis. Drug susceptibility testing (DST) was then performed for the identified NTMs against STR, INH, and RIF using microplate Alamar blue assay. The results were analyzed against patients’ biodata for statistical associations.
Results
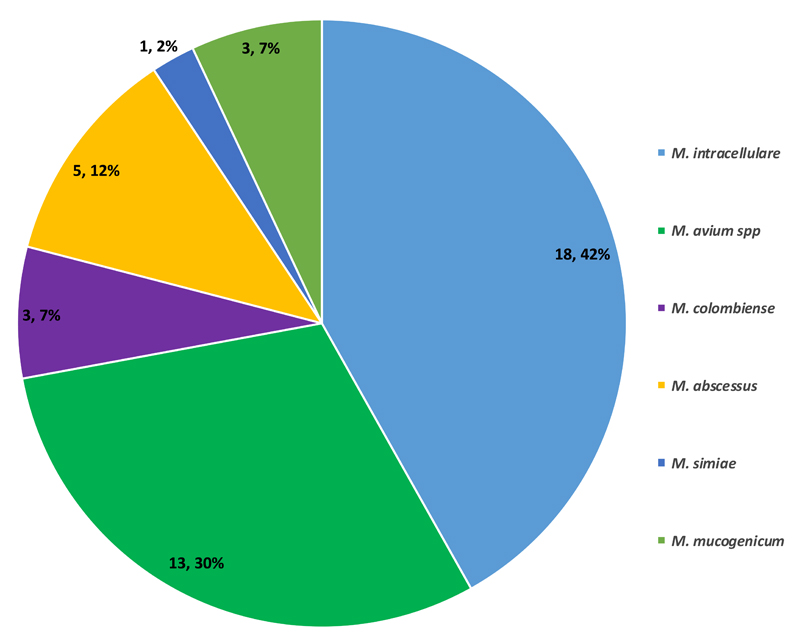
Of the 1755 analyzed isolates, we identified 43 (2.5%) NTMs, which included 18 (41.9%) Mycobacterium intracellulare, 13 (30.2%) Mycobacterium avium subs. paratuberculosis, 5 (11.3%) Mycobacterium abscessus, 3 (7.0%) each of Mycobacterium mucogenicum and Mycobacterium colombiense, and 1 (2.3%) Mycobacterium simiae. Patients infected with NTMs (52.0%) were more likely to be human immunodeficiency virus-positive (P = 0.001, odds ratio = 6.6, 95% confidence interval = 2.7–16.2) than those infected with M. tuberculosis complex (5.8%). All the 43 (100%) NTMs were resistant to INH, whereas 32 (74%) and 19 (44%) were resistant to RIF and STR, respectively. Furthermore, 16 (37.2%) NTMs were resistant to all three drugs, 20 were resistant to INH and RIF, and 3 were resistant to STR and INH. All the M. abscessus isolates were resistant to all the three drugs, whereas all the M. avium isolates were resistant to INH and RIF, but only three were resistant to STR. Among the M. intracellulare isolates, 8, 18, and 15 isolates were resistant to STR, INH, and RIF, respectively.
Conclusion
The observed high-resistance level to INH and RIF supports the need for rapid species identification and DST of nonresponding TB cases before retreatment.
1. Background
Nontuberculous mycobacteria (NTMs) species are mycobacteria other than members of the Mycobacterium tuberculosis complex (MTBC) and Mycobacterium leprae. There are over 170 species identified to date, and unlike M. tuberculosis, NTMs are generally free-living, ubiquitous organisms in the environment.[1] The ecology of NTMs makes it easier for human exposure.[2] Until recently, NTMs were not considered clinically important because they were not found to cause diseases.[2] However, lately, the prevalence of clinical NTMs, especially pulmonary NTMs, has been on the increase worldwide.[1,3] The prevalence rates have been shown to vary among countries globally,[1] and in regions of high tuberculosis (TB) endemicity, NTMs are often unrecognized, misdiagnosed, and/or under diagnosed due to overreliance on smear microscopy.[4–6] Immune suppression (due to factors, such as human immunodeficiency virus [HIV] infection, and chemotherapy) and underlying lung diseases, such as cystic fibrosis, have been identified as contributing factors for the increasing global incidence of NTMs-associated infections.[1,7] Although Africa is a TB/HIV-endemic region,[8] information concerning isolation and characterization of pulmonary NTMs are scanty. This lack of knowledge regarding the contribution of NTMs inevitably may lead to an overly high rate of misdiagnosis of NTMs as MTBC since smear microscopy, the most widely used diagnostic tool, cannot differentiate NTMs from MTBC, which could have dire consequence on the control of TB. [4] Ghana has TB and HIV coinfection rate of approximately 21%;[8] however, there is not a single report regarding isolation and characterization of pulmonary NTMs.
Most NTMs are innately resistant to the conventional anti-TB drugs, [9] leading to treatment failure of the standard TB regimen. Therefore, NTMs infections are likely to be misdiagnosed as multidrug-resistant TB; therefore, patients are put on second-line anti-TB drugs,[6,9] which may also fail. Unlike MTBC infections, there is no single treatment regimen for NTMs because of the diverse spectrum of drug resistance influenced by the genetic background of the infecting species/ strain.[10–12] Therefore, it is imperative that proper identification of NTMs is performed to advice an appropriate treatment regimen.[10–12]
Although smear microscopy is limited by its low sensitivity and discriminatory power, it is the most widely used diagnostic tool for mycobacterial infections, especially in resource-limited countries.[13] Classical identification of NTMs by culture-based biochemical tests has an improved sensitivity and discrimination; however, it is tedious, requires expertise, and the results are sometimes not reproducible.[14] The use of nucleic acid-based typing tools, such as 16S rRNA and hspt65 encoding a 65 kDa protein conserved in all Mycobacteria species with species-specific polymorphic loci have revolutionized the diagnosis of NTM.[15,16] The DNA-based assays are rapid, very reproducible, and highly discriminatory for differential diagnosis to advice on species specific therapy.[14]
We set out to use hsp65 gene sequencing to identify NTMs isolated from pulmonary TB cases from Ghana and to phenotypically test the identified NTMs against isoniazid (INH), rifampicin (RIF), and streptomycin (STR). Our findings suggest that NTMs infections may play a vital role in causing lung diseases, thereby impacting the management of TB in endemic settings such as Ghana.
2. Methodology
2.1. Recruitment of study participants
Sputum smear-positive patients reporting to selected health facilities in the Accra Metropolitan Authority of the Greater-Accra Region and in the northern region (Mamprusi East District and Tamale Metropolis) of Ghana between August 2012 and July 2014 were recruited for the study. The procedures used for sample collection were as routinely employed by the National Tuberculosis Program. The structured standard questionnaire was employed to gather patients’ demographic and clinical data. The questionnaire and protocols for this work were reviewed and approved by the Institutional Review Board of the Noguchi Memorial Institute for Medical Research with federal assurance number FWA00001824.
2.2. Cultivation of mycobacteria from sputa
All specimens were decontaminated using the 5% oxalic acid decontamination method,[13] inoculated on Lowenstein–Jensen medium supplemented with glycerol, incubated at 37°C, and observed for macroscopic growth until 12 weeks. The isolates obtained were suspended in nuclease-free water, deactivated at 98°C for 60 min and spun at 14,000 rpm to pellet cells for DNA extraction.
2.3. Isolation of genomic DNA
The protocol used for DNA extraction was a hybrid of the protocols as outlined by van Soolingen et al.[17] and Käser et al.[18] Briefly, the mycobacterial cell wall was disrupted by the addition of lysozyme solution (50 μL lysozyme of 10 mg/mL), vortexed, and incubated overnight. Next, 75 μL of 10% SDS and 10 μL of proteinase K (20 mg/mL) was added to the suspension, vortexed, and incubated at 65°C for 15 min. After incubation, 100 μL of 5M NaCl was added to the suspension, followed by CTAB/NaCl which was prewarmed at 65°C, and incubated at 65°C for 10 min. After vortexing, the extracted DNA was purified using the chloroform/isoamyl alcohol extraction method. The DNA contained in the upper phase was precipitated with isopropanol, washed with ethanol, dried, resuspended in 100 μl of Tris-EDTA buffer, and stored at 4°C until use.
4.2. Polymerase chain reaction detection of nontuberculous mycobacteria
All isolates obtained were typed as MTBC or NTM by polymerase chain reaction (PCR) amplification of the insertion sequence 6110 (IS6110) with MTBC-specific primers as previously described.[19] All IS6110-negative strains were suspected as NTMs and were subsequently used for further analysis.
2.5. Hsp65 gene amplification, DNA sequencing, and blast analysis
Oligonucleotide primers used for the mycobacterial hsp65 gene were Tb11 (5′-CAACGATGGTGTGTCCCAT-3′) and Tb12 (5′-CTTGTCGAACCGCATACCCT-3′). Briefly, the PCR reactions contained 3 μL of 10X buffer, 1.8 μL of 15 mM MgCl2, 3 μL of Q solution, 0.6 μL of 10 mM dNTP mix, 1.8 μL of each primer, 0.2 μL of hot-start Taq polymerase from Qiagen, 14.8 μL of nuclease-free water, and 3 μL of template DNA. Cycling conditions were (a) initial denaturation at 95°C for 15 min and 35 cycles of denaturation at 96°C for 1 min, (b) annealing at 60°C for 1 min, (c) extension at 68°C for 1 min, and (d) final extension at 72°C for 10 min. Amplification of the 441-bp product was confirmed by 1.5% agarose gel electrophoresis and UV transillumination. PCR products were purified and sequenced by outsourcing (Macrogen Europe). Vector sequences in the sequenced hsp65 genes were removed, the sequences were processed into gap experimental files with pregap4, edited with gap4 of the Staden package,[20] and the corresponding consensus sequence was saved in FASTA format for further analysis. The consensus hsp65 gene sequence of each isolate was analyzed by NCBI nucleotide blast against the microbial database of representative genomes only and selecting highly similar sequences (megablast), whereas leaving all other settings to default.
2.6. Drug susceptibility testing
Microplate Alamar blue assay (MABA) for STR, INH, and RIF drug susceptibility testing was performed in clear-bottomed, 96-well microplates (Nunc International, Rochester, NY, US). Outer perimeter wells were filled with sterile water to prevent dehydration in experimental wells. The drug concentrations used were prepared, and the assay was set up by following the standard method,[21,22] with few modifications as described by Otchere et al.[23] and reduced incubation time from 7 to 4 days. All manipulations were conducted in an appropriate, safe working area in the P3 laboratory of the Noguchi Memorial Institute for Medical Research.
2.7. Statistical analysis
The Fisher’s exact test was used to assess the associations between patient characteristics and species identified. All statistical analyses were performed in STATA 10.1 (Stata Corp., College Station, TX, USA)[24] with significant threshold set to P < 0.05 at 95% confidence interval (CI).
3.0. Results
3.1. Patients with nontuberculous mycobacteria infection
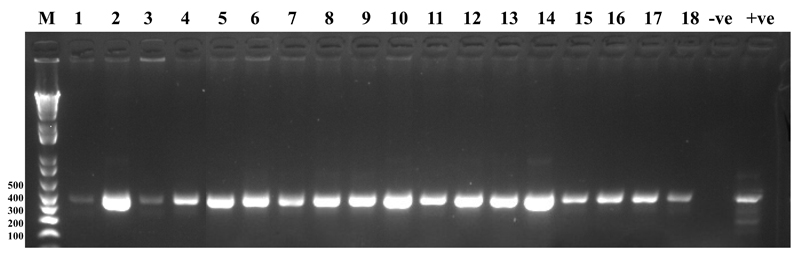
In total, 2036 smear-positive patients were recruited in the study. We excluded 278 from further analysis because no macroscopic cultures were obtained from their sputa after 12 weeks of observation; the remaining 1755 (86.2%) unique isolates were used for further analyses. The characteristics of the study population are summarized in Table 1. Out of 43 patients, comprising 27 (62.8%) male and 16 (37.2%) female patients of average age 39.4 (range, 10–75) years, 43 (2.5%) isolates were identified as NTMs after unsuccessful PCR amplification of IS6110, but positive hsp65 gene amplification [Figure 1]. Furthermore, 25 patients consented to HIV testing, and13 (52.0%) of them were seropositive. Univariate analysis comparing patients with HIV and NTMs coinfection (52%) and those with HIV and MTBC coinfection (5.8%) showed a statistical association of NTMs infection with HIV (P = 0.001, odd ratio [OR] =15.6, 95% CI = 7.7–32.4). Among 10 patients who consented to diabetic testing, only 1 (10%) was diabetic. Out of the 25 patients with information on Bacillus Calmette–Guerin, 16 (64%) had the scar.
Table 1. Biodata of Patients from Whom NTM were Isolated.
| Feature (Number of Patients) | Number (%) |
|---|---|
| Average Age (43) | 39.4 years |
| Gender (43) | |
| Male | 27(62.8) |
| Female | 16(37.2) |
| HIV available (25) | |
| Positive | 13(52.0) |
| Negative | 12(48.0) |
| HIV Not Available (18) | |
| Diabetes Available (10) | |
| Positive | 1(10.0) |
| Negative | 9(90.0) |
| Diabetes Not Available (33) | |
| BCG Scar Available (25) | |
| Present | 16(64.0) |
| Not Present | 9(36.0) |
| BCG Scar Not Available (18) |
Figure 1. Agarose Gel Picture of PCR Amplified hsp65 Gene of Suspected NTM Strains.
Lane M: DNA size marker, lane 1 to 18 are the 441 bp hsp65 gene amplified from candidate NTM strains. Lane -ve is the negative pcr control, Lane +ve is the positive control amplified from Mycobacterium tuberculosis H37Rv
3.2. Species identity of the isolated nontuberculous mycobacteria
The DNA sequencing hsp65 gene identified 13 (30.2%) Mycobacterium avium subs. paratuberculosis, 3 (7.0%) each of Mycobacterium colombiense and Mycobacterium mucogenicum, 18 (41.9%) Mycobacterium intracellulare, 5 (11.3%) Mycobacterium abscessus, and 1 (2.3%) Mycobacterium simiae [Figure 2]. No species-specific statistical association was found between the identified NTMs and any of the patient’s clinical data.
Figure 2. Distribution of Identified NTM.
3.3. Drug susceptibility profiling of nontuberculous mycobacteria against streptomycin, isoniazid, and rifampicin
All the 43 NTMs identified were screened for their susceptibility profiles against INH, STR, and RIF using MABA. All 43 (100%) identified NTMs were resistant to INH, 26 (60.5%) were resistant to all the 3 drugs tested, 11 (25.6%) were resistant to both INH and RIF, and 3 (6.9%) were resistant to RIF and STR in addition to INH resistance. Moreover, 3 (6.9%) of the isolates were susceptible to STR and RIF, 6 (13.9%) were susceptible to RIF irrespective of INH and STR resistance, and none was susceptible to INH [Table 2].
Table 2. Distribution of Drug Resistance Among the NTMs.
| Resistance | Number (Percentage) |
|---|---|
| INH + RIF + STR | 26 (60.5%) |
| INH + STR | 3 (6.9%) |
| RIF + INH | 11 (25.6%) |
Stratifying drug profile by species, all the 13 M. avium subs. paratuberculosis strains were resistant to INH and RIF irrespective of INH resistance, whereas 7 (53.8%) were additionally resistant to STR. All the 5 M. abscessus (100%) strains were resistant to all drugs, whereas 12 (66.7%), 18 (100%), and 16 (88.9%) of the M. intracellulare strains were resistant to STR, INH, and RIF, respectively. All the 3 M. colombiense strains were resistant to INH, whereas 2 (66.7%) and 1 (33.3%) were resistant to STR and RIF, respectively. While all the 3 M. mucogenicum strains were additionally resistant to STR, 2 (66.7%) out of the 3 were additionally resistant to RIF. Last, the single M. simiae strain was susceptible to STR and RIF, but resistant to INH [Table 3].
Table 3. Drug Susceptibility Profile of Identified NTM.
| Species (Number) | Resistance | ||
|---|---|---|---|
| STR | INH | RIF | |
| M. avium paratuberculosis (13) | 7 (53.8%) | 13 (100%) | 13 (100%) |
| M. columbiense (3) | 2 (66.7%) | 3 (100%) | 1(33.3%) |
| M. intracellulare (18) | 12 (66.7%) | 18 (100%) | 16 (88.9%) |
| M. abscessus (3) | 5 (100%) | 5 (100%) | 5 (100%) |
| M. mucogenicum (3) | 3 (100%) | 3 (100%) | 2 (66.7%) |
| M. simiae (1) | - | 1 (100%) | - |
| Total Number of isolates | 29 (67.4%) | 43 (100%) | 37 (86.0%) |
4.0. Discussion
Pulmonary infections caused by NTMs are often misdiagnosed as conventional TB caused by members of the MTBC because of the nonspecificity of sputum smear microscopy.[4–6] In addition, because of their intrinsic resistance to most first-line anti-TB drugs,[9,10] NTMs-associated TB may be normally diagnosed as TB and managed as MDR with its associated public health implications.[25–28] In Ghana, the prevalence and epidemiology of NTMs are largely unknown.[25] We sought to (1) determine the proportion and identity of NTMs among our prospective collection of mycobacteria isolates from smear-positive pulmonary TB cases reporting to selected health facilities in Ghana; (2) establish their susceptibility to the anti-TB drugs INH, RIF, and STR; and (3) analyzed for possible association of NTMs infection with patient characteristics. We found that (1) 2.5% of our mycobacterial isolates were NTMs and were dominated by M. intracellulare; (2) most of the identified NTMs were resistant to the two backbone drugs (INH and RIF) of the Directly Observed Treatment Short-course (DOTS) regimen; and (3) there was a significant association between NTMs-infected patients and HIV. Compared with the global incidence and a recent report on NTMs from Ghana, which identified 8% of NTMs among clinical mycobacteria from presumptive TB cases, our report of 2.5% is relatively small.[29–32] However, the previous study in Ghana involved only HIV-positive presumptive TB patients, which may explain the high rate of NTMs from that study. Nevertheless, for individual patient care and public health purposes, 2.5% is still high enough to reiterate the importance of rapid identification of the infecting species to facilitate effective treatment of pulmonary TB cases.
Our data set was dominated by M. intracellulare and M. avium subs. paratuberculosis [Figure 2], which constitute the M. avium complex (MAC). This finding compares with earlier reports of the dominance of MAC among pulmonary NTM infections.[1,29,32] This result is suggestive of the possible dominance of the MAC species in the environmental settings of the country. In addition, our results give more credit to the use of hsp65 gene sequence analysis for rapid identification of NTMs [Figure 2]. Compared with the earlier report from Ghana that could identify 17/38 (44.7%) of the NTMs using genotyping tools other than hsp65 gene sequencing,[32] we were able to identify all the isolated NTMs using the hsp65 gene sequencing. Our study, therefore, highlights the discriminatory power of the use of hsp65 gene sequencing for mycobacterial speciation.
Individuals that are immunocompromised due to HIV infection are at a risk of NTMs infection as evidenced by our study. We found a significant association between NTMs-infected patients and HIV (P = 0.001, OR = 15.6, 95% CI = 7.7–32.4). This finding supports earlier reports implicating the global HIV pandemic on the increasing global cases of NTMs-associated infections[33,34] and also compares with earlier works done in Tanzania and South Africa,[29,35] where HIV coinfection was implicated as a predictor of pulmonary NTMs infections. It is, therefore, imperative that TB treatment failures on the standard DOTS, especially among HIV-coinfected patients, are properly investigated to exclude the possibility of treating NTMs infection as an MDR-TB case.
NTMs pose a major challenge for TB treatment programs since such patients are managed mainly on the basis of smear microscopy,[9] which cannot determine their drug susceptibity profiles leading to their misdiagnoses as MDR-TB cases. Most of the NTMs (60.5%) identified were resistant to all the three drugs (INH, STR, and RIF), whereas 11 (25.6%) were resistant to the two most important drugs of the DOTS regimen (INH and RIF). None of the 43 isolates was sensitive to INH. This result is in agreement with the already established high resistance of NTMs to the drugs commonly used in the management of pulmonary TB cases[36,37] and reinforces the need to correctly identify the causative agent (to the species level) of such patients before they are put on an appropriate treatment regimen. Comparing independent resistance to the three drugs (43 INH, 29 STR, and 37 RIF) suggests that STR was the most active drug against our identified NTMs [Table 3] even though the proportion of identified NTMs resistant to STR (67.4%) was still high. Among the identified species, M. abscessus was the only species that had all strains resistant to all the three drugs [Table 3] and had the highest minimal inhibitory concentrations (S1). This finding supports earlier reports of resistance of M. abscessus to various antimicrobials, hence, making the treatment of M. abscessus infections very difficult.[9,38,39] This extreme resistance of M. abscessus has been partly attributed to the unique genetic constitution, probably due to horizontal gene transfer, a trait that is largely missing in the MTBC.[39–42]
5.0. Conclusion
Our results emphasize the importance of NTMs in pulmonary infections, which is dominated by the M. intracelluare complex. The high level of resistance of the identified species to INH and RIF calls for rapid identification and susceptibility testing of the causative agents of nonconverting TB patients or treatment failures for proper management.
Supplementary Material
Bibliography
- 1.Nunes-Costa D, Alarico S, Dalcolmo MP, Correia-Neves M, Empadinhas N. The looming tide of nontuberculous mycobacterial infections in Portugal and Brazil. Tuberculosis (Edinb) 2016;96:107–19. doi: 10.1016/j.tube.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 2.Falkinham JO., 3rd Environmental sources of nontuberculous mycobacteria. Clin Chest Med. 2015;36:35–41. doi: 10.1016/j.ccm.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Shinnick TM, Good RC. Mycobacterial taxonomy. Eur J Clin Microbiol Infect Dis. 1994;13:884–901. doi: 10.1007/BF02111489. [DOI] [PubMed] [Google Scholar]
- 4.Desikan P. Sputum smear microscopy in tuberculosis: Is it still relevant? Indian J Med Res. 2013;137:442–4. [PMC free article] [PubMed] [Google Scholar]
- 5.Steingart KR, Ng V, Henry M, Hopewell PC, Ramsay A, Cunningham J, et al. Sputum processing methods to improve the sensitivity of smear microscopy for tuberculosis: A systematic review. Lancet Infect Dis. 2006;6:664–74. doi: 10.1016/S1473-3099(06)70602-8. [DOI] [PubMed] [Google Scholar]
- 6.Perkins MD, Cunningham J. Facing the crisis: Improving the diagnosis of tuberculosis in the HIV era. J Infect Dis. 2007;196(Suppl 1):S15–27. doi: 10.1086/518656. [DOI] [PubMed] [Google Scholar]
- 7.Falkinham JO., 3rd Surrounded by mycobacteria: Nontuberculous mycobacteria in the human environment. J Appl Microbiol. 2009;107:356–67. doi: 10.1111/j.1365-2672.2009.04161.x. [DOI] [PubMed] [Google Scholar]
- 8.World Health Organization. Global Tuberculosis Report 2014. WHO/HTM/ TB/2014.08. Geneva, Switzerland: World Health Organization; 2014. [Google Scholar]
- 9.Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, et al. American Thoracic Society Documents An Official ATS/IDSA Statement: Diagnosis, Treatment, and Prevention of Nontuberculous Mycobacterial Diseases. 2007 doi: 10.1164/rccm.200604-571ST. [DOI] [PubMed] [Google Scholar]
- 10.Griffith DE. Nontuberculous mycobacterial lung disease. Curr Opin Infect Dis. 2010;23:185–90. doi: 10.1097/QCO.0b013e328336ead6. [DOI] [PubMed] [Google Scholar]
- 11.Lee MR, Sheng WH, Hung CC, Yu CJ, Lee LN, Hsueh PR. Mycobacterium abscessus complex infections in humans. Emerg Infect Dis. 2015;21:1638–46. doi: 10.3201/2109.141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffith DE. Mycobacterium abscessus subsp abscessus lung disease: ‘Trouble ahead, trouble behind…’. F1000Prime Rep. 2014;6:107. doi: 10.12703/P6-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yeboah-Manu D, Bodmer T, Mensah-Quainoo E, Owusu S, Ofori-Adjei D, Pluschke G. Evaluation of decontamination methods and growth media for primary isolation of Mycobacterium ulcerans from surgical specimens. J Clin Microbiol. 2004;42:5875–6. doi: 10.1128/JCM.42.12.5875-5876.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu TS, Lu CC, Lai HC. Current situations on identification of nontuberculous mycobacteria. J Biomed Lab Sci. 2009;21:1–6. [Google Scholar]
- 15.Saifi M, Jabbarzadeh E, Bahrmand AR, Karimi A, Pourazar S, Fateh A, et al. HSP65-PRA identification of non-tuberculosis mycobacteria from 4892 samples suspicious for mycobacterial infections. Clin Microbiol Infect. 2013;19:723–8. doi: 10.1111/j.1469-0691.2012.04005.x. [DOI] [PubMed] [Google Scholar]
- 16.Galagan JE. Genomic insights into tuberculosis. Nat Rev Genet. 2014;15:307–20. doi: 10.1038/nrg3664. [DOI] [PubMed] [Google Scholar]
- 17.van Soolingen D, Hermans PW, de Haas PE, Soll DR, van Embden JD. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: Evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol. 1991;29:2578–86. doi: 10.1128/jcm.29.11.2578-2586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Käser M, Ruf MT, Hauser J, Marsollier L, Pluschke G. Optimized method for preparation of DNA from pathogenic and environmental mycobacteria. Appl Environ Microbiol. 2009;75:414–8. doi: 10.1128/AEM.01358-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Asante-Poku A, Otchere ID, Danso E, Mensah DD, Bonsu F, Gagneux S, et al. Evaluation of GenoType MTBDRplus for the rapid detection of drug-resistant tuberculosis in Ghana. Int J Tuberc Lung Dis. 2015;19:954–9. doi: 10.5588/ijtld.14.0864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Staden R, Beal KF, Bonfield JK. The Staden package, 1998. Methods Mol Biol. 2000;132:115–30. doi: 10.1385/1-59259-192-2:115. [DOI] [PubMed] [Google Scholar]
- 21.Palomino JC, Martin A, Camacho M, Guerra H, Swings J, Portaels F. Resazurin microtiter assay plate: Simple and inexpensive method for detection of drug resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2002;46:2720–2. doi: 10.1128/AAC.46.8.2720-2722.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Franzblau SG, Witzig RS, McLaughlin JC, Torres P, Madico G, Hernandez A, et al. Rapid, low-technology MIC determination with clinical Mycobacterium tuberculosis isolates by using the microplate Alamar Blue assay. J Clin Microbiol. 1998;36:362–6. doi: 10.1128/jcm.36.2.362-366.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otchere ID, Asante-Poku A, Osei-Wusu S, Baddoo A, Sarpong E, Ganiyu AH, et al. Detection and characterization of drug-resistant conferring genes in Mycobacterium tuberculosis complex strains: A prospective study in two distant regions of Ghana. Tuberculosis (Edinb) 2016;99:147–54. doi: 10.1016/j.tube.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomz M, Wittenberg J, King G. Clarify: Software for interpreting and presenting statistical results. J Stat. 2003;8:1–28. [Google Scholar]
- 25.Couto I, Machado D, Viveiros M, Rodrigues L, Amaral L. Identification of nontuberculous mycobacteria in clinical samples using molecular methods: A 3-year study. Clin Microbiol Infect. 2010;16:1161–4. doi: 10.1111/j.1469-0691.2009.03076.x. [DOI] [PubMed] [Google Scholar]
- 26.Adjemian J, Olivier KN, Prevots DR. Nontuberculous mycobacteria among patients with cystic fibrosis in the United States: Screening practices and environmental risk. Am J Respir Crit Care Med. 2014;190:581–6. doi: 10.1164/rccm.201405-0884OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fusco da Costa AR, Lopes ML, de Sousa MS, Suffys PN, Sales LHM, Lima KVB. In: Pulmonary Nontuberculous Mycobacterial Infections in the State of Para, an Endemic Region for Tuberculosis in North of Brazil, Pulmonary Infection. Amer Amal., editor. InTech; 2012. [Last accessed on 27-May-2016]. ISBN: 978-953-51-0286-1, Available from: http://www.intechopen.com/books/pulmonaryinfection/pulmonary-nontuberculous-mycobacterial-infections-in-the-state-of-para-an-endemic-region-fortuberculosis. [Google Scholar]
- 28.da Costa AR, Lopes ML, Leão SC, Schneider MP, de Sousa MS, Suffys PN, et al. Molecular identification of rapidly growing mycobacteria isolates from pulmonary specimens of patients in the State of Pará, Amazon region, Brazil. Diagn Microbiol Infect Dis. 2009;65:358–64. doi: 10.1016/j.diagmicrobio.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 29.Hoza AS, Mfinanga SG, Rodloff AC, Moser I, König B. Increased isolation of nontuberculous mycobacteria among TB suspects in Northeastern, Tanzania: Public health and diagnostic implications for control programmes. BMC Res Notes. 2016;9:109. doi: 10.1186/s13104-016-1928-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zamarioli LA, Coelho AG, Pereira CM, Ferrazoli L, Bammann RH. Laboratory identification of mycobacteria in respiratory samples from HIV-positive patients suspected of tuberculosis. Rev Soc Bras Med Trop. 2009;42:290–7. doi: 10.1590/s0037-86822009000300010. [DOI] [PubMed] [Google Scholar]
- 31.Lima CA, Gomes HM, Oelemann MA, Ramos JP, Caldas PC, Campos CE, et al. Nontuberculous mycobacteria in respiratory samples from patients with pulmonary tuberculosis in the state of Rondônia, Brazil. Mem Inst Oswaldo Cruz. 2013;108:457–62. doi: 10.1590/0074-0276108042013010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bjerrum S, Oliver-Commey J, Kenu E, Lartey M, Newman MJ, Addo KK, et al. Tuberculosis and non-tuberculous mycobacteria among HIV-infected individuals in Ghana. Trop Med Int Health. 2016;21:1181–90. doi: 10.1111/tmi.12749. [DOI] [PubMed] [Google Scholar]
- 33.Ferreira RM, Saad MH, Silva MG, Fonseca Lde S. Non-tuberculous mycobacteria I: One year clinical isolates identification in Tertiary Hospital Aids Reference Center, Rio de Janeiro, Brazil, in pre highly active antiretroviral therapy era. Mem Inst Oswaldo Cruz. 2002;97:725–9. doi: 10.1590/s0074-02762002000500024. [DOI] [PubMed] [Google Scholar]
- 34.Senna SG, Marsico AG, Vieira GB, Sobral LF, Suffys PN, Fonseca Lde S. Identification of nontuberculous mycobacteria isolated from clinical sterile sites in patients at a university hospital in the city of Rio de Janeiro, Brazil. J Bras Pneumol. 2011;37:521–6. doi: 10.1590/s1806-37132011000400015. [DOI] [PubMed] [Google Scholar]
- 35.Sookan L, Coovadia YM. A laboratory-based study to identify and speciate non-tuberculous mycobacteria isolated from specimens submitted to a central tuberculosis laboratory from throughout KwaZulu-Natal Province, South Africa. S Afr Med J. 2014;104:766–8. doi: 10.7196/samj.8017. [DOI] [PubMed] [Google Scholar]
- 36.World Health Organization. Global Tuberculosis Report. Geneva, Switzerland: World Health Organization; 2013. p. 306. [Google Scholar]
- 37.Viveiros M, Leandro C, Amaral L. Mycobacterial efflux pumps and chemotherapeutic implications. Int J Antimicrob Agents. 2003;22:274–8. doi: 10.1016/s0924-8579(03)00208-5. [DOI] [PubMed] [Google Scholar]
- 38.Cândido PH, Nunes Lde S, Marques EA, Folescu TW, Coelho FS, de Moura VC, et al. Multidrug-resistant nontuberculous mycobacteria isolated from cystic fibrosis patients. J Clin Microbiol. 2014;52:2990–7. doi: 10.1128/JCM.00549-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nessar R, Cambau E, Reyrat JM, Murray A, Gicquel B. Mycobacterium abscessus: A new antibiotic nightmare. J Antimicrob Chemother. 2012;67:810–8. doi: 10.1093/jac/dkr578. [DOI] [PubMed] [Google Scholar]
- 40.Nobre A, Alarico S, Maranha A, Mendes V, Empadinhas N. The molecular biology of mycobacterial trehalose in the quest for advanced tuberculosis therapies. Microbiology. 2014;160(Pt 8):1547–70. doi: 10.1099/mic.0.075895-0. [DOI] [PubMed] [Google Scholar]
- 41.Shallom SJ, Gardina PJ, Myers TG, Sebastian Y, Conville P, Calhoun LB, et al. New rapid scheme for distinguishing the subspecies of the Mycobacterium abscessus group and identifying Mycobacterium massiliense isolates with inducible clarithromycin resistance. lin Microbiol. 2013;51:2943–9. doi: 10.1128/JCM.01132-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Macheras E, Roux AL, Bastian S, Leão SC, Palaci M, Sivadon Tardy V, et al. Multilocus sequence analysis and rpoB sequencing of Mycobacterium abscessus (sensu lato) strains. J Clin Microbiol. 2011;49:491–9. doi: 10.1128/JCM.01274-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.