Abstract
Tannerella forsythia is an anaerobic, Gram-negative periodontal pathogen. A unique O-linked oligosaccharide decorates the bacterium's cell surface proteins and was shown to modulate the host immune response. In our study, we investigated the biosynthesis of the nonulosonic acid (NulO) present at the terminal position of this glycan. A bioinformatic analysis of T. forsythia genomes revealed a gene locus for the synthesis of pseudaminic acid (Pse) in the type strain ATCC 43037 while strains FDC 92A2 and UB4 possess a locus for the synthesis of legionaminic acid (Leg) instead. In contrast to the NulO in ATCC 43037, which has been previously identified as a Pse derivative (5-N-acetimidoyl-7-N-glyceroyl-3,5,7,9-tetradeoxy-l-glycero-l-manno-NulO), glycan analysis of strain UB4 performed in this study indicated a 350-Da, possibly N-glycolyl Leg (3,5,7,9-tetradeoxy-d-glycero-d-galacto-NulO) derivative with unknown C5,7 N-acyl moieties. We have expressed, purified and characterized enzymes of both NulO pathways to confirm these genes’ functions. Using capillary electrophoresis (CE), CE–mass spectrometry and NMR spectroscopy, our studies revealed that Pse biosynthesis in ATCC 43037 essentially follows the UDP-sugar route described in Helicobacter pylori, while the pathway in strain FDC 92A2 corresponds to Leg biosynthesis in Campylobacter jejuni involving GDP-sugar intermediates. To demonstrate that the NulO biosynthesis enzymes are functional in vivo, we created knockout mutants resulting in glycans lacking the respective NulO. Compared to the wild-type strains, the mutants exhibited significantly reduced biofilm formation on mucin-coated surfaces, suggestive of their involvement in host-pathogen interactions or host survival. This study contributes to understanding possible biological roles of bacterial NulOs.
Keywords: bacterium, biofilm, biosynthesis pathway, pseudaminic and legionaminic acid
Introduction
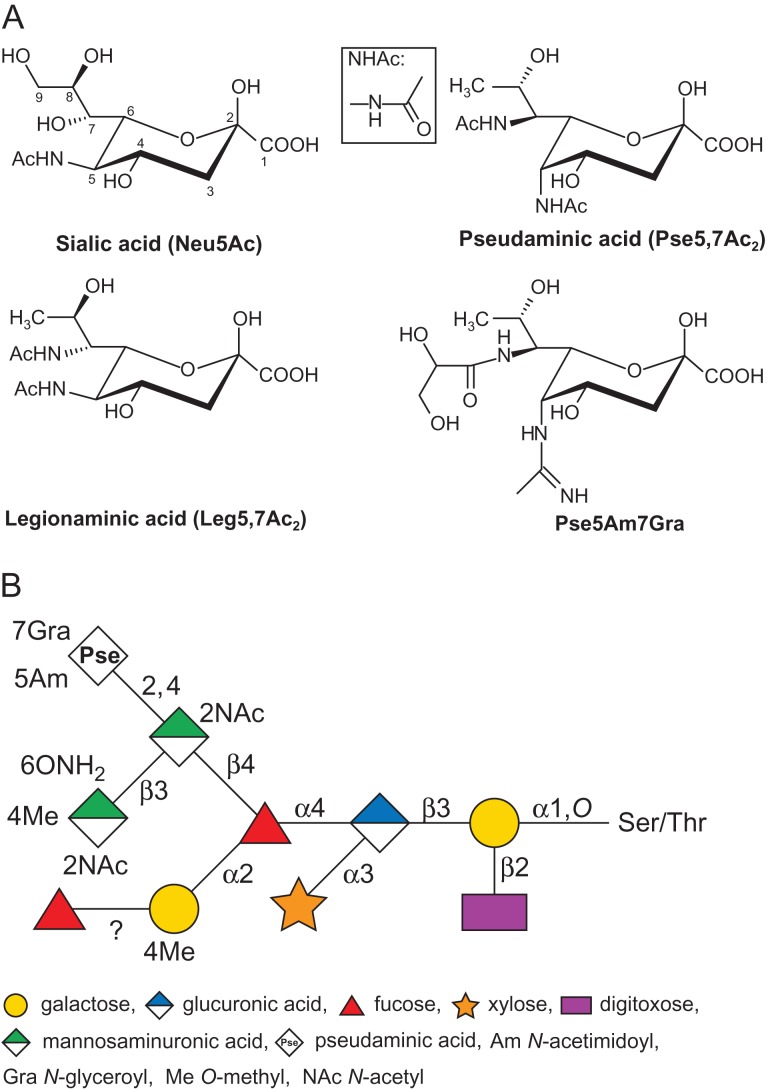
Nonulosonic acids (NulOs) are a class of 9-carbon sugars that are α-keto acids. The most abundant naturally occurring NulOs are the sialic acids (Sia or neuraminic acids, Neu) and their derivatives (Varki 1992; Traving and Schauer 1998). It is established knowledge that Sias play crucial roles in the life-cycles of various organisms from eukaryotes to prokaryotes (Varki et al. 2009). While Sias are commonly present as terminal residues of many eukaryotic glycoconjugates and have also been identified in several prokaryotic polysaccharides, a number of NulOs appear to be unique to bacterial species (Knirel et al. 2003; Morrison and Imperiali 2014; Zunk and Kiefel 2014; Kenyon et al. 2015; Schäffer and Messner 2016). Due to their structural and biosynthetic similarities to Sia, these NulOs are also often referred to as Sia-like sugars (Figure 1A), the most studied of which are pseudaminic acids (such as 5,7-diacetamido-3,5,7,9-tetradeoxy-l-glycero-l-manno-NulO, Pse5,7Ac2) and legionaminic acids (such as 5,7-diacetamido-3,5,7,9-tetradeoxy-d-glycero-d-galacto-NulO, Leg5,7Ac2). For both pseudaminic acid (Pse) and legionaminic acid (Leg), several naturally occurring derivatives have been identified. These most commonly include substitutions of the N-acyl groups at the C-5 and C-7 positions, e.g., with N-acetyl or acetamido (Ac), N-acetimidoyl or acetamidino (Am), N-formyl (Fo) and N-hydroxybutyryl (Hb) groups (Zunk and Kiefel 2014).
Fig. 1.
(A) Structures of Sia and Sia-like sugars (NulOs). Sialic acid (Neu5Ac), legionaminic acid (Leg5,7Ac2) and pseudaminic acids (Pse5,7Ac2 and Pse5Am7Gra) are shown. To note, Pse5Am7Gra is found as the terminal sugar of the S-layer glycan in T. forsythia ATCC 43037. For reference, the nine carbon atoms of Sia are numbered, and the structure of the NHAc group is shown in the boxed inset. (B) Schematic drawing of the structure of the S-layer O-glycan in T. forsythia ATCC 43037 (amended from Posch et al. (2011)). This figure is available in black and white in print and in color at Glycobiology online.
Pse was first identified in the O-polysaccharide chain of the lipopolysaccharide (LPS) from Pseudomonas aeruginosa and Shigella boydii (Knirel et al. 1984). Since then, Pse and derivatives have been frequently found as constituents of cell surface glycoconjugates such as pili (Horzempa et al. 2006), flagella (Thibault et al. 2001; Schirm et al. 2003), LPS (Knirel et al. 2003) and capsular polysaccharides (Kiss et al. 2001; Kenyon et al. 2014). In Campylobacter jejuni, Helicobacter pylori and Aeromonas spp., heavy glycosylation of the flagellin protein with Pse has been shown to be an essential requirement for flagellar assembly and, thus, motility and virulence of these pathogens (Guerry et al. 2006; Wilhelms et al. 2012; Lowry et al. 2014).
Leg—a C5,7,8 isomer of Pse (Figure 1A)—was first characterized in a repeating homopolymer of the LPS O-antigen from Legionella pneumophila, the causative agent of Legionnaires’ disease (Knirel et al. 1994). Since then, Leg and its derivatives have also been found in the LPS of various other Gram-negative bacteria such as Vibrio salmonicida (Edebrink et al. 1996), Pseudomonas fluorescens (Knirel et al. 1996), Acinetobacter baumannii (Haseley and Wilkinson 1997), Cronobacter turicensis (MacLean et al. 2011), Salmonella arizonae (Vinogradov et al. 1992) and certain serotypes of Escherichia coli (Li et al. 2010). Leg has also been found as a major component in the O-linked glycosylation of flagellins in Campylobacter spp. (McNally et al. 2007; Schoenhofen et al. 2009) and Clostridium botulinum (Twine et al. 2008), the latter marking the first occurrence of a Sia-like sugar in the glycosylation of a Gram-positive flagellar protein. While modification of C. jejuni flagellins with a NulO is required for flagellin assembly and motility, the specific NulO present may impart particular biological consequences, such as seen in a strain lacking the ability to display a 5-N-acetimidoyl Leg derivative (Leg5Am7Ac). This strain exhibited decreased cell hydrophobicity, reduced biofilm formation, and reduced ability to colonize the chicken intestine (Howard et al. 2009).
Apart from some evidence for involvement in bacterial fitness and pathogenicity of distinct organisms, the function of Sia-like sugars as part of cell surface components is still largely unknown. It was suggested that bacterial NulOs could play a role as molecular mimic of eukaryotic Sia, thereby contributing to the pathogen's ability to evade or modulate the host's immune responses (Carlin et al. 2009). A recent study proposes that Pse derivatives on the flagella of C. jejuni interact with host Siglec-10 to increase IL-10 expression, thus promoting an anti-inflammatory response and host persistence (Stephenson et al. 2014). In contrast to the scarcity of evidence for their function, an increasing number of studies suggests that Leg and Pse derivatives are widespread among prokaryotes, where more than 20% of 1000 microbial genomes examined were found to encode a predicted NulO biosynthesis pathway (Lewis et al. 2009).
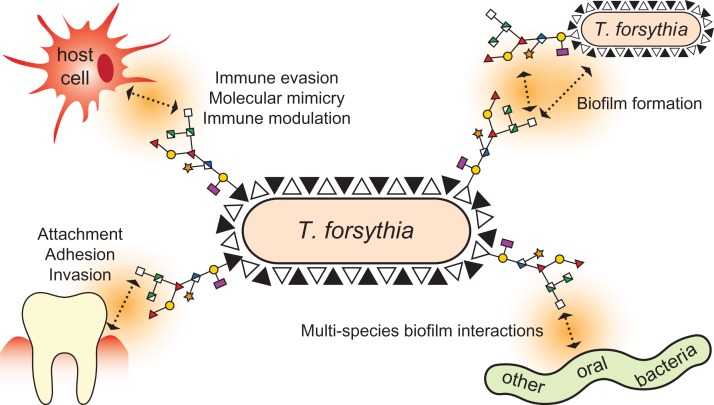
In our group, the ATCC 43037 type strain of the periodontal pathogen Tannerella forsythia was found to display a Pse derivative as the terminal residue of its abundant cell surface (S-) layer O-glycans (Figure 1B) (Posch et al. 2011). T. forsythia is a Gram-negative anaerobe that inhabits subgingival plaque biofilms in the human oral cavity (Tanner et al. 1986). Together with Porphyromonas gingivalis and Treponema denticola it forms the so-called “red complex” consortium of oral pathogens, which is strongly associated with periodontal diseases in humans (Holt and Ebersole 2005). Periodontitis is a multifactorial inflammatory disease which progresses as a result of the direct effects of bacterial virulence factors on host tissues, as well as self-damaging host responses to the colonizing bacteria (Socransky et al. 1998; Hajishengallis and Lamont 2012). Apart from being the most common cause of tooth loss worldwide and affecting millions of people (Darveau 2010), periodontitis is increasingly linked to systemic diseases such as cardiovascular disease (Leishman et al. 2010). Among the identified virulence factors of T. forsythia is its S-layer (Lee et al. 2006; Sakakibara et al. 2007; Sharma 2010). S-Layers are found as the outermost cell envelope component of many bacteria and archaea (Sára and Sleytr 2000); the constituting (glyco)proteins are predominantly water-insoluble and endowed with the intrinsic capability of self-assembling into 2D crystalline arrays (Schäffer and Messner 2004). Apart from their role as virulence factors, S-layers can have many functions in bacterial growth and survival, including the maintenance of cell integrity, enzyme display and, in pathogens and commensals, interaction with the host and its immune system (Fagan and Fairweather 2014).
Compared to other S-layer-carrying bacteria, T. forsythia's status is unique in two ways: it is the only Gram-negative bacterium that is known to possess a glycosylated S-layer and its S-layer is comprised of two S-layer proteins (named TfsA and TfsB) instead of just one (Sekot et al. 2012). After the T. forsythia S-layer had been found to delay the host immune response, at least at the early stage of infection, (Sekot et al. 2011), a detailed investigation of the bacterium's cell surface architecture (Sekot et al. 2013) and glycosylation ensued (Posch et al. 2011). As previously mentioned, the S-layer of the ATCC 43037 type strain was shown to be modified with an O-linked oligosaccharide containing several rare sugar residues (Figure 1B), including a Pse derivative with an N-acetimidoyl (Am) group at C-5 and an N-glyceroyl (Gra) group at C-7 (Pse5Am7Gra; Figure 1A; (Sekot et al. 2011)). The original publication by our group correctly identified the C-7 modification by NMR spectroscopy but erroneously described it as an “N-glycolyl (Gc)” group. The correct term for this modification is “N-glyceroyl”, with “Gra” as the proposed abbreviation, although the designation “N-2,3-dihydroxypropionyl”, “glycerate group” or “glyceric acid” is also in use for this substitution (Thibault et al. 2001; Vinogradov et al. 2009). Interestingly, the S-layer glycan is also present on several other proteins, residing mainly in the outer membrane, indicative of the presence of a general protein O-glycosylation system in T. forsythia that gives rise to a rich glycoproteome. Considering the prominent location of the modified Pse5Am7Gra as a terminal non-reducing residue of the outermost cell surface decoration of T. forsythia, it is likely to participate in bacterium-host interactions and impact the bacterium's life-style. First indications of this role can be inferred from immunologic studies showing that the O-glycan's terminal trisaccharide branch containing the Pse5Am7Gra residue dampens T-helper 17 differentiation, mitigates neutrophil infiltration to the gingival tissue, and increases periodontal bone loss in mice (Settem et al. 2013).
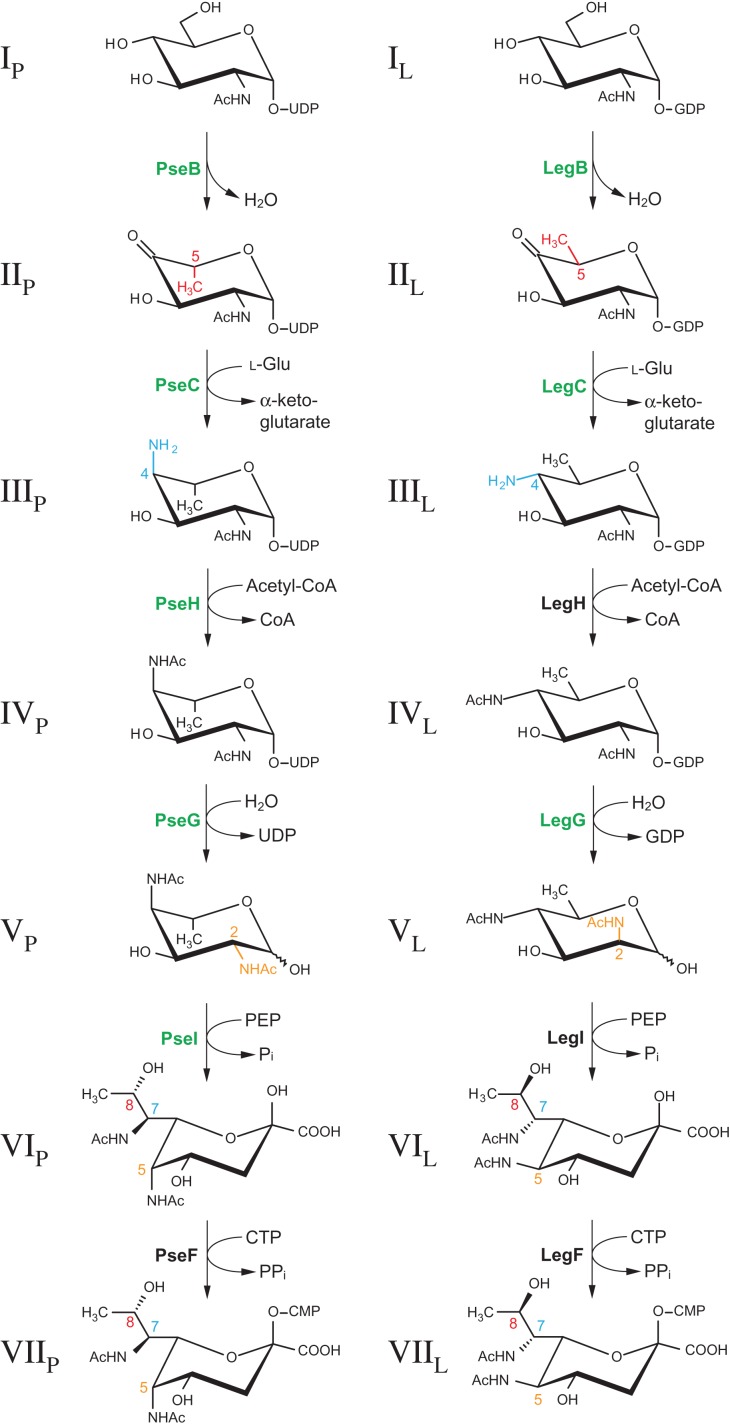
In light of these findings, we launched an investigation into the NulO biosynthetic pathway of T. forsythia, which relied primarily on knowledge of the Pse and Leg biosynthesis pathways in H. pylori (Schoenhofen et al. 2006b) and C. jejuni (Schoenhofen et al. 2009), respectively. In both cases, CMP-NulO, representing the activated form of NulO required for its incorporation into glycoconjugates, is formed by the subsequent action of six enzymes starting from nucleotide-activated N-acetylglucosamine. While the pathway to Pse utilizes UDP-GlcNAc as a precursor, Leg biosynthesis in C. jejuni was shown to involve GDP-GlcNAc. Apart from using different sugar nucleotides, the biosynthetic routes for Pse and Leg differ in the reactions performed at the C-2, C-4 and C-5 positions of the hexose intermediates, which results in stereochemical differences at C-5, C-7 and C-8 positions in the final NulO (Figure 2). In the first enzymatic step of the Leg pathway, the dehydratase LegB catalyzes a C-4,6 dehydration, while the corresponding enzyme of the Pse pathway, PseB, additionally performs a 5-epimerization reaction. The subsequent amino transfer at C-4 introduces another stereochemical distinction as an axial (PseC) vs. equatorial (LegC) addition of the amino group is performed. After N-4 acetylation (PseH/LegH), NDP removal catalyzed by LegG results in C-2 epimerization, whereas the corresponding enzyme of the Pse biosynthesis pathway, PseG, only possesses NDP-sugar hydrolase activity. In both NulO routes, the formation of the 9-carbon-backbone sugar is catalyzed by homologous enzymes (PseI/LegI) via the condensation of a 6-carbon hexose intermediate with the 3-carbon molecule pyruvate. The final enzymatic step, accomplished by PseF/LegF, CMP-activates the free NulO residue using cytidine triphosphate.
Fig. 2.
Experimentally confirmed enzymatic steps (in green) in T. forsythia strains ATCC 43037 (left) and FDC 92A2/UB4 (right) corresponding to the CMP-Pse and CMP-Leg biosynthetic pathways as elucidated in H. pylori and C. jejuni, respectively (Schoenhofen et al. 2006b, Schoenhofen et al. 2009). In T. forsythia, pathways will deviate at some point to produce the unique NulO derivatives found in our strains, such as Pse5Am7Gra. These deviations would be anticipated to occur either within the NulO biosynthetic pathway or post CMP-NulO biosynthesis. Red, blue and orange highlight enzymatic steps that introduce the stereochemical differences between the two pathways and also indicate the positions altered for both hexose intermediates and final NulO. The assignment of roman numerals to each compound is consistent with label designations found throughout the text. Subscripts “P” and “L” indicate intermediates from the CMP-Pse and CMP-Leg biosynthesis pathway, respectively. For simplicity, all sugars except for the NulOs are shown in 4C1 form. This figure is available in black and white in print and in color at Glycobiology online.
A previous bioinformatic analysis of the T. forsythia ATCC 43037 type strain genome revealed the presence of a protein O-glycosylation gene cluster encoding genes involved in sugar biosynthesis, glycan assembly and transport (Posch et al. 2011). The identification of a dedicated NulO biosynthesis gene locus of T. forsythia—as can be found in H. pylori (Schoenhofen et al. 2006b) and C. jejuni (Schoenhofen et al. 2009)—was complicated by the fact that the publicly available genome thought to match the ATTC 43037 type strain, was in fact derived from T. forsythia strain FDC 92A2 (or ATCC BAA-2717). After the discovery of this strain misattribution, we obtained the correct genome sequence of the type strain (Friedrich et al. 2015). In this current work, a bioinformatic analysis of both genomes as well as that of a new T. forsythia isolate, strain UB4, predicted a gene locus for Pse biosynthesis in the type strain ATCC 43037, while the genes in strain FDC 92A2 and UB4 pointed towards a Leg biosynthetic route. To confirm the function of the predicted NulO enzymes encoded by these loci and hence the resulting NulO type present in each organism, we have biochemically characterized five enzymes for Pse synthesis (homologs of PseB, PseC, PseH, PseG and PseI) in the type strain and the three stereochemically defining enzymes (homologs of LegB, LegC and LegG) of strains FDC 92A2/UB4. By deleting one of the NulO biosynthetic genes in either pathway, we created knockout mutants displaying O-glycans deficient in their respective terminal NulO, thus demonstrating that the NulO biosynthesis genes are indeed transcribed and the enzymes are functional in vivo. As a first indication of a biological function, we assessed biofilm formation in wild-type and NulO-deficient mutants of T. forsythia. Our findings provide a basis for determining the roles of Pse and Leg in the pathogenicity of T. forsythia and may contribute to understanding the influence of NulOs on the physiology and life-style of this bacterium.
Results
Bioinformatic analysis of NulO biosynthetic gene loci in three T. forsythia strains
The presence of terminal Pse5Am7Gra on the S-layer glycan of T. forsythia ATCC 43037 type strain prompted an investigation of the genes involved in the biosynthesis of this NulO. For a comparative genomic analysis, we selected enzymes of the flagellin O-linked glycosylation systems of C. jejuni 11168 and H. pylori. These organisms possess well-characterized biosynthetic pathways to Pse, as well as Leg and Neu in the former (Schoenhofen et al. 2006b; Schoenhofen et al. 2009).
An investigation of the T. forsythia ATCC 43037 genome (JUET00000000) recently obtained by our laboratory (Friedrich et al. 2015) revealed the presence of candidate genes for all six dedicated Pse biosynthesis genes (Supplementary Figure 1). They cluster in a locus downstream of a proposed wzx flippase gene (Tanf_01180) and upstream of the predicted T. forsythia protein O-glycosylation gene locus (Posch et al. 2011). All NulO genes showed much higher similarity to the C. jejuni/H. pylori Pse biosynthetic genes than to the Leg counterparts of C. jejuni (Table I). The presence of a homolog to PseB (Tanf_01185), in particular, was a strong indication towards Pse, as this enzyme is unique among other dehydratases in its ability to additionally perform a C-5 epimerization (McNally et al. 2008).
Table I.
Bioinformatic analysis of candidate genes for CMP-Pse and CMP-Leg biosynthesis in T. forsythia
| Strain locus tag | Best BLASTP match | Identity (%) | Bitscore | Known activity | Comments and references |
|---|---|---|---|---|---|
| ATCC 43037 | |||||
| Tanf_01185 | H. pylori pseB (HP0840) | 56 | 355 | 4,6-Dehydratase/5-epimerase | McNally et al. (2008), Schoenhofen et al. (2006c) |
| Tanf_01190 | C. jejuni pseC (Cj1294) | 35 | 201 | Aminotransferase | Schoenhofen et al. (2006a, 2006c) |
| Tanf_01215a | C. jejuni pseH (Cj1313) | 34 | 60 | N-acetyltransferase | Schoenhofen et al. (2006b) |
| Tanf_01235a | C. jejuni pseG (Cj1312) | 27 | 70 | UDP-sugar hydrolase | Contains PseG consensus sequence (Rangarajan et al. 2009, Schoenhofen et al. 2006b) |
| Tanf_01240 | H. pylori pseI (HP0178) | 49 | 332 | Pseudaminic acid synthase | Chou et al. (2005) |
| Tanf_01200 | C. jejuni pseF (Cj1311) | 37 | 37 | CMP-pseudaminic acid synthetase | Schoenhofen et al. (2006b) |
| 92A2 / UB4 | |||||
| BFO_1074 | C. jejuni legB (Cj1319) | 62 | 408 | 4,6-Dehydratase | Schoenhofen et al. (2009) |
| BFO_1073 | C. jejuni legC (Cj1320) | 49 | 385 | Aminotransferase | Schoenhofen et al. (2009) |
| BFO_1067 a | C. jejuni pglD (Cj1123c) | 30 | 83 | N-acetyltransferase | Olivier et al. (2006), Schoenhofen et al. (2009) |
| BFO_1065 | C. jejuni legG (Cj1328) | 46 | 350 | NDP-sugar hydrolase/2-epimerase | Member of NDP-N-acetylglucosamine 2-epimerase family (PFAM) (Schoenhofen et al. 2009) |
| BFO_1066 | C. jejuni legI (Cj1327) | 56 | 391 | Legionaminic acid synthase | Schoenhofen et al. (2009) |
| BFO_1063 | C. jejuni legF (Cj1331) | 32 | 102 | CMP-legionaminc acid synthetase | Schoenhofen et al. (2009) |
| BFO_1064 | C. jejuni ptmE (Cj1329) | 29 | 162 | GlcN-1-P guanylyltransferase | Specific for GTP (Schoenhofen et al. 2009) |
aEarlier start codon than in NCBI annotation.
A homology search of C. jejuni NulO biosynthesis amino acid sequences against the genomes of strains FDC 92A2 (Genbank CP003191) and UB4 (Stafford et al. in preparation) revealed major differences with respect to the genome of strain ATCC 43037. The putative flippase gene (Tanf_01180/BFO_1075) is identical in FDC 92A2, UB4 and the type strain along almost its entire length, however, alignment breaks down over the last 30 base pairs (Supplementary Figure 1). Downstream of this ORF is a locus of NulO biosynthetic genes that is identical in FDC 92A2 and UB4 but exhibits no significant similarity to the putative Pse biosynthesis locus found in strain ATCC 43037, regarding both the arrangement and types of genes. The first gene that shows homology between the strains again is an ORF at the 5′-end of the locus which encodes a hypothetical protein (BFO_1060), the C-terminal half of which is identical to the corresponding gene in the type strain (Tanf_01245). A closer inspection of the NulO biosynthesis genes found in this genomic locus suggested that strains FDC 92A2 and UB4 possess the biosynthetic route towards Leg (Table I). Firstly, for the dehydratase (LegB/PseB), aminotransferase (LegC/PseC), NulO synthase (LegI/PseI) and CMP-activating synthetase (LegF/PseF), strains FDC 92A2/UB4 genes display higher similarity to the Leg biosynthetic enzymes of C. jejuni than to those of the Pse pathway. Secondly, no PseG or UDP-sugar hydrolase consensus sequence could be identified, while there is a good candidate for the hydrolyzing 2-epimerase LegG. This strongly supported Leg synthesis as 2-epimerization does not occur in the Pse pathway. Lastly, the presence of a homolog to the nucleotidyl transferase PtmE points towards a biosynthetic pathway utilizing a GDP-linked intermediate. With respect to activator NTP donors, PtmE has been shown to be absolutely specific for GTP in reactions involving the abundant hexosamine metabolite GlcN-1-P, converting it to GDP-GlcN where GDP-GlcNAc is the LegB substrate in C. jejuni (Schoenhofen et al. 2009). The Pse pathway, on the other hand, would likely require UDP-GlcNAc as a precursor. Even though there are no good matches for the N-acetyltransferase LegH, the gene BFO_1067 encodes an amino acid sequence that is similar to PglD, an N-acetyltransferase involved in UDP-2,4-diacetamido-bacillosamine biosynthesis in C. jejuni (Olivier et al. 2006). PglD has been shown to accept both GDP- and UDP-linked substrates, and as there is no evidence of bacillosamine occurring in T. forsythia, we propose that BFO_1067 is a dedicated part of the Leg biosynthesis pathway (i.e., LegH).
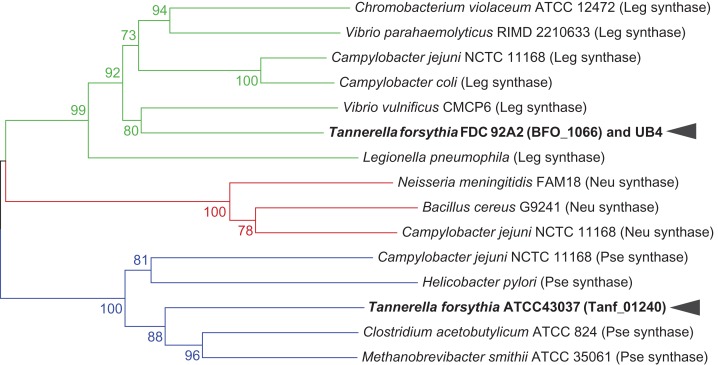
A phylogenetic comparison of NulO biosynthetic enzyme sequences by Lewis et al. (2009) indicated that NulO synthase sequences are better conserved than other NulO biosynthetic enzymes and, therefore, form the most conclusive basis for prediction of specific NulO sugar types in different organisms. Including T. forsythia sequences in this analysis, Tanf_01240 of the type strain ATCC 43037 is clustered in the phylogenetic clade of PseI enzymes, whereas BFO_1066 of strains FDC 92A2/UB4 shares ancestry with LegI sequences (Figure 3).
Fig. 3.
Phylogenetic clusters of selected microbial NulO synthase homologs. Based on the prediction of NulO sugar type by Lewis et al. (2009), a distance-based neighbor joining tree calculated from the amino acid sequences of NulO synthase enzymes places Tanf_01240 of T. forsythia type strain ATCC 43037 in a phylogenetic clade of pseudaminic acid synthases, while BFO_1066 of strain FDC 92A2/UB4 is clustered with Leg synthases (bootstrap values shown at relevant nodes). Green denotes Leg synthases, red Neu synthases and blue Pse synthases. This figure is available in black and white in print and in color at Glycobiology online.
Biochemical characterization of a CMP-Pse biosynthetic locus in T. forsythia ATCC 43037
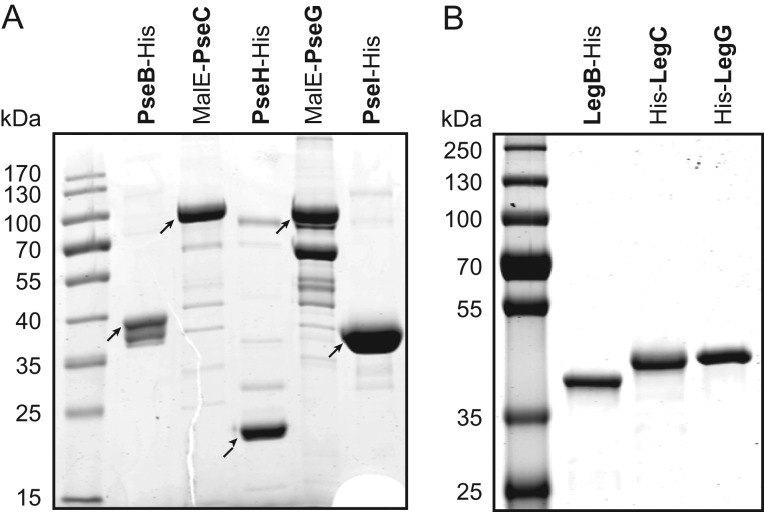
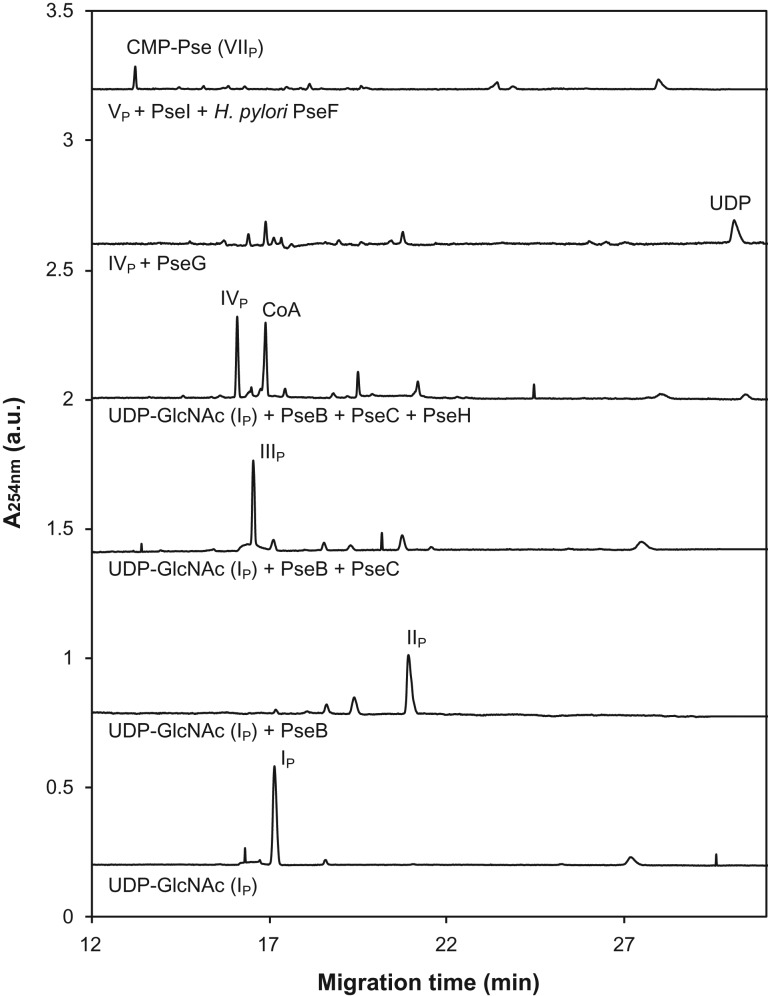
To confirm the role of the predicted Pse biosynthesis enzymes in the ATCC 43037 type strain, we recombinantly produced and characterized these proteins. PseB, PseH and PseI were expressed as His6-tagged proteins in E. coli and purified by nickel-affinity chromatography (Figure 4A). In all three cases, C-terminal tags resulted in better protein expression and solubility than N-terminal tags. For PseC and PseG, a maltose-binding protein (MBP) fusion tag followed by purification over an amylose affinity column was used since, in these cases, the His6-tagged constructs were found to be inactive. Subsequently, the enzymes were assayed for their ability to catalyze the various steps in the formation of CMP-Pse. The activity and product of each enzymatic step was confirmed by either capillary electrophoresis (CE) (Figure 5), CE–mass spectrometry (MS) (Supplementary Table I) or NMR spectroscopy (Table II). Starting with UDP-GlcNAc (IP) as a precursor, PseB-His6 was found to exhibit both C-4,6 dehydratase and C-5 epimerase activity that resulted in the production of UDP-2-acetamido-2,6-dideoxy-β-l-arabino-hexos-4-ulose (IIP). When the subsequent aminotransferase enzyme PseC-MBP was not present and this metabolite was left to build up in overnight reactions, the formation of UDP-2-acetamido-2,6-dideoxy-α-d-xylo-hexos-4-ulose could be observed (data not shown), resulting from a further C5 epimerization of the arabino-4-keto Pse intermediate (IIP). This is characteristic of the PseB enzymes studied to date. Also, the addition of exogenous NAD+ or NADP+ did not improve reaction speed, indicating that sufficient cofactor was already tightly bound to the enzyme and recycled throughout catalysis. Identical properties have already been described for the C. jejuni PseB enzyme Cj1293 (Schoenhofen et al. 2006b). In the next step, a C-4 amino transfer of the 4-keto intermediate (IIP) forming UDP-4-amino-4,6-dideoxy-β-L-AltNAc (IIIP) was accomplished by the aminotransferase PseC-MBP. PseH-His6 was found to perform N-4 acetylation of IIIP, forming UDP-2,4-diacetamido-2,4,6-trideoxy-β-l-altropyranose (IVP). Next, 2,4-diacetamido-2,4,6-trideoxy-β-l-altropyranose (VP) was formed by the removal of UDP from IVP by the sugar hydrolase PseG-MBP. The condensation of VP with pyruvate by PseI-His6 created Pse5,7Ac2 (VIP). For the last enzyme of the pathway, CMP-Pse synthetase PseF, neither the His-tagged nor the MBP-fusion construct showed any activity on VIp. However, using PseF from H. pylori, we were able to catalyze the activation of VIP with CMP, forming CMP-Pse5,7Ac2 (VIIP) (Figure 5; Table II). This finding might be indicative of the requirement of the 5Am7Gra modifications of Pse for activity of the T. forsythia PseF enzyme, or may simply indicate inactive enzyme due to in vitro tagging strategies (i.e., MalE- or His-tags). The former would also suggest possible relaxed specificity of enzymes PseB/C/H/G/I, as we were able to produce Pse5,7Ac2 with enzymes ultimately responsible for making Pse5Am7Gra. Importantly, the CMP-Pse5,7Ac2 produced using Pse5,7Ac2 prepared with T. forsythia enzymes was found to be identical to CMP-Pse5,7Ac2 previously characterized by NMR (Table II; Supplementary Figure 2). Our findings suggest that Pse biosynthesis in T. forsythia ATCC 43037 follows essentially the same biosynthetic route as in C. jejuni and H. pylori (Schoenhofen et al. 2006b).
Fig. 4.
SDS-PAGE gels (12%) of purified recombinant enzymes of the (A) CMP-Pse biosynthesis pathway from T. forsythia ATCC 43037 and (B) CMP-Leg biosynthesis pathway from T. forsythia FDC 92A2/UB4.
Fig. 5.
CE analysis of the reaction products obtained by the incubation of UDP-GlcNAc (IP) upon sequential addition of PseB-His6, forming UDP-2-acetamido-2,6-dideoxy-β-l-arabino-hexos-4-ulose (IIP); PseC-MBP, forming UDP-4-amino-4,6-dideoxy-β-L-AltNAc (IIIP); PseH-His6, forming UDP-2,4-diacetamido-2,4,6-trideoxy-β-l-altro-pyranose (IVP) and PseG-MBP, releasing UDP. Lastly, the combined action of PseI-His6 and H. pylori PseF convert 2,4-diacetamido-2,4,6-trideoxy-β-l-altro-pyranose (VP) via Pse5,7Ac2 (VIP) to CMP-Pse5,7Ac2 (VIIP). Subscript “P” indicates intermediates from the CMP-Pse biosynthesis pathway.
Table II.
NMR chemical shifts δ (ppm) for the pyranose rings of T. forsythia NulO biosynthesis intermediates analyzed in this study
| Compound | 1H | Literature δH (ppm) | Measured δH (ppm) | 13C | Literature δC (ppm) | Measured δC (ppm) | |
|---|---|---|---|---|---|---|---|
| VIIP | CMP-pseudaminic acid (CMP-Pse5,7Ac2) | H3ax | 1.60 | 1.60 | |||
| H3eq | 2.22 | 2.21 | C3 | 36.7 | 37.1 | ||
| H4 | 4.23 | 4.23 | C4 | 65.5 | 66.1 | ||
| H5 | 4.28 | 4.28 | C5 | 49.5 | 50.0 | ||
| H6 | 4.30 | 4.29 | C6 | 73.4 | 73.9 | ||
| H7 | 4.02 | 4.02 | C7 | 54.4 | 55.0 | ||
| H8 | 4.11 | 4.11 | C8 | 69.4 | 69.9 | ||
| H9 | 1.20 | 1.19 | C9 | 17.9 | 18.4 | ||
| IIL | GDP-2-acetamido-2,6-dideoxy-α-d-xylo-hexos-4-ulose | H1 | 5.45 | 5.46 | C1 | 95.3 | 95.5 |
| H2 | 4.10 | 4.09 | C2 | 53.5 | 53.7 | ||
| H3 | 3.82 | 3.83 | C3 | 72.4 | 72.5 | ||
| H5 | 4.11 | 4.11 | C5 | 70.9 | 71.1 | ||
| H6 | 1.21 | 1.21 | C6 | 12.4 | 12.5 | ||
| IIIL | GDP-4-amino-4,6-dideoxy-α-d-GlcNAc | H1 | 5.50 | 5.50 | C1 | 95.6 | 95.5 |
| H2 | 4.06 | 4.06 | C2 | 55.0 | 55.1 | ||
| H3 | 3.87 | 3.89 | C3 | 69.1 | 69.0 | ||
| H4 | 2.96 | 2.99 | C4 | 58.6 | 58.7 | ||
| H5 | 4.21 | 4.22 | C5 | 68.0 | 67.9 | ||
| H6 | 1.32 | 1.32 | C6 | 18.0 | 18.4 | ||
| VL | 2,4-diacetamido-2,4,6-trideoxy-d-mannopyranose | α/β | α/β | α/β | α/β | ||
| H1 | 5.11/4.96 | 5.11/4.97 | C1 | 94.0/94.0 | 93.8/93.8 | ||
| H2 | 4.30/4.46 | 4.30/4.47 | C2 | 53.8/54.8 | 53.8/54.7 | ||
| H3 | 4.06/3.84 | 4.08/3.85 | C3 | 67.7/71.7 | 67.6/71.0 | ||
| H4 | 3.77/3.66 | 3.78/3.67 | C4 | 54.8/54.5 | 54.6/54.3 | ||
| H5 | 3.97/3.51 | 3.98/3.52 | C5 | 68.1/72.6 | 68.2/72.4 | ||
| H6 | 1.19/1.21 | 1.19/1.22 | C6 | 18.0/18.0 | 18.0/18.0 |
Biochemical characterization of the CMP-Leg biosynthesis locus in T. forsythia strains FDC 92A2/UB4
In contrast to the type strain, protein homology searches of the NulO biosynthesis gene locus in strains FDC 92A2/UB4 suggested a biosynthetic route towards Leg. To support this prediction, we conducted a series of functional characterizations of those biosynthetic enzymes that determine the stereochemical differences between the Pse and Leg pathway (Figure 2). For this purpose, BFO_1074 (legB), BFO_1073 (legC) and BFO_1065 (legG) were cloned and expressed in E. coli, and purified as His6-tagged proteins (Figure 4B). Enzyme activity and reaction products were analyzed as described for strain ATCC 43037 above.
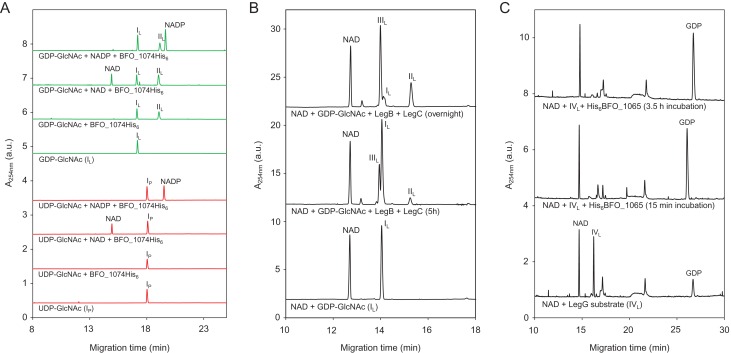
To substantiate the hypothesis that NulO biosynthesis in T. forsythia FDC 92A2/UB4 uses a GDP-linked precursor, we first assessed the substrate specificity of the first enzyme in the pathway, the LegB homolog BFO_1074 (Figure 6A). Since NAD(P)+ is a necessary cofactor for the C-4,6 dehydratase reaction, both NAD+ and NADP+ were added in separate reactions. Of all different nucleotide-activated GlcNAc-substrates that were used in our study (i.e., GDP-GlcNAc, UDP-GlcNAc (IP), CDP-GlcNAc, dTDP-GlcNAc and IDP-GlcNAc), only GDP-GlcNAc (IL) was converted by BFO_1074-His6 (data only shown for UDP/GDP). Moreover, enough cofactor was bound to the purified enzyme to facilitate substrate conversion without any additional NAD(P)+ present, albeit with slightly lower yields. In subsequent studies, we were able to determine that the reaction requires NAD+ and does not use NADP+ (data not shown), although increased conversion can even be seen for NAD-containing samples vs. NADP-containing ones (Figure 6A). The identity of the LegB-His6 reaction product was confirmed by NMR (Table II) to be identical to the LegB product described in C. jejuni, which is GDP-2-acetamido-2,6-dideoxy-α-d-xylo-hexos-4-ulose (IIL). This finding marked a substantial difference to the Pse pathway as found in the type strain, where, in addition to the C-4,6 dehydration, epimerization at C-5 takes place.
Fig. 6.
CE analysis of the reaction products obtained by incubation of (A) BFO_1074-His6 with UDP-GlcNAc (IP, red traces) and GDP-GlcNAc (IL, green traces) as substrate. Formation of product (GDP-2-acetamido-2,6-dideoxy-α-d-xylo-hexos-4-ulose (IIL)) could only be observed with GDP-GlcNAc (IL), confirming that BFO_1074-His6 is a LegB homolog that utilizes GDP-linked substrates. (B) GDP-GlcNAc (IL) with both the dehydratase BFO_1074-His6 (LegB) and the aminotransferase His6-BFO_1073 (LegC), resulted in the formation of GDP-4-amino-4,6-dideoxy-α-d-GlcNAc (IIIL). In the overnight reaction, a conversion of roughly 75% could be achieved. (C) GDP-2,4-diacetamido-2,4,6-trideoxy-α-d-glucopyranose (IVL) with the hydrolyzing 2-epimerase BFO_1065 (LegG). Complete release of GDP from the substrate could be observed after 15 min of incubation. Subscripts “P” and “L” indicate intermediates from the CMP-Pse and CMP-Leg biosynthesis pathway, respectively. This figure is available in black and white in print and in color at Glycobiology online.
Previous studies in C. jejuni have shown that the aminotransferases of the Pse, Leg and bacillosamine pathways display no cross-talk with their respective substrates, demonstrating the stereospecificity of each enzyme (Morrison and Imperiali 2014). Most importantly, C. jejuni LegC is absolutely specific for the GDP-4-keto intermediate formed by LegB. Overnight incubation of GDP-GlcNAc (IL) with both BFO_1074-His6 (LegB) and His6-BFO_1073 (LegC) resulted in approximately 75% substrate conversion to GDP-4-amino-4,6-dideoxy-α-d -GlcNAc (IIIL) (Figure 6B), the identity of which was again confirmed by NMR spectroscopy (Table II). Therefore, BFO_1073 can be considered a true LegC homolog that performs an equatorial addition of the amino group as opposed to the axial addition that PseC would catalyze (Figure 2). Lastly, the hydrolyzing 2-epimerase activity of BFO_1065 (putative LegG) was assayed. As with most of the enzymes of the C. jejuni-Leg pathway, C. jejuni LegG does not efficiently use the UDP-sugar precursor from the bacillosamine pathway, further reinforcing the specificity for a GDP-activated sugar (Schoenhofen et al. 2009; Morrison and Imperiali 2014). Since our attempts to express the putative N-acetyltransferase BFO_1067 failed, we used GDP-2,4-diacetamido-2,4,6-trideoxy-α-d -glucopyranose (IVL) produced by C. jejuni LegH in our assay (Schoenhofen et al. 2009). Complete GDP removal and C-2 epimerization by His6-BFO_1065 could be observed after just 15 min of incubation (Figure 6C; Table II), forming 2,4-diacetamido-2,4,6-trideoxy-d-mannopyranose (VL). To note, due to lability of biosynthetic intermediates, a small amount of GDP is observed in control reactions lacking LegG. Our findings suggest that the NulO found in strains FDC 92A2/UB4 is a Leg derivative, and the biosynthetic pathway closely resembles that found in C. jejuni (Schoenhofen et al. 2009). This is in contrast to the Pse5Am7Gra NulO found in the type strain ATCC 43037 (Figure 1).
Construction and characterization of NulO-deficient T. forsythia strains
After having characterized both Pse and Leg biosynthesis pathways in vitro, the next step was the construction of knockout mutants deficient in the respective terminal NulO, thus facilitating the investigation of the enzymes’ in vivo function and providing a valuable tool for future studies (Supplementary Figure 3). Since it was impossible to grow strain FDC 92A2 reliably, we used only strain UB4 for the creation of a Leg-deficient deletion mutant. The amplification and sequencing of various NulO biosynthesis genes indicated that UB4 possesses the same Leg biosynthesis gene locus as strain FDC 92A2 (Supplementary Figure 4). This was confirmed by an alignment of the FDC 92A2 genome with the recently published genome of strain UB4, which revealed a nucleotide identity of 99.8% in this cluster (Stafford et al. in preparation). The aminotransferase genes pseC and legC were selected as targets for gene knockouts in the ATCC 43037 type strain and UB4, respectively. These were predicted to be specific and crucial to NulO synthesis and, therefore, result in a NulO-deficient background when deleted. To allow the identification of possible polar effects from the insertion of the knockout cassette, complementation mutants were also created.
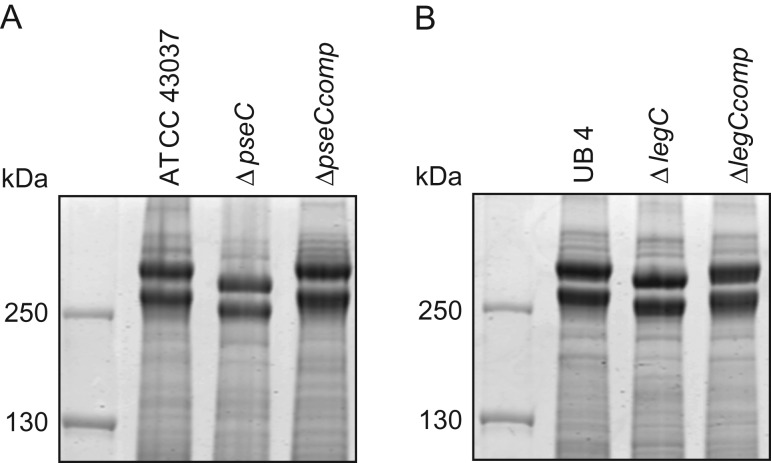
As a first indication of altered S-layer glycan structure, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis of T. forsythia ATCC 43037, UB4, and the respective NulO-deficient deletion mutants ATCC 43037 ΔpseC and UB4 ΔlegC showed a clear downshift of the glycosylated S-layer proteins TfsA and TfsB (Figure 7). To analyze these changes in detail, in-gel reductive β-elimination was performed to release O-glycans for subsequent analysis by LC–electrospray ionization (ESI)–MS/MS (Supplementary Figure 5). With the ATCC 43037 wild-type glycan (1897.7 Da) serving as a reference, data from the ATCC 43037 ΔpseC knockout mutant were consistent with the loss of the terminal 379 Da Pse5Am7Gra residue (Table III). Moreover, we could observe the loss of a methyl group on the terminal N-acetylmannosaminuronic acid residue, while the MS/MS profile of the rest of the glycan remained unchanged (data not shown). Additional mass peaks corresponding to glycans lacking digitoxose, fucose, N-acetyl residues or combinations thereof are all in agreement with the previously published microheterogeneity of the T. forsythia oligosaccharide (Posch et al. 2011). In terms of overall sugar composition and glycan structure, the O-glycan of strain UB4 was found to be identical to that of ATCC 43037 apart from a −43 Da mass difference in the terminal trisaccharide branch. MS/MS data and analysis of glycan fragments indicated that the methyl group on the terminal N-acetyl-mannosaminuronic acid residue was missing (−14 Da), while the remaining mass defect of 29 Da could be attributed to a change in the terminal NulO (Table III; Supplementary Figure 5). Importantly, the S-layer glycan from a UB4 ΔlegC knockout mutant was also found to be lacking a terminal NulO residue (Table III), specifically, a Leg derivative with a mass of 350 Da. In addition, we could consistently observe another glycan species with an additional +16 Da at the position of the digitoxose, indicating the presence of a deoxyhexose instead of a dideoxyhexose in some forms of the glycan.
Fig. 7.
SDS-PAGE analysis of T. forsythia parent, NulO-deficient, and complemented strains. (A) CBB stained SDS-PAGE (7.5% gel) of whole cell extracts from T. forsythia ATCC 43037, ATCC 43037 ΔpseC and the complemented strain ATCC 43037 ΔpseCcomp, and (B) T. forsythia UB4 wild-type, UB4 ΔlegC and the complemented strain UB4 ΔlegCcomp. For both T. forsythia ATCC 43037 ΔpseC and UB4 ΔlegC, a downshift of the S-layer protein bands (TfsA and TfsB) could be observed, which was reverted in the complemented strains.
Table III.
Overview of T. forsythia glycan species, detected m/z values, neutral masses and the anticipated mass of respective NulOs
| Strain | Structure | m/z [M+H+]+ | m/z [M+2 H+]2+ | Neutral mass [Da] | Neutral NulO mass [Da] | |
|---|---|---|---|---|---|---|
| with linkage (-18 Da) | Full mass | |||||
| ATCC 43037 |
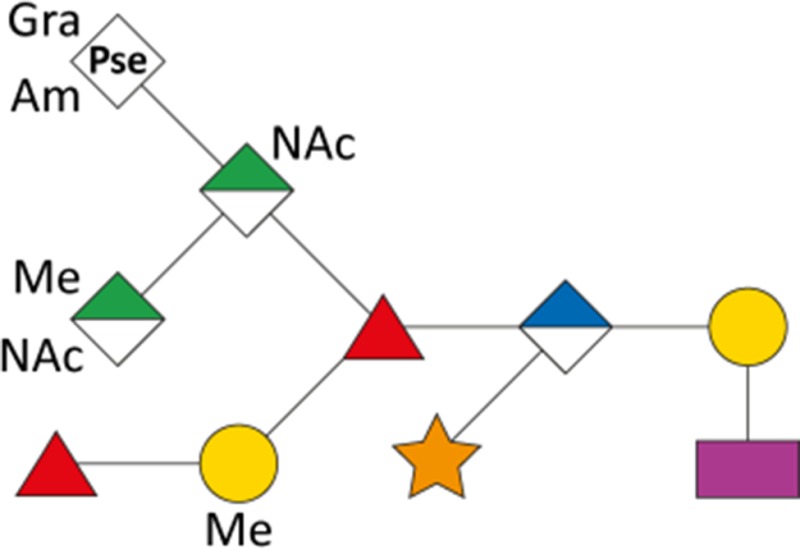
|
1898.7 | 949.9 | 1897.7 | 361.14 | 379.15 |
| ATCC 43037 ΔpseC |
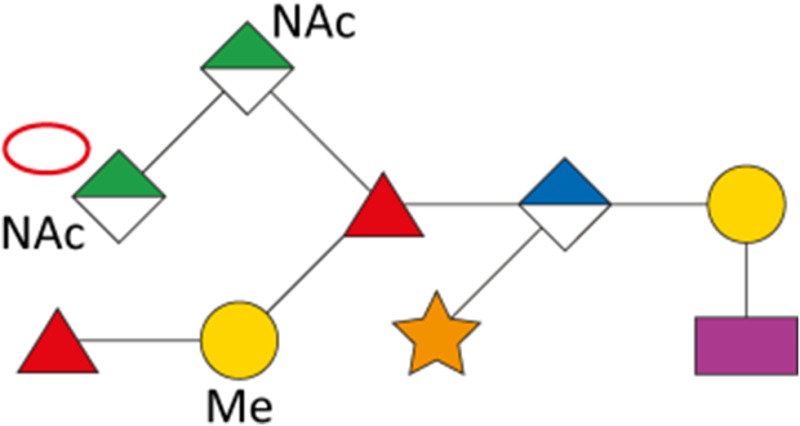
|
1523.4 | 762.3 | 1522.6 | – | – |
| UB4 |
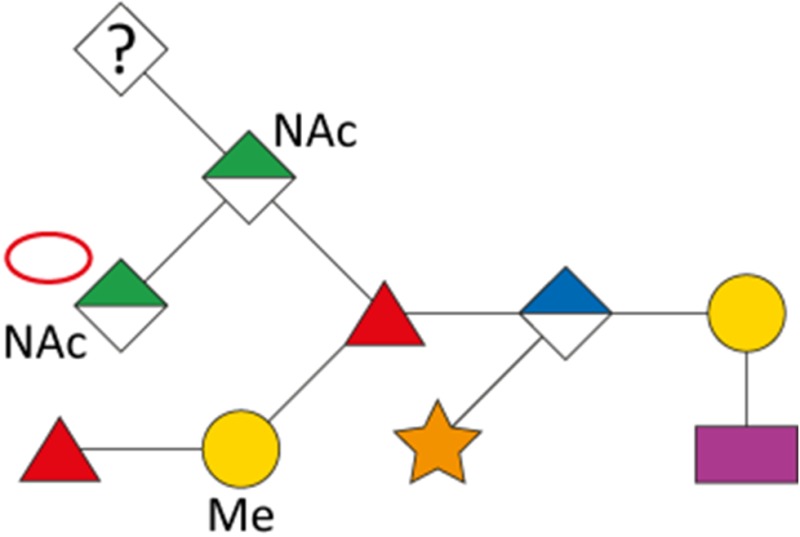
|
1855.6 | 928.3 | 1854.6 | 332.12 | 350.13 |
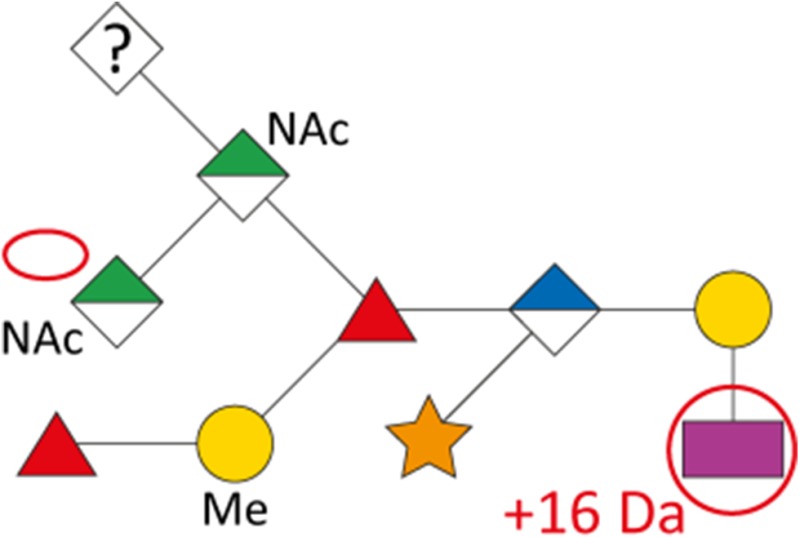
|
1871.6 | 936.3 | 1870.6 | 332.12 | 350.13 | |
| UB4 ΔlegC |
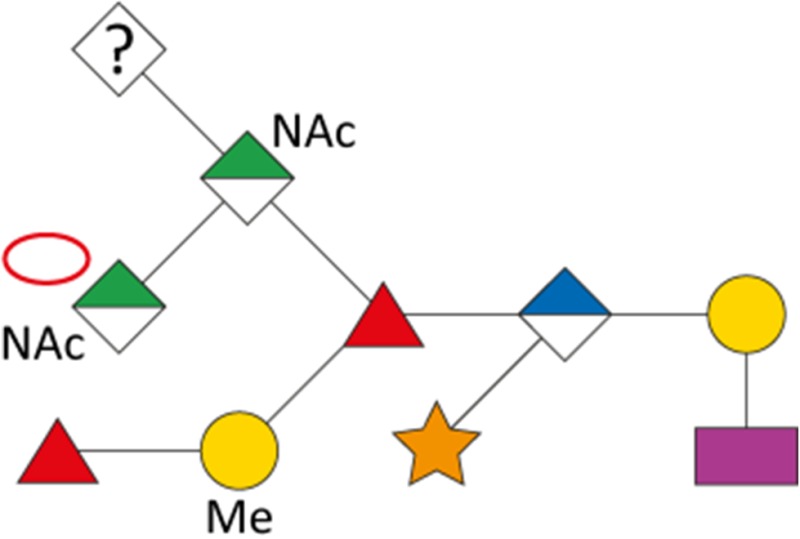
|
1523.4 | 762.3 | 1522.6 | – | – |
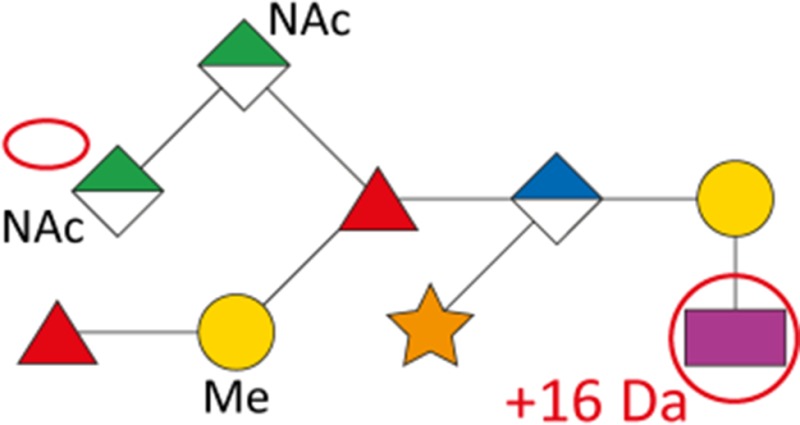
|
1539.4 | 770.3 | 1538.6 | – | – | |
Our data indicate that the Pse and Leg acid biosynthesis gene loci are functional and active in vivo and that strain UB4 carries a Leg residue with a different modification than the Pse in strain ATCC 43037, accounting for a mass difference of 29 Da. By way of calculation this could be explained by a change of the glyceroyl for a glycolyl and the acetimidyl for an acetyl group.
Analysis of biofilm formation of T. forsythia wild-type and NulO-deficient strains
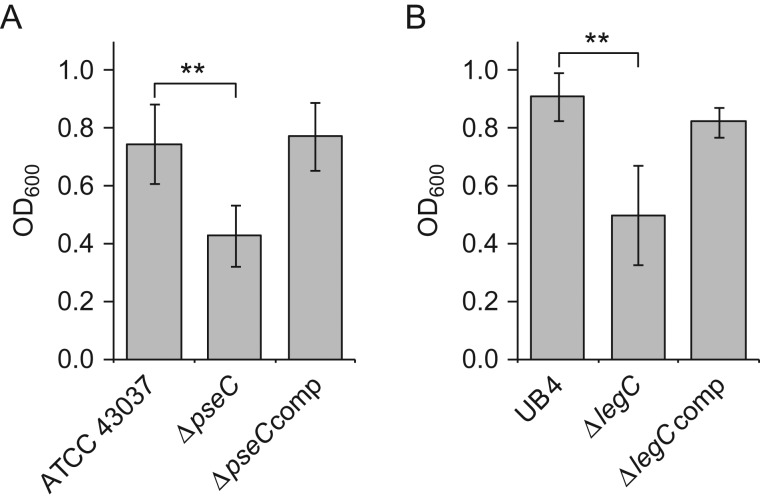
To test a possible influence of the terminal NulO residue on biofilm growth, liquid cultures of T. forsythia parent strains, deletion mutants, and complemented strains were incubated in mucin-coated 24-well polysterene plates. The salivary glycoprotein mucin is heavily sialylated and has been shown to facilitate attachment and promote growth in the initial stages of biofilm formation in various oral bacteria including T. forsythia (Roy et al. 2011). After removal of planktonic cells, the OD600 of the resuspended biofilm was determined. Biofilm formation in the NulO-deficient mutants showed a 1.7-fold and 1.8-fold decrease for strain ATCC 43037 ΔpseC (Figure 8 A) and for UB4 ΔlegC (Figure 8B), respectively, when compared to the corresponding parent strain. In the complemented strains, biofilm formation was restored to the parent strains’ levels, indicating that the decrease in biofilm growth is attributable to the loss of NulO biosynthesis rather than the insertion of a knockout cassette or the disruption of an operon.
Fig. 8.
Knockout of pseC and legC decreases biofilm formation of T. forsythia ATCC 43037 and T. forsythia UB4 cells, respectively. (A) Biofilm formation in T. forsythia ATCC 43037 wild-type compared to ATCC 43037 ΔpseC and the complemented strain ATCC 43037 ΔpseCcomp. (B) Biofilm formation in T. forsythia UB4 wild-type compared to UB4 ΔlegC and the complemented strain T. forsythia UB4 ΔlegCcomp. Data represent mean values ± SD of at least four independent experiments with four replicates each and were analyzed by the unpaired Student's t-test. Asterisks indicate significant differences (**P < 0.01).
Discussion
The purpose of this study was to examine the genetic and biochemical basis for the biosynthesis of the terminal, non-reducing end Pse derivative (Pse5Am7Gra) found on the S-layer O-glycan of T. forsythia ATCC 43037 (Posch et al. 2011). After discovering a strain misattribution in the publicly available genome of the type strain, we extended our investigation to strains FDC 92A2 and UB4. We have identified in three strains of T. forsythia distinct gene loci involved in the synthesis of the structurally distinct NulOs Pse or Leg. Through the subsequent characterization of the ATCC 43037 NulO biosynthetic enzymes, we could show their capability of producing CMP-Pse following the well-characterized UDP-sugar pathway of H. pylori, while the enzymes of strains FDC 92A2 and UB4 can follow the GDP-sugar route leading to CMP-Leg in C. jejuni. Considering the unique modifications of the NulOs in T. forsythia strains, however, it is likely that there might be deviations from the said pathways in vivo (Figure 2). Interestingly, a preliminary analysis of unpublished genomes found that a significantly higher number of the T. forsythia strains available to us contain the biosynthetic gene cluster for Leg than for Pse, suggesting that this type of NulO might be more prevalent than the one found in the type strain ATCC 43037. Whether the “equipment” of T. forsythia strains with Leg or Pse may reflect adaptations to specific oral microenvironments or is linked to the health status of the patients from which the isolates were derived remains elusive.
Despite testing both His6-tagged and MBP-fusion protein constructs, we were unable to detect activity for the last enzyme of the Pse pathway, the CMP-Pse synthetase PseF. While this can have several reasons, it is conceivable that at this stage of the pathway, PseF can only process Pse that carries one or both of the modifications observed in the ATCC 43037 type strain (i.e., 5Am7Gra). Among NulO biosynthesis genes, sequence conservation between LegF and PseF homologs, respectively, is relatively low, which could indicate high specificity for a particularly modified substrate. Since knowledge about the identity and function of NulO-modifying enzymes is scarce, the elucidation of the biochemical mechanism and genetic basis for the N-acetimidoyl (Am) and N-glyceroyl (Gra) substitutions observed in the ATCC 43037 type strain remains a major challenge. In Campylobacter spp., the Am- modifications of Pse and Leg appear to occur at the CMP-sugar stage (McNally et al. 2007). The genes involved in these substitutions (pseA and ptmG (legA), respectively) have been identified, but have no homolog in T. forsythia (data not shown). However, it may not be surprising that there is low similarity between a T. forsythia and a C. jejuni Am-modifying enzyme, since in C. jejuni the Am group is added to the C7 position of Pse, whereas in T. forsythia it is added to the C5 position. With regard to the Gra modification of Pse, the substitution of an N-acetyl (Ac) group of Pse by a 2,3-di-O-methyl glyceric acid residue has been described on the flagellin of C. jejuni 11168 (Logan et al. 2009). In C. jejuni, this Gra modification is found at C5, whereas in T. forsythia it is found at C7, and, again, may explain the inability to identify clear homologs between the two organisms. A glyceric acid residue at the C7 position of Pse has also been found in the LPS of Vibrio vulnificus (Vinogradov et al. 2009), but the responsible genes have yet to be identified.
While there are numerous reports of one distinct bacterium being capable of producing both Leg and Pse, variations of the NulO-type between strains of the same species have, to the best of our knowledge, so far only been described in C. jejuni (strain 81-176 only displays Pse derivatives, whereas strain 11168 displays Pse and Leg), Campylobacter concisus (Kaakoush et al. 2011), Acinetobacter spp. (Scott et al. 2014) and Photobacterium profundum (Lewis et al. 2009). The evolutionary purpose for such structural variation remains a matter of speculation. In T. forsythia, a likely driving force would be the development of sophisticated strategies for host evasion and virulence. It is conceivable that different NulO types play distinct roles in the molecular mimicry of host tissues, thus enabling the pathogen to avoid immune detection, modulate the host immune response and enter into host cells to escape immune surveillance (Amano et al. 2014). This may also suggest that some T. forsythia strains have a special “fit” with only certain hosts since NulO display is not a matter of turning gene content on or off but is fixed in their genomes—they are either displaying Pse or Leg at the cell surface. Owing to T. forsythia's slow growth and recalcitrance to genetic manipulation, insights into the possible immunological function of the modified Pse in T. forsythia could so far only be derived from an ATCC 43037 ΔwecC deletion mutant (ED1 (Honma et al. 2007)) lacking the terminal trisaccharide motif of the S-layer glycan (comprised of two ManNAcA residues and the terminal Pse5Am7Gra; compare with Figure 1B; (Posch et al. 2011)). The ΔwecC mutant was found to induce significantly higher amounts of the pro-inflammatory cytokines IL-6, IL-1β and IL-23 secretion in mouse bone-marrow-derived dendritic cells and macrophages compared with T. forsythia parent cells. Moreover, the loss of that glycan motif rendered the pathogen increasingly prone to capture and killing by dendritic cells (Settem et al. 2013, 2014). In vivo, mice infected with the T. forsythia strain showed higher inflammatory alveolar bone loss than the ΔwecC mutant-infected mice, a consequence that could be attributed primarily to the parent-type's ability to activate TLR2, drive CD4+ T-cell differentiation toward Th2 whilst blocking Th17 and innate neutrophil responses (Myneni et al. 2011; Settem et al. 2013). Although the ΔwecC mutant strain lacks three sugars instead of only just the Pse5Am7Gra residue, we speculate that the terminal NulO derivative plays the major role in the observed immunomodulatory effects, possibly through interactions with Siglec (sialic acid binding Ig-like lectin) receptors on immune cells. The deletion mutant ΔpseC created in the course of this study lacks only the terminal modified Pse and, therefore, represents a valuable tool for future studies in this direction. The analysis of T. forsythia UB4, a strain possessing the biosynthetic pathway for Leg, and its deletion mutant ΔlegC revealed the presence of a NulO of unknown structure with a mass of 350 Da. A possible candidate that matches this mass requires an N-glycolyl (Gc) substitution at either C-5 or C-7, resulting in Leg5Ac7Gc or Leg5Gc7Ac. This modification is commonly found in the Sias of mammals in the form of the hydroxylated derivative of N-acetylneuraminic acid, Neu5Gc, but has, to the best of our knowledge, not yet been described in bacteria. The elucidation of the exact structure of the unknown NulO in T. forsythia UB4 using NMR spectroscopic methods will therefore be the main focus of future studies. Moreover, having identified different NulOs in the strains ATCC 43037 and UB4, in conjunction with our respective ΔpseC and ΔlegC mutants, it is now possible to investigate the respective influence of Pse and Leg on Toll-like receptor 2 signaling and T-helper 17 responses.
As shown by our analysis of growth on mucin-coated microtiter plates, the knockout of pseC and legC in T. forsythia ATCC 43037 and UB4 cells, respectively, resulted in impaired biofilm formation as evidenced by a 1.7- (ΔpseC) and 1.8-fold (ΔlegC) decrease compared to the parent strains. In either case biofilm formation could be fully restored in the respective complemented mutant (Figure 8), indicating that NulO loss directly correlates with reduced biofilm formation. For comparison, the T. forsythia ATCC 43037 ΔwecC mutant for which enhanced biofilm formation had been reported previously (Honma et al. 2007), was included in our study. Interestingly, in our microtiter assay, this mutant exhibited a 3.6-fold decrease in biofilm formation compared to the parent strain (data not shown). This discrepancy of our and previous data might be explained by the different surface characteristics of the plates used in the assays. In our study, mucin was used to foster the biofilm formation (Roy et al. 2011, 2012), since apart from facilitating initial attachment and promoting growth, coating of the plates with this glycoprotein also confers highly hydrophilic properties to the inherent hydrophobic polystyrene surfaces (Crouzier et al. 2013) and simultaneously mirrors the native habitat of the bacterium. Thus, it is conceivable that a ΔwecC mutant (lacking three charged sugar residues) exhibits reduced biofilm formation when grown on hydrophilic surface, while the comparably more hydrophobic, non-coated polystyrene plates might have favored the opposite. Furthermore, a study of a deletion mutant of the OmpA-like protein (Tanf_10935/TF1331) showed that this knockout resulted in a significant reduction of cell surface hydrophobicity and biofilm-forming activity on hydrophobic polystyrene plates (Abe et al. 2011). Both these findings are in agreement with our results and support the hypothesis that the surface of the plates, when rendered from hydrophobic to hydrophilic, causes a decrease of biofilm formation in strains such as ΔwecC (Honma et al. 2007; Posch et al. 2011), ΔpseC and ΔlegC (this study) that lack one or more charged residues.
In summary, our findings contribute to the understanding of the biosynthesis of two distinct NulOs in two strains of the same species and provide valuable tools for the investigation of the biofilm life-style and the immunogenic potential of T. forsythia. Considering the likely involvement of this pathogen's NulOs in the modulation of immune responses and the increasing reports of Sia-like sugars being linked to bacterial pathogenesis (Figure 9), future experiments will be aimed at elucidating the mechanisms that govern the interactions of T. forsythia with the host as well as other bacteria of the oral microflora. Ultimately, these insights will facilitate the development of future strategies to control periodontal disease.
Fig. 9.
Possible biological roles of the NulOs of the T. forsythia S-layer glycan. Representing the terminal sugars on the outermost surface structure of T. forsythia strains, Pse or Leg could have several functions. As previous studies have found the terminal trisaccharide of the S-layer glycan to be involved in the modulation of cytokine release and the suppression of Th17 responses, we hypothesize that the different NulOs displayed by T. forsythia strains are implicated in this phenomenon. Apart from the effect on monospecies biofilm formation described in this paper, the terminal NulOs are likely to play a role in the bacterium's interactions with other species of the subgingival microbiota. Furthermore, they could exert an influence on how the bacterium attaches to periodontal surfaces and how it invades host tissue. The variation of the NulO type among various T. forsythia strains may reflect a special ‘fit’ with only certain hosts and/or microenvironments. This figure is available in black and white in print and in color at Glycobiology online.
Materials and methods
Strains and culture conditions
T. forsythia ATCC 43037 (American Type Culture Collection, Manassas, VA), T. forsythia FDC 92A2 (obtained from Dr. Graham Stafford, University of Sheffield, UK) and T. forsythia UB4 (obtained from Dr. Ashu Sharma, University of Buffalo, NY) and the knockout mutants were grown anaerobically at 37 °C for 4–7 days in brain heart infusion (BHI) broth or 0.8% (w/v) BHI agar, supplemented with N-acetylmuramic acid and horse serum as described previously (Tomek et al. 2014). And 5 g/ml erythromycin (Erm), 10 µg/ml chloramphenicol (Cat) and 50 µg/ml gentamycin (Gent) were added when appropriate. Escherichia coli DH5α (Invitrogen), E. coli BL21 (DE3) (Invitrogen) and E. coli BL21-CodonPlus[DE3]-RIL (Novagen) cells were cultivated at 37°C and 200 rpm in lysogeny broth (LB) medium or 2xYT medium (16 g/l tryptone, 10 g/l yeast extract, 5 g/L NaCl) optionally supplemented with 100 µg/ml ampicillin (Amp), 50 µg/ml kanamycin (Km) or 30 µg/ml chloramphenicol (Cm), respectively. All strains used in this study are listed in Supplementary Table II.
DNA techniques and plasmid construction
All enzymes were purchased from Thermo Scientific. Genomic DNA of T. forsythia was isolated from 2 ml of bacterial suspension as described previously (Cheng and Jiang 2006). The GeneJET™ Gel Extraction Kit (Fermentas) was used to purify DNA fragments from agarose gels and to purify digested plasmids and oligonucleotides. Plasmid DNA from transformed cells was isolated with the GeneJET™ Plasmid Miniprep kit (Fermentas). Primers for PCR and DNA sequencing were purchased from Invitrogen (Supplementary Table III). PCR was performed using the Phusion® High-Fidelity DNA Polymerase (Fermentas) and the thermal cycler My CyclerTM (Bio-Rad). For recombinant expression in E. coli BL21 (DE) cells, plasmids encoding PseB, PseH and PseI were created as C-terminal His6-tag fusion constructs by PCR using T. forsythia ATCC 43037 genomic DNA as a template and the primers 11/12, 28/22 and 33/34, respectively. The His6-tagged amplification products were digested with NcoI/XhoI (pseB and pseH) and BsaI/XhoI (pseI), and all were cloned into NcoI/XhoI-linearized pET28a(+) (Novagen). PseC, PseG and PseH were produced as N-terminal MBP fusion proteins. After amplification using the primers 80/86, 81/105 and 101/102, respectively, the amplicons were digested with BamHI/HindIII and cloned into BamHI/HindIII-linearized pMAL_c2E vector (NEB). Plasmids encoding the Leg biosynthetic enzymes of strain FDC 92A2 were constructed as C-terminal (LegB) and N-terminal (LegC and LegG) His6-tagged fusion constructs, after having the genes synthesized (Life Technologies). To overcome problems with low expression, legG was codon-optimized for E. coli. The synthesized gene sequences were amplified using the primers V83/84, V133/134 and V114/115, used for blunt-ended ligations into pCR2.1 and subsequently cloned into pET22b via NdeI/XhoI.
Transformation of chemically competent E. coli DH5α, E. coli BL21 (DE3) and E. coli BL21-CodonPlus[DE3]-RIL (Novagen) cells was performed according to the manufacturer's protocols. Transformants were screened by PCR using RedTaq ReadyMix PCR mix (Sigma-Aldrich), and recombinant clones were analyzed by restriction mapping and confirmed by sequencing (Microsynth).
His6-tagged protein expression and purification
The plasmids encoding the Pse biosynthesis enzymes (pET28a_pseB, pMAL_2cE_pseC, pET28a_pseH, pMAL_2cE_pseG, pET28a_pseI and pMAL_2cE_pseH) were transformed into E. coli BL21 (DE3) cells, while those encoding biosynthesis enzymes of the Leg pathway (TF2074N-pet22b, TF2075C-pet22b and BFO_1065 N-pet22b) were expressed in E. coli BL21-CodonPlus[DE3]-RIL. Freshly transformed cells were grown in LB or 2xYT medium to the mid-exponential growth phase (OD600 ~0.6) and protein expression was induced with a final concentration of 0.5 mM (Pse enzymes) or 0.1 mM (Leg enzymes) isopropyl-β-d-thiogalactopyranosid (IPTG). Cultures were grown for an additional 4 h at 37 °C and 200 rpm (PseG and PseH), were put on ice for 20 min and incubated overnight at 22 °C (PseB, PseC, PseI, PseF) or were incubated for 3 h at 30 °C (LegB, LegC and LegG). Cells were harvested by centrifugation (4500 × g, 30 min, 4 °C).
For purification of His6-tagged enzymes, the cell pellets were resuspended in lysis buffer (50 mM sodium phosphate, pH 7.3, 400 mM NaCl, 10 mM β-mercaptoethanol) containing 10 mM imidazole, 10 µg/ml of DNaseI (Roche Applied Science) and EDTA-free protease inhibitor mixture (cOmplete, Roche Applied Science). The cells were disrupted by two passes through a French pressure cell (Amicorp) or an emulsiflex C5 (20,000 psi). Lysates were centrifuged at 100,000 × g for 50 min at 4 °C, and the supernatant fraction was applied to a 3-ml nickel–nitrilotriacetic acid (Qiagen) column equilibrated in lysis buffer using a flow rate of 1 ml/min. After sample application, the column was washed with eight column volumes of lysis buffer. To elute the protein of interest, a linear gradient from 10 to 100 mM imidazole in lysis buffer was applied over 14 column volumes prior to a final pulse of 14 column volumes of 200 mM imidazole in lysis buffer. MBP-fusion proteins were purified according to the manufacturer's protocol using 2-ml amylose resin (NEB) per litre of culture. Fractions containing the purified protein of interest, as determined by SDS-PAGE (12.5%) upon Coomassie Brilliant Blue G250 (CBB) staining, were pooled and dialyzed against dialysis buffer (25 mM sodium phosphate, 25 mM NaCl, pH 7.3) overnight at 4 °C. Protein concentration was measured spectrophotometrically and calculated using A280 0.1% values obtained from the exPASy ProtParam tool (http://web.expasy.org/protparam).
Enzymatic assays
Enzymatic assays and metabolite/product purification was performed similarly to methods described previously (Schoenhofen et al. 2006b; Schoenhofen et al. 2009; Gulati et al. 2015). Typically, NulO biosynthetic pathway reactions were performed at 37 °C in 25 mM sodium phosphate pH 7.3–7.8, 25 mM NaCl with 1 mM NDP-GlcNAc, 0.5 mM NAD+ or NADP+, 10 mM L-Glu, 0.5 mM PLP, 1.2 mM acetyl-CoA, 1.2 mM PEP and 2 mM MgCl2 as necessary. CMP-activation of Pse5,7Ac2 was performed using a CMP-pseudaminic acid synthetase, PseF, from H. pylori (Schoenhofen et al. 2006a Glycobiology). Reactions contained 50 mM Tris/HCl pH 9, 50 mM MgCl2, 0.5 mM CTP, ~0.5 mM Pse5,7Ac2, and sufficient quantities of pyrophosphatase as well as PseF.
Detection of Pse/Leg pathway biosynthetic genes in T. forsythia ATCC 43037, FDC 92A2 and UB4
To check for the presence of selected genes from the Pse or Leg biosynthesis pathway, genomic DNA of T. forsythia ATCC 43037, T. forsythia FDC 92A2 and T. forsythia UB4 was used as a template in six separate PCRs. Primers 11/12, 80/86 and 23/24 were used to amplify pseB, pseC and pseF of the Pse biosynthesis pathway. Primers V83/84, V26/27 and V63/64 were used to amplify legB, legC and legF of the Leg biosynthesis pathway. Amplification products of the UB4 strain were sequenced to confirm identity to the sequenced genome of strain FDC 92A2.
Gene knockouts
The gene knockout cassette used for constructing the T. forsythia ATCC 43037 ∆pseC and UB4 ∆legC mutants consisted of a 1093-base-pair (bp) Erm resistance gene P(ermF)_ermF flanked by homologous upstream and downstream regions. The ~1000-bp upstream and ~1000-bp downstream homology regions of the pseC gene (Tanf_01190) and of the legC gene (BFO_1073) were amplified from genomic DNA of T. forsythia ATCC 43037 and T. forsythia UB4, respectively, using the primers 5/6 (ATCC upstream), 7/8 (ATCC downstream), 88/89 (UB4 upstream) and 90/91 (UB4 downstream), respectively. The 1093-bp Erm resistance gene P(ermF)_ermF was amplified from genomic DNA of T. forsythia ΔwecC (obtained from A. Sharma, State University of New York at Buffalo, NY) using the primers 1 and 2. The three fragments were joined by overlap extension (OE-) PCR and sub-cloned into the blunt-end cloning vector pJET1.2 (Thermo Scientific). The corresponding plasmids were named pJET1.2_pseC_ermF and pJET1.2_legC_ermF. Transformation of T. forsythia ATCC and T. forsythia UB4 was done as described previously (Tomek et al. 2014, Megson et al. 2015). Cells were regenerated in BHI medium for 24 h before plating on BHI agar plates containing Erm as selection marker. Single colonies were picked and used for inoculation of liquid BHI medium. Once bacterial growth was visible, genomic DNA was isolated to confirm the integration of the knockout cassette via PCR amplification. The upstream and downstream integration as well as the loss of the gene were checked, using the primers 38/3 (ATCC upstream), 4/10 (ATCC downstream), 15/18 (ATCC loss of gene), 92/3 (UB4 upstream), 4/93 (UB4 downstream) and V26/V27 (UB4 loss of gene), respectively.
The complementation cassette for T. forsythia mutants consisted of a 651-bp Cm resistance gene flanked by a homologous ~1000-bp upstream region, the gene of interest and a ~1000-bp downstream region. The Cm resistance gene was amplified containing KpnI/SphI restriction sites at the C-terminus from the pEXALV vector (Zarschler et al. 2009) using the primers 75/77. The ~1000-bp upstream region plus the gene of interest, and the ~1000-bp downstream homology regions containing also the two restriction sites were amplified from genomic DNA of T. forsythia ATCC 43037 or T. forsythia UB4, using the primers 5/46 (ATCC upstream), 78/79 (ATCC downstream), 88/86 (UB4 upstream) and 94/95 (UB4 downstream), respectively. The ~1000-bp upstream region plus the gene of interest and the Cm resistance gene were joined by overlap extension (OE-) PCR and sub-cloned into the blunt-end cloning vector pJET1.2 (Thermo Scientific) resulting in plasmids pJET1.2_pseC_up+gene_cat and pJET1.2_legC_up+gene_cat. The ~1000-bp downstream homology regions were cloned into pJET1.2_pseC_up+gene_cat and pJET1.2_ legC_up+gene_cat using the KpnI and SphI restriction sites. The corresponding plasmids were named pJET1.2_pseC_cat and pJET1.2_legC_cat. Transformation of T. forsythia ATCC ∆pseC and T. forsythia UB4 ∆legC was done as described previously (Tomek et al. 2014; Megson et al. 2015).
CE analysis
CE analysis was performed using a P/ACE MDQ system (Beckman Instruments) with diode array detection. The running buffer was 25 mM sodium tetraborate, pH 9.4. The capillary was bare silica 50 µm × 50 cm, with a detector at 50 cm. The capillary was conditioned before each run by washing with 0.2 M NaOH for 2 min, water for 2 min, and running buffer for 2 min. Samples were introduced by pressure injection for 6 s, and the separation was performed at 18 kV for 20–30 min. Peak integration was done using the Beckman P/ACE station software.
Capillary electrophoresis–mass spectrometry
Separation of ions was achieved by CE (Prince CE system (Prince Technologies, Netherlands)) in a 90-cm long bare fused-silica capillary (365 μm OD × 50 μm ID). The 30 mM morpholine aqueous running CE buffer (adjusted to pH 9 with formic acid) was coupled with the capillary sheath fluid (isopropanol:methanol [2:1]) at their interface prior to MS (API3000 mass spectrometer [Applied Biosystems/Sciex, Concord, ON, Canada]).
NMR spectroscopy
For structural characterization of NulO pathway intermediates, filtered enzymatic reactions (Amicon Ultra-4 centrifugal filter unit with 3000 kDa molecular weight cut-off) or material purified according to previously described methods was exchanged into 100% D2O (Gulati et al. 2015). Structural analysis was performed using either a Varian Inova 500 MHz (1H) spectrometer with a Varian Z-gradient 3-mm probe or a Varian 600 MHz (1H) spectrometer with a Varian 5 mm Z-gradient probe. All spectra were referenced to an internal acetone standard (δH 2.225 ppm and δC 31.07 ppm).
Mass spectrometric glycan structure analysis
For the analysis of the glycan structure in T. forsythia strains and mutants, S-layer O-glycans were released from purified TfsA S-layer protein by in-gel reductive β-elimination (Posch et al. 2011). The excised gel slices were covered with 1.0 M NaBH4 in 0.5 M NaOH and incubated at 50 °C overnight. Excess salt was removed with porous graphitic carbon (PGC) solid phase extraction cartridges (HyperSep-96- Hypercarb, 25 mg, Thermo Scientific). Briefly, the cartridges were equilibrated with 500 μl of 60% acetonitrile and washed three times with 500 µl water. Sample was added and washed twice with water. Reduced glycans were eluted using 350 μl 60% acetonitrile, vacuum dried and taken up in water. O-glycans were then analyzed by LC–ESI–MS/MS on a Hypercarb PGC column (100 × 0.32 mm, 5 μm pore size, Thermo Scientific) with a 20-min gradient from 1 to 28% acetonitrile in formate buffer of pH 3.0 at a flow rate of 6 μl/min using an Ultimate 3000 capillary flow LC (Dionex). Eluted glycans were registered on a Bruker amaZon ion trap using ESI and data dependent acquisition. Qualitative evaluation was done using Bruker's Data Analysis 4.0, where the all spectra within the relevant elution range were summed up to generate an average spectrum. The glycan nature of the main peaks was confirmed by the identification of specific reporter ions in MS/MS spectra, which was done by the help of Glycoworkbench 2.0 (Ceroni et al. 2008).
Biofilm microtiter plate assay
The ability of the different T. forsythia strains and NulO mutants to form biofilm on an abiotic surface was evaluated according to Roy et al. (2011). Cultures were grown for 2–3 d in half-concentrated BHI medium, diluted to an OD600 ~0.05 with sterile nutrient medium, and 1-ml aliquots were transferred into 24-well polystyrene plates (STARLAB) that were coated overnight at 37 °C with 500 µl of mucin (from bovine submaxillary gland, Sigma-Aldrich) solution (0.5 mg/ml in 0.1 M sodium acetate buffer pH 4.5). As a control, sterile nutrient medium was used. Cultures were incubated under anaerobic conditions at 37°C for 3 d (T. forsythia ATCC 43037, ATCC 43037 ∆pseC and ATCC 43037 ∆pseCcomp) or 6 d (T. forsythia UB4, T. forsythia UB4 ∆legC and T. forsythia UB4 ∆legCcomp) without shaking. Two wells of each sample were used to determine the total optical density of the strains for later normalization. Subsequently, planktonic cells were gently removed with a pipette and the plates were washed once with PBS buffer. Cells were resuspended in 1 ml of PBS buffer and the OD600 of the biofilm was determined. Biofilm values were normalized to the corresponding absorbance of the total biofilm. Data represent mean values ± SD of at least four independent experiments with four replicates each and were analyzed by the unpaired Student's t-test. Asterisks indicate significant differences (*P < 0.05; **P < 0.01; ***P < 0.001).
Supplementary data
Funding
Financial support came from the Austrian Science Fund FWF, projects P24317-B22 (to C.S.) and the Doctoral Program “Biomolecular Technology of Proteins” W1224. I.C.S. acknowledges support by the National Research Council of Canada. A.S. acknowledges the support from the U.S. Public Health grants DE14749 and DE22870.
Supplementary Material
Acknowledgements
The authors gratefully acknowledge Amanda Lewis for sharing NulO-synthase sequences for phylogenetic analysis and Graham Stafford for providing T. forsythia strains FDC 92A2 and UB4. We would also like to thank Melissa Schur and Jacek Stupak for CE and CE–MS analysis, respectively.
Conflict of interest statement
None declared.
Abbreviations
Ac, N-acetyl or acetamido; Am, N-acetimidoyl or acetamidino; Amp, ampicillin; AltNAc, N-acetyl-β-l-altrosamine; BLASTP, protein-protein Basic Local Alignment Search Tool; BHI, brain heart infusion; Cat, chloramphenicol; CBB, Coomassie Brilliant Blue G250; CDP, cytidine-5′-diphosphate; CE, capillary electrophoresis; Cm, chloramphenicol; CMP, cytidine-5′-monophosphate; CoA, coenzyme A; CTP, cytidine-5′-triphosphate; dTDP, 2′-deoxythymidine-5′-diphosphate; Erm, erythromycin; ESI, electrospray ionization; Fo, N-formyl; Gc, N-glycolyl; GDP, guanosine-5′-diphosphate; Gent, gentamycin; GlcN, glucosamine; GlcNAc, N-acetyl glucosamine; Glu, glutamate; Gra, N-glyceroyl or N-2,3-dihydroxypropionyl or glycerate group; Hb, N-hydroxybutyryl; IDP, inosine-5′-diphosphate; IL, interleukin; IPTG, isopropyl-β-d-thiogalactopyranosid; Km, kanamycin; LB, lysogeny broth; LC, liquid chromatography; Leg, legionaminic acid (Leg5,7Ac2), 5,7-diacetamido-3,5,7,9-tetradeoxy-d-glycero-d-galacto-nonulosonic acid; LPS, lipopolysaccharide; MBP, maltose-binding protein; MS, mass spectrometry; NAD+, nicotinamide adenine dinucleotide; NADP+, nicotinamide adenine dinucleotide phosphate; NDP, nucleotide diphosphate; Neu, neuraminic acid; NMR, nuclear magnetic resonance; NulO, nonulosonic acid; ORF, open reading frame; PBS, phosphate-buffered-saline; PEP, phosphoenolpyruvate; PGC, porous graphitic carbon; Pi, inorganic phosphate; PLP, pyridoxal-5′-phosphate; PPi, pyrophosphate; Pse, pseudaminic acid (Pse5,7Ac2), 5,7-diacetamido-3,5,7,9-tetradeoxy-l-glycero-l-manno-nonulosonic acid; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis; Sia, sialic acid (Neu5Ac), 5-acetamido-3,5-dideoxy-d-glycero-d-galacto-nonulosonic acid; Siglec, sialic acid binding Ig-like lectin; Th, T helper cell; TLR2, toll-like receptor 2; UDP, uridine-5′-diphosphate
References
- Abe T, Murakami Y, Nagano K, Hasegawa Y, Moriguchi K, Ohno N, Shimozato K, Yoshimura F. 2011. OmpA-like protein influences cell shape and adhesive activity of Tannerella forsythia. Mol Oral Microbiol. 26:374–387. [DOI] [PubMed] [Google Scholar]
- Amano A, Chen C, Honma K, Li C, Settem RP, Sharma A. 2014. Genetic characteristics and pathogenic mechanisms of periodontal pathogens. Adv Dent Res. 26:15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlin AF, Uchiyama S, Chang YC, Lewis AL, Nizet V, Varki A. 2009. Molecular mimicry of host sialylated glycans allows a bacterial pathogen to engage neutrophil Siglec-9 and dampen the innate immune response. Blood. 113:3333–3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceroni A, Maass K, Geyer H, Geyer R, Dell A, Haslam SM. 2008. GlycoWorkbench: A tool for the computer-assisted annotation of mass spectra of glycans. J Proteome Res. 7:1650–1659. [DOI] [PubMed] [Google Scholar]
- Cheng HR, Jiang N. 2006. Extremely rapid extraction of DNA from bacteria and yeasts. Biotechnol Lett. 28:55–59. [DOI] [PubMed] [Google Scholar]
- Chou WK, Dick S, Wakarchuk WW, Tanner ME. 2005. Identification and characterization of NeuB3 from Campylobacter jejuni as a pseudaminic acid synthase. J Biol Chem. 280:35922–35928. [DOI] [PubMed] [Google Scholar]
- Crouzier T, Jang H, Ahn J, Stocker R, Ribbeck K. 2013. Cell patterning with mucin biopolymers. Biomacromolecules. 14:3010–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darveau RP. 2010. Periodontitis: A polymicrobial disruption of host homeostasis. Nat Rev Microbiol. 8:481–490. [DOI] [PubMed] [Google Scholar]
- Edebrink P, Jansson PE, Bogwald J, Hoffman J. 1996. Structural studies of the Vibrio salmonicida lipopolysaccharide. Carbohydr Res. 287:225–245. [DOI] [PubMed] [Google Scholar]
- Fagan RP, Fairweather NF. 2014. Biogenesis and functions of bacterial S-layers. Nat Rev Microbiol. 12:211–222. [DOI] [PubMed] [Google Scholar]
- Friedrich V, Pabinger S, Chen T, Messner P, Dewhirst FE, Schäffer C. 2015. Draft genome sequence of Tannerella forsythia type strain ATCC 43037. Genome Announc. 3: e00660–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerry P, Ewing CP, Schirm M, Lorenzo M, Kelly J, Pattarini D, Majam G, Thibault P, Logan S. 2006. Changes in flagellin glycosylation affect Campylobacter autoagglutination and virulence. Mol Microbiol. 60:299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulati S, Schoenhofen IC, Whitfield DM, Cox AD, Li J, St Michael F, Vinogradov EV, Stupak J, Zheng B, Ohnishi M, et al. 2015. Utilizing CMP-sialic acid analogs to unravel Neisseria gonorrhoeae lipooligosaccharide-mediated complement resistance and design novel therapeutics. PLoS Pathog. 11:e1005290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Lamont RJ. 2012. Beyond the red complex and into more complexity: The polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol. 27:409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseley SR, Wilkinson SG. 1997. Structural studies of the putative O-specific polysaccharide of Acinetobacter baumannii O24 containing 5,7-diamino-3,5,7,9-tetradeoxy-l-glycero-d-galacto-nonulosonic acid. Eur J Biochem. 250:617–623. [DOI] [PubMed] [Google Scholar]
- Holt SC, Ebersole JL. 2005. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: The “red complex”, a prototype polybacterial pathogenic consortium in periodontitis. Periodontol 2000. 38:72–122. [DOI] [PubMed] [Google Scholar]
- Honma K, Inagaki S, Okuda K, Kuramitsu HK, Sharma A. 2007. Role of a Tannerella forsythia exopolysaccharide synthesis operon in biofilm development. Microb Pathog. 42:156–166. [DOI] [PubMed] [Google Scholar]
- Horzempa J, Dean CR, Goldberg JB, Castric P. 2006. Pseudomonas aeruginosa 1244 pilin glycosylation: Glycan substrate recognition. J Bacteriol. 188:4244–4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard SL, Jagannathan A, Soo EC, Hui JP, Aubry AJ, Ahmed I, Karlyshev A, Kelly JF, Jones MA, Stevens MP, et al. 2009. Campylobacter jejuni glycosylation island important in cell charge, legionaminic acid biosynthesis, and colonization of chickens. Infect Immun. 77:2544–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaakoush NO, Deshpande NP, Wilkins MR, Raftery MJ, Janitz K, Mitchell H. 2011. Comparative analyses of Campylobacter concisus strains reveal the genome of the reference strain BAA-1457 is not representative of the species. Gut Pathog. 3:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon JJ, Marzaioli AM, De Castro C, Hall RM. 2015. 5,7-di-N-acetyl-acinetaminic acid: A novel non-2-ulosonic acid found in the capsule of an Acinetobacter baumannii isolate. Glycobiology. 25:644–654. [DOI] [PubMed] [Google Scholar]
- Kenyon JJ, Marzaioli AM, Hall RM, De Castro C. 2014. Structure of the K2 capsule associated with the KL2 gene cluster of Acinetobacter baumannii. Glycobiology. 24:554–563. [DOI] [PubMed] [Google Scholar]
- Kiss E, Kereszt A, Barta F, Stephens S, Reuhs BL, Kondorosi A, Putnoky P. 2001. The rkp-3 gene region of Sinorhizobium meliloti Rm41 contains strain-specific genes that determine K antigen structure. Mol Plant Microbe Interact. 14:1395–1403. [DOI] [PubMed] [Google Scholar]
- Knirel YA, Helbig JH, Zähringer U. 1996. Structure of a decasaccharide isolated by mild acid degradation and dephosphorylation of the lipopolysaccharide of Pseudomonas fluorescens strain ATCC 49271. Carbohydr Res. 283:129–139. [DOI] [PubMed] [Google Scholar]
- Knirel YA, Rietschel ET, Marre R, Zähringer U. 1994. The structure of the O-specific chain of Legionella pneumophila serogroup 1 lipopolysaccharide. Eur J Biochem. 221:239–245. [DOI] [PubMed] [Google Scholar]
- Knirel YA, Shashkov AS, Tsvetkov YE, Jansson PE, Zähringer U. 2003. 5,7-diamino-3,5,7,9-tetradeoxynon-2-ulosonic acids in bacterial glycopolymers: Chemistry and biochemistry. Adv Carbohydr Chem Biochem. 58:371–417. [DOI] [PubMed] [Google Scholar]
- Knirel YA, Vinogradov EV, L'Vov V L, Kocharova NA, Shashkov AS, Dmitriev BA, Kochetkov NK. 1984. Sialic acids of a new type from the lipopolysaccharides of Pseudomonas aeruginosa and Shigella boydii. Carbohydr Res. 133:C5–C8. [DOI] [PubMed] [Google Scholar]
- Lee SW, Sabet M, Um HS, Yang J, Kim HC, Zhu W. 2006. Identification and characterization of the genes encoding a unique surface (S-) layer of Tannerella forsythia. Gene. 371:102–111. [DOI] [PubMed] [Google Scholar]
- Leishman SJ, Do HL, Ford PJ. 2010. Cardiovascular disease and the role of oral bacteria. J Oral Microbiol. 21:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis AL, Desa N, Hansen EE, Knirel YA, Gordon JI, Gagneux P, Nizet V, Varki A. 2009. Innovations in host and microbial sialic acid biosynthesis revealed by phylogenomic prediction of nonulosonic acid structure. Proc Natl Acad Sci U S A. 106:13552–13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Perepelov AV, Wang Q, Senchenkova SN, Liu B, Shevelev SD, Guo X, Shashkov AS, Chen W, Wang L, et al. 2010. Structural and genetic characterization of the O-antigen of Escherichia coli O161 containing a derivative of a higher acidic diamino sugar, legionaminic acid. Carbohydr Res. 345:1581–1587. [DOI] [PubMed] [Google Scholar]
- Logan SM, Hui JP, Vinogradov E, Aubry AJ, Melanson JE, Kelly JF, Nothaft H, Soo EC. 2009. Identification of novel carbohydrate modifications on Campylobacter jejuni 11168 flagellin using metabolomics-based approaches. FEBS J. 276:1014–1023. [DOI] [PubMed] [Google Scholar]
- Lowry RC, Parker JL, Kumbhar R, Mesnage S, Shaw JG, Stafford GP. 2014. The Aeromonas caviae AHA0618 gene modulates cell length and influences swimming and swarming motility. Microbiologyopen. doi:1002/mbo3.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean LL, Vinogradov E, Pagotto F, Perry MB. 2011. Characterization of the lipopolysaccharide O-antigen of Cronobacter turicensis HPB3287 as a polysaccharide containing a 5,7-diacetamido-3,5,7,9-tetradeoxy-d-glycero-d-galacto-non-2-ulosonic acid (legionaminic acid) residue. Carbohydr Res. 346:2589–2594. [DOI] [PubMed] [Google Scholar]
- McNally DJ, Aubry AJ, Hui JP, Khieu NH, Whitfield D, Ewing CP, Guerry P, Brisson JR, Logan SM, Soo EC. 2007. Targeted metabolomics analysis of Campylobacter coli VC167 reveals legionaminic acid derivatives as novel flagellar glycans. J Biol Chem. 282:14463–14475. [DOI] [PubMed] [Google Scholar]
- McNally DJ, Schoenhofen IC, Houliston RS, Khieu NH, Whitfield DM, Logan SM, Jarrell HC, Brisson JR. 2008. CMP-pseudaminic acid is a natural potent inhibitor of PseB, the first enzyme of the pseudaminic acid pathway in Campylobacter jejuni and Helicobacter pylori. ChemMedChem. 3:55–59. [DOI] [PubMed] [Google Scholar]
- Megson ZA, Koerdt A, Schuster H, Ludwig R, Janesch B, Frey A, Naylor K, Wilson IB, Stafford GP, Messner P, et al. 2015. Characterization of an α-l-fucosidase from the periodontal pathogen Tannerella forsythia. Virulence. 6:282–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison MJ, Imperiali B. 2014. The renaissance of bacillosamine and its derivatives: Pathway characterization and implications in pathogenicity. Biochemistry. 53:624–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myneni SR, Settem RP, Connell TD, Keegan AD, Gaffen SL, Sharma A. 2011. TLR2 signaling and Th2 responses drive Tannerella forsythia-induced periodontal bone loss. J Immunol. 187:501–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier NB, Chen MM, Behr JR, Imperiali B. 2006. In vitro biosynthesis of UDP-N,N′-diacetylbacillosamine by enzymes of the Campylobacter jejuni general protein glycosylation system. Biochemistry. 45:13659–13669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posch G, Pabst M, Brecker L, Altmann F, Messner P, Schäffer C. 2011. Characterization and scope of S-layer protein O-glycosylation in Tannerella forsythia. J Biol Chem. 286:38714–38724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangarajan ES, Proteau A, Cui Q, Logan SM, Potetinova Z, Whitfield D, Purisima EO, Cygler M, Matte A, Sulea T, et al. 2009. Structural and functional analysis of Campylobacter jejuni PseG: A UDP-sugar hydrolase from the pseudaminic acid biosynthetic pathway. J Biol Chem. 284:20989–21000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Honma K, Douglas CW, Sharma A, Stafford GP. 2011. Role of sialidase in glycoprotein utilization by Tannerella forsythia. Microbiology. 157:31953–31202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Phansopa C, Stafford P, Honma K, Douglas CW, Sharma A, Stafford GP. 2012. Beta-hexosaminidase activity of the oral pathogen Tannerella forsythia influences biofilm formation on glycoprotein substrates. FEMS Immunol Med Microbiol. 65:116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara J, Nagano K, Murakami Y, Higuchi N, Nakamura H, Shimozato K, Yoshimura F. 2007. Loss of adherence ability to human gingival epithelial cells in S-layer protein-deficient mutants of Tannerella forsythensis. Microbiology. 153:866–876. [DOI] [PubMed] [Google Scholar]
- Sára M, Sleytr UB. 2000. S-layer proteins. J Bacteriol. 182:859–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäffer C, Messner P. 2004. Surface-layer glycoproteins: An example for the diversity of bacterial glycosylation with promising impacts on nanobiotechnology. Glycobiology. 14:31R–42R. [DOI] [PubMed] [Google Scholar]
- Schäffer C, Messner P. Emerging facets of prokaryotic glycosylation. FEMS Microbiol Rev. doi:10.1033/femsre/fuw036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirm M, Soo EC, Aubry AJ, Austin J, Thibault P, Logan SM. 2003. Structural, genetic and functional characterization of the flagellin glycosylation process in Helicobacter pylori. Mol Microbiol. 48:1579–1592. [DOI] [PubMed] [Google Scholar]
- Schoenhofen IC, Lunin VV, Julien JP, Li Y, Ajamian E, Matte A, Cygler M, Brisson JR, Aubry A, Logan SM, et al. 2006. a. Structural and functional characterization of PseC, an aminotransferase involved in the biosynthesis of pseudaminic acid, an essential flagellar modification in Helicobacter pylori. J Biol Chem. 281:8907–8916. [DOI] [PubMed] [Google Scholar]
- Schoenhofen IC, McNally DJ, Brisson JR, Logan SM. 2006. b. Elucidation of the CMP-pseudaminic acid pathway in Helicobacter pylori: Synthesis from UDP-N-acetylglucosamine by a single enzymatic reaction. Glycobiology. 16:8C–14C. [DOI] [PubMed] [Google Scholar]
- Schoenhofen IC, McNally DJ, Vinogradov E, Whitfield D, Young NM, Dick S, Wakarchuk WW, Brisson JR, Logan SM. 2006. c. Functional characterization of dehydratase/aminotransferase pairs from Helicobacter and Campylobacter: Enzymes distinguishing the pseudaminic acid and bacillosamine biosynthetic pathways. J Biol Chem. 281:723–732. [DOI] [PubMed] [Google Scholar]
- Schoenhofen IC, Vinogradov E, Whitfield DM, Brisson JR, Logan SM. 2009. The CMP-legionaminic acid pathway in Campylobacter: Biosynthesis involving novel GDP-linked precursors. Glycobiology. 19:715–725. [DOI] [PubMed] [Google Scholar]
- Scott NE, Kinsella RL, Edwards AV, Larsen MR, Dutta S, Saba J, Foster LJ, Feldman MF. 2014. Diversity within the O-linked protein glycosylation systems of acinetobacter species. Mol Cell Proteomics. 13:2354–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekot G, Posch G, Messner P, Matejka M, Rausch-Fan X, Andrukhov O, Schäffer C. 2011. Potential of the Tannerella forsythia S-layer to delay the immune response. J Dent Res. 90:109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekot G, Posch G, Oh YJ, Zayni S, Mayer HF, Pum D, Messner P, Hinterdorfer P, Schäffer C. 2012. Analysis of the cell surface layer ultrastructure of the oral pathogen Tannerella forsythia. Arch Microbiol. 194:525–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekot G, Schuster D, Messner P, Pum D, Peterlik H, Schäffer C. 2013. Small-angle X-ray scattering for imaging of surface layers on intact bacteria in the native environment. J Bacteriol. 195:2408–2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settem RP, Honma K, Nakajima T, Phansopa C, Roy S, Stafford GP, Sharma A. 2013. A bacterial glycan core linked to surface (S)-layer proteins modulates host immunity through Th17 suppression. Mucosal Immunol. 6:415–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settem RP, Honma K, Sharma A. 2014. Neutrophil mobilization by surface-glycan altered Th17-skewing bacteria mitigates periodontal pathogen persistence and associated alveolar bone loss. PLoS One. 9:e108030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A. 2010. Virulence mechanisms of Tannerella forsythia. Periodontol 2000. 54:106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL Jr. 1998. Microbial complexes in subgingival plaque. J Clin Periodontol. 25:134–144. [DOI] [PubMed] [Google Scholar]
- Stafford GP, Chaudhuri RR, Haraszthy V, Friedrich V, Schäffer C, Ruscitto A, Honma K, Sharma A in preparation. Draft genome sequences of three clinical isolates of Tannerella forsythia isolated from sub-gingival plaque from periodontitis patients in the USA. Genome Announc. 4:pii: e01286–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson HN, Mills DC, Jones H, Milioris E, Copland A, Dorrell N, Wren BW, Crocker PR, Escors D, Bajaj-Elliott M. 2014. Pseudaminic acid on Campylobacter jejuni flagella modulates dendritic cell IL-10 expression via Siglec-10 receptor: A novel flagellin-host interaction. J Infect Dis. 210:1487–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner ACR, Listgarten MA, Ebersole JL, Strezempko MN. 1986. Bacteroides forsythus sp. nov., a slow-growing, fusiform Bacteroides sp. from the human oral cavity. Int J Syst Bacteriol. 36:213–221. [Google Scholar]
- Thibault P, Logan SM, Kelly JF, Brisson JR, Ewing CP, Trust TJ, Guerry P. 2001. Identification of the carbohydrate moieties and glycosylation motifs in Campylobacter jejuni flagellin. J Biol Chem. 276:34862–34870. [DOI] [PubMed] [Google Scholar]
- Tomek MB, Neumann L, Nimeth I, Koerdt A, Andesner P, Messner P, Mach L, Potempa JS, Schäffer C. 2014. The S-layer proteins of Tannerella forsythia are secreted via a type IX secretion system that is decoupled from protein O-glycosylation. Mol Oral Microbiol. 29:307–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traving C, Schauer R. 1998. Structure, function and metabolism of sialic acids. Cell Mol Life Sci. 54:1330–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twine SM, Paul CJ, Vinogradov E, McNally DJ, Brisson JR, Mullen JA, McMullin DR, Jarrell HC, Austin JW, Kelly JF, et al. 2008. Flagellar glycosylation in Clostridium botulinum. FEBS J. 275:4428–4444. [DOI] [PubMed] [Google Scholar]
- Varki A. 1992. Diversity in the sialic acids. Glycobiology. 2:25–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A, Cummings RD, Esko JD, Hudson FH, Stanley P, Bertozzi CR, Hart GW, Etzler ME. 2009. Essentials of glycobiology, 2nd ed Cold Spring Harbor (NY), Cold Spring Harbor Laboratory Press. [PubMed] [Google Scholar]
- Vinogradov E, Wilde C, Anderson EM, Nakhamchik A, Lam JS, Rowe-Magnus DA. 2009. Structure of the lipopolysaccharide core of Vibrio vulnificus type strain 27562. Carbohydr Res. 344:484–490. [DOI] [PubMed] [Google Scholar]
- Vinogradov EV, Shashkov AS, Knirel YA, Kochetkov NK, Dabrowski J, Grosskurth H, Stanislavsky ES, Kholodkova EV. 1992. The structure of the O-specific polysaccharide chain of the lipopolysaccharide of Salmonella arizonae O61. Carbohydr Res. 231:1–11. [DOI] [PubMed] [Google Scholar]
- Wilhelms M, Fulton KM, Twine SM, Tomas JM, Merino S. 2012. Differential glycosylation of polar and lateral flagellins in Aeromonas hydrophila AH-3. J Biol Chem. 287:27851–27862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarschler K, Janesch B, Zayni S, Schäffer C, Messner P. 2009. Construction of a gene knockout system for application in Paenibacillus alvei CCM 2051T, exemplified by the S-layer glycan biosynthesis initiation enzyme WsfP. Appl Environ Microbiol. 75:3077–3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zunk M, Kiefel MJ. 2014. The occurrence and biological significance of the alpha-keto-sugars pseudaminic acid and legionaminic acid within pathogenic bacteria. RSC Adv. 4:3413–3421. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.