Abstract
Rationale: Acute kidney injury is a common and severe complication of critical illness and cardiac surgery. Despite significant attempts at developing treatments, therapeutic advances to attenuate acute kidney injury and expedite recovery have largely failed.
Objectives: Identifying genetic loci associated with increased risk of acute kidney injury may reveal novel pathways for therapeutic development.
Methods: We conducted an exploratory genome-wide association study to identify single-nucleotide polymorphisms associated with genetic susceptibility to in-hospital acute kidney injury.
Measurements and Main Results: We genotyped 609,508 single-nucleotide polymorphisms and performed genotype imputation in 760 acute kidney injury cases and 669 controls. We then evaluated polymorphisms that showed the strongest association with acute kidney injury in a replication patient population containing 206 cases with 1,406 controls. We observed an association between acute kidney injury and four single-nucleotide polymorphisms at two independent loci on metaanalysis of discovery and replication populations. These include rs62341639 (metaanalysis P = 2.48 × 10−7; odds ratio [OR], 0.64; 95% confidence interval [CI], 0.55–0.76) and rs62341657 (P = 3.26 × 10−7; OR, 0.65; 95% CI, 0.55–0.76) on chromosome 4 near APOL1-regulator IRF2, and rs9617814 (metaanalysis P = 3.81 × 10−6; OR, 0.70; 95% CI, 0.60–0.81) and rs10854554 (P = 6.53 × 10−7; OR, 0.67; 95% CI, 0.57–0.79) on chromosome 22 near acute kidney injury–related gene TBX1.
Conclusions: Our findings reveal two genetic loci that are associated with acute kidney injury. Additional studies should be conducted to functionally evaluate these loci and to identify other common genetic variants contributing to acute kidney injury.
Keywords: genetics, genotype, intensive care unit, cardiac, surgery
At a Glance Commentary
Scientific Knowledge on the Subject
Acute kidney injury significantly contributes to morbidity and mortality during hospitalization, particularly in critically ill patients. However, patients with similar renal insults can have remarkably different outcomes, suggesting genetic risk factors that influence the development of acute kidney injury. A genome-wide association study represents an unbiased search for links between common genetic markers, such as single-nucleotide polymorphisms, and a common disease, such as acute kidney injury. Identifying these genetic markers will enhance the understanding of acute kidney injury pathophysiology and guide future therapeutic strategies and patient management.
What This Study Adds to the Field
After performing a genome-wide association study on one of the largest in-hospital acute kidney injury case–control populations assembled, we identified and validated single-nucleotide polymorphisms in two genetic loci on chromosomes 4 and 22 that may represent risk factors for development of acute kidney injury. These markers are found near APOL1-regulator IRF2 and acute kidney injury–related TBX1 genes, respectively, though additional research is necessary to determine underlying molecular mechanisms. Our work also represents a basis for future investigations using similar patient populations that may confirm current findings and identify additional genetic markers associated with acute kidney injury.
Acute kidney injury (AKI) is increasingly common among hospitalized patients (1). Even minor injury has been associated with increased mortality (2) and may contribute to increased risk of chronic kidney disease (CKD) (3). Unfortunately, no clinical treatments have been developed for AKI, partially because of the complex pathophysiology of the disease.
Identifying genetic risk factors for AKI could help elucidate novel pathways for therapeutic development and important subtypes of AKI. Although investigators have begun identifying genetic variants that predispose patients to AKI under minimal physiologic stress (e.g., during exercise), patients can experience markedly different renal outcomes during severe physiologic stress (e.g., during sepsis or surgery), when AKI is more commonly encountered (4). This suggests that there may be an underlying genetic predisposition to AKI.
Previous studies using a candidate gene approach to identify genetic risk factors for AKI have produced mixed results (5). Genome-wide association studies (GWASs), which allow for an unbiased search for correlation between single-nucleotide polymorphisms (SNPs) and phenotypes, may be better suited to the complexity of AKI. Thus far, only one GWAS of AKI has been completed, which identified two novel loci associated with AKI (6). However, given the heterogeneous and complex nature of AKI and the relatively small sample size of this prior study, there are likely other important genetic variants. Thus, we undertook a prospective multipopulation GWAS investigating the association of SNPs with the development of AKI in critically ill patients.
Methods
Study Populations
Cases and controls for the discovery population were derived from two independent populations of critically ill patients. The first population was assembled from patients in medical, surgical, trauma, and cardiac intensive care units (ICUs) from the VALID (Validation of Biomarkers in Acute Lung Injury Diagnosis) study (7). The second population enrolled patients who underwent cardiac surgery from the TRIBE (Translational Research Investigating Biomarker Endpoints in AKI) study (8). In addition, we performed genotyping for replication of GWAS signals in a case–control population consisting of patients who underwent cardiac surgery from the CABG Genomics (Coronary Artery Bypass Graft Genomics) study (9) and the BWH CSS (Brigham and Women’s Hospital Cardiac Surgery Study) (10).
AKI cases for discovery and replication were defined as patients with at least a 0.3-mg/dl or 50% increase in serum creatinine from baseline for at least two consecutive days. These criteria are stricter than currently accepted definitions and improve the specificity for AKI detection (11). The criterion for an at least 2-day duration of AKI selects patients with AKI due to intrinsic renal damage as opposed to those with hemodynamic changes, who often have transient AKI. Duration of AKI has been strongly associated with short- and long-term outcomes after hospitalization (12–15). Non-AKI controls were defined as patients with an increase in serum creatinine not exceeding 25% relative to baseline, to enrich for patients with minimal alterations in kidney function despite an insult (e.g., renal ischemia with cardiopulmonary bypass). Detailed descriptions of patient populations and selection criteria are included in the online supplement.
Genotyping, Imputation, and Association Analysis in the AKI Discovery Population
Genotyping was performed in three different batches, using the HumanOmni1-Quad v1.0 BeadChip, HumanOmniExpress v1.0 BeadChip, and HumanOmniExpress v1.1 BeadChip platforms (Illumina, San Diego, CA) without any evidence of batch bias. The final discovery population after quality control contained 709 AKI cases and 619 non-AKI controls, representing 93% of enrolled patients in both settings. Samples with a call rate less than 97%, sex mismatch, or cryptic relatedness were excluded. After quality control, 609,508 SNPs were checked for alignment against the 1000 Genomes Project phase I integrated reference panel (released June 16, 2014) (16) and used for haplotype phasing with SHAPEIT2 software (17). Genome-wide SNP imputation using inferred haplotypes was performed with IMPUTE2, generating data for 9,237,809 SNPs (18). In total, 161,689 SNPs with poor imputation quality (info score < 0.5) were removed.
Association testing for AKI was performed on 9,076,120 SNPs, using the additive model in SNPTEST v2.4.1 (19) with conditioning on the following covariates: patient age, sex, recruitment site (TRIBE vs. ICU), genotype batch, and the first three principal components for ethnicity. Further models adjusted for other important clinical covariates, including the presence or absence of diabetes mellitus (DM), hypertension, and CKD. Because AKI definitions in our discovery populations varied, we completed a sensitivity analysis by defining cases as patients with an increase of at least 25, 50, or 100% in serum creatinine relative to baseline or the lowest in-hospital level. Additional details are available in the online supplement.
Genomic Functional Annotation and Selection of SNPs for Validation
We used a two-tiered approach to select SNPs for validation from the discovery cohort. First, we included SNPs that had a P value less than 5 × 10−6. Second, we used GenoWAP (Genome-Wide Association Prioritizer) (20), which is a post-GWAS prioritization approach based on integrative analysis of genomic regions with functional annotation, to identify any SNPs with marginal P values that were more likely to be functional in the disease process. GenoWAP uses a mixture model of multiple layers and empirical Bayes techniques to assign a posterior score to each SNP in the data set. Up to three SNPs per gene or locus of interest were selected for additional study. Further details regarding GenoWAP and selection of SNPs for validation are provided in the online supplement.
Genotyping and Association Analysis in the AKI Replication Populations
Genotyping was performed on a MassARRAY system using matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry with iPLEX Gold chemistry (Agena Bioscience, San Diego, CA). After primer design, genotyping was performed for 22 SNPs representing 13 loci. AKI association testing was completed with adjustment for patient age and sex, using SNPTEST v2.4.1 (19). Detailed descriptions are available in the online supplement.
Variant Database Query for SNPs Associated with Renal Disease
Apart from the SNPs selected for validation, we also examined whether any SNPs previously associated with AKI or other kidney diseases were associated with the phenotype. These SNPs were identified by detailed review of published reports on GWASs of renal diseases (21–26) and the BioMart portal (27) to examine publicly available human variant databases (including ClinVar, dbSNP, ESP, HGMD-Public, and PhenCode). Expression quantitative trait loci (eQTLs) without tissue specification were searched using the Genotype-Tissue Expression (GTEx) release v4 portal (28) for matches to the 38 SNPs of interest found in our study.
Statistical Analysis
Patient demographic variables are reported as either mean and standard deviation or as median and quartiles. The primary end point for our GWAS was the development of AKI. Odds ratios (ORs) are reported with 95% confidence intervals. To make our analysis more comparable to the previously published GWAS of AKI (6), we also used the percent change in serum creatinine as an alternative end point for association.
In the discovery phase, P < 5 × 10−6 was considered the initial cutoff for statistical significance to guide SNP selection for the replication phase, and P < 5 × 10−8 indicated genome-wide significance. Power calculations were performed with QUANTO software (29). The discovery panel had 80% power to identify variants with odds ratios greater than 1.6 at P < 5 × 10−6, assuming a minor allele frequency not less than 30%. GenoWAP (20) was used to prioritize and select SNPs for replication testing. In the replication phase, P < 0.05 was considered significant. For the metaanalysis, we applied a random-effects model to estimate heterogeneity and a fixed-effect model to estimate odds ratios and P values. Detailed descriptions are available in the online supplement.
Results
Study Design and Characteristics of Patient Populations
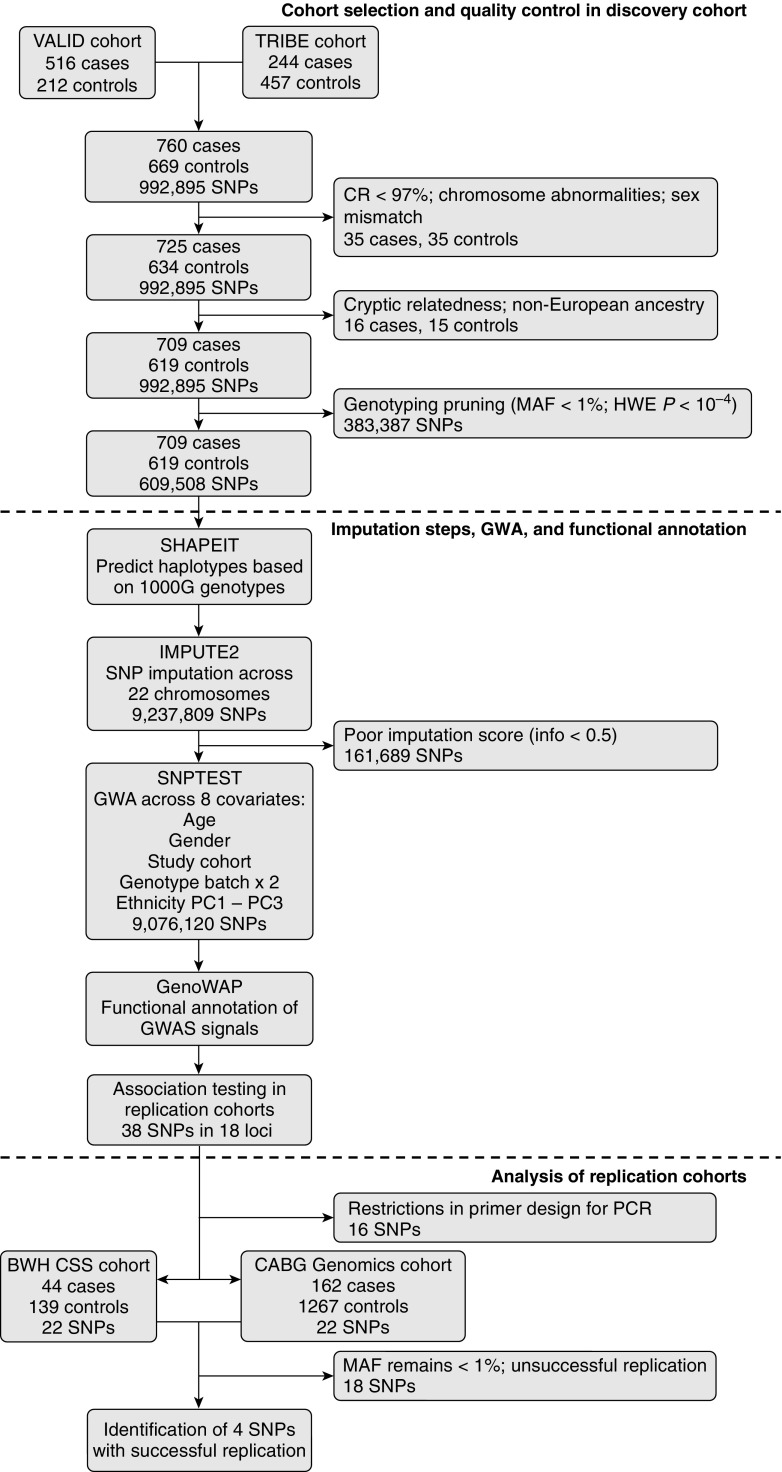
Figure 1 describes the cases and controls in the discovery population. We identified 709 AKI cases (474 from VALID, 235 from TRIBE) and 619 non-AKI controls (198 from VALID, 421 from TRIBE) for the combined discovery population that underwent SNP genotyping (Table 1). Using the same definitions as in the discovery population, 206 cases along with 1,406 controls from the CABG Genomics and BWH CSS studies, respectively, were included as the combined replication population (Table E1 in the online supplement).
Figure 1.
Flow diagram of study design, quality control, and imputation steps for selection and analysis of acute kidney injury (AKI) cases and non-AKI controls. CR = call rate; GWA = genome-wide association; GWAS = genome-wide association study; HWE P = Hardy–Weinberg equilibrium deviation P value; MAF = minor allele frequency; PC = principal component; PCR = polymerase chain reaction; SNP = single-nucleotide polymorphism. See text for abbreviations of study population names.
Table 1.
Patient Demographics of the Acute Kidney Injury Discovery Case–Control Population
| VALID Population |
TRIBE Population |
|||
|---|---|---|---|---|
| Cases (n = 474) | Controls (n = 198) | Cases (n = 235) | Controls (n = 421) | |
| Males, n (%) | 286 (60) | 109 (55) | 160 (68) | 277 (66) |
| Age, yr | ||||
| Mean (SD) | 55 (17) | 59 (15) | 73 (9) | 73 (9) |
| p25, median, p75 | 43, 56, 68 | 50, 62, 69 | 69, 74, 79 | 70, 74, 79 |
| Diabetes, n (%) | 122 (26) | 37 (19) | 116 (49) | 142 (34) |
| Hypertension, n (%) | 239 (50) | 100 (51) | 205 (87) | 337 (80) |
| CKD,* n (%) | 91 (19) | 58 (29) | 118 (50) | 209 (50) |
| Baseline Cr, mg/dl, mean (SD) | 0.86 (0.53) | 1.08 (0.50) | 1.19 (0.4) | 1.12 (0.32) |
| ICU setting (VALID only), n (%) | ||||
| Medical | 207 (44) | 86 (43) | — | — |
| Trauma | 143 (30) | 19 (10) | — | — |
| Surgical | 96 (20) | 88 (44) | — | — |
| Cardiothoracic | 28 (6) | 5 (3) | — | — |
| Sepsis (VALID only), n (%) | 216 (46) | 63 (32) | — | — |
| Surgery details (TRIBE only) | ||||
| Valve only, n (%) | — | — | 65 (28) | 132 (31) |
| CPB time, mean (SD) | — | — | 129 (66) | 118 (52) |
| Outcomes | ||||
| Acute dialysis, n (%) | 60 (13) | 0 (0) | 10 (6) | 0 (0) |
| Length of ventilation, d, mean (SD) | 8.7 (9.2) | 3.2 (5.2) | 1.0 (1.3) | 0.6 (0.7) |
| Length of ICU stay, d, mean (SD) | 12.9 (10.9) | 5.8 (5.5) | 6.1 (14.7) | 2.4 (3.1) |
| Length of hospital stay, d, mean (SD) | 21.1 (15) | 10.2 (7.6) | 13.1 (18.6) | 7.6 (6.8) |
| In-hospital mortality, n (%) | 90 (19) | 16 (8) | 9 (6) | 0 (0) |
Definition of abbreviations: CPB = cardiopulmonary bypass; Cr = creatinine; ICU = intensive care unit; p25 = 25th percentile; p75 = 75th percentile; TRIBE = Translational Research Investigating Biomarker Endpoints in AKI study; VALID = Validation of Biomarkers in Acute Lung Injury Diagnosis study.
CKD (chronic kidney disease) was defined as baseline estimated glomerular filtration rate less than 60 ml/min/1.73 m2.
GWAS Analysis in the AKI Discovery Population
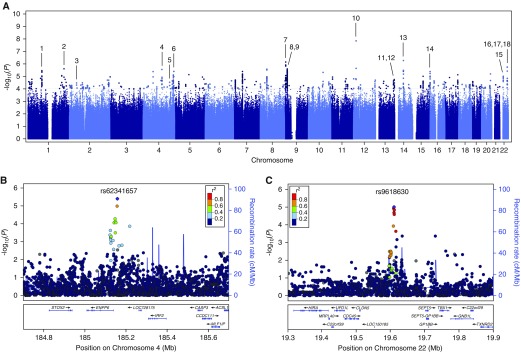
Association testing in the discovery population revealed 74 SNPs with P < 5 × 10−6 for association with AKI, including 1 SNP with P < 5 × 10−8 and 3 additional SNPs with P < 1 × 10−6 (Figure 2). The genome-wide inflation factor (λ) was 1.01, indicating negligible variation in population structure between cases and controls. The quantile–quantile plot showed overall adherence to expected P values (Figure E1). One SNP with a genome-wide significant P value (rs148018420, P = 1.43 × 10−8 and P = 1.02 × 10−8, before and after adjustment for clinical covariates, respectively) is located in an intergenic region on chromosome 12 and has an estimated OR of 0.017 (95% confidence interval [CI], 0.0042–0.068), but exhibits less than 1% minor allele frequency after imputation. The three additional SNPs closest to reaching genome-wide significance include rs72681624 (P = 5.41 × 10−7 and P = 4.41 × 10−7; OR, 0.34; 95% CI, 0.22–0.52), located in the promoter sequence of the SAV1 gene on chromosome 14, as well as two neighboring SNPs in the first intron of IL33 on chromosome 9: rs10815380 (P = 7.06 × 10−7 and P = 6.93 × 10−7; OR, 0.60; 95% CI, 0.49–0.74) and rs10815381 (P = 7.11 × 10−7 and P = 6.96 × 10−7; OR, 0.60; 95% CI, 0.49–0.74). All measured and imputed SNPs were subsequently subjected to functional annotation.
Figure 2.
Manhattan plots generated from genome-wide association study analysis of the acute kidney injury cases and controls. Plots include (A) all markers or single-nucleotide polymorphisms (SNPs) at loci of interest on (B) chromosome 4 and (C) chromosome 22 at higher magnification. Linkage disequilibrium (r2) is indicated relative to the SNP with lowest P value (purple marker) by colored scale. Recombination rates at nearby positions are shown as the background blue lines. Genes located near each locus are displayed in the bottom panels. Loci of interest, corresponding to those in Table E2, are indicated by black lines labeled with the numbers 1 through 18.
Functional Annotation to Reveal Candidate SNPs for Validation
Of the 74 SNPs with P values less than 5 × 10−6 for association with AKI in the discovery GWAS, 31 SNPs reside in 9 protein-coding genes. GenoWAP (20) was applied to prioritize SNPs with marginal significance in coding regions as well as in the noncoding genome. This prioritization algorithm successfully converged in our study, confirming the enrichment of GWAS signals in genomic regions containing functional annotations compared with regions without (P = 0.007 by one-sided Kolmogorov–Smirnov test). GenoWAP revealed nine additional intra- and intergenic loci. In total, 38 SNPs representing 18 candidate loci were selected for additional testing (Figure 2A; Table E2). Of these, 24 SNPs are found in the coding regions of 13 genes. Among the 38 SNPs, 18 showed an OR greater than 1 and were designated as risk alleles, whereas the remaining 20 SNPs had an OR less than 1 and were designated as protective alleles. There were four SNPs with P values near our selection threshold (P < 5 × 10−6) that no longer met this criterion after adjustment for the presence of DM, hypertension, or CKD (Table E2). However, because these resulting P values were only slightly above the original selection threshold value of P < 5 × 10−6, these SNPs remained candidates for further analysis. Of the 38 SNPs, 22 SNPs in 13 loci passed the primer design stage (5 loci failed) and were successfully genotyped in the replication population (Table E3).
Sensitivity analysis of 4 AKI definitions revealed that nearly all 38 SNPs of interest retained their odds ratios regardless of the definition of AKI (Table E4). Additional covariates, such as presence of DM, CKD, or hypertension, also did not significantly affect the odds ratios of these SNPs (Table E2).
Association Analysis in the AKI Replication Populations
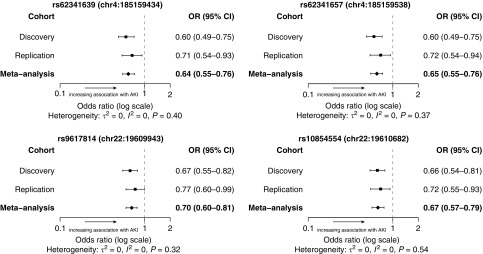
For 22 SNPs of interest, their association with AKI with adjustment for covariates was tested in a replication case–control population, followed by metaanalysis of the effect size of each SNP across the populations (Table E3). For additional quality control, three SNPs (rs148018420, rs111732708, and rs202139590) representing two loci were removed from the analysis because their minor allele frequencies remained less than 1% after replication testing (Table E3). No SNPs reached the genome-wide significance threshold of 5 × 10−8 in the metaanalysis. However, of the 22 SNPs selected for replication, 4 SNPs in 2 genetic loci showed significant association with AKI (P < 0.05) in the replication population (Table 2; Figure 3). The first locus includes two proximate intergenic SNPs on chromosome 4, rs62341639 (discovery P = 4.01 × 10−6, metaanalysis P = 2.48 × 10−7) and rs62341657 (discovery P = 3.98 × 10−6, metaanalysis P = 3.26 × 10−7), with ORs of 0.64 (95% CI, 0.55–0.76) and 0.65 (95% CI, 0.55–0.76), respectively. The second locus includes two close intergenic SNPs on chromosome 22, rs9617814 (discovery P = 2.04 × 10−5, metaanalysis P = 3.81 × 10−6) and rs10854554 (discovery P = 1.44 × 10−5, metaanalysis P = 6.53 × 10−7), with ORs of 0.70 (95% CI, 0.60–0.81) and 0.67 (95% CI, 0.57–0.79), respectively. All four SNPs in the two loci on chromosomes 4 and 22 showed ORs less than 1, suggesting that the SNPs have a protective effect (Table 2). Furthermore, both signals are supported by additional SNPs in high linkage disequilibrium with P < 10−4 for AKI association in the discovery population (Figures 2B and 2C).
Table 2.
Summary of Acute Kidney Injury Genome-Wide Association Study Signals in Discovery and Replication Case–Control Populations
| SNP | Chr | Position | Discovery Population |
Replication Populations |
Metaanalysis |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P Value* | Minor Allele | MAF Cases | MAF Ctrls | OR*† | 95% CI | P Value | MAF Cases | MAF Ctrls | OR† | 95% CI | P Value | OR† | 95% CI | |||
| rs62341639 | 4 | 185159434 | 3.16 × 10−6 | T | 0.18 | 0.24 | 0.60 | 0.49–0.75 | 0.014 | 0.14 | 0.19 | 0.71 | 0.54–0.93 | 2.48 × 10−7 | 0.64 | 0.55–0.76 |
| rs62341657 | 4 | 185159538 | 3.09 × 10−6 | A | 0.18 | 0.24 | 0.60 | 0.49–0.75 | 0.018 | 0.14 | 0.19 | 0.72 | 0.54–0.94 | 3.26 × 10−7 | 0.65 | 0.55–0.76 |
| rs9617814 | 22 | 19609943 | 3.31 × 10−5 | G | 0.22 | 0.29 | 0.67 | 0.55–0.82 | 0.040 | 0.20 | 0.25 | 0.77 | 0.60–0.99 | 3.81 × 10−6 | 0.70 | 0.60–0.81 |
| rs10854554 | 22 | 19610682 | 4.46 × 10−5 | G | 0.19 | 0.26 | 0.66 | 0.54–0.81 | 0.012 | 0.17 | 0.22 | 0.72 | 0.55–0.93 | 6.53 × 10−7 | 0.67 | 0.57–0.79 |
Definition of abbreviations: Chr = chromosome; CI = confidence interval; Ctrls = controls; MAF = minor allele frequency; OR = odds ratio; SNP = single-nucleotide polymorphism.
P value and odds ratio after adjustment for the presence or absence of clinical covariates (diabetes mellitus, hypertension, and chronic kidney disease) in cases and controls are shown for the discovery population.
The odds ratio was calculated as eβ, where β represents the regression coefficient of the additive model in SNPTEST (19).
Figure 3.
Forest plots of odds ratios (ORs) for four single-nucleotide polymorphisms supported by acute kidney injury (AKI) association testing in the replication population. Odds ratios from each tested population and resulting metaanalysis based on a fixed-effect model are shown with error bars indicating the 95% confidence interval (CI). Heterogeneity variance (τ2) and statistic (I2) were calculated from a random-effects model as described in the online supplement. The P value is reported for Cochran’s Q test. A nonsignificant Q test result suggests that contributing samples are statistically represented by one underlying distribution.
Query of Public Databases for Variations Implicated in Kidney Disease
Review of publicly available genetic databases and reported GWAS of renal diseases (21–26) failed to reveal any shared SNPs or genetic loci between the literature and our discovery population. In addition, there were no shared SNPs or loci when using the change in serum creatinine as a continuous outcome for association testing, which was the approach used by the only previously reported GWAS of AKI (6) (Table E5). Finally, examination of the GTEx project database (28) revealed eQTLs for several of the 38 SNPs identified in the discovery cohort (Table E6).
Discussion
We present the largest GWAS of AKI to date, which identified four promising and novel SNPs that have not been found in prior genetic studies of AKI. Although neither locus contained SNPs that showed genome-wide significance in the metaanalysis of the replication populations, each locus included SNPs that were independently significant in a replication population. The modest differences between discovery and replication results may be due to reduced statistical power as there were limited numbers of available cases in the replication population. Nonetheless, both loci showed consistent odds ratios with improved P values after replication testing relative to the discovery phase, suggesting that these SNPs reliably associate with the development of AKI but fail to meet traditional genome-wide significance due to limited power.
Because the SNPs supported by replication study are located in intergenic regions on chromosomes 4 and 22, we examined all nearby genes for previously reported relevance to kidney disease. The first locus on chromosome 4 is located 150 kb upstream of IRF2, a transcription factor involved in innate immunity pathways. IRF2 regulates expression of the kidney disease risk gene APOL1 (30), in which variation has been found to be a strong genetic risk factor for focal segmental glomerulosclerosis and kidney failure in African Americans (31). Disruption of IRF-2 has been found to up-regulate the inflammatory response to infection in murine models (32). In addition, the IRF2-antagonizing interferon regulatory factor IRF1 has been implicated in AKI exacerbation (33, 34). The second locus on chromosome 22 is found 140 kb upstream of TBX1, a T-box transcription factor involved in embryonic renal development (35). TBX1 expression was correlated with activation of transforming growth factor-β and with kidney injury in a gentamicin-induced AKI model (36). Although proximity does not guarantee genetic interaction, these intergenic regions could represent enhancer or mediator elements for nearby genes, and both loci may affect pathways that contribute to AKI pathophysiology.
Comparison of our results with publicly available databases and previous reports of GWASs of kidney diseases, including one in the setting of postoperative AKI (6), did not reveal overlapping SNP markers or genetic loci. This discrepancy may be partially due to different study designs and sample selection—in particular, the prior study did not exclude transient AKI of short duration. Our GWAS also failed to confirm findings from a previously published candidate gene study using mouse models (37). This is not unexpected as there is a growing body of literature suggesting that the overlap between murine and human transcriptomes is minimal (38–40). Moreover, candidate gene studies have a low replication rate (5). Regardless, AKI as defined by changes in serum creatinine is a complex heterogeneous clinical phenotype, analogous to schizophrenia—a heterogeneous psychiatric disorder. Ultimately, 108 loci for schizophrenia have been definitively identified, but only after combining multiple large cohorts (41). Thus, we anticipate that data from multiple patient populations, including from the previously published GWAS (6) and the current study, will need to be considered before all genetic variants associated with AKI can be definitively identified.
Because our discovery case–control population was composed of patients from two large heterogeneous settings of AKI, we performed sensitivity analysis that used various cutoffs for the change in serum creatinine as the definition of AKI cases. Nearly all SNPs of interest continued to have consistent odds ratios, suggesting that our AKI definitions were adequately sensitive for detecting GWAS signals across AKI settings. Furthermore, we performed association testing with conditioning on covariates that represent AKI risk factors and comorbidities. Association P values and odds ratios for SNPs of interest were nearly identical across all covariate adjustments, suggesting that these SNPs associate independently with the risk of AKI.
The present study has multiple strengths. Cases and non-AKI controls were strictly defined, using definitions more likely to be specific for “true” AKI (11, 42). We used the latest genomic platforms with rigorous quality control measures and GenoWAP software to identify SNPs with the highest potential for functional consequences. Given the heterogeneous nature of AKI in the postsurgical and medical ICU settings, combining these two patient populations would be expected to bias our findings toward the null. Nonetheless, we identified and found additional support for two loci in the discovery and replication phase, respectively.
However, our study has important limitations as well. Although we undertook extensive effort to identify a replication case–control population that was similar to our discovery population, there are still some differences between the two studies combined to make the final replication population that may have contributed to varying results. For example, the BWH CSS population selected a substantially sicker population than the TRIBE population as evidenced by the longer cardiopulmonary bypass time and higher incidence of CKD. Despite these differences, the confidence intervals for all of the populations overlapped substantially. An additional limitation of our study was the unavailability of urine output data in our populations, which could have been used with serum creatinine values to more sensitively detect AKI. Moreover, because of primer design limitations, we were able to study only 13 of 18 genetic loci of interest in the replication populations. We were unable to condition the replication population on patient ethnicity because there were no genome-wide data available, which can lead to inaccuracies in estimations of SNP frequencies. We also could not condition the replication population on clinical covariates because of missing data and differences in patient selection (and therefore in covariate distribution in cases and controls) among the replication cohorts. Thus, future studies using alternative sequencing techniques are necessary to complete the validation of our discovery population findings. In addition, our study included only white patients and may not be generalizable to other ethnicities. Extremely rare, segmental (e.g., copy number variants, microsatellites), or somatic genetic variants are not well assayed in GWASs and instead require whole-exome or whole-genome sequencing in the tissue of interest to be identified. Indeed, such screens have discovered genetic risk factors for complex traits including cholesterol levels, insulin metabolism, type 2 diabetes, and AKI (43–46). Future studies may use such sequencing as a complement to GWAS analysis.
In conclusion, we have performed the largest GWAS to date evaluating genetic susceptibility for AKI and have identified two intergenic loci, each containing at least two SNPs of interest, that have suggestive association with AKI development. Our results provide the framework for future investigations of genetic associations with AKI, which can illuminate biological pathways involving genes located near the identified SNPs.
Acknowledgments
Acknowledgment
The authors thank Garcia Milan Ronaldo (Yale University) for assistance with the identification of kidney disease genes in the literature across several databases.
Members of the TRIBE-AKI Consortium are as follows: Dr. Jay Koyner (University of Chicago), Dr. Steven Coca (Yale University), Dr. Prasad Devarajan (University of Cincinnati Children’s Hospital), Dr. Cary Passik (Danbury Hospital), Dr. Madhav Swaminathan and Dr. Uptal Patel (Duke University), and Dr. Michael Zappitelli (Montreal Children’s Hospital).
Footnotes
Supported by the National Institutes of Health (NIH) grant R01HL085757 (C.R.P.) to fund the TRIBE-AKI Consortium to study novel biomarkers of AKI in cardiac surgery, and by P30DK079310 (O’Brien Center Grant). C.R.P. was supported by the NIH (K24DK090203). A.X.G. and C.R.P. are also members of the NIH-sponsored Assess, Serial Evaluation and Subsequent Sequelae in Acute Kidney Consortium (U01DK082185). B.Z. was supported by NIH Medical Scientist Training Program Training Grant T32GM007205. The VALID study investigators (E.D.S., T.A.I., and L.B.W.) were supported by NIH grants UO1HL081332, R21HL112656-02, K24HL103836, K24DK62849, and Clinical Translational Science Award 1 UL-1RR024975 from the National Center for Research Resources. E.D.S. was also supported by NIH grant K23DK088964-03 from the National Institute of Diabetes and Digestive and Kidney Diseases and the Vanderbilt Center for Kidney Diseases.
Author Contributions: B.Z., Q.L., Y.C., and M.C. analyzed genome-wide association study data. C.R.P. and R.P.L. designed the study. C.R.P., R.P.L., and H.Z. supervised the study. E.D.S., D.E.L., S.C.B., A.A.F., S.S.W., C.D.C., T.A.I., L.B.W., C.L.E., and A.X.G. acquired DNA samples. B.Z., Q.L., J.M.B., and C.R.P. wrote the manuscript. All authors contributed equally to revising the manuscript.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.201603-0518OC on August 30, 2016
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Waikar SS, Liu KD, Chertow GM. Diagnosis, epidemiology and outcomes of acute kidney injury. Clin J Am Soc Nephrol. 2008;3:844–861. doi: 10.2215/CJN.05191107. [DOI] [PubMed] [Google Scholar]
- 2.Linder A, Fjell C, Levin A, Walley KR, Russell JA, Boyd JH. Small acute increases in serum creatinine are associated with decreased long-term survival in the critically ill. Am J Respir Crit Care Med. 2014;189:1075–1081. doi: 10.1164/rccm.201311-2097OC. [DOI] [PubMed] [Google Scholar]
- 3.Coca SG, Singanamala S, Parikh CR. Chronic kidney disease after acute kidney injury: a systematic review and meta-analysis. Kidney Int. 2012;81:442–448. doi: 10.1038/ki.2011.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaito H, Ishimori S, Nozu K, Shima Y, Nakanishi K, Yoshikawa N, Iijima K. Molecular background of urate transporter genes in patients with exercise-induced acute kidney injury. Am J Nephrol. 2013;38:316–320. doi: 10.1159/000355430. [DOI] [PubMed] [Google Scholar]
- 5.Lu JC, Coca SG, Patel UD, Cantley L, Parikh CR Translational Research Investigating Biomarkers and Endpoints for Acute Kidney Injury (TRIBE-AKI) Consortium. Searching for genes that matter in acute kidney injury: a systematic review. Clin J Am Soc Nephrol. 2009;4:1020–1031. doi: 10.2215/CJN.05411008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stafford-Smith M, Li YJ, Mathew JP, Li YW, Ji Y, Phillips-Bute BG, Milano CA, Newman MF, Kraus WE, Kertai MD, et al. Duke Perioperative Genetics and Safety Outcomes (PEGASUS) Investigative Team. Genome-wide association study of acute kidney injury after coronary bypass graft surgery identifies susceptibility loci. Kidney Int. 2015;88:823–832. doi: 10.1038/ki.2015.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siew ED, Ware LB, Gebretsadik T, Shintani A, Moons KG, Wickersham N, Bossert F, Ikizler TA. Urine neutrophil gelatinase–associated lipocalin moderately predicts acute kidney injury in critically ill adults. J Am Soc Nephrol. 2009;20:1823–1832. doi: 10.1681/ASN.2008070673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parikh CR, Coca SG, Thiessen-Philbrook H, Shlipak MG, Koyner JL, Wang Z, Edelstein CL, Devarajan P, Patel UD, Zappitelli M, et al. TRIBE-AKI Consortium. Postoperative biomarkers predict acute kidney injury and poor outcomes after adult cardiac surgery. J Am Soc Nephrol. 2011;22:1748–1757. doi: 10.1681/ASN.2010121302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fox AA, Muehlschlegel JD, Body SC, Shernan SK, Liu KY, Perry TE, Aranki SF, Cook EF, Marcantonio ER, Collard CD. Comparison of the utility of preoperative versus postoperative B-type natriuretic peptide for predicting hospital length of stay and mortality after primary coronary artery bypass grafting. Anesthesiology. 2010;112:842–851. doi: 10.1097/ALN.0b013e3181d23168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leaf DE, Rajapurkar M, Lele SS, Mukhopadhyay B, Rawn JD, Frendl G, Waikar SS. Increased plasma catalytic iron in patients may mediate acute kidney injury and death following cardiac surgery. Kidney Int. 2015;87:1046–1054. doi: 10.1038/ki.2014.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin J, Fernandez H, Shashaty MG, Negoianu D, Testani JM, Berns JS, Parikh CR, Wilson FP. False-positive rate of AKI using consensus creatinine-based criteria. Clin J Am Soc Nephrol. 2015;10:1723–1731. doi: 10.2215/CJN.02430315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coca SG, King JT, Jr, Rosenthal RA, Perkal MF, Parikh CR. The duration of postoperative acute kidney injury is an additional parameter predicting long-term survival in diabetic veterans. Kidney Int. 2010;78:926–933. doi: 10.1038/ki.2010.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown JR, Kramer RS, Coca SG, Parikh CR. Duration of acute kidney injury impacts long-term survival after cardiac surgery. Ann Thorac Surg. 2010;90:1142–1148. doi: 10.1016/j.athoracsur.2010.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoste EA, Kellum JA. AKI severity class doesn’t tell all: the case for transient AKI. Nephrol Dial Transplant. 2010;25:1738–1739. doi: 10.1093/ndt/gfq133. [DOI] [PubMed] [Google Scholar]
- 15.Uchino S, Bellomo R, Bagshaw SM, Goldsmith D. Transient azotaemia is associated with a high risk of death in hospitalized patients. Nephrol Dial Transplant. 2010;25:1833–1839. doi: 10.1093/ndt/gfp624. [DOI] [PubMed] [Google Scholar]
- 16.Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delaneau O, Marchini J 1000 Genomes Project Consortium. Integrating sequence and array data to create an improved 1000 Genomes Project haplotype reference panel. Nat Commun. 2014;5:3934. doi: 10.1038/ncomms4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu Q, Yao X, Hu Y, Zhao H. GenoWAP: GWAS signal prioritization through integrated analysis of genomic functional annotation. Bioinformatics. 2016;32:542–548. doi: 10.1093/bioinformatics/btv610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorski M, Tin A, Garnaas M, McMahon GM, Chu AY, Tayo BO, Pattaro C, Teumer A, Chasman DI, Chalmers J, et al. Genome-wide association study of kidney function decline in individuals of European descent. Kidney Int. 2015;87:1017–1029. doi: 10.1038/ki.2014.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Köttgen A, Glazer NL, Dehghan A, Hwang SJ, Katz R, Li M, Yang Q, Gudnason V, Launer LJ, Harris TB, et al. Multiple loci associated with indices of renal function and chronic kidney disease. Nat Genet. 2009;41:712–717. doi: 10.1038/ng.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Köttgen A, Pattaro C, Böger CA, Fuchsberger C, Olden M, Glazer NL, Parsa A, Gao X, Yang Q, Smith AV, et al. New loci associated with kidney function and chronic kidney disease. Nat Genet. 2010;42:376–384. doi: 10.1038/ng.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pezzolesi MG, Poznik GD, Mychaleckyj JC, Paterson AD, Barati MT, Klein JB, Ng DP, Placha G, Canani LH, Bochenski J, et al. DCCT/EDIC Research Group. Genome-wide association scan for diabetic nephropathy susceptibility genes in type 1 diabetes. Diabetes. 2009;58:1403–1410. doi: 10.2337/db08-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sandholm N, Forsblom C, Mäkinen VP, McKnight AJ, Osterholm AM, He B, Harjutsalo V, Lithovius R, Gordin D, Parkkonen M, et al. SUMMIT Consortium; FinnDiane Study Group. Genome-wide association study of urinary albumin excretion rate in patients with type 1 diabetes. Diabetologia. 2014;57:1143–1153. doi: 10.1007/s00125-014-3202-3. [DOI] [PubMed] [Google Scholar]
- 26.Stanescu HC, Arcos-Burgos M, Medlar A, Bockenhauer D, Kottgen A, Dragomirescu L, Voinescu C, Patel N, Pearce K, Hubank M, et al. Risk HLA-DQA1 and PLA2R1 alleles in idiopathic membranous nephropathy. N Engl J Med. 2011;364:616–626. doi: 10.1056/NEJMoa1009742. [DOI] [PubMed] [Google Scholar]
- 27.Smedley D, Haider S, Durinck S, Pandini L, Provero P, Allen J, Arnaiz O, Awedh MH, Baldock R, Barbiera G, et al. The BioMart community portal: an innovative alternative to large, centralized data repositories. Nucleic Acids Res. 2015;43:W589–W598. doi: 10.1093/nar/gkv350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.GTEx Consortium. Human genomics: the Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gauderman WJ. Sample size requirements for matched case–control studies of gene–environment interaction. Stat Med. 2002;21:35–50. doi: 10.1002/sim.973. [DOI] [PubMed] [Google Scholar]
- 30.Nichols B, Jog P, Lee JH, Blackler D, Wilmot M, D’Agati V, Markowitz G, Kopp JB, Alper SL, Pollak MR, et al. Innate immunity pathways regulate the nephropathy gene Apolipoprotein L1. Kidney Int. 2015;87:332–342. doi: 10.1038/ki.2014.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Genovese G, Friedman DJ, Ross MD, Lecordier L, Uzureau P, Freedman BI, Bowden DW, Langefeld CD, Oleksyk TK, Uscinski Knob AL, et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329:841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsuyama T, Kimura T, Kitagawa M, Pfeffer K, Kawakami T, Watanabe N, Kündig TM, Amakawa R, Kishihara K, Wakeham A, et al. Targeted disruption of IRF-1 or IRF-2 results in abnormal type I IFN gene induction and aberrant lymphocyte development. Cell. 1993;75:83–97. [PubMed] [Google Scholar]
- 33.Wang Y, John R, Chen J, Richardson JA, Shelton JM, Bennett M, Zhou XJ, Nagami GT, Zhang Y, Wu QQ, et al. IRF-1 promotes inflammation early after ischemic acute kidney injury. J Am Soc Nephrol. 2009;20:1544–1555. doi: 10.1681/ASN.2008080843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winterberg PD, Wang Y, Lin KM, Hartono JR, Nagami GT, Zhou XJ, Shelton JM, Richardson JA, Lu CY. Reactive oxygen species and IRF1 stimulate IFNα production by proximal tubules during ischemic AKI. Am J Physiol Renal Physiol. 2013;305:F164–F172. doi: 10.1152/ajprenal.00487.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fu Y, Li F, Zhao DY, Zhang JS, Lv Y, Li-Ling J. Interaction between Tbx1 and HoxD10 and connection with TGFβ–BMP signal pathway during kidney development. Gene. 2014;536:197–202. doi: 10.1016/j.gene.2012.06.069. [DOI] [PubMed] [Google Scholar]
- 36.Jiang H, Li L, Li-Ling J, Qiu G, Niu Z, Jiang H, Li Y, Huang Y, Sun K. Increased Tbx1 expression may play a role via TGFβ–Smad2/3 signaling pathway in acute kidney injury induced by gentamicin. Int J Clin Exp Pathol. 2014;7:1595–1605. [PMC free article] [PubMed] [Google Scholar]
- 37.Chousterman BG, Boissonnas A, Poupel L, Baudesson de Chanville C, Adam J, Tabibzadeh N, Licata F, Lukaszewicz AC, Lombès A, Deterre P, et al. Ly6Chigh monocytes protect against kidney damage during sepsis via a CX3CR1-dependent adhesion mechanism. J Am Soc Nephrol. 2016;27:792–803. doi: 10.1681/ASN.2015010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin S, Lin Y, Nery JR, Urich MA, Breschi A, Davis CA, Dobin A, Zaleski C, Beer MA, Chapman WC, et al. Comparison of the transcriptional landscapes between human and mouse tissues. Proc Natl Acad Sci USA. 2014;111:17224–17229. doi: 10.1073/pnas.1413624111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, Richards DR, McDonald-Smith GP, Gao H, Hennessy L, et al. Inflammation and Host Response to Injury, Large Scale Collaborative Research Program. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci USA. 2013;110:3507–3512. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burns TC, Li MD, Mehta S, Awad AJ, Morgan AA. Mouse models rarely mimic the transcriptome of human neurodegenerative diseases: a systematic bioinformatics-based critique of preclinical models. Eur J Pharmacol. 2015;759:101–117. doi: 10.1016/j.ejphar.2015.03.021. [DOI] [PubMed] [Google Scholar]
- 41.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palevsky PM, Liu KD, Brophy PD, Chawla LS, Parikh CR, Thakar CV, Tolwani AJ, Waikar SS, Weisbord SD. KDOQI US commentary on the 2012 KDIGO clinical practice guideline for acute kidney injury. Am J Kidney Dis. 2013;61:649–672. doi: 10.1053/j.ajkd.2013.02.349. [DOI] [PubMed] [Google Scholar]
- 43.Morrison AC, Voorman A, Johnson AD, Liu X, Yu J, Li A, Muzny D, Yu F, Rice K, Zhu C, et al. Cohorts for Heart and Aging Research in Genetic Epidemiology (CHARGE) Consortium. Whole-genome sequence–based analysis of high-density lipoprotein cholesterol. Nat Genet. 2013;45:899–901. doi: 10.1038/ng.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steinthorsdottir V, Thorleifsson G, Sulem P, Helgason H, Grarup N, Sigurdsson A, Helgadottir HT, Johannsdottir H, Magnusson OT, Gudjonsson SA, et al. Identification of low-frequency and rare sequence variants associated with elevated or reduced risk of type 2 diabetes. Nat Genet. 2014;46:294–298. doi: 10.1038/ng.2882. [DOI] [PubMed] [Google Scholar]
- 45.Huyghe JR, Jackson AU, Fogarty MP, Buchkovich ML, Stančáková A, Stringham HM, Sim X, Yang L, Fuchsberger C, Cederberg H, et al. Exome array analysis identifies new loci and low-frequency variants influencing insulin processing and secretion. Nat Genet. 2013;45:197–201. doi: 10.1038/ng.2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leaf DE, Body SC, Muehlschlegel JD, McMahon GM, Lichtner P, Collard CD, Shernan SK, Fox AA, Waikar SS. Length polymorphisms in heme oxygenase-1 and AKI after cardiac surgery. J Am Soc Nephrol. 2016;27:3291–3297. doi: 10.1681/ASN.2016010038. [DOI] [PMC free article] [PubMed] [Google Scholar]