Abstract
Mechanistic understanding of how the brain gives rise to complex behavioral and cognitive functions is one of science’s grand challenges. The technical challenges that we face as we attempt to gain a systems-level understanding of the brain are manifold. The brain’s structural complexity requires us to push the limit of imaging resolution and depth, while being able to cover large areas, resulting in enormous data acquisition and processing needs. Furthermore, it is necessary to detect functional activities and ‘map’ them onto the structural features. The functional activity occurs at multiple levels, using electrical and chemical signals. Certain electrical signals are only decipherable with sub-millisecond timescale resolution, while other modes of signals occur in minutes to hours. For these reasons, there is a wide consensus that new tools are necessary to undertake this daunting task. Optical techniques, due to their versatile and scalable nature, have great potentials to answer these challenges. Optical microscopy can now image beyond the diffraction limit, record multiple types of brain activity, and trace structural features across large areas of tissue. Genetically encoded molecular tools opened doors to controlling and detecting neural activity using light in specific cell types within the intact brain. Novel sample preparation methods that reduce light scattering have been developed, allowing whole brain imaging in rodent models. Adaptive optical methods have the potential to resolve images from deep brain regions. In this roadmap article, we showcase a few major advances in this area, survey the current challenges, and identify potential future needs that may be used as a guideline for the next steps to be taken.
Keywords: neurophotonics, brain, imaging, biophotonics, microscopy, neuroscience, spectroscopy
1. Introduction
Yong Ku Cho and Guoan Zheng
University of Connecticut
The BRAIN initiative, an acronym for ‘Brain Research through Advancing Innovative Neurotechnologies’, was launched in 2013 [1]. As the name suggests, the project is heavily driven by the tools available to study the brain. Over the past two years, the goal of the project crystallized into the following focal areas [2]: (1) cataloguing all cell types in the brain and identifying their roles in brain health and disease; (2) generating a structural map of the brain at length scales from synapses to the whole brain; (3) capturing the functional dynamics of the brain activity at a large scale; (4) demonstrating the causality of neural circuit dynamics in behavior; (5) identifying theoretical principles of the basis of mental processes; and (6) developing tools to understand the human brain and its disorders. Overall, the project goals listed above aim to understand how the dynamics of neural activity is transformed into the human cognition, emotion, and behavior.
Optical techniques are in a unique and central position in addressing these ambitious goals. Optical methods are scalable, allowing the analysis of sub-cellular compartments to the whole brain. They are also highly versatile, enabling the detection of essentially any signal that can be converted to light. Moreover, they provide high temporal accuracy, which is essential in deciphering the millisecond-timescale signals that neuronal cells utilize. For a long time, the field of neuroscience has benefited from optical imaging approaches by utilizing chemically synthesized dyes that report the dynamics of signaling queues tied closely to neuronal and glial function. Optical approaches have also been central in studying the vasculature and energetics of the central nervous system. However, the capability of optical techniques has been greatly expanded in recent years. Over the past decade, several technical breakthroughs using light have provided us with unprecedented abilities to study the brain. This roadmap article is an effort to highlight these new developments, brainstorm their full potential, and project future needs in analyzing the brain.
The first set of sections addresses the development of genetically encoded tools (proteins expressed in genetically defined population of cells) that enable cell-type specific imaging and manipulation of neural activity. These techniques are referred to as optogenetics. As emphasized by Augustine (section 2), this relatively new development provides the ability to functionally map the circuit activity with single cell resolution, complementing existing low resolution approaches. In order to take full advantage of optogenetics, it is critical to develop proteins with optimal performance. Section 3 by Hochbaum and Cohen focuses on optogenetic tools that function as reporters of neuronal activity, while the following section by Knöpfel (section 4) covers tools to control circuit activity. These tools can be expressed in genetically defined cell types in the intact brain, to monitor and manipulate a specific cell population among the myriad of cells, all with high temporal and spatial resolution. The optogenetic tools need to be accompanied by ways to interface the brain with light. Section 5 by Pisanello discusses micro- and nano-fabricated optical interfaces that can be used to manipulate and monitor neural activity.
The next set of sections discusses approaches to overcome the critical issue of light scattering in the brain. Section 6 by Pavone covers new methods to process brain tissue samples to reduce light scattering. Section 7 by Vellekoop discusses the use of wavefront shaping for deep-tissue imaging. In this case, the imaging depth is not limited by the exponential decrease of ballistic light; instead, it is determined by the collection and control of scattering components in turbid tissue. The following section by Booth (section 8) discusses the recent developments in the field of adaptive optics (AO). Similar to wavefront shaping, AO will play an important role in tackling challenges in deep-tissue fluorescence and multi-photon imaging. Section 9 by Hu focuses on photoacoustic imaging (PAI) techniques that can operate at distinct spatiotemporal scales. It is pointed out that, two major embodiments—photoacoustic microscopy (PAM) and photoacoustic computed tomography (PACT) can operate across the optical ballistic and diffusive regimes. Section 10 by Zhu and Chen discusses the recent progresses in the field of optical coherent tomography. This technology is able to provide information on tissue structure, vascular and neuronal dynamics as well as other physiological information simultaneously, making it a powerful tool for brain imaging. Finally, section 11 by Hoshi covers the near-infrared spectroscopy (NIRS) technology, where diffused optical tomography can be used for image reconstruction in the photon-diffusive regime.
Even though we attempted to gain a meaningful snapshot of current technologies relevant in this area, by no means the approaches and techniques covered here are complete. The editors are responsible for any major developments that are not included here, and emphasize that these sections are intended to create a constructive dialogue in the optics community, and to break existing barriers that we might be currently bound by.
2. Functional brain mapping via optogenetics
George J Augustine
Nanyang Technological University
Status
The brain processes and stores information via its synaptic circuits. In the human brain there are approximately 100 billion neurons and 100 trillion synaptic connections. This enormous complexity makes it very challenging to understand how the brain works.
Several well-publicized efforts are addressing this challenge by defining the ‘connectome’ of the brain. These efforts are largely focused on determining the number and spatial organization of synaptic connections, defined either by their appearance in the electron microscope [3, 4], their labeling with immuno-cytochemical markers of synapses [5], or by modern updates on classical pathway-tracing approaches [6]. While such efforts are valuable, ultimately they cannot answer questions about brain function because they yield only structural information.
Until recently, functional mapping of brain circuitry could only be achieved at the ‘mesoscopic’ scale, namely at the level of connections between different brain areas. This has been done via electro-physiological analysis of neuronal activity [7] or by functional imaging approaches such as MRI [8] or intrinsic signal imaging [9]. Such efforts have been highly informative, but ultimately do not offer sufficient precision or spatial resolution to tell us how information is processed at the level of discrete synaptic circuits or neuronal populations. It is clear that this higher level of resolution will be required to comprehend how neural circuits process information and generate behavior.
Here I advocate the use of optogenetic technology as a powerful and promising means of mapping the functional connectome of the brain. Optogenetics is based on the use of genetically-targetable optical probes, such as the light-sensitive cation channel channelrhodopsin-2 (ChR2), that photosensitizes neurons and thereby allows them to be stimulated when exposed to light [10; see also section 4 below].
Current and future challenges
The key benefit of optogenetics for mapping brain circuits is cellular specificity: by using cell-specific promoters to target expression of optogenetic probes exclusively to a genetically defined population of neurons, it is possible to side-step much of the cellular complexity of the brain. The experiment shown in figure 1 demonstrates this advantage and illustrates how it can be leveraged to map the functional organization of brain circuits with high resolution.
Figure 1.
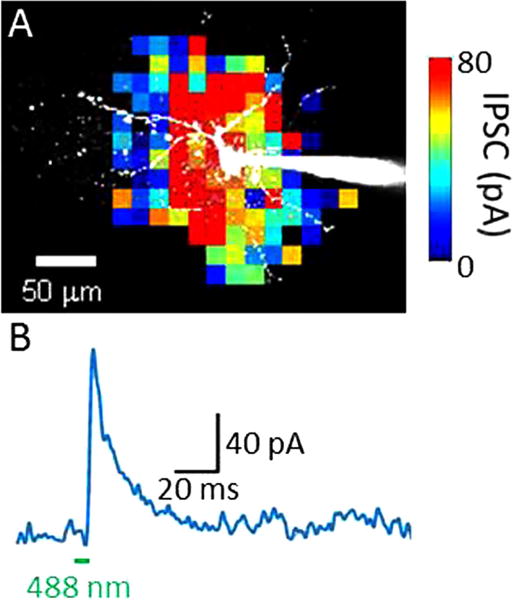
Optogenetic mapping of local inhibitory circuit in mouse claustrum. (A) Patch clamp recording from postsynaptic projection neuron (white). Colored pixels indicate laser spot locations that evoked inhibitory postsynaptic currents (IPSCs); IPSC amplitude indicated by pseudocolor scale shown at lower left. (B) Example of IPSC evoked by 488 nm light spot at time indicated by green bar. Unpublished results of Dr Yanxia Tang.
In this experiment, a slice of tissue was prepared from the brain of a transgenic mouse genetically engineered to express ChR2 in interneurons within the claustrum, a part of the brain that may be involved in higher-order brain functions. The ChR2 allowed these neurons to be selectively photostimulated by small spots of laser light (488 nm). While scanning the laser light spot throughout the brain slice, a patch-clamp pipette (at right in figure 1(A)) was used to detect postsynaptic electrical responses in a projection neuron (white cell) within the claustrum. Whenever the scanned laser spot was positioned over the cell body of a ChR2-expressing interneuron that innervated the projection neuron, an inhibitory postsynaptic current (IPSC) could be observed in the projection neuron (figure 1(B)). This defines the function of the circuit: in this case, it is inhibitory. By correlating the location of the light spot with the amplitude of the IPSC evoked at that location (colored pixels in figure 1(A)), it was possible to generate a map of the spatial distribution of presynaptic interneurons innervating the postsynaptic projection neuron.
The significance of this result is that it provides novel information about the spatial organization of this circuit while simultaneously defining the function of the circuit, thereby neatly merging function with structure. With suitable calibration, it can also determine the number of presynaptic neurons converging on a given postsynaptic target [11].
This optogenetic circuit mapping approach has many advantages. First, it is the only approach that provides both structural and functional information at a cellular level of resolution, a level of resolution required to advance our understanding of brain function beyond what has already been provided by MRI and related imaging approaches. In addition, this approach is very general and can be applied to any circuit within the brain. It is particularly valuable for mapping local circuits, though it can also be applied to long-range circuits as well. Also important is its high speed: this approach is approximately 10 000 times faster than would be possible with conventional electro-physiological measurements [11].
While representing a very substantial improvement over more conventional techniques, optogenetic circuit mapping is constrained by the need for patch clamp recording to detect postsynaptic responses. Learning patch-clamping skills is time-consuming and experimental implementation is slow and tedious.
Advances in science and technology to meet challenges
A further substantial improvement in the throughput of functional circuit mapping will come by applying technologies that bypass electrophysiology altogether. In particular, optical technologies for imaging neuronal activity seem the best way forward. Existing cell-level imaging technologies—such as voltage-sensitive dyes, fluorescent calcium reporters—are already starting to be applied for such purposes. However, optogenetic sensors of membrane potential will be the best option for detecting postsynaptic activity. Like all optogenetic approaches, these benefit from the advantages of cellular targeting strategies and major strides are being made in the development of such probes [12; see also section 3 below]. Focusing future development on optimizing optogenetic voltage sensors for detection of subthreshold postsynaptic responses will make it possible to combine optogenetic photostimulation of defined presynaptic neurons with optogenetic identification of their postsynaptic actions in defined targets.
Having suitable optogenetic sensors will greatly accelerate mapping of the brain functional connectome at the cellular level of resolution. This, in turn, will catalyze development of automated systems for such all-optogenetic functional mapping, further accelerating progress. Bioinformatics will then need to tackle challenges such as defining what sort of information is required and how such information will be processed and assimilated. The final challenge will be at the policy level, namely deciding how to best utilize this new information. Resolution of the functional connectome of the brain not only will help us to understand how the brain works, but also will have considerable value for commercial applications such as medical diagnostics and therapeutics.
Concluding remarks
In conclusion, all-optogenetic mapping of the functional connectome of the brain has tremendous potential and requires only a few technological advances to fully deliver on its promise.
3. Genetically encoded optical sensors of brain function
Daniel Hochbaum1 and Adam Cohen1,2
1Harvard University
2Howard Hughes Medical Institute
Status
We dream of peering into the brain of a mouse or a rat to see individual neurons receiving inputs, signaling to each other and then, collectively, inducing behavior. Neuronal activity is encoded in dynamic fluctuations of the transmembrane potential: subthreshold events and action potentials. Thus we need to see membrane voltage, or a proxy. The voltages range from ~1 mV (a postsynaptic potential from a single vesicle release) to 100 mV (an action potential), and over times from 100 μs (axonal conduction) to 10 s (plateau potentials). The brain contains hundreds of neuronal subtypes (nobody knows precisely how many) with distinct functions and patterns of gene expression. A mouse brain contains approximately 7.5 × 107 neurons in a volume of ~0.5 ml. While the goal of recording all activity in all neurons is still years away, recent progress has brought us closer.
Genetically encoded optical sensors are a promising route toward this goal. The most highly developed are genetically encoded calcium indicators (GECIs), with the GCaMP6 family most widely used [13]. Action potentials trigger an increase in intracellular calcium concentration from an initial 50–100 nM to a peak concentration of 5–10 μM, which modulates the GECI fluorescence. Recent innovations include calcium integrators which report cumulative calcium activity during an illumination-gated window [14]; and red-shifted indicators which might enable multiplex imaging or combination with optogenetic stimulation [15].
GECI fluorescence is a good proxy for mean action potential firing rate, but GECIs miss many aspects of neuronal activity. Calcium transients are ~100-fold slower than the underlying electrical waveforms. Most sub-threshold changes in membrane potential do not elicit changes in calcium concentration (figure 2). Thus a longstanding challenge has been to develop a robust genetically encoded voltage indicator (GEVI). Voltage imaging in vivo has been limited by the modest sensitivity of existing GEVIs, and the technical challenges associated with kHz frame-rate imaging in scattering tissues.
Figure 2.
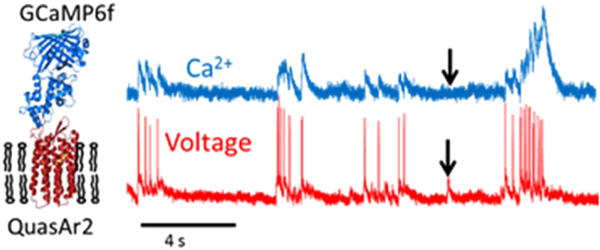
Comparison of fluorescence signals recorded simultaneously from a GECI, GCaMP6f, and a GEVI, QuasAr2, expressed as a fusion construct in a rat hippocampal neuron. Sub-threshold depolarizations, indicated by the arrows, do not have a correlate in the Ca2+ signal. Rapid spike trains manifest as a broad increase in Ca2+ concentration.
Existing GEVIs are based either on fusions of fluorescent proteins (FPs) to voltage sensitive transmembrane domains [16]; or on microbial rhodopsins which position a voltage-sensitive chromophore directly in the membrane [17]. Until recently, FP-based GEVIs suffered from either low time resolution or low sensitivity, though the recently reported ASAP1 protein has good performance by both measures [16]. However, photobleaching remains a challenge for all FP-based GEVIs. Rhodopsin-based GEVIs have sub-millisecond response times and excellent voltage sensitivity, yet they are dim, requiring ~30-fold more intense illumination than FP-based GEVIs.
Current and future challenges
One would like a palette of GEVIs that are bright, photostable, respond quickly and sensitively to voltage changes, traffic efficiently to the plasma membrane in most cell types, and come in a wide range of colors. These improvements may come from modifications to existing GEVI scaffolds, but advances attained in this way are likely to be incremental.
It will also be important to screen novel GEVI backbones, which could have dramatically different (and better) properties. Any transmembrane protein could, in principle, undergo voltage-dependent motions which could modulate an attached chromophore. There is also merit in exploring transmembrane proteins with naturally associated chromophores, including genes from the electron transport chain and plant photosynthetic proteins.
Optical sensors will also need to be optimized for specialized functions. These include:
Absolute measurements. Existing GECIs and GEVIs report relative level changes, but not absolute Ca2+ concentration or absolute voltage. Uncontrolled variations in fluorescence background and reporter concentration prevent direct calibration. Nascent efforts to encode information in temporal dynamics or ratiometric signals show promise but require further work.
Integrators. No existing data recording system could handle kilohertz rate recordings from all 7.5 × 107 neurons in a mouse brain. Light-gated activity reporters promise to deliver a snapshot of this activity during a defined time interval. Early calcium integrators [14] and voltage integrators [18] show promise. Variants of these molecules which convert the signal into activation of gene expression will be particularly powerful because one could then modify the function of neurons that were activated at a particular moment.
Multiple colors. GEVIs, GECIs, and other functional reporters are needed in a wide range of colors. Far red-shifted reporters will be particularly important to avoid autofluorescence, phototoxicity, and scatter in brain tissue.
More modalities. Novel genetically encoded reporters are also needed for neurotransmitters and neuropeptides, which play crucial modulatory roles in the nervous system. Reporters for glutamate [19] show promise. Much can be learned from reporters of vesicle release [20], but measures of vesicle content would be even more useful.
Advances in science and technology to meet challenges
At present we cannot predict the effects of mutations on GEVI function, so progress is largely through trial and error, with low-throughput manual characterization of GEVI mutants. We need high-throughput, multi-parameter screens. Mammalian cell lines capable of mimicking action potential transients provide a possible substrate for such a screen [21]. Technologies for in situ identification of novel mutants or scaffolds within a mixed population would further increase screening throughput. Ultimately this work will require integration of novel hardware and software with protein engineering.
An often-underappreciated aspect of GEVI performance is the requirement that the protein traffic to the plasma membrane. Quantitative assessments of membrane trafficking will assist in engineering efforts. Targeting GEVIs to different sub-cellular compartments will be useful. For example, neurons in the brain are often packed tightly and surrounded by neuropil whose fluorescence contributes background noise. Localized trafficking of GEVIs to somatic plasma membranes may allow for highly multiplexed measurements within large networks.
Advances in imaging must accompany advances in reporter technology. Two-photon raster scan imaging is widely applied for GECI imaging. One can record at cellular resolution from up to ~1000 neurons at near video frame rates (10–30 Hz), and whole-brain imaging has been demonstrated in the worm, fly larva, and fish. But this time resolution is inadequate for GEVI imaging. Development of imaging systems that can achieve kHz frame rates, over hundreds of cells, in tissue, remains an unresolved challenge. 2P illumination with sufficient intensity to elicit kHz-resolvable signals results in rapid photobleaching of all known FP-based GEVIs, and rhodopsin-based GEVIs are too dim for 2P imaging. Simply increasing the scan rate and laser power of 2P imaging systems will not address the photobleaching problem.
Thus there is a need to explore alternate approaches to depth-resolved imaging in tissue. Optical sectioning can be provided by one-photon light sheet microscopy or structured illumination techniques. Photothermal, photoacoustic, and optical coherence tomographies are possible approaches, but it is not yet clear how to produce activity-dependent contrast for these methods.
Concluding remarks
As genetically encoded sensors and the instruments to probe them improve, we will require better data analysis algorithms. Existing instruments for optical electrophysiology [17] or volumetric calcium imaging [22] generate data at ~1 GByte s−1. Segmenting these movies to identify cells (figure 3), and then interpreting the cellular dynamics, will require advances in signal processing and close collaboration with computational and experimental neuroscientists. Ultimately, advances in genetically encoded sensors of neural activity must be tightly coupled to the instrumentation and the software for all parts to function harmoniously.
Figure 3.
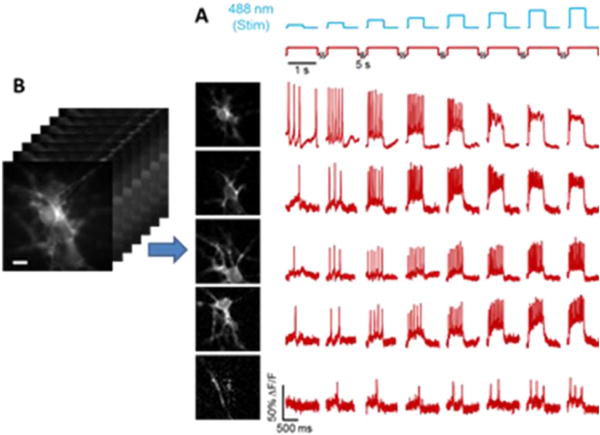
Activity-based image segmentation. (A) Neurons expressing a channelrhodopsin, CheRiff, and a GEVI, QuasAr2, are stimulated with pulses of blue light and imaged with red excitation and near infrared fluorescence. (B) The cells form a clump, but pixels associated with each cell vary in a unique temporal pattern, allowing a decomposition of the image into a sum of single-cell images, each with an associated firing pattern.
4. Optical control of neural activity
Thomas Knöpfel
Imperial College London
Status
Understanding how cognitive and emotional behavior emerges from the brain’s electrical activity is one of the ultimate goals of neuroscience. The experimental approach to decipher neuronal codes and algorithms is, in principle, simple: observe the neuronal activity, correlate it with brain function and deduce causality between activity and function by perturbation of the presumed causal activity patterns.
To achieve the latter we require methods for activation and inhibition of specific neurons and of circuits formed by their synaptic connections. Electrical stimulation lacks spatial precision and—in densely packed brain tissue composed of heterogeneous cell populations—specificity. Optical stimulation methods capitalize on the relative ease of confining light delivery spatially and temporally (relevant scales are μm and ms) and, therefore, often outperform microelectrode-based techniques.
For many years neurophysiologists have used light to systematically manipulate neuronal activity using tools such as caged glutamate [24]. Optical control of neuronal activity has evolved earlier in nature to enable vision. Naturally evolved light-sensitive proteins, such as microbial opsins, can be expressed in neurons which then can be excited or inhibited by illumination [25, 26]. This approach involves methods from photonics, protein engineering and genetics and has been associated with the term optogenetics [27]. Optogenetic tools are proteins (or DNA coding for such proteins) designed for manipulating and monitoring neuronal circuits. This fast-growing toolbox consists of three classes: (1) actuators, proteins that transduce light into neuronal signals for manipulation (prominent representative: ChR2 that mediates an excitatory cation current when activated with blue light, figure 4 upper panel), (2) silencers, such as halorhodopsin (NpHR) that generates an inhibitory current when illuminated with yellow light, figure 4 lower panel, and (3) indicators, proteins that transduce neuronal activity into optical signals for monitoring (prominent representatives: GCaMP calcium indicators and VSFP voltage indicators) [28, 29]. The core experimental design, paraphrased as an ‘optogenetic approach’, utilizes light-based (‘optical’) interventions and recordings of natural neural activity in genetically-defined cell classes and connectivity-defined projections to elucidate the time-resolved role of specified pathways in mammalian behavior. The term genetically defined cell classes refers to the use of genetic methods to target the optogenetic tool (the expressed protein) to cells defined by their phenotype, activity patterns or long-range connections.
Figure 4.
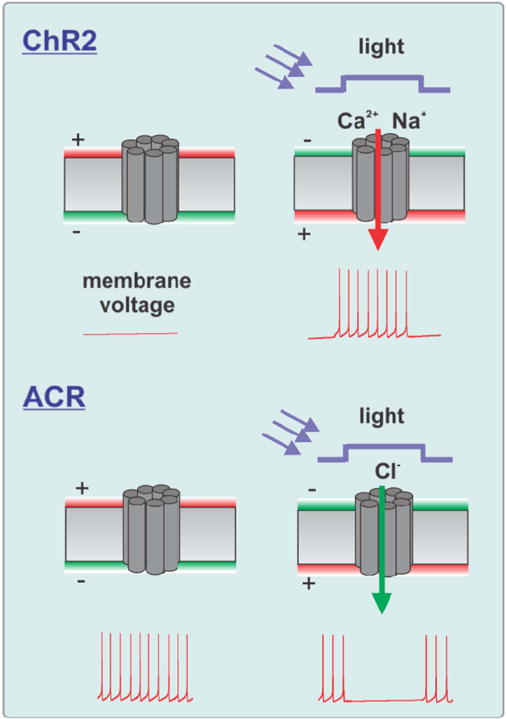
Genetically encoded and targetable actuator proteins that allow the use of light to either excite or inhibit action potential generation of neurons. Excitation and Inhibition is exemplified by the light-gated ion channel channelrhodopsin 2 (ChR2) and anion channel rhodopsin (ACR) [23].
Alternative emerging stimulation technologies include chemogenetics and magnetogenetics [30].
Current and future challenges
During recent years, optical methods have developed to enable neuronal actuation and monitoring at the single cell level in awake, task-performing animals. The number of neurons that can be simultaneously activated or inhibited and at the same time monitored is, however, many orders of magnitude smaller than the cells involved in cognitive and emotional functions of the brain.
Currently evolving methods seek to address this issue on two fronts: (i) development of instrumentation increasing the number of neurons controlled and monitored in cognitive and emotionally active experimental animals; and (ii) defining sets of neurons of similar (if not redundant) functional properties (encoding the same information) and treating them as a single functional unit.
Advances in science and technology to meet challenges
Further technical advances are need on several fronts:
- Refining the toolbox of optogenetic actuators
- Opsins that produce photocurrents with higher unitary conductance that respond faster to onset and offset of illumination.
- Opsins (or other genetically encoded light activated channels) with permeability more selective either for Ca2+, K+, Na+ or Cl−.
- Opsins (or other genetically encoded light activated channels) with increased two-photon cross-section.
- Opsins with more narrow absorption spectra, covering a larger spectral range, in particular in the infrared region.
- Opsins with activation spectra that do not overlap with the spectral range occupied by optogenetic monitoring approaches.
- Improved optical instrumentation
- Instruments for optogenetic monitoring in combination with two-photon activation of many (thousands) of individual opsin-expressing cells in living animals.
- Implantable micro-optoelectronic devices for activation of many individual opsin-expressing cells (or several genetically defined cell classes) in living animals.
- Refined genetic targeting
- Development of regulatory sequences (e.g. combination of naturally occurring sequences) that drive optogenetic tool expression in functionally homogenous subsets of neurons.
Along with technical advances, conceptual advances are needed, such as:
New concepts and associated analysis methods for brain activity data obtained while optogenetically exciting (or inhibiting) many individual opsin expressing cells (or several genetically defined cell classes) in living animals.
Concluding remarks
There are worldwide efforts towards improved optogenetic actuation technologies, indicating that the importance of these efforts is well recognized. Given the multidisciplinary and modular nature of these technologies, it will be important to define standards to interface components (e.g. optical instruments optimized for the same range of wavelength as optogenetic tools) and standards for benchmarking and comparing alternative technological solutions to the same problem (e.g. information content of data obtained with different imaging approaches). Standards in the format of experimental data are yet to be established. Rigorous open source dissemination of best technical solutions and experimental data would further advance the application of these exciting new technologies.
5. Micro- and nano-neurophotonics for next generation of optical neural interfaces
Ferruccio Pisanello
Istituto Italiano di Tecnologia
Status
In recent years the ability to optically control and simultaneously monitor neural activity in the mammalian brain has allowed an unprecedented picture of the brain in action. In particular, optogenetics [25]—the use of genetically encoded, optically-active proteins for manipulating or monitoring neural activity—has emerged as a powerful research tool. New optical approaches have in turn stimulated the development of new technologies to optically interface with the central nervous system [31]. New classes of implantable nano- and micro-photonic devices have emerged for investigating causal connections in brain microcircuits [32, 33], employing a combination of multipoint optical control and multipoint readout of brain activity. The first optrode for simultaneous optogenetic manipulation and electrical readout of neural activity was reported in 2007 [34], and much progress has been subsequently made toward increasing the density of stimulation/readout points. A promising strategy for multisite modulation is implanting multiple solid-state light sources directly into the brain region(s) of interest. For example, multiple micro light-emitting diodes (μLEDs) have been used for site-selective optical stimulation in neocortex [35], or have been integrated in multifunctional probes with electrodes for extracellular recording, temperature sensing, or microfluidic drug delivery (figures 5(A) and (B)) [36, 37]. Although these devices are convenient for operation in freely-moving animals, a major limitation is that the μLEDs can cause tissue heating with long illumination periods. To avoid this problem, an alternative approach is to have an external light source coupled to the brain with implanted waveguides, tethered to the animal with an extension fiber. Examples are multipoint-emitting tapered optical fibers [38] (figure 5(C)) and multifunctional optical fibers [39] (figure 5(D)), both of which can support multipoint extracellular recording of neural activity [38, 39].
Figure 5.
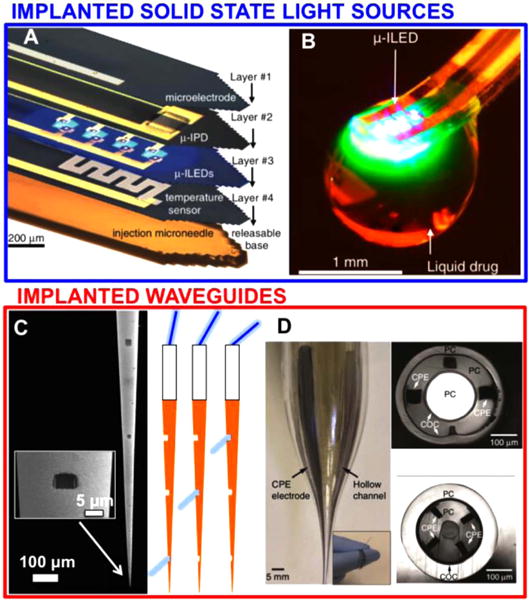
State of the art for user-selectable light delivery. Panels (A) and (B): implanted μLEDs with integrated functionalities. Panels (C) and (D): optical fibers for multiplipoint interface and/or integrated drug delivery, Panel (A), reproduced from [36], copyright 2015, with permission from Elsevier. Panel (B), from [37], reproduced with permission from AAAS. Panel (C), reproduced by permission from Macmillan Publishers Ltd: [39], copyright 2015.
Current and future challenges
For optical neural interfaces to reach their full potential, a crucial point will be the integration of multiple functionalities in a single implanted device (see schematic representation in figure 6). Such devices would allow simultaneous control and monitoring of neural activity in multiple brain regions, with spatial configurations customized to conform to specific experimental needs. Moreover, while recent work has focused on multi-point light delivery, new techniques are also needed for light collection, for multi-point monitoring of neural activity, including from deep-brain structures. Although these methods are still at their embryonic stage (e.g., flat-cleaved and large core optical fibers [40, 41]), they allow for cell-type specific readout when used in combination with genetically encoded fluorescent indicators of neural activity, such as Ca2+ indicators or voltage-sensitive dyes. The ability to both modulate and read out neural activity with cell-type specificity could provide powerful new approaches for investigating and manipulating brain function, such as closed-loop optogenetics [32].
Figure 6.
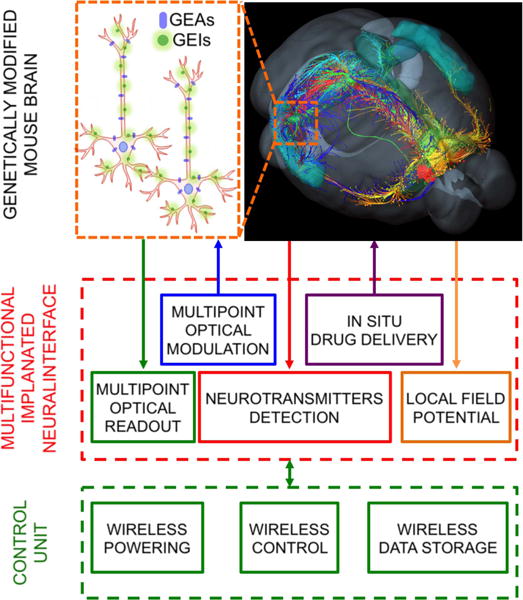
A schematic representation of the components of optogenetic neural interfaces and expected functionalities to be integrated in next devices generation.
Optical methods could also be applied to monitor in vivo release of neurotransmitters, in real time (tens to hundreds of ms). Combined with optical modulation of specific cell types, such an approach could help to unravel the complexity of chemical communication in brain circuits. A promising strategy for optical interrogation of released neurotransmitters is localized surface-plasmon enhanced Raman spectroscopy, which is compatible with both planar and fiber-optic based devices [42, 43].
Finally, optical neural interfaces could also be complemented with key non-optical components, such as microfluidics for drug delivery, or integrated electrodes for extracellular recording of neural activity. Integrated electrodes allow access to neuronal signals that currently cannot be measured optically (e.g., local field potentials).
Advances in science and technology to meet challenges
Current all-optical neural interfaces are constituted by three main components (figure 6): (i) an implant carrying nano- and micro-photonic devices into brain tissue; (ii) genetically encoded light-driven actuators (GEAs) and/or fluorescent indicators (GEIs) of neural activity; and (iii) a control/powering unit. Future advances in optical interfacing will involve improvements in all three components.
The photonic implant should integrate multiple functions, but with as small a profile as possible to minimize damage to brain tissue. Deep brain regions are especially difficult to access with currently available optical technologies, due to their blunt insertional cross-section and overall invasiveness. On the other hand, smaller optical devices generally deliver less light power and have lower efficiency for photon collection. Overcoming this trade-off will require the synergy of material science and optical and photonics engineering. For example, new methods are needed to multiplex and de-multiplex light used for excitation and detection, with the overall goal of reducing the number of implants. Furthermore, a key point for next negation of multifunctional devices will be their compatibility with existing high-resolution or whole brain imaging and stimulation techniques. Both two-photon microscopy [44, 45] and functional magnetic resonance imaging [46] will, indeed, greatly benefit of low-invasive endoscopes that can combine cell-type specificity to a set of discrete points to control and monitor neural activity in deep brain regions.
Cell-type specificity of optogenetic neural interfaces is obtained using GEAs and GEIs of neural activity (see sections 3 and 4, respectively). The optical efficiency of these reagents depends on the absorption cross-section of GEAs and the fluorescence intensity of GEIs. Increasing their efficiency will mitigate some of the issues discussed in point (i). Two recent steps in this direction are the introduction of ChR2-XXL for low-intensity applications [47] and the development of constructs for all-optical ‘electrophysiology’ [17]. At the same time, alternative methods to optogenetics are currently being investigated, for both optical control and optical readout of neural activity. Carvalho-de-Souza et al [48] recently proposed an interesting strategy to obtain nanoparticles-mediated photothermal neural stimulation. They succeeded in functionalizing gold nanoparticles on the cellular membrane, and used short pulses of green light resonant with the plasmonic band to induce local heating and therefore a variation of membrane capacitance, which lead to cell depolarization and action potentials generation. Marshal et al [49] instead proposed to exploit the dependence of the emission wavelength and fluorescence intensity of ultra-bright inorganic colloidal nanoparticles to optically monitor action potentials. Although this latter is, at the moment, only a theoretical proposal, nanoparticles-based methods can open new perspectives for next generation of neural activity indicators and actuators.
To link brain function to behavior, optical approaches should also be compatible with untethered and free-moving animal models. This will require the development of integrated wireless systems for spatiotemporal control of multiple stimuli (e.g., light and drug delivery), as well as methods for on-board storage of recorded signals for post processing. Experiments in feely moving animals would also be facilitated by the development of wireless powering systems, to mitigate the weight and undependability of on-board batteries [50, 51].
Concluding remarks
Application of nano- and micro-technologies have provided powerful new possibilities for optical neural interfaces. New implantable devices envision the integration of multiple functionalities in a single device that can simultaneously access multiple points of the brain. The next generation of optical neural interfaces will allow multipoint optical modulation and control of neural activity, as well as in situ drug delivery and specific monitoring of neurotransmitter release.
Acknowledgments
The author acknowledges helpful conversations with M De Vittorio, J A Assad, M Pisanello and L Sileo.
6. Novel sample preparation methods for brain imaging
Francesco S Pavone
University of Florence
Status
In the last few years a field called connectomics has aroused general interest. It consists of mapping each neuron at the level of synaptic connections through the whole brain. The structure of the nervous system is extraordinarily complicated since individual neurons are interconnected to hundreds or even thousands of other cells in networks that can extend over large volumes. Estimating the functional interactions between brain regions and mapping those connections are central pursuits for understanding how the brain works. A deeper comprehension of the brain would have an enormous impact for the whole society. Bridging the gap from the macro-scale to the micro-scale will allow the understanding of the neural basis of cognition, behavior and memory. Moreover, a complete reconstruction of the neuronal network will allow a better understanding of diseases and open the way to new treatments. In recent years, technological innovations have been proposed to achieve high resolution while allowing three-dimensional (3D) reconstruction of large tissue volume. The most common are confocal microscopy, multiphoton microscopy, and light sheet microscopy. All of them are optical techniques based on fluorescence detection to obtain a high contrast. Confocal and two-photon fluorescence microscopy (TPFM) can achieve, for example, very high resolution; however, to image large specimens, both of them need to be coupled with a serial sectioning of the tissue (STP) [53]. Light-sheet fluorescence microscopy (LSFM) [54], instead, does not need sample cutting because it uses optical sectioning at the price of lower resolution. In this case, the sample is illuminated with a thin sheet of light confined into the focal plane of the detection objective which collects the fluorescence emission along an axis that is perpendicular to the plane of illumination. All of these imaging techniques require a specific sample preparation, which should be adapted to the peculiar features of analyzed samples. Some of them require embedding the sample in hard resin [55] with specific fixation of the tissue [52]. Anyway, during the past few years, the technologies for making the tissue transparent to improve the imaging depth achievable with optical microscopy, such as LSM, have become very powerful.
Current and future challenges
Normally, biological tissues are opaque due to light scattering caused by the inhomogeneity of the refractive index (RI) distribution of macromolecules, which is different from the RI of water, surrounding them. To reduce the scattering and to make the tissue almost transparent, the RI has to be homogenized inside and outside the sample. During recent years, different approaches have been used to obtain optimal transparency. The oldest techniques involve the use of high RI organic solvents as benzyl alcohol/benzyl benzoate or dibenzylether [56]. These methods guarantee high transparency, but lead to tissue shrinking and fast protein fluorescence quenching, therefore preventing long-term measurement. Subsequently, other approaches have been introduced involving water-based optical clearing agents, such as SeeDB [57] and CUBIC [58]. Aqueous solutions improve fluorescence preservation, however, they present other non-negligible limitations such as long incubation times, structural alteration and incompatibility with immunostaining. In 2013, Chung et al [59] introduced a new approach based on tissue transformation called CLARITY. This method transforms tissue into a nano-porous, hydrogel-hybridized, lipid-free form. By removing membrane lipid bilayers, CLARITY allows high transparency and whole brain immunolabeling with structural and molecular preservation. This method, however, requires an expensive RI matching solution (FocusClear™) whose composition is unknown because of patent protection, limiting its practical applicability. Recently, a new clearing method was published [60]: it utilizes a simple, rapid and inexpensive clearing agent, the 2,2′-thiodiethanol (TDE). TDE does not quench protein fluorescence, is compatible with immunostaining and does not introduce tissue deformation at the sub-cellular level (table 1). Because of the non-viscous nature of the agent, it is a suitable agent to perform serial two-photon tomography, and has allowed a 3D reconstruction of the whole hippocampus of a fluorescent transgenic mouse, with high resolution and sensitivity (figure 7(a)). Furthermore, since the RI of TDE can be finely adjusted, it can also be used as an optical clearing medium for CLARITY allowing whole mouse brain imaging with LSFM (figure 7(b)). Finally, TDE clearing can be coupled with immunohistochemistry to stain large volume of human dysplastic brain tissue. It represents an innovative translational tool for fine characterization of macroscopic and microscopic circuit alterations in diseases.
Table 1.
Comparison of clearing techniques.
| Method | Clearing composition | Refractive index | Protein fluorescence quencing | Clearing capability | Immunostaining |
|---|---|---|---|---|---|
| Et + BABB | Ethanol Benzyl alcohol Benzyl Benzoate | 1.54 | Yes | Good | No |
| THF + DBE | Thetrahydrofuran Dibenzyl ether | 1.56 | Yes | Good | No |
| SeeDB | Fructose saturated α thioglycerol | 1.49 | No | Moderate | No |
| CUBIC | Urea Aminoalcohols | 1.47 | No | Good | Yes |
| CLARITY | Focus Cl ear™ | 1.45 | No | Good | Yes |
| TDE | 2,2′ Thiodioethanol | Tunable | No | Good | Yes |
Figure 7.
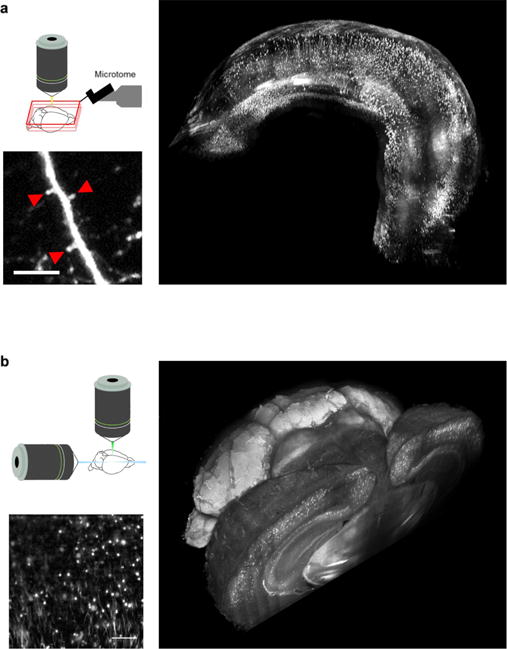
(a) Reconstruction of entire Thy1-GFP-M mouse hippocampus imaged with STP. Magnified insets scale bar = 10 μm. (b) Whole mouse parvalbumin-tdTomato brain tomography obtained with LSFM brain. High resolution insert scale bar = 100 μm. From Costantini et al [60].
Advances in science and technology to meet challenges
In the last few years great advances in fluorescent light microscopy have occurred. Different optical techniques are commercially available, leading to the necessity of having useful techniques for sample preparation. In particular, clearing procedures are crucial to obtain high transparency while preserving sample fluorescence. However, some methods are developed for a specific imaging technique, which therefore limits their applicability. A single imaging technique can reveal only a small part of the complex machinery that is the brain. To obtain a more comprehensive view of brain functionality complementary imaging approaches should be used. Versatile clearing techniques will make multi-modal acquisition available (such as correlative STP/EM, as demonstrated with the TDE [60]). Such approaches will also expand the application to mechanical and optical sectioning imaging and provide a novel and useful method for quantitative morphological analysis of the brain. Furthermore, the main goal for the future will be the standardization of clearing protocols, maybe through the use of commercial kit, to remove errors and obtain more reliable results. Moreover, it could be interesting to combine a clearing procedure with expansion microscopy [61]. This approach could allow to obtain tomographies of large samples with superresolution. Another big challenge concerns the use of fluorescent samples. Transgenic animal models are widely used to study brain structures. However, the use of genetically encoded fluorescent indicators is limited to a few animal models and requires complex genetic engineering procedures. To expand fluorescence microscopy to different types of samples, such as pathological models and human brains, it is necessary to develop new techniques for labeling. Antibodies and nucleic acids conjugated with fluorescent indicators may be used to access all different kinds of specimen. They should be optimized to better access epitopes deep in a sample in a short period of time. Finally, an important aspect that will require many efforts concerns the analysis of large datasets and the reconstruction of 3D images of whole specimen from raw data. It will be necessary to develop novel technologies for massive data collection, data sharing, and multimodal integration to achieve a global organization of the information.
Concluding remarks
Recently, different imaging methods have been developed to reconstruct neuronal connections in the brain. Several clearing protocols have been proposed to reduce light scattering, and achieve higher imaging depths and resolution during acquisition. Some of them combine different approaches, such as TPFM and LSFM, and make it possible to overcome the inherent limitations of the single technique, providing a more comprehensive and solid view of the brain. Together with advances in microscopy (new objectives with long working distance and high numerical aperture), computational analysis and sample labeling, tissue clearing contributes to enhance the understanding of the anatomic structure of the brain, facilitating the opening of new doors to many discoveries.
Acknowledgments
I would like to acknowledge Irene Costantini for the contribution to this paper.
7. The end of turbidity
Ivo M Vellekoop
University of Twente
Status
High-resolution optical imaging of neurons and their activity is currently only possible up to a depth of about a millimetre. Beyond this point, scattering prevents the formation of a sharp focus. As a consequence, neuroscientists either have to limit their attention to superficial structures, or resort to invasive techniques such as endoscopy, surgery, or even the use of acute brain slices.
Over recent decades, there has been an intense research effort to develop methods to look deeper while maintaining microscopic resolution. Highlights along this road include multiphoton microscopy, optical coherence tomography (OCT) (see section 10), PAM (section 9), chemical clearing protocols (section 6), and even animals that are genetically engineered for transparency.
So far, however, all microscopes have in common that they rely on non-scattered (ballistic) light for imaging. As the amount of ballistic light decreases exponentially with depth, looking deeper while maintaining microscopic resolution is exponentially hard. As a result, at depths exceeding ~1 mm, typically only mesoscopic imaging methods, such as NIRS (section 11) or computed photoacoustic tomography (section 9) can be used.
The exponential decrease of ballistic light has long been considered the fundamental limitation for microscopy. However, with the introduction of complex wavefront shaping [62] it became clear that scattering is not an insurmountable obstacle. By shaping the wavefront of the incident light with a spatial light modulator (SLM), light can be focused through or inside even the most turbid materials, and subsequently be used for imaging (see figures 8(a) and (b)).
Figure 8.
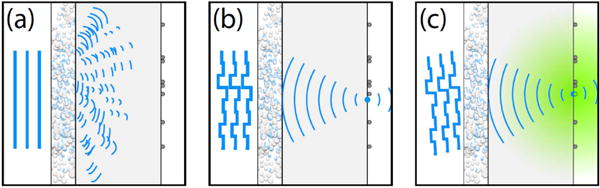
Principle of scattered light fluorescence microscopy (SLFM). (a) A turbid layer blocks a fluorescent structure from sight; all incident light is scattered. (b) The wavefront is shaped with a high-resolution spatial light modulator. For a unique configuration of the modulator, the scattered light interferes constructively at a chosen point; giving rise to a sharp focus. (c) Imaging: the focus follows rotations of the incident beam. The hidden structure is imaged. Reproduced with permission from [66], copyright 2010 OSA Publishing.
The possibility to use scattered light for high resolution imaging has a profound impact on tissue microscopy. If current trends continue, researchers will soon be able to perform fluorescence microscopy in turbid tissue at a depth of several millimetres.
Finding the required wavefront for focusing to a given point is far from trivial. So far, over a dozen approaches have been demonstrated, all based on feedback-based wavefront shaping, or optical phase conjugation (see [63, 64] for recent reviews on these respective topics).
Most approaches use some kind of marker that the light will be focused onto. The nature of the mechanism varies: second harmonic generation, fluorescence, photoacoustics, moving particles, two-photon fluorescence, etc [63, 64]. Alternatively, it is possible to use a marker-free method called time-reversal of ultrasound encoded light (TRUE) [64].
Current and future challenges
Research in the field of complex wavefront shaping revolves around three key challenges.
Scanning the focus
Marker-based approaches only focus light onto the marker itself. For imaging, however, the focus should be scanned over a region of interest. In general, scanning is not possible since a completely different wavefront is needed for shifting the focus by even half a wavelength.
Fortunately, there are two known special cases in which scanning is possible. The first case is when the structure of interest lies far behind a thin scattering layer. In this case, the optical memory effect [65] allows tilting of the transmitted wave by applying a tilt to the incident wave (see figure 8). This effect was exploited for scattered light fluorescence microscopy (SLFM), first through a layer of white paint [66], and recently through the intact skull of a living mouse [67].
When the medium mainly scatters light in the forward direction, as is the case with most biological tissue, a complementary type of memory effect occurs, which allows scanning a focus inside a scattering medium [68].
The use of the memory effect extends beyond wavefront shaping: it can be exploited to image sparse structures through scattering layers even without shaping the incident wave [69]. Unfortunately, the scan range of both memory effects is limited (e.g. 26 μm for the mouse skull), and decreases rapidly with the thickness of the scattering layer [65, 68].
Contrast
TRUE solves the challenge of focusing light at arbitrary positions. However, it suffers from the problem that the focus is relatively large. The size is roughly the size of the ultrasound focus: tens of micrometers in diameter. Since the peak intensity in the focus is inversely proportional to the number of independent speckles in the focus [63, 64] the signal-to-background contrast of TRUE is only about 20:1. Even though this contrast is sufficient for imaging isolated fluorescent structures, a problem arises when the background is even weakly fluorescent: the signal easily gets swamped by the signal from the much larger background volume.
Speed
The first wavefront shaping experiments needed minutes to form a focus [62]. Since the sample must be completely static during this time, in vivo applications were considered infeasible. At the moment, however, it is already possible to construct an appropriately shaped wavefront in 5.3 ms and focus light through 2.3 mm of perfused mouse skin [70]. In addition, for imaging through static materials such as bone, the setup need not be fast at all: for in vivo brain imaging the wavefront could remain static for over half an hour [67]. These results demonstrate that, even though a further speed increase is always desirable, many applications are already feasible at current speeds.
Advances in science and technology to meet challenges
At the moment of writing, there is no single method that solves all three challenges at once. For mesoscopic-resolution imaging at ~20 μm resolution, ultrasound-based methods seem to be the most promising so far, especially when the contrast can be enhanced. This contrast enhancement could be achieved by shrinking the focus through variance encoding [71], or perhaps by using nonlinear ultrasound propagation or ultrasound contrast agents, to create a smaller tagging volume. Also, the ultrasound frequency may be increased, at the cost of a steep increase in the attenuation coefficient. Finally, the contrast may be increased by using a nonlinear contrast mechanism, such as multi-photon fluorescence.
Since the contrast scales linearly with the number of pixels on the light modulator [62], the contrast can be increased by simply having more pixels on the SLM. Here lies a challenge for industry to manufacture light modulators with tens of megapixels. The development of 4K video projectors is a step in the right direction, and higher resolutions may become available when holographic television is developed. At these resolutions, fast data transfer from the camera, and pixel-perfect alignment between the camera and the SLM becomes increasingly hard. Conceptually, a simple solution would be to construct a specialized device where the image array is integrated with the SLM, i.e. where each SLM pixel has its own integrated photodetector and control logic.
For microscopic imaging, memory-based scanning techniques [66–69] are very promising. The work by Park et al [67] demonstrates that relevant in vivo applications are already possible for structures lying far behind a thin layer of bone. A scientific challenge here lies in increasing the fundamental understanding of wave propagation in order to extend the range over which the focus can be scanned, or to find ways to extrapolate or interpolate shaped wavefronts for scanning. A particularly interesting area is the cross-over regime between AO and wavefront shaping: combining ideas from both fields may increase the penetration depth of existing microscopes significantly.
Concluding remarks
The promise of complex wavefront shaping is clear: imaging deep inside turbid objects at a microscopic resolution. The traditional depth/resolution trade-off (see figure 9) can finally be broken by using focused, multiply scattered light for imaging. The ideal ‘all purpose’ technique has to simultaneously solve the challenges of scanning, contrast, and speed. Innovative solutions to these challenges are currently being demonstrated in rapid succession, and the first in vivo imaging results are already a fact.
Figure 9.
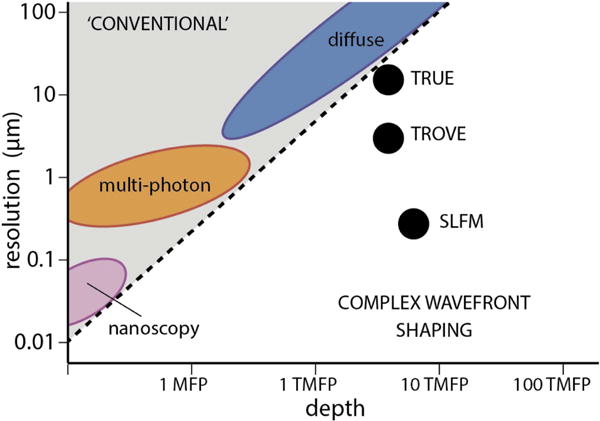
Complex wavefront shaping versus ‘conventional’ fluorescence imaging. Wavefront shaping techniques are breaking through the typical depth/resolution trade-off that is seen from super resolution imaging to diffuse tomography. MFP, scattering mean free path; TMFP, transport mean free path. Note: SLFM and TROVE required a physical distance between the scattering layer and the imaged object. Approximate indications, loosely based on [72].
Acknowledgments
I thank Roarke Horstmeyer and Gerwin Osnabrugge for proof-reading the manuscript and for useful discussions.
8. Adaptive optics for neuroscience
Martin J Booth
University of Oxford
Status
The ability to focus light on a microscopic scale inside tissue means that neurons can be observed or manipulated with 3D sub-micrometre resolution in a minimally invasive way. Whether for imaging or for manipulation, optical microscopes are central to a wide range of neuroscientific research. For functional imaging, two-photon laser scanning fluorescence microscopy has emerged as the dominant method, due to its ability to produce 3D images from deep inside specimens. Other fluorescence methods, such as confocal or widefield microscopy, are still widely employed for routine structural imaging. Super-resolution methods are also being employed to elucidate fine structural details, such as synaptic configurations. The same microscope optics provides the route for lasers in photo-activation and stimulation applications.
The major advantage of all of these methods is the ability to access deep within neural tissue. However, this presents one of the big challenges, namely that the light must pass through the volume of the specimen on the way to or from the focus. Tissue is not uniform, but rather a complex arrangement of diverse structures, each of which exhibits variations in optical properties. When light propagates through regions of differing RI, the wavefronts become distorted, suffering phase aberrations. These aberrations have detrimental effects in both focusing and imaging systems: a laser focus becomes dimmer and blurred by aberrations thus reducing the precision and efficiency of optical targeting; aberrated microscope images have lower resolution and contrast due to distortion of the point spread function. Furthermore, the aberrations change at different positions within the specimen, as light then passes through different structures to reach the focus.
AO technology has been introduced into microscopes to compensate aberrations and adapt to variations within and between specimens. In AO systems, a dynamically reconfigurable optical element, such as a deformable mirror (DM) or a liquid crystal SLM, corrects the specimen-induced aberrations (figure 10). A variety of AO schemes has been implemented in a wide range of microscopes [73]. In this roadmap article, we discuss the specific challenges relevant to optics in neuroscience.
Figure 10.
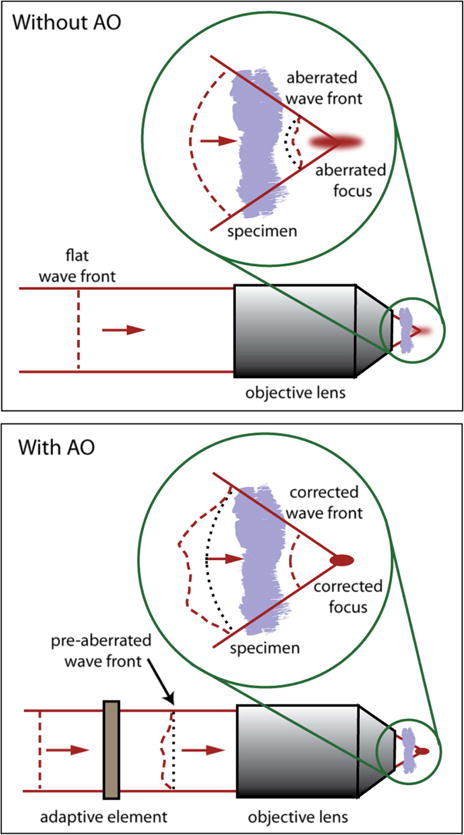
Aberration effects in a microscope focussing system and the principle of adaptive optics (AO) aberration correction.
Current and future challenges
Fundamentally, an AO system consists of three components: a method of aberration measurement, a wavefront corrector, and a control system. In the traditional paradigm, as commonly implemented in astronomical telescopes, aberrations are measured using a wavefront sensor, such as the Hartmann Shack sensor, and are in turn corrected using a DM. This approach has been used to good effect in some adaptive microscopes and has the advantage of fast closed-loop correction (figure 11(a)) [74]. In other microscopes, where wavefront sensors are not easily incorporated, indirect image-based optimization has been employed. In these systems, the specimen aberrations are inferred from a sequence of images taken with different aberrations added intentionally using the correction element. Information about the specimen aberrations is in effect encoded in the sequence of images. These indirect methods encompass modal sensing [75], pupil-segmentation [76] and scattering compensation (figure 11(b); see also section 7) [67].
Figure 11.
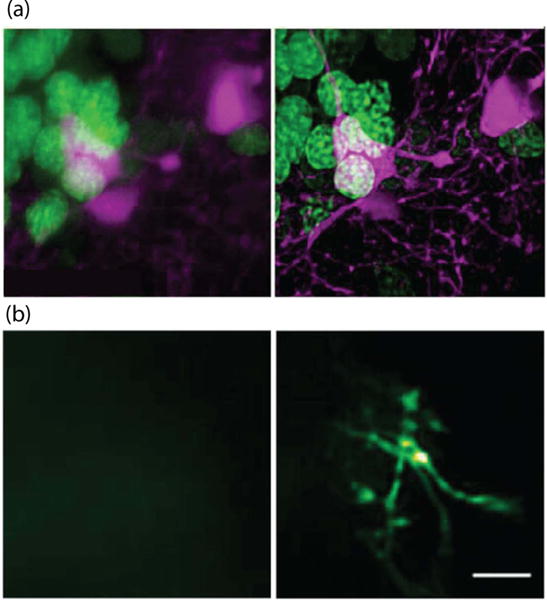
Adaptive microscopy demonstrations. (a) Two-color fluorescence confocal images of living zebrafish brain at depth ~200 μm: left, no aberration correction; right, with AO and deconvolution. Image width 40 μm (reprinted by permission from Macmillan Publishers Ltd [74], copyright 2014). (b) Two-photon fluorescence image of microglia taken through intact mouse skull: left, no compensation; right, with adaptive scattering compensation. Scale bar 5 μm (reproduced with permission from [67], copyright 2015 National Academy of Sciences, USA).
Like most biological specimens, neural tissue, whether intact brains or sections, consists of a mixture of complex structures. Different types of tissue can also have considerably different optical properties, such that aberrations can vary significantly between specimens. It is however possible to make some generalized comments about the aberrating effects of such tissue.
First, there is an effect caused by the average change in RI between the immersion medium and the tissue. If the interface between the two media is planar, then the dominant mode will be spherical aberration. Of course, most specimens will not have planar interfaces, but more complex structures that contribute more varied aberrations.
For shallow focusing (~10 s of μm), the profile of the aberrations tends to be smooth, and as such can be described by a small number of low order aberration modes, such as spherical, astigmatism and coma. In principle, these might be fully corrected by a simple adaptive element, such as a DM with a few actuators.
As one focuses deeper (~100 s of μm), the aberrations tend to become more complicated in form, leading to more complex, higher order aberrations. Compensation of these effects would typically require a more advanced correction element, such as a DM with tens or hundreds of actuators, or a SLM with thousands of pixels. Some of the light would be scattered strongly and in effect lost from the system, but reasonable imaging quality can be maintained.
Focusing even further into the specimen (~1 mm or more), multiple scattering can occur. This leads not only to phase aberrations, but also to significant amplitude variations, which cannot be recovered by the simple phase compensation provided by a single adaptive element. In this highly scattering regime, phase compensation can recover imaging performance, but usually only at low efficiency. Nevertheless, many interesting advances are being made in this area (see section 7).
Advances in science and technology to meet challenges
For applications requiring shallower focusing (10 s–100 s μm), where confocal, two-photon and even super-resolution methods might be employed, the challenges in neuroscience are similar to those in other applications of microscopy. The advances required here concern the efficiency (time and exposure) required for image-based AO and the effectiveness of direct sensing methods in scattering specimens
At greater focusing depths, aberrations are more complex, which means that more information about the phase aberration is required in order to determine the correction setting for the adaptive element. This requires more measurements and hence more time, although development of parallelized sensing methods could address this issue [77, 78].
For deep imaging, one route to mitigating the effects of scattering is to use longer wavelength lasers and three-photon excitation [79]. Although the magnitude and complexity of phase distortions is reduced through the longer wavelength, this method is more sensitive than two-photon microscopy to a drop in focal intensity due to the higher order nonlinear effects —whereas in two-photon fluorescence, the emission is proportional to the square of the focal intensity, in three-photon it depends upon the intensity cubed. Aberration correction will therefore still be necessary to achieve optimal performance.
When imaging wider areas, it will be necessary to address the issue of spatially variant aberrations. Most AO implementations so far have made implicit assumptions about aberrations being approximately constant over the field of view. This may in many scenarios be valid, but it is known that aberrations can change across the field. For full correction, it will be necessary either to change aberration correction rapidly during scanning using a fast adaptive element, or to use advanced AO configurations such as ‘conjugate’ or ‘multi-conjugate’ AO, where one or more correction elements are used in different positions in the optical train [67, 80].
Although this discussion has concentrated on neural imaging, the methods of AO are also applicable to optical targeting for optogenetic and other light activation applications. Indeed, SLMs have already been employed to sculpt light for the targeting of individual cells and there is plenty of scope here for inclusion of aberration compensation [81].
An important observation to make is that the diversity of microscopes in use and the variety of specimen types, even within neuroscience, are broad. Hence there is unlikely to be a single, universal, AO method that is applicable across all applications. A suite of technologies must therefore be developed that will permit improvements across the gamut.
Concluding remarks
Expectations about the performance of future optical microscopes for neuroscientific investigations are extremely demanding, requiring high speed, high spatial resolution 3D imaging at hundreds of μm (even mm) inside aberrating and scattering neural tissue. Furthermore, this would ideally be performed in living and mobile animals. Achieving these goals will require an impressive combination of many cutting-edge technologies, but AO will certainly play an important role.
9. Photoacoustic imaging of brain activity
Song Hu
University of Virginia
Status
Capitalizing on the elegant marriage of light and sound, PAI fills a critical niche in neuroimaging—bridging the gap between pre-clinical and clinical techniques that operate at distinct spatiotemporal scales and rely on different contrasts.
The technology envelope of PAI spans a seamless combination of spatiotemporal scales and a diverse collection of functional and molecular contrasts [82]. Spatially, by confining optical excitation and/or acoustic detection, the two major embodiments—PAM and PACT—can operate across the optical ballistic and diffusive regimes with either cellular-level optical resolution or tissue-level acoustic resolution [82]. This unique scalability, by trading off the spatial resolution against the tissue penetration, well poises PAI to translate novel basic findings from bench to bedside. Temporally, recent advances in laser technology, scan mechanism, and image reconstruction strategy have led to real-time PAM and PACT of the brain activity [83, 84]. In parallel, minimally invasive PAI has demonstrated long-term monitoring of neurodegeneration. The broad temporal coverage from rapid neurovascular coupling to chronic disease progression makes PAI a versatile tool for brain research. In terms of contrast, PAI has demonstrated functional brain imaging—including the concentration and oxygen saturation of hemoglobin, blood flow, oxygen metabolism, and hydrocephalus—using the endogenous optical absorption of hemoglobin and water. Relying on the endogenous lipid contrast, high-resolution PAI of large-scale neuronal networks in the brain is possible. Equally exciting, emerging molecular probes based on organic dyes, inorganic nanoparticles, and reporter genes have expanded the scope of PAI to image specific molecular and pathway activities in the brain—including calcium dynamics, glucose uptake, and aberrant protein aggregation.
Integrating the rich contrasts, PAI holds the potential to comprehensively characterize the brain at multiple spatiotemporal scales—a perspective that is otherwise impossible to achieve with agglomerated observations from a collection of different techniques.
Current and future challenges
As the envelope of neuro-PAI expands, new challenges emerge. Based on my view of this burgeoning field, a few prominent technical challenges are highlighted below.
Anisotropic resolution in (quasi)ballistic regime
Within the optical diffusion limit, the lateral resolution of PAI can be refined to the cellular level through tight optical focusing. In contrast, the axial resolution is usually limited by the acoustic detection bandwidth. In current multi-parametric PAM [85], for example, the axial resolution is 46.4 μm—one order of magnitude coarser than the lateral resolution (2.7 μm). This anisotropy impairs the 3D imaging capability of optical-resolution PAM. With a wideband piezoelectric transducer (6 dB:100 MHz), the axial resolution can be refined to 7.6 μm. However, the broad coverage is accompanied with a high central frequency (125 MHz), which compromises its sensitivity to the low-frequency component. New detection strategies providing broad and flat frequency responses are highly desired.
Fluence compensation in (quasi)diffusive regime
Beyond the optical diffusion limit, the impact of tissue scattering and absorption on optical fluence is non-negligible. The lack of means to accurately compensate for wavelength-dependent fluence is a major obstacle to quantitative and spectroscopic PAI at depth. The inability of acoustic-resolution PAI to assess the total concentration and oxygen saturation of hemoglobin (CHb and sO2, respectively) in absolute values prevents quantification of the cerebral metabolic rate of oxygen (CMRO2), thereby limiting its impact in deep brain imaging. To address this challenge, numerical models and invasive approaches are developed; however, both have significant limitations.
High-resolution mapping of CMRO2
As a direct measure of the brain energetics, CMRO2 is inherently a better surrogate of neural activity than CHb, sO2, and cerebral blood flow (CBF), particularly under disease states where neurovascular uncoupling might happen. Also, expanding the contrast of functional-connectivity PAI [84] from current CHb to CMRO2 may provide new insights into disease mechanisms. To this end, exciting progresses have been made. In particular, PAM has enabled the measurement of relative CMRO2 changes, but only in selected regions with closed circulation [83]. To overcome this limitation, advances in both system instrumentation and data processing are required.
Advances in science and technology to meet challenges
Intensive efforts have been undertaken to address the challenges and led to substantial progress. However, further advances are required.
Optical detection of ultrasound
Recently, novel optical approaches—including micro-ring resonator [86], Fabry–Perot etalon [87], and surface plasmon resonance [88]—have emerged for broadband high-sensitivity acoustic detection in PAI. In particular, the micro-ring resonator provides high sensitivity (6.8 Pa) and large bandwidth (6 dB:280 MHz), leading to all-optical PAM with near isotropic spatial resolution (i.e., 2.0 μm laterally and 5.3 μm axially) [86]. Future improvements in the long-term stability of this novel detector will excite numerous in vivo applications and open a new avenue for convenient integration of PAM and other optical microscopy techniques to perform multi-modal neuroimaging.
Acoustic spectral analysis for quantitative PAI at depth
In light of that the acoustic spectrum of the PAI-detected vascular signal depends on blood absorption but not optical fluence, an acoustic spectral method has recently been developed to quantify absolute CHb and sO2 in the optical diffusive regime without the need for fluence compensation [89]. Although encouraging, this method only applies to vessels with diameters much larger than the lateral resolution of PAI. Extending it to other types of vessels is a prerequisite for imaging CMRO2 at depth. Alternatively, the tissue-induced fluence variation can be potentially compensated for using an acousto-optic modulation approach [90], which however requires in vivo validation.
Multi-parametric PAI
High-resolution mapping of CMRO2 requires quantification of absolute CHb, sO2, and CBF at the same spatiotemporal scale. To this end, we have demonstrated simultaneous PAM of vascular anatomy, oxygen saturation, and blood flow down to the capillary level in the mouse skin [85], and recently extended it to the mouse brain (figure 12) [91]. Statistical, spectroscopic, and correlation analysis of the same PAM dataset allows quantifying absolute CHb, sO2, and CBF at the microscopic level. Development of algorithms to extend these vascular parameters to the tissue level will ultimately enable us to derive the microscopic CMRO2 using the Fick’s law. High-resolution mapping of CMRO2 in the diffusive regime is more challenging. Integrating the acoustic spectral analysis and ultrasound-induced thermal encoding approach [92] into PACT might be a potential solution.
Figure 12.
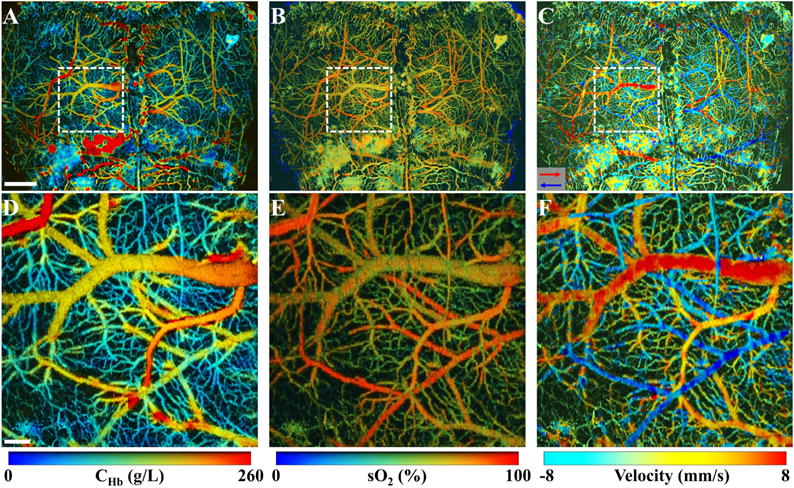
Simultaneous multi-parametric PAM of (A) CHb, (B) sO2, and (C) CBF (both flow speed and direction) in the mouse brain through the intact skull. (D)–(F) are the blow-up views of the boxed regions in (A)–(C), showing the microvasculature resolved by PAM. Scale bars: 1 mm in (A) and 200 μm in (D). The red and blue arrows in panel (C) indicate the directions of the blood flow.
Concluding remarks
Recent advances in the spatiotemporal coverage and contrast mechanism of PAI are rapidly expanding our understanding of the brain. As the technology envelope evolves, new challenges arise—particularly in the anisotropic spatial resolution in the ballistic regime, the optical fluence compensation in the diffusive regime, and the high-resolution mapping of CMRO2. Current research efforts have led to highly promising solutions; however, breakthroughs in both system instrumentation and data analysis are required to fully address these challenges.
10. Optical coherence tomography for brain imaging
Jiang Zhu and Zhongping Chen
University of California, Irvine
Status
OCT is a high-resolution (~1–10 μm) and non-invasive biomedical imaging modality which provides 3D structural and functional information of biological tissue. Since the first imaging of retina, the OCT imaging technique has played an increasingly important role for brain imaging.
From early time-domain OCT to current spectral domain OCT and swept source OCT (SS-OCT), OCT technology has rapidly advanced in both imaging speed (from a few Hz to a few MHz A-line speed) and resolution (from more than 10 μm to a few micrometers). These advancements enable a number of basic science and clinical applications in ophthalmology, cardiovascular diseases, cancer biology, and neuroscience.
Recently, there has been a growing interest in OCT brain imaging [93–95]. OCT can differentiate white matter and gray matter, and may provide surgical guidance in laser treatment of an epilepsy lesion. Layered organization of a rat olfactory bulb can be visualized using in vivo 3D OCT imaging. Single myelin fibers in living rodents can be imaged, and the RI in the rat somatosensory cortex can be measured using OCT without labeling. OCT signal change has also been used to measure and map neural activation [93–95].
In addition to morphological structure imaging, functional OCT, such as Doppler OCT (D-OCT) and polarization sensitive OCT (PS-OCT), can also image and quantify physiological parameters. D-OCT combines the Doppler principle with OCT to obtain high resolution tomographic images of moving constituents in highly scattering biological tissues [96, 97]. D-OCT has been successfully used to image cortical blood flow and map the blood vessel network for brain research [93–95]. Quantitative measurement of flow velocities and qualitative analysis of vascular networks from OCT brain imaging can expose changes of CBF induced by drugs, diseases and stimulation, all of which may result in functional changes of the brain cortex. PS-OCT, which incorporates polarization sensitive detection to determine tissue birefringence, has been used for the localization of nerve fiber bundles and the mapping of micrometer-scale fiber pathways in the brain [98].
The first D-OCT image of blood flow in the rat cerebral cortex was reported in 1997 with time domain OCT [96]. Recent advances in Fourier domain phase-resolved D-OCT have significantly improved imaging speed and velocity sensitivity, which make imaging of a full map of the neurovascular cortex down to capillary level possible [93–95]. Figure 13 shows D-OCT images of rat cortex blood vessels that reveal the detailed blood perfusion network over the cortex at capillary level resolution [93].
Figure 13. En face.
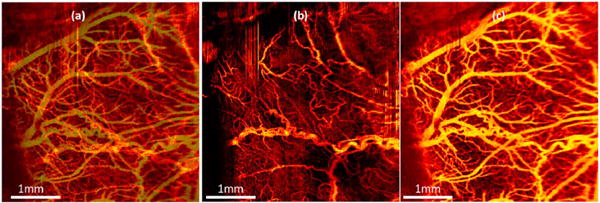
projection view of rat cortex blood perfusion using D-OCT. (a) Projection view from the full imaging depth; (b) projection view from skull; (c) projection view from cortex [93].
D-OCT has been used to measure stimulus-induced changes in blood flow, evaluate and monitor brain ischemia and trauma, quantify blood circulation and flow resistance before and after a localized ischemic stroke in a mouse model, and map the density of the vascular network in the brain of diseased animal models [93–95]. Figure 14 compares cerebrovascular networks in a 20 month old control and in a triple transgenic Alzheimer’s mouse model (3xTg-AD) [100].
Figure 14.
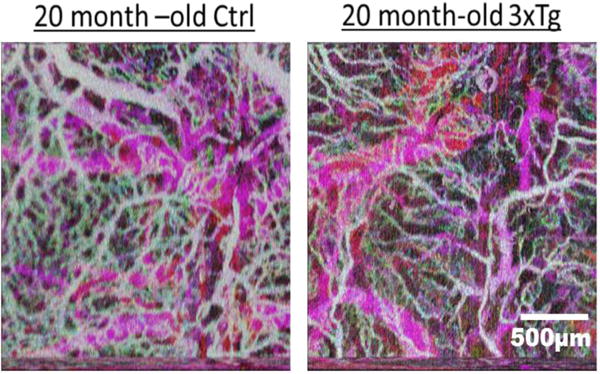
Doppler OCT images of cerebrovascular network in a 20 month old control and a 3xTg-AD mouse. Superficial vessels and deeper vessels are in white and in pink, respectively. There was a 29% decrease in the superficial cortical vessel volume in the 3xTg-AD mouse than in the control.
Current and future challenges
For brain studies, an ideal imaging method should provide high-resolution images of microvascular networks in a wide field of view, cover deep imaging depth, enable broad dynamic range for flow quantification, and possess high speed for real-time flow analysis in each vessel of a brain cortex. However, while each individual goal has been achieved with recent advances in technology, achievement of all target performances simultaneously remains a challenge.
First, the imaging speed is a major challenge for micro-vascular angiography and quantitative D-OCT imaging over a large field of view. In order to image the microvascular network in a wider field of view, A-line speed in the order of tens of MHz will be necessary. However, since the flow velocity sensitivity depends on the time window used for correlation calculation, the high imaging speed may reduce velocity sensitivity if inter-A-line correlation is used for flow quantification. Therefore, inter-2D-frame or inter-3D-frame correlation may be necessary in order to increase velocity sensitivity, which will put further requirements on the stability of the imaging system and the light source.
Second, D-OCT can provide an accurate flow velocity and flow direction after calculation of a Doppler phase shift and a Doppler angle. Real-time calculation of a Doppler angle is required at each location of each vessel, and the distortion due to phase shift wrapping should be corrected automatically during Doppler phase calculation. Efficiency of automatic algorithms and speed of data processing are challenges for real-time flow calculation.
Finally, due to strong optical scattering of tissues, OCT penetration depth is limited to 1–2 mm for brain imaging. Therefore, OCT can only detect microvascular networks near the surface of a cortex, which may not be enough for studies of deeper cerebral cortices. The penetration depth is also a challenge for brain imaging.
Advances in science and technology to meet challenges
In order to image and quantify vascular and neuronal dynamics occurring in the brain over a larger field of view with higher spatial resolution and higher velocity sensitivity and broader dynamic range, a number of scientific and technological advancements are necessary. These advancements include developing better light sources, OCT system hardware, and data acquisition cards to increase imaging speed and spatial resolution, as well as developing better algorithms for volumetric vessel reconstruction and flow quantification.
Benefiting from the development of swept sources, OCT imaging speed has significantly increased in SS-OCT systems over the last ten years. A swept source with an A-line speed of 200 kHz has been widely used in OCT systems, and a FDML swept source with an A-line speed of 1.6 MHz has also been available recently. The 1.6 MHz OCT system can image the microvascular network of a mouse brain at a rate of 4.7 volumes s−1 (512 × 200 × 720 voxels) [101]. However, a light source with tens of MHz swept rate is necessary to cover the full rat cortex at a video rate. Although this light source has been demonstrated, the noise and coherence length remain a problem. In addition, A/D digitizing speed needs to be significantly increased.
In addition, a full-field OCT (FF-OCT) is another potential solution for increasing the imaging speed. FF-OCT increases the frame rate by parallel acquisition of A-scans using a charge-coupled device or complementary metaloxide-semiconductor camera. The Fourier-domain FF-OCT with an equivalent A-line rate of 1.5 MHz has been developed, which can record a volume of 640 × 24 × 512 voxels in 10 ms. A modified dark-field FF-OCT can provide an isotropical resolution of 1.5 μm, which is much higher than that of conventional OCT. FF-OCT may have great potential to promote the studies of cerebral vasculature due to high resolution and super-fast imaging speed. However, FF-OCT has limited imaging depth. Innovation in system design and signal processing to overcome this limitation will greatly enhance the potential of FF-OCT for brain imaging.
Furthermore, for real-time calculation of flow velocities, automatic and robust processing algorithms for 3D visualization of vascular networks and for correction of phase shift wrapping can significantly improve the efficiency of the data analysis [102]. Parallel graphics processing unit based data processing can accelerate calculation of Doppler angles and correction of Doppler shift wrapping. Faster and automatic data processing will enable real-time volumetric rendering and blood flow analysis.
Finally, to overcome the limitation of penetration depth in OCT imaging, an OCT system that incorporates AO may be used. In addition, the combination of Doppler ultrasound imaging or PAI with D-OCT has great potential to extend the imaging depth for quantifying vascular hemodynamics in the brain.
Concluding remarks
OCT is a rapidly developing imaging technology with many potential applications in the field of brain imaging. D-OCT not only can map cerebrovascular networks with high resolution, but also can accurately measure flow velocities. Given the noninvasive and label-free nature with exceptionally high spatial resolution and velocity sensitivity, functional OCT can simultaneously provide tissue structure, vascular and neuronal dynamics as well as other physiological information. It has great potential to become a powerful brain imaging tool for basic biomedical research and clinical medicine.
11. Near-infrared spectroscopy for brain imaging
Yoko Hoshi
Hamamatsu University School of Medicine
Status
NIRS utilizes light in the near-infrared region, from 700 to 2500 nm, and is a non-invasive technique for analysis of various substances including agricultural products and food. Biological tissue is relatively transparent from 650 to 1300 nm, the so-called optical window, where absorption spectra vary with the oxygenation–deoxygenation states of hemoglobin (Hb) and myoglobin and the redox state of cytochrome c oxidase. These spectroscopic characteristics were the background for biomedical applications of NIRS in 1977 [103]: the technique was originally designed for clinical monitoring of tissue oxygenation. Since the early 1990s, it has also been developed into a useful tool in neuroimaging studies, with so-called functional NIRS [104, 105].
A wide range of NIRS instruments have been developed, including commonly commercially available instruments for continuous wave (CW) measurements based on the modified Beer–Lambert law [106] (CW-type instruments). Although NIRS provides only information of the functioning of the cerebral cortex facing the skull convexity, NIRS has become widely applied in a variety of neuroimaging studies because of its advantages: (1) measurements are non-invasive and can be performed with fewer motion restrictions, (2) the temporal resolution is high, and (3) simultaneous measurements with other neuroimaging modalities are possible. In addition, the introduction of multichannel CW-type instruments and wearable systems is facilitating neuroimaging studies on children, the elderly, and patients with psychoneurological complaints, where it is difficult to conduct measurements by other neuroimaging techniques. It may also be expected that NIRS will allow development of other new neuroimaging studies different from those performed with fMRI and PET.
Current and future challenges
During the past 39 years, difficulties in selective and quantitative measurements of cerebral Hb have been central issues in the NIRS field. To overcome the problems with quantification, time-resolved spectroscopy (TRS) and frequency-domain spectroscopy (FDS), which can determine mean total optical path lengths (t-PL), have been developed. However, functional brain activation is accompanied by localized Hb concentration changes within cerebral tissue. This requires the determination of partial mean optical path lengths (p-PL) in the cerebral tissue for the quantification, something which is not feasible at present. Since the mean p-PL varies with measurement position, as does the mean t-PL, direct comparisons of amplitudes of NIRS signals across regions cannot be used in comparisons of regional differences in Hb concentration changes.
Changes in the scalp blood flow are not homogeneous but heterogeneous and can influence NIRS signals. Currently, resting-state fMRI is a focus of research interests, and NIRS has also been employed in investigating functional connectivity. However, the scalp blood flow fluctuates during rest, as is the case for the CBF, making it necessary to distinguish the NIRS signals originating in the cerebral tissue from those coming from the scalp. Several approaches have been tried to achieve this, and in recent years, various methods with a multi-distance probe arrangement have been proposed. Here short source-detector (SD) separation measurements are used as regressors to eliminate the scalp blood flow signals from the longer SD measurements [107]. These methods have been reported to improve quality of NIRS signals, and further refining the algorithms for routine applications to NIRS measurements is also required.
In TRS, the tissue is irradiated by ultrashort (picosecond order) laser pulses, and the intensity of the emerging light is detected as a function of time (the temporal point spread function, TPSF) with picosecond resolution (figure 15). The mean t-PL is determined by multiplying the light speed in the media by the mean transit time of the scattered photons, which is calculated with the TPSF. TRS carries information about depth-dependent attenuation based on the correlation of detection time to penetration depth of photons. Changes in cerebral Hb concentrations can be more selectively determined by analyzing TPSF and several different methods of analysis have been proposed [108, 109]. However, the selective and quantitative accuracy is insufficient and further investigation must be continued to apply these methods to human head measurement.
Figure 15.

Temporal profiles of incident light and intensity of detected light. tm, mean transit time; 1, 2, 3 denote detection times of photons traveling through the optical paths 1, 2, and 3 (inserted figure), respectively. Reproduced with permission from [113]. Copyright 2016 SPIE.
Advances in science and technology to meet challenges
A very promising approach to the above-mentioned issues is diffuse optical tomography (DOT). This technique reconstructs 3D images of the optical properties from boundary measurements, and could enable a quantitative detection of regional cerebral Hb (figure 16). Except for extremely low birth weight infants, whose head circumference is very small as is shown in figure 16(b), little light illuminated on the human head is detected in a transmittance mode. Thus, DOT rarely provides information of the subcortical structures. The DOT algorithm essentially consists of two parts. One is a forward model to calculate the light propagation and the resultant outward reemissions at the boundary of the tissue. The other is an inverse model to search for the distribution of optical properties by minimizing the differences between the calculated and the measured data.
Figure 16.

DOT image of total Hb concentrations of the brain in an extremely low birth weight infant under hypocapnic conditions [112]. (a) Sixteen coaxial optical fibers, consisting of a single fiber for illumination in the center and outer bundle fibers for detection, were placed on the scalp of the infant. (b) Image reconstruction was performed by using the PDE-based algorithm. The axial image shows the total Hb concentration changes cause by hypocapnia in comparison with normocapnic conditions.
The DOT image reconstruction can be approximately divided into two: one is a linear-single step image reconstruction; the other is a nonlinear iterative image reconstruction. The former was first proposed in 1995 [110], and can now provide high quality images with multichannel CW-type instruments [111]. This approach reconstructs only qualitative images of measured changes, which is, however, sufficient for neuroimaging studies. In clinical usage, quantitative images of steady-state Hb are further useful with diagnostic optical imaging, and DOT based on the latter approach can be performed with CW measurements, FDS, and TRS.
DOT has been applied to imaging of organs and tissue, including in the brain and breasts. However, image quality is still inadequate. The photon diffusion equation (PDE) is widely used as a forward model, but the PDE is invalid in low-scattering regions, highly absorbing regions and in the vicinity of light sources. The radiative transfer equation (RTE) correctly describes photon propagation in living tissue, making development of RTE-based algorithms attractive; however, the computational load in solving the RTE is very heavy. Further, the inverse model is an ill-posed and ill-conditioned problem, and the initial values of the optical properties are critical for image reconstruction. However, these are still unknown quantities. For this reason, it is crucial to develop accurate and efficient algorithms for DOT image reconstruction and also to determine optical properties in biological tissue.
Concluding remarks
Temporal resolution of conventional NIRS (CW measurements) is very high, whereas even linear-single step CW-DOT cannot yet reconstruct images in real-time. Although TRS provides more information required for image reconstruction, such as depth information, it takes a few seconds to obtain a temporal profile with the detected photons. When high-quality DOT images (the desired spatial resolution of 3–5 mm) are achieved, the next step should focus on improving the temporal resolution. Development of DOT is very challenging, but can be extended to fluorecsence tomography (FDOT). With FDOT, molecular imaging in living human subjects is possible. Thus, combining DOT and FDOT will make dynamic multi-level, from molecular to whole organism level, bio-optical imaging possible.
Table 2.
Functional OCT methods for vascular imaging.
Footnotes
(Some figures may appear in colour only in the online journal)
References
- 1.Alivisatos AP, et al. Nanotools for neuroscience and brain activity mapping. ACS Nano. 2013;7:1850–66. doi: 10.1021/nn4012847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jorgenson LA, et al. The BRAIN Initiative: developing technology to catalyse neuroscience discovery. Phil Trans R Soc B. 2015;370:20140164. doi: 10.1098/rstb.2014.0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Denk W, Briggman KL, Helmstaedter M. Structural neurobiology: missing link to a mechanistic understanding of neural computation. Nat Rev Neurosci. 2012;13:351–8. doi: 10.1038/nrn3169. [DOI] [PubMed] [Google Scholar]
- 4.Kasthuri N, et al. Saturated reconstruction of a volume of neocortex. Cell. 2015;162:648–61. doi: 10.1016/j.cell.2015.06.054. [DOI] [PubMed] [Google Scholar]
- 5.Collman F, Buchanan J, Phend KD, Micheva KD, Weinberg RJ, Smith SJ. Mapping synapses by conjugate light-electron array tomography. J Neurosci. 2015;35:5792–807. doi: 10.1523/JNEUROSCI.4274-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oh SW, et al. A mesoscale connectome of the mouse brain. Nature. 2014;508:207–14. doi: 10.1038/nature13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nicolelis MA, Dimitrov D, Carmena JM, Crist R, Lehew G, Kralik JD, Wise SP. Chronic, multisite, multielectrode recordings in macaque monkeys. Proc Natl Acad Sci USA. 2003;100:11041–6. doi: 10.1073/pnas.1934665100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calhoun VD, Miller R, Pearlson G, Adalı T. The chronnectome: time-varying connectivity networks as the next frontier in fMRI data discovery. Neuron. 2014;84:262–74. doi: 10.1016/j.neuron.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malonek D, Grinvald A. Vascular regulation at submillimeter range, sources of intrinsic signals for high resolution optical imaging. Adv Exp Med Biol. 1997;413:215–20. [PubMed] [Google Scholar]
- 10.Deisseroth K. Optogenetics. Nat Methods. 2011;8:26–9. doi: 10.1038/nmeth.f.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J, Lee S, Tsuda S, Zhang X, Asrican B, Gloss B, Feng G, Augustine GJ. Optogenetic mapping of cerebellar inhibitory circuitry reveals spatially biased coordination of interneurons via electrical synapses. Cell Rep. 2014;7:1601–13. doi: 10.1016/j.celrep.2014.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piao HH, Rajakumar D, Kang BE, Kim EH, Baker BJ. Combinatorial mutagenesis of the voltage-sensing domain enables the optical resolution of action potentials firing at 60 Hz by a genetically encoded fluorescent sensor of membrane potential. J Neurosci. 2015;35:372–85. doi: 10.1523/JNEUROSCI.3008-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen T, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fosque BF, et al. Neural circuits, labeling of active neural circuits in vivo with designed calcium integrators. Science. 2015;347:755–60. doi: 10.1126/science.1260922. [DOI] [PubMed] [Google Scholar]
- 15.Inoue M, et al. Rational design of a high-affinity, fast, red calcium indicator R-CaMP2. Nat Methods. 2015;12:64–70. doi: 10.1038/nmeth.3185. [DOI] [PubMed] [Google Scholar]
- 16.St-Pierre F, et al. High-fidelity optical reporting of neuronal electrical activity with an ultrafast fluorescent voltage sensor. Nat Neurosci. 2014;17:884–9. doi: 10.1038/nn.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hochbaum DR, et al. All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nat Methods. 2014;11:825–33. doi: 10.1038/nmeth.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Venkatachalam V, et al. Flash memory: photochemical imprinting of neuronal action potentials onto a microbial rhodopsin. J Am Chem Soc. 2014;136:2529–37. doi: 10.1021/ja411338t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marvin JS, et al. An optimized fluorescent probe for visualizing glutamate neurotransmission. Nat Methods. 2013;10:162–70. doi: 10.1038/nmeth.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miesenbock G, De Angelis DA, Rothman JE. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature. 1998;394:192–5. doi: 10.1038/28190. [DOI] [PubMed] [Google Scholar]
- 21.Park J, et al. Screening fluorescent voltage indicators with spontaneously spiking HEK cells. PLoS One. 2013;8:e85221. doi: 10.1371/journal.pone.0085221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahrens MB, Orger MB, Robson DN, Li JM, Keller PJ. Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nat Methods. 2013;10:413–20. doi: 10.1038/nmeth.2434. [DOI] [PubMed] [Google Scholar]
- 23.Govorunova EG, Sineshchekov OA, Janz R, Liu X, Spudich JL. Natural light-gated anion channels: a family of microbial rhodopsins for advanced optogenetics. Science. 2015;349:6248. doi: 10.1126/science.aaa7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wieboldt R, Gee KR, Niu L, Ramesh D, Carpenter BK, Hess GP. Photolabile precursors of glutamate: synthesis, photochemical properties, and activation of glutamate receptors on a microsecond time scale. Proc Natl Acad Sci USA. 1994;91:8752–6. doi: 10.1073/pnas.91.19.8752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–8. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 26.Li X, Gutierrez DV, Hanson MG, Han J, Mark MD, Chiel H, Hegemann P, Landmesser LT, Herlitze S. Fast noninvasive activation and inhibition of neural and network activity by vertebrate rhodopsin and green algae channelrhodopsin. Proc Natl Acad Sci USA. 2005;102:17816–21. doi: 10.1073/pnas.0509030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deisseroth K, Feng G, Majewska AK, Miesenbock G, Ting A, Schnitzer MJ. Next-generation optical technologies for illuminating genetically targeted brain circuits. J Neurosci. 2006;26:10380–6. doi: 10.1523/JNEUROSCI.3863-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knopfel T. Genetically encoded optical indicators for the analysis of neuronal circuits. Nat Rev Neurosci. 2012;13:687–700. doi: 10.1038/nrn3293. [DOI] [PubMed] [Google Scholar]
- 29.Deisseroth K, Schnitzer MJ. Engin structure and dynamics. Neuron. 2013;80:568–77. doi: 10.1016/j.neuron.2013.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knopfel T, Akemann W. Nanobiotechnology: Remote control of cells. Nat Nanotechnol. 2010;5:560–1. doi: 10.1038/nnano.2010.163. [DOI] [PubMed] [Google Scholar]
- 31.Warden MR, Cardin JA, Deisseroth K. Optical neural interfaces. Annu Rev Biomed Eng. 2014;16:103–29. doi: 10.1146/annurev-bioeng-071813-104733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grosenick L, Marshel James H, Deisseroth K. Closed-loop and activity-guided optogenetic control. Neuron. 2015;86:106–39. doi: 10.1016/j.neuron.2015.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pisanello F, Sileo L, De Vittorio M. Micro- and nanotechnologies for optical neural interfaces. Frontiers Neurosci. 2016;10:70. doi: 10.3389/fnins.2016.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gradinaru V, et al. Targeting and readout strategies for fast optical neural control in vitro and in vivo. J Neurosci. 2007;27:14231–8. doi: 10.1523/JNEUROSCI.3578-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McAlinden N, Gu E, Dawson MD, Sakata S, Mathieson K. Optogenetic activation of neocortical neurons in vivo with a sapphire-based micro-scale LED probe. Frontiers Neural Circuits. 2015;9:25. doi: 10.3389/fncir.2015.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jeong J-W, et al. Wireless optofluidic systems for programmable in vivo pharmacology and optogenetics. Cell. 2015;162:662–74. doi: 10.1016/j.cell.2015.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim T-I, et al. Injectable, cellular-scale optoelectronics with applications for wireless optogenetics. Science. 2013;340:211–6. doi: 10.1126/science.1232437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pisanello F, et al. Multipoint-emitting optical fibers for spatially addressable in vivo optogenetics. Neuron. 2014;82:1245–54. doi: 10.1016/j.neuron.2014.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Canales A, et al. Multifunctional fibers for simultaneous optical, electrical and chemical interrogation of neural circuits in vivo. Nat Biotechnol. 2015;33:277–84. doi: 10.1038/nbt.3093. [DOI] [PubMed] [Google Scholar]
- 40.Cui G, et al. Concurrent activation of striatal direct and indirect pathways during action initiation. Nature. 2013;494:238–42. doi: 10.1038/nature11846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gunaydin Lisa A, et al. Natural neural projection dynamics underlying social behavior. Cell. 2014;157:1535–51. doi: 10.1016/j.cell.2014.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smythe EJ, Dickey MD, Bao J, Whitesides GM, Capasso F. Optical antenna arrays on a fiber facet for in situ surface-enhanced raman scattering detection. Nano Lett. 2009;9:1132–8. doi: 10.1021/nl803668u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dipalo M, et al. 3D plasmonic nanoantennas integrated with MEA biosensors. Nanoscale. 2015;7:3703–11. doi: 10.1039/c4nr05578k. [DOI] [PubMed] [Google Scholar]
- 44.Bovetti S, Fellin T. Optical dissection of brain circuits with patterned illumination through the phase modulation of light. J Neurosci Methods. 2015;241:66–77. doi: 10.1016/j.jneumeth.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 45.Packer AM, Russell LE, Dalgleish HWP, Hausser M. Simultaneous all-optical manipulation and recording of neural circuit activity with cellular resolution in vivo. Nat Methods. 2015;12:140–6. doi: 10.1038/nmeth.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ferenczi EA, et al. Prefrontal cortical regulation of brainwide circuit dynamics and reward-related behavior. Science. 2016;351:6268. doi: 10.1126/science.aac9698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nagel G, et al. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci. 2003;100:13940–5. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carvalho-de-Souza João L, Treger Jeremy S, Dang B, Kent Stephen BH, Pepperberg David R, Bezanilla F. Photosensitivity of neurons enabled by cell-targeted gold nanoparticles. Neuron. 2015;86:207–17. doi: 10.1016/j.neuron.2015.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marshall JD, Schnitzer MJ. Optical strategies for sensing neuronal voltage using quantum dots and other semiconductor nanocrystals. ACS Nano. 2013;7:4601–9. doi: 10.1021/nn401410k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park SI, et al. Ultraminiaturized photovoltaic and radio frequency powered optoelectronic systems for wireless optogenetics. J Neural Eng. 2015;12:056002. doi: 10.1088/1741-2560/12/5/056002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Montgomery KL, et al. Wirelessly powered, fully internal optogenetics for brain, spinal and peripheral circuits in mice. Nat Methods. 2015;12:969–74. doi: 10.1038/nmeth.3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ragan T, et al. Serial two-photon tomography for automated ex vivo mouse brain imaging. Nat Methods. 2012;9:255–8. doi: 10.1038/nmeth.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Economo MN, et al. A platform for brain-wide imaging and reconstruction of individual neurons. Elife. 2016;5:1–22. doi: 10.7554/eLife.10566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huisken J, Stainier DY. Selective plane illumination microscopy techniques in developmental biology. Development. 2009;136:1963–75. doi: 10.1242/dev.022426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li A, et al. Micro-optical sectioning tomography to obtain a high-resolution atlas of the mouse brain. Science. 2010;330:1404–8. doi: 10.1126/science.1191776. [DOI] [PubMed] [Google Scholar]
- 56.Dodt HU, et al. Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat Methods. 2007;4:331–6. doi: 10.1038/nmeth1036. [DOI] [PubMed] [Google Scholar]
- 57.Ke MT, Fujimoto S, Imai T. SeeDB: a simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat Neurosci. 2013;16:1154–61. doi: 10.1038/nn.3447. [DOI] [PubMed] [Google Scholar]
- 58.Susaki EA, et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell. 2014;157:726–39. doi: 10.1016/j.cell.2014.03.042. [DOI] [PubMed] [Google Scholar]
- 59.Chung K, et al. Structural and molecular interrogation of intact biological systems. Nature. 2013;497:332–7. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Costantini I, et al. A versatile clearing agent for multimodal brain imaging. Sci Rep. 2015;5:9808. doi: 10.1038/srep09808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen F, et al. Optical imaging, expansion microscopy. Science. 2016;347:543–8. doi: 10.1126/science.1260088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vellekoop IM, Mosk AP. Focusing coherent light through opaque strongly scattering media. Opt Lett. 2007;32:2309–11. doi: 10.1364/ol.32.002309. [DOI] [PubMed] [Google Scholar]
- 63.Vellekoop IM. Feedback-based wavefront shaping. Opt Express. 2015;23:12189–206. doi: 10.1364/OE.23.012189. [DOI] [PubMed] [Google Scholar]
- 64.Horstmeyer R, Ruan H, Yang C. Guidestar-assisted wavefront-shaping methods for focusing light into biological tissue. Nat Photon. 2015;9:563–71. doi: 10.1038/nphoton.2015.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Feng S, Kane C, Lee PA, Stone AD. Correlations and fluctuations of coherent wave transmission through disordered media. Phys Rev Lett. 1988;61:834–7. doi: 10.1103/PhysRevLett.61.834. [DOI] [PubMed] [Google Scholar]
- 66.Vellekoop IM, Aegerter CM. Scattered light fluorescence microscopy: imaging through turbid layers. Opt Lett. 2010;35:1245–7. doi: 10.1364/OL.35.001245. [DOI] [PubMed] [Google Scholar]
- 67.Park J-H, Sun W, Cui M. High-resolution in vivo imaging of mouse brain through the intact skull. Proc Natl Acad Sci USA. 2015;112:9236–41. doi: 10.1073/pnas.1505939112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Judkewitz B, Horstmeyer R, Vellekoop IM, Papadopoulos IN, Yang C. Translation correlations in anisotropically scattering media. Nat Phys. 2015;11:684–9. [Google Scholar]
- 69.Bertolotti J, van Putten EG, Blum C, Lagendijk A, Vos WL, Mosk AP. Non-invasive imaging through opaque scattering layers. Nature. 2012;491:232–4. doi: 10.1038/nature11578. [DOI] [PubMed] [Google Scholar]
- 70.Wang D, Zhou EH, Brake J, Ruan H, Jang M, Yang C. Focusing through dynamic tissue with millisecond digital optical phase conjugation. Optica. 2015;2:728–35. doi: 10.1364/OPTICA.2.000728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Judkewitz B, Wang YM, Horstmeyer R, Mathy A, Yang C. Speckle-scale focusing in the diffusive regime with time reversal of variance-encoded light (TROVE) Nat Photon. 2013;7:300–5. doi: 10.1038/nphoton.2013.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ntziachristos V. Going deeper than microscopy: the optical imaging frontier in biology. Nat Methods. 2010;7:603–14. doi: 10.1038/nmeth.1483. [DOI] [PubMed] [Google Scholar]
- 73.Booth MJ. Adaptive optical microscopy: the ongoing quest for a perfect image. Light: Sci Appl. 2014;3:e165. [Google Scholar]
- 74.Wang K, Milkie DE, Saxena A, Engerer P, Misgeld T, Bronner ME, Mumm J, Betzig E. Rapid adaptive optical recovery of optimal resolution over large volumes. Nat Methods. 2014;11:625–8. doi: 10.1038/nmeth.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Débarre D, et al. Image-based adaptive optics for two-photon microscopy. Opt Lett. 2009;34:2495–7. doi: 10.1364/ol.34.002495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ji N, Sato TR, Betzig E. Characterization and adaptive optical correction of aberrations during in vivo imaging in the mouse cortex. Proc Natl Acad Sci. 2012;109:22–7. doi: 10.1073/pnas.1109202108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang C, et al. Multiplexed aberration measurement for deep tissue imaging in vivo. Nat Methods. 2014;11:1037–40. doi: 10.1038/nmeth.3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fiolka R, Se K, Cui M. Parallel wavefront measurements in ultrasound pulse guided digital phase conjugation. Opt Express. 2012;20:24827–34. doi: 10.1364/OE.20.024827. [DOI] [PubMed] [Google Scholar]
- 79.Horton NG, Xu C. Dispersion compensation in three-photon fluorescence microscopy at 1700 nm. Biomed Opt Express. 2015:1392–7. doi: 10.1364/BOE.6.001392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li J, et al. Conjugate adaptive optics in widefield microscopy with an extended-source wavefront sensor. Optica. 2015;2:682–8. [Google Scholar]
- 81.Papagiakoumou E, et al. Functional patterned multiphoton excitation deep inside scattering tissue. Nat Photon. 2013;7:274–8. [Google Scholar]
- 82.Wang LV, Hu S. Photoacoustic tomography: in vivo imaging from organelles to organs. Science. 2012;335:1458–62. doi: 10.1126/science.1216210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yao J, Wang L, Yang J-M, Maslov KI, Wong TTW, Li L, Huang C-H, Zou J, Wang LV. High-speed label-free functional photoacoustic microscopy of mouse brain in action. Nat Methods. 2015;12:407–10. doi: 10.1038/nmeth.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nasiriavanaki M, Xia J, Wan H, Bauer AQ, Culver JP, Wang LV. High-resolution photoacoustic tomography of resting-state functional connectivity in the mouse brain. Proc Natl Acad Sci USA. 2014;111:21–6. doi: 10.1073/pnas.1311868111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ning B, et al. Simultaneous photoacoustic microscopy of microvascular anatomy, oxygen saturation, and blood flow. Opt Lett. 2015;40:910. doi: 10.1364/OL.40.000910. [DOI] [PubMed] [Google Scholar]
- 86.Li H, Dong B, Zhang Z, Zhang HF, Sun C. A transparent broadband ultrasonic detector based on an optical micro-ring resonator for photoacoustic microscopy. Sci Rep. 2014;4:4496. doi: 10.1038/srep04496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jathoul AP, et al. Deep in vivo photoacoustic imaging of mammalian tissues using a tyrosinase-based genetic reporter. Nat Photon. 2015;9:239–46. [Google Scholar]
- 88.Wang T, Cao R, Ning B, Dixon AJ, Hossack JA, Klibanov AL, Zhou Q, Wang A, Hu S. All-optical photoacoustic microscopy based on plasmonic detection of broadband ultrasound. Appl Phys Lett. 2015;107:153702. [Google Scholar]
- 89.Guo Z, Favazza C, Garcia-Uribe A, Wang L. Quantitative photoacoustic microscopy of optical absorption coefficients from acoustic spectra in the optical diffusive regime. J Biomed Opt. 2012;17:066011. doi: 10.1117/1.JBO.17.6.066011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Daoudi K, Hussain A, Hondebrink E, Steenbergen W. Correcting photoacoustic signals for fluence variations using acousto-optic modulation. Opt Express. 2012;20:14117–29. doi: 10.1364/OE.20.014117. [DOI] [PubMed] [Google Scholar]
- 91.Ning B, Sun N, Cao R, Chen R, Kirk Shung K, Hossack JA, Lee J-M, Zhou Q, Hu S. Ultrasound-aided multiparametric photoacoustic microscopy of the mouse brain. Sci Rep. 2015;5:18775. doi: 10.1038/srep18775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang L, Xia J, Yao J, Maslov KI, Wang L. Ultrasonically encoded photoacoustic flowgraphy in biological tissue. Phys Rev Lett. 2013;111:204301. doi: 10.1103/PhysRevLett.111.204301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu G, Chen Z. Optical coherence tomography for brain imaging. In: Madsen SJ, editor. Optical Methods and Instrumentation in Brain Imaging. New York: Springer Science +Business Media; 2013. pp. 157–72. [Google Scholar]
- 94.Lee J, Boas DA. OCT and coherence imaging for the neurosciences. In: Drexler W, Fujimoto JG, editors. Optical Coherence Tomography: Technology and Applications. 2. Switzerland: Springer; 2015. p. 2025. [Google Scholar]
- 95.Chen Z, Zhang J. Doppler optical coherence tomography. In: Drexler W, Fujimot J, editors. Optical Coherence Tomography: Technology and Applications. Switzerland: Springer; 2015. p. 1289. [Google Scholar]
- 96.Chen Z, et al. Noninvasive imaging of in vivo blood flow velocity using optical doppler tomography. Opt Lett. 1997;22:1119–21. doi: 10.1364/ol.22.001119. [DOI] [PubMed] [Google Scholar]
- 97.Zhao Y, Chen Z, Saxer C, Xiang S, de Boer JF, Nelson JS. Phase-resolved optical coherence tomography and optical Doppler tomography for imaging blood flow in human skin with fast scanning speed and high veocity sensitivity. Opt Lett. 2000;25:114–6. doi: 10.1364/ol.25.000114. [DOI] [PubMed] [Google Scholar]
- 98.Nakaji H, Kouyama N, Muragaki Y, Kawakami Y, Iseki H. Localization of nerve fiber bundles by polarization-sensitive optical coherence tomography. J Neurosci Methods. 2008;174:82–90. doi: 10.1016/j.jneumeth.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 99.Zhao Y, et al. Doppler standard deviation imaging for clinical monitoring of in vivo human skin blood flow. Opt Lett. 2000;25:1358–60. doi: 10.1364/ol.25.001358. [DOI] [PubMed] [Google Scholar]
- 100.Lin AJ, et al. Optical imaging in an Alzheimer’s mouse model reveals amyloid-β-dependent vascular impairment. Neurophotonics. 2014;1:011005. doi: 10.1117/1.NPh.1.1.011005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhi Z, Qin W, Wang J, Wei W, Wang RK. 4D optical coherence tomography-based micro-angiography achieved by 1.6 MHz FDML swept source. Opt Lett. 2015;40:1779–82. doi: 10.1364/OL.40.001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Qi L, et al. Fully distributed absolute blood flow velocity measurement for middle cerebral arteries using Doppler optical coherence tomography. Biomed Opt Express. 2016;7:601–15. doi: 10.1364/BOE.7.000601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jöbsis FF. Noninvasive infrared monitoring of cerebral and myocardial oxygen sufficiency and circulatory parameters. Science. 1977;198:1264–7. doi: 10.1126/science.929199. [DOI] [PubMed] [Google Scholar]
- 104.Hoshi Y, Tamura M. Detection of dynamic changes in cerebral oxygenation coupled to neuronal function during mental work in man. Neurosci Lett. 1993;50:5–8. doi: 10.1016/0304-3940(93)90094-2. [DOI] [PubMed] [Google Scholar]
- 105.Villringer A, Plank J, Hock C, Schleikofer L, Dirnagl U. Near-infrared spectroscopy (NIRS): a new tool to study hemodynamic changes during activation of brain function in human adults. Neurosci Lett. 1993;154:101–4. doi: 10.1016/0304-3940(93)90181-j. [DOI] [PubMed] [Google Scholar]
- 106.Delpy DT, Cope M, van der Zee P, Arridge S, Wray S, Wyatt J. Estimation of optical pathlength through tissue from direct time of flight measurement. Phys Med Biol. 1988;33:1433–42. doi: 10.1088/0031-9155/33/12/008. [DOI] [PubMed] [Google Scholar]
- 107.Saager RB, Berger AJ. Direct characterization and removal of interfering absorption trends in two-layer turbid media. J Opt Soc Am A. 2005;22:1874–82. doi: 10.1364/josaa.22.001874. [DOI] [PubMed] [Google Scholar]
- 108.Hoshi Y. Towards the next generation of near-infrared spectroscopy. Phil Trans R Soc A. 2011;369:4425–39. doi: 10.1098/rsta.2011.0262. [DOI] [PubMed] [Google Scholar]
- 109.Torricelli A, Contini D, Pifferi A, Caffini M, Re R, Zucchelli L, Spinelli L. Time domain functional NIRS imaging for human brain mapping. NeuroImage. 2014;85:28–50. doi: 10.1016/j.neuroimage.2013.05.106. [DOI] [PubMed] [Google Scholar]
- 110.O’Leary MA, Boas DA, Chance B, Yodh AG. Experimental images of heterogenous turbid media by frequency-domain diffusing-photon tomography. Opt Lett. 1995;20:426–8. doi: 10.1364/ol.20.000426. [DOI] [PubMed] [Google Scholar]
- 111.Eggebrecht AT, Ferradal SL, Robichaux-Viehoever A, Hassanpour MS, Dehghani H, Snyder AZ, Hershey T, Culver JP. Mapping distributed brain function and networks with diffuse optical tomography. Nat Photon. 2014;8:448–54. doi: 10.1038/nphoton.2014.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fukuzawa R. Master Thesis. The University of Electro-Communications; Tokyo, Japan: 2009. Diffuse optical tomography for premature neonate head. [Google Scholar]
- 113.Hoshi Y, Yamada Y. Overview of diffuse optical tomography and its clinical applications. J Biomed Opt. 2016;21:91312. doi: 10.1117/1.JBO.21.9.091312. [DOI] [PubMed] [Google Scholar]


